Abstract
Nickel-dependent ureases are activated by a multiprotein complex that includes the GTPase UreG. Prior studies showed that nucleotide-free UreG from Klebsiella aerogenes is monomeric and binds one nickel or zinc ion with near equivalent affinity using an undefined binding site, whereas nucleotide-free UreG from Helicobacter pylori selectively binds one zinc ion per dimer via a universally conserved Cys-Pro-His motif in each protomer. Iodoacetamide-treated K. aerogenes UreG was nearly unaffected in nickel binding compared to non-treated sample, suggesting the absence of thiolate ligands to the metal. X-ray absorption spectroscopy of nickel-bound UreG showed the metal possessed four-coordinate geometry with all O/N donor ligands including one imidazole, thus confirming the absence of thiolate ligation. The nickel site in Strep-tag II-modified protein possessed six-coordinate geometry, again with all O/N donor ligands, but now including two or three imidazoles. An identical site was noted for the Strep-tag II-modified H74A variant, substituted in the Cys-Pro-His motif, ruling out coordination by this His residue. These results are consistent with metal binding to both His6 and a His residue of the fusion peptide in Strep-tagged K. aerogenes UreG. We conclude that the nickel and zinc binding site in nucleotide-free K. aerogenes UreG is distinct from that of nucleotide-free H. pylori UreG and does not involve the Cys-Pro-His motif. Further, we show the Strep-tag II can perturb metal coordination of this protein.
Keywords: Urease, Metallocenter assembly, Nickel binding, X-ray absorption Spectroscopy
Introduction
For many microorganisms, the incorporation of Ni into the urease active site requires four accessory proteins: UreD (called UreH in some microorganisms), UreE, UreF, and UreG [1–3]. In our working model for activation [4], UreE functions as a metallochaperone to deliver Ni to the multiprotein complex of urease-apoprotein·UreD/UreH·UreF·UreG, UreG accepts this metal and couples GTP hydrolysis to subsequent Ni transfer, with UreF plus UreD serving as a conduit to the nascent active site and altering the conformation of urease apoprotein. Synthesis of a functional dinuclear metallocenter in the holoenzyme is followed by dissociation of the accessory proteins. This study focuses on UreG, a metal-binding GTPase that plays a central role in this elaborate process.
UreG proteins have been characterized from several sources, with marked differences in their properties (Table 1). Klebsiella aerogenes UreG (KaUreG) is monomeric as purified or in the presence of Ni [5, 6], whereas the proteins from Sporosarcina (formerly Bacillus) pasteurii (SpUreG) [7] and Mycobacterium tuberculosis (MtUreG) [8] are predominantly dimers stabilized by intermolecular disulfides involving Cys residues in a conserved Cys-Pro-His motif. Additional studies have revealed a monomer-dimer equilibrium for UreG proteins from S. pasteurii [9], Helicobacter pylori (HpUreG) [10], Methanocaldococcus jannaschii (MjUreG) [11], Metallosphaera sedula (MsUreG) [11], and Glycine max (soybean; GmUreG) [12], with the dimers stabilized by Zn coordination. All UreG samples examined bind Zn and Ni, but the number of equivalents bound and the measured affinities differ in each case [6, 7, 9–14]. The single Zn site in the nucleotide-free dimeric H. pylori protein is coordinated by two His and two Cys according to X-ray absorption spectroscopy (XAS) [15]. Homology models show close positioning of two Cys66 and two His74 residues in this dimer [10, 16], and a ~6-fold decreased affinity for Zn is observed in the C66A and H68A variants [10]. These results are consistent with Zn coordination to the symmetric Cys-Pro-His motifs in the HpUreG dimer lacking nucleotide. Two equivalents of Ni bind to this nucleotide-free monomeric protein with weak affinity, but a single Ni binds per dimer with high affinity when GTP is present [13, 14]. Notably, the sites of Ni binding are not established for GTP-bound HpUreG although the aforementioned motif is very likely to be involved. All UreG sequences contain the Cys-Pro-His motif; however, only a single copy of this motif (involving residues Cys72 and His74) is available in monomeric KaUreG, thus raising the question of whether it coordinates Zn or Ni in the nucleotide-free protein. Perhaps significantly, KaUreGStr (a Strep-tag II form of the protein) exhibits near equivalent affinity for binding Zn and Ni, and 63Ni binding studies show these metals compete for the same site [6]. Notably, the KdNi/ KdZn ratio is much lower for KaUreG than for other UreG samples examined (Table 1), perhaps indicating a unique Ni-binding site.
Table 1.
Quaternary structure and metal binding properties of UreG proteins
| Proteina | Quaternary structure |
Equiv. Zn bound | Zn Kd (µM) | Equiv. Ni bound | Ni Kd (µM) | KdNi/KdZn | Refs |
|---|---|---|---|---|---|---|---|
| KaUreG | Monomer | NRb, added Zn causes precipitation |
NR | 1.0/monomer | 16.0 ± 3.1 | NR | [5, 6] |
| KaUreGStrc | Monomer | 1.10 ± 0.08 /monomer |
7.2 ± 1.6 | 0.95 ± 0.09 /monomer |
5.0 ± 1.8 | 0.7 | [6] |
| SpUreG | Monomer/dimer equilibriumd |
2/dimer | 0.0030 ± 0.0003 and 0.53 ± 0.006e |
3/monomerf | 14 ± 1, 270 ± 22, and 160 ± 13f |
4670 | [7, 9] |
| MtUreG | Dimerd | NR | NR | NR | NR | NR | [8] |
| HpUreG | Monomer/dimer equilibrium |
1/dimer | 0.33 ± 0.03 | 1.8/monomer | 10 ± 1 | 30 | [10] |
|
HpUreG (with GTP) |
Monomer/dimer equilibrium |
NR | NR | 1.2 or 1.0/dimer | 0.36 ± 0.05 | NR | [13, 14] |
| MjUreG | Monomer/dimer equilibrium |
1.0 and 1.6/dimer | 0.08 ±0.03 and 3.7 ± 0.9 |
0.74 ± 0.09 /monomer |
11 ±1 | 138 | [11] |
| MsUreG | Monomer/dimer equilibrium |
1.0 and 2.0/dimer | 0.22 ± 0.07 and 60 ±20 |
1.9/monomer | 37 ± 7 | 168 | [11] |
| GmUreG | Monomer/dimer equilibrium; Zn stabilizes dimer (Ni leads to precipitation) |
1.0, 2, and 5.2/dimer |
0.02 ± 0.01, 1.31 ± 0.05, 0.16 ± 0.09 |
2.15/monomer | 4.8 ± 0.3 | 240 | [12] |
UreG proteins derived from Klebsiella aerogenes, Sporosarcina pasteurii, Mycobacterium tuberculosis, Methanocaldococcus jannaschii, Metallosphaera sedula, Helicobacter pylori, and Glycine max.
Not reported,
Strep-tag II fusion protein.
Dimer is stabilized by an intermolecular disulfide involving the Cys residue of the Cys-Pro-His motif.
Initially reported as 42 ± 3 µM for two sites/dimer.
Initially reported as 360 ± 30 µM for four sites/dimer.
The studies reported here examine the Ni-binding site of nucleotide-free KaUreG and demonstrate that it is distinct from the Cys-Pro-His motif used to bind Zn in nucleotide-free HpUreG (and probably used to bind Ni when GTP is provided). In addition, the presence of Strep-tag II is shown to profoundly affect the Ni coordination geometry when compared to that of wild-type UreG.
Materials and methods
Preparation of protein samples
KaUreG, KaUreGStr (containing Strep-tag II fused at the C-terminus), C72A KaUreGStr, and H74A KaUreGStr were purified as previously described [5, 6]. A construct for synthesizing H6A KaUreGStr was created using the same approach as for the C72A and H74A variants; however, the H6A protein was insoluble for all conditions tested. Iodoacetamide-treated KaUreG and KaUreGStr were prepared by incubating 30 µM protein with 20-fold excess of iodoacetamide for 1 h in 50 mM Tris-HCl (pH 8.0), quenching with β-mercaptoethanol, and dialyzing into 50 mM HEPES (pH 7.4) containing 200 mM NaCl. For XAS analysis, proteins were first incubated in 50 mM HEPES (pH 7.4) buffer containing 200 mM NaCl, 1 mM EDTA, and 1 mM dithiothreitol, dialyzed into the same buffer lacking EDTA and dithiothreitol, exchanged into the same buffer containing 200 µM NiCl2, and concentrated to 0.5 mL. The final protein concentrations ranged from 0.885 mM to 1.9 mM, with substoichiometric levels of nickel (0.77 to 1.8 mM) in all cases. These samples were desalted by Sephadex G25 chromatography in the same buffer containing 25% glycerol and concentrated by use of Amicon centrifugal filters (10,000 MWCO).
Equilibrium dialysis analysis
Ni binding interactions with iodoacetamide-treated KaUreG and KaUreGStr samples were assessed by using an equilibrium microvolume dialyzer (Hoefer Scientific Instruments). Purified protein (400 µL of 10 µM) was dialyzed against 400 µL of various concentrations of NiCl2 overnight at 4 °C by using a 3500 Da molecular mass cutoff membrane (Spectra-Por). Ni concentrations on both sides of the membrane were determined by adding 100 µL of these solutions to 900 µL of 100 µM 4-(2-pyridylazo)resorcinol made in 50 mM HEPES (pH 7.4) with 200 mM NaCl, incubating for 10 min, and monitoring the absorbance at 500 nm [17]. The data were plotted and analyzed in Sigma Plot (Systat Software, Inc.) according to the following equation:
where Y is the number of Ni bound per UreG, Bmax is the maximum number of Ni bound per UreG, [Ni]f is the concentration of free Ni, and Kd is the dissociation constant.
XAS data collection and analysis
Data were collected at the Brookhaven National Laboratory – National Synchrotron Light Source (BNL-NSLS, 2.8 GeV ring) on beam line X3B equipped with a Canberra 31 element Germanium detector and a helium displex cryostat attached to a vacuum chamber allowing the samples to be run at ~50 K. A Si(111) crystal monochromator was used and a 3 µm Z-1 element filter was placed between the sample and the detector to reduce scattering. Internal energy calibration was performed by collecting spectra simultaneously in transition mode on a Ni metal foil. Samples of frozen protein solutions (0.885–1.9 mM protein containing 0.5–0.95 equivalents of Ni) were placed in polycarbonate holders. X-ray absorption near-edge spectroscopy (XANES) data were collected from ±200 eV relative to the nickel K-edge.
Data analysis was performed as previously described [18, 19] by using the SixPack software package [20] and the Horae (Athena, Artemis) software package [21]. Data were loaded, averaged, background corrected and calibrated in Athena or Sixpack with the Ni-K-edge calibration energy set to 8331.6 eV. Extended X-ray absorption fine structure (EXAFS) data were extracted by using an Rbkg of 1 and a spline series from k = 2 to 12.5 Å−1 with a strong clamp at high k-values. The k3-weighted data were fit in R-space over the k = 2 to 12.5 Å−1 region with E° for nickel set to 8340 eV. All data sets were processed using a Kaiser-Bessel window with a dk = 1 (window sill).
Both Artemis and SixPack fitting software packages were built on the IFEFFIT engine and use iterative FEFF calculations to fit EXAFS data during model refinement [22]. Single-scatter and multiple-scatter fits were performed and fit using the general EXAFS equation below:
Average values and bond lengths obtained from crystallographic data were used to construct rigid imidazole rings (imid) to fit His residues [23]. The distance of the imidazole rings from the metal center was fit in terms of the metal-ligand bond distance (Reff) and a rotation angle α, around an axis perpendicular to the plane of the ring and going through the coordinating nitrogen [18, 24].
To assess the goodness of fit from different fitting models, the fit parameters χ2, reduced χ2(χv2), and the Rfactor were minimized. Increasing the number of adjustable parameters is generally expected to improve the Rfactor, however χv2 (equivalent to χ2/(Nidp − Nvar) where Nvar is the number of refining parameters) may go through a minimum then increase indicating the model is over-fitting the data. The parameters are defined as follows [25]:
where Nidp is the number of independent data points defined as ; Δr is the fitting range in r-space; Δk is the fitting range in k-space; Npts is the number of points in the fitting range; ε is the measurement of uncertainty; Re() is the real part of the EXAFS Fourier transformed data and theory functions; Im() is the imaginary part of the EXAFS Fourier transformed data and theory functions; χ(Ri) is the Fourier transformed data or theory function; and
RESULTS
Ni binding to nucleotide-free UreG from K. aerogenes is unaffected by Cys chemical modification
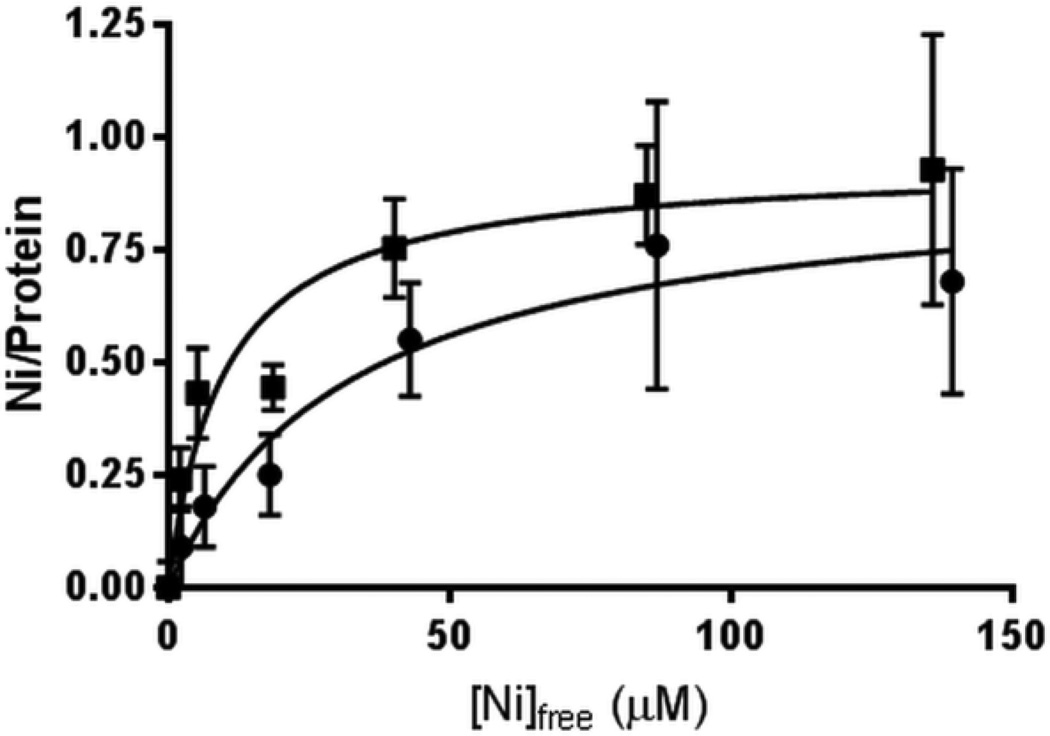
Ni and Zn were previously shown to compete for binding to a single metal-binding site in monomeric KaUreGStr [6]. As one approach to test whether a Cys residue coordinates the metal in the nucleotide-free protein, equilibrium dialysis studies were carried out for Ni using KaUreG and KaUreGStr samples for which the available surface thiol groups were modified by reaction with iodoacetamide (Figure 1). The chemically-derivatized samples bound 0.92 ± 0.11 Ni and 0.94 ± 0.09 Ni, respectively, equivalent to the untreated samples (Table 1). The modified samples provided Ni Kd values of 32 ± 11.6 µM and 9.7 ± 4.0 µM, respectively, each exhibiting about twice the values reported for untreated samples [6]. These small differences between alkylated and untreated samples are consistent with the metal binding site of KaUreG lacking thiol ligation. Further support for this conclusion is available from prior equilibrium dialysis results of variants affecting the two Cys residues in KaUreGStr; the C28A and C72A variants bound near-stoichiometric amounts of Ni although with some perturbation of the Kd [6]. The iodoacetamide-treated KaUreG and KaUreGStr samples also were examined for their interactions with Zn, but precipitation proved to be a problem with this metal (data not shown) and the site of Zn(II) binding to the alkylated protein is not established.
Fig. 1.
Equilibrium dialysis analysis of Ni binding to iodoacetamide-treated KaUreG (●) and KaUreGStr (■).
XANES Analysis
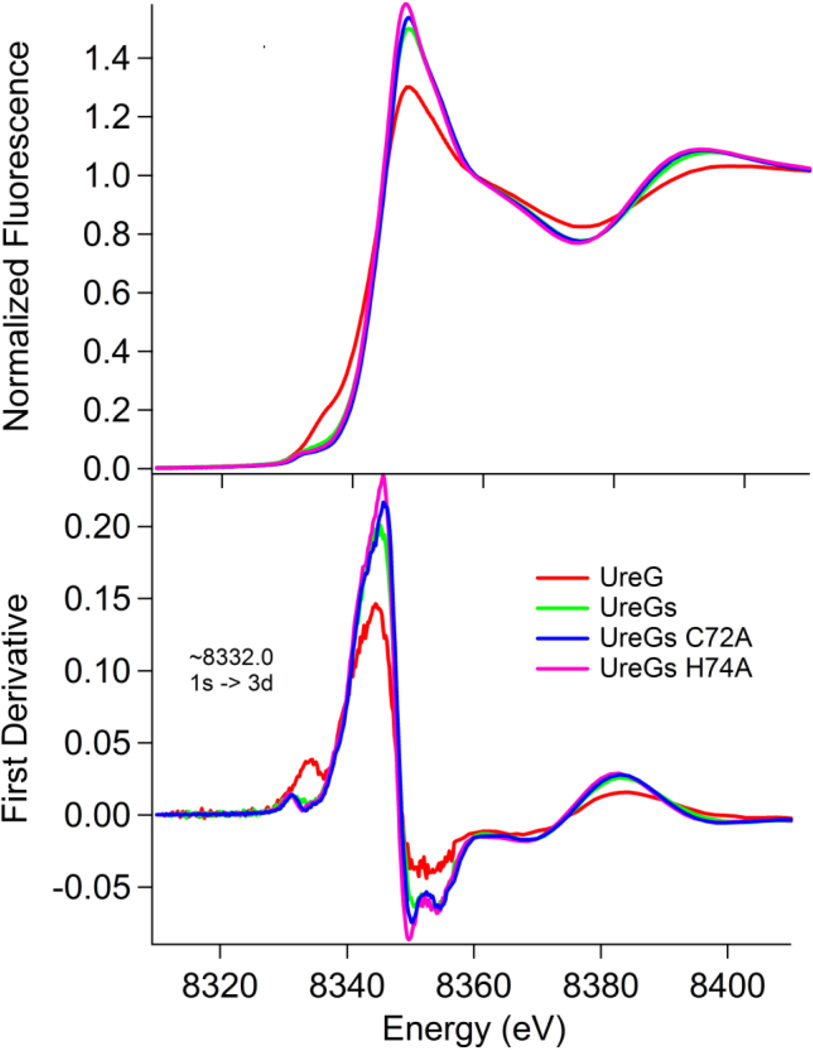
XAS was used to directly examine the Ni coordination environment of KaUreG, KaUreGStr, C72A KaUreGStr, and H74A KaUreGStr. The XAS spectrum of KaUreG exhibits a well-resolved shoulder in the pre-edge region centered around 8334.8 eV consistent with the presence of 1s → 4p transitions and shake down contributions. First derivative analysis of this region coupled with peak fitting (Supplementary material Figure S1) reveal an additional feature at ~8332 eV from 1s → 3d transitions having an intensity of 8.9 normalized units (Figure 2). The intensity of the 1s → 3d feature and the profile of the rising edge are consistent with either a five-coordinate trigonal pyramidal or a four-coordinate tetrahedral environment, the latter being consistent with EXAFS analysis (vide infra) [26]. In contrast, the pre-edge features of the three Strep-tag II species are distinct from that of KaUreG, having closely overlapping spectra indicating very similar coordination geometries and ligand environments, with weak transitions at ~8332 eV from the dipole forbidden 1s → 3d transition. The weak intensity of these features (Table 2) are consistent with a centrosymmetric arrangement of ligands, and the absence of a peak near 8338 eV, diagnostic of four-coordinate planar coordination, indicates a six-coordinate nickel center for these species [26]. Unfortunately, the Zn coordination could not be examined due to protein precipitation problems.
Fig. 2.
Ni K-edge XANES spectra of KaUreG (red), KaUreGStr (abbreviated UreGs; green), C72A KaUreGStr (blue) and H74A KaUreGStr (pink).
Table 2.
Ni K-edge XANES analysisa
| Sample | 1s → 3d Peak Area (×102 eV) |
Coordination Number |
|---|---|---|
| KaUreG | 8.9(1) | 4 |
| KaUreGStr | 3.9(1) | 6 |
| C72A KaUreGStr | 4.6(1) | 6 |
| H74A KaUreGStr | 4.8(1) | 6 |
1s→3d pre-edges were fit with pseudo-Voight peak functions having 75% Gaussian character.
EXAFS Analysis
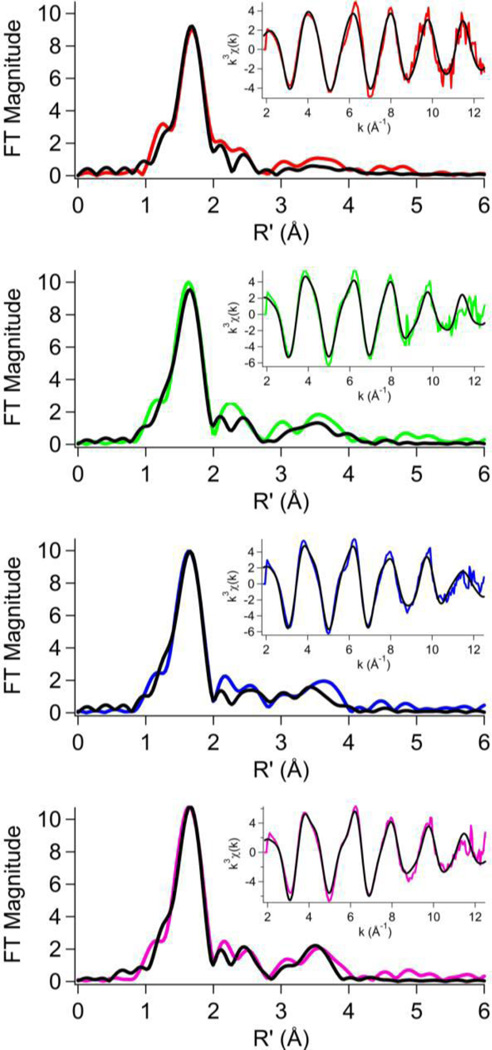
The EXAFS region of the XAS spectrum provides insightful information regarding the metal center’s coordination environment, such as the number of donor atoms, the types of donor atoms, and the bonding distance. The Fourier-transformed spectrum of KaUreG exhibited an intense feature at ~1.7 Å with some additional intensity at 2.0 – 4.0 Å in R-space (uncorrected for phase shifts, Figure 3). Best fit analysis indicated solely O/N donor ligands that included a single imidazole in a four coordinate model (Table 3, Supplementary material Tables S1–S4), consistent with the XANES analysis. The spectra obtained for the Strep-tag II series of KaUreGStr, C72A KaUreGStr, and H74A KaUreGStr proteins are very similar with an intense feature at ~1.7 Å and relatively greater density in the 2.0 – 4.0 Å range of R-space (Figure 3). The best fits indicate exclusively O/N donors with the presence of multiple imidazole scatterers, accounting for the additional intensity in the 2.0–4.0 range of R-space, in six coordinate models (Table 3), consistent with the octahedral environment suggested by XANES analysis.
Fig. 3.
Fourier-transformed extended X-ray absorption fine structure spectra (colored lines) and fits (black lines) from Table 3 for KaUreG (red), KaUreGStr (green), C72A KaUreGStr (blue) and H74A KaUreGStr (pink). Insets: k3-Weighted unfiltered EXAFS spectra and fits.
Table 3.
Ni K-edge EXAFS Analysis best fits
| Sample | N | R (Å) |
σ2 (× 103 Å2) |
ΔE° (eV) |
Rfactor |
|---|---|---|---|---|---|
| KaUreG | 3 N/O 1 imid |
2.08(1) 2.08(1) |
2.4(1) 2.4(2) |
4(1) | 0.051 |
| KaUreGStr | 3 N/Oa 3 imida |
2.09(1) 2.09(1) |
5.1(1) 5.1(1) |
2(1) | 0.061 |
| C72A KaUreGStr |
4 N/O 2 imid |
2.11(1) 2.06(2) |
5(1) 4(2) |
2(1) | 0.042 |
| H74A KaUreGStr |
4 N/O 2 imid |
2.10(1) 2.06(2) |
4(1) 4(2) |
2(1) | 0.048 |
Two imid and 4 N/O cannot be excluded for this sample.
Discussion
Nucleotide-free UreG from K. aerogenes binds competitively one Zn or Ni per monomer with similar affinities [6], whereas homologs isolated from other species and lacking nucleotides exhibit a clear preference for Zn over Ni (Table 1). The Zn site is best characterized for HpUreG where XAS studies demonstrated coordination by two Cys and two His [15], and structural models show close juxtaposition of two Cys-Pro-His motifs in the dimer [10, 16]. This motif is present in all UreG proteins; however, one would expect metal coordination by a single copy of this motif to be greatly attenuated in monomeric KaUreG. We demonstrate that nucleotide-free KaUreG uses a distinct site to bind Ni and, by implication [6], Zn.
Thiolate ligation of Ni is absent in KaUreG according to both XAS analysis and the near lack of effect on Ni binding during equilibrium dialysis studies when using samples treated with iodoacetamide, a Cys-specific chemical reagent. A prior study by us had noted the generation of a thiolate-to-Ni ligand-to-metal charge-transfer (LMCT) absorption band in KaUreGStr at high concentrations of Ni whereas this transition was absent in C72A KaUreGStr, leading to the suggestion that Cys72 was a Ni ligand [6]; however, we wish to emphasize that the concentration dependence of LMCT formation was inconsistent with the measured Ni binding thermodynamics. For example, equimolar levels of Ni and protein (58 µM) led to only very small increases in the absorption at 330 nm, with the difference spectra continuing to increase up to the largest amount of Ni added (300 µM). We conclude that the spectral changes reflect very weak binding to a second site, likely involving the Cys-Pro-His motif. The C72A variant of KaUreGStr was similar to the non-variant KaUreGStr in binding stoichiometric amounts of Ni, although the substituted protein exhibited a 12-fold increase in Kd for the binding of Ni [6], likely attributed to conformational changes. In conclusion, the metal-binding site of nucleotide-free KaUreG is distinct from the reported Zn-binding site of HpUreG.
Wild-type KaUreG binds Ni in a site with tetrahedral geometry using all nitrogen/oxygen ligands including one imidazole, whereas each of the Strep-tag II species binds Ni in a six-coordinate site that includes two or three imidazole ligands. Significantly, KaUreGStr and the C72A and H74A KaUreGStr variants yielded equivalent EXAFS fits, demonstrating that His74 is not a ligand to Ni in these proteins. The only other His residue in the wild-type protein is His6, which must coordinate the metal. We were unable to examine the Ni binding properties of H6A KaUreGStr because the variant protein was insoluble. HpUreG, MjUreG, and MsUreG do not possess a His residue at this position, but SpUreG possesses a His residue in the same region and GmUreG possesses an N-terminal His-rich extension. In addition to the likely metal coordination by the His6 residue in all KaUreG samples, the Strep-tag II (Trp-Ser-His-Pro-Gln-Phe-Glu-Lys) includes a His residue that we propose also participates in Ni binding. A homology model of KaUreG (Supplementary material Figure S2) positions His6 and His74 with a separation of more than 20 Å, so simultaneous coordination of the same Ni by both of these residues would require a large conformational change that is unlikely. By contrast, the very flexible C-terminal region containing the fusion tag is very likely to be positioned near His6 allowing for their joint participation in a Ni binding site. The presence of one His ligand in non-tagged samples and two His ligands in tagged samples is congruent with the observed decrease in Kd for KaUreGStr and iodoacetamide-treated KaUreGStr compared to their non-tagged counterparts [6]. This finding highlights the non-innocent nature of the Strep-tag II with regard to Ni binding, even though it often is used in preference to His tags to avoid interaction with metals. Of interest, the change in metal coordination geometry for tagged and non-tagged forms of the protein does not affect its function in urease activation [6].
Supplementary Material
Acknowledgments
These studies were supported by the National Institutes of Health (Grant DK045686, RPH, and GM069696, MJM). XAS data collection at the National Synchrotron Light Source at Brookhaven National Laboratory was supported by the U.S. Department of Energy, Division of Materials Science and Division of Chemical Sciences. Beamline X3B at NSLS was supported by the NIH Grant P30-EB-009998 from the National Institute of Biomedical Imaging and Bioengineering.
Abbreviations
- EXAFS
extended x-ray absorption fine structure
- LMCT
ligand-to-metal charge-transfer
- XANES
x-ray absorption near edge spectroscopy
- XAS
x-ray absorption spectroscopy
References
- 1.Zambelli B, Musiani F, Benini S, Ciurli S. Acc Chem Res. 2011;44:520–530. doi: 10.1021/ar200041k. [DOI] [PubMed] [Google Scholar]
- 2.Farrugia MA, Macomber L, Hausinger RP. J Biol Chem. 2013;288:13178–13185. doi: 10.1074/jbc.R112.446526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higgins KA, Carr CE, Maroney MJ. Biochemistry. 2012;51:7816–7832. doi: 10.1021/bi300981m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farrugia MA, Wang B, Feig M, Hausinger RP. Biochemistry. 2015;54:6392–6401. doi: 10.1021/acs.biochem.5b00942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moncrief MBC, Hausinger RP. J Bacteriol. 1997;179:4081–4086. doi: 10.1128/jb.179.13.4081-4086.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boer JL, Quiroz-Valenzuela S, Anderson KL, Hausinger RP. Biochemistry. 2010;49:5859–5869. doi: 10.1021/bi1004987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zambelli B, Stola M, Musiani F, De Vriendt K, Samyn B, Devreese B, Van Beeumen J, Dikiy A, Bryant DA, Ciurli S. J Biol Chem. 2005;280:4684–4695. doi: 10.1074/jbc.M408483200. [DOI] [PubMed] [Google Scholar]
- 8.Zambelli B, Musiani F, Savini M, Tucker P, Ciurli S. Biochemistry. 2007;46:3171–3182. doi: 10.1021/bi6024676. [DOI] [PubMed] [Google Scholar]
- 9.D'Urzo A, Santambrogio C, Grandori R, Ciurli S, Zambelli B. J Biol Inorg Chem. 2014;19:1341–1354. doi: 10.1007/s00775-014-1191-9. [DOI] [PubMed] [Google Scholar]
- 10.Zambelli B, Turano P, Musiani F, Neyroz P, Ciurli S. Proteins. 2009;74:222–239. doi: 10.1002/prot.22205. [DOI] [PubMed] [Google Scholar]
- 11.Miraula M, Ciurli S, Zambelli B. J Biol Inorg Chem. 2015;20:739–755. doi: 10.1007/s00775-015-1261-7. [DOI] [PubMed] [Google Scholar]
- 12.Real-Guerra R, Staniscuaski F, Zambelli B, Musiani F, Ciurli S, Carlini CR. Plant Molec Biol. 2012;78:461–475. doi: 10.1007/s11103-012-9878-1. [DOI] [PubMed] [Google Scholar]
- 13.Fong YH, Wong HC, Yuen MH, Lau PH, Chen YW, Wong K-B. PLoS Biol. 2013;11:e1001678. doi: 10.1371/journal.pbio.1001678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X, Li H, Lai T-P, Sun H. J Biol Chem. 2015;290:12474–12485. doi: 10.1074/jbc.M114.632364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin-Diaconescu V, Bellucci M, Musiani F, Ciurli S, Maroney MJ. J Biol Inorg Chem. 2011;17:353–361. doi: 10.1007/s00775-011-0857-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bellucci M, Zambelli B, Musiani F, Turano P, Ciurli S. Biochem J. 2009;422:91–100. doi: 10.1042/BJ20090434. [DOI] [PubMed] [Google Scholar]
- 17.Hunt JB, Neece SH, Ginsburg A. Anal Biochem. 1985;146:150–157. doi: 10.1016/0003-2697(85)90409-9. [DOI] [PubMed] [Google Scholar]
- 18.Banaszak K, Martin-Diaconescu V, Bellucci M, Zambelli B, Rypniewski W, Maroney MJ, Ciurli S. Biochem J. 2012;441:1017–1026. doi: 10.1042/BJ20111659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryan K, Johnson O, Cabelli DE, Brunold T, Maroney MJ. J Biol Inorg Chem. 2010;15:795–807. doi: 10.1007/s00775-010-0645-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webb SM. Physical Scripta. 2005;T115:1011–1014. [Google Scholar]
- 21.Ravel B, Newville M. J Synchrotron Radiat. 2005;12:537–541. doi: 10.1107/S0909049505012719. [DOI] [PubMed] [Google Scholar]
- 22.Newville M. J Synchrotron Radiat. 2001;8:322–324. doi: 10.1107/s0909049500016964. [DOI] [PubMed] [Google Scholar]
- 23.Engh RA, Huber R. Acta Crystallogr. 1991;A47:392–400. [Google Scholar]
- 24.Blackburn NJ, Hasnain SS, Pettingill TM, Strange rW. J Biol Chem. 1991;266:23120–23127. [PubMed] [Google Scholar]
- 25.Ferreira GC, Franco R, Mangravita A, George GN. Biochemistry. 2002;41:4809–4818. doi: 10.1021/bi015814m. [DOI] [PubMed] [Google Scholar]
- 26.Colpas GJ, Maroney MJ, Bagyinka C, Kumar M, Willis WS, Suib SL, Baidya N, Mascharak PK. Inorg Chem. 1991;30:920–928. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.