Abstract
In higher plants, the Ndh complex reduces plastoquinones and is involved in cyclic electron flow around photosystem I, supplying extra-ATP for photosynthesis, particularly under environmental stress conditions. Based on plastid genome sequences, the Ndh complex would contain 11 subunits (NDH-A to -K), but homologies with bacterial complex indicate the probable existence of additional subunits. To identify missing subunits, tobacco (Nicotiana tabacum) NDH-H was His tagged at its N terminus using plastid transformation. A functional Ndh subcomplex was purified by Ni2+ affinity chromatography and its subunit composition analyzed by mass spectrometry. Five plastid encoded subunits (NDH-A, -H, -I, -J, and -K) were identified as well as three new subunits (NDH-M, -N, and -O) homologous to cyanobacterial and higher plant proteins. Arabidopsis thaliana mutants missing one of these new subunits lack a functional Ndh complex, and NDH-M and NDH-N are not detected in a tobacco transformant lacking the Ndh complex. We discuss the involvement of these three nuclear-encoded subunits in the functional integrity of the plastidial complex.
INTRODUCTION
Thylakoid membranes of higher plant chloroplasts contain two photosynthetic complexes (photosystem II and photosystem I [PSI]) and several electron carriers, including the cytochrome b6f complex, plastocyanin, and ferredoxin, involved in water photolysis and electron transport reactions leading to NADP+ reduction. In addition to these major components of the Z scheme of oxygenic photosynthesis, higher plant chloroplasts contain electron carriers, components of a respiratory electron transport chain (Nixon, 2000; Peltier and Cournac, 2002). A NAD(P)H dehydrogenase complex (Ndh complex), whose existence in thylakoid membranes was first assumed based on the characterization of plastid genes (ndh genes) encoding homologs of mitochondrial complex I subunits (Shinozaki et al., 1986; Ohyama et al., 1988), was identified and partly purified (Berger et al., 1993; Guedeney et al., 1996; Burrows et al., 1998; Sazanov et al., 1998). More recently, a plastid terminal oxidase was characterized (Carol et al., 1999; Wu et al., 1999; Cournac et al., 2000; Joët et al., 2002b). Studies of transplastomic tobacco (Nicotiana tabacum) plants inactivated in ndh genes were realized in different laboratories (Burrows et al., 1998; Kofer et al., 1998; Shikanai et al., 1998; Horvath et al., 2000) and indicate the existence of a functional Ndh complex implicated in the nonphotochemical reduction of plastoquinones. Besides its involvement in chlororespiration, the Ndh complex also participates in cyclic electron transport reactions around PSI (Burrows et al., 1998; Shikanai et al., 1998; Horvath et al., 2000; Joët et al., 2002a). These reactions allow optimal functioning of photosynthesis by increasing the pH gradient and supplying extra-ATP for CO2 fixation. This function would be particularly important in environmental conditions, such as drought, in which the ATP demand is increased (Horvath et al., 2000). Two cyclic electron transport pathways have been identified around PSI on the basis of their sensitivity to antimycin A (AA), and the involvement of the Ndh complex in the AA-insensitive pathway has been clearly demonstrated (Joët et al., 2001; Munekage et al., 2004). Although electron carriers of the AA-sensitive pathway have not been identified yet, a new thylakoid component (PGR5) recently has been shown to be a required partner (Munekage et al., 2002).
Subunit composition of the plastid Ndh complex is not elucidated yet and is still a matter of debate. In bacteria, minimal Ndh complex contains 14 subunits, 11 of them being homologous to chloroplast ndh genes products. Interestingly, the remaining three subunits, present in bacterial complexes but not encoded by plastid genomes (75-, 51-, and 24-kD subunits), are key components of complex I involved in NADH binding and oxidation. Electron microscopy has shown that bacterial or mitochondrial complex I is formed of two main parts: a peripheral arm mediating NADH oxidation and a membrane embedded arm (Friedrich and Böttcher, 2004). The nature of missing subunits involved in electron input associated to the peripheral arm as well as the nature of chloroplast electron donors to the complex remain unknown. It has been proposed that ferredoxin-NADP reductase (FNR) could interact with the Ndh complex to supply electrons from NADPH, thanks to its diaphorase activity (Guedeney et al., 1996), but this possibility has been a subject of controversy (Quiles and Cuello, 1998; Sazanov et al., 1998; Funk et al., 1999; Quiles et al., 2000). Genes encoding missing subunits could have been relocated to the nucleus, as is the case for major photosynthetic complexes (PSI, photosystem II, cytochrome b6f, and ATPase), which are partly encoded by nuclear genes (Rochaix, 2002). The Arabidopsis thaliana nuclear genome contains genes homologous to the 75-, 51-, and 24-kD complex I subunits encoding genes, but corresponding gene products are predicted to be targeted to mitochondria and likely participate in mitochondrial complex I (Grohmann et al., 1996). Another possibility would be that the plastidial complex contains a new unidentified electron input module, as recently proposed in the case of cyanobacteria (Prommeenate et al., 2004). Several attempts to purify the plastidial complex of higher plants did not identify missing subunits involved in electron input (Sazanov et al., 1998; Funk et al., 1999).
His tagging has been successfully used for the purification of photosynthetic complexes (Bricker et al., 1998; Sugiura et al., 1998). Very recently, this strategy was used in Synechocystis PCC6803 to purify the cyanobacterial Ndh complex and elucidate the diversity of Ndh complexes in this species (Prommeenate et al., 2004). Based on their copurification with other NDH subunits, the existence of the two new subunits, designated NDH-M and NDH-N, was proposed (Prommeenate et al., 2004). In this work, we used a strategy based on His tagging and plastid transformation of tobacco to fuse a hepta-His (7His) sequence to the N terminus or C terminus of NDH subunits (NDH-B and NDH-H) and purify the plastid Ndh complex. We report on the identification of three nuclear-encoded subunits (NDH-M, -N, and -O) as essential components of the plastid Ndh complex of higher plants.
RESULTS
Engineering a PolyHis-Tag at the N Terminus of NDH-H
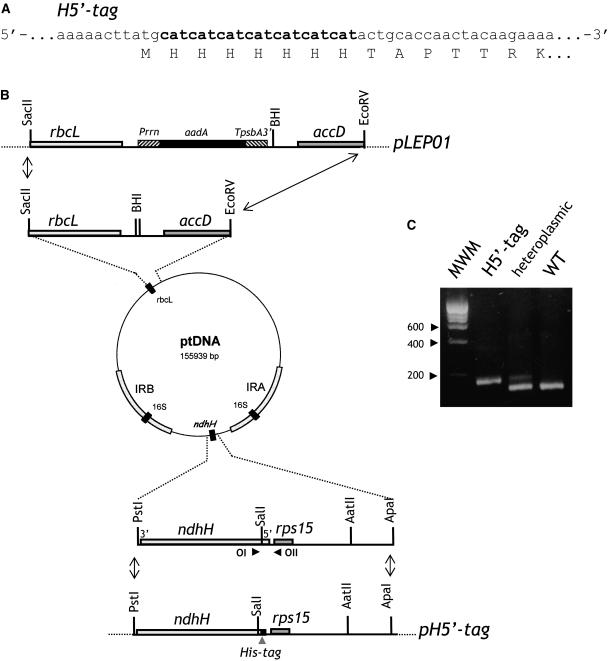
In an attempt to develop an efficient purification protocol of the Ndh complex from tobacco thylakoids, we engineered a polyHis-tag at the C terminus or N terminus of plastid-encoded NDH subunits. In a first attempt, a 7His-tag was engineered at the C terminus or N terminus of the hydrophobic NDH-B subunit (see Supplemental Figure 1 online). This approach proved to be unsuccessful (see below), the tagged complex being undetectable in transformant plants. In a second attempt, a 7His-sequence was engineered at the N terminus of the hydrophilic NDH-H subunit. 7His-encoding sequence was inserted downstream of the ATG codon of ndhH (Figure 1A). Because ndhH is part of a transcription unit including different ndh genes, ndhH tagging was performed by cotransformation, which allows targeted plastid genome modification without physical linkage to the marker gene (Carrer and Maliga, 1995; Rumeau et al., 2004). Cotransformation was performed by bombardment with a mix of two vectors, one containing the engineered ndhH gene (pH5′-tag, Figure 1B), the other the selection cassette targeted to a silent region of the plastid chromosome (LEP01; Svab and Maliga, 1993). As shown in Figure 1B, pLEP01 carries a chimeric spectinomycin resistance cassette (aadA) targeted for insertion between the accD and the rbcL genes. Bombardment was followed by selection for spectinomycin resistant clones, identified as green shoots on the bleached leaf sections. Plantlets were regenerated and characterized. The presence of the 7His-tagged ndh gene in spectinomycin resistant lines was analyzed by PCR using OI and OII as primers (Figure 1B) and high-resolution agarose gel electrophoresis (Figure 1C; see Methods). Two clones appeared to contain the 7His-tagged ndh gene. Because one single band was detected on the agarose gel after PCR reaction with the DNA extracted from one of them, H5′-tag (Figure 1C), it was considered homoplasmic. This plant was backcrossed twice with wild-type plants and used for further characterization.
Figure 1.
Engineering a 7His-Tag Sequence at the 5′ End of ndhH in the Tobacco Plastid Genome.
(A) Nucleotide and amino acid sequences in the vicinity of the introduced His tag. H5′-tag, mutant carrying the 7His-tag at the 5′ end of ndhH, immediately downstream from the start codon. Added nucleotides are in bold.
(B) Plastid cotransformation vectors. Plasmid pLEP01 carries an aadA cassette (i.e., the aadA gene controlled by a modified 16SrDNA promoter [Prrn] and psbA terminator [TpsbA3′]). AadA cassette integration into the ptDNA is targeted between rbcL and accD. Plasmid pH5′-tag contains an ApaI-PstI fragment of plastid DNA encompassing the engineered 5′ end of ndhH. OI and OII indicate the position of the oligonucleotides used for PCR characterization. Dotted lines represent the vector portion of pBSII. BHI, BamHI.
(C) PCR analysis. DNA from wild-type or selected tobacco plants (H5′-tag) was extracted and used as template for PCR with OI and OII. After amplification, products were separated by electrophoresis on a high-resolution agarose gel. MWM, molecular weight marker.
7His-Tagged NDH-H Subunits Are Integrated in the Ndh Complex without Impairing Its Function
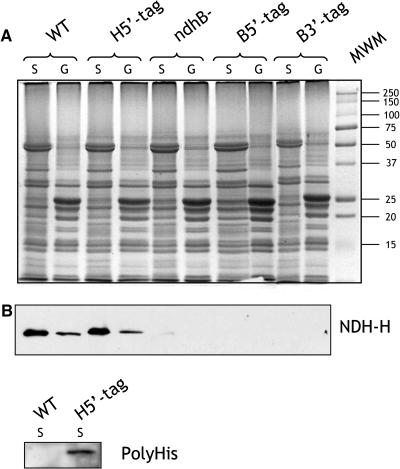
To investigate the presence of a stable Ndh complex in thylakoid membranes of transplastomic plants, immunodetection experiments were first performed using antibodies against the NDH-H subunit. Thylakoid membranes were extracted from wild-type, H5′-tag, B5′-tag, and B3′-tag plants and from ndhB− tobacco plants. B5′-tag and B3′-tag are transplastomic plants containing a 5′ end and 3′ end His-tagged ndhB gene, respectively (see Supplemental Figure 1 online); ndhB− plants are plastid transformant plants in which the ndhB gene was translationally inactivated (Horvath et al., 2000). Granal and stromal lamellae were separated by detergent treatment, and protein extracts were analyzed by SDS-PAGE (Figure 2A). Coomassie blue staining did not reveal any difference between protein patterns of wild-type and transplastomic plants. Protein gel blot analysis with an NDH-H antiserum (Horvath et al., 2000; Figure 2B) indicated the absence of NDH-H in ndhB− plants as well as in the B5′-tag and B3′-tag plastid transformants. In H5′-tag plants as in wild-type plants, the NDH-H subunit was mainly located in stromal lamellae and was present in similar amounts (Figure 2B). A slight molecular weight increase because of the 7His-tag could be detected. The presence of a 7His-tag on NDH-H was confirmed with commercial antibodies directed against a polyHis motif (Figure 2B).
Figure 2.
Biochemical Characterization of H5′-Tag Transplastomic Plants.
(A) Coomassie blue staining of proteins extracted from granal (G) and stromal (S) lamellae purified from wild-type, H5′-tag, B5′-tag, B3′-tag, and ndhB− tobacco leaves and separated by SDS-PAGE.
(B) Protein gel blot analysis using anti-NDH-H and anti-polyHis antibodies.
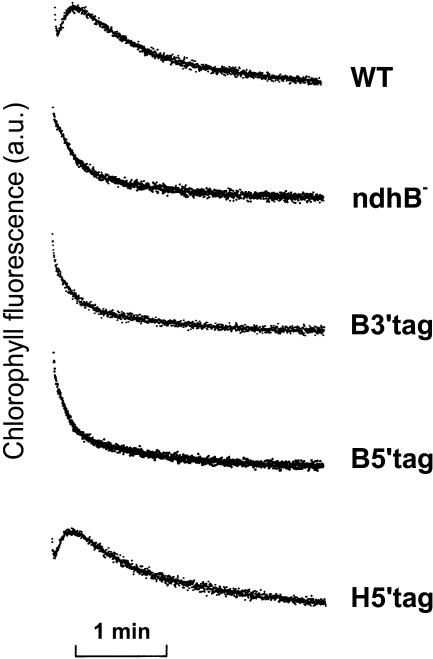
Plastid transformants were also characterized using chlorophyll fluorescence measurements (Figure 3). The transient increase in chlorophyll fluorescence measured in the dark after actinic illumination has been previously reported to be related to the functioning of the Ndh complex (Burrows et al., 1998; Joët et al., 2001). In B5′-tag, B3′-tag, and ndhB− tobacco plants, the transient increase was absent, clearly demonstrating that no functional Ndh complex is present in these plants. In wild-type and H5′-tag transplastomic leaves, the transient increase in chlorophyll fluorescence was similar. We conclude from these experiments that NDH-B tagging prevented assembly of the Ndh complex, whereas NDH-H tagging did not affect its assembly and functioning.
Figure 3.
In Vivo Detection of Ndh Activity by Chlorophyll Fluorescence Measurements.
Leaf chlorophyll fluorescence of wild-type and transplastomic tobacco plants was monitored with a pulse-amplitude modulated fluorometer. After a 5-min actinic illumination (250 μmol photons·m−2·s−1), Fs levels reached similar levels in wild-type and transplastomic leaves. After switching the light off, transient increases in chlorophyll fluorescence were recorded under low nonactinic light. a.u., arbitrary units.
Purification of a His-Tagged Ndh Complex Preparation
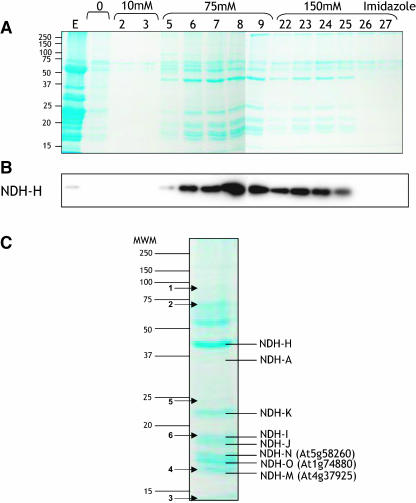
Thylakoid membranes were prepared from leaves of H5′-tag transplastomic plants. Stromal lamellae proteins were selectively solubilized by dodecyl maltoside treatment and loaded onto a Ni2+-NTA column. After extensive washing with a buffer containing a low imidazole concentration (10 mM), polypeptides fixed on the affinity column were eluted with 75 and 100 mM imidazole (Figure 4). SDS-PAGE and protein gel blotting were used to examine the protein composition of collected fractions. Coomassie blue staining (Figure 4A) revealed that the majority of polypeptides present in the stromal lamellae extract (Figure 4A, lane E) did not bind to the column. Most of the polypeptides eluted with 75 mM imidazole were also detected in the 150 mM imidazole fractions but at a lower intensity. A protein gel blot performed with anti-NDH-H antibodies demonstrated that the NDH-H subunit was retained onto the column and eluted mostly by the 75 mM imidazole wash (Figure 4B). Collected fractions (lanes 5 to 9 and 23 to 25, Figure 4A) were pooled and analyzed for NADH and NADPH:ferricyanide oxidoreductase activity. Specific activities of ∼30 and ∼0.02 μmol·min−1·mg−1 of protein were measured in the presence of NADH and NADPH, respectively.
Figure 4.
Purification of a His-Tagged Ndh Complex by Affinity Chromatography on Ni2+-NTA.
(A) Coomassie blue–stained SDS-PAGE of protein from collected fractions. Lane E shows the stromal lamellae protein extract loaded onto the column, lane 0 shows wash fractions without imidazole, lane 10 is the wash fraction with 10 mM imidazole, lanes 75 and 150 are successive collected fractions eluted with 75 and 150 mM imidazole, respectively. The number above each lane refers to the fraction number. Fraction volume is 400 μL.
(B) Protein gel blot analysis using anti-NDH-H antibodies.
(C) Coomassie blue–stained SDS-PAGE of protein from pooled fractions 5 to 9 and 23 to 25 eluted with 75 and 150 mM imidazole, respectively. Stained bands whose MS/MS characterization was performed are indicated.
Subunit Composition of the Ndh Complex Preparation
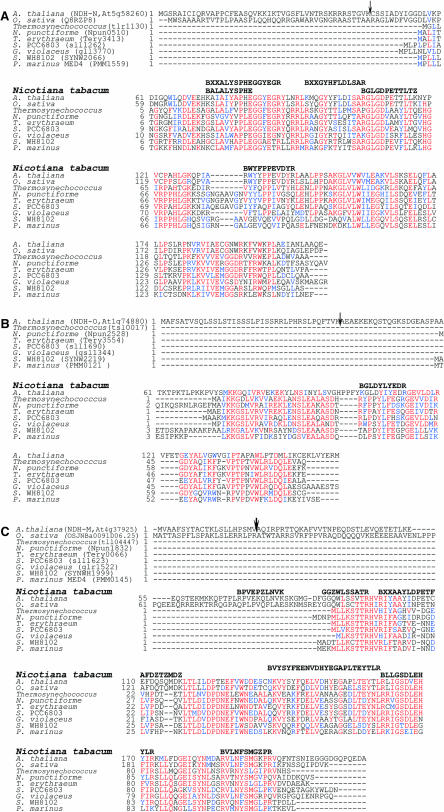
To assess the subunit composition of the complex, the His-tagged Ndh complex preparation was subjected to SDS-PAGE (Figure 4C). Approximately 15 bands were identified by Coomassie blue staining. After excising stained polypeptides, tandem mass spectrometry (MS/MS) sequencing was performed. Five out of the 11 plastid-encoded Ndh complex subunits were identified as NDH-A, -H, -I, -J, and -K (Figures 4C and 5A). Peptides identified by MS/MS from protein bands numbered from 1 to 6 in Figure 4C could not be assigned to known proteins, likely because of low homologies with Arabidopsis proteins (Figure 5B). On the other hand, peptides issued from three other bands (Figures 4C and 6) revealed sequence similarities with hypothetical proteins of three Arabidopsis predicted genes, At5g58260, At1g74880, and At4g37920, described in The Arabidopsis Information Resource (TAIR) database as of September, 2004 (http://www.arabidopsis.org/; Rhee et al., 2003). These three genes harbored many EST and full-length cDNA (Riken Arabidopsis Full Length cDNA [RAFL]; Seki et al., 2002) records in the TAIR database and Arabidopsis cDNA database from the Riken BioResource Center (http://pfgweb.gsc.riken.go.jp/index.html, September, 2004; see Supplemental Figure 2 online), respectively, indicating that they are expressed in vivo. From the RAFL cDNA sequences we confirmed the genomic annotation of At1g74880 and At5g58260 (see Supplemental Figure 2 online) and deduced the full sequence of the encoded peptides. For At4g37920, two distinct RAFL cDNAs were retrieved (see Supplemental Figure 2 online), demonstrating that the gene numbered At4g37920 in the TAIR database must be split into two, At4g37925 and At4g37920, as suggested in the Riken Bioresource Center database and Munich Information Center for Protein Sequences Arabidopsis database (MATDB; http://mips.gsf.de/proj/thal/db/index.html, September, 2004). The MS/MS-identified peptides matched the At4g37925 sequence (Figure 6). These three polypeptides are proposed to be new NDH subunits designated NDH-M (At4g37925), NDH-N (At5g58260), and NDH-O (At1g74880).
Figure 5.
Peptide Sequences Identified by Mass Spectrometry.
(A) Identification of plastid-encoded NDH subunits by mass spectrometry. Predicted amino acid sequences of tobacco plastid encoded NDH subunits are presented. Peptides identified by mass spectrometry analysis are in bold.
(B) The polypeptide bands stained and numbered from 1 to 6 in Figure 4C were excised and submitted to MS/MS analysis after trypsin digestion.
Figure 6.
Identification of Three New NDH Subunits, NDH-M, NDH-N, and NDH-O, by Mass Spectrometry.
Alignment of the amino acid sequences of NDH-N (A), NDH-O (B), and NDH-M (C) homologs. The tobacco peptides identified by MS/MS analysis and sequencing are indicated in bold on upper lines. Similarity searches were performed using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). Multiple alignments were performed with ClustalW (http://www.infobiogen.fr/services/analyseq/cgi-bin/clustalw_in.pl) and the alignment output in red and blue letters with the Boxshade program (http://www.ch.embnet.org/software/BOX_form.html). The first amino acid of the tobacco peptides is B:R or K according to trypsin specificity. In the peptides BXXALYSPHEGGYEGR, BXXAAYLDPETFAFDZTZMDK, and BXXGYHFLDLSAR, XX could be LA, AL, SP, or PS; YL, LY, FE, or EF; YQ, QY, KY, or YK, respectively. In the peptides, L is I or L and Z is K or Q. The predicted cleavage site using ChloroP 1.1 is indicated by an arrow.
The deduced amino acid sequences of the three Arabidopsis proteins was then compared with those of known proteins of plants and organisms in various public databases using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). Homologous proteins were found in other plants and in cyanobacteria (Figure 6). Using TBLASTN, EST encoding homologous proteins were identified in maize (Zea mays) and Solanaceae databases (http://www.maizegdb.org/blast.php and http://www.sgn.cornell.edu/cgi-bin/tools/blast/simple.pl; data not shown). No homologous protein was found in bacteria, Chlamydomonas, or animal databases.
The program ChloroP 1.1 (http:// www.cbs.dtu.dk/services/ChloroP/) predicted chloroplast targeting for NDH-M, -N, and -O. The putative cleavage site of the transit peptide is indicated by an arrow in Figure 6. The other target prediction programs, Target P (http://www.cbs.dtu.dk/services/TargetP/) and Predotar (http://www.inra.fr/predotar/), predicted chloroplast targeting for NDH-N and NDH-O but mitochondria targeting for NDH-M. Based on predicted cleavage sites, NDH-M, -N, and -O molecular masses were estimated to 22, 18, and 13 kD, respectively. This prediction is in agreement with NDH-N measured size (∼18 kD), but not consistent with NDH-M and -O sizes (Figure 4C) of ∼16.5 and 17.5 kD, respectively. This discrepancy might be either attributed to an abnormal electrophoretic migration or to an erroneous cleavage site prediction (note, for instance, that the predicted NDH-M sequence harbors an N terminus extension as compared with cyanobacterial orthologs).
Computer aided analysis did not reveal a transmembrane or a specific domain suggesting a putative function for NDH-N and NDH-M peptides. For NDH-O, two transmembrane domains were predicted by TmPred (http://www.ch.embnet.org/software/TMPRED_form.html). One of them was encompassed within the putative targeting sequence, and the other, from amino acid 121 to 142, was not found in NDH-O cyanobacterial orthologs.
Newly Characterized Ndh Complex Subunits Are Required for NAD(P)H:Plastoquinone Oxidoreductase Activity
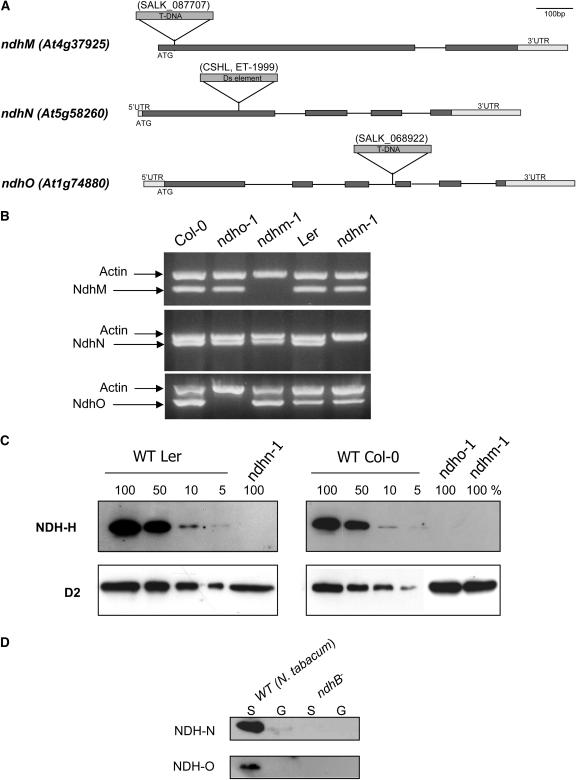
To characterize the role of the three newly identified proteins, Arabidopsis mutant lines were obtained from insertion mutants collections (Figure 7A). Mutants SALK_068922 and SALK_087707, in which NdhO and NdhM were disrupted, respectively, were provided by the Salk Institute Genomic Analysis Laboratory (Alonso et al., 2003). Mutant ET-1999, in which NdhN was disrupted, was obtained from the Cold Spring Harbor Laboratory (Martienssen, 1998). Arabidopsis mutant lines were named ndhm-1, ndhn-1, and ndho-1, respectively. For each mutant line, PCR amplifications and sequencing were performed to confirm T-DNA or Ds element insertion. Single insertion was also confirmed by thermal asymmetric interlaced PCR and DNA gel blotting (data not shown). Full gene disruptions by the T-DNA or Ds insertions were confirmed by RT-PCR (Figure 7B). No specific amplification band was detected in each ndh mutant, indicating that both alleles are null. Biochemical and physiological investigations were performed with homozygous lines. Protein gel blot analyses were performed to detect NDH-H subunits. NDH-H could not be detected in the thylakoid membranes of the three different mutants (Figure 7B), suggesting that the three different peptides are required for Ndh complex assembly. From diluted loading of wild-type proteins, we estimated that NDH-H level was much less than 5%.
Figure 7.
Biochemical Characterization of Arabidopsis Mutants ndhm-1, ndhn-1, and ndho-1 Lacking Peptides Encoded by Nuclear Genes NdhM (At4g37925), NdhN (At5g58260), and NdhO (At1g74880), Respectively.
(A) Positions of T-DNA or Ds insertions in gene sequences. Solid boxes, exons; thin lines, introns; UTR, untranslated region.
(B) RT-PCR analysis of NdhM, NdhN, and NdhO transcripts. Total RNA was extracted from wild-type and Arabidopsis mutants. RT-PCR was performed using gene-specific primers for the nuclear Ndh genes and actin gene as an internal control. After agarose gel electrophoresis, amplified frgments were visualized with ethidium bromide.
(C) Protein gel blot analysis. Thylakoids were extracted from wild-type (ecotype Columbia [Col-0] or Landsberg erecta [Ler]) and mutant Arabidopsis leaves. Proteins were separated by SDS-PAGE, transferred onto nitrocellulose, and analyzed using anti-NDH-H antibodies. Immunodetection of D2 was used as a loading control.
(D) Protein gel blot analysis. Proteins extracted from granal (G) and stromal (S) lamellae purified from wild-type and ndhB− tobacco leaves were separated by SDS-PAGE transferred onto nitrocellulose and analyzed using anti-NDH-N and anti-NDH-O antibodies (aliquots were taken from the same protein samples as in Figure 2; see loading controls on Figure 2A).
Antibodies directed against NDH-N and against a well-conserved region of NDH-O (from amino acid 134 to 149) were produced and used for protein gel blotting of stromal lamellae proteins from wild-type and ndhB− tobacco plants (Figure 7C). Although homologous proteins to NDH-N or NDH-O could be detected in wild-type tobacco stromal thylakoids, no signal was observed in ndhB− transplastomic plants, suggesting that these peptides are components of the plastid Ndh complex. Unfortunately, these antibodies did not detect any protein in Arabidopsis wild-type thylakoids, probably because of the relatively low sensitivity of the antibodies and to the fact that peptides encoded by NdhN and NdhO are likely present in low amounts in thylakoid membranes.
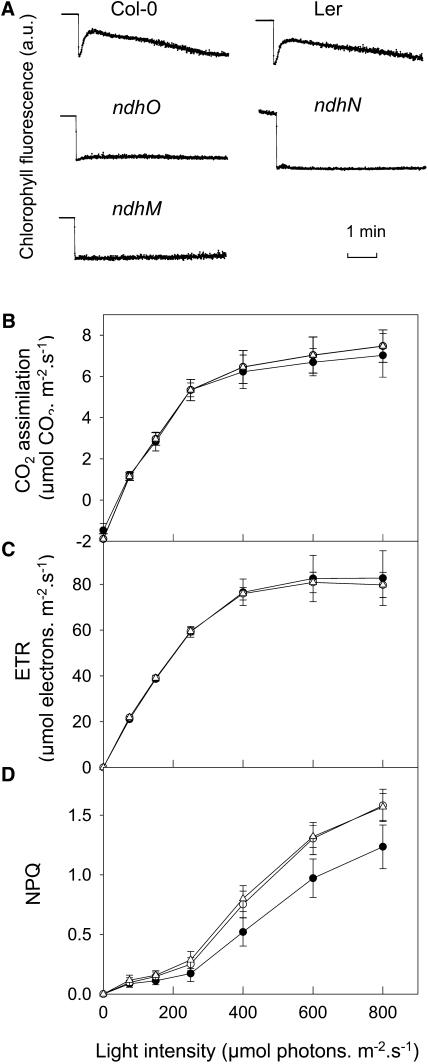
Chlorophyll fluorescence analysis was performed to probe in vivo the Ndh complex activity in the Arabidopsis mutants. The transient increase in chlorophyll fluorescence measured after illumination could not be detected in any of the three mutants, demonstrating the absence of functional Ndh complex (Figure 8A). Photosynthetic activity was also assessed by following basic chlorophyll fluorescence parameters, CO2 assimilation, and growth. Whereeas electron transport rate (ETR; Figure 8B), CO2 assimilation (Figure 8C), and growth were not affected under various illuminations, a slight but significant increase in nonphotochemical quenching (NPQ) was observed in ndho-1 and ndhm-1 when compared with the Columbia-0 (Col-0) wild-type control (Figure 8D). Similar effects, including the increase in NPQ, were observed in the ndhn-1 mutant in comparison with the Landsberg erecta (Ler) wild-type control (data not shown).
Figure 8.
Physiological Characterization of ndhm-1, ndhn-1, and ndho-1 Arabidopsis Mutants.
(A) Transient chlorophyll fluorescence rise measured under nonactinic light after a 5-min illumination in wild-type (Col-0 or Ler) and ndhm-1, ndhn-1, and ndho-1 mutant lines. a.u., arbitrary units.
(B) CO2 assimilation as a function of light intensity measured in the presence of 21% O2 and 250 μL·L−1 CO2 in wild-type Col (closed circles), ndhm-1 (open circles), and ndho-1 (triangles) mutants.
(C) and (D) ETR and NPQ calculated from chlorophyll fluorescence parameters; experimental conditions and symbols are the same as in (B).
DISCUSSION
The Ndh complex of plant thylakoid membranes is present in very low amounts estimated to 0.2% of the thylakoid protein (Sazanov et al., 1996). Because of its low abundance, its low functional stability (Berger et al., 1993; Matsuo et al., 1998; Sazanov et al., 1998), and possible contamination with the mitochondrial analogous complex I, isolation of Ndh complex from plant thylakoids has remained a challenge, requiring the development of efficient and rapid purification procedures. Although attempts to immunopurify plant thylakoid Ndh complexes have remained unsuccessful (Funk and Steinmüller, 1995; Sazanov et al., 1998), purification procedures involving multiple steps have been performed for isolating pea (Pisum sativum) (Sazanov et al., 1998) and maize plastidial Ndh complex (Funk et al., 1999). We have used a different strategy involving His tagging of a complex subunit for purifying the tobacco plastidial Ndh complex. This technique, based on the high affinity interaction between a genetically engineered polyHis-tag and a commercially available Ni2+ resin, has been proven to be highly effective for purifying membrane protein complexes from bacteria and microalgae (Bricker et al., 1998; Sugiura et al., 1998). In a first approach, NDH-B was selected for His tagging (see Supplemental Figure 1 online). NDH-B with its 14 putative transmembrane domains (according to TmPred) is probably the most hydrophobic subunit of the Ndh complex. In bacteria and mitochondria, NDH-B homologs have been attributed to the membrane arm of the complex I that anchors the enzyme in the membrane and takes part in proton transport (Friedrich and Böttcher, 2004). Addition of the tag to the most intrinsic subdomain was expected to favor full native Ndh complex isolation. However, the absence of a functional Ndh complex in the His-tagged NDH-B plants indicates that His-tag fusion hampered either thylakoid membrane integration of the His-tagged subunit or complex assembly or stability. Disruption of one of the ndh genes in tobacco plastid transformants resulted in disappearance of the other NDH subunits investigated (Burrows et al., 1998; Kofer et al., 1998; Horvath et al., 2000). The increased susceptibility of NDH subunits to degradation, observed when their assembly partners are absent, is a general feature of eukaryotic photosynthetic proteins and is considered as a posttranslational regulation step in chloroplastic protein biogenesis (Wollman et al., 1999).
In a second attempt, ndhH was His tagged at its 5′ end. In bacteria and mitochondria, NDH-H homologs are located in the connecting fragment that links the NADH-oxidizing unit to the membrane arm (Friedrich and Böttcher, 2004). Much less hydrophobic than NDH-B, NDH-H, whose bacterial ortholog is involved in the quinone binding site (Dupuis et al., 2001), appeared as an attractive target to His tagging. The Ndh complex containing a His-tagged NDH-H subunit was fully active, allowing its purification using a Ni2+-NTA chromatography. The purified Ndh complex harbored a high NADH-specific ferricyanide oxidoreductase activity when compared with other plant thylakoid Ndh complex preparations (Sazanov et al., 1998; Funk et al., 1999), which likely results from the use of a single short purification step. In previous studies, Sazanov et al. (1998) used two anion-exchange chromatography steps followed by one size-exclusion chromatography step to purify pea Ndh complex, and Funk et al. (1999) performed maize Ndh complex purification through a sucrose gradient centrifugation followed by an anion-exchange chromatography. The low stability of the plant thylakoid Ndh complex has been well documented (Berger et al., 1993; Sazanov et al., 1998). Therefore, any purification protocol involving extensive manipulation of the plastid complex leads to activity lost partially because of extensive delipidation as demonstrated by Sazanov et al. (1998).
Five of the plastid-encoded NDH subunits were identified by sequence analysis as NDH-A, -H, -I, -J, and -K. The other subunits of the membrane arm NDH-B, -C, -D, -E, -F, and -G, more hydrophobic, were not identified either because they could not be detected by staining or because they were lost during the purification as a result of the dissociation of the membrane arm. In the mitochondrial and bacterial complex I, NDH-A is a membrane-associated subunit involved in quinone binding (Dupuis et al., 2001). NDH-A was present in the tobacco Ndh subcomplex, suggesting a direct interaction with the other identified subunits, NDH-I, -J, or -K, whose homologs in complex I constitute the interconnecting domain. NDH-A was much less represented in the tobacco Ndh complex preparation than hydrophilic NDH subunits (Figure 5A), suggesting a partial loss of the membrane arm during the purification. Similar results were obtained by Sazanov et al. (1998) or Funk et al. (1999), who did not characterize the most hydrophobic NDH subunits in pea and maize Ndh complex preparations, respectively. The existence of a ferricyanide oxidoreductase activity in an Ndh subcomplex lacking hydrophobic subunits has been previously reported (Funk et al., 1999). This activity likely results from the ability of artificial acceptors to receive electrons from an accessible reduced component of the hydrophilic arm.
Our sequencing work allowed the identification of three nuclear-encoded NDH subunits called NDH-M, -N, and -O, whose Arabidopsis orthologs are respectively designated in MATDB as At4g37925, At5g58260, and At1g74880. In higher plants, 11 ndh genes (ndhA to ndhK) are present in the plastid genome. In cyanobacteria, although it was not confirmed by further studies, the existence of an additional ndh gene (ndhL) has been reported (Ogawa, 1992). Very recently, Prommeenate et al. (2004) His tagged ndhJ from Synechocystis and isolated an Ndh complex preparation in which they could also identify At4g37925 and At5g58260 orthologs (slr1623 and sll1262) that they suggested to name NDH-M and NDH-N, respectively. So, to avoid any possible confusion and in agreement with published results, we designated the three tobacco subunits NDH-M, -N, and -O.
Steinmüller and colleagues first identified the N terminus sequence of NDH-M in a Ndh complex preparation of Synechocystis PCC6803 (Berger et al., 1993) and the N terminus sequence of NDH-N in a maize Ndh complex preparation (Funk et al., 1999) but could not assign these sequences to known gene products. Neither Berger et al. (1993) nor Prommeenate et al. (2004) demonstrated that these newly identified peptides are NDH subunits. Here, using antibodies directed against Arabidopsis NDH-N and NDH-O, we show that both peptides are restricted to lamellae thylakoid membranes in tobacco, as expected for Ndh complex subunits. We also show that NDH-H could not be detected in Arabidopsis mutants affected in NdhM, N, and O, supporting the existence of a close interaction between nuclear- and plastid-encoded NDH subunits. Moreover, we show that Arabidopsis insertional mutants of NdhM, N, or O are devoid of a functional Ndh complex, clearly demonstrating that these peptides are essential for functional integrity of the Ndh complex.
If sequence similarity searches led to the identification of NdhM, NdhN, and NdhO orthologs in plants and cyanobacteria, interestingly, no homologous sequence could be detected in Chlamydomonas reinhardtii, although the nuclear genome of this alga has nearly been fully sequenced (http://genome.jgi-psf.org/chlre2/chlre2.home.html). This absence is consistent with the fact that Chlamydomonas lacks the 11 ndh plastid genes (Maul et al., 2002) and is recognized to lack a plastidial Ndh complex (Peltier and Cournac, 2002).
The localization of NDH-M and NDH-O homologs (slr1623 and ssl1690) in cyanobacterial thylakoid membranes was demonstrated by bidimensional electrophoresis analysis and peptide sequencing a few years ago (http://www.kazusa.or.jp/cyanobase/Synechocystis/cyano2D/thylakoid.html). Surprisingly, Prommeenate et al. (2004) when purifying the Synechocystis Ndh complex did not identify NDH-O in their preparation, although they noticed the presence of a small peptide present in a very low abundance of molecular mass ∼7 kD close to that of sll1690 ∼8.3 kD.
The nature of missing subunits of the plastid Ndh complex of higher plants has been a subject of controversy for many years. FNR (Guedeney et al.,1996; Quiles and Cuello, 1998; Funk et al., 1999; Quiles et al., 2000) and nuclear-encoded homologs of NuoE, F, and G (Rasmusson et al., 1998; Quiles et al., 2003) have been suggested to be possible candidates, but no genetic or biochemical confirmation of their involvement was obtained. An alternative strategy, using a genetic approach based on Arabidopsis mutant screening and chlorophyll fluorescence imaging, has been used in recent years to identify missing Ndh complex subunits (Hashimoto et al., 2003). Nuclear genes (crr) involved in the control of plastid ndh gene expression (Hashimoto et al., 2003; Munekage et al., 2004) were characterized, but until now this strategy failed to identify new subunits of the Ndh complex. crr Arabidopsis mutants were shown to contain low amounts of NDH subunits (∼6% of NDH-H levels; Hashimoto et al., 2003) because of the fact that expression of ndh genes, although affected in the mutant, was still possible but at lower rates (Hashimoto et al., 2003; Munekage et al., 2004). In NdhM, NdhN, and NdhO mutants, the NDH-H subunit was never detected even when the detection limit in wild-type plant thylakoids was as low as 5% (Figure 7). Based on the copurification with hydrophilic NDH subunits and on the fact that corresponding mutants are affected in the Ndh complex assembly and functioning, NDH-M, -N, and -O are concluded to be essential subunits of this complex. Therefore, we interpret the disappearance of the NDH-H subunit as a result of a failure of the whole complex to assemble when one of these subunits is missing, as typically observed for photosynthetic complexes (Rochaix, 2002) and previously reported in transplastomic tobacco plants inactivated in one or several ndh genes (Burrows et al., 1998; Horvath et al., 2000). To our knowledge, these three mutants are the first Arabidopsis fully deficient ndh mutants reported so far. Interestingly, whereas photosynthetic ETR, CO2 assimilation, and growth were not affected in the three ndh mutants, as previously reported for tobacco ndhB− mutants (Joët et al., 2001) and for the crr2 Arabidopsis mutant (Hashimoto et al., 2003), a slight but significant increase in NPQ was detected in NdhM, N, and O deficient mutants. This effect, which was not reported in crr mutants (Hashimoto et al., 2003; Munekage et al., 2004), likely indicates that the complete loss of the Ndh complex affects photosynthetic reactions linked to the establishment of qE. Additional experiments will be needed to fully understand this phenomenon.
Although three new NDH subunits have been identified, the nature of the electron input device still remains to be determined. Indeed, none of the new identified subunits contains an NADH or NADPH binding domain. Clearly, because the Ndh complex isolated here showed a high and specific NADH:ferricyanide oxidoreductase activity, a functional association to FNR can be ruled out. Interestingly, homologs of the Escherichia coli NuoE, F, and G subunits were not identified in purified plastid Ndh complex preparations, although it should be noted that all peptides have not been sequenced yet. The existence of a different electron input module, involving an archaebacterial-type dehydrogenase, appears as an interesting hypothesis as recently proposed by Prommeenate et al. (2004). Future efforts will aim to identify components of the electron input device of the plastid Ndh complex.
METHODS
Plant Material and Culture Conditions
Tobacco wild plants (Nicotiana tabacum cv Petit Havana) and transplastomic plants were grown in soil under controlled conditions in a growth chamber (14-h photoperiod at 25°C followed by a 10-h night at 20°C) and watered with a complete nutrient solution (Rumeau et al., 2004). Arabidopsis (ecotype Col-0 or Ler) was grown in soil in a growth chamber with an 8-h-light at 22°C/16-h-dark at 18°C cycle for a short day condition. T-DNA insertion mutant lines N_068922 and N_087707 were provided by the Salk Institute Genomic Analysis Laboratory. In N_068922 and N_087707 mutant lines, T-DNA is inserted in NdhO (At1g74880) and NdhM (At4g37920), respectively. The mutant line ET-1999, which contains a Ds insertion in NdhN (At5g58260), was provided by the Cold Spring Harbor Laboratory.
Plasmid Construction for Plastid Transformation
Plasmid pLEP01 (Figure 1B; Kay et al., 2002) is similar to pSZ197 described by Svab and Maliga (1993). It is a pBSII-SK− (Stratagene, La Jolla, CA) derivative containing the aadA gene (gift from Michel Goldschmidt-Clermont, University of Geneva), conferring spectinomycin and streptomycin resistance and controlled by a modified plastid rRNA operon promoter (Prrn) and the 3′ region of the plastid psbA gene (TpsbA). The aadA gene is inserted at a BamHI site (nucleotide 59,313) in the plastid sequence encompassing the rbcL and accD genes (nucleotides 57,758 to 60,571).
From pTBa2, a tobacco ptDNA subclone (Shinozaki et al., 1986), a PstI-ApaI restriction fragment (nucleotides 123,765 to 126,745) was generated and cloned into pBSII-SK− (Stratagene). The generated vector carries the ndhH gene (Figure 1B). A 7His-tag was engineered at the N terminus of NDH-H by PCR (Figure 1A). Two separate PCRs were performed with two oligonucleotide couples, OIII-OIV and OV-OVI, using the vector carrying ndhH as template. The PCR products were mixed and used as template for a new PCR with OVI and OIV. The PCR product was digested with SalI and AatII and exchanged with its homologous counterpart in the ndhH vector. This yielded the plastid transformation vector designated pH5′-tag (Figure 1B).
Oligonucleotides
Oligonucleotide sequences are as follows: OI (5′-ACTAAATCAATAAAGTATCGAT-3′), OII (5′-TGCATCATTCGCATACCGGT-3′), OIII (5′-GGTGCAGTATGATGATGATGATGATGATGCATAAGTTTTTTACCGATTC-3′), OIV (5′-TTATGGTCTAGACGTCGTAGAGAATTGA-3′), OV (5′-CATCATCATACTGCACCAACTACAAGAAAAGACCTCATG-3′), and OVI (5′-ATATTGGTTCGCAGTCGACAACAT-3′).
Molecular Analysis
Plasmids were isolated from Escherichia coli cells and purified using anion-exchange resin columns (Tip-500; Qiagen, Courteboeuf, France). General molecular cloning techniques (restriction digests, cloning, growth, and transformation of bacterial strains) were performed according to Sambrook et al. (1989). High-resolution agarose gel electrophoresis was performed in 3% (w/v) agarose (high resolution, Sigma A4718; Sigma-Aldrich, Saint-Quentin, France) in Tris-borate-Na2-EDTA (44 mM/44 mM/1 mM) buffer. Sequencing was performed using the BigDye Terminator cycle sequencing ready reaction kit (PE-Applied Biosystems, Foster City, CA). Sequencing products were analyzed using an ABI Prism 377 DNA sequencer (PE Applied Biosystems).
Total DNA was extracted from regenerating plants by a rapid minipreparation procedure (DNeasy; Qiagen).
Total RNA was extracted by a rapid minipreparation procedure (RNeasy; Qiagen). Reverse transcription was performed with 1 μg of RNA and reverse transcriptase (Omniscript; Qiagen) using oligo(dT) primers. Specific transcript amplifications were performed using Taq polymerase (Qiagen) and the following specific gene primers: for NdhM, 5′-ATGGTTGCAGCATTCTCTTACA-3′ and 5′-TCAAACTCACAAGTCTCTGGAT-3′; for NdhN, 5′-ATGGGAAGCCGTGCAATATGTATA-3′ and 5′-TTACTCTTGTGCAGCTAGATTTGCAA-3′, and for NdhO, 5′-ATGGCTTTCTCTGCGACTGTGTCT-3′ and 5′-GTAAGCGAGCATTTGTTCAAA-3′. As internal control, actin-specific primers were also included in the NdhN and NdhM PCR mix: 5′-AAAATGGCCGATGGTGAGGATAT-3′ and 5′-CAATACCGGTTGTACGACCACT-3′. As internal control in the NdhO mix, the following actin-specific primers were included: 5′-AAAATGGCCGATGGTGAGGATAT-3′ and 5′-GATGGTTATGACTTGTCCAT-3′.
Generation of Transplastomic Plants
Tobacco leaves were transformed using the biolistic method essentially as described by Svab and Maliga (1993). Tobacco leaves were bombarded with a mix (1:1, 25 μg each) of both vectors pLEP01 and pH5′-tag. Green calli were selected on growth media containing spectinomycin (1 mg·mL−1) until they formed shoots. The shoots were rooted on MS medium also containing spectinomycin.
Preparation of Thylakoid Membranes
Thylakoid membrane extraction and stromal and granal lamellae separation were performed as described by Sazanov et al. (1998). Briefly, after grinding in a Waring blender, chloroplasts were collected by three successive low-speed centrifugations to avoid mitochondria contamination. Chloroplasts were osmotically lysed and thylakoid membranes collected by centrifugation. Stromal and grana lamellae were separated by centrifugation after stacking in a buffer containing 20 mM Mes, pH 6, 15 mM NaCl, 5 mM MgCl2, and dodecyl maltoside (1% final concentration).
Affinity Purification of His-Tagged Ndh Complex
Stromal lamellae fractions extracted from H5′-tag transplastomic plants were loaded onto a Ni2+ metal affinity column (HiTrap chelating; Amersham-Pharmacia Biotech, Uppsala, Sweden), which had been pre-equilibrated with buffer A (phosphate buffer, pH 7.4, 0.15 M NaCl, and 0.1% dodecyl maltoside). The column was washed with buffer A and with buffer A containing 10 mM imidazole. The bound His-tagged Ndh complex was eluted with the same buffer containing 75 mM imidazole. The eluted fractions (400 μL) were pooled and either acetone (75%) precipitated before electrophoresis analysis or directly used for activity determination.
PAGE and Immunodetection
Denaturing SDS-PAGE was performed as described by Laemmli (1970) using 13% (w/v) acrylamide gels. Proteins were either stained with Bio-Safe Coomassie (Bio-Rad, Hercules, CA) or electrotransferred onto 0.45-μm nitrocellulose membranes (Biotrace NT; Pall, Fontenay-sous-Bois, France) and probed with antibodies. Immunocomplexes were detected using peroxidase-conjugated antibodies and SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL) as peroxidase substrate. Antibodies against NDH-H are described in Horvath et al. (2000). Antibodies against polyHis-tag were purchased at Roche (Meylan, France). NDH-O was detected using rabbit antibodies prepared against a peptide antigen, GEVLDLRVFETGEYALV, conjugated with keyhole limpet hemocyanin (Agro-Bio, La Ferté Saint-Aubin, France). The Arabidopsis gene encoding NDH-N (At5g58260) was cloned into an E. coli expressing vector (pQE; Qiagen). Protein synthesis was induced with isopropylthio-β-galactoside for 6 h at 37°C. Protein purification was performed using Ni2+-NTA affinity chromatography according to the manufacturer's instructions. Antiserum was raised against the protein in chicken (Agro-Bio).
Chlorophyll Fluorescence
Post-illumination chlorophyll fluorescence rises were measured on attached leaves using a PAM-2000 modulated fluorometer (Walz, Effeltrich, Germany). The basal chlorophyll fluorescence level measured under low nonactinic light was recorded after a 5-min actinic illumination (250 μmol photons·m−2·s−1).
Photosynthetic gas exchange and chlorophyll fluorescence parameter measurements were simultaneously performed on detached leaves using an LI-6400 portable photosynthesis system equipped with a 6400-40 fluorometer (LI-COR Biosciences, Lincoln, NE). To limit variations in photosynthetic activity because of stomatal regulations, experiments were performed under lowered CO2 concentration (250 μmol·mol−1 CO2) favoring stomatal opening. The gas flow rate at the leaf level was 250 mol air·s−1, and the temperature was 25°C. Air humidity was controlled by a dew point generator (LI-COR Biosciences; LI-610) set at 17°C. Gas exchange and cholorophyll fluorescence measurements were performed at different actinic light intensities supplied with light emitting diodes (90% red light, 10% blue light) after a 20-min equilibration period under a light intensity of 150 μmol photons·m−2·s−1. Fluorescence levels Fs (stationary fluorescence level in the light), Fm′ (maximal fluorescence in the light measured after a saturating pulse), and Fm (maximal fluorescence in the dark), were used to calculate NPQ [NPQ = (Fm−Fm′)/Fm′]. Apparent photosynthetic ETRs, expressed in μmol electrons·m−2·s−1, were estimated as (Fm′−Fs)/Fm′ × PFFDi × LA × 0.5, where PFFDi is the incident photosynthetic photon flux density, LA is the leaf absorbance (0.84), and 0.5 the factor accounting for the light partition between the two photosystems.
NADH and NADPH Dehydrogenase Assays
NADH and NADPH:ferricyanide oxidoreductase activities were assayed at room temperature according to Sazanov et al. (1998). The assay buffer consisted of 50 mM Tris-HCl, pH 7.5, 0.1% dodecyl maltoside, 0.1 mM NADH or NADPH, and 0.5 mM FeCN. Activity was recorded on a spectrophotometer after reduction of FeCN at 420 nm.
Mass Spectrometry and Protein Identification
After SDS-PAGE, discrete bands were excised from the Coomassie Brilliant Blue–stained gel. The in-gel digestion was performed as previously described (Ferro et al., 2000). Gel pieces were then extracted with 5% (v/v) formic acid solution and acetonitrile. After drying, tryptic peptides were resuspended in 2% aqueous trifluoroacetic acid. The samples were injected into an LC-Packings (Dionex, Sunnyvale, CA) nanoLC system and first preconcentrated on a 300 μm × 5 mm PepMap C18 precolumn. The peptides were then eluted onto a C18 column (75 μm × 150 mm). The chromatographic separation used a gradient from solution A (2% acetonitrile:98% water:0.1% formic acid) to solution B (80% acetonitrile:20% water:0.1% formic acid) over 60 min at a flow rate of 200 nL·min−1. The liquid chromatography system was directly coupled to a QTOF Ultima mass spectrometer (Waters, Milford, MA). MS and MS/MS data were acquired and processed automatically using MassLynx 3.5 software. Database searching was performed using the MASCOT 1.7 program available intranet. Two protein databases were used: an updated compilation of SwissProt and Trembl (ftp://us.expasy.org/databases/sp_tr_nrdb/) and the Arabidopsis Genome Initiative protein database (ftp://ftp.arabidopsis.org/home/tair/Sequences/blast_datasets/). Using MASCOT, proteins, which were identified with at least two peptides both showing a score higher than 40, were validated without any manual validation. For proteins identified by only one peptide having a score higher than 40, the peptide sequence was checked manually. Peptides, with scores higher than 20 and lower than 40, were systematically checked and/or interpreted manually to confirm or cancel the MASCOT suggestion. The remaining unassigned peptides were interpreted manually, and internet MS-Pattern (http://128.40.158.151/ucsfhtml3.4/mspattern.htm) and Blast (http://www.ncbi.nlm.nih.gov/BLAST/) were used for database searching.
Supplementary Material
Acknowledgments
The authors thank Michel Goldschmidt-Clermont (University of Geneva) for providing the aadA gene and Elena Baena Gonzalez (University of Turku, Finland) for providing the anti-D2 antibodies. We also thank S. Cuiné (Commissariat à l'Energie Atomique Cadarache, France) for his help with the fast protein liquid chromatography and T. Shikanai (Kyushu University, Japan), Y. Munekage, M. Havaux, and B. Genty (Commissariat à l'Energie Atomique Cadarache, France) for helpful discussions. We also acknowledge the ABRC, the Salk Institute Genomic Analysis Laboratory, the Cold Spring Harbor Laboratory, and the Max-Planck-Institut für Züchtungsforschung for supplying the Arabidopsis mutants. This work was supported by the European Community Biotechnology program (Grant BIO4-CT-97-2245).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Dominique Rumeau (dominique.rumeau@cea.fr).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.028282.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Berger, S., Ellersiek, U., Kinzelt, D., and Steinmüller, K. (1993). Immunopurification of a subcomplex of the NAD(P)H-plastoquinone-oxidoreductase from the cyanobacterium Synechocystis sp. PCC6803. FEBS Lett. 326, 246–250. [DOI] [PubMed] [Google Scholar]
- Bricker, T.M., Morvant, J., Masri, N., Sutton, H.M., and Frankel, L.K. (1998). Isolation of a highly active photosystem II preparation from Synechocystis 6803 using a histidine-tagged mutant of CP47. Biochim. Biophys. Acta 1409, 50–57. [DOI] [PubMed] [Google Scholar]
- Burrows, P.A., Sazanov, L.A., Svab, Z., Maliga, P., and Nixon, P. (1998). Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J. 17, 868–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carol, P., Stevenson, D., Bisanz, C., Breitenbach, J., Sandmann, G., Mache, R., Coupland, G., and Kuntz, M. (1999). Mutations in the Arabidopsis gene immutans cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell 11, 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrer, H., and Maliga, P. (1995). Targeted inactivation of foreign genes into the tobacco plastid genome without physical linkage to the selectable marker gene. Biotechniques 13, 791–794. [Google Scholar]
- Cournac, L., Redding, K., Ravenel, J., Rumeau, D., Josse, E.M., Kuntz, M., and Peltier, G. (2000). Electron flow between photosystem II and oxygen in chloroplasts of photosystem I-deficient algae is mediated by a quinol oxidase involved in chlororespiration. J. Biol. Chem. 275, 17256–17262. [DOI] [PubMed] [Google Scholar]
- Dupuis, A., Prieur, I., and Lunardi, J. (2001). Toward a characterization of the connecting module of Complex I. J. Bioenerg. Biomembr. 25, 367–375. [DOI] [PubMed] [Google Scholar]
- Ferro, M., Seigneurin-Berny, D., Rolland, N., Chapel, A., Salvi, D., Garin, J., and Joyard, J. (2000). Organic solvent extraction as a versatile procedure to identify hydrophobic chloroplast membrane proteins. Electrophoresis 21, 3517–3526. [DOI] [PubMed] [Google Scholar]
- Friedrich, T., and Böttcher, B. (2004). The gross structure of the respiratory complex I: A Lego system. Biochim. Biophys. Acta 1608, 1–9. [DOI] [PubMed] [Google Scholar]
- Funk, E., Schafer, E., and Steinmüller, K. (1999). Characterization of the complex I-homologous NAD(P)H- plastoquinone-oxidoreductase (NDH-complex) of maize chloroplasts. J. Plant Physiol. 154, 16–23. [Google Scholar]
- Funk, E., and Steinmüller, K. (1995). Photosynthesis: From Light to Biosphere, Vol. II. P. Mathis, ed (Boston: Kluwer), pp. 701–704.
- Grohmann, L., Rasmusson, A.G., Heiser, V., Thieck, O., and Brennicke, A. (1996). The NADH-binding subunit of respiratory chain complex I is nuclear-encoded in plants and identified only in mitochondria. Plant J. 10, 793–803. [DOI] [PubMed] [Google Scholar]
- Guedeney, G., Corneille, S., Cuiné, S., and Peltier, G. (1996). Evidence for an association of ndhB, ndhJ gene products and ferredoxin-NADP-reductase as components of a chloroplastic NAD(P)H dehydrogenase complex. FEBS Lett. 378, 277–280. [DOI] [PubMed] [Google Scholar]
- Hashimoto, M., Endo, T., Peltier, G., Tasaka, M., and Shikanai, T. (2003). A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J. 36, 541–549. [DOI] [PubMed] [Google Scholar]
- Horvath, E., Peter, S.O., Joët, T., Rumeau, D., Cournac, L., Horvath, G.V., Kavanagh, T.A., Schäfer, C., Peltier, G., and Medgyesy, P. (2000). Targeted inactivation of the plastid ndhB gene in tobacco results in an enhanced sensitivity of photosynthesis to moderate stomatal closure. Plant Physiol. 123, 1337–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joët, T., Cournac, L., Horvath, E., Medgyesy, P., and Peltier, G. (2001). Increased sensitivity of photosynthesis to antimycin A induced by inactivation of the chloroplast ndhB gene. Evidence for a participation of the NADH dehydrogenase complex to cyclic electron flow around photosystem I. Plant Physiol. 125, 1919–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joët, T., Cournac, L., Peltier, G., and Havaux, M. (2002. a). Cyclic electron flow around photosystem I in C3 plants. In vivo control by the redox state of chloroplasts and involvement of the NADH-dehydrogenase complex. Plant Physiol. 128, 760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joët, T., Genty, B., Josse, E.M., Kuntz, M., Cournac, L., and Peltier, G. (2002. b). Involvement of a plastid terminal oxidase in plastoquinone oxidation as evidenced by expression of the Arabidopsis thaliana enzyme in tobacco. J. Biol. Chem. 277, 31623–31630. [DOI] [PubMed] [Google Scholar]
- Kay, E., Vogel, T.M., Bertolla, F., Nalin, R., and Simonet, P. (2002). In situ transfer of antibiotic resistance genes from transgenic (transplastomic) tobacco plants to bacteria. Appl. Environ. Microbiol. 68, 3345–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofer, W., Koop, H.U., Wanner, G., and Steinmüller, K. (1998). Mutagenesis of the genes encoding subunits A, C, H, I, J and K of the plastid NAD(P)H-plastoquinone-oxidoreductase in tobacco by polyethylene glycol-mediated plastome transformation. Mol. Gen. Genet. 258, 166–173. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Martienssen, R. (1998). Functional genomics: Probing plant gene function and expression with transposons. Proc. Natl. Acad. Sci. USA 95, 2021–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo, M., Endo, T., and Asada, K. (1998). Isolation of a novel NAD(P)H-quinone oxidoreductase from cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol. 39, 751–755. [DOI] [PubMed] [Google Scholar]
- Maul, J.E., Lilly, J.W., Cui, L., dePamphilis, C.W., Miller, W., Harris, E.H., and Stern, D.B. (2002). The Chlamydomonas reinhardtii plastid chromosome: Islands of genes in a sea of repeats. Plant Cell 14, 2659–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munekage, Y., Hashimoto, M., Miyake, C., Tomizawa, K.I., Endo, T., Tasaka, M., and Shikanai, T. (2004). Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429, 579–582. [DOI] [PubMed] [Google Scholar]
- Munekage, Y., Hojo, M., Meurer, J., Endo, T., Tasaka, M., and Shikanai, T. (2002). PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110, 361–371. [DOI] [PubMed] [Google Scholar]
- Nixon, P.J. (2000). Chlororespiration. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 1541–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, T. (1992). Identification and characterization of the ictA/ndhL gene product essential to inorganic carbon transport of Synechocystis PCC6803. Plant Physiol. 99, 1604–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama, K., Kohchi, T., Sano, T., and Yamada, Y. (1988). Newly identified groups of genes in chloroplasts. Trends Biochem. Sci. 13, 19–22. [DOI] [PubMed] [Google Scholar]
- Peltier, G., and Cournac, L. (2002). Chlororespiration. Annu. Rev. Plant Biol. 53, 523–550. [DOI] [PubMed] [Google Scholar]
- Prommeenate, P., Lennon, A.M., Markert, C., Hippler, M., and Nixon, P.J. (2004). Subunit composition of NDH-1 complexes of Synechocystis sp. PCC 6803: Identification of two new ndh gene products with nuclear-encoded homologues in the chloroplast Ndh complex. J. Biol. Chem. 279, 28165–28173. [DOI] [PubMed] [Google Scholar]
- Quiles, M.J., and Cuello, J. (1998). Association of ferredoxin-NADP oxidoreductase with the chloroplastic pyridine nucleotide dehydrogenase complex in barley leaves. Plant Physiol. 117, 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiles, M.J., Garcia, A., and Cuello, J. (2000). Separation by blue-native PAGE and identification of the whole NAD(P)H dehydrogenase complex from barley stroma thylakoids. Plant Physiol. Biochem. 38, 225–232. [Google Scholar]
- Quiles, M.J., Garcia, A., and Cuello, J. (2003). Comparison of the thylakoidal NAD(P)H dehydrogenase complex and the mitochondrial complex I separated from barley leaves by blue-native PAGE. Plant Sci. 164, 541–547. [Google Scholar]
- Rasmusson, A.G., Heiser, V., Irrgang, K.D., Brennicke, A., and Grohmann, L. (1998). Molecular characterisation of the 76 kDa iron-sulphur protein subunit of potato mitochondrial complex I. Plant Cell Physiol. 39, 373–381. [DOI] [PubMed] [Google Scholar]
- Rhee, S.Y., et al. (2003). The Arabidopsis Information Resource (TAIR): A model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 31, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix, J.D. (2002). Chlamydomonas, a model system for studying the assembly and dynamics of photosynthetic complexes. FEBS Lett. 529, 34–38. [DOI] [PubMed] [Google Scholar]
- Rumeau, D., Bécuwe-Linka, N., Beyly, A., Carrier, P., Cuiné, S., Genty, B., Medgyesy, P., Horvath, E., and Peltier, G. (2004). Increased zinc content in transplastomic tobacco plants expressing a polyhistidine-tagged Rubisco large subunit. Plant Biotechnol. J. 2, 389–399. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sazanov, L.A., Burrows, P.A., and Nixon, P. (1996). Detection and characterization of a complex I-like NADH-specific dehydrogenase from pea thylakoids. Biochem. Soc. Trans. 24, 739–743. [DOI] [PubMed] [Google Scholar]
- Sazanov, L.A., Burrows, P.A., and Nixon, P. (1998). The plastid ndh genes code for an NADH-specific dehydrogenase: Isolation of a complex I analogue from pea thylakoid membranes. Proc. Natl. Acad. Sci. USA 95, 1319–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, M., et al. (2002). Functional annotation of a full-length Arabidopsis cDNA collection. Science 296, 141–145. [DOI] [PubMed] [Google Scholar]
- Shikanai, T., Endo, T., Hashimoto, T., Yamada, Y., Asada, K., and Yokota, A. (1998). Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc. Natl. Acad. Sci. USA 95, 9705–9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki, K., et al. (1986). The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 5, 2043–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura, M., Inoue, Y., and Minagawa, J. (1998). Rapid and discrete isolation of oxygen-evolving His-tagged photosystem II core complex from Chlamydomonas reinhardtii by Ni2+ affinity column chromatography. FEBS Lett. 426, 140–144. [DOI] [PubMed] [Google Scholar]
- Svab, Z., and Maliga, P. (1993). High frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl. Acad. Sci. USA 90, 913–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman, F.A., Minai, L., and Nechushtai, R. (1999). The biogenesis and assembly of photosynthetic proteins in thylakoid membranes. Biochim. Biophys. Acta 1411, 21–85. [DOI] [PubMed] [Google Scholar]
- Wu, D.Y., Wright, D.A., Wetzel, C., Voytas, D.F., and Rodermel, S. (1999). The immutans variegation locus of Arabidopsis defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis. Plant Cell 11, 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.