Abstract
Membrane proteins are the gateway to the cell. These proteins are also a control center of the cell as information from the outside is passed through membrane protein signaling networks to the cellular machinery. The design of membrane proteins seeks to harness the power of these gateways and signaling networks. This review will focus on the design of the membrane proteins that are in the outer membrane, a membrane which only exists for gram negative bacteria, mitochondria and chloroplasts. Unlike other membrane proteins, outer membrane proteins are uniquely shaped as β-barrels. Herein, I describe most known examples of outer-membrane, β-barrel design to date, focusing particularly on categorizing designs as 1) structural deconstruction, 2) structural changes, 3) chemical function design, and 4) the creation of new folds.
Introduction
Membrane proteins are classified by backbone configuration which determines their function and location. In a quirk of biology, due to the mechanisms of their respective insertions through the Sec translocon [1,2], inner membrane proteins are all α-helical, and outer membrane proteins are almost [3] all β-sheets.
The field of helical (inner) membrane protein design is much more developed than that of its β-barrel (outer) membrane protein counterpart. This is evident in the comprehensive reviews of membrane protein design that focus exclusively on helical membrane protein design [4–8]. However, there is growing interest in outer membrane proteins (OMPs) because of their role in antibiotic resistance, their potential applications as biosensors, and their location which makes them accessible to the exterior of the cell.
OMP Anatomy
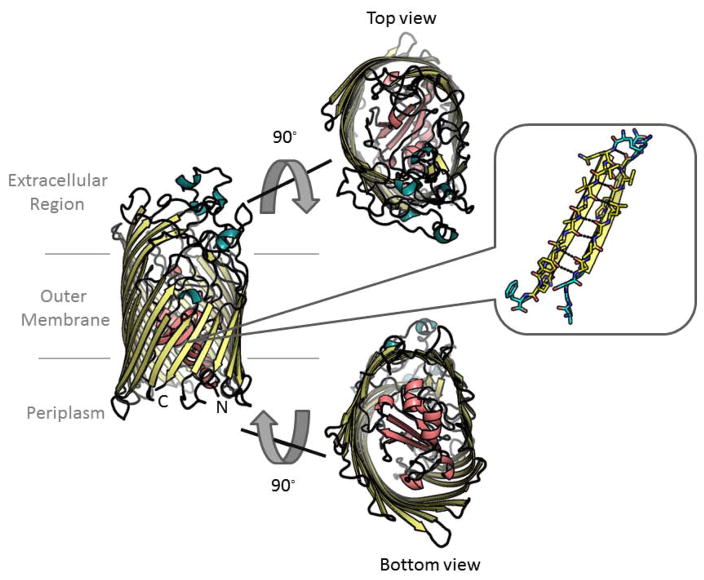
The anatomy of OMPs is fundamental to their design. The known anatomic patterns of these proteins are illustrated by the ~100 non-homologous structures of OMPs in the PDB. β-barrel strands are amphipathic and are generally oriented antiparallel to each other. The architecture of the strands which are hydrogen bound through the backbone causes the side chains to alternate in direction between those facing the pore and those facing the membrane. Loops connect the strands, with larger loops in the extracellular space and smaller turns in the periplasmic space. The loops sometimes create plugs in the barrels before connecting to the next strand (figure 1).
Figure 1. Anatomy of an outer membrane protein using FhuA [35] for illustration.
Strands shown in yellow. Longer extracellular loops and shorter periplasmic turns shown in teal/black. The plug domain is shown in pink. Side view at left, top view and bottom view in the center, and strand view at right.
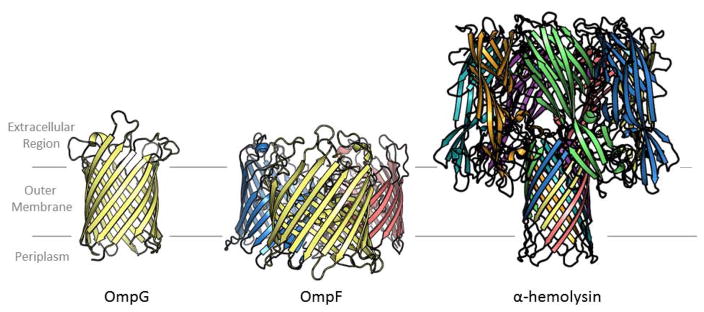
Most structurally characterized outer membrane β-barrels are monomeric with one chain making up one barrel. However, some of the barrels oligomerize. When they do it’s most often as trimers although dimers are seen in some cases. More complicated topologies also exist where multiple protein chains contribute strands to a single barrel (figure 2).
Figure 2. The three oligomeric configurations of outer membrane barrels.
Each chain is colored differently. On the left is a single chain making a single barrel, OmpG [63]. In the center are three chains making three barrels, OmpF [64]. On the right is seven chains making one barrel, α-hemolysin [65].
This review focuses on how amino acids create OMP conformations. Synthetic modifications of OMPs are not within the scope of this review.
Outer Membrane Protein Design
There are four types of protein design: (1) Structural deconstruction—removing parts of proteins to see if structural or functional elements can be maintained (2) structural transformation – intentionally changing a structural characteristic of a protein (3) chemical function design—adding a new chemical feature to a protein, and (4) creating a new fold—making a fold topology that has not yet been found in nature.
1) Structural deconstruction
A primary stage of protein design is determining the relationship of the protein’s anatomy to its structure, function, and stability. To do this, protein designers have determined the extent to which strand-strand interactions, loops, and sequence content can be altered before changing these three characteristics.
Strand-strand interactions
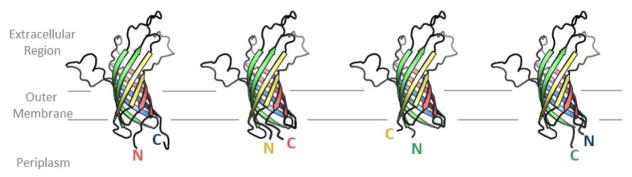
In β-barrels, side chains don’t point towards each other but rather alternately point towards the interior of the pore and towards the membrane. This makes their interactions more oblique. Given the indirectness of the interaction, we need to ask the question: how important are strand-strand interactions in β-barrels? To address this question, 8-stranded β-barrel, OmpA was divided into four hairpins—i.e. two strands which are antiparallel to each other. These four hairpins were then permuted in almost all possible manners [9]. As might be expected, very few of the constructs folded into the outer membrane in vivo. The ones that did fold were the three circular permutations, i.e. the permutations that preserved all of the relationships between all of the strands although they did not maintain the covalent connectivity between the strands (figure 3). This suggests that although the side chains do not point towards each other they interact sufficiently that changing hairpin-hairpin interactions can abrogate folding. Moreover, these circular permutations change the location of the β-signature that is conserved on the C-terminus [10] of barrels and which is known to be used for insertion into the membrane through interactions with BamA [11]. That these circular permutations fold with little loss of efficiency into the outer membrane suggests that either the β-signature sequence on the C-terminus is not required for folding or that it can be recognized at a location other than the terminus.
Figure 3. Schematic of circular permutations of OmpA.
The four hairpins of OmpA have each been colored differently. The permutations shift the strands on which the N- and C-termini are located, but all strands still maintain their relationship relative to each other. N and C are colored relative to the strands on which the termini are located. These permutations were found to fold and most engendered phage sensitivity. Drawn using the first pose of the OmpA NMR structure [66].
Loops
In the previously described study, not only did circular permutations fold, but the OmpA conformation remained recognizable to phages which target them. Unintuitively, the periplasmic turns were found to be important in facilitating phage recognition even though the phage targets the extracellular side of the barrel. Particularly important for phage recognition was the periplasmic turn between strand 6 and strand 7. A separate study experimented with deleting the extracellular loops and found that OmpA would fold even if all extracellular loops were deleted. [12] However in order to make the deletion constructs, amino acids were added to the periplasmic turns. These additions decreased OmpA’s stability. The subsequent deletion of the extracellular loops however did not further decrease the protein’s stability. Thus periplasmic turns are shown to be important for stability and maintenance of the fold.
Sequence content
Focusing then on the sequence content of the strands, the interior facing residues and the exterior facing residues of strands 4, 6, and 8 of OmpA were subjected to genetic randomization. [13] Despite the use of the β-signature sequence for insertion, strand 8 which contains the β-signature sequence was the most permissive to randomization as measured by conferring the phage sensitive phenotype of native-OmpA. Conversely, strand 6 was the strand least permissive to sequence randomization. Overall, a key determinant of successful randomization of membrane-facing residues was maintaining hydrophobicity. Although it has since been shown evolutionarily that membrane-facing residues are more amenable to substitution than interior facing residues [14], this work indicated that outward-facing mutants were not necessarily more successful than inward-facing mutants.
Later that same year, drastic mutation to the exotoxin α-hemolysin resulted in the maintenance of a multi-chain β-barrel fold [15]. By reversing the sequence of the transmembrane barrel region of α-hemolysin, the barrel still folded though it did not maintain function. The sequence reversal employed meant that the N to C sequence was encoded in the C to N direction. Although the reverse sequenced barrel still oligomerized, the role of the barrel in the formation in the oligomerization is unclear as the cap region has been shown to oligomerize without the transmembrane barrel [16]. Importantly, function of the altered α-hemolysin returned with the return of the N to C sequence of the barrel’s turns in the otherwise reversed hairpin sequence. This experiment further underscores previous results demonstrating the important role of the turn region in OMP interactions.
The most radical sequence change for a transmembrane β-barrel has been the surface redesign of OmpA [17]. This redesign was a computational design using the previous creation of a statistical potential for each residue’s membrane-depth preference [18]. The statistical potential coupled with an entropy term was implemented to change almost the entire surface of the 8-stranded barrel. Although the initial design did not demonstrate function, reverting one-third of the mutations created a construct whose surface was still 60% designed which folded and conferred phage sensitivity. Notably, strand 6 required the most reversion to wild type, consistent with the difficulty in mutating strand 6 in the genetic randomization of strands study [13].
2) Structural transformation
Structural transformation design consists of intentionally changing a structural feature or characteristic of a protein. The following is a discussion of outer membrane β-barrels that have been stabilized, lengthened/shortened, widened/narrowed, and unblocked/occluded.
Stabilizing the barrel
The computational program, TmSIP uses amino acid interactions and amino acid position to predict unstable regions of proteins [19]. These predictions of instability have been used to find positions in β-barrels that are bound to other parts of proteins (plugs, clamps, or other barrels) to increase stability [20]. TmSIP can therefore predict unstable positions to be mutated that make the β-barrel more stable. This methodology was used for a variety of proteins [21,22], and is best illustrated with the case of eukaryotic OMP Tom40 where two of the three predicted mutations were stabilizing, despite not having an experimentally determined structure of the protein[23].
While the above examples of stabilizing strands affected unfolding temperature, stabilization has also been utilized to stop gating, notably with OmpG. A non-gating “quietOmpG” was created by stabilizing a loop with a disulphide bond and deleting the bulge of a strand [24]. The gating of OmpG has since been further stabilized by dramatically removing all of the loops [25].
Lengthening/shortening the barrel
The hydrophobic region of the membrane and the hydrophobic surface of a protein should be of similar height in order to facilitate optimal insertion of the protein into the membrane [26,27]. In order to make the 22-stranded FhuA barrel long enough to incorporate into a thicker membrane, a ‘copy and paste’ approach was used by repeating the last five residues of each strand in the 22-stranded protein. This elongation was successful and the protein was demonstrated to allow insertion and pore formation in thicker membranes compared to an un-elongated FhuA.[28] Conversely, even when more than half of the barrel portion of α-hemolysin is taken off from the bottom of the barrel, the shortened construct can still create membrane pores, possibly by assisting in the organization of the lipids beneath the cut-off pore [29].
Increase/decrease pore size
In light of the utility of β-barrels for biosensors and the dependence of biosensors on pore size, attempts were made to increase the pore size of beta-barrels by duplicating native strands. Although an attempt to widen the barrel of OmpG using two strands at its C-terminus was not successful [30], duplication of two strands at the N-terminus of FhuA successfully increased both its pore size and diffusion kinetics[31]. Conversely, removing one and two strands from the middle of the VDAC1 barrel resulted in a smaller barrel [32].
Unblocking/occluding the pore
In line with widening the pore of FhuA by adding strands, there have also been efforts to widen the pore of FhuA by removing blockages. Before the crystal structure of FhuA was published there was a proposed structure of FhuA with 32 strands [33]. Using this structure, the predicted largest loop was pruned resulting in an FhuA with a much higher conductance [34]. When the structure was determined [35,36] it was noted that there was a large N-terminal plug and only 22 strands. Moreover, the predicted loop pruning which led to higher conductance was actually a deletion of half of strand 8. Neither did deletion of the plug give similar significant increase in high conductance channels, nor did pruning the loop do the same without pruning strand 8 [37]. Ultimately for FhuA, the more extreme the loop-pruning, the higher the conductance [38,39]. However, as would be expected, it has also been shown that in some proteins deletion of the plug does in fact lead to higher conductance. Such is the case for PapC [40].
Protein occlusions have been engineered in the cavity of the extracellular cap region of the pore of α-hemolysin. This cavity accommodated loops of serines and glycines totaling 175 amino acids.[41]. Temperature sensitive loops were also engineered in that cavity region, specifically, elastin-like polypeptides that order at higher temperatures [42]. By adding these elastin-like polypeptides to the extracellular cavity of one of the chains of the α-hemolysin heptamer, a temperature sensitive gate of the α-hemolysin pore was created.[43]
3) Chemical function design
The third type of protein design is engineering a new chemical function. Because the outer membrane is the outermost part of the cell, developing chemical functions in OMPs allows for cells to change their local environment. In the outer membrane, this type of design has had three major successes thus far: changing the ion selectivity of a pore, the engineering of metal binding sites and the change in specificity of an endopeptidase.
Changing ion selectivity of a pore
The porin OmpF is a non-selective channel that natively prefers cations to anions. By changing the charge balance of the residues at the constriction site of the pore the preference for cations or anions can be shifted [44]. Overall, the greater the shift in the charge ratio of the constriction site, the greater the selectivity of the channel [45]. Removing all positive charges at the constriction site transforms OmpF into a Ca2+ selective channel [46]. Replacing positive residues with negative residues at the constriction site of P. denitrificans porin was also effective in creating cation selectivity [47]. A similar approach was also attempted by neutralizing the negatively charged residues on the loops of FhuA [39].
Metal binding sites
Successes in metal binding sites may be found to be useful for biosensing or even future manipulation towards catalysis. The first instance of OMP metal binding site design was a divalent-metal binding site engineered into the pore of α-hemolysin. This was accomplished by adding four histidine residues to one of the seven chains[48]. The functional design is ultimately a 6:1 WT: His-mutant heptamer, a combination that is isolated by mixing the two components together and then separation of the desired combination from all other combinations using SDS PAGE.
A Tb3+ binding site was engineered into the loop region of OmpA to facilitate NMR characterization. This was accomplished by shortening extracellular loops of OmpA and then putting an EF-hand domain [49] surrounded by glycines in one of the previously cut loop positions [50].
Evolution of an enzyme
OmpT is an outer membrane endopeptidase that natively cleaves an Arg-Arg peptide bond. Its active site was evolved to cleave new substrate. Through mutagenesis of the binding pocket and the development of a clever FACS screen that screens for fluorophores that have been cleaved from a quencher [51], six new enzymes were created with high selectivity and catalytic efficiency. OmpT’s substrate specificity was changed to Ala-Arg[51], Glu-Arg, Tyr-Arg, Thr-Arg, Arg-Val, and Glu-Ala.[52] Since then the process has been optimized for modified-Tyr-Arg peptides as well. [53,54]
4) Novel fold design
Given the rigid requirements of OMPs as β-barrels imposed by the mechanism of the Sec translocon, and the rigidity of the β-barrel hydrogen bonding as imposed by the water depleted outer membrane, it might not be easy to create significantly new topologies for β-barrels. However, we have seen novel multi-chain β-barrels with new chain numbers.
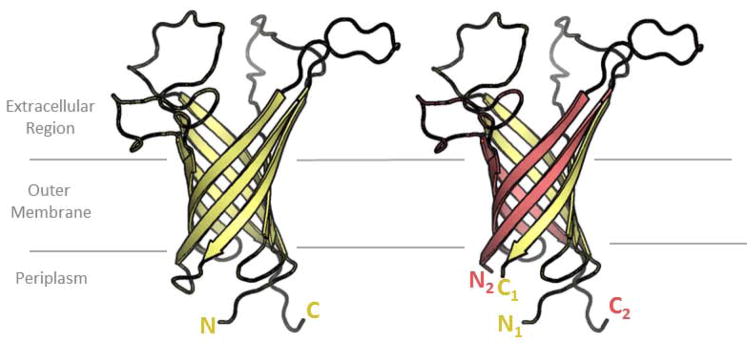
Split variants of OmpA offer the tantalizing possibility of multi-chain β-barrels that can be designed with the specificity of interaction encoded in the side chains. OmpA was divided in half to make two separate constructs. It was found that the N-terminal four-strands of OmpA could pair with a non-covalently attached C-terminal four-strands of OmpA to make a native-like, eight-stranded OmpA (figure 4). This multi-chain OmpA is susceptible to phage, is heat modifiable and is protected from subtilisin degradation. [55] Other co-expressed groupings of hairpins showed somewhat native-like characteristics though it’s notable that in both this study and in the circular permutation study [9] maintaining the turn between hairpin 3 and hairpin 4 of OmpA (turn between strands 6 and 7) is correlated with more phage susceptibility.
Figure 4. A split variant of OmpA.
OmpA is transformed from a single-chain barrel (left) to a dual-chain barrel (right). Dual chain barrels have not yet been seen in nature. drawn using the first pose of the OmpA NMR structure [66]
Other designs of multi-chain β-barrels were created through iterative combinatorial library screening to find new β-stranded, pore-forming peptides. [56] Although the structure or oligomeric state of these hairpin multimers has not yet been characterized, future studies may reveal them to have new folds as well.
Methods for future design
There is strong reason to believe that we will see substantial increase in OMP design in the coming years, particularly via computational design. The recent doubling of solved, outer-membrane protein structures will yield a more general understanding of this fold which can then be utilized for design. For example, a generalized feature has been found in the charge-out rule i.e. that there is a ring of charged residues in the extracellular side of the barrel that interacts with the phosphate region of the membrane [57]. Rosetta, the widespread computational protein design software, has recently been rebooted for membrane proteins. In the new RosettaMP (membrane protein), β-barrels were benchmarked alongside their alpha helical cousins. The energy functions provided produce reasonable correlation for predicted vs. experimental ΔΔG values for mutations in OmpLA though the calculations produce a less strong correlation for predicted vs. experimental ΔΔG values for mutations in OmpA. [58] Finally, TmSIP which was used to stabilize β-barrels [20,23] has been updated. This computational potential is now more powerful and includes intrastrand side-chain interactions and an asymmetric membrane. TmSIP’s results excellently correlate with ΔΔG values for mutations in OmpLA and it also predicts the membrane insertion direction. [59]
With respect to experimental techniques, improvement of directed evolution [60] is poised to result in more optimization of designs as well as further engineering of native OMPs. As has been seen for inner membrane (helical) proteins, directed evolution can lead to membrane proteins that are more detergent-stable [61] or more fluorescent [62]. Thus the evolution of the function of OmpT discussed above will likely be the first of many evolved OMPs and evolved OMP designs.
Conclusion
Overall, OMP design has clarified the structural roles of the components of the β-barrel and has also created useful biomolecules. Structural designs have shown that the β-barrel fold is extremely robust to mutation. There are important strand-strand interactions. The larger, extracellular loops may matter for function—if the function is at the exterior of the cell—but the smaller, periplasmic turns matter more for stability and conformation. Finally, particular strands may carry more responsibility than others in creating the overall conformation of the barrel. Designers have yet to figure out why this might be or how to accommodate for it in their designs. With respect to structural transformation, bulging residues and residues of the wrong hydrophobicity can be mutated or deleted in order to stabilize a β-barrel fold. Lengthening and shortening a barrel is possible. Additions to the N-terminus have been successful in expanding the width of a barrel; additions to the C-terminus have not. Pores are sometimes cleared by trimming loops, sometimes cleared by deleting pores, and occlusion of pores can be engineered to create desirable gating conditions. Chemical function design in beta barrels is at its infancy. Chemical designs to date include: change in ion selectivity of porins, two instances of metal binding sites added to OMPs, and the evolution of an OMP’s enzymatic function. Finally, it appears that fold design in this field is possible and will most frequently exploit multi-chain constructs.
As the field further utilizes the increasing power of computational design and directed evolution, I anticipate more useful designs that fulfill current needs for dealing with antibiotic resistance, providing biosensors, and using extracellular enzymatic capacity to alter bacteria’s environment.
Highlights.
Functional roles for β-barrel components have been elucidated
Outer membrane enzymes have been evolved for new substrates
New β-barrel folds have been created
Acknowledgments
I thank Dr. Pinakin Sukthankar, Meghan Franklin, and Cyril Cook for their critical reading of the manuscript. This work was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20 GM103418. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health. All figures were drawn using PyMol.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hagan CL, Silhavy TJ, Kahne D. β-barrel membrane protein assembly by the Bam complex. Annual Review of Biochemistry. 2011;80(1):189–210. doi: 10.1146/annurev-biochem-061408-144611. [DOI] [PubMed] [Google Scholar]
- 2.Osborne AR, Rapoport TA, Berg Bvd. Protein translocation by the Sec61/SecY channel. Annual Review of Cell and Developmental Biology. 2005;21(1):529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- 3.Dong C, Beis K, Nesper J, Brunkan-LaMontagne AL, Clarke BR, Whitfield C, Naismith JH. Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature. 2006;444(7116):226–229. doi: 10.1038/nature05267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Senes A. Computational design of membrane proteins. Current Opinion in Structural Biology. 2011;21(4):460–466. doi: 10.1016/j.sbi.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Perez-Aguilar Jose M, Saven Jeffery G. Computational design of membrane proteins. Structure. 2012;20(1):5–14. doi: 10.1016/j.str.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghirlanda G. Design of membrane proteins: Toward functional systems. Current Opinion in Chemical Biology. 2009;13(5–6):643–651. doi: 10.1016/j.cbpa.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 7.Simms J, Booth PJ. Membrane proteins by accident or design. Current Opinion in Chemical Biology. 2013;17(6):976–981. doi: 10.1016/j.cbpa.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Scott DJ, Kummer L, Tremmel D, Plückthun A. Stabilizing membrane proteins through protein engineering. Current Opinion in Chemical Biology. 2013;17(3):427–435. doi: 10.1016/j.cbpa.2013.04.002. [DOI] [PubMed] [Google Scholar]
- ∘∘9.Koebnik R, Krämer L. Membrane assembly of circularly permuted variants of the E. coli outer membrane protein OmpA. Journal of Molecular Biology. 1995;250(5):617–626. doi: 10.1006/jmbi.1995.0403. The authors divide OmpA’s eight strands into hairpins and make OmpA variants of all possible orders of the hairpins. They find that only the circularly permuted variants fold and most of the circularly permuted variants retain the phage sensitivity phenontype. [DOI] [PubMed] [Google Scholar]
- 10.Struyvé M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. Journal of Molecular Biology. 1991;218(1):141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- 11.Robert V, Volokhina EB, Senf F, Bos MP, Gelder PV, Tommassen J. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS biology. 2006;4(11):e377. doi: 10.1371/journal.pbio.0040377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koebnik R. Structural and functional roles of the surface-exposed loops of the beta-barrel membrane protein OmpA from Escherichia coli. J Bacteriol. 1999;181(12):3688–3694. doi: 10.1128/jb.181.12.3688-3694.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koebnik R. Membrane assembly of the escherichia coli outer membrane protein OmpA: Exploring sequence constraints on transmembrane [beta]-strands. Journal of Molecular Biology. 1999;285(4):1801–1810. doi: 10.1006/jmbi.1998.2405. [DOI] [PubMed] [Google Scholar]
- 14.Jimenez-Morales D, Liang J. Pattern of amino acid substitutions in transmembrane domains of β-barrel membrane proteins for detecting remote homologs in bacteria and mitochondria. PLoS ONE. 2011;6(11):e26400. doi: 10.1371/journal.pone.0026400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheley S, Braha O, Lu X, Conlan S, Bayley H. A functional protein pore with a “retro” transmembrane domain. PRS. 1999;8(06):1257–1267. doi: 10.1110/ps.8.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheley S, Malghani MS, Song L, Hobaugh M, Gouaux JE, Yang J, Bayley H. Spontaneous oligomerization of a staphylococcal alpha-hemolysin conformationally constrained by removal of residues that form the transmembrane beta-barrel. Protein Engineering. 1997;10(12):1433–1443. doi: 10.1093/protein/10.12.1433. [DOI] [PubMed] [Google Scholar]
- ∘∘17.Stapleton JA, Whitehead TA, Nanda V. Computational redesign of the lipid-facing surface of the outer membrane protein OmpA. Proceedings of the National Academy of Sciences. 2015;112(31):9632–9637. doi: 10.1073/pnas.1501836112. The authors resurface the entirety of OmpA using a computational potential that encodes residue depth preference and sequence entropy. They find a series of cross back mutations that allow for OmpA folding and for the phage sensitivity phenotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh D, Davis A, Nanda V. A knowledge-based potential highlights unique features of membrane α-helical and β-barrel protein insertion and folding. Protein Science. 2012;21(1):50–62. doi: 10.1002/pro.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackups R, Jr, Liang J. Interstrand pairing patterns in β-barrel membrane proteins: The positive-outside rule, aromatic rescue, and strand registration prediction. Journal of Molecular Biology. 2005;354(4):979–993. doi: 10.1016/j.jmb.2005.09.094. [DOI] [PubMed] [Google Scholar]
- 20.Naveed H, Jackups R, Liang J. Predicting weakly stable regions, oligomerization state, and protein–protein interfaces in transmembrane domains of outer membrane proteins. Proceedings of the National Academy of Sciences. 2009;106(31):12735–12740. doi: 10.1073/pnas.0902169106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naveed H, Jimenez-Morales D, Tian J, Pasupuleti V, Kenney LJ, Liang J. Engineered oligomerization state of ompf protein through computational design decouples oligomer dissociation from unfolding. Journal of Molecular Biology. 2012;419(1–2):89–101. doi: 10.1016/j.jmb.2012.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geula S, Naveed H, Liang J, Shoshan-Barmatz V. Structure-based analysis of vdac1 protein: Defining oligomer contact sites. Journal of Biological Chemistry. 2012;287(3):2179–2190. doi: 10.1074/jbc.M111.268920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gessmann D, Mager F, Naveed H, Arnold T, Weirich S, Linke D, Liang J, Nussberger S. Improving the resistance of a eukaryotic β-barrel protein to thermal and chemical perturbations. Journal of Molecular Biology. 2011;413(1):150–161. doi: 10.1016/j.jmb.2011.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ∘24.Chen M, Khalid S, Sansom MSP, Bayley H. Outer membrane protein g: Engineering a quiet pore for biosensing. Proceedings of the National Academy of Sciences. 2008;105(17):6272–6277. doi: 10.1073/pnas.0711561105. The authors stabilize the OmpG pore by deleting a bulge and cysteine crosslinking a loop. The resultant pore demonstrates very little gating. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grosse W, Psakis G, Mertins B, Reiss P, Windisch D, Brademann F, Bürck J, Ulrich A, Koert U, Essen L-O. Structure-based engineering of a minimal porin reveals loop-independent channel closure. Biochemistry. 2014;53(29):4826–4838. doi: 10.1021/bi500660q. [DOI] [PubMed] [Google Scholar]
- 26.Mouritsen OG, Bloom M. Mattress model of lipid-protein interactions in membranes. Biophysical Journal. 1984;46(2):141–153. doi: 10.1016/S0006-3495(84)84007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramakrishnan M, Qu J, Pocanschi CL, Kleinschmidt JH, Marsh D. Orientation of β-barrel proteins OmpA and FhuA in lipid membranes. Chain length dependence from infrared dichroism. Biochemistry. 2005;44(9):3515–3523. doi: 10.1021/bi047603y. [DOI] [PubMed] [Google Scholar]
- 28.Muhammad N, Dworeck T, Fioroni M, Schwaneberg U. Engineering of the e. Coli outer membrane protein FhuA to overcome the hydrophobic mismatch in thick polymeric membranes. Journal of Nanobiotechnology. 2011;9(1):1–9. doi: 10.1186/1477-3155-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoddart D, Ayub M, Höfler L, Raychaudhuri P, Klingelhoefer JW, Maglia G, Heron A, Bayley H. Functional truncated membrane pores. Proceedings of the National Academy of Sciences. 2014;111(7):2425–2430. doi: 10.1073/pnas.1312976111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korkmaz F, van Pee K, Yildiz Ö. IR-spectroscopic characterization of an elongated OmpG mutant. Archives of Biochemistry and Biophysics. 2015;576:73–79. doi: 10.1016/j.abb.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 31.Krewinkel M, Dworeck T, Fioroni M. Engineering of an E. coli outer membrane protein FhuA with increased channel diameter. Journal of Nanobiotechnology. 2011;9(1):1–8. doi: 10.1186/1477-3155-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reina S, Magrì A, Lolicato M, Guarino F, Impellizzeri A, Maier E, Benz R, Ceccarelli M, De Pinto V, Messina A. Deletion of β-strands 9 and 10 converts VDAC1 voltage-dependence in an asymmetrical process. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2013;1827(6):793–805. doi: 10.1016/j.bbabio.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 33.Koebnik R, Braun V. Insertion derivatives containing segments of up to 16 amino acids identify surface- and periplasm-exposed regions of the FhuA outer membrane receptor of Escherichia coli k-12. Journal of Bacteriology. 1993;175(3):826–839. doi: 10.1128/jb.175.3.826-839.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Killmann H, Benz R, Braun V. Conversion of the fhua transport protein into a diffusion channel through the outer membrane of Escherichia coli. The EMBO Journal. 1993;12(8):3007–3016. doi: 10.1002/j.1460-2075.1993.tb05969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferguson AD, Hofmann E, Coulton JW, Diederichs K, Welte W. Siderophore-mediated iron transport: Crystal structure of FhuA with bound lipopolysaccharide. Science. 1998;282(5397):2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 36.Locher KP, Rees B, Koebnik R, Mitschler A, Moulinier L, Rosenbusch JP, Moras D. Transmembrane signaling across the ligand-gated FhuA receptor: Crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell. 1998;95(6):771–778. doi: 10.1016/s0092-8674(00)81700-6. [DOI] [PubMed] [Google Scholar]
- 37.Braun M, Killmann H, Maier E, Benz R, Braun V. Diffusion through channel derivatives of the Escherichia coli FhuA transport protein. Eur J Biochem. 2002;269 doi: 10.1046/j.1432-1033.2002.03195.x. [DOI] [PubMed] [Google Scholar]
- 38.Mohammad MM, Howard KR, Movileanu L. Redesign of a plugged β-barrel membrane protein. Journal of Biological Chemistry. 2011;286(10):8000–8013. doi: 10.1074/jbc.M110.197723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolfe AJ, Mohammad MM, Thakur AK, Movileanu L. Global redesign of a native β-barrel scaffold. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2016;1858(1):19–29. doi: 10.1016/j.bbamem.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mapingire OS, Henderson NS, Duret G, Thanassi DG, Delcour AH. Modulating effects of the plug, helix, and n- and c-terminal domains on channel properties of the PapC usher. Journal of Biological Chemistry. 2009;284(52):36324–36333. doi: 10.1074/jbc.M109.055798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jung Y, Cheley S, Braha O, Bayley H. The internal cavity of the staphylococcal α-hemolysin pore accommodates ~175 exogenous amino acid residues. Biochemistry. 2005;44(25):8919–8929. doi: 10.1021/bi0473713. [DOI] [PubMed] [Google Scholar]
- 42.Urry DW, Long MM, Sugano H. Cyclic analog of elastin polyhexapeptide exhibits an inverse temperature transition leading to crystallization. Journal of Biological Chemistry. 1978;253(18):6301–6302. [PubMed] [Google Scholar]
- 43.Jung Y, Bayley H, Movileanu L. Temperature-responsive protein pores. Journal of the American Chemical Society. 2006;128(47):15332–15340. doi: 10.1021/ja065827t. [DOI] [PubMed] [Google Scholar]
- 44.Saint N, Lou K-L, Widmer C, Luckey M, Schirmer T, Rosenbusch JP. Structural and functional characterization of OmpF porin mutants selected for larger pore size. Journal of Biological Chemistry. 1996;271(34):20676–20680. [PubMed] [Google Scholar]
- 45.Phale PS, Philippsen A, Widmer C, Phale VP, Rosenbusch JP, Schirmer T. Role of charged residues at the OmpF porin channel constriction probed by mutagenesis and simulation. Biochemistry. 2001;40(21):6319–6325. doi: 10.1021/bi010046k. [DOI] [PubMed] [Google Scholar]
- 46.Miedema H, Meter-Arkema A, Wierenga J, Tang J, Eisenberg B, Nonner W, Hektor H, Gillespie D, Meijberg W. Permeation properties of an engineered bacterial OmpF porin containing the EEEE-locus of Ca2+ channels. Biophysical Journal. 2004;87(5):3137–3147. doi: 10.1529/biophysj.104.041384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saxena K, Drosou V, Maier E, Benz R, Ludwig B. Ion selectivity reversal and induction of voltage-gating by site-directed mutations in the Paracoccus denitrificans porin. Biochemistry. 1999;38(7):2206–2212. doi: 10.1021/bi982296f. [DOI] [PubMed] [Google Scholar]
- ∘∘48.Braha O, Walker B, Cheley S, Kasianowicz JJ, Song L, Gouaux JE, Bayley H. Designed protein pores as components for biosensors. Chemistry & Biology. 1997;4(7):497–505. doi: 10.1016/s1074-5521(97)90321-5. The authors engineer a divalent metal binding site into α-hemolysin using four histidines on a single beta hairpin. They combine the mutated monomer with six wild type α-hemolysins in order to create the successful metal binding heptamer. This is the first metal binding site engineered into an outer membrane protein. [DOI] [PubMed] [Google Scholar]
- 49.Ye Y, Lee H-W, Yang W, Shealy SJ, Wilkins AL, Liu Z-r, Torshin I, Harrison R, Wohlhueter R, Yang JJ. Metal binding affinity and structural properties of an isolated EF-loop in a scaffold protein. Protein Engineering. 2001;14(12):1001–1013. doi: 10.1093/protein/14.12.1001. [DOI] [PubMed] [Google Scholar]
- 50.Johansson MU, Alioth S, Hu K, Walser R, Koebnik R, Pervushin K. A minimal transmembrane β-barrel platform protein studied by nuclear magnetic resonance. Biochemistry. 2007;46(5):1128–1140. doi: 10.1021/bi061265e. [DOI] [PubMed] [Google Scholar]
- 51.Varadarajan N, Gam J, Olsen MJ, Georgiou G, Iverson BL. Engineering of protease variants exhibiting high catalytic activity and exquisite substrate selectivity. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(19):6855–6860. doi: 10.1073/pnas.0500063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ∘∘52.Varadarajan N, Rodriguez S, Hwang B-Y, Georgiou G, Iverson BL. Highly active and selective endopeptidases with programmed substrate specificities. Nat Chem Biol. 2008;4(5):290–294. doi: 10.1038/nchembio.80. I verson and colleagues invented a screen for endopeptidase OmpT utilizing a ligand that contains a positive charged fluorophore linked by a peptide to a quencher. If OmpT cleaves the peptide, the fluorophore sticks to the negatively charged surface of E. coli and the cell can be isolated through FACS. In this paper they changed the peptide and heavily mutated the binding site to screen for OmpT variants that would cleave the peptides used. This paper details five endopeptidases which they created with cleavage specificity for an impressive variety of peptides. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Varadarajan N, Georgiou G, Iverson BL. An engineered protease that cleaves specifically after sulfated tyrosine. Angewandte Chemie. 2008;120(41):7979–7981. doi: 10.1002/anie.200800736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varadarajan N, Pogson M, Georgiou G, Iverson BL. Proteases that can distinguish among different post-translational forms of tyrosine engineered using multicolor flow cytometry. Journal of the American Chemical Society. 2009;131(50):18186–18190. doi: 10.1021/ja907803k. [DOI] [PubMed] [Google Scholar]
- ∘∘55.Koebnik R. In vivo membrane assembly of split variants of the E. coli outer membrane protein ompa. EMBO J. 1996;15(14):3529–3537. Here, Koebnik creates the first novel topology for an outer membrane protein. By splitting OmpA’s eight strands into two separate four-stranded constructs, he finds that these constructs bind together to make an eight stranded protein that is heat modifiable and that confers phage sensitivity phenotype. Tantalizingly, he also finds that a six-stranded construct co-expressed with a four-stranded construct functions moderately well. [PMC free article] [PubMed] [Google Scholar]
- 56.Krauson AJ, He J, Wimley AW, Hoffmann AR, Wimley WC. Synthetic molecular evolution of pore–forming peptides by iterative combinatorial library screening. ACS chemical biology. 2013;8(4):823–831. doi: 10.1021/cb300598k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Slusky JSG, Dunbrack RL. Charge asymmetry in the proteins of the outer membrane. Bioinformatics. 2013;29(17):2122–2128. doi: 10.1093/bioinformatics/btt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ∘58.Alford RF, Koehler Leman J, Weitzner BD, Duran AM, Tilley DC, Elazar A, Gray JJ. An integrated framework advancing membrane protein modeling and design. PLoS Comput Biol. 2015;11(9):e1004398. doi: 10.1371/journal.pcbi.1004398. This paper describes a membrane focused update of the widely successful protein design software ROSETTA. The update allows for the storing of membrane specific information such as membrane potentials, and preferred movements in the membrane. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ∘59.Lin M, Gessmann D, Naveed H, Liang J. Outer membrane protein folding and topology from a computational transfer free energy scale. Journal of the American Chemical Society. 2016;138(8):2592–2601. doi: 10.1021/jacs.5b10307. This paper describes an update to the membrane potential, TmSIP. This computational potential now includes intrastrand side-chain interactions and an asymmetric membrane. It can be used to calculate ΔΔG values for mutations in OmpLA and can predict the membrane insertion direction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Packer MS, Liu DR. Methods for the directed evolution of proteins. Nat Rev Genet. 2015;16(7):379–394. doi: 10.1038/nrg3927. [DOI] [PubMed] [Google Scholar]
- 61.Scott DJ, Plückthun A. Direct molecular evolution of detergent-stable g protein-coupled receptors using polymer encapsulated cells. Journal of Molecular Biology. 2013;425(3):662–677. doi: 10.1016/j.jmb.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 62.McIsaac RS, Engqvist MKM, Wannier T, Rosenthal AZ, Herwig L, Flytzanis NC, Imasheva ES, Lanyi JK, Balashov SP, Gradinaru V, Arnold FH. Directed evolution of a far-red fluorescent rhodopsin. Proceedings of the National Academy of Sciences. 2014;111(36):13034–13039. doi: 10.1073/pnas.1413987111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yildiz Ö, Vinothkumar KR, Goswami P, Kühlbrandt W. Structure of the monomeric outer-membrane porin OmpG in the open and closed conformation. The EMBO Journal. 2006;25(15):3702–3713. doi: 10.1038/sj.emboj.7601237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cowan SW, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit RA, Jansonius JN, Rosenbusch JP. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358(6389):727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 65.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science. 1996;274(5294):1859–1865. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 66.Cierpicki T, Liang B, Tamm LK, Bushweller JH. Increasing the accuracy of solution NMR structures of membrane proteins by application of residual dipolar couplings. High-resolution structure of outer membrane protein A. Journal of the American Chemical Society. 2006;128(21):6947–6951. doi: 10.1021/ja0608343. [DOI] [PMC free article] [PubMed] [Google Scholar]