Abstract
The endoplasmic reticulum (ER) of eukaryotic cells serves as a checkpoint tightly monitoring protein integrity and channeling malformed proteins into different rescue and degradation routes. The degradation of several ER lumenal and membrane-localized proteins is mediated by ER-associated protein degradation (ERAD) in yeast (Saccharomyces cerevisiae) and mammalian cells. To date, evidence for the existence of ERAD-like mechanisms in plants is indirect and based on heterologous or artificial substrate proteins. Here, we show that an allelic series of single amino acid substitution mutants of the plant-specific barley (Hordeum vulgare) seven-transmembrane domain mildew resistance o (MLO) protein generates substrates for a postinsertional quality control process in plant, yeast, and human cells, suggesting conservation of the underlying mechanism across kingdoms. Specific stabilization of mutant MLO proteins in yeast strains carrying defined defects in protein quality control demonstrates that MLO degradation is mediated by HRD pathway-dependent ERAD. In plants, individual aberrant MLO proteins exhibit markedly reduced half-lives, are polyubiquitinated, and can be stabilized through inhibition of proteasome activity. This and a dependence on homologs of the AAA ATPase CDC48/p97 to eliminate the aberrant variants strongly suggest that MLO proteins are endogenous substrates of an ERAD-related plant quality control mechanism.
INTRODUCTION
Both lumenal and integral membrane proteins enter the secretory pathway via the endoplasmic reticulum (ER). Impairment or delay of protein folding by mutation, perturbation of protein–protein interactions, or stress stimuli can induce a detrimental accumulation and/or aggregation of unfolded proteins (Brodsky and McCracken, 1999). Malformed proteins retained in the ER can either be assisted in maturation by the upregulation of molecular chaperones (unfolded protein response) or targeted for destruction (Casagrande et al., 2000; Friedlander et al., 2000; Travers et al., 2000). Such proteins can be either transported to the vacuole, where they are subsequently degraded by vacuolar proteases (Hong et al., 1996), or they are disposed of in the cytosol by a mechanism described as ER-associated protein degradation (ERAD). The quality control of most yeast (Saccharomyces cerevisiae) and mammalian lumenal and membrane-localized ERAD substrates has been shown to involve retrotranslocation into the cytosol, substrate ubiquitination, and degradation by the proteasome (Brodsky and McCracken, 1999; Plemper and Wolf, 1999; Hampton, 2002; Jarosch et al., 2002; Tsai et al., 2002). Key constituents of this mechanism were identified in yeast by genetic screens for mutants impaired in protein quality control (Hampton et al., 1996; Knop et al., 1996; Swanson et al., 2001). A subset of yeast ERAD substrates is degraded via the HRD pathway, involving polyubiquitination by the membrane-associated ubiquitin ligase Hrd1p/Der3p together with Hrd3p and the ubiquitin-conjugating enzyme Ubc7p (Bordallo et al., 1998; Gardner et al., 2001). An alternative ERAD-associated ubiquitination cascade is mediated by the ubiquitin ligase Doa10p in cooperation with a dimer of the ubiquitin-conjugating enzymes Ubc6p and Ubc7p (Swanson et al., 2001).
Comparably little is known about protein quality control in the secretory pathway of plants. Rescue reactions induced by accumulation of malformed proteins similar to the unfolded protein response in yeast and mammalian cells have been described (Pedrazzini et al., 1997; Jelitto-Van Dooren et al., 1999; Leborgne-Castel et al., 1999; Vitale and Denecke, 1999; Martínez and Chrispeels, 2003). Recently, studies with heterologous and artificial substrate proteins have demonstrated that components of the ERAD pathway exist in plants. The ability of the ribosome-inactivating ricin A chain to gain access to the cytosol of mammalian cells by subverting the ERAD machinery is paralleled by a similar retrograde transport and proteasome-dependent degradation in tobacco (Nicotiana tabacum) cells (Di Cola et al., 2001). Furthermore, Brandizzi et al. (2003) described that a presumably misfolded fusion of the calreticulin P-region to secreted green fluorescent protein (GFP) is recognized as aberrant in tobacco cells, retained inside the cell, and slowly degraded by a proteasome-independent mechanism.
Barley (Hordeum vulgare) powdery mildew resistance o (MLO) is the founder of a sequence-diversified protein family with seven-transmembrane (7-TM) helices that is unique to plants. MLO accumulates at low levels in the plasma membrane and acquires a conformation with its N- and C-terminal ends located extracellularly and intracellularly, respectively (Büschges et al., 1997; Devoto et al., 1999, 2003). Barley MLO interacts with the Ca2+ sensor calmodulin and appears to inhibit a vesicle-associated and soluble NSF attachment protein receptor protein-dependent resistance reaction to the widespread powdery mildew pathogen, Blumeria graminis f. sp hordei (Bgh; Kim et al., 2002; Collins et al., 2003; Panstruga and Schulze-Lefert, 2003). Barley plants lacking functional MLO (mlo mutants) terminate fungal pathogenesis of all known Bgh strains during the transition from surface to invasive growth.
A collection of chemical- or radiation-induced barley mlo mutant alleles conferring Bgh resistance has been previously described (Büschges et al., 1997; Piffanelli et al., 2002). In this study, we show that a series of mlo alleles leading to single amino acid replacements generates mainly destabilized mutant MLO forms. We noted that various reporter proteins were consistently codestabilized in plant cells upon fusion to the C termini of MLO mutant variants. This phenomenon was exploited to adopt a facile reporter gene–based dual luciferase assay for quantification of MLO proteins in different cell types. We demonstrate that destabilizing amino acid replacements in MLO generate universal signals for protein quality control in barley, Arabidopsis thaliana, yeast, and human cells. By taking advantage of numerous quality control components previously identified in yeast, we could assign the underlying process in this organism to HRD-dependent ERAD. This, in turn, provided leads to investigate the mechanism in planta. Reporter protein–based and biochemical analysis indicated that MLO proteins are endogenous substrates of an ERAD-like protein quality control surveillance system in plant cells.
RESULTS
Single Amino Acid Substitutions in Different Regions of Barley MLO Impair Accumulation of the Protein
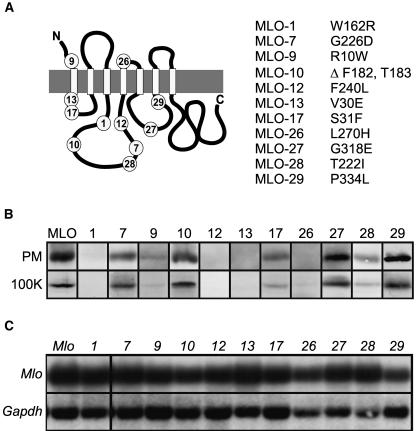
We selected for this study mlo alleles that result in either single amino acid substitutions (mlo-1, -7, -9, -12, -13, -17, -26, -27, -28, and -29) or a two–amino acid deletion (mlo-10) in lumenal and cytoplasmic regions of the 7-TM protein (Figure 1A). Protein gel blot analysis of leaf plasma membrane vesicles and microsomal pellets isolated from leaf tissue of the barley mutants revealed allele-specific differences in MLO protein accumulation, ranging from unaltered abundance to undetectable levels in comparison with wild-type plants (Figure 1B). The ratio of MLO protein detected in enriched plasma membrane vesicles and in total microsomal fractions was similar in all tested mutants, suggesting that the aberrant forms were not significantly retained in intracellular membrane compartments (Figure 1B). Because mlo transcript abundance in wild-type and all tested mutant plants was indistinguishable (Figure 1C), a posttranscriptional event must impair protein accumulation in an allele-specific manner.
Figure 1.
MLO Amino Acid Substitution Sites and mlo mRNA Levels.
(A) Schematic representation of the MLO protein. The C terminus is located in the cytosol, whereas the N terminus faces the ER lumen/extracellular space. White boxes represent transmembrane domains, and the gray boxes represent the lipid bilayer. Positions of the amino acid substitutions/deletions are indicated by numbers corresponding to the listed protein variants.
(B) Protein gel blot analysis of barley mlo plants. Enriched leaf plasma membrane fractions (PM) and microsomal fractions (100K) were probed with the MLO-specific MYP antibody as described previously (Devoto et al., 1999). Equal protein amounts were loaded in each lane. This blot is a representative example of three independent experiments.
(C) Analysis of poly(A)+-enriched RNA fractions prepared from barley wild-type and mlo mutants hybridized with Mlo (top) and Gapdh (bottom) cDNA probes.
Half-Lives of MLO Variants -1, -7, and -12 Are Significantly Reduced
To test whether differences in protein accumulation were because of a translational or posttranslational process, we expressed fusions of various reporter genes (Neomycin phosphotransferase II, β-glucuronidase, GFP, and Renilla luciferase) to the 3′-ends of mutant mlo cDNAs in planta under control of the constitutive 35S promoter of Cauliflower mosaic virus (CaMV). We noted that the accumulation of the reporter proteins was determined by amino acid substitutions in the MLO portion (data not shown). We exploited this phenomenon and adopted a reporter gene–based dual luciferase assay as a tool for quantification of proteins in different eukaryotic cell types. Luminescence generated by translational fusions of MLO proteins to Renilla luciferase was normalized against the activity of coexpressed firefly luciferase.
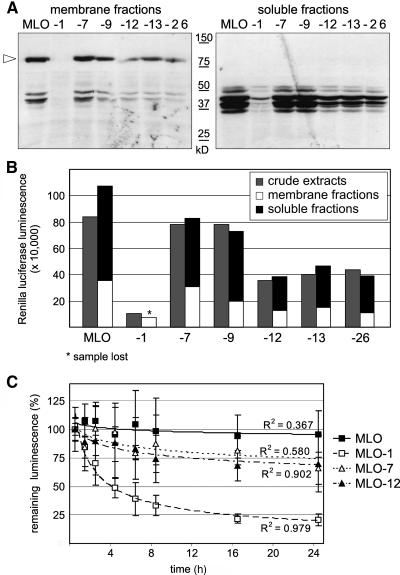
To test whether Renilla luciferase luminescence is a reliable indicator of MLO abundance, we introduced a triple hemagglutinin (3xHA) tag to the C terminus of a subset of MLO-Renilla luciferase fusion constructs. Extracts of protoplasts expressing MLO, MLO-1, -7, -9, -12, -13, and -26 fusions to the Renilla luciferase-3xHA tag were prepared under the conditions usually applied for dual luciferase assays and separated by ultracentrifugation (100,000g). Soluble and membrane fractions were subsequently analyzed by protein gel blot hybridization with an antiserum directed against the HA epitope (Figure 2A) and by Renilla luciferase assays (Figure 2B). In membrane fractions, we detected predominantly the full-length fusion protein of ∼80 kD. C-terminal degradation products ranging from 30 to 50 kD were mainly detected in the soluble fractions. The relative abundance of these fragments correlated roughly to the amounts of the corresponding full-length proteins. The same peptides were also detected in samples containing the stable wild-type MLO protein, suggesting that these are allele-independent cleavage products possibly resulting from the sample preparation. Renilla luciferase measurements of both soluble and membrane fractions confirmed these results. Importantly, the sum of Renilla luciferase luminescence detected in pellet and supernatant corresponded approximately to the luminescence detected in crude extracts for each tested allele (Figure 2B). We conclude that luminescence originating from MLO-Renilla luciferase fusion proteins is a valid means to determine relative protein amounts.
Figure 2.
MLO Protein Stability.
(A) and (B) Crude extracts of Arabidopsis protoplasts expressing MLO proteins translationally fused to combined C-terminal Renilla luciferase-3xHA tags were separated in membrane pellets and supernatants by ultracentrifugation (100,000g).
(A) Membrane (left) and soluble (right) fractions were analyzed by SDS-PAGE and protein gel blot hybridization with an antiserum directed against the HA epitope. The arrowhead indicates the position of full-length MLO fusion proteins. Sizes of a molecular weight standard are indicated.
(B) Corresponding fractions of the same crude extracts (gray bars), membrane (white bars), and soluble (black bars) fractions were analyzed by Renilla luciferase measurements.
(C) CHX chase was performed with MLO, MLO-1, MLO-7, and MLO-12 using dual luciferase assays in Arabidopsis protoplasts. CHX was added at a concentration of 200 μg mL−1 at 7 h after transfection. Samples were taken at the indicated time points. Relative protein accumulation is expressed as the percentage of the value determined at time point zero. R2 indicates the R2 value for the shown trend lines. Each data point represents the mean ± sd of three to five independent experiments, including two samples per construct and time point.
We first applied the dual luciferase technology for assessing half-lives of wild-type MLO, MLO-1, -7, and -12 by cycloheximide (CHX) chase in cell culture–derived Arabidopsis protoplasts. Whereas the relative amount of luminescence obtained from the MLO-Renilla luciferase fusion protein decreased only slightly during the 24-h chase period, the MLO-1-Renilla luciferase fusion revealed a half-life of ∼3.5 h (Figure 2C). Tested MLO-12 and -7 stabilities were similar to each other and ranged between the half-lives determined for wild-type MLO and MLO-1 (Figure 2C). Allele-specific MLO decay variation in Arabidopsis was similar to the reduction of MLO steady state levels detected in the corresponding barley mlo plants (Figure 1B). We conclude that the reduced MLO accumulation in mlo mutant plants is mediated by allele-specific protein degradation and that the underlying mechanism may have been conserved because the lineages of the monocot barley and dicot Arabidopsis diverged 200 million years ago (Wolfe et al., 1989).
MLO Quality Control Occurs after Insertion into the ER Membrane
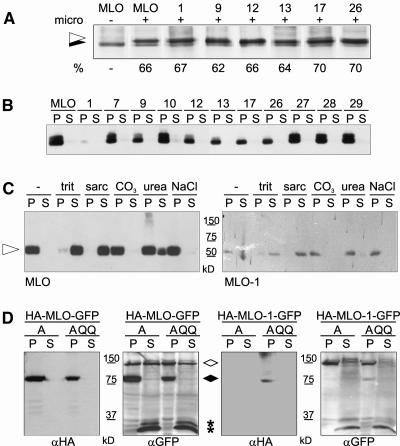
To find out whether mutant and wild-type MLO proteins differ in the ability to insert into the ER membrane, we tested a subset of the variants for in vitro membrane insertion efficiency. Because N-glycosylation of polypeptides is initiated by an oligosaccharyltransferase enzyme complex at the lumenal side of the ER membrane (Trombetta and Helenius, 2000), glycan attachment indicates localization of a protein/domain in the ER lumen. The native MLO protein lacks endogenous N-glycosylation consensus sequences. Upon introduction of an N-glycosylation acceptor site into the first extracellular loop, wild-type and mutant proteins were expressed in an in vitro transcription/translation system in the presence of canine ER microsomal membranes. As previously demonstrated by Devoto et al. (1999), the slower migrating band corresponds to N-glycosylated MLO proteins. We observed almost identical glycosylation efficiencies for wild-type MLO and each tested derivative (MLO-1, -9, -12, -13, -17, and -26; Figure 3A). Exposure of the first extracellular loop to the ER lumen suggested that at least in vitro transmembrane domains 1 and 2 of the tested MLO variants adopted their correct membrane topology.
Figure 3.
In Vitro and in Vivo Membrane Association of Wild-Type and Mutant MLO Proteins.
(A) In vitro membrane insertion efficiencies of wild-type MLO and six mutant derivatives engineered to carry an N-glycosylation acceptor site in the first extracellular loop. Presence or absence of canine microsomes (micro) is denoted by (+) or (−). The efficiency of N-glycosylation (%) indicated below the signals in each lane was calculated from the integrated areas corresponding to the N-glycosylated (indicated by the open arrowhead) and nonglycosylated (closed arrowhead) full-length products.
(B) In vitro membrane insertion competence of all MLO protein variants was verified in vivo. Extracts of Arabidopsis protoplasts expressing MLO proteins with C-terminal 3xHA tags were separated into membrane (P) and soluble (S) fractions by ultracentrifugation at 100,000g and analyzed by 10% SDS-PAGE and protein gel blot hybridization with an antiserum directed against the HA epitope.
(C) Integral membrane association of full-length MLO (left) and MLO-1 (right) in Arabidopsis protoplasts. C-terminally 3xHA-tagged MLO and MLO-1 were transiently expressed in Arabidopsis protoplasts under control of the CaMV 35S promoter. Membrane fractions were resuspended in 100 mM Na2CO3 (CO3) or 8 M Urea or in extraction buffer supplemented with 2% Triton X-100 (trit), 2% Sarcosyl (sarc), or 2 M NaCl and cleared by ultracentrifugation at 125,000g. Equal fractions of the resulting supernatants and pellets were analyzed by 12% SDS-PAGE and protein gel blot hybridization with an αHA antiserum. The arrowhead indicates full-length MLO fusion proteins. Sizes of a molecular weight standard are indicated.
(D) The entire MLO-1 protein is destabilized by the single amino acid substitution. Wild-type MLO and MLO-1 were engineered to carry a single HA tag between Lys 5 and Gly 6 of the cytosolic N terminus in addition to a C-terminal GFP moiety. Crude extracts of Arabidopsis protoplasts transiently expressing the double-labeled proteins in the presence of either cyan fluorescent protein (CFP)-AtCDC48A (A) or CFP-AtCDC48A QQ (AQQ) under control of the CaMV 35S promoter were separated in membrane (P) and soluble (S) fractions by centrifugation at 100,000g. Equal amounts of both fractions were analyzed by 10% SDS-PAGE and protein gel blot hybridization with αHA and αGFP (recognizing GFP and CFP) antisera. The open rhombus indicates CFP-tagged AtCDC48 A/AtCDC48A QQ, the closed rhombus indicates the position of full-length HA/GFP double-labeled MLO proteins, and asterisks indicate the position of small soluble C-terminal degradation products.
To verify efficient in vivo membrane insertion in our experimental model system, we fused the complete collection of MLO protein variants to a C-terminal 3xHA tag and transfected corresponding constructs into Arabidopsis protoplasts for transient expression. Cell extracts were separated in soluble and 100,000g microsomal fractions and analyzed by SDS-PAGE and protein gel blot hybridization (Figure 3B). αHA cross-reacting polypeptides of an apparent molecular mass of ∼60 kD were detected exclusively in membrane fractions. The consistent presence of two similarly sized, prominent bands upon expression of MLO wild-type and some mutant variants (e.g., MLO-7, -10, -27, -28, and -29) in independent protein gel blot experiments is indicative of a potential posttranslational modification during passage through the secretory pathway. Consistent absence of the upper signals in a subset of mutant variants (e.g., MLO-17 and MLO-26) may be evidence for differential progression of these variants within the endomembrane system. This is especially obvious for MLO-1 that accumulated to significantly lower levels than the other tested MLO proteins. Proper insertion of MLO and variants -1, -7, -9, and -12 in membranes rather than their peripheral association was further corroborated by the finding that the proteins remained attached to microsomal membranes in the presence of 100 mM Na2CO3, 8 M Urea, or 2 M NaCl, whereas detergent treatment caused their release into the soluble fraction (shown for MLO and MLO-1 in Figure 3C; similar data were obtained for MLO-7, -9, and -12; data not shown). Full-length wild-type or mutant MLO proteins were undetectable in the soluble fraction of membrane preparations or in immunoprecipitates of such fractions (data not shown), thus excluding the existence of a substantial soluble pool of MLO proteins in the cytosol. We conclude that the tested amino acid substitutions do not interfere with membrane insertion and that the suspected quality control process is activated subsequently to or coincidentally with membrane insertion of MLO proteins.
The Entire Unstable MLO-1 Protein Is Degraded
To rule out the possibility that C-terminal tags/reporter proteins may be degraded independently of the remaining major part of MLO proteins, we introduced a single HA tag in the extracellular/ER-lumenal N terminus of C-terminally GFP-tagged MLO and MLO-1 (between Lys 5 and Gly 6). Crude extracts derived from Arabidopsis protoplasts transfected with corresponding constructs were separated in soluble and 100,000g microsomal fractions and analyzed by protein gel blot hybridization with antibodies directed against both the N-terminal HA and the C-terminal GFP tag. Membrane fractions of MLO-expressing protoplasts revealed signals of ∼80 kD, corresponding to the full-length fusion protein (Figure 3D, left panels). MLO-1 was detectable only upon stabilization by the dominant negative variant of the Arabidopsis AAA ATPase Cdc48/p97 (AtCDC48A QQ), whose mammalian and yeast homologs are known to contribute to the retrotranslocation of ERAD substrates preceding proteasomal protein degradation (Figure 3D, right panels; Braun et al., 2002; Jarosch et al., 2002; see also below). The absence of major N- or C-terminal protein fragments suggested that the suspected quality control mechanism results in the complete removal of MLO-1 rather than a selective cleavage and degradation of the cytosolic C terminus.
Conservation of MLO Quality Control in Eukaryotic Cells
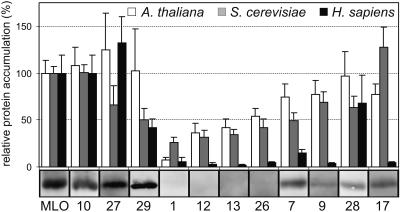
To test a potential conservation of protein quality control in plants and other eukaryotes, steady state protein levels were determined for all MLO mutants by dual luciferase assays of cell lysates obtained from Arabidopsis protoplasts, yeast, and human umbilical vein endothelial cells (HUVEC) transfected or transformed with suitable DNA constructs (Figure 4). Whereas the two–amino acid deletion derivative MLO-10 retained wild-type–like stability in all tested organisms, MLO-27 and -29 were stable in barley and Arabidopsis but exhibited variable accumulation in yeast and mammalian cells. Remarkably, all variants that were essentially undetectable in the barley mutant plants (MLO-1, -12, -13, and -26) were highly unstable in Arabidopsis, yeast, and HUVEC cells. Especially the MLO-1 variant was detected at consistently low levels in the tested cell types. Likewise, variants accumulating to intermediate levels in barley (MLO-7, -17, -28, and -9) showed, in general, a similar pattern in the heterologous test cells, though with exceptions (efficient removal of variants MLO-7, -9, and -17 in HUVEC cells and apparent stability of MLO-17 in S. cerevisiae). The ability of plants, yeast, and human cells to discriminate wild-type MLO from most aberrant forms is suggestive of a common recognition and/or degradation mechanism. We propose that destabilizing amino acid replacements in MLO generate universal signals recognized across kingdom borders.
Figure 4.
Relative Accumulation of Wild-Type and Mutant MLO Proteins in Barley Leaves, Arabidopsis Protoplasts, Yeast, and Mammalian Cells.
The relative accumulation of MLO protein variants in Arabidopsis protoplasts (white bars), yeast (gray bars), and HUVEC cells (black bars) was determined by dual luciferase assays. The protein gel blot analysis of plasma membrane fractions of the corresponding barley mutants is shown below the graph for comparison (identical but reassembled data of Figure 1B, top). Arabidopsis protoplasts and HUVEC cells were incubated for 24 to 27 h after transfection, whereas stably transformed yeast cells were grown to early stationary phase. For each cell lysate, luminescence values generated by the MLO-Renilla luciferase fusion proteins were divided by the corresponding firefly luciferase values. Relative protein accumulation was expressed as the percentage of the value determined for the wild-type MLO fusion. Each data point represents the mean + sd of at least three independent experiments including two to four samples.
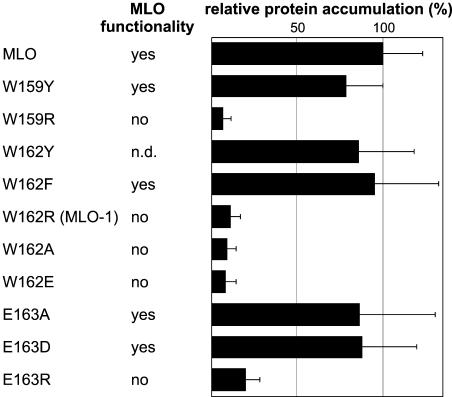
The Physico-Chemical Characteristics of Conserved Amino Acids Determine MLO Stability
In the least stable MLO mutant form, MLO-1, Trp 162 is substituted by Arg. Site-directed mutagenesis was employed next to this residue, which is highly conserved in all known plant MLO family members, to identify potential destabilization determinants. Replacements of Trp 159 and 162 as well as Glu 163, each also conserved among known MLO proteins (Devoto et al., 1999; Elliott et al., 2005), were tested for their impact on MLO accumulation by dual luciferase assays in Arabidopsis protoplasts. In parallel, we assessed the biological activity of each of the resulting MLO variants by single cell complementation assays in barley leaves upon Bgh challenge (Shirasu et al., 1999). Conservative substitutions of Trp 159 and 162 to nonpolar aromatic residues like Tyr or Phe affected neither protein stability nor activity relative to wild-type MLO (Figure 5). However, replacements by charged or small hydrophobic residues destabilized the protein to levels comparably low as MLO-1 and abolished its function. Similarly, conservative exchange of Glu 163 to Asp or replacement to the small uncharged residue Ala was tolerated, whereas substitution by positively charged Arg caused instability and loss of function (Figure 5). We conclude that the physico-chemical properties of conserved amino acids in this region are critical for MLO stability.
Figure 5.
Site-Directed Mutagenesis Next to MLO Residue W162.
Dual luciferase assays were performed with different MLO protein variants upon transient expression in Arabidopsis protoplasts as described in Figure 4. Relative protein accumulation was expressed as the percentage of the value determined for the wild-type MLO fusion. Each data point represents the mean + sd of at least three independent experiments, including two to four samples. Functionality of the respective MLO variants was determined by single cell complementation assays in detached mlo barley leaves (Shirasu et al., 1999). Transformed cells supporting invasive growth of Bgh sporelings indicate MLO functionality. n.d., not determined.
MLO Quality Control in Yeast Is Dependent on Proteasome Function
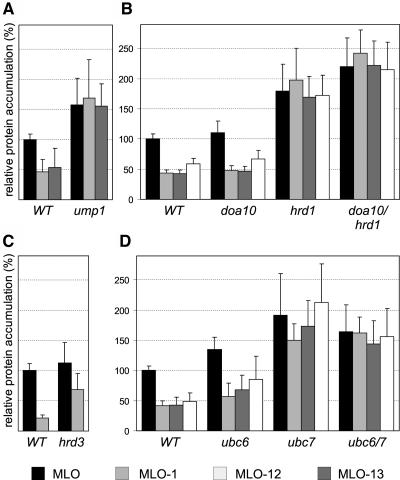
We took advantage of the apparent cross-kingdom conservation of MLO quality control to examine the underlying mechanism in yeast. We determined steady state accumulation of MLO and unstable variants MLO-1, -12, and -13 using dual luciferase experiments in mutant yeast strains with well-defined defects in protein degradation or quality control. MLO-1 and -12 were stabilized to wild-type levels in cells lacking the proteasome maturation factor Ump1p (Figure 6A; Ramos et al., 1998) and partially stabilized in a yeast strain carrying the pre1-1 and pre2-2 alleles that encode functionally impaired proteasome subunits (data not shown). Enhanced stability of MLO in the ump1 deletion strain (Figure 6A) may indicate that a fraction of the wild-type protein is also degraded in wild-type yeast. MLO-1 stability was unaffected in a yeast strain deficient in vacuolar proteolysis (Δpep4; data not shown), indicating that MLO quality control is mediated by a proteasome-dependent degradation mechanism rather than by vacuolar proteolysis.
Figure 6.
Relative MLO Protein Accumulation in Yeast Strains That Are Impaired in Proteasomal Protein Degradation and ERAD.
Dual luciferase assays were performed with different MLO protein variants, as described in Figure 4, in yeast strains deficient for Ump1p (A), Doa10p and/or Hrd1p (B), Hrd3p (C), and Ubc6p and/or Ubc7p (D). Accumulation of MLO variants was expressed as the percentage of the luminescence generated by wild-type MLO in the respective isogenic wild-type strains. Each data point represents the mean + sd of four to six independent experiments, including two samples per construct.
MLO Quality Control in Yeast Is Mediated by HRD-Dependent ERAD
Degradation of most known ERAD substrates in yeast is dependent on either Doa10p or Der3/Hrd1p, two integral ER ubiquitin ligases (Hampton, 2002). Relative to wild-type MLO accumulation, MLO-1, -12, and -13 were stabilized in hrd1 and hrd1/doa10 deletion strains but were efficiently degraded in a doa10 strain (Figure 6B; Swanson et al., 2001). We also detected a partial stabilization of MLO-1 in a yeast strain deficient for Hrd3p, an integral ER membrane protein physically interacting with Der3/Hrd1p and required for HRD-mediated ERAD (Figure 6C; Bays et al., 2001; Deak and Wolf, 2001). Furthermore, we could show that the ubiquitin-conjugating enzyme Ubc7p but not Ubc6p or Ubc1p was necessary for efficient disposal of the tested MLO variants (Figure 6D; data not shown). Ubc7p mediates Doa10p-dependent ubiquitination of some quality control substrates cooperatively with Ubc6p but acts independently of Ubc6p in the HRD pathway (Bays et al., 2001; Hampton, 2002). Enhanced accumulation of wild-type MLO was observed in ubc7, ubc6/7, and hrd1 cells and was reminiscent of the stabilization seen in an ump1 background (cf. Figure 6A). This indicated that a portion of the wild-type protein is subject to ERAD, possibly because of an overflow of the ER-folding capacity resulting from high-level expression of the heterologous protein. We conclude that in yeast, the mutant MLO proteins are preferred ERAD substrates that are recognized and degraded via the HRD pathway but are unaffected by Doa10p-dependent degradation.
MLO-1 Quality Control in Plants Requires Proteasome Function
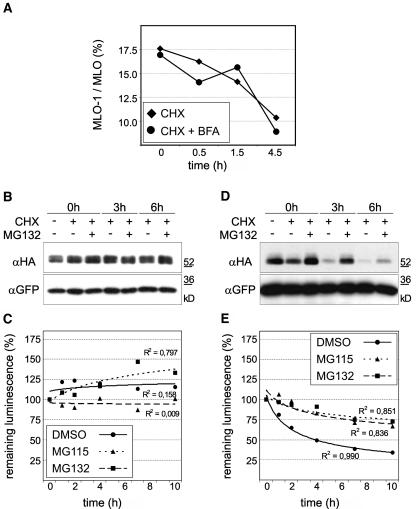
To investigate the suspected quality control mechanism in planta, we determined the effect of chemical inhibitors (Brefeldin A [BFA] and proteasome inhibitors) on the accumulation and stability of MLO and MLO-1 in Arabidopsis protoplasts. BFA impedes ER to Golgi vesicle transport in plant cells (Nebenführ et al., 2002). To test whether BFA affects the accumulation of MLO-1 relative to the wild-type protein, we performed protein accumulation assays (data not shown) and CHX chase experiments in the presence of 20 μg mL−1 BFA (Figure 7A). Collectively, the ratio of MLO-1 relative to the wild-type protein was not affected by BFA, suggesting that MLO-1 quality control does not require vesicle transport beyond the ER (Figure 7A).
Figure 7.
In Planta MLO Quality Control Is Unaffected by BFA Treatment but Requires Proteasome Function.
(A) CHX chase experiments in the presence of BFA were performed with Arabidopsis protoplasts expressing MLO and MLO-1 dual luciferase constructs. CHX (200 μg mL−1) and BFA (20 μg mL−1) were added after 12 h of incubation, and samples were taken after the indicated time points. Data points represent the percentage of MLO-1 with respect to wild-type MLO for each time point and treatment as determined by dual luciferase measurements. Each data point represents the mean of two independent experiments, including two samples per time point and treatment.
(B) to (E) CHX chase experiments with MLO ([B] and [C]) or MLO-1 ([D] and [E]) in Arabidopsis protoplasts were qualitatively evaluated by protein gel blot analysis ([B] and [D]) and quantified in independent experiments by dual luciferase assays ([C] and [E]; XY plots). CHX (200 μg mL−1) and the proteasome inhibitor MG115 (50 μM) or MG132 (50 μM) were added to the protoplasts at 7 h after transfection. Samples were taken at the indicated time points. The 3xHA-tagged MLO or MLO-1 was transiently coexpressed with GFP from the same plasmid. Crude extracts were analyzed by protein gel blot analysis using antibodies directed against the HA epitope (αHA; [B] and [D], top panels). The same blots were probed with an antiserum against GFP (αGFP; [B] and [D], bottom panels) as loading control. In independent experiments, MLO and MLO-1 stabilities were quantified by dual luciferase assays. Relative protein accumulation is expressed as the percentage of the value determined at the time of inhibitor addition (time point zero). Each data point represents the mean of two independent experiments, including two samples per time point and treatment. R2 indicates the R2 values for the shown trend lines.
We further performed CHX chase experiments in the presence of proteasome inhibitors. Relative amounts of full-length proteins were visualized by protein gel blot analysis of 3xHA-tagged MLO proteins (Figures 7B and 7D) and independently tested by quantitative dual luciferase assays (Figures 7C and 7E). Decay of MLO-1 was significantly delayed in the presence of proteasome inhibitors MG115 and MG132 in both assays (Figures 7D and 7E), indicating that proteasomal protein degradation is either directly or indirectly required for MLO-1 quality control in planta. By contrast, wild-type MLO levels, though variable, showed no clear evidence of stabilization or destabilization (Figures 7B and 7C).
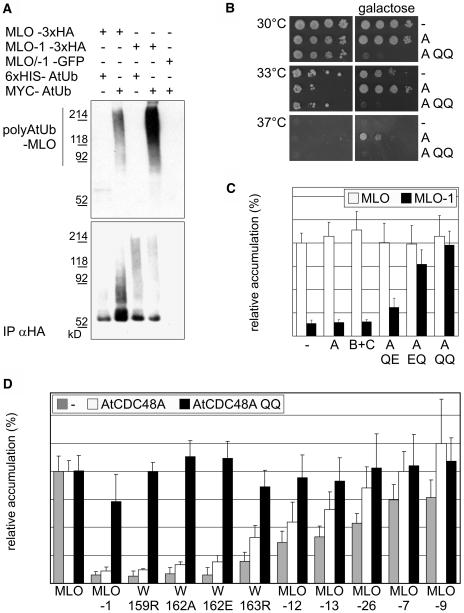
MLO-1 Is Polyubiquitinated in Arabidopsis Cells
To reveal a potential polyubiquitination of MLO-1, we coexpressed 3xHA-tagged MLO or MLO-1 with either c-myc proto-oncogene epitope (MYC)-tagged or 6xHIS-tagged Arabidopsis ubiquitin (AtUb), respectively, in Arabidopsis protoplasts. Immunoprecipitations from detergent-treated and cleared cell lysates were performed with antiHA affinity matrix and analyzed by SDS-PAGE and protein gel blot hybridization. A high molecular weight signal corresponding to multiubiquitinated protein was detected with a MYC-specific antiserum in the assay containing MLO-1 coexpressed with MYC-tagged ubiquitin (Figure 8A, top panel). This result is consistent with a direct degradation of MLO-1 by the proteasome. Polyubiquitinated MLO was also detected but at greatly reduced levels, suggesting that a small fraction of the wild-type protein is subject to degradation. This finding is reminiscent of the increased stability of wild-type MLO observed in ERAD-deficient yeast strains (cf. Figures 6A, 6B, and 6D). Thus, overexpression in the Arabidopsis protoplast system may exceed the ER-folding capacity and could also explain variation of wild-type MLO levels seen in these experiments (cf. Figure 7C).
Figure 8.
MLO-1 Is Polyubiquitinated and Can Be Stabilized by Dominant Negative Mutants of the AAA ATPase AtCDC48A in Arabidopsis Cells.
(A) Immunoprecipitates of detergent-treated and cleared Arabidopsis protoplast lysates with an antiHA affinity matrix were analyzed by SDS-PAGE and protein gel blot hybridization with αHA (bottom) and αMYC (top) specific antibodies. We coexpressed 3xHA-tagged MLO or MLO-1 with either MYC-tagged or 6xHIS-tagged AtUb, respectively. The proteasome inhibitor MG115 was added to the samples at a concentration of 50 μM 90 min before protoplast harvesting. No multiubiquitinated proteins were detected with the αMYC antibody in immunoprecipitates from extracts containing 3xHA-tagged MLO proteins in combination with 6xHIS-tagged AtUb. Likewise, no multiubiquitinated proteins were detected in αHA immunoprecipitates from protoplasts expressing a 1:1 mixture of GFP-tagged MLO and MLO-1 in combination with MYC-tagged AtUb.
(B) The cdc48 temperature-sensitive yeast strain KFY197 was transformed with an empty vector (−) or with plasmids encoding either AtCDC48A (A) or the AtCDC48A QQ (E308Q E581Q; indicated as A QQ) variant under the control of a galactose-inducible promoter. Cells were grown on media containing either glucose (left) or galactose (right) as carbon source at the permissive temperature (30°), at a temperature leading to impaired growth (33°C), or at nonpermissive temperature (37°C).
(C) Wild-type MLO (white bars) or MLO-1 (black bars) dual luciferase constructs were coexpressed in Arabidopsis protoplasts with wild-type AtCDC48A (A) or AtCDC48B plus AtCDC48C (B+C). Steady state MLO accumulation was determined as described in Figure 4. The same MLO dual luciferase constructs were coexpressed with mutant variants of AtCDC48A containing single amino acid substitutions in the Walker B motifs of either one or both ATPase domains (E308Q [A QE], E581Q [A EQ], and E308Q E581Q [A QQ]). Samples were incubated for 20 to 24 h post-transfection before lysis. Each data point represents the mean + sd of four to five independent experiments, including two samples per time point and treatment.
(D) Accumulation of MLO variants MLO-1, -7, -9, -12, -13, and -26 and single amino acid replacement variants W159R, W162A, W162E, and W163R (compare Figures 4 and 5) in the absence (gray bars) and presence of wild-type AtCDC48A (white bars) or AtCDC48A QQ (black bars) was determined as described in Figure 4. Relative protein accumulation was expressed as the percentage of the value determined for the wild-type MLO fusion. Each data point represents the mean + sd of at least three independent experiments, including two samples per time point and treatment.
Dominant Negative Mutants of the AAA ATPase AtCDC48A Impair MLO Quality Control in Arabidopsis Cells
One cellular function of the AAA ATPase Cdc48/p97 is a direct contribution to the retrotranslocation of ERAD substrates at an intermediate step preceding proteasomal protein degradation in mammalian cells and yeast (Braun et al., 2002; Jarosch et al., 2002). Three close sequence homologs of this AAA ATPase are present in the Arabidopsis genome, of which one (AtCDC48A) has been shown to functionally complement a yeast cdc48 mutant (Feiler et al., 1995; Rancour et al., 2002). Replacement of the conserved Glu of the Walker B motifs of either one or both ATPase domains in Cdc48p and p97 to Gln (E308Q [QE], E581Q [EQ], and E308Q E581Q [QQ]) leads to a dominant negative inhibition of retrotranslocation and consequently ERAD-mediated degradation. The mutations also exert a strong dominant negative effect on cell growth (Ye et al., 2003). We introduced the corresponding mutations in AtCDC48A and transformed a cdc48 temperature-sensitive yeast strain (KFY197) with expression vectors carrying AtCDC48A or the AtCDC48A QQ variant under control of a galactose-inducible promoter. At a nonpermissive temperature of 37°C, KFY197 cells were unable to grow on glucose as carbohydrate source, irrespective of the introduced plasmid (Figure 8B). Upon expression of the plasmid-encoded AtCDC48 variants on a galactose-containing culture medium, yeast growth was rescued in the presence of wild-type AtCDC48A but neither by an empty vector control nor by the AtCDC48A QQ variant. This finding confirms the ability of AtCDC48A to complement a cdc48 yeast mutant (Feiler et al., 1995) and shows that the AtCDC48A QQ modification abolishes this function. At 33°C, KFY197 growth was significantly reduced. On galactose-containing medium, AtCDC48A expression enhanced viability compared with the vector control, whereas AtCDC48A QQ expression completely terminated cell growth. This dominant negative effect of AtCDC48A QQ was further corroborated by the observation that it impaired cell growth at the permissive temperature of 30°C.
The effect of AtCDC48A and AtCDC48A QQ expression on the accumulation of wild-type MLO and a subset of mutant MLO variants in Arabidopsis protoplasts was subsequently analyzed by dual luciferase assays. Coexpression of wild-type AtCDC48A or AtCDC48B and AtCDC48C did not specifically alter the accumulation of individual MLO variants but seemed to cause a general increase in the Renilla luciferase:firefly luciferase ratio (Figures 8C and 8D). MLO-1 levels relative to wild-type MLO were slightly enhanced in the presence of AtCDC48A QE and significantly increased by AtCDC48A EQ (Figure 8C). Remarkably, the variant impaired in both ATPase domains (AtCDC48A QQ) stabilized MLO-1 and other variants carrying destabilizing replacements of conserved residues neighboring the MLO-1 mutation to levels that were comparable to the wild-type MLO protein (Figures 8C and 8D). Whereas MLO variants -12 and -13 accumulated to moderately increased levels in the presence of AtCDC48A QQ, the slight impact on variants -7, -9, and -26 was not significant. This may indicate variant-specific differences in the dependence on AtCDC48A activity or may be because of insufficient sensitivity of the protein accumulation assay. Protein gel blots of microsomal membrane fractions isolated from Arabidopsis protoplasts coexpressing 3xHA-tagged MLO protein variants and HA-/GFP- double tagged MLO and MLO-1 with AtCDC48A QQ corroborated that the stabilization deduced from dual luciferase assays is indeed because of an increased accumulation of membrane-bound full-length MLO proteins (Figure 3D; data not shown). We conclude that in planta quality control of several unstable MLO variants shares CDC48 ATPase activity requirement for ERAD-mediated protein degradation with yeast and human cells (Ye et al., 2003).
DISCUSSION
Single Amino Acid Replacements Target MLO for Postinsertional Quality Control
Allele-specific impairment of MLO protein accumulation was observed in a series of barley mlo mutants. We show that single amino acid replacements target the plant-specific 7-TM protein for degradation by a posttranscriptional and posttranslational quality control process. Glycosylation at an engineered N-glycosylation site indicated that at least transmembrane domains 1 and 2, the first intracellular, and a substantial part of the first extracellular loop of the tested MLO variants retained the ability to insert in membranes in vitro (Figure 3A). Although we have not tested whether the remaining protein portion adopts also a wild-type–like membrane topology, exclusive detection of tagged-MLO variants in microsomal fractions upon transient expression in Arabidopsis protoplasts (Figures 3B to 3D) indicated that MLO quality control is initiated by membrane-inserted substrates. CHX chase experiments with 3xHA- and Renilla luciferase–tagged MLO proteins (Figures 2C and 7) confirmed in vivo that reduced accumulation is not attributable to attenuation of translation because detection of C-terminal tags requires translation of the entire fusion protein. Furthermore, experiments using N-terminally tagged MLO and MLO-1 demonstrated that the quality control mechanism mediates degradation of the entire protein (Figure 3D).
The analysis of an allelic series of mutant MLO variants allows several conclusions. First, single amino acid substitutions in both lumenal and cytoplasmic regions of the protein suffice to trigger degradation. This is reminiscent of the distributed degron (multiple distinct regions of primary sequence) responsible for regulated, ERAD-mediated degradation of Hmg2p (Gardner and Hampton, 1999) and multiple degradation signals identified on the unassembled Na,K-ATPase α subunit (Béguin et al., 2000). Second, the remarkable variation in the degree of allele-specific destabilization implies that individual MLO variants may be recognized either at different maturation stages, with different efficiencies, in different subcellular compartments, or by different components of the cellular quality control machinery. Individual single amino acid replacements may affect the extent or the kinetics of protein folding by reducing accessibility to interacting partners or prolonging association with maturation factors, which in turn may stimulate cellular quality control. This was demonstrated for folding-incompetent N-glycosylated proteins that are retained in the calnexin cycle and targeted for ERAD-mediated degradation (Trombetta and Helenius, 2000). Third, we could show that the physico-chemical properties rather than the identity of particular amino acid side chains of a region in intracellular loop 2 are decisive for overall MLO stability (Figure 5). Bulky hydrophobic amino acids next to Trp 162 may be crucial in enabling proper protein folding, possibly creating an interaction surface for molecular chaperones that are highly conserved across kingdoms (Flynn et al., 1991; Béguin et al., 2000). Thus, one intriguing hypothesis is that inappropriate chaperone binding to MLO-1 mutant proteins might promote their rapid degradation, as reported for Hsp90/CFTR in humans (Loo et al., 1998).
Recognition and Degradation of Mutated MLO Proteins Is Conserved in Eukaryotic Cells
Protein accumulation of the complete series of variants relative to wild-type MLO was tested in barley mutants, Arabidopsis protoplasts, yeast, and mammalian HUVEC cells (Figure 4). We observed striking similarities in relative protein accumulation profiles for MLO variants -1, -12, -13, and -26, which were immunologically undetectable in barley membrane fractions and relatively unstable in each of the other tested cell types. The comparably high levels of MLO-12, -13, and -26 observed in Arabidopsis and yeast cells may be explained by elevated expression levels under the respective experimental conditions, which may saturate the cellular quality control capacity. However, this does not appear to be the case for MLO-1. Consistently low accumulation levels in all tested cell types may reflect exceptionally efficient removal of this variant (Figure 4). The reasons for this remain unclear. A subset of the variants that accumulated to wild-type or intermediate levels in barley and Arabidopsis exhibited a different accumulation pattern in yeast and mammalian cells (e.g., MLO-7, -9, -17, -27, and -29; Figure 4). These deviations could reflect quantitative differences that are the result of particular physiological conditions inflicted on the cells by the experimental setup or may result from species- or cell type–specific differences in protein quality control. It is known that yeast integral membrane proteins contain usually longer transmembrane domains than mammalian proteins. Short transmembrane helices promote vacuolar targeting and subsequent degradation (Rayner and Pelham, 1997). Thus, the increased stability of MLO-17 in yeast may be caused by the extension of the hydrophobic amino acid stretch forming transmembrane helix 1 through replacement of Ser 31 by Phe. ERAD-mediated degradation of mutated MLO proteins may be superimposed on this independent vacuolar quality control mechanism, thereby leading to a generally low basal accumulation of MLO proteins in yeast cells.
We conclude that the specificity of substrate recognition appears largely conserved in monocot and dicot plant species, yeast, and human cells, although MLO-like proteins are absent in animals and yeast. Thus, the discrimination between malformed and native MLO variants is most likely based on an intrinsic property of the proteins and independent of their functional integrity or the necessity to associate with specific interaction partners not expected to be present in distantly related organisms. A corollary of this is that the flexibility of eukaryotic membrane protein evolution is not only limited by structural constraints associated with the formation of TM helices (Leabman et al., 2003) but also by a universal ERAD recognition mechanism monitoring cytoplasmic and lumenal regions.
MLO Quality Control Is Mediated by HRD Pathway–Dependent ERAD in Yeast
We have exploited the cross-species conservation of MLO quality control to identify HRD-mediated ERAD as the underlying mechanism in yeast. Factors required for the degradation of several endogenous and heterologous ERAD substrates have been characterized biochemically and genetically in this unicellular eukaryote. The accumulation of mutated MLO variants relative to the wild-type protein was specifically increased in yeast strains impaired in proteasome function (Ump1p deletion strain; Figure 6A). Furthermore, stabilization of MLO variants was observed in strains lacking an ERAD-specific ubiquitin-conjugating enzyme (Ubc7p) and components of an ubiquitin ligase complex (Hrd1/Der3p and Hrd3p; Figures 6B to 6D). Elevated levels of the wild-type protein in these yeast strains indicate that overexpression of a heterologous polytopic membrane protein can trigger the cellular quality control, though at reduced stringency compared with aberrant variants of the same protein. Collectively, it appears that the HRD pathway of ERAD can discriminate between wild-type and unstable MLO proteins and is necessary to drive their proteasome-dependent degradation in yeast. This is corroborated by the finding that neither wild-type MLO nor unstable variants are stabilized in yeast strains deficient for components of an alternative Doa10p- and Ubc6p-dependent ERAD pathway (Figures 6B and 6D).
Arabidopsis sequence homologs of yeast Ump1p, Hrd1p, Hrd3p, and Ubc7p are potential determinants of in planta MLO stability (Table 1). We therefore tested the effect of overexpression as well as double stranded RNA interference–mediated gene silencing of such candidates on MLO-1 stability by dual luciferase assays in Arabidopsis protoplasts. With the exception of an unspecific stabilization of MLO-1 and several cytosolic proteasome substrates (Worley et al., 1998, 2000) upon overexpression of Arabidopsis homologs of the ubiquitin conjugating enzymes Ubc1p and Ubc7p, we failed to detect a consistent impairment of MLO quality control (data not shown). This finding may indicate that the selected genes do not encode functional orthologs of yeast ERAD components in spite of their sequence relatedness. Analysis of Arabidopsis T-DNA insertion mutants will provide an alternative approach to evaluate the role of such candidate genes in plant protein quality control.
Table 1.
Arabidopsis Homologs of Yeast Genes Involved in ERAD
| Yeast Gene | Arabidopsis Homologs |
|---|---|
| Ump1 | At1g67250, At5g38650 |
| Ubc7 | At3g46460, At3g55380, At5g59300 |
| Ubc6 | At1g17280, At5g50430, At3g17000 |
| Ubc1 | At5g50870 |
| Der3/Hrd1 | At1g65040, At3g16090, At4g25230, At5g51450 |
| Hrd3 | At1g18260 |
| Cdc48 | At3g09840, At3g53230, At5g03340 |
ERAD in Plants: A Novel Role for AtCDC48
We have shown for a subset of MLO variants that in planta, MLO quality control occurs posttranscriptionally and postinsertionally (Figures 1C and 3), is dependent on proteasome function (Figures 7B to 7E), involves substrate polyubiquitination (Figure 8A), and can be quantitatively abolished by overexpression of dominant negative mutants of the AAA ATPase AtCDC48A (Figures 3D, 8C, and 8D)—features typically associated with ERAD in yeast and mammalian cells. In these cells, the AAA ATPase Cdc48/p97 participates in diverse cellular processes, including membrane fusion and the dislocation of ERAD substrates (Kondo et al., 1997; Ye et al., 2003). Three close sequence homologs of this AAA ATPase are present in the Arabidopsis genome (AtCDC48A, B, and C; Rancour et al., 2002), sharing 91 to 96% sequence identity among each other and 65 and 76% identity with yeast Cdc48p and human p97, respectively. It has been shown that AtCDC48A functionally complements S. cerevisiae cdc48 mutants. Similar to their yeast and mammalian counterparts, AtCDC48 proteins assemble mainly in homo-hexameric and higher order hetero-oligomeric complexes (Feiler et al., 1995; Rancour et al., 2002). It is likely that AtCDC48 participates in at least two distinct membrane fusion pathways during plant cytokinesis (Rancour et al., 2002). Our data are strongly indicative of an additional AtCDC48 function in plant ERAD. This demonstrates the conservation of multiple functions of this protein family in eukaryotes.
ERAD in Plants: Are All Eukaryotic Cells Alike?
Aberrant variants of barley MLO are the first described examples of endogenous polytopic membrane proteins monitored by an ERAD-like quality control mechanism in plant cells. In accordance with the previously reported degradation of the small soluble ricin A chain upon heterologous expression in tobacco cells, our results strengthen the evolving picture of protein quality control in plants (Di Cola et al., 2001; Kostova and Wolf, 2003). Though characteristic ERAD events such as substrate recognition, retrograde transport, and proteasomal degradation have been maintained from yeast to multicellular eukaryotes, including plants, particular cellular and environmental contexts may demand alternative/complementary facets of protein quality control. Stimulus-dependent regulation of protein surveillance and folding capacity may be especially important for sessile plants obliged to adapt to varying environmental conditions. The finding that feedback regulation of sterol synthesis in mammalian cells uses the ERAD machinery (Hampton, 2002) illustrates co-option of the basic quality control mechanism for regulatory processes and reveals potential functions in cell-to-cell signaling. The identification of further plant degradation substrates and genetic screens similar to those previously performed in S. cerevisiae (Hampton et al., 1996; Knop et al., 1996; Swanson et al., 2001) will be instrumental in the identification of conserved as well as plant-specific aspects of protein quality control.
METHODS
RNA Gel Blot Analysis
Total RNA was isolated from 3-week-old seedlings using the TriReagent (Sigma-Aldrich Chemie, Taufkirchen, Germany) protocol, and poly(A)+-enriched RNA fractions prepared using Oligotex columns (Qiagen, Hilden, Germany). RNA gel blot analysis was performed with 500 ng of poly(A)+ RNA per lane using standard procedures. Probes consisted of the full-length Mlo cDNA (Büschges et al., 1997) and the full-length cDNA-encoding barley (Hordeum vulgare) GAPDH (Chojecki, 1986).
Barley Microsomal and Plasma Membrane Vesicle Preparation
Plasma membrane vesicle purification by two-phase aqueous partitioning and protein gel blot analysis were performed as described previously (Devoto et al., 1999) using the MLO-specific affinity-purified antiserum raised against a peptide of the MLO C terminus (MYP antiserum). Immunostained protein amounts were quantified with a Fujix BAS 1000 phosphor imaging plate scanner (Fuji, Tokyo, Japan) and MacBas software (version 2.31).
In Vitro Membrane Insertion Analysis
We engineered six single amino acid substitution mutants in an MLO derivative containing an N-glycosylation acceptor site in the first extracellular loop (plasmid pEC1B; Devoto et al., 1999). Supercoiled-plasmid DNA was used directly for in vitro transcription/translation with the TNT SP6-coupled reticulocyte lysate system (Promega, Madison, WI) following the manufacturer's instructions. Occurrence of N-linked glycosylation was tested in the presence of canine pancreatic microsomal membranes (Promega). Membrane integration of translation products was determined by extraction with 150 μL of ice-cold 0.1 M Na2CO3, pH 11, followed by recovery of the stripped microsomes by centrifugation (245,000g for 10 min). All samples were analyzed by 10% SDS-PAGE. Gels were scanned using a Fujix BAS 1000 phosphor imaging plate scanner and images analyzed using the MacBas software (version 2.31).
Plasmid Constructions
DNA manipulations were performed according to standard methods. All plant expression constructs were based on the binary plant expression vector pAMPAT-MCS (GenBank accession number AY436765). For dual luciferase constructs, in-frame fusions of Renilla luciferase–encoding DNA fragments to the respective Mlo cDNAs were inserted between the double CaMV 35S promoter and terminator of the vector, and a second expression cassette consisting of a single CaMV 35S promoter, the firefly luciferase cDNA, and the 35S terminator was introduced into the NotI and PmeI sites of the same construct. For protein gel blot analysis, the Renilla luciferase cDNA of these constructs was replaced by a 3xHA-encoding fragment and the firefly luciferase by the cDNA encoding chloramphenicol acetyltransferase GFP (kindly provided by Guido Jach). The combined Renilla luciferase-3xHA tag was obtained by ligating PCR-amplified stop codonless Renilla luciferase between the MLO and the HA portion of these constructs. The N-terminal HA tag was introduced after amino acid five of MLO and MLO-1 by PCR mutagenesis. cDNAs corresponding to Arabidopsis thaliana AtCDC48A-C (Rancour et al., 2002) were PCR amplified from a flower cDNA library (kindly provided by Laurent Deslandes). The mutations E308Q and/or E581Q were introduced into the AtCDC48A construct by PCR mutagenesis. For CFP fusions of AtCDC48A and AtCDC48A QQ, the PCR-amplified stop codonless CFP cDNA was ligated between the 35S promoter and the start codon of corresponding AtCDC48A expression constructs. AtUb was PCR amplified from a plasmid carrying the first repeat of the Arabidopsis UBQ11 gene (kindly provided by Hans-Peter Stuible).
Vectors of the pRS series (Sikorski and Hieter, 1989) carrying the GAL1 promoter and CYC1 terminator were used for experiments in yeast (Saccharomyces cerevisiae). MLO-Renilla luciferase fusions and firefly luciferase were expressed from a bicistronic-mRNA molecule. Introduction of the internal ribosomal-entry site from Potato leafroll virus (Jaag et al., 2003) between the stop codon of the MLO-Renilla luciferase fusion protein and the ATG of firefly luciferase allowed an efficient cap-dependent translation of the MLO-fusion protein and a less efficient internal cap-independent translation initiation of firefly luciferase.
For expression in mammalian cells, cDNAs encoding MLO-Renilla luciferase fusions were cloned between the cytomegalovirus promoter and the bovine growth hormone polyadenylation site of pcDNA3 (Invitrogen, Carlsbad, CA). Firefly luciferase was coexpressed from pCLuc (Plank et al., 1992).
Protein Expression and Detection in Arabidopsis Protoplasts, Yeast, and Mammalian Cells
Arabidopsis cells (ecotype Columbia-0, line At7) were propagated as reported in Trezzini et al. (1993). Protoplast preparations and transfections were performed according to the protocol described previously (Sprenger-Haussels and Weisshaar, 2000) with a modified-enzyme solution containing 12 units mL−1 Cellulase Onuzoka R-10 (Merck, Darmstadt, Germany) and 1.5 units mL−1 Mazerozyme R-10 (Serva, Heidelberg, Germany). Chemical treatments with CHX (Sigma-Aldrich Chemie), BFA (Sigma-Aldrich Chemie), and proteasome inhibitors MG115 (Z-Leu-Leu-Nva-H) and MG132 (Z-Leu-Leu-Leu-H; Peptides International, Louisville, KY) were performed as indicated. For protein gel blot analysis, Arabidopsis protoplasts were harvested in W5 medium (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM Mes, pH 5.7). Protoplast pellets were lysed in 20 mM Hepes, pH 7.5, 12% (w/v) sucrose, 1 mM EDTA, pH 8.0, 1 mM DTT, and complete protease inhibitor mixture (Roche Diagnostics, Mannheim, Germany).
For microsome isolation, protoplasts were harvested in W5 medium. Protoplast pellets were resuspended in resuspension buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 10 mM EDTA, 5 mM Ascorbat, 5 mM DTT, and protease inhibitor mixture [Roche Diagnostics]). Solubilization experiments were performed as indicated in the text. For immunoprecipitations, protoplasts were supplemented with 50 μM MG115 or MG132 (Peptides International) 8 h after transfection, incubated for another 90 min, and harvested in W5 medium supplemented with 1 mM N-ethyl-maleimide (Sigma-Aldrich Chemie). Protoplast pellets were resuspended in resuspension buffer containing 20 mM N-ethyl-maleimide, 2% Sarcosyl, and 1% SDS by sonication, incubated for 1 h at 30°C, and centrifuged at 4500g for 1 h. The supernatant was cleared by ultracentrifugation at 100,000g for 1 h and diluted 1:1 with resuspension buffer without detergents. Immunoprecipitation was performed with antiHA affinity matrix (Roche Diagnostics) overnight at 4°C. Seven washings were performed with resuspension buffer containing 0.2% Sarcosyl. Samples were supplemented with SDS-PAGE loading buffer and denatured at 55°C for 15 min before SDS-PAGE and protein gel blot analysis.
Antibodies directed against the following epitopes were used in this study: αHA and αGFP (Roche Diagnostics) and αMYC (Santa Cruz Biotechnology, Santa Cruz, CA). Corresponding secondary antisera and detection kits were purchased from Sigma-Aldrich Chemie, Amersham (Freiburg, Germany [ECL]), or Bio-Rad Laboratories (Hercules, CA [Immun Star AP]).
For dual luciferase reporter assays, protoplast suspensions were diluted in 240 mM CaCl2, harvested by centrifugation (15,000g), shock-frozen in liquid nitrogen, and extracted in the lysis buffer supplied with the dual luciferase kit (Promega). Measurements were performed according to the manufacturer's instructions.
Yeast strains used in this study are listed in Table 2. Standard techniques were used for transformation and propagation of yeast cells (Ausubel et al., 1989). Expression of the reporter constructs was induced in galactose-containing culture media. For stationary chase experiments (cf. Bays et al., 2001), cells were grown to early stationary phase (12 to 36 h, depending on the yeast strain), harvested by centrifugation, and shock-frozen in liquid nitrogen. Cell extracts were prepared by vigorous vortexing in lysis buffer in the presence of glass beads (425 to 600 μm; Sigma-Aldrich Chemie). HUVEC were transfected with the Nucleofector in cooperation with Amaxa (Cologne, Germany) and lysed for dual luciferase assays following the standard protocol.
Table 2.
Yeast Strains Used in This Study
| Strain | Genotype | Reference |
|---|---|---|
| BBY61 | MATa, pep4::His3, ura3-52, his3Δ200, leu2-3,112, lys2-801, gal- | Bartel et al. (1990) |
| JD47-13C | MATa, his3-Δ200, leu2-3,112, lys2-801, trp1-Δ63, ura3-52 | Ramos et al. (1998) |
| JD59 | MATa, his3-Δ200, leu2-3,112, lys2-801, trp1-Δ63, ura3-52, ump1Δ::HIS3 | Ramos et al. (1998) |
| BR1 | MATa, ura3Δ5, his3-11,15, GAL+ | Heinemeyer et al. (1993) |
| BR4 | MATα, pre1-1, pre2-2, ura3Δ5, his3-11,15, Leu2-3,112, GAL+ | Heinemeyer et al. (1993) |
| MHY501 | MATα, ura3-52, leu2-3,-112, his3-Δ200, trp1-1, lys2-801 | Chen et al. (1993) |
| MHY552 | MATα, his3-Δ200, ura3-52, leu2-3112, lys2-801, trp1-1, ubc6- 1::HIS3, ubc7::LEU2 | Chen et al. (1993) |
| MHY495 | MATα, ura3-52, leu2-3,-112, his3-Δ200, trp1-1, lys2-801, ubc6::HIS3 | Chen et al. (1993) |
| MHY507 | MATα, ura3-52, leu2-3,-112, his3-Δ200, trp1-1, lys2-801, ubc7::LEU2 | Chen et al. (1993) |
| MHY501 | MATα, ura3-52, leu2-3,-112, his3-Δ200, trp1-1, lys2-801, gal2 | Chen et al. (1993) |
| MHY1631 | MATα, ura3-52, leu2-3,-112, his3-Δ200, trp1-1, lys2-801, doa10Δ1::HIS3 | Swanson et al. (2001) |
| MHY1669 | MATα, ura3-52, leu2-3,-112, his3-Δ200, trp1-1, lys2-801, hd1Δ::LEU2 | Bays et al. (2001) |
| MHY1703 | MATα, ura3-52, leu2-3,-112, his3-Δ200, trp1-1, lys2-801, hd1Δ::LEU2, doa10Δ::HIS3 | Swanson et al. (2001) |
| RHY717 | leu2Δ, trp1Δ, URA3::6MYC HMG2 (Ura+6myc), hmg1Δ::LYS2, hmg2Δ::HIS3, MATa | Bays et al. (2001) |
| RHY749 | hrd3Δ::URA3, URA3::6MYC HMG2 (Ura+6myc), hmg1Δ::LYS2, hmg2Δ::HIS3, MATα | Bays et al. (2001) |
| KFY197 | KFY197 is a temperature-sensitive revertant of the cold-sensitive cdc48-1 strain and carries an intragenic suppressor mutation in cdc48. | Rabinovich et al. (2002); K.-U. Fröhlich, personal communication |
Acknowledgments
We thank R.J. Dohmen, R.Y. Hampton, and M. Hochstrasser for providing yeast strains; L. Deslandes, G. Jach, and H.P. Stuible for providing DNA constructs; A. Reinstädler, M. Decker, and T. Turbanski for technical assistance; and L. Bollenbach for maintenance of cell cultures. A. Bachmair and T.A. Rapoport are acknowledged for constructive discussions. We thank N. Collins and T. Langer for critical reading of the manuscript. J.M. was supported by a postdoctoral fellowship of the Max Planck Society.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Judith Müller (jmueller@mpiz-koeln.mpg.de), Paul Schulze-Lefert (schlef@mpiz-koeln.mpg.de), and Ralph Panstruga (panstrug@mpiz-koeln.mpg.de).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.026625.
References
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1989). Current Protocols in Molecular Biology. (New York: John Wiley & Sons).
- Bartel, B., Wunning, I., and Varshavsky, A. (1990). The recognition component of the N-end rule pathway. EMBO J. 9, 3179–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bays, N.W., Gardner, R.G., Seelig, L.P., Joazeiro, C.A., and Hampton, R.Y. (2001). Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat. Cell Biol. 3, 24–29. [DOI] [PubMed] [Google Scholar]
- Béguin, P., Hasler, U., Staub, O., and Geering, K. (2000). Endoplasmic reticulum quality control of oligomeric membrane proteins: Topogenic determinants involved in the degradation of the unassembled Na,K-ATPase alpha subunit and in its stabilization by beta subunit assembly. Mol. Biol. Cell 11, 1657–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordallo, J., Plemper, R.K., Finger, A., and Wolf, D.H. (1998). Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol. Biol. Cell 9, 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi, F., Hanton, S., DaSilva, L.L., Boevink, P., Evans, D., Oparka, K., Denecke, J., and Hawes, C. (2003). ER quality control can lead to retrograde transport from the ER lumen to the cytosol and the nucleoplasm in plants. Plant J. 34, 269–281. [DOI] [PubMed] [Google Scholar]
- Braun, S., Matuschewski, K., Rape, M., Thoms, S., and Jentsch, S. (2002). Role of the ubiquitin-selective CDC48(UFD1/NPL4) chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 21, 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, J.L., and McCracken, A.A. (1999). ER protein quality control and proteasome-mediated protein degradation. Semin. Cell Dev. Biol. 10, 507–513. [DOI] [PubMed] [Google Scholar]
- Büschges, R., et al. (1997). The barley Mlo gene: A novel control element of plant pathogen resistance. Cell 88, 695–705. [DOI] [PubMed] [Google Scholar]
- Casagrande, R., Stern, P., Diehn, M., Shamu, C., Osario, M., Zuniga, M., Brown, P.O., and Ploegh, H. (2000). Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol. Cell 5, 729–735. [DOI] [PubMed] [Google Scholar]
- Chen, P., Johnson, P., Sommer, T., Jentsch, S., and Hochstrasser, M. (1993). Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell 74, 357–369. [DOI] [PubMed] [Google Scholar]
- Chojecki, J. (1986). Identification and characterization of a cDNA clone for cytosolic glyceraldehyde-3-phosphate dehydrogenase in barley. Carlsberg Res. Commun. 51, 203–210. [Google Scholar]
- Collins, N.C., Thordal-Christensen, H., Lipka, V., Bau, S., Kombrink, E., Qiu, J.L., Hückelhoven, R., Stein, M., Freialdenhoven, A., Somerville, S.C., and Schulze-Lefert, P. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425, 973–977. [DOI] [PubMed] [Google Scholar]
- Deak, P.M., and Wolf, D.H. (2001). Membrane topology and function of Der3/Hrd1p as a ubiquitin-protein ligase (E3) involved in endoplasmic reticulum degradation. J. Biol. Chem. 276, 10663–10669. [DOI] [PubMed] [Google Scholar]
- Devoto, A., Andreas Hartmann, A.H., Piffanelli, P.H., Elliott, C.H., Simmons, C.H., Taramino, G.H., Goh, C.S., Cohen, F.E., Emerson, B.C., Schulze-Lefert, P.C., and Panstruga, R.C. (2003). Molecular phylogeny and evolution of the plant-specific seven-transmembrane MLO family. J. Mol. Evol. 56, 77–88. [DOI] [PubMed] [Google Scholar]
- Devoto, A., Piffanelli, P., Nilsson, I., Wallin, E., Panstruga, R., von Heijne, G., and Schulze-Lefert, P. (1999). Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J. Biol. Chem. 274, 34993–35004. [DOI] [PubMed] [Google Scholar]
- Di Cola, A., Frigerio, L., Lord, J.M., Ceriotti, A., and Roberts, L.M. (2001). Ricin A chain without its partner B chain is degraded after retrotranslocation from the endoplasmic reticulum to the cytosol in plant cells. Proc. Natl. Acad. Sci. USA 98, 14726–14731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, C., Müller, J., Miklis, M., Bhat, R.A., Schulze-Lefert, P., and Panstruga, R. (2005). Conserved extracellular cysteines and cytoplasmic loop-loop interplay are required for functionality of the heptahelical MLO protein. Biochem. J., in press. [DOI] [PMC free article] [PubMed]
- Feiler, H.S., Desprez, T., Santoni, V., Kronenberger, J., Caboche, M., and Traas, J. (1995). The higher plant Arabidopsis thaliana encodes a functional CDC48 homologue which is highly expressed in dividing and expanding cells. EMBO J. 14, 5626–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn, G.C., Pohl, J., Flocco, M.T., and Rothman, J.E. (1991). Peptide-binding specificity of the molecular chaperone BiP. Nature 353, 726–730. [DOI] [PubMed] [Google Scholar]
- Friedlander, R., Jarosch, E., Urban, J., Volkwein, C., and Sommer, T. (2000). A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat. Cell Biol. 2, 379–384. [DOI] [PubMed] [Google Scholar]
- Gardner, R.G., and Hampton, R.Y. (1999). A ‘distributed degron’ allows regulated entry into the ER degradation pathway. EMBO J. 18, 5994–6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, R.G., Shearer, A.G., and Hampton, R.Y. (2001). In vivo action of the HRD ubiquitin ligase complex: Mechanisms of endoplasmic reticulum quality control and sterol regulation. Mol. Cell. Biol. 21, 4276–4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton, R. (2002). ER-associated degradation in protein quality control and cellular recognition. Curr. Opin. Cell Biol. 14, 476–482. [DOI] [PubMed] [Google Scholar]
- Hampton, R.Y., Gardner, R.G., and Rine, J. (1996). Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell 7, 2029–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer, W., Gruhler, A., Mohrle, V., Mahe, Y., and Wolf, D.H. (1993). PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chrymotryptic activity and degradation of ubiquitinated proteins. J. Biol. Chem. 268, 5115–5120. [PubMed] [Google Scholar]
- Hong, E., Davidson, A.R., and Kaiser, C.A. (1996). A pathway for targeting soluble misfolded proteins to the yeast vacuole. J. Cell Biol. 135, 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaag, H.M., Kawchuk, L., Rohde, W., Fischer, R., Emans, N., and Prüfer, D. (2003). An unusual internal ribosomal entry site of inverted symmetry directs expression of a potato leafroll polerovirus replication-associated protein. Proc. Natl. Acad. Sci. USA 100, 8939–8944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosch, E., Taxis, C., Volkwein, C., Bordallo, J., Finley, D., Wolf, D.H., and Sommer, T. (2002). Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat. Cell Biol. 4, 134–139. [DOI] [PubMed] [Google Scholar]
- Jelitto-Van Dooren, E.P., Vidal, S., and Denecke, J. (1999). Anticipating endoplasmic reticulum stress. A novel early response before pathogenesis-related gene induction. Plant Cell 11, 1935–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M.C., Panstruga, R., Elliott, C., Müller, J., Devoto, A., Yoon, H.W., Park, H.C., Cho, M.J., and Schulze-Lefert, P. (2002). Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416, 447–451. [DOI] [PubMed] [Google Scholar]
- Knop, M., Finger, A., Braun, T., Hellmuth, K., and Wolf, D.H. (1996). Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J. 15, 753–763. [PMC free article] [PubMed] [Google Scholar]
- Kondo, H., Rabouille, C., Newman, R., Levine, T.P., Pappin, D., Freemont, P., and Warren, G. (1997). p47 is a cofactor for p97-mediated membrane fusion. Nature 388, 75–78. [DOI] [PubMed] [Google Scholar]
- Kostova, Z., and Wolf, D.H. (2003). Waste disposal in plants: Where and how? Trends Plant Sci. 8, 461–462. [DOI] [PubMed] [Google Scholar]
- Leabman, M.K., et al. (2003). Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc. Natl. Acad. Sci. USA 100, 5896–5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leborgne-Castel, N., Jelitto-Van Dooren, E.P., Crofts, A.J., and Denecke, J. (1999). Overexpression of BiP in tobacco alleviates endoplasmic reticulum stress. Plant Cell 11, 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo, M.A., Jensen, T.J., Cui, L., Hou, Y., Chang, X.B., and Riordan, J.R. (1998). Perturbation of Hsp90 interaction with nascent CFTR prevents its maturation and accelerates its degradation by the proteasome. EMBO J. 17, 6879–6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez, I.M., and Chrispeels, M.J. (2003). Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15, 561–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ, A., Ritzenthaler, C., and Robinson, D.G. (2002). Brefeldin A: Deciphering an enigmatic inhibitor of secretion. Plant Physiol. 130, 1102–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panstruga, R., and Schulze-Lefert, P. (2003). Corruption of host seven-transmembrane proteins by pathogenic microbes: A common theme in animals and plants? Microbes Infect. 5, 429–437. [DOI] [PubMed] [Google Scholar]
- Pedrazzini, E., Giovinazzo, G., Bielli, A., de Virgilio, M., Frigerio, L., Pesca, M., Faoro, F., Bollini, R., Ceriotti, A., and Vitale, A. (1997). Protein quality control along the route to the plant vacuole. Plant Cell 9, 1869–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piffanelli, P., Zhou, F., Casais, C., Orme, J., Jarosch, B., Schaffrath, U., Collins, N.C., Panstruga, R., and Schulze-Lefert, P. (2002). The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 129, 1076–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank, C., Zatloukal, K., Cotten, M., Mechtler, K., and Wagner, E. (1992). Gene transfer into hepatocytes using asialoglycoprotein receptor mediated endocytosis of DNA complexed with an artificial tetra-antennary galactose ligand. Bioconjug. Chem. 3, 533–539. [DOI] [PubMed] [Google Scholar]
- Plemper, R.K., and Wolf, D.H. (1999). Retrograde protein translocation: ERADication of secretory proteins in health and disease. Trends Biochem. Sci. 24, 266–270. [DOI] [PubMed] [Google Scholar]
- Rabinovich, E., Kerem, A., Fröhlich, K.U., Diamant, N., and Bar-Nun, S. (2002). AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 22, 626–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos, P.C., Hockendorff, J., Johnson, E.S., Varshavsky, A., and Dohmen, R.J. (1998). Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell 92, 489–499. [DOI] [PubMed] [Google Scholar]
- Rancour, D.M., Dickey, C.E., Park, S., and Bednarek, S.Y. (2002). Characterization of AtCDC48. Evidence for multiple membrane fusion mechanisms at the plane of cell division in plants. Plant Physiol. 130, 1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner, J.C., and Pelham, H.R. (1997). Transmembrane domain-dependent sorting of proteins to the ER and plasma membrane in yeast. EMBO J. 16, 1832–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu, K., Nielsen, K., Piffanelli, P., Oliver, R., and Schulze-Lefert, P. (1999). Cell-autonomous complementation of mlo resistance using a biolistsic transient expression system. Plant J. 17, 293–299. [Google Scholar]
- Sikorski, R.S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger-Haussels, M., and Weisshaar, B. (2000). Transactivation properties of parsley proline-rich bZIP transcription factors. Plant J. 22, 1–8. [DOI] [PubMed] [Google Scholar]
- Swanson, R., Locher, M., and Hochstrasser, M. (2001). A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 15, 2660–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers, K.J., Patil, C.K., Wodicka, L., Lockhart, D.J., Weissman, J.S., and Walter, P. (2000). Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101, 249–258. [DOI] [PubMed] [Google Scholar]
- Trezzini, G.F., Horrichs, A., and Somssich, I.E. (1993). Isolation of putative defense-related genes from Arabidopsis thaliana and expression in fungal elicitor-treated cells. Plant Mol. Biol. 21, 385–389. [DOI] [PubMed] [Google Scholar]
- Trombetta, E.S., and Helenius, A. (2000). Conformational requirements for glycoprotein reglucosylation in the endoplasmic reticulum. J. Cell Biol. 148, 1123–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, B., Ye, Y., and Rapoport, T.A. (2002). Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell Biol. 3, 246–255. [DOI] [PubMed] [Google Scholar]
- Vitale, A., and Denecke, J. (1999). The endoplasmic reticulum-gateway of the secretory pathway. Plant Cell 11, 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K.H., Gouy, M., Yang, Y.W., Sharp, P.M., and Li, W.H. (1989). Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc. Natl. Acad. Sci. USA 86, 6201–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley, C.K., Ling, R., and Callis, J. (1998). Engineering in vivo instability of firefly luciferase and Escherichia coli beta-glucuronidase in higher plants using recognition elements from the ubiquitin pathway. Plant Mol. Biol. 37, 337–347. [DOI] [PubMed] [Google Scholar]
- Worley, C.K., Zenser, N., Ramos, J., Rouse, D., Leyser, O., Theologis, A., and Callis, J. (2000). Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J. 21, 553–562. [DOI] [PubMed] [Google Scholar]
- Ye, Y., Meyer, H.H., and Rapoport, T.A. (2003). Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: Dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol. 162, 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]