Abstract
Recently, transfer learning has been successfully applied in early diagnosis of Alzheimer’s Disease (AD) based on multi-domain data. However, most of existing methods only use data from a single auxiliary domain, and thus cannot utilize the intrinsic useful correlation information from multiple domains. Accordingly, in this paper, we consider the joint learning of tasks in multi-auxiliary domains and the target domain, and propose a novel Multi-Domain Transfer Learning (MDTL) framework for early diagnosis of AD. Specifically, the proposed MDTL framework consists of two key components: 1) a multi-domain transfer feature selection (MDTFS) model that selects the most informative feature subset from multi-domain data, and 2) a multidomain transfer classification (MDTC) model that can identify disease status for early AD detection. We evaluate our method on 807 subjects from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database using baseline magnetic resonance imaging (MRI) data. The experimental results show that the proposed MDTL method can effectively utilize multi-auxiliary domain data for improving the learning performance in the target domain, compared with several state-of-the-art methods.
Keywords: Transfer learning, Multi-domain, Alzheimer’s disease (AD), Feature selection
Introduction
Alzheimer’s Disease (AD) is characterized by the progressive impairment of neurons and their connections, which leads to the loss of cognitive function and the ultimate death. It is reported that an estimated 700,000 elderly Americans will die with AD, and many of them will die from complications caused by AD in 2014 (Association, A.s 2014). Mild cognitive impairment (MCI) is a prodromal stage of AD, where some MCI patients will convert to AD over time, i.e., progressive MCI (pMCI), and other MCI patients remain stable, i.e., stable MCI (sMCI). Thus, for timely therapy that might be effective to slow the disease progression, it is important for early diagnosis of AD and its early stage (i.e., MCI). For the last decades, neuroimaging has been successfully used to investigate the characteristics of neurodegenerative progression in the spectrum between AD and normal controls (NC). In recent years, magnetic resonance imaging (MRI) data are widely applied to early diagnosis of AD, which can measure the structural brain atrophy (Fan et al. 2008; Misra et al. 2009; Risacher et al. 2009). For instance, several studies have shown that AD patients exhibited significant decrease of gray matter volume (Chao et al. 2010; Chetelat et al. 2005; Guo et al. 2010).
Recently, many machine learning methods based on MRI biomarkers have been used for early diagnosis of AD (Cho et al. 2012; Coupé et al. 2012; Cuingnet et al. 2011; Eskildsen et al. 2013; Gaser et al. 2013; Li et al. 2014; Liu et al. 2014; Liu et al. 2016a, b; Ota et al. 2014; Wee et al. 2013; Westman et al. 2013; Westman et al. 2012; Zhang et al. 2016). According to the point of view in the machine learning field, the number of training samples available to build a generalized model is often overwhelmed by the feature dimensionality. In other words, the number of training samples is usually very limited, while the feature dimensionality is very high. This so-called small-sample-size problem has been one of the main challenges in neuroimaging data analysis, which may lead to over-fitting issue (Zhu et al. 2012). To overcome the small-sample-size problem, feature selection has been commonly used in many neuroimaging based studies (Cheng et al. 2015b; Eskildsen et al. 2013; Jie et al. 2015; Liu et al. 2014; Ye et al. 2012; Zhu et al. 2014), where various feature selection methods have been developed to select informative feature subset for reducing the feature dimensionality. Especially, in neuroimaging data analysis for disease diagnosis and therapy, features may be corresponding to brain regions. In such a case, feature selection can detect the regions with brain atrophy, thus potentially useful for timely therapy of brain diseases.
Besides feature selection, many studies have used multimodal data to improve classification performance (Jie et al. 2015; Liu et al. 2014; Ye et al. 2012; Zhang et al. 2012; Zhu et al. 2014). For example, to enhance the generalization of classifiers, some studies have used multi-task learning for multimodal feature selection (Jie et al. 2015; Liu et al. 2014; Zhang et al. 2012; Zhu et al. 2014). In all these studies using multimodal data, different biomarkers are regarded as different modalities, and each modality data is regarded as a learning task (Jie et al. 2015; Liu et al. 2014). On the other hand, several studies have considered each learning approach as a learning task (Zhang et al. 2012; Zhu et al. 2014). All these studies suggest that the use of multimodal data for multi-task learning of features can significantly improve classification performance and enhance generalization performance of classifiers. However, in the clinical practice of AD/MCI diagnosis, the collection of complete multimodal biomarkers from each subject is expensive and time-consuming; on the other hand, it is relatively easy to get single modality data (e.g., MRI) for different categories of subjects. Therefore, in this paper, we address the latter case to build respective classification models for early diagnosis of AD.
According to the pathology of AD, it is the progressive impairment of neurons, and MCI and advanced AD are thus highly related. In this way, several studies suggested that the learning domain of AD diagnosis is related to the learning domain of MCI diagnosis (Cheng et al. 2015b; Coupé et al. 2012; Da et al. 2014; Filipovych et al. 2011; Westman et al. 2013; Young et al. 2013). Also in machine learning community, transfer learning aims to extract the knowledge from one or more auxiliary domains and applies the extracted knowledge to a target domain (Duan et al. 2012; Pan and Yang 2010; Yang et al. 2007), where the auxiliary domain is assumed to be related to the target domain. However, in recent years, several transfer learning methods were developed just for AD/MCI diagnosis (Cheng et al. 2015a; Cheng et al. 2015b; Filipovych et al. 2011; Schwartz et al. 2012; Young et al. 2013). Although these studies suggested that the data from the auxiliary domain can improve the classification performance of target domain, the training data are often from just a single auxiliary domain. Actually, there are multiple auxiliary domain data that can be available in clinical practice. According to the principle of transfer learning, the application of multiple auxiliary domain data could further promote the performance of the target domain.
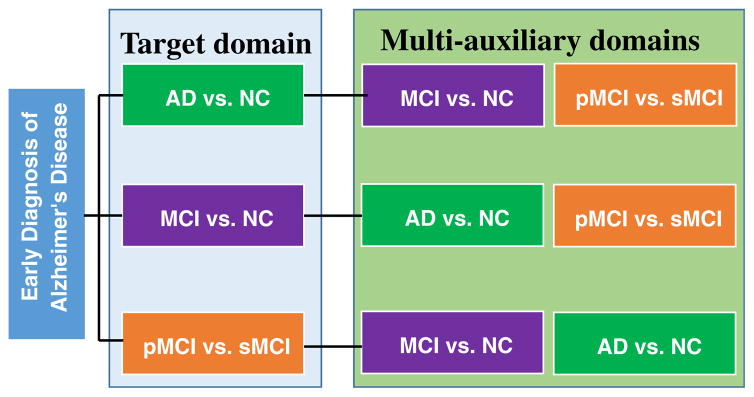
In addition, in our previous works (Cheng et al. 2015a; Cheng et al. 2015b; Cheng et al. 2012), we mainly consider the prediction of MCI conversion based on a single auxiliary domain data, to construct the respective transfer learning model. Although in our work (Cheng et al. 2015b) we proposed a domain transfer learning method for classification groups such as MCI vs. NC and MCI vs. AD, our proposed method still cannot acquire the deep structured information between the target domain and the auxiliary domain. Furthermore, few studies considered the heterogeneity of MCI to construct semi-supervised classification or regression models (where MCI subjects are regarded as unlabeled samples), which shows that using information of MCI diagnosis can help improve performance of classifying or estimating AD patients from NCs (Cheng et al. 2013; Filipovych et al. 2011; Zhang and Shen 2011). Inspired by the aforementioned issues and successes, in this paper, we assume that there is underlying relationship between each binary classification problem in the early diagnosis of AD, where each binary classification problem can be regarded as target domain, with the other binary classification problems as auxiliary domains. In Fig. 1, we illustrate this novel description of relationships between target domain and corresponding multi-auxiliary domains for early diagnosis of AD. Then, those single modal data that contain multiple data categories can be regarded as multiple related-learning-domains.
Fig. 1.
Our proposed relationships between target domain and auxiliary domains
In particular, we develop a novel multi-domain transfer learning (MDTL) method for early diagnosis of AD, where training data from multiple auxiliary domains are jointly learned with the target domain. Specifically, we first develop a multidomain transfer feature selection (MDTFS) model by using the training data from multiple auxiliary domains and target domain to select a subset of discriminative features. Then, we build a multi-domain transfer classifier (MDTC) that can conjointly apply the training data from multi-auxiliary domains and target domain to construct the classifier. The proposed method is evaluated on the baseline Alzheimer’s Disease Neuroimaging Initiative (ADNI) database of 807 subjects with MRI data. The experimental results demonstrate that the proposed method can further improve the performance of early diagnosis of AD, compared with several state-of-the-art methods.
Materials
ADNI Database
The data used in the preparation of this paper were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (http://adni.loni.usc.edu/). ADNI researchers collect, validate and utilize data such as MRI and positron emission tomography (PET) images, genetics, cognitive tests, cerebrospinal fluid (CSF) and blood biomarkers as predictors for Alzheimer’s disease. Data from the North American ADNI’s study participants, including Alzheimer’s disease patients, mild cognitive impairment subjects and elderly controls, are available in this database. In addition, the ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies, and non-profit organizations, as a $60 million, 5-year public-private partnership. The primary goal of ADNI has been to test whether the serial MRI, PET, other biological markers, and clinical and neuropsychological assessments can be combined to measure the progression of MCI and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials.
The ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 adults, aged 55 to 90, to participate in the research approximately 200 cognitively normal older individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years, and 200 people with early AD to be followed for 2 years (see www.adni-info.org for up-to-date information). The research protocol was approved by each local institutional review board, and the written informed consent is obtained from each participant.
Subjects
The ADNI general eligibility criteria are described at www.adniinfo.org. Briefly, subjects are between 55 and 90 years of age, having a study partner able to provide an independent evaluation of functioning. Specific psychoactive medications will be excluded. General inclusion/ exclusion criteria are as follows: 1) healthy subjects:MMSE scores between 24 and 30, a Clinical Dementia Rating (CDR) of 0, non-depressed, non-MCI, and non-demented; 2) MCI subjects: MMSE scores between 24 and 30, a memory complaint, having objective memory loss measured by education adjusted scores on Wechsler Memory Scale Logical Memory II, a CDR of 0.5, absence of significant levels of impairment in other cognitive domains, essentially preserved activities of daily living, and an absence of dementia. MCI is a prodromal stage of AD, where some MCI patients will convert to AD, i.e., progressive MCI (pMCI), and other MCI patients remain stable, i.e., stable MCI (sMCI); and 3) Mild AD: MMSE scores between 20 and 26, CDR of 0.5 or 1.0, and meets the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s Disease and Related Disorders Association (NINCDS/ADRDA) criteria for probable AD.
In this work, we focus on using the baseline ADNI database with MRI data. Specifically, the structural MR scans were acquired from 1.5 Tscanners. We downloaded raw Digital Imaging and Communications in Medicine (DICOM) MRI scans from the public ADNI website (www.loni.ucla.edu/ADNI), reviewed for quality, and corrected spatial distortion caused by gradient nonlinearity and B1 field inhomogeneity. More detailed description can be found in (Zhang et al. 2011).
Method
In this section, we first briefly introduce our proposed learning method, and then present our proposed multi-modal transfer feature selection (MDTFS) model, as well as an optimization algorithm for solving the proposed objective function. Finally, we elaborate the proposed multi-domain transfer classification (MDTC) model.
Overview
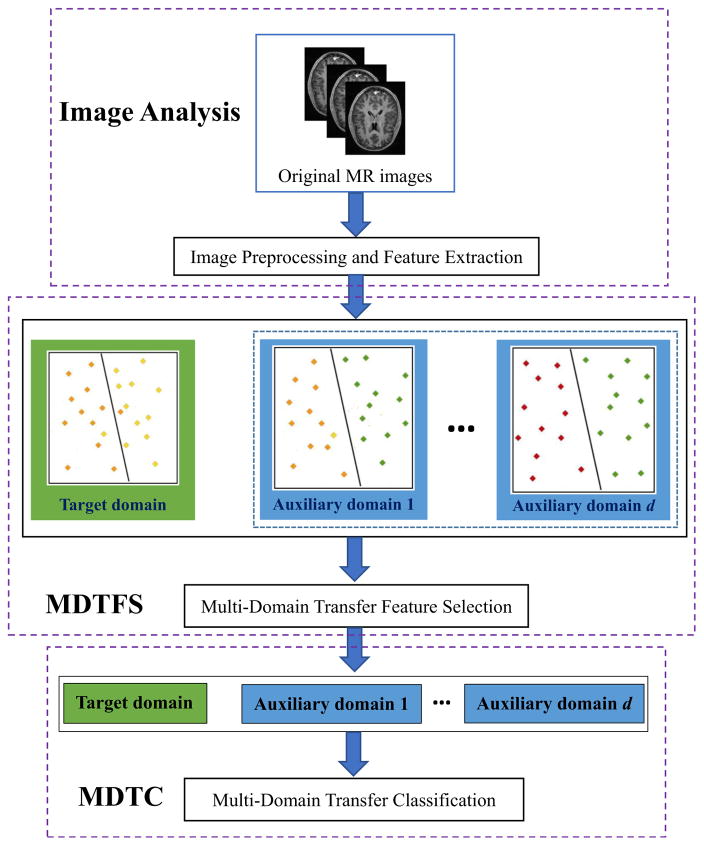
In Fig. 2, we illustrate the proposed framework for early diagnosis of AD. Specifically, our framework consists of three main components, i.e., (1) image pre-processing and feature extraction, (2) multi-domain transfer feature selection (MDTFS), and (3) multi-domain transfer classification (MDTC). As shown in Fig. 2, we first pre-process all MR images, and extract features from MR images as described in the Image Preprocessing and Feature Extraction section below. Then, we select informative features via the proposed MDTFS method. We finally build a multi-domain transfer classifier using both the target domain and multi-auxiliary domains data for the classification of AD and MCI.
Fig. 2.
Summary of our proposed framework for early diagnosis of AD using multi-domain transfer learning (MDTL) method
Image Preprocessing and Feature Extraction
All MR images were pre-processed by first performing an anterior commissure-posterior commissure (AC-PC) correction using the MIPAV software (CIT 2012). The AC-PC corrected images were resampled to 256 × 256 × 256, and the N3 algorithm (Sled et al. 1998) was used to correct intensity inhomogeneity. Then, a skull stripping method (Wang et al. 2011) was performed, and the skull stripping results were manually reviewed to ensure cleaning of skull and dura. The cerebellum was removed by first registering the skull-stripped image to a manually-labeled cerebellum template, and then removing all voxels within the labeled cerebellum mask. FAST in FSL (Zhang et al. 2001) was then used to segment human brain into three different tissues: grey matter (GM), white matter (WM), and cerebrospinal fluid (CSF). We used HAMMER (Shen and Davatzikos 2002) for registration. After registration, each subject image was labeled using the Jacob template (Kabani et al. 1998) with 93 manually-labeled regions-of-interests (ROIs). Then, for each of 93 ROIs, we computed its GM tissue volume as a feature. As a result, for each subject, we have a 93-dimensional feature vector for representing it.
Multi-Domain Transfer Feature Selection (MDTFS)
Unlike previous methods that only considered a single auxiliary domain in model training, in this work, we use samples of target domain as well as multi-auxiliary domains to build a generalized model for feature selection. Hereafter, we denote D as the number of different domains with an index d ∈ {1, 2, ⋯, D} throughout the whole paper. Assume that we have one target domain T, with NT samples xT,i and the class labels yT,i, denoted as , where xT, i ∈ RF is the i-th sample with F features, and yT, i ∈ {+1, −1} is its corresponding class label. Also, assume that we have D − 1 auxiliary domains A, with samples and the class labels for each auxiliary domain, denoted as , where is the j-th sample with F features of the d-th auxiliary domain, and is the corresponding class label for the d-th auxiliary domain. Therefore, by adding up one target domain and D − 1 auxiliary domains, we have D domains in total.
In this work, we use a traditional multi-task feature selection method (Obozinski et al. 2006) to design our model for feature selection, and use all the available domain data from the multi-auxiliary domains as well as the target domain to build a more generalized model. Since they are related between the target domain and each auxiliary domain, we need to utilize the intrinsic useful correlation information from multi-auxiliary domain, and introduce an L2-norm regularizer based on weight vectors (i.e., ) for different learning domains, which can capture the correlation information between the target domain and multi-auxiliary domains. To learn the common subset of features from all domains (i.e., target domain and all auxiliary domains), we also introduce an L2/L1-norm regularizer (i.e., ) based on the weight matrix W, where wf is the f-th row vector of weight matrix W and is associated with the f-th feature weight across all domains). In addition, to keep the useful decision information of itself, we also use the ‘group sparsity’ regularization of weight matrix for all domains (i.e., ). Accordingly, the proposed multi-domain transfer feature selection (MDTFS) model H(W) can be written as follows:
| (1) |
where y(d) ∈ RN(d)×1 is the class label vector of the d-th domain (including target domain and all auxiliary domains), and x(d) ∈ RN(d)×F is the training dataset of the d-th domain. The ‘group sparsity’ regularizer matrix||W||1,1 (W ∈ RF×D) can select a discriminative subset of features relevant to self-domain, and can select a common subset of features relevant to all domains. The regularization term can control the similarity of multiple weight vectors between the target domain and each auxiliary domain, thus keeping each weight vector of auxiliary domain close to the target domain (Zhou et al. 2013). The column vector is the d-th auxiliary-domain weight vector, and the column vector wT is the target-domain weight vector. In addition, λ1, λ2, λ3 > 0 are the regularization parameters that control the relative contributions of the three regularization terms. By minimizing Eq. (1), we can learn a converged W from the target domain and multi-auxiliary domain. It is worth noting that, because of using ‘group sparsity’, the elements of the weight matrix W will be zero. For feature selection, we just keep those features with non-zero weights.
To solve the optimization problem of Eq. (1), we employ an accelerated gradient descent (AGD) method (Chen et al. 2009; Nemirovski 2005). To be specific, we decompose the objective function of H(W) in Eq. 1 into two parts, i.e., a smooth term S(W) and a non-smooth term G(W), as follows:
| (2) |
| (3) |
Then, we define the generalized gradient update rule as follows:
| (4) |
where ∇S(Wt) denotes the gradient of S(W) at the point Wt at the t-th iteration, h is a step size, 〈W − Wt, ∇S(Wt)〉 = tr((W−Wt) ∇S(Wt)) is the matrix inner product, ||·||F denotes a Frobenius norm for matrix, and tr(·) denotes a trace of a matrix. According to (Chen et al. 2009), the generalized gradient update rule of Eq. (4) can be further decomposed into N separate subproblems with a gradient mapping update approach. We summarize the details of AGD algorithm in Algorithm 1.
Algorithm 1.
AGD algorithm for MDTFS in Eq. (1)
| 1: | Initialization: h0 > 0, η > 1, W0 ∈ RF×D, W̄0 = W0, h=h0 and α0 = 1. |
| 2: | for t = 0,1,2, … until convergence of Wt do: |
| 3: | Set h = ht |
| 4: | while H(qh(W̄t)) > Qh(qh(W̄t), W̄t), h = ηh |
| 5: | Set ht+1 = h and compute |
| Wt+1 = arg minWQht+1(W, W̄t), | |
| , βt+1 = Wt+1 − Wt, | |
| end-while | |
| 6: | end-for |
Multi-Domain Transfer Classification (MDTC)
After performing MDTFS, we can obtain the most discriminative common features, upon which we will build a multidomain transfer classifier (MDTC) for final classification. Denote and as the new d-th auxiliary and target domains, with the corresponding labels denoted as and , respectively. Here, , xq∈RF̄, and F̄ denote the number of features for new d-th auxiliary and target domain after feature selection (via MDTFS). However, since we use the regularizer of ||W||1,1 in the MDTFS step, the selected features from each domain are different. For simplicity, we roughly select same feature subset F̄ for each auxiliary domain as the target domain.
Unlike our previous work (Cheng et al. 2015b) that only considered a single auxiliary domain in model training of classification. In this work, we will use multi-auxiliary domains for aiding the learning task of target domain. Due to the domain distribution relatedness between the target domain and each auxiliary domain, Yang et al. (Yang et al. 2007) consider that learning a multi-domain transfer classifier f(x) is to learn the “delta function Δf(x)” between the target and auxiliary classifiers using an objective function similar to SVMs. To combine multi-auxiliary domain classifiers , we construct an “ensemble” of auxiliary classifiers . Then, we employed the A-SVMs model of Yang et al. (Yang et al. 2007) to get the multi-domain transfer classifier, which has the following form:
| (5) |
where vd ∈ (0, 1) is the weight of each auxiliary classifier , which sums to one as . Also, Φ(x) is a kernel-based mapping function, and u is the weight vector of target domain classifier. In addition, u′ denotes the transpose of u.
To learn the weight vector u in Eq. 5, we use the following objective function, similar to the SVM (Yang et al. 2007):
| (6) |
where l is the l-th sample in the target domain training subset (xl, yl) ∈ XT, and βl is the slack variable that represents the prediction error of objective function of Eq. 6, thus it can be used for nonlinear classification. The parameter C balances contributions between auxiliary classifier and target-domain training samples. According to (Yang et al. 2007), we can solve this objective function in Eq. 6 to obtain the solution for the weight vector u. Then, we can obtain the final solution for f(x). In this study, is trained by SVM, and Δf(x) is solved by Eq.5 using the method of kernel learning.
Results
In this section, we first describe experimental settings in our experiments. Then, we show the classification results on the ADNI database by comparing our proposed method with several state-of-the-art methods. In addition, we illustrate the most discriminative brain regions identified by our proposed method.
Experimental Settings
We use the samples of 807 subjects (186 AD, 395 MCI, and 226 NC), for whom the baseline MRI data were all available. It is worth noting that, for all 395 MCI subjects, during the 24- month follow-up period, 167 MCI subjects converted to AD (pMCI for short) and 228 MCI subjects remained stable (sMCI for short). In addition, we consider three binary classification problems, i.e., AD vs. NC classification, MCI vs. NC classification, and pMCI vs. sMCI classification. For our proposed multi-modal transferring method, we explicitly list the target domain and the corresponding auxiliary domains for each classification task in Table 1.
Table 1.
Target domains and corresponding multiple auxiliary domains used in three binary classification tasks. The number in each bracket denotes the class label, where +1 denotes positive class and −1 represents negative class.
| Classification problem | Target domain | Auxiliary domain |
|---|---|---|
| AD vs. NC | AD(+1) vs. NC(−1) | 1) MCI(+1) vs. NC(−1), 2) pMCI(+1) vs. sMCI(−1) |
| MCI vs. NC | MCI(+1) vs. NC(−1) | 1) AD(+1) vs. NC(−1), 2) pMCI(+1) vs. sMCI(−1) |
| pMCI vs. sMCI | pMCI(+1) vs. sMCI(−1) | 1) AD(+1) vs. NC(−1), 2) MCI(+1) vs. NC(−1) |
In the experiments, we adopt a 10-fold cross-validation strategy to partition the target domain data into training and testing subsets. In particular, the target domain samples of each classification problem is partitioned into 10 subsets (each subset with a roughly equal size), and then one subset was successively selected as the testing samples and all the remaining subsets were used for training. To avoid the possible bias occurred during sample partitioning, we repeat this process 10 times. We report the average performances in terms of area under the receiver operating characteristic curve (AUC), accuracy (ACC), sensitivity (SEN), and specificity (SPE).
We compared the proposed method with a standard SVM (SVM for short), Lasso (Tibshirani 1996),MTFS (Zhang et al. 2012), and M2TFS (Jie et al. 2015). These methods are listed as follows.
SVM: training samples only from the target domain, and without any feature selection before classification stage;
Lasso: training samples only from the target domain, and the Lasso method conducted for feature selection before using SVM for classification;
MTFS and M2TFS: training samples from the target and multi-auxiliary domains, and the MTFS and M2TFS methods conducted for feature selection before using the selected classification method in the literatures (Jie et al. 2015; Zhang et al. 2012).
The SVM method is implemented using the LIBSVM toolbox (Chang and Lin 2001) with a linear kernel and a default value for the parameter C. Also, other competing methods with the SVM for classification are implemented using the LIBSVM toolbox, with the same settings of parameters as the SVM method. For the Lasso and MTFS methods, we adopt the SLEP toolbox (Liu et al. 2009) to solve the optimization problem. In addition, we employ the accelerated proximal gradient (APG) method in the literature (Chen et al. 2009) to solve the optimization problem of M2TFS. There are multiple regularization parameters of these methods (including Lasso, MTFS, M2TFS, and proposed MDTL) to be optimized. All regularization parameters of these methods are chosen from the range of P1 by a nested 10-fold cross-validation on the training data. In the proposed MDTL frame, the weight vd of auxiliary classifier for MDTC is learned within a nested 10-fold cross-validation via a grid search in the range of 0 and 1 at a step size of 0.1, and adopted the SVM based linear kernel for training the target-domain and auxiliary-domain classifiers. Before training models, we normalized features following (Zhang et al. 2011).
Comparison between MDTL and Other Methods
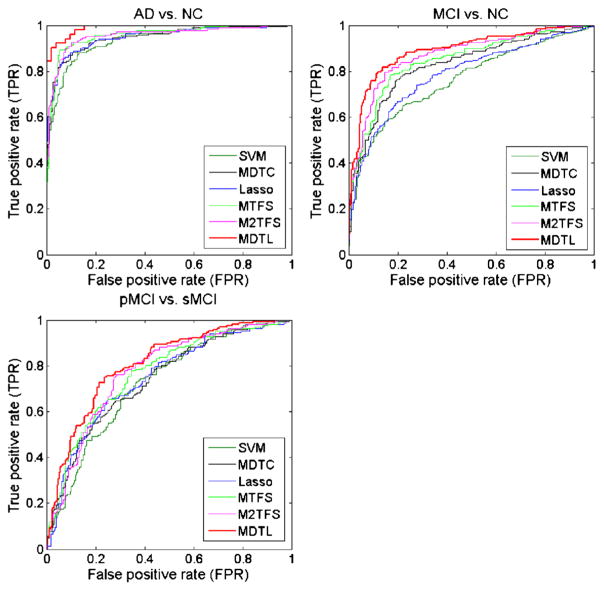
To investigate the effectiveness of the proposed method, we compare the proposed method with several state-of-the-art methods. Table 2 shows the classification results achieved by six methods, including SVM (traditional SVM), Lasso, MTFS (Zhang et al. 2012), M2TFS (Jie et al. 2015), and the proposed method (i.e., MDTL and MDTC). In Table 2, the proposed ‘MDTL’ method first performs the MDTFS for feature selection and then adopts MDTC for classification, while the ‘MDTC’ method performs only MDTC for classification. Also, note that each value in Table 2 is the averaged result of the 10-fold cross validation, which was performed for ten different times. In addition, we plot the ROC curves achieved by these six methods in Fig. 3.
Table 2.
Comparison of our proposed methods (MDTL and MDTC) and other 4 state-of-the-art methods (SVM, Lasso, MTFS and M2TFS) in three binary classification problems.
| AD vs. NC Classification | |||||
|---|---|---|---|---|---|
| Method | Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC | p-value |
| SVM | 85.8 | 84.6 | 85.9 | 0.933 | <0.0001 |
| MDTC | 88.4 | 87.2 | 88.5 | 0.950 | <0.0001 |
| Lasso | 87.9 | 87.8 | 88.1 | 0.951 | <0.0001 |
| MTFS | 90.7 | 89.5 | 90.8 | 0.966 | <0.001 |
| M2TFS | 91.5 | 91.4 | 91.6 | 0.979 | <0.005 |
| MDTL | 94.7 | 94.1 | 94.8 | 0.988 | - |
| MCI vs. NC Classification | |||||
| Method | Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC | p-value |
| SVM | 72.5 | 78.8 | 60.4 | 0.769 | <0.0001 |
| MDTC | 75.8 | 81.4 | 65.1 | 0.810 | <0.0001 |
| Lasso | 74.1 | 80.1 | 62.7 | 0.787 | <0.0001 |
| MTFS | 78.1 | 83.2 | 68.5 | 0.849 | <0.0005 |
| M2TFS | 78.6 | 83.6 | 69.1 | 0.870 | <0.005 |
| MDTL | 81.5 | 85.8 | 73.3 | 0.882 | - |
| pMCI vs. sMCI Classification | |||||
| Method | Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC | p-value |
| SVM | 67.1 | 61.0 | 71.5 | 0.728 | <0.0001 |
| MDTC | 69.2 | 63.5 | 73.3 | 0.742 | <0.0001 |
| Lasso | 69.4 | 63.8 | 73.5 | 0.744 | <0.0001 |
| MTFS | 71.0 | 62.7 | 74.9 | 0.757 | <0.0005 |
| M2TFS | 71.4 | 65.8 | 77.8 | 0.768 | <0.001 |
| MDTL | 73.8 | 69.0 | 77.4 | 0.796 | - |
Fig. 3.
ROC curves of different methods for three binary classification problems
As can be seen from Table 2 and Fig. 3, for three binary classification problems, the proposed MDTL method consistently outperforms SVM, Lasso, MDTC, MTFS and M2TFS in terms of the classification accuracy, sensitivity, and AUC measures. We also perform DeLong’s method (DeLong et al. 1988) on the AUC between the proposed method and each of other five competing methods, with the corresponding p-values shown in Table 2. The DeLong’s test is a nonparametric statistical test for comparing AUC between two ROC curves, which can be employed to assess statistical significance by computing z-scores for the AUC estimate (Robin et al. 2011; Sabuncu et al. 2015). For both AD vs. NC and MCI vs. NC classification tasks, the proposed MDTL method consistently outperforms the competing methods in all classification measures. In pMCI vs. sMCI classification, the proposed MDTL method outperforms the competing methods except for the specificity. Also, in Fig. 3, we can see from the ROC shown for pMCI vs. sMCI classification, which implies that the MDTL method can achieve better diagnostic performance in recognizing pMCI and sMCI patients than the competing methods. From the results in Table 2 and Fig. 3, it is clear that the proposed MDTL method can effectively integrate information of target domain and multi-auxiliary domains, which can achieve more significant performance improvement than the methods that use samples only from the target domain for training.
On the other hand, in Table 2 and Fig. 3, the proposed MDTC method consistently outperforms the SVM method in all classification measures for three binary classification problems. Also, there are slight differences of performance between the MDTC and Lasso method for three classification problems. These results imply that, compared with the case of only using SVM for performing classification, using MDTC can also improve the diagnostic performance, similar to the case of using the Lasso method for feature selection. We can see from Table 2 and Fig. 3, Lasso, MTFS, M2TFS, and MDTL methods also outperform the SVM method in all classification measures for three classification problems, which suggest that using feature selection on the high-dimensional features before performing classification can effectively improve the classification performance. In addition, from Table 2 and Fig. 3, MTFS, M2TFS, and MDTL methods can achieve better classification performance than the Lasso method, and the MDTL method also outperforms both the MTFS and M2TFS methods. These results also suggest that the inclusion of multi-auxiliary domains can improve the classification performance compared to the case of only using target domain, and that our proposed regularization factor based on multidomain weight vector is more suitable than the manifold regularization factor for the transfer learning problem.
In addition, there is an interesting observation from Table 2 and Fig. 3. Specifically, different from conventional studies (Cheng et al. 2012; Coupé et al. 2012; Da et al. 2014; Young et al. 2013), using pMCI and sMCI subjects as auxiliary domain can also help improve the performance of AD and NC classification. The main reason for this observation is that we proposed a regularizer (i.e., ) in step of MDTFS, which can use the weight vector from each of the multi-auxiliary domains to adjust the weight vector of target domain, and combine L2/L1-norm and L1/L1-norm regularizers to select features relevant to all domains (including self-domain), followed by using the MDTC based linear kernel SVM to keep these selected helpful features for classification. Furthermore, our proposed MDTFS model can also keep the target domain as the most important task in classification. Therefore, our proposed MDTL method can effectively use related multi-auxiliary domain data to improve the performance of target learning domain in early diagnosis of AD.
Comparison with MDTL and Other Variants
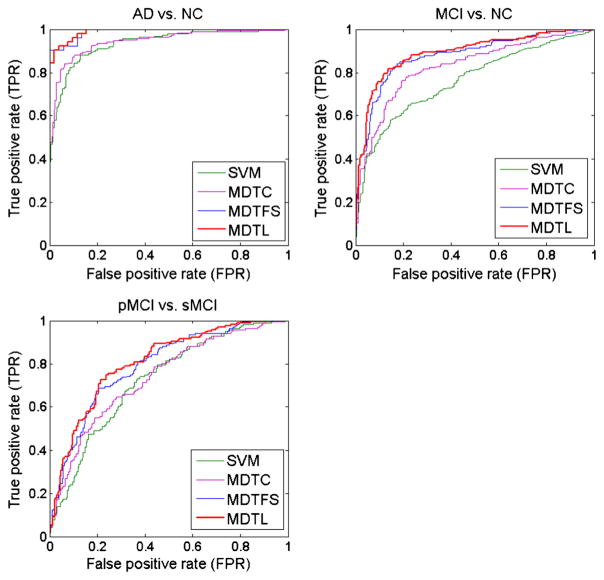
To investigate the relative contributions of the two components (i.e., MDTC and MDTFS) in our proposed method, we compare our method with its two variant methods. In Table 3, we give the classification measures by our proposed MDTL method, its variant methods (MDTC and MDTFS), and SVM (as a baseline method). Note that the proposed ‘MDTL’ method first performs feature selection using MDTFS model and then adopts MDTC for classification (i.e., MDTFS + MDTC, while the ‘MDTC’ method only performs classification using the proposed MDTC model. The ‘MDTFS’ method first performs feature selection using MDTFS model and then adopts SVM for classification. In Fig. 4, we also plot the ROC curves achieved by different methods. In addition, we also report the p-values, which are computed by DeLong’s method (DeLong et al. 1988) on the AUC between the proposed method and its two variant methods, as well as baseline method, in Table 3. From Table 3 to Fig. 4, we can observe that each component can boost the classification performance compared with SVM method. However, using feature selection method (i.e., MDTFS) can achieve better improvement than the MDTC method for classification. In general, our proposed MDTL method that integrates all the two components together achieves the best performance.
Table 3.
The comparison of our proposed methods (MDTL), its two variant methods (MDTC and MDTFS), and SVM(as a baseline method) in three binary classification problems.
| AD vs. NC Classification | |||||
|---|---|---|---|---|---|
| Method | Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC | p-value |
| SVM | 85.8 | 84.6 | 85.9 | 0.933 | <0.0001 |
| MDTC | 88.4 | 87.2 | 88.5 | 0.950 | <0.0001 |
| MDTFS | 93.4 | 93.3 | 93.5 | 0.982 | <0.05 |
| MDTL | 94.7 | 94.1 | 94.8 | 0.988 | - |
| MCI vs. NC Classification | |||||
| Method | Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC | p-value |
| SVM | 72.5 | 78.8 | 60.4 | 0.769 | <0.0001 |
| MDTC | 75.8 | 81.4 | 65.1 | 0.810 | <0.0001 |
| MDTFS | 80.2 | 84.8 | 71.4 | 0.876 | <0.05 |
| MDTL | 81.5 | 85.8 | 73.3 | 0.882 | - |
| pMCI vs. sMCI Classification | |||||
| Method | Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC | p-value |
| SVM | 67.1 | 61.0 | 71.5 | 0.728 | <0.0001 |
| MDTC | 69.2 | 63.5 | 73.3 | 0.742 | <0.0001 |
| MDTFS | 72.8 | 67.8 | 76.4 | 0.792 | <0.05 |
| MDTL | 73.8 | 69.0 | 77.4 | 0.796 | - |
Fig. 4.
ROC curves of different methods for three binary classification problems
Discriminative Brain Regions Detection
To evaluate the efficacy of our proposed multi-domain transfer feature selection (MDTFS) method in detecting the discriminative brain regions, we compare our proposed MDTFS method with the single-domain based feature selection method (i.e., Lasso) and the commonly used multi-domain based feature selection methods (i.e., MTFS and M2TFS). Table 4 shows the classification performances of four different methods, including Lasso, MTFS (Zhang et al. 2012), M2TFS (Jie et al. 2015), and the proposed MDTFS, using classification accuracy, sensitivity, specificity and AUC measures. In addition, we also compute p-values on the AUC between the MDTFS method and other three methods via DeLong’s method (DeLong et al. 1988), as also shown in Table 4. It is worth noting that, for fair comparison, we use SVM on the target domain in the classification step for our method and competing methods. Also, each value in Table 4 is the averaged result of 10-fold cross-validation strategy in 10 independent runs. As shown in Table 4, MDTFS, MTFS and M2TFS methods can achieve better classification performance than the Lasso method. The possible reason could be that MDTFS, MTFS and M2TFS use data from multi-auxiliary domains. On the other hand, our proposed MDTFS method outperforms MTFS and M2TFSmethods, suggesting that our method can better capture useful information between the target domain and multi-auxiliary domains.
Table 4.
Comparison of our proposed feature selection method (MDTFS) and other state-of-the-art feature selection methods (Lasso, MTFS and M2TFS) in three binary classification problems.
| AD vs. NC Classification | |||||
|---|---|---|---|---|---|
| Method | Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC | p-value |
| Lasso | 87.9 | 87.8 | 88.1 | 0.951 | <0.0001 |
| MTFS | 90.7 | 89.5 | 90.8 | 0.966 | <0.001 |
| M2TFS | 91.5 | 91.4 | 91.6 | 0.979 | <0.01 |
| MDTFS | 93.4 | 93.3 | 93.5 | 0.982 | - |
| MCI vs. NC Classification | |||||
| Method | Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC | p-value |
| Lasso | 74.1 | 80.1 | 62.7 | 0.787 | <0.0001 |
| MTFS | 78.1 | 83.2 | 68.5 | 0.849 | <0.0005 |
| M2TFS | 78.6 | 83.6 | 69.1 | 0.870 | <0.01 |
| MDTFS | 80.2 | 84.8 | 71.4 | 0.876 | - |
| pMCI vs. sMCI Classification | |||||
| Method | Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC | p-value |
| Lasso | 69.4 | 63.8 | 73.5 | 0.744 | <0.0001 |
| MTFS | 71.0 | 62.7 | 74.9 | 0.757 | <0.0005 |
| M2TFS | 71.4 | 65.8 | 77.8 | 0.768 | <0.001 |
| MDTFS | 72.8 | 67.8 | 76.4 | 0.792 | - |
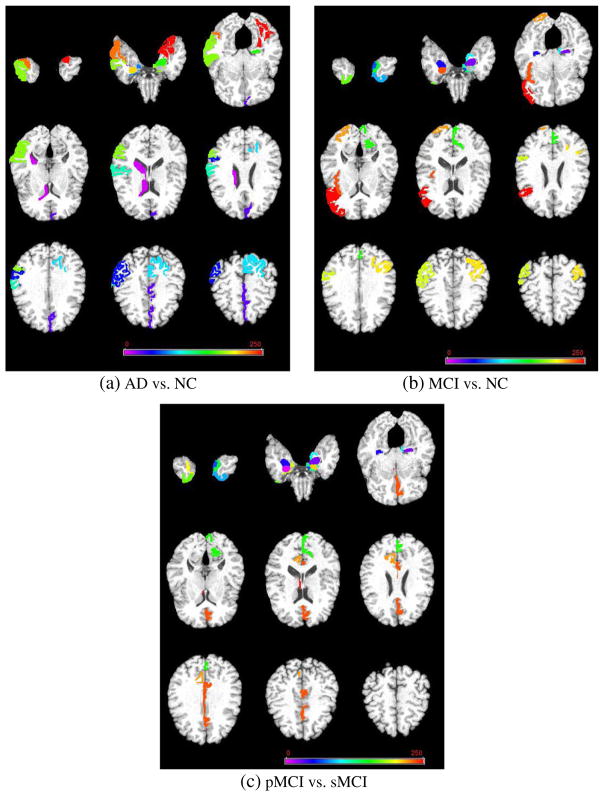
Furthermore, we also investigate the most discriminative regions identified by the proposed feature selection method. Since the feature selection in each fold was performed only based on the training set, the selected features could vary across different cross-validations. We thus defined the most discriminative brain regions based on the selected frequency of each region over the cross-validations. In Fig. 5, for three classification problems, we list all selected brain regions with the highest frequency of occurrence (i.e., each feature and selected across all folds and all runs) by MDTL (i.e., MDTFS + MDTC) on template MR image. As can be seen from Fig. 5, our proposed MDTL method successfully finds out the most discriminative brain regions (e.g., amygdala, hippocampal formation, entorhinal cortex, temporal pole, uncus, perirhinal cortex, cunecus, and temporal pole) that are known to be related to Alzheimer’s disease (Davatzikos et al. 2011; Eskildsen et al. 2013; Jie et al. 2015; Ye et al. 2012; Zhang et al. 2012; Zhang et al. 2011; Zhu et al. 2014).
Fig. 5.
The most discriminative brain regions identified by the proposed MDTL method for three classification tasks. Note that different colors indicate different brain regions
Discussion
In this paper, we propose a multi-domain transfer learning (MDTL) method for early diagnosis of AD, in which we combine the data from multi-auxiliary domains and target domain in both feature selection and classification steps. We evaluate the performance of our method on 807 subjects from the publicly available ADNI database and compare our method with the state-of-the-art methods. The experimental results show that the proposed method consistently and substantially improved the performance of early diagnosis of AD.
Learning with Multi-Domain Data
In the field of neuroimaging-based early diagnosis of AD, multimodal biomarker is widely used for the model design of feature selection and classification (Cheng et al. 2015a; Cheng et al. 2015b; Hinrichs et al. 2011; Jie et al. 2015; Liu et al. 2014; Suk et al. 2014; Ye et al. 2012; Zhang et al. 2012; Zhang et al. 2011; Zhu et al. 2014), which can achieve better performance than the case of using single-modal biomarker. However, in clinical practice, the collection of multimodal biomarker from subject is expensive and time-consuming, and hence the size of collected complete multimodal biomarker dataset is often small. On the other hand, it is relatively easy to get more single-modal biomarker data (e.g., MRI) that contain different categories of subjects. Because of the characteristic of AD cohorts and the requirement of the clinical diagnosis of AD, these single modal data are classified as multiple learning domains that are related to each other. Some studies also show the effectiveness of transfer learning or semi-supervised learning technique in combining these data from related learning domains (Cheng et al. 2012; Da et al. 2014; Filipovych et al. 2011; Young et al. 2013; Zhang and Shen 2011). However, auxiliary data from a single related learning domain is often used in the aforementioned studies. In this paper, we developed MDTL method to enhance the generalization and accuracy of classifiers for the case of singlemodal data with multiple related learning domains,.
To integrate these data with multiple categories of subjects, the transfer learning can be used to build the learning model as done in our previous works (Cheng et al. 2015a; Cheng et al. 2015b; Cheng et al. 2012). However, in our previous works, we only adopted a single related domain data as auxiliary domain to help design classification model for MCI conversion prediction. For example, in our work (Cheng et al. 2015b), we gave an explanation that the domain of classifying pMCI and sMCI subjects was related to the domain of classifying AD and NC subjects, but it was only used for MCI conversion prediction. According to the pathology of AD and its progression, we extended our previous work (Cheng et al. 2015b), by assuming that the classification problem in the target domain is related to each classification problem in the auxiliary domains. To validate this assumption, we have listed results of our proposed method (MDTL), as well as SDTL1, SDTL2, and Lasso methods, in Table 5. For purpose of comparison, we used the Lasso as a baseline method. Also, SDTL1 and SDTL2 methods are the variants of our proposed MDTL method performed on single auxiliary domain and target domain. As we can see from Table 5, using multiauxiliary domain data in the MDTL method can achieve better performance than using single auxiliary domain data in both SDTL1 and SDTL2 methods, and the baseline method (i.e., Lasso) is inferior to MDTL, SDTL1 and SDTL2 methods. These results make it clear that the use of multi-auxiliary domain can effectively improve the performance of early diagnosis of AD.
Table 5.
Comparison of our proposed method (MDTL), SDTL1, SDTL2, and Lasso methods in three binary classification problems. The ‘SDTL1’ is the MDTL method using one auxiliary domain data, which actually is the first one in the auxiliary domains of Table 1. The ‘SDTL2’ is the MDTL method using one auxiliary domain data, which is the second one in the auxiliary domains of Table 1.
| Method | AD vs. NC
|
MCI vs. NC
|
pMCI vs. sMCI
|
|||
|---|---|---|---|---|---|---|
| Accuracy (%) | AUC | Accuracy (%) | AUC | Accuracy (%) | AUC | |
| Lasso | 87.9 | 0.951 | 74.1 | 0.787 | 69.4 | 0.744 |
| SDTL1 | 93.0 | 0.978 | 80.2 | 0.876 | 72.6 | 0.782 |
| SDTL2 | 92.2 | 0.969 | 79.0 | 0.857 | 72.1 | 0.786 |
| MDTL | 94.7 | 0.988 | 81.5 | 0.882 | 73.8 | 0.796 |
Multi-Domain Transfer Learning Model
To effectively integrate the domain knowledge between target domain and multi-auxiliary domain, we introduced the regularizer based on weight vector (i.e., ), which can keep the similarity of multi-weight vectors between target domain and each auxiliary domain. In addition, we introduced two regularizations of weight matrix for all domains (i.e., ||W||1,1 and ||W||2,1) simultaneously, which can select a common feature subset relevant to all domains and also keep useful features relevant to self-domain. In this paper, we combined the three regularizers for feature selection from target-domain and multi-auxiliary-domain data, namely Multi- Domain Transfer Feature Selection (MDTFS). To evaluate the efficacy of each regularizer, we performed some experiments for testing the contribution of each regularizer. In Table 6, we give results of the proposed MDTL method for three classification problems using different setting of regularization parameters.
Table 6.
Comparison of our proposed method (MDTL) in three binary classification problems using different setting of regularization parameters.
| Regularization parameter | AD vs. NC
|
MCI vs. NC
|
pMCI vs. sMCI
|
|||
|---|---|---|---|---|---|---|
| Accuracy (%) | AUC | Accuracy (%) | AUC | Accuracy (%) | AUC | |
| λ1 = 0 | 93.3 | 0.985 | 79.7 | 0.871 | 72.1 | 0.786 |
| λ2 = 0 | 93.7 | 0.986 | 80.6 | 0.875 | 73.1 | 0.792 |
| λ3 = 0 | 92.7 | 0.984 | 79.6 | 0.871 | 72.3 | 0.787 |
| λ1, λ2, λ3 > 0 | 94.7 | 0.988 | 81.5 | 0.882 | 73.8 | 0.796 |
In the MDTFS model, the regularization parameters (i.e., λ1, λ2, λ3) can control the relative contribution of the three regularizers. In Table 6, we investigate the contribution of each regularizer by setting the respective parameter to zero. For example, we set the regularization parameter λ1 to zero (i.e., λ1 = 0), which is used for evaluating contribution of the first regularizer. As can be seen from Table 6, combining three regularizers (i.e., λ1, λ2, λ3 ≠ 0) can achieve better performance for early diagnosis of AD. Specifically, for three classification problems, the minimum reduction of classification performance is without use of the second regularizer (i.e., λ2 = 0), while the reduction of classification performance is small compared to the case without using the first regularizer (i.e., λ1 = 0) and the case without the third regularizer (i.e., λ3 = 0). These results suggest the importance of selecting the common feature subset relevant to all domains and keeping the similarity of weight vectors between target domain and each auxiliary domain, which also confirms the efficacy of using multi-auxiliary domain data.
Strategy for Selecting Feature Subset
Recently, many studies in early diagnosis of AD focus on designing feature selection methods to overcome the small-sample- size problem in neuroimaging data analysis (Cheng et al. 2015b; Li et al. 2014; Liu et al. 2014; Moradi et al. 2015; Ota et al. 2015; Ye et al. 2012; Zhu et al. 2014). In this paper, we develop a multi-domain transfer learning (MDTL) method that can simultaneously utilize approaches of related multi-domain data and feature selection to improve generalization ability of classifiers.
In the MDTL framework, the multi-domain transfer feature selection (MDTFS) is developed, which can use the optimal weight matrix W to select informative feature subset. Since we use the regularizer of ||W||1,1 in the MDTFS step, the selected features from each domain are different. For simplicity, in the current work, we select the same feature subset for each auxiliary domain as the target domain (i.e., Strategy1 in Table 7). Actually, we should consider the target domain more than the auxiliary domains in classification task.
Table 7.
Comparison of two different strategies in selecting feature subset from each domain in our proposed MDTL method. AUC: area under the receiver operating characteristic curve; ACC: accuracy.
| Strategy | AD vs. NC
|
MCI vs. NC
|
pMCI vs. sMCI
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| ACC (%) | AUC | p-value | ACC (%) | AUC | p-value | ACC (%) | AUC | p-value | |
| Strategy1 | 94.7 | 0.988 | >0.05 | 81.5 | 0.882 | >0.05 | 73.8 | 0.796 | >0.05 |
| Strategy2 | 94.8 | 0.989 | - | 82.0 | 0.886 | - | 74.2 | 0.798 | - |
To validate the above assumption, we also adopt another strategy for selecting feature subset, i.e., keeping just features with non-zero weights from each column weight vector of W for all domains (i.e., Strategy2 in Table 7). In Table 7, we list classification results obtained by two different strategies. As we can see from Table 7, the MDTL method using Strategy2 for feature subset selection can achieve slight improvement, compared with the case of using Strategy1. Generally, it is believed that using Strategy2 should be more effective than using Strategy1; but, when using the DeLong’s test (DeLong et al. 1988) to assess the statistical difference between AUC values of two strategies, we found no statistical difference between these two strategies.
To further investigate the difference of using these two strategies, we do a statistical analysis on selected features of each domain. Specifically, we count the number of features selected across all folds and all runs (i.e., a total of 100 times for 10-fold cross-validation with 10 independent runs) on the training set. Then, those features with frequency of 100 (i.e., always selected in all folds and all runs) are regarded as stable features. Accordingly, we compute the average percentage of stable features in the target domain and also those stable features in each auxiliary domain, using MDTL method with the Strategy2 for selecting feature subset. For AD vs. NC, MCI vs. NC and pMCI vs. sMCI classification tasks, their corresponding mean ratios are 83 %, 88 % and 92 %. Similarly, we adopt the Strategy1 to select feature subset, and obtained the results that are slightly inferior to the case of the Strategy2. This implies that the target domain plays a critical role in the classification performance, compared with the auxiliary domain.
Comparison with Previous Methods
To further evaluate the efficacy of our proposed multi-domain transfer learning (MDTL) method for early diagnosis of AD, we list a comparison between the MDTL and some representative state-of-the-art methods in the recent 5 years (Cho et al. 2012; Coupé et al. 2012; Cuingnet et al. 2011; Duchesne and Mouiha 2011; Eskildsen et al. 2013; Hu et al. 2016; Khedher et al. 2015; Liu et al. 2014; Moradi et al. 2015; Ota et al. 2015; Westman et al. 2013; Wolz et al. 2011; Zhu et al. 2014), and show them in Table 8. Here, we provide two performance measurements (i.e., ACC: Accuracy; and AUC: Area Under the receiver operating characteristic Curve) in Table 8. Since it is not available the ACC and AUC from the paper of Cuingnet et al. 2011, we just list measurements of sensitivity (SEN) and specificity (SPE). Note that, in Table 8, for several studies using multimodal biomarker (Liu et al. 2014; Zhu et al. 2014), we report their results using only MRI data if available; otherwise, we report their results using multimodal data. Although feature extraction method is different for the comparison methods, this comparison also can show the efficacy of MDTL method at certain level. In most cases, the AUC and ACC of MDTL method are better than those of the comparison methods, indicating that MDTL has better diagnostic performance in early diagnosis of AD.
Table 8.
Comparison with the state-of-the-art methods in early diagnosis of AD. AUC: area under the receiver operating characteristic curve; ACC: accuracy; SEN: sensitivity; SPE: specificity.
| Reference | Data | Feature extraction | Result |
|---|---|---|---|
| Our proposed method | 186 AD, 226 NC 167 pMCI, 228 sMCI |
ROIs of GM | AD vs. NC ACC = 94.7 %, AUC = 0.988 MCI vs. NC ACC = 81.5 %, AUC = 0.882 pMCI vs. sMCI ACC = 73.8 %, AUC = 0.796 |
| Hu et al. 2016 | 188 AD, 228 NC 71 pMCI, 62 sMCI |
VBM of GM, WM, and CSF | AD vs. NC ACC = 84.13 %, AUC = 0.9 pMCI vs. sMCI ACC = 76.69 %, AUC = 0.79 |
| Moradi et al. 2015 | 200 AD, 231 NC 164 pMCI, 100 sMCI |
VBM of GM VBM of GM, age and cognitive measures |
pMCI vs. sMCI ACC = 74.74 %, AUC = 0.7661 ACC = 82 %, AUC = 0.9 |
| Khedher et al. 2015 | 188 AD, 229 NC 401 MCI |
VBM of GM, and WM | AD vs. NC ACC = 88.49 % MCI vs. NC ACC = 81.89 % ACC = 77.57 % (only GM) |
| Ota et al. 2015 | 40 pMCI, 40 sMCI | VBM of whole brain | pMCI vs. sMCI AUC = 0.75 |
| Zhu et al. 2014 | 51 AD, 52 NC 43 pMCI, 56 sMCI |
ROIs of GM | AD vs. NC (Only MRI) ACC = 93.8 %, AUC = 0.979 MCI vs. NC (Only MRI) ACC = 79.7 %, AUC = 0.852 pMCI vs. sMCI (Only MRI) ACC = 70.8 %, AUC = 0.756 |
| Liu et al. 2014 | 51 AD, 52 NC 43 pMCI, 56 sMCI |
ROIs of GM | AD vs. NC (PET + MRI) ACC = 94.37 %, AUC = 0.9724 MCI vs. NC (PET + MRI) ACC = 78.8 %, AUC = 0.8284 pMCI vs. sMCI (PET + MRI) ACC = 67.83 %, AUC = 0.6957 |
| Eskildsen et al. 2013 | 194 AD, 226 NC 161 pMCI, 227 sMCI |
Cortical Thickness | AD vs. NC ACC = 86.7 %, AUC = 0.917 pMCI vs. sMCI ACC = 73 %, AUC = 0.803 |
| Westman et al. 2013 | 187 AD, 225 NC 87 pMCI, 200 sMCI |
Regional Volume and Cortical Thickness | AD vs. NC ACC = 91.5 %, AUC = 0.96 pMCI vs. sMCI (Conversion time of 24 months) ACC = 69.3 %, AUC = 0.748 |
| Coupé et al. 2012 | 198 AD, 231 NC 167 pMCI, 238 sMCI |
Nonlocal Image Patch of ROIs | AD vs. NC ACC = 89 % pMCI vs. sMCI ACC = 71 % |
| Cho et al. 2012 | 128 AD, 160 NC 72 pMCI, 131 sMCI |
Cortical Thickness | AD vs. NC ACC = 86 % pMCI vs. sMCI ACC = 71 % |
| Cuingnet et al. 2011 | 137 AD, 162 NC 76 pMCI, 134 sMCI |
Various (Voxel-based, hippocampus, and Cortical thickness) | AD vs. NC SEN = 81 %, SPE = 95 % pMCI vs. sMCI SEN = 62 %, SPE = 69 % |
| Duchesne and Mouiha 2011 | 75 AD, 75 NC 20 pMCI, 29 sMCI |
Volume of ROIs | AD vs. NC ACC = 90 %, AUC = 0.9444 pMCI vs. sMCI ACC = 72.3 %, AUC = 0.794 |
| Wolz et al. 2011 | 198 AD, 231 NC 167 pMCI, 238 sMCI |
Various (Hippocampal volume, Cortical thickness, MBL, TBM and CTH) | AD vs. NC ACC = 89 % pMCI vs. sMCI ACC = 68 % |
Limitations
The current study is limited by several factors. First, our proposed method is based on the single modal (i.e., MRI) biomarker from the ADNI database. In the ADNI database, many subjects also have multimodal biomarkers. Also, many status-unlabeled subjects can be used to extend our current method. In the future work, we will investigate whether adding more auxiliary domain (e.g., multimodal biomarker, and status-unlabeled data) can further improve the performance.
Second, considering the small number of training samples, as well as the sensitivity of those very local features (i.e., thickness, and tissue density) to noises as well as errors in processing pipeline (including skull stripping, tissue segmentation, image registration, and regions-of-interest (ROI) labeling), our current study considers only using ROI features, and no surface-based cortical thickness features are extracted and used although some studies already show the sensitivity of cortical thickness in early diagnosis of AD (Cho et al. 2012; Cuingnet et al. 2011; Eskildsen et al. 2013; Querbes et al. 2009; Wee et al. 2013; Wolz et al. 2011). In the future work, we will consider extracting cortical thickness features from MR images and combine with volume-based features for early diagnosis of AD.
Finally, due to the small number of training samples, we adopted only the volume of gray matter (GM) tissue in each ROI as a feature and input the MDTL model for early diagnosis of AD. However, the study of Cuingnet et al. (Cuingnet et al. 2011) showed that other tissue volumetric feature (i.e., white matter (WM) and CSF) also contributed to AD and NC classification. Accordingly, we also used all types of volumetric features (i.e., GM + WM + CSF) to test the classification performance of our proposed MDTL model, by comparison with the MDTL model using only the GM features. In Table 9, we list their respective ACC and AUC, and further perform the DeLong’s test (DeLong et al. 1988) on the AUC to test their statistical difference, with p-values provided. The results in Table 9 suggest that using three types of volumetric features can improve the performance, but the corresponding p-values show no statistical significant improvement by using three types of volumetric features. In future work, we will improve the MDTL model and combine with the improvement of neuroimaging pre-processing pipeline to enhance the final classification results.
Table 9.
Effects of using different imaging features in our proposed MDTL method. AUC: area under the receiver operating characteristic curve; ACC: accuracy.
| Feature | AD vs. NC
|
MCI vs. NC
|
pMCI vs. sMCI
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| ACC (%) | AUC | p-value | ACC (%) | AUC | p-value | ACC (%) | AUC | p-value | |
| GM | 94.7 | 0.988 | >0.05 | 81.5 | 0.882 | >0.05 | 73.8 | 0.796 | >0.05 |
| GM + WM + CSF | 95.1 | 0.988 | - | 82.1 | 0.892 | - | 74.0 | 0.797 | - |
Conclusion
In this paper, we propose a novel multi-domain transfer learning (MDTL) method for early diagnosis of AD, which consists of multi-domain transfer feature selection (MDTFS) and multi-domain transfer classifier (MDTC). The main idea of our multi-domain transfer learning based method is to exploit the multi-auxiliary domain data to improve classification performance (e.g., AD vs. NC, MCI vs. NC and pMCI vs. sMCI) in the target domain. Also, we further combine the source data from multi-auxiliary domain and target domain to guide both feature selection and classification steps. We evaluate our method on the baseline ADNI database with MRI data, and the experimental results demonstrate the efficacy of our method by comparison with several state-of-the-art methods.
Acknowledgments
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson and Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co., Inc., Novartis AG, Pfizer Inc., F. Hoffman-La Roche, Schering-Plough, Synarc, Inc., as well as non-profit partners the Alzheimer’s Association and Alzheimer’s Drug Discovery Foundation, with participation from the U.S. Food and Drug Administration. Private sector contributions to ADNI are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuron Imaging at the University of California, Los Angeles. This work was supported in part by the National Natural Science Foundation of China (Nos. 61602072, 61422204 and 61473149), the Chongqing Cutting-edge and Applied Foundation Research Program (Nos. cstc2016jcyjA0063, cstc2014jcyjA1316, and cstc2014jcyjA40035), the Scientific and Technological Research Program of Chongqing Municipal Education Commission (Nos. KJ1501014, KJ1401010, and KJ1601003), the NUAA Fundamental Research Funds (No. NE2013105), and NIH grants (AG041721, AG049371, AG042599, AG053867).
Footnotes
P ∈ {0.000001, 0.00001, 0.0001, 0.0003, 0.0007, 0.001, 0.003, 0.005, 0.007, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9}
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (http://adni.loni.usc.edu/). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report.
Information Sharing Statement
The dataset used in this paper are from the Alzheimer’s Disease Neuroimaging Initiative (ADNI, RRID:SCR_003007) which are available at http://adni.loni.usc.edu/. Source code and binary programs developed in this paper are available via our website (http://ibrain.nuaa.edu.cn/, RRID:SCR_014691), cb729@nuaa.edu.cn.
References
- Association As. 2014 Alzheimer’s disease facts and figures. Alzheimers Dement. 2014;10:47–92. doi: 10.1016/j.jalz.2014.02.001. [DOI] [PubMed] [Google Scholar]
- Chang CC, Lin CJ. LIBSVM: a library for support vector machines. 2001 http://www.csie.ntu.edu.tw/~cjlin/libsvm/
- Chao LL, Buckley ST, Kornak J, Schuff N, Madison C, Yaffe K, Miller BL, Kramer JH, Weiner MW. ASL perfusion MRI predicts cognitive decline and conversion from MCI to dementia. Alzheimer Disease and Associated Disorders. 2010;24:19–27. doi: 10.1097/WAD.0b013e3181b4f736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Pan W, Kwok JT, Carbonell JG. Accelerated gradient method for multi-task sparse learning problem. Proceeding of ninth IEEE international conference on data mining and knowledge discovery; 2009. pp. 746–751. [Google Scholar]
- Cheng B, Zhang D, Shen D. Domain transfer learning for MCI conversion prediction. Proceeding of International Conference on Medical Image Computing and Computer-Assisted Intervention-MICCAI 2012 7510; 2012. pp. 82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B, Zhang D, Chen S, Kaufer DI, Shen D ADNI. Semi-supervised multimodal relevance vector regression improves cognitive performance estimation from imaging and biological biomarkers. Neuroinformatics. 2013;11:339–353. doi: 10.1007/s12021-013-9180-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B, Liu M, Suk H, Shen D, Zhang D. Multimodal manifold-regularized transfer learning for MCI conversion prediction. Brain Imaging and Behavior. 2015a;9:913–926. doi: 10.1007/s11682-015-9356-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B, Liu M, Zhang D, Munsell BC, Shen D. Domain transfer learning for MCI conversion prediction. IEEE Transactions on Biomedical Engineering. 2015b;62:1805–1817. doi: 10.1109/TBME.2015.2404809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetelat G, Landeau B, Eustache F, Mezenge F, Viader F, de la Sayette V, Desgranges B, Baron JC. Using voxel-based morphometry to map the structural changes associated with rapid conversion in MCI: a longitudinal MRI study. NeuroImage. 2005;27:934–946. doi: 10.1016/j.neuroimage.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Cho Y, Seong JK, Jeong Y, Shin SY ADNI. Individual subject classification for Alzheimer’s disease based on incremental learning using a spatial frequency representation of cortical thickness data. NeuroImage. 2012;59:2217–2230. doi: 10.1016/j.neuroimage.2011.09.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CIT. Medical Image Processing, Analysis and Visualization (MIPAV) 2012 http://mipav.cit.nih.gov/clickwrap.php.
- Coupé P, Eskildsen SF, Manjón JV, Fonov VS, Pruessner JC, Allard M, Collins DL. Scoring by nonlocal image patch estimator for early detection of Alzheimer’s disease. NeuroImage: Clinical. 2012;1:141–152. doi: 10.1016/j.nicl.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehericy S, Habert MO, Chupin M, Benali H, Colliot O. Automatic classification of patients with Alzheimer’s disease from structural MRI: a comparison of ten methods using the ADNI database. NeuroImage. 2011;56:766–781. doi: 10.1016/j.neuroimage.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Da X, Toledo JB, Zee J, Wolk DA, Xie SX, Ou Y, Shacklett A, Parmpi P, Shaw L, Trojanowski JQ, Davatzikos C. Integration and relative value of biomarkers for prediction of MCI to AD progression: spatial patterns of brain atrophy, cognitive scores, APOE genotype and CSF biomarkers. NeuroImage: Clinical. 2014;4:164–173. doi: 10.1016/j.nicl.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davatzikos C, Bhatt P, Shaw LM, Batmanghelich KN, Trojanowski JQ. Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiology of Aging. 2011;32:2322.e2319–2322.e2327. doi: 10.1016/j.neurobiolaging.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- Duan LX, Tsang IW, Xu D. Domain transfer multiple kernel learning. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2012;34:465–479. doi: 10.1109/TPAMI.2011.114. [DOI] [PubMed] [Google Scholar]
- Duchesne S, Mouiha A. Morphological factor estimation via high-dimensional reduction: prediction of MCI conversion to probable AD. International Journal of Alzheimer’s Disease. 2011;2011:914085. doi: 10.4061/2011/914085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskildsen SF, Coupé P, García-Lorenzo D, Fonov V, Pruessner JC, Collins DL. Prediction of Alzheimer’s disease in subjects with mild cognitive impairment from the ADNI cohort using patterns of cortical thinning. NeuroImage. 2013;65:511–521. doi: 10.1016/j.neuroimage.2012.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Batmanghelich N, Clark CM, Davatzikos C, Initia ADN. Spatial patterns of brain atrophy in MCI patients, identified via high-dimensional pattern classification, predict subsequent cognitive decline. NeuroImage. 2008;39:1731–1743. doi: 10.1016/j.neuroimage.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipovych R, Davatzikos C, Initia ADN. Semi-supervised pattern classification of medical images: application to mild cognitive impairment (MCI) NeuroImage. 2011;55:1109–1119. doi: 10.1016/j.neuroimage.2010.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaser C, Franke K, Kloppel S, Koutsouleris N, Sauer H. Brain AGE in mild cognitive impaired patients: predicting the conversion to Alzheimer’s disease. PloS One. 2013;8:e67346. doi: 10.1371/journal.pone.0067346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Wang Z, Li K, Li Z, Qi Z, Jin Z, Yao L, Chen K. Voxel-based assessment of gray and whitematter volumes in Alzheimer’s disease. Neuroscience Letters. 2010;468:146–150. doi: 10.1016/j.neulet.2009.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs C, Singh V, Xu GF, Johnson SC, Neuroimaging AD. Predictive markers for AD in a multi-modality framework: an analysis of MCI progression in the ADNI population. NeuroImage. 2011;55:574–589. doi: 10.1016/j.neuroimage.2010.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K, Wang Y, Chen K, Hou L, Zhang X. Multi-scale features extraction from baseline structure MRI for MCI patient classification and AD early diagnosis. Neurocomputing. 2016;175:132–145. [Google Scholar]
- Jie B, Zhang D, Cheng B, Shen D. Manifold regularized multitask feature learning for multimodality disease classification. Human Brain Mapping. 2015;36:489–507. doi: 10.1002/hbm.22642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani N, MacDonald D, Holmes CJ, Evans A. A 3D atlas of the human brain. NeuroImage. 1998;7:S717. [Google Scholar]
- Khedher L, Ramírez J, Górriz JM, Brahim A, Segovia F. Early diagnosis of Alzheimer’s disease based on partial least squares, principal component analysis and support vector machine using segmented MRI images. Neurocomputing. 2015;151:139–150. [Google Scholar]
- Li H, Liu Y, Gong P, Zhang C, Ye J. Hierarchical interactions model for predicting mild cognitive impairment (MCI) to Alzheimer’s disease (AD) conversion. PloS One. 2014;9:e82450. doi: 10.1371/journal.pone.0082450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ji S, Ye J. SLEP: sparse learning with efficient projections. Arizona State University; 2009. http://www.public.asu.edu/~jye02/Software/SLEP. [Google Scholar]
- Liu F, Wee CY, Chen HF, Shen DG ADNI. Intermodality relationship constrained multi-modality multi-task feature selection for Alzheimer’s disease and mild cognitive impairment identification. NeuroImage. 2014;84:466–475. doi: 10.1016/j.neuroimage.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang D, Shen D. Inherent structure based multiview learning with multi-atlas feature representation for alzheimer’s disease diagnosis. IEEE Transactions on Biomedical Engineering. 2016a;63:1473–1482. doi: 10.1109/TBME.2015.2496233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang D, Shen D. Relationship induced multitemplate learning for diagnosis of alzheimer’s disease and mild cognitive impairment. IEEE Transactions on Medical Imaging. 2016b;35:1463–1474. doi: 10.1109/TMI.2016.2515021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra C, Fan Y, Davatzikos C. Baseline and longitudinal patterns of brain atrophy in MCI patients, and their use in prediction of short-term conversion to AD: results from ADNI. NeuroImage. 2009;44:1415–1422. doi: 10.1016/j.neuroimage.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradi E, Pepe A, Gaser C, Huttunen H, Tohka J. Machine learning framework for early MRI-based Alzheimer’s conversion prediction in MCI subjects. NeuroImage. 2015;104:398–412. doi: 10.1016/j.neuroimage.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemirovski A. Efficient methods in convex programming. 2005. [Google Scholar]
- Obozinski G, Taskar B, Jordan MI. Statistics Department, UC Berkeley: Technical report. 2006. Multi-task feature selection. [Google Scholar]
- Ota K, Oishi N, Ito K, Fukuyama H, Grp SJS. A comparison of three brain atlases for MCI prediction. Journal of Neuroscience Methods. 2014;221:139–150. doi: 10.1016/j.jneumeth.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Ota K, Oishi N, Ito K, Fukuyama H. Effects of imaging modalities, brain atlases and feature selection on prediction of Alzheimer’s disease. Journal of Neuroscience Methods. 2015;256:168–183. doi: 10.1016/j.jneumeth.2015.08.020. [DOI] [PubMed] [Google Scholar]
- Pan SJ, Yang Q. A survey on transfer learning. IEEE Transactions on Knowledge and Data Engineering. 2010;22:1345–1359. [Google Scholar]
- Querbes O, Aubry F, Pariente J, Lotterie JA, Demonet JF, Duret V, Puel M, Berry I, Fort JC, Celsis P ADNI. Early diagnosis of Alzheimer’s disease using cortical thickness: impact of cognitive reserve. Brain: A Journal of Neurology. 2009;132:2036–2047. doi: 10.1093/brain/awp105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risacher SL, Saykin AJ, West JD, Shen L, Firpi HA, McDonald BC. Baseline MRI predictors of conversion from MCI to probable AD in the ADNI cohort. Current Alzheimer Research. 2009;6:347–361. doi: 10.2174/156720509788929273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011:12. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabuncu MR, Konukoglu E ADNI. Clinical prediction from structural brain MRI scans: a large-scale empirical study. Neuroinformatics. 2015;13:31–46. doi: 10.1007/s12021-014-9238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y, Varoquaux G, Pallier C, Pinel P, Poline J, Thirion B. Improving accuracy and power with transfer learning using a meta-analytic database. Proceeding of International Conference on Medical Image Computing and Computer-Assisted Intervention- MICCAI 2012 7512; 2012. pp. 248–255. [DOI] [PubMed] [Google Scholar]
- Shen D, Davatzikos C. HAMMER: hierarchical attribute matching mechanism for elastic registration. IEEE Transactions on Medical Imaging. 2002;21:1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Transactions on Medical Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Suk H, Lee SW, Shen D ADNI. Hierarchical feature representation and multimodal fusion with deep learning for AD/ MCI diagnosis. NeuroImage. 2014;101:569–582. doi: 10.1016/j.neuroimage.2014.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani RJ. Regression shrinkage and selection via the LASSO. Journal of the Royal Statistical Society, Series B. 1996;58:267–288. [Google Scholar]
- Wang Y, Nie J, Yap P-T, Shi F, Guo L, Shen D. Robust Deformable-Surface-Based Skull-Stripping for Large-Scale Studies. In: Fichtinger G, Martel A, Peters T, editors. Medical Image Computing and Computer-Assisted Intervention. Springer; Berlin / Heidelberg, Toronto, Canada: 2011. pp. 635–642. [DOI] [PubMed] [Google Scholar]
- Wee CY, Yap PT, Shen DG ADNI. Prediction of Alzheimer’s disease and mild cognitive impairment using cortical morphological patterns. Human Brain Mapping. 2013;34:3411–3425. doi: 10.1002/hbm.22156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westman E, Muehlboeck JS, Simmons A. Combining MRI and CSF measures for classification of Alzheimer’s disease and prediction of mild cognitive impairment conversion. NeuroImage. 2012;62:229–238. doi: 10.1016/j.neuroimage.2012.04.056. [DOI] [PubMed] [Google Scholar]
- Westman E, Aguilar C, Muehlboeck JS, Simmons A. Regional magnetic resonance imaging measures for multivariate analysis in Alzheimer’s disease and mild cognitive impairment. Brain Topography. 2013;26:9–23. doi: 10.1007/s10548-012-0246-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolz R, Julkunen V, Koikkalainen J, Niskanen E, Zhang DP, Rueckert D, Soininen H, Lotjonen J. Multi-method analysis of MRI images in early diagnostics of Alzheimer’s disease. PloS One. 2011;6:e25446. doi: 10.1371/journal.pone.0025446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yan R, Hauptmann AG. Cross-domain video concept detection using adaptive SVMs. Proceedings of the 15th international conference on Multimedia; 2007. pp. 188–197. [Google Scholar]
- Ye J, Farnum M, Yang E, Verbeeck R, Lobanov V, Raghavan N, Novak G, DiBernardo A, Narayan VA ADNI. Sparse learning and stability selection for predicting MCI to AD conversion using baseline ADNI data. BMC Neurology. 2012;12:1471-2377-1412-1446. doi: 10.1186/1471-2377-12-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J, Modat M, Cardoso MJ, Mendelson A, Cash D, Ourselin S. Accurate multimodal probabilistic prediction of conversion to Alzheimer’s disease in patients with mild cognitive impairment. NeuroImage: Clinical. 2013;2:735–745. doi: 10.1016/j.nicl.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Shen D. Semi-supervised multimodal classification of Alzheimer’s disease. Proceeding of IEEE International Symposium on Biomedical Imaging; 2011. pp. 1628–1631. [Google Scholar]
- Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation maximization algorithm. IEEE Transactions on Medical Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- Zhang J, Gao Y, Munsell BC, Shen D. Detecting anatomical landmarks for fast alzheimer’s disease diagnosis. IEEE Transactions on Medical Imaging. 2016 doi: 10.1109/TMI.2016.2582386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Wang Y, Zhou L, Yuan H, Shen D ADNI. Multimodal classification of Alzheimer’s disease and mild cognitive impairment. NeuroImage. 2011;55:856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Shen D ADNI. Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer’s disease. NeuroImage. 2012;59:895–907. doi: 10.1016/j.neuroimage.2011.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu J, Narayan VA, Ye J ADNI. Modeling disease progression via multi-task learning. NeuroImage. 2013;78:233–248. doi: 10.1016/j.neuroimage.2013.03.073. [DOI] [PubMed] [Google Scholar]
- Zhu X, Huang Z, Shen HT, Cheng J, Xu C. Dimensionality reduction by mixed kernel canonical correlation analysis. Pattern Recognition. 2012;45:3003–3016. [Google Scholar]
- Zhu X, Suk H, Shen D. A novel matrix-similarity based loss function for joint regression and classification in AD diagnosis. NeuroImage. 2014;100:91–105. doi: 10.1016/j.neuroimage.2014.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]