Abstract
Studies have suggested mosaic loss of chromosome Y (mLOY) in blood-derived DNA is common in older men. Cohort studies investigating mLOY and mortality have reported contradictory results. Previous work found a 1.6 Mb deletion of the AZFc region on the Y chromosome (the “gr/gr” deletion) is associated with both male infertility and increased risk of testicular germ cell tumors (TGCT). We investigated whether mosaic loss across the entire Y chromosome was associated with TGCT. We obtained blood and buccal-derived DNA from two case-control studies: the NCI Familial Testicular Cancer Study (FTC; cases=172, controls=163) and the NCI US Servicemen's Testicular Tumor Environmental and Endocrine Determinants Study (STEED; cases=506, controls=611). We utilized 15 quantitative polymerase chain reactions (qPCR) spanning the Y chromosome to assess mLOY. Multivariate logistic regression models adjusted for study batch effects detected no significant overall relationship between mean chromosome Y T/R ratio and TGCT (OR=0.34, 95% CI=0.10–1.17, P=0.09). When restricted to familial TGCT cases, a significantly lower T/R ratio was observed in cases compared with controls (0.993 vs 1.014, P-value=0.01). Our study suggests mLOY, as measured by 15 probes spanning the Y chromosome, could be associated with familial TGCT, but larger studies are required to confirm this observation.
Keywords: Mosaicism, chromosome Y loss, testicular germ cell tumors, association
Introduction
In the United States, testicular germ cell tumors (TGCTs) affect approximately 5.3 out of 100,000 men with a peak incidence occurring among men 15 to 40 years of age(1, 2). Established TGCT risk factors include family history of TGCT, undescended testis, contralateral TGCT, impaired fertility and 25 GWAS susceptibility loci(3–5). Evidence also suggests a 1.6 Mb deletion in the AZFc region (known as the gr/gr deletion) of chromosome Y, which has previously been associated with male infertility, could be associated with TGCT risk(6). Men harboring a gr/gr deletion are estimated to have a two- and three-fold increased risk of sporadic and familial TGCT, respectively. It remains to be determined if other regions of the Y chromosome are important contributors to TGCT risk.
Genetic mosaicism is the presence of two or more populations of cells with acquired genetic differences in an individual who developed from a single zygote. The phenotypic consequences of mosaicism can be quite broad, ranging from no apparent health effects to life-threatening disorders such as cancer, and are dependent on: (1) where in the genome the alteration occurs, (2) the developmental timing of the alteration, (3) the tissue type involved and (4) the overall percentage of cells affected in a tissue(7, 8). Large scale (>2Mb) genetic mosaicism is relatively rare in autosomes (<1% frequency in most populations), making it difficult to perform sufficiently powered studies to adequately asses associations with cancer risk(9–11). Mosaicism of the sex chromosomes is more common than that observed in autosomes, with frequencies of X mosaicism observed at four times the autosomal rate, and frequencies of Y mosaicism reaching ≥15 percent in smokers and in men over 75 years old(12–14). Thus, mosaic loss of chromosome Y (mLOY) is a relatively frequent event which may be a useful biomarker and potentially a metric of overall genomic maintenance capacity or stem cell diversity. While one group has suggested that the presence of mLOY could be associated with risk of cancer mortality(12), a larger subsequent study has not confirmed this hypothesis(14). These studies were performed looking at a range of cancers and did not focus specifically on TGCT.
In this study, we characterized the frequency of mLOY in TGCT cases and controls. Our goal was to investigate whether mosaic loss across the entire Y chromosome could be associated with risk for TGCT.
Materials and Methods
Blood- or buccal-derived DNA samples for this analysis originated from two TGCT retrospective case-control studies: the National Cancer Institute (NCI) Familial Testicular Cancer Study (FTC)(15, 16) and the NCI United States Servicemen's Testicular Tumor Environmental and Endocrine Determinants Study (STEED)(17, 18). The FTC contributed 172 familial TGCT cases and 163 controls for analysis, while STEED contributed 506 sporadic TGCT cases and 611 controls resulting in a combined study sample size of 678 TGCT cases and 774 controls. Both studies were reviewed and approved by the Institutional Review Board of the NCI, and all participants provided written informed consent prior to study participation.
A previously-validated quantitative polymerase chain reaction (qPCR) panel with markers spanning the Y chromosome was used to evaluate potential mLOY(14). Mosaic chromosome Y loss across the entire Y chromosome was assessed using a ratio of Y chromosome marker signal to an autosomal single copy gene signal, RPPH1. Ratios below 1 are considered evidence of mosaic chromosome Y loss, whereas ratios above 1 suggest evidence of mosaic chromosome Y gain. Fifteen qPCR gene assays spanning the p and q arms of the Y chromosome were included in the qPCR panel to enable assessment of loss across the Y chromosome (Supplemental Table 1). This was a pre-designed assay to detect mLOY and did not include any markers for the gr/gr deletion. Each target assay was run in duplex with RPPH1 as the reference gene, known to be present in a single copy in the human genome(19). A serial dilution of pooled male gDNA with no detectable Y chromosome loss and pooled female gDNA was made across 7 target ratios to use as an internal standard curve and to guide the analysis. Assay control samples (3 target ratios, prepared similarly to the standard curve) were applied to the assay plates to indicate overall quality of assay performance.
A total of 5 ng of sample DNA as measured by Quant-iT PicoGreen dsDNA quantitation (Life Technologies, Grand Island, NY), was transferred to LightCycler-compatible 384-well plates (Roche, Indianapolis, IN) and dried down. Duplex qPCR was performed using 5 uL reaction volumes consisting of: 2.5 uL of LightCycler 480 Probes Master Mix (Roche, Indianapolis, IN), 2.0 uL of MBG Water, 0.25 uL of VIC-dye labeled 20X TaqMan® Copy Number Reference Assay, human, RNase P (Life Technologies, Grand Island, NY), and 0.25 uL of FAM-dye labeled target-specific 20X TaqMan® Copy Number Assay (Life Technologies, Grand Island, NY). Thermal cycling was performed with a LightCycler 480 (Roche), in which PCR conditions consisted of: 95°C hold for 5 minutes, denature at 95°C for 15 seconds, anneal at 60°C for 30 seconds, and fluorescence data collection for 45 cycles. All TGCT case and control samples were assayed in triplicate on each plate.
LightCycler software (Release 1.5.0) was used for initial analysis of the raw data. Utilizing absolute quantification analysis with the second derivative maximum method and high confidence detection algorithm, single target sequences were quantified and expressed as a target to reference (T/R) ratio, based on the internal standard curve, for each well. Final probe-specific T/R ratios for each individual were calculated as the average across the triplicate results. The resulting overall chromosome Y T/R ratio was calculated as the average across all 15 final probe-specific T/R ratios. Variability across the 15 probe T/R ratios within an individual was low with an overall average standard deviation of 0.088. Samples were run in 3 batches, with the FTC study in one batch and the STEED study split between two batches. Inter-batch variability was adjusted for in multivariate analyses. All statistical tests and regression modeling was carried out using R statistical software (R Foundation for Statistical Computing, Vienna, Austria). All reported P-values are two-tailed and are presented without correction for multiple testing.
Results
Chromosome Y qPCR assays for 15 genes spanning the Y chromosome were completed for 678 TGCT cases and 774 cancer-free controls (Table 1). Control T/R ratios were observed to significantly differ among batches, with a significant mean T/R ratio difference noted between controls in batch 1 and batch 3 (1.014 vs 1.077, P-value=1.24×10−9) and controls in batch 2 and batch 3 (1.000 vs 1.077, P-value=6.52 x10−18), but no significant mean T/R difference was observed between controls in batch 1 and batch 2 (1.014 vs 1.000, P-value=0.09) (Figure 1A).
Table 1.
Chromosome Y qPCR mean assay target to reference (T/R) ratios
| Batch 1 (FTC) | Batch 2 (STEED) | Batch 3 (STEED) | ||||
|---|---|---|---|---|---|---|
| Assay | Cases | Controls | Cases | Controls | Cases | Controls |
| (N=172) | (N=163) | (N=308) | (N=301) | (N=198) | (N=310) | |
| SRY | 0.9968 | 1.0131 | 1.0005 | 1.0000 | 1.0913 | 1.1098 |
| RPS4Y1 | 1.0035 | 1.0097 | 1.0005 | 1.0004 | 1.0447 | 1.0433 |
| ZFY | 0.9874 | 1.0257 | 1.0011 | 0.9994 | 1.0592 | 1.0754 |
| AMELY | 1.0284 | 0.9676 | 0.9997 | 0.9991 | 1.1093 | 1.1317 |
| TBL1Y | 0.9990 | 1.0047 | 1.0060 | 0.9957 | 1.1039 | 1.1240 |
| PRKY | 0.9960 | 1.0100 | 0.9954 | 1.0063 | 1.1394 | 1.1403 |
| USP9Y | 0.9703 | 1.0461 | 0.9978 | 1.0037 | 1.0652 | 1.0699 |
| DDX3Y | 0.9896 | 1.0197 | 0.9992 | 1.0014 | 1.0866 | 1.0860 |
| UTY | 1.0070 | 0.9904 | 1.0007 | 0.9978 | 1.1729 | 1.1702 |
| TMSB4Y | 1.0221 | 0.9823 | 0.9933 | 1.0022 | 0.9574 | 0.9694 |
| NLGN4Y | 0.9740 | 1.0369 | 0.9978 | 1.0034 | 1.0375 | 1.0445 |
| CYorf15A | 0.9754 | 1.0356 | 1.0007 | 0.9962 | 1.0397 | 1.0519 |
| CYorf15B | 0.9792 | 1.0313 | 0.9994 | 1.0008 | 1.0827 | 1.0920 |
| KDM5D | 1.0094 | 0.9911 | 0.9978 | 1.0019 | 0.9265 | 0.9568 |
| EIF1AY | 0.9688 | 1.0439 | 1.0089 | 0.9938 | 1.0898 | 1.0863 |
| Combined Mean | 0.9937 | 1.0140 | 1.0000 | 1.0002 | 1.0670 | 1.0767 |
Figure 1.
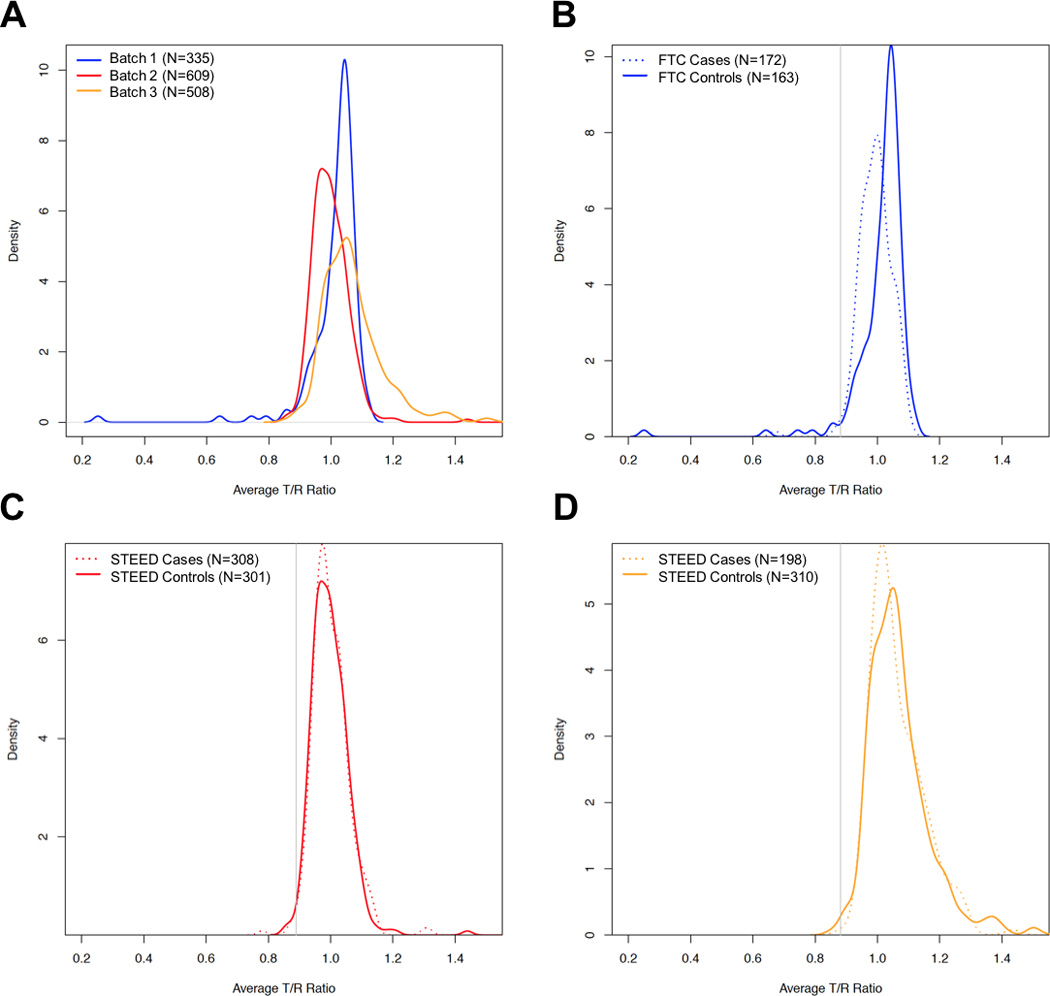
Plots of average T/R ratios across all 15 chromosome Y qPCR markers for (A) cancer-free controls across all three batches, (B) FTC familial TGCT cases and controls in batch 1, (C) STEED sporadic TGCT cases and controls in batch 2 and (D) STEED sporadic TGCT cases and controls in batch 3. TGCT case kernel density distributions are plotted as dashed lines and cancer-free controls are plotted as solid lines. The solid gray line indicates 1.5 standard deviations below the control mean T/R ratio and is used as the calling threshold for categorizing men as mosaic chromosome Y loss.
When comparing average T/R ratios between TGCT cases and controls, a significant difference in mean T/R ratio was observed in the FTC samples that comprised batch 1 (0.993 vs 1.014, P-value=0.01; Figure 1B). No significant difference in average T/R ratio between TGCT cases and controls was observed for the STEED samples in either batch 2 (1.000 vs 1.000, P-value=0.97; Figure 1C) or batch 3(1.067 vs 1.077, P-value=0.34; Figure 1D).
An analysis of individual qPCR markers found mean differences in T/R ratio between TGCT cases and controls for 9 of the 15 markers in FTC (ZFY, AMELY, USP9Y, DDX3Y, TMSB4Y, NLGN4Y, CYorf15A, CYorf15B, and EIF1AY) and only 1 marker (KDM5D) in STEED batch 2 (Supplementary Table 2). For the 9 statistically significant qPCR markers in the FTC study, 7 showed lower T/R ratios in TGCT cases when compared with controls. Merging all batches of case and control samples together and statistically adjusting for batch effects indicated that TGCT cases (on average) had lower T/R ratios compared with controls (1.018 vs. 1.034), a difference that was not statistically significant (OR=0.34, 95% CI=0.10–1.17, P=0.09).
Prior studies have used a threshold approach for calling mLOY in which samples falling below a defined threshold can be categorized as having mLOY(12–14). We chose a conservative threshold of 1.5 standard deviations less than the mean T/R ratio in the control samples. Applying this thresholding approach for calling mLOY resulted in 8 men in batch 1, 7 in batch 2 and 1 man in batch 3 with evidence of mLOY. We did not observe any statistically significant differences in frequency of mLOY in TGCT cases samples when compared with control samples, with P-values equal to 0.16, 1.00, and 1.00 for batch 1, 2, and 3, respectively.
Evidence in our study suggested men with familial TGCT in the FTC study, but not men with sporadic TGCT in the STEED study, may have elevated rates of mosaic Y loss compared with controls. We therefore investigated 123 TGCT cases in the FTC with available clinical data to examine whether any TGCT-related clinical characteristics were associated with mean T/R ratio. There was no statistically significant relationship between age-at-first-TGCT diagnosis and mean T/R ratio (P-value=0.54). A modest association was observed between the occurrence of bilateral TGCT (commonly thought of as a marker of familial risk(20, 21)) and T/R ratio, with lower T/R ratios being associated with increased risk of bilateral TGCT (P-value=0.02). We found no association between surgery or exposure to chemotherapy or radiation associated with T/R ratio across the Y chromosome (P-values=0.27, 0.80 and 0.69, respectively).
Discussion
Our analysis of 15 qPCR markers spanning the Y chromosome detected lower mean T/R ratios in familial TGCT cases compared with controls. We did not observe a significant difference in T/R ratios between sporadic TGCT cases and controls. These findings suggest higher frequencies of mLOY may be present in men at risk of familial TGCT risk, although replication of these findings is needed.
There is an ongoing controversy whether mLOY is associated with increased overall cancer mortality(12, 14). While cancer mortality is a composite of both cancer incidence and cancer survival, it remains to be established whether mLOY has an influence on cancer risk, cancer survival or a combination of both. A recent investigation on mLOY and cancer risk has provided limited support for the relationship between cancer risk and mLOY(14). Interestingly, both studies grouped together a range of cancer diagnoses, where in our study, we have only examined TCGT. Our analysis suggests that if mLOY has an effect on combined familial and sporadic TGCT risk, its effects are likely minimal and therefore would explain only a limited proportion of TGCT risk, since only about 2% of TGCT cases have a positive family history.
A small deletion of chromosome Y, known as the gr/gr deletion, has been associated previously with TGCT risk(6), with a stronger effect size for familial TGCT (OR=3.2) than sporadic forms (OR=2.1). Our study employed a previously-validated qPCR assay for detecting mLOY that did not include an assay for the gr/gr deletion. Although our study did not capture variation specifically in the gr/gr deletion region, we observed a similar differential in T/R ratios for familial TGCT cases versus controls than that observed for the sporadic TGCT cases. We also observed an interesting association in bilateral TGCT cases, which tend to have a higher degree of heritability, in which bilateral TGCT cases were more likely to have lower T/R ratios than non-bilateral TGCT cases. Future studies targeted at better understanding the role of the gr/gr deletion in TGCT risk are needed to isolate whether the effect observed is specific to deletions of the gr/gr region of the Y chromosome or if the gr/gr deletion serves as a surrogate haplotype for larger chromosome Y deletions that have greater relevance for TGCT risk. Such a study would be challenged by the low absolute frequencies of gr/gr in familial (3%) and sporadic cases (2%) and in controls (1.3%), which would mandate large sample sizes to achieve adequate statistical power.
Our investigation studied a unique collection of 678 cases of TGCT, a relatively rare solid tumor. We had the advantage of investigating both familial as well as sporadic forms of TGCT. While we were unable to obtain prospectively collected blood or buccal DNA samples collected prior to TGCT diagnosis, we found no association with potential confounders in the familial TGCT cases such as surgery, chemotherapy or radiation exposure that may have influenced T/R ratio measurements. Our study offers preliminary findings on the relationship between mLOY and TGCT that presents association P-values that are unadjusted for multiple testing. A biological connection linking detected mLOY in blood or buccal tissue DNA to carcinogenesis in the testes is currently unavailable, but a hypothetical link could include mLOY acting as a surrogate for the overall capacity to maintain one’s genome against endogenous and exogenous agents capable of inducing DNA damage or preventing appropriate repair(8). Future studies with large sets of prospectively-collected TGCT cases are needed for more definitively establishing the relationship between mLOY in blood- or buccal-derived DNA and TGCT risk.
In conclusion, we used a previously validated panel of 15 qPCR markers spanning the Y chromosome to investigate a potential relationship between mLOY and TGCT risk. We report a significantly lower mean T/R ratio only in familial TGCT cases compared with controls. While further replication is needed, these findings suggest mLOY could be associated with familial TGCT risk.
Supplementary Material
Acknowledgments
This project was funded by the Intramural Research Program of the United States National Cancer Institute. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government.
Footnotes
Conflict of Interest Disclosure
The authors have no relevant conflicts of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Ghazarian AA, Trabert B, Graubard BI, Schwartz SM, Altekruse SF, McGlynn KA. Incidence of testicular germ cell tumors among US men by census region. Cancer. 2015;121(23):4181–4189. doi: 10.1002/cncr.29643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajpert-De Meyts E, McGlynn KA, Okamoto K, Jewett MA, Bokemeyer C. Testicular germ cell tumours. Lancet. 2016;387(10029):1762–1774. doi: 10.1016/S0140-6736(15)00991-5. [DOI] [PubMed] [Google Scholar]
- 4.Chung CC, Kanetsky PA, Wang Z, Hildebrandt MA, Koster R, Skotheim RI, et al. Meta-analysis identifies four new loci associated with testicular germ cell tumor. Nat Genet. 2013;45(6):680–685. doi: 10.1038/ng.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Litchfield K, Holroyd A, Lloyd A, Broderick P, Nsengimana J, Eeles R, et al. Identification of four new susceptibility loci for testicular germ cell tumour. Nat Commun. 2015;6:8690. doi: 10.1038/ncomms9690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nathanson KL, Kanetsky PA, Hawes R, Vaughn DJ, Letrero R, Tucker K, et al. The Y deletion gr/gr and susceptibility to testicular germ cell tumor. Am J Hum Genet. 2005;77(6):1034–4043. doi: 10.1086/498455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Youssoufian H, Pyeritz RE. Mechanisms and consequences of somatic mosaicism in humans. Nat Rev Genet. 2002;3(10):748–758. doi: 10.1038/nrg906. [DOI] [PubMed] [Google Scholar]
- 8.Machiela MJ, Chanock SJ. Detectable clonal mosaicism in the human genome. Semin Hematol. 2013;50(4):348–359. doi: 10.1053/j.seminhematol.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobs KB, Yeager M, Zhou W, Wacholder S, Wang Z, Rodriguez-Santiago B, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44(6):651–658. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laurie CC, Laurie CA, Rice K, Doheny KF, Zelnick LR, McHugh CP, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet. 2012;44(6):642–650. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Machiela MJ, Zhou W, Sampson JN, Dean MC, Jacobs KB, Black A, et al. Characterization of large structural genetic mosaicism in human autosomes. Am J Hum Genet. 2015;96(3):487–497. doi: 10.1016/j.ajhg.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forsberg LA, Rasi C, Malmqvist N, Davies H, Pasupulati S, Pakalapati G, et al. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat Genet. 2014;46(6):624–628. doi: 10.1038/ng.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dumanski JP, Rasi C, Lonn M, Davies H, Ingelsson M, Giedraitis V, et al. Mutagenesis. Smoking is associated with mosaic loss of chromosome Y. Science. 2015;347(6217):81–83. doi: 10.1126/science.1262092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou W, Machiela MJ, Freedman ND, Rothman N, Malats N, Dagnall C, et al. Mosaic loss of chromosome Y is associated with common variation near TCL1A. Nat Genet. 2016;48(5):563–568. doi: 10.1038/ng.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korde LA, Premkumar A, Mueller C, Rosenberg P, Soho C, Bratslavsky G, et al. Increased prevalence of testicular microlithiasis in men with familial testicular cancer and their relatives. Br J Cancer. 2008;99(10):1748–1753. doi: 10.1038/sj.bjc.6604704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greene MH, Mai PL, Loud JT, Pathak A, Peters JA, Mirabello L, et al. Familial testicular germ cell tumors (FTGCT) - overview of a multidisciplinary etiologic study. Andrology. 2015;3(1):47–58. doi: 10.1111/andr.294. [DOI] [PubMed] [Google Scholar]
- 17.McGlynn KA, Sakoda LC, Rubertone MV, Sesterhenn IA, Lyu C, Graubard BI, et al. Body size, dairy consumption, puberty, and risk of testicular germ cell tumors. Am J Epidemiol. 2007;165(4):355–363. doi: 10.1093/aje/kwk019. [DOI] [PubMed] [Google Scholar]
- 18.Cook MB, Chia VM, Berndt SI, Graubard BI, Chanock SJ, Rubertone MV, et al. Genetic contributions to the association between adult height and testicular germ cell tumors. Int J Epidemiol. 2011;40(3):731–739. doi: 10.1093/ije/dyq260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baer M, Nilsen TW, Costigan C, Altman S. Structure and transcription of a human gene for H1 RNA, the RNA component of human RNase P. Nucleic Acids Res. 1990;18(1):97–103. doi: 10.1093/nar/18.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindor NM MM, Lindor CJ, Greene MH. Concise handbook of familial cancer susceptibility syndromes - Second Edition. J Natl Cancer Inst Monogr. 2008:1–93. doi: 10.1093/jncimonographs/lgn001. [DOI] [PubMed] [Google Scholar]
- 21.Mai PL, Friedlander M, Tucker K, Phillips KA, Hogg D, Jewett MA, et al. The International Testicular Cancer Linkage Consortium: a clinicopathologic descriptive analysis of 461 familial malignant testicular germ cell tumor kindred. Urol Oncol. 2010;28(5):492–499. doi: 10.1016/j.urolonc.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


