Abstract
Isoprenoid biosynthesis is an important area for anti-infective drug development and one target is IspH, (E)-1-hydroxy-2-methyl-but-2-enyl 4-diphosphate (HMBPP) reductase, which forms isopentenyl diphosphate and dimethylallyl diphosphate from HMBPP in a 2H+/2e− reduction. IspH contains a 4Fe-4S cluster and here, we first investigated how small molecules can bind to the cluster using HYSCORE and NRVS spectroscopies. The results of these as well as other structural and spectroscopic investigations led to the conclusion that in most cases, ligands bind to IspH 4Fe-4S clusters via η1 coordination, forming tetrahedral geometries at the unique 4th Fe, ligand side-chains preventing further ligand (e.g. H2O, O2) binding. Based on these ideas, we sought using in silico methods to find drug-like inhibitors that might occupy the HMBPP substrate binding pocket and bind to Fe, leading to the discovery of a barbituric acid analog having a Ki ~ 500 nM against Pseudomonas aeruginosa IspH.
Keywords: Isoprenoid, IspH, NRVS, HYSCORE, in silico
Introduction
The enzymes IspG ((E)-1-hydroxy-2-methyl-but-2-enyl 4-diphosphate synthase, also known as GcpE) and IspH ((E)-1-hydroxy-2-methyl-but-2-enyl 4-diphosphate reductase, also known as LytB) are the last two enzymes of the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway of isoprenoid biosynthesis in many bacteria, as well as in some protozoa, and in plants[1]. They are both 4Fe-4S cluster-containing proteins that are involved in 2H+/2e reductions: IspG converts 2-C-methyl-D-erythritol-2,4-cyclo-diphosphate (MEcPP, 1, Scheme 1) to (E)-1-hydroxy-2-methyl-but-2-enyl 4-diphosphate (HMBPP; 2), and IspH converts 2 to dimethylallyl diphosphate (3) and isopentenyl diphosphate (4), the building blocks of isoprenoid biosynthesis. Both enzymes are not produced by humans (who use the mevalonate pathway for isoprenoid biosynthesis), but are essential in plants and in many bacteria and protozoa, so IspG and IspH are of interest as potential herbicide or drug targets. In earlier work[2] we reported the first structure (PDB ID 3DNF) of an IspH, from Aquifex aeolicus, finding that one Fe atom in the cluster was lost during crystallization, and similar results (PDB ID 3F7T) were reported for IspH from E. coli[3]. However, using computational modeling to add back the unique 4th Fe, together with computational docking of the HMBPP substrate 1, we were able to produce[1–2] ligand-bound structures that were very similar to later 4Fe-4S IspH/1 X-ray structures[4–5], and these structures have led to detailed mechanism of action models[6–8] for IspH catalysis. In other early work we discovered, based on previous reports that alkynes could bind to and be reduced by 4Fe-4S clusters[9–10], that alkyne diphosphates such as 5 (propargyl diphosphate, PPP) were low μM inhibitors of IspH (as well as IspG, which also contains a 4Fe-4S cubane-like structure)[1]. Plus, the 1-amino (6) and 1-thio (7) analogs of HMBPP have been found to inhibit IspH with Ki ~20–50 nM[11–12], and we[13] and others (PDB ID codes: 3ZGL, 3ZGN) have reported their X-ray structures which are basically the same as found with the HMBPP substrate, with N,S binding to the unique, 4th Fe. We also reported[14] several other IspH-ligand complex structures with, in each case, a single O-containing ligand (alcoholate or enolate) bound to the unique, 4th Fe atom with Fe-O bond lengths of ~2 Å. What has been (and still is) missing is the X-ray structure of a “ligand free” IspH-either reduced ([Fe4S4]+) or oxidized ([Fe4S4]2+), the problem being that it has not been possible to crystallize the 4-Fe containing protein. However, the results of 57Fe Mössbauer spectroscopy[15] indicated the presence of a 5 or 6-coordinate 4th iron in “ligand-free” oxidized IspH, with three cluster S and most likely three additional N/O ligands bound to the 4th Fe. More recently, Faus et al.[16] reported a NRVS (nuclear resonant vibrational spectroscopy) investigation of oxidized IspH and suggested that the three non-cluster ligands were H2O molecules (or presumably OH−, or a mixture of both). This structure is surprisingly labile, leading under crystallization conditions to loss of the 4th Fe. “Ligand-free” IspH is also very sensitive to O2 while HMBPP, PPP as well as an enolate-liganded species are much less sensitive to cluster degradation by O2[17], suggesting that the presence of relatively bulky ligand side-chains might block the H2O/O2 binding that leads to lability. For example, in the PPP (5) structure (crystallized from oxidized IspH), there is a single H2O (or OH−) bound to the 4th Fe[14] while the acetylene group is ~3.6 Å from the Fe, acting perhaps as a barrier to water and oxygen ligands.
Scheme 1.
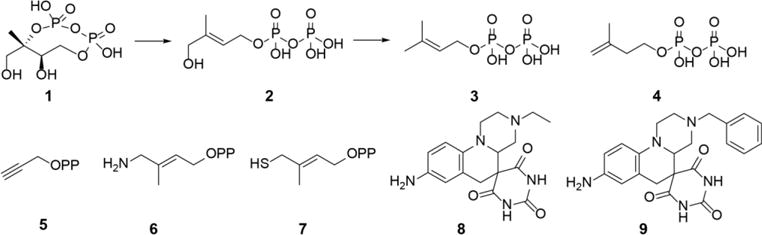
Structures of IspG (GcpE) and IspH (LytB) substrates and reaction products, and some ligands/inhibitors of interest. OPP in 5–7 is diphosphate.
In our EPR work on reduced IspH-5, we concluded (based in part on early observations of Fe4S4–acetylene interactions) that there could be π-bonding with the 4th Fe in the reduced cluster, but with oxidized IspH, S = 0, so the system is not accessible via EPR and it is not clear if there is any alkyne interaction with the cluster. We thus used NRVS to investigate the IspH-5 system. In addition, we used HYSCORE (hyperfine sublevel correlation spectroscopy) to investigate how another small ligand, CN−, might bind to reduced IspH, of interest since CN− can be involved in π-back-bonding with d-orbitals. The results of these experiments together with an examination of numerous IspH and IspG structures suggested the importance of η1 σ-bonding of ligands to the 4th Fe, together with the presence of a bulky side-chain, for IspH inhibition, so we then used in silico screening of possible inhibitors, finding interesting new drug-like leads that, we propose, bind in this manner.
Results and Discussion
In the following we first investigated the interactions of IspH with the alkyne diphosphate 5, as well as with a small molecule ligand, CN−, to see if there were any interactions of the alkyne with the cluster, and whether the anionic species CN− bound since in principle, both might be involved in metal-ligand π-bonding/back-bonding. Then, using information from these and other related studies, we used in silico screening to try and find new, drug-like inhibitor leads.
NRVS spectroscopy of the IspH-5 complex
NRVS provides 57Fe-specific information on the partial vibrational density of states (PVDOS) for Fe-X vibrational modes in a molecule[18]. For systems containing 4Fe-4S cubane-like clusters, such as IspH, there are typically four main NRVS features, as shown schematically in Figure 1A[18–21]: cluster torsional modes occur at <100 cm−1; δ (S-Fe-X) bending modes at ~150 cm−1; ν (Fe-S) (cluster) stretching modes at ~280 cm−1 and ν (Fe-Ster) (terminal) stretching modes in the ~320–370 cm−1 range. Although there are of course additional mixed modes, these NRVS spectral features are present in most if not all species containing 4Fe-4S as well as 4Fe-3S clusters.[18–21] For example, the NRVS spectrum of oxidized Pyrococcus furiosus ferredoxin containing a D14C mutation (which therefore has 4 Cys ligands; Figure 1B, black line)[20], is very similar to that seen with the model compound [Fe4S4Cl4](Ph4P)2, which contains a [Fe4S4Cl4]2− cluster in which the four terminal ligands are Cl (Figure 1B, grey line)[20]. Basically the same features are also seen in oxidized IspH with HMBPP (2), the amino-analog (6) or the thiol analog (7) as ligands [16], in which O, N or S are directly bonded to the 4th Fe. In sharp contrast, these features are all less obvious (or absent) in the NRVS spectrum of IspH in the absence of 2, 6 or 7[16]. In the presence of NO, the 4Fe-4S cluster in P. furiosus D14C ferredoxin[22] is converted to Roussin’s black salt, [Fe4S3(NO)7] − and the same set of peaks (at ~150, 280 and 370 cm−1) as seen in the 4-Cys liganded protein are present. However, there are in addition strong peaks at ~540, 610 cm−1, Figure 1B (red line), suggesting a contribution from Fe-N-O and/or N-Fe-N vibrational modes, due in part to metal-ligand pi-bonding. We thus investigated the NRVS spectra of the IspH-5 complex to see whether there might be any evidence for interaction between the alkyne group and the oxidized 4Fe-4S cluster (Fe-C bonding) that we previously proposed to be important in the reduced protein.
Figure 1.
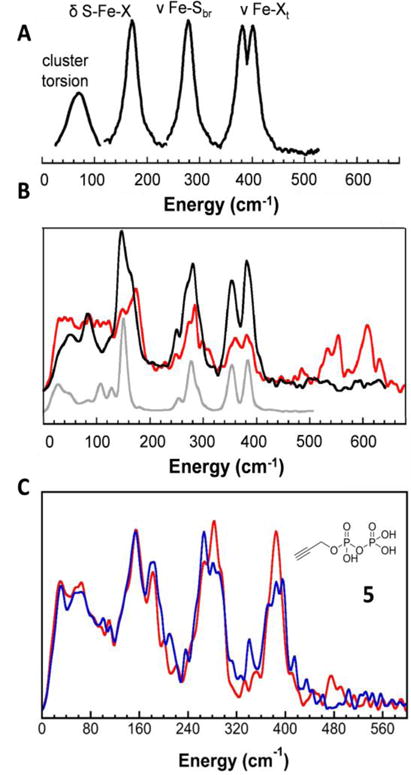
NRVS spectra. A, Cartoon representation of a NRVS spectrum of a 4Fe-4S cluster-containing σ-bonding ligands (e.g. OR, NHR, SR, Cl) at a 4th, unique Fe-site. B, Experimental spectra of oxidized P. furiosus D14C ferredoxin (PfFd; black), PfFd+NO (red) and [Fe4S4Cl4](Ph4P)2 (grey). C, EcIspH + 5 (red) and [13C]-5 (black).
We show in Figure 1C the NRVS spectrum of EcIspH in the oxidized state ([Fe4S4]2+) bound to 5 (red line). The spectrum is very similar to that seen with HMBPP (2) as well as the amino 6 and thiol 7 analogs of HMBPP, bound to IspH,[16] and the [Fe4S4Cl4]2− model compound[16], consistent with each Fe being bound to 3 cluster sulfurs and a single 4th atom. We also found no spectral shifts when a uniformly 13C-labeled analog of PPP (5) was bound to the protein (blue line, Figure 1C). These results are consistent with the presence of a single water molecule [14] binding to the 4th Fe—the tetrahedral geometry seen in the other IspH structures with 2, 6 and 7 [16]—with no significant bonding between the alkyne group and the cluster. A compilation of the NRVS spectra of all of the variously ligated protein and model compound 4Fe-4S cluster-containing systems discussed above, highlighting their similarities, is shown in Supporting Information Figure S1. The spectra with 5 are also dissimilar to that observed with IspH containing 3 H2O molecules bound to the 4th Fe, again consistent with the lack of multiple (alkyne, water) interactions with the 4th Fe.
We next sought to see how other small molecules/ions might bind to the 4Fe-4S cluster. Attempts to bind CO were unsuccessful, as determined by UV-VIS and EPR spectroscopy. However, in previous work[23] we showed that CN− bound to reduced IspH, yielding an EPR spectrum characterized by g-values of 2.08, 1.94 and 1.93 (with a small shoulder at g = 2.05[23]), but the number of ligands as well as the binding mode were unknown. We thus next collected HYSCORE spectra using [13C15N]− bound to E. coli IspH. As can be seen in Figure 2A, there are clearly HYSCORE features that are consistent with binding of a single CN− to the unique, 4th Fe in the cluster. Using the EasySpin program[24] we simulated hyperfine coupling tensors of A(13C) = [−3.9, −3.8, 0.1] MHz, Figure 2B, and A(15N) = [1.1, 1.1, 2.3] MHz, Figure 2C. Interestingly, the g-values observed[23] in the EPR spectrum of IspH•CN (g=2.08, 1.94, 1.93) are virtually identical to those found (g = 2.09, 1.94, 1.93) for CN- bound to Shewanella oneidensis HydG (minus the “dangler Fe”)[25] involved in formation of the [Fe(CO)2CN] synthon in hydrogenase function. Moreover, the HYSCORE spectrum of [13CN]-SoHydG has a 13C isotropic hyperfine coupling Aiso = −2.7 MHz, similar to the Aiso = −2.9 MHz in PfFd with bound 13CN[26] and the Aiso ~ 2.5 MHz we find here. Plus, the 15N HYSCORE result for [13C15N] − bound to SoHydG [25] is extremely similar to that we observe with IspH. So, IspH, SoHydG (the 4Fe-4S cluster) as well as the (wild type) ferredoxin all appear to bind CN− to the 4th Fe, forming a tetrahedral species. It should be noted, however, that CN− is actually a very poor IspH inhibitor (IC50>1 mM) and, as noted by Suess et al.[25], CN− binds only weakly to other biological[26] as well as synthetic[27] 4Fe-4S clusters, and cysteine displaces CN− from HydG[25].
Figure 2.
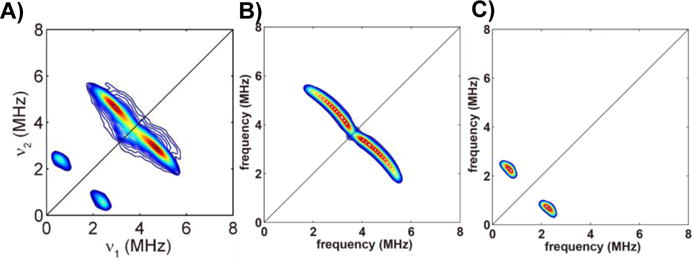
HYSCORE spectra of IspH•CN. A, Experimental HYSCORE spectrum of EcIspH•13C15N. Sample is reduced ([Fe4S4]+) with dithionite. B, Simulation of 13C hyperfine coupling with A = [−3.9, −3.8, 0.1] MHz, Euler angle = [0, 40±5, 0]°. C, Simulation of 15N hyperfine coupling with A = [1.1, 1.1, 2.3] MHz, Euler angle = [0, 30±10, 0]°. Euler angle follows zyz convention.
Ligand binding to IspH and IspG: Clues for inhibitor discovery?
When taken together, the results shown above together with other reported work[1], show that there are several ways that ligands can bind to IspH, and suggest a potential route to finding new inhibitors. First, most ligands bind with η1 coordination with O, N and S binding via σ-interactions with the 4th Fe. Second, potential π-bonding species (CN−, CO, propargyl alcohol) do not bind strongly. Third, while η2 as well as η3 coordination is possible[1], these cases are rare. Fourth, when alkyne diphosphates bind to oxidized IspH, the main cluster interaction is with a 4th H2O (or OH−), the alkyne fragment blocking addition of further H2O molecules. Fifth, the diphosphate moiety must contribute in a major way to IspH inhibition, because propargyl alcohol itself is a very weak inhibitor (Ki>10 mM). Sixth, these general patterns of ligand binding are very similar in IspH and in IspG. This is shown in Figure 3A in which we compare PPP (5) binding to IspH and IspG. In both cases, there is a 4th H2O bound to the unique 4th Fe (dFe-O = 1.9 Å), while the alkyne is more distant (d ~3.6 Å). The IspH•5 structure is very similar to the structures found with the amino (6) and thiol (7) HMBPP analogs, the IspH•5 water being in the same position as the 6, 7 NH2 and SH groups, and the diphosphate groups overlap, Figure 3B. The IspH•5 water also co-locates with the ligand-bound oxygen in the IspG-MEcPP (1) 1st reaction intermediate[28], Figure 3C, and the IspG•5 bound water is in the same position as a carboxylate oxygen in E307, in IspG (Figure 3D). So, in the vast majority of cases the 4th Fe has a tetrahedral coordination geometry with O, N or S binding to Fe. Unfortunately, these potent diphosphate-containing inhibitors are not active in cells, presumably because the diphosphate groups are very highly charged, reducing cell penetration. These observations led us to try and find more lipophilic, drug-like species that might bind to Fe, while also occupying the relatively large substrate-binding pocket.
Figure 3.
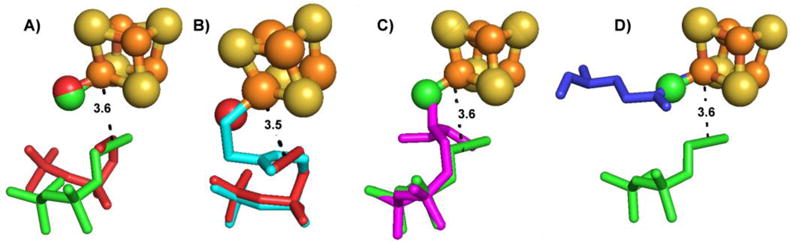
Comparisons between IspH/IspG structures with various ligands binding to the unique, 4th Fe site. A, PPP (5) bound to EcIspH (red) and EcIspG (green) (PDB ID codes 3URK, 4S3E). B, Superposition of EcIspH PPP (5)/H2O (red) and thiolate (cyan, 7) X-ray structues (PDB ID codes 3URK, 4H4E). C, Superposition of PPP (5)/H2O IspG (green) structures with PPP (5)/H2O-GcPE reaction intermediate structures (magenta) (PDB ID codes 4S3B, 4S3E). All structures contain a σ-bonding ligand (H2O, RNH2, RSH, enolate) at the unique, 4th Fe structure. Three water molecules binding to 4th Fe lead to low stability; a H2O and a “bulky” ligand (that prevents binding of additional water molecules) leads to more stable species. D, Superposition of E307 from AaIspG (blue) and PPP (5)/H2O-GcPE structures (green) (PDB ID codes 3NOY, 4S3E).
We thus next used an in silico approach to screen a library of drug-like compounds from the ZINC and NCI libraries against both Aquifex aeolicus IspH (PDB ID 3DNF) and E. coli IspH (PDB ID 3F7T). In both cases, the 4th Fe was reconstituted computationally as described previously[2], and we used Glide docking[29], again as described previously[30]. Using this approach we obtained and tested 15 potential hits (Figure S2) that were commercially available, for IspH inhibition. 14 out of the 15 compounds were inactive (IC50 > 1 mM), but 8 was active. We then obtained and tested 11 commercially available analogs of 8 (Figure S3) and tested them against E. coli IspH and Pseudomonas aeruginosa IspH, both organisms (unlike A. aeolicus) being important pathogens. The most active compounds were 8 and 9 with 8 having a Ki = 500 nM against P. aeruginosa IspH, Figure 4A, and the benzyl analog 9 having a Ki = 3 μM, against E. coli IspH, Figure 4B. AaIspH was also inhibited by 9 with a Ki=700 nM. These compounds are barbituric acid analogs that have some structural similarity to anti-infectives developed by Pharmacia[31], and are predicted to bind to the 4th Fe of the 4Fe-4S cluster. A view of the AaIspH•9 complex (obtained by docking) from the electron-transfer side of the protein is shown in Figure 4C; a bottom-view (substrate side) in Figure 4D, and a side-view in Figure 4E. A close up view of the ligand interacting with the 4th Fe in the cluster (dFe-N = 2.6 Å; dFe-O = 2.7 Å) is shown in Figure 4F. The ligand can clearly occupy the large substrate-binding site seen in the early X-ray structures, with the enolate form of the barbiturate reacting with the Fe, similar to the binding of related barbiturate enolates to Zn2+ in other metalloproteins [32–33].
Figure 4.
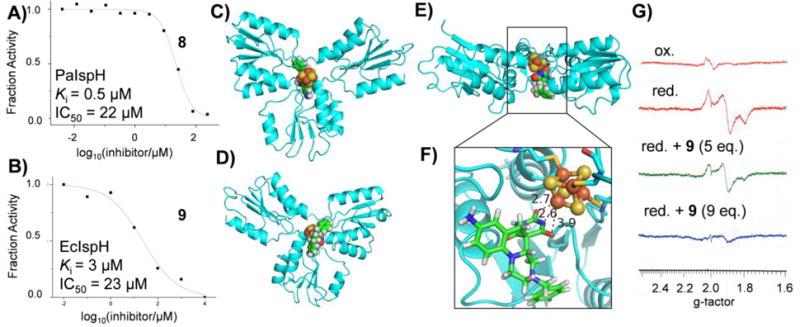
IspH inhibition by barbiturate analogs. A,B. Dose-response curves for EcIspH inhibition. C, Computational docking structure of 9 binding to oxidized AaIspH. View from electron transfer side. D, View from substrate binding side. E, Side view. F, Proposed binding of the barbiturate enolate group in 9 to the unique, 4th Fe in the 4Fe-4S cluster in oxidized AaIspH. G, 9 GHz EPR spectra of PaIspH. Oxidized protein (top, orange); dithionite reduce (red); dithionite reduced plus 5 eq. 9 (green); dithionote reduced plus 9 eq. 9 (blue).
Interestingly, we were unable to obtain EPR spectra of 9 bound to IspH (from the pathogen Pseudomonas aeruginosa), ligand addition resulting in loss of signal intensity, Figure 4G, due perhaps to a shift in redox potential on ligand binding, or a change in relaxation behavior but in either case, 8, 9 appear to represent interesting new IspH inhibitor leads for further development since they are far more lipophilic than the diphosphates, they do not violate Lipinski’s rules[34] and they are not PAINS compounds[35].
Conclusions
The results we have shown above are of interest for a number of reasons. First, we find that NRVS spectra of the alkyne diphosphate inhibitor 5 bound to oxidized IspH are very similar to those found for binding of HMBPP 2, as well as the amino and thiol analogs of HMBPP, 6 and 7. There is no evidence for any alkyne-cluster interaction. Second, we show (using HYSCORE) that CN− binds to IspH (but is a very weak inhibitor), and that the EPR/HYSCORE spectra of the CN− bound protein are very similar to those found with CN− binding to SoHydG and P. furiosus ferredoxin, consistent with binding of a single cyanide in all three cases. Third, when compared with all known IspH and IspG structures, it is clear that in most cases, O, N and S-bonding ligands bind to the unique 4th Fe in the cluster forming tetrahedral geometries. Fourth, based on the results noted above, we sought to find novel IspH inhibitors that might bind to Fe in the active site and be more drug-like than the diphosphate inhibitors. Using in silico screening we found that the barbiturate analogs 8, 9 had Ki ~ 0.5–3 μM, binding we propose, via the barbiturate enolate moiety to the 4th Fe, the hydrophobic domains occupying the substrate-binding site.
Experimental Section
Chemical Synthesis: General Methods
8 and its analogs were purchased from Vitas-M Laboratory (Hong Kong) and used without further purification. Compounds were analyzed by LC/MS and were >97% pure. Other compounds were from Aldrich, Asinex, Enamine, NCI/Developmental Therapeutics Program Open Chemical Repository (dtp.cancer.gov/), or TimTec. All chemicals for the re-synthesis of 9 were purchased from Sigma-Aldrich. 1H NMR spectra were obtained on Varian Unity spectrometers at 400 and 500 MHz. High resolution MS and elemental analyses were carried out in the University of Illinois Mass Spectrometry and Microanalytical Laboratories.
8-amino-3-benzyl-2,3,4,4a-tetrahydro-1H,2′H,6H-spiro[pyrazino[1,2-a]quinoline-5,5′-pyrimidine]-2′,4′,6′ (1′H,3′H)-trione (9)
The synthesis of 9 was based on the synthesis of a morpholine analog[36], and is illustrated in Scheme 1. 1-benzylpiperazine (9a). Piperazine (59 mmol, 5.09 g) was dissolved in 26 mL of THF by heating. Benzyl bromide (8.4 mmol, 1mL) was added dropwise to a refluxing solution of piperazine in THF. After stirring overnight at reflux, the reaction was cooled, and THF removed by evaporation. The resulting residue was washed with aq. K2CO3 (20 mL), extracted with EtOAc (10 mL × 3), washed with satd. NaCl (10 mL × 1), dried with Na2SO4, then evaporated to dryness under vacuum (9a, 1.29 g, 87%). 9a (207 mg, 1.174 mmol) was dissolved in 3 mL MeCN and 1 mL Et3N. Then, 2-fluoro-5-nitrobenzaldehyde (200 mg, 1.18 mmol) was added and the solution stirred at reflux overnight. The reaction mixture was then diluted with EtOAc (10 mL) and washed with water (10 mL). The aqueous layer was extracted with EtOAc (10 mL × 2), dried with Na2SO4, then solvent removed under vacuum. The crude mixture was purified by column chromatography (2:1 Hex:EtOAc, 9b, 272 mg, 71% yield). 9b (192.77 mg, 0.592 mmol) was dissolved in 10 mL MeOH. To this solution, barbituric acid (79.76 mg, 0.623 mmol) was added and the mixture heated to reflux and stirred overnight. The crude reaction mixture was loaded onto a column and purified with 2:3 EtOAc:Tol (9c, 77.33 mg, 30%). 9c (30 mg, 0.069) was dissolved in 8 mL AcOH and Zn dust (52 mg, 0.795 mmol) added. The reaction mixture was stirred at room temperature for 2 hours and then quenched with K2CO3 (20 mL). Next, the mixture was diluted with EtOAc (20 mL) and washed with aq. K2CO3 (20 mL), satd. NaCl, dried with Na2SO4, then solvent was removed under vacuum. The crude mixture was purified by preparative TLC (100% EtOAc) to yield an orange powder 9 (3.92 mg, 9%). ESI-HRMS: Calc: 406.1879, found: 406.1868 C22H24N5O3. Compound purity determined by HPLC (Phenomenex C6-Phenyl 110A, 100×2 mm, 3μM, 250 nm, retention time = 1.5 min): 99.7%.
Scheme 1.
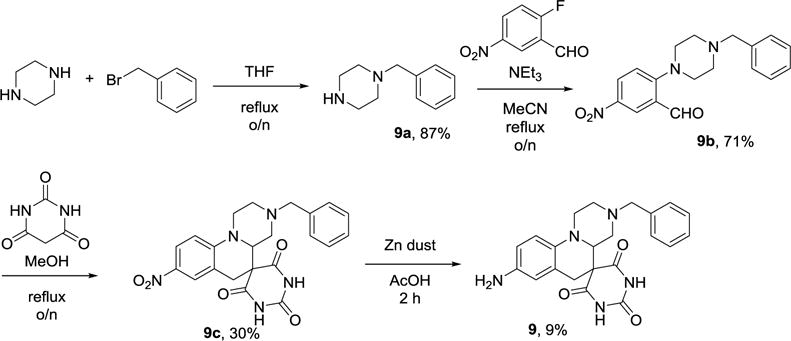
Synthesis of 9.
Sample preparation
57Fe IspHs were prepared as described elsewhere.[5]. Compound 5 was described previously [23]. For EPR spectroscopy, EcIspH and PaIspH in the oxidized state ([Fe4S4]2+) and in the presence of a 20-fold excess of 5 were concentrated to ~0.3 mM by using an Amicon Ultra centrifugal device (EMD Millipore Corporation, Billerica, MA, USA), then glycerol was added to 20% (v/v) as a glassing agent. NRVS EcIspH samples were loaded into a Lucite cuvette (internal dimensions = 10 × 2.5 × 1 mm), then frozen in liquid nitrogen.
NRVS Measurements
NRVS samples (6 mM EcIspH) were attached to a cryogenic sample base connected to a liquid helium (LHe) cryostat maintained at 10K. Spectra were recorded using published procedures at 03-ID at the Advanced Photon Source (APS) [37]. Photon flux was ~2.5 × 109 photons/s in a 1.1 eV energy resolution. Delayed nuclear fluorescence Fe K fluorescence were recorded with a single 1 cm2 square avalanche photodiode. Total data aquisition time was 19 hours. Data reduction was performed by using the PHOENIX software package[38] where the observed raw NRVS spectra were calibrated (aligned) to the nuclear resonant peak, normalized to the I0, then summed and converted to the 57Fe partial vibrational density of states (PVDOS). The spectral conversion was optimized when the observed Stokes/anti-Stokes imbalance matched the imbalance calculated using the entered temperature as a variable. The real sample temperature obtained by using this procedure was ~60 K.
CW-EPR/ENDOR/HYSCORE spectroscopy
All CW (continuous wave)-EPR experiments were performed on a Varian E-line 122 X-band spectrometer with an Air Products helium cryostat. Typical data acquisition parameters were: microwave frequency = 9.05 GHz; field center = 3250 G; field sweep = 1000 G; modulation frequency = 100 kHz; modulation amplitude = 5 Gauss; time constant = 32 ms; temperature = 8 – 20 K. HYSCORE spectra were obtained on a Bruker ElexSys E-580–10 FT-EPR EPR spectrometer equipped with an Oxford Instruments CF935 cryostat. HYSCORE used a four-pulse sequence π/2mw − τ − π/2mw − t1 − πmw − t2 − π/2mw − echo; π/2mw = 16 ns and πmw = 32 ns, 128 points for both t1 and t2, each using 24 ns steps. Time-domain data were baseline corrected using a 3rd order polynomial, then Hamming windowed, followed by zero-filling, 2D-Fourier transformation, and symmetrization. Parameters were typically: microwave frequency = 9.65 – 9.72 GHz, temperature = 8 – 15 K, microwave power attenuation = 6.5 – 9 dB.
Spectral simulations
HYSCORE spectra were simulated by using the EasySpin program[24].
In silico screening
In order to find new inhibitors, we carried out in silico screens of AaIspH and EcIspH using ZINC and NCI libraries and Glide docking, basically as described previously[30].
Supplementary Material
Acknowledgments
This work was supported by the United States Public Health Service (NIH grants GM065307 to EO and GM-65440 to SPC) and by the Department of Energy Office of Biological and Environmental Research (SPC). The NRVS measurements were performed at APS (proposals 39192/ 43032) and at SPring-8 (2015B1134). The APS is supported by the DOE Office of Basic Energy Sciences. The EPR instrumentation was supported by NIH Grants S10-RR15878 and S10-RR025438. Work at UCSD was supported by NIH, NSF, HHMI, NBCR, and SDSC. W.W. was supported by a Predoctoral Fellowship from the American Heart Association, Midwest Affiliate (award 10PRE4430022). We would like to thank Drs. J. Zhao, M. Hu and E. E. Alp at APS for assistance with the NRVS measurements, and Mark J. Nilges for assistance with the EPR measurements.
References
- 1.Wang W, Oldfield E. Angew Chem Int Ed. 2014;53:4294–4310. doi: 10.1002/anie.201306712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rekittke I, Wiesner J, Rohrich R, Demmer U, Warkentin E, Xu W, Troschke K, Hintz M, No JH, Duin EC, Oldfield E, Jomaa H, Ermler U. J Amer Chem Soc. 2008;130:17206–17207. doi: 10.1021/ja806668q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gräwert T, Rohdich F, Span I, Bacher A, Eisenreich W, Eppinger J, Groll M. Angew Chem Int Ed. 2009;48:5756–5759. doi: 10.1002/anie.200900548. [DOI] [PubMed] [Google Scholar]
- 4.Grawert T, Span I, Eisenreich W, Rohdich F, Eppinger J, Bacher A, Groll M. Proc Nat Acad Sci USA. 2010;107:1077–1081. doi: 10.1073/pnas.0913045107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Span I, Grawert T, Bacher A, Eisenreich W, Groll M. J Mol Biol. 2012;416:1–9. doi: 10.1016/j.jmb.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 6.Wang W, Wang K, Span I, Jauch J, Bacher A, Groll M, Oldfield E. J Amer Chem Soc. 2012;134:11225–11234. doi: 10.1021/ja303445z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Citron CA, Brock NL, Rabe P, Dickschat JS. Angew Chem Int Ed. 2012;51:4053–4057. doi: 10.1002/anie.201201110. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Wang K, Smirnova TI, Khade RL, Zhang Y, Oldfield E. Angew Chem Int Ed. 2013;52:6522–6525. doi: 10.1002/anie.201302343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McMillan RS, Renaud J, Reynolds JG, Holm RH. J Inorg Biochem. 1979;11:213–227. doi: 10.1016/s0162-0134(00)80019-7. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka K, Nakamoto M, Tsunomori M, Tanaka T. Chem Lett. 1987:613–616. [Google Scholar]
- 11.Janthawornpong K, Krasutsky S, Chaignon P, Rohmer M, Poulter CD, Seemann M. J Amer Chem Soc. 2013;135:1816–1822. doi: 10.1021/ja309557s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahrens-Botzong A, Janthawornpong K, Wolny JA, Tambou EN, Rohmer M, Krasutsky S, Poulter CD, Schünemann V, Seemann M. Angew Chem Int Ed. 2011;50:11976–11979. doi: 10.1002/anie.201104562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Span I, Wang K, Wang W, Jauch J, Eisenreich W, Bacher A, Oldfield E, Groll M. Angew Chem Int Ed. 2013;52:2118–2121. doi: 10.1002/anie.201208469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Span I, Wang K, Wang W, Zhang Y, Bacher A, Eisenreich W, Li K, Schulz C, Oldfield E, Groll M. Nat Comm. 2012;3:1042. doi: 10.1038/ncomms2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seemann M, Janthawornpong K, Schweizer J, Bottger LH, Janoschka A, Ahrens-Botzong A, Tambou EN, Rotthaus O, Trautwein AX, Rohmer M. J Amer Chem Soc. 2009;131:13184–13185. doi: 10.1021/ja9012408. [DOI] [PubMed] [Google Scholar]
- 16.Faus I, Reinhard A, Rackwitz S, Wolny JA, Schlage K, Wille HC, Chumakov A, Krasutsky S, Chaignon P, Poulter CD, Seemann M, Schunemann V. Angew Chem Int Ed. 2015 doi: 10.1002/anie.201502494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rao G, Oldfield E. Biochemistry. 2016;55:4119–4129. doi: 10.1021/acs.biochem.6b00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, Alp EE, Yoda Y, Cramer SP. In: Metalloproteins: Methods and Protocols. Fontecilla-Camps JC, Nicolet Y, editors. Humana Press; Totowa, NJ: 2014. pp. 125–137. [Google Scholar]
- 19.Lauterbach L, Wang H, Horch M, Gee LB, Yoda Y, Tanaka Y, Zebger I, Lenz O, Cramer SP. Chem Sci. 2015;6:1055–1060. doi: 10.1039/c4sc02982h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitra D, Pelmenschikov V, Guo Y, Case DA, Wang H, Dong W, Tan ML, Ichiye T, Jenney FE, Adams MWW, Yoda Y, Zhao J, Cramer SP. Biochemistry. 2011;50:5220–5235. doi: 10.1021/bi200046p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamali S, Wang H, Mitra D, Ogata H, Lubitz W, Manor BC, Rauchfuss TB, Byrne D, Bonnefoy V, Jenney FE, Adams MWW, Yoda Y, Alp E, Zhao J, Cramer SP. Angew Chem Int Ed. 2013;52:724–728. doi: 10.1002/anie.201204616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tonzetich ZJ, Wang H, Mitra D, Tinberg CE, Do LH, Jenney FE, Adams MWW, Cramer SP, Lippard SJ. J Amer Chem Soc. 2010;132:6914–6916. doi: 10.1021/ja101002f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K, Wang W, No JH, Zhang Y, Zhang Y, Oldfield E. J Amer Chem Soc. 2010;132:6719–6727. doi: 10.1021/ja909664j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoll S, Schweiger A. J Mag Res. 2006;178:42–55. doi: 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 25.Suess DLM, Bürstel I, Paz L De La, Kuchenreuther JM, Pham CC, Cramer SP, Swartz JR, Britt RD. Proc Nat Acad Sci USA. 2015;112:11455–11460. doi: 10.1073/pnas.1508440112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Telser J, Smith ET, Adams MWW, Conover RC, Johnson MK, Hoffman BM. J Amer Chem Soc. 1995;117:5133–5140. [Google Scholar]
- 27.Zhou C, Holm RH. Inorg Chem. 1997;36:4066–4077. [Google Scholar]
- 28.Quitterer F, Frank A, Wang K, Rao G, O’Dowd B, Li J, Guerra F, Abdel-Azeim S, Bacher A, Eppinger J, Oldfield E, Groll M. J Mol Biol. 2015;427:2220–2228. doi: 10.1016/j.jmb.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. J Med Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 30.Lindert S, Zhu W, Liu YL, Pang R, Oldfield E, McCammon JA. Chem Biol Drug Des. 2013;81:742–748. doi: 10.1111/cbdd.12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller AA, Bundy GL, Mott JE, Skepner JE, Boyle TP, Harris DW, Hromockyj AE, Marotti KR, Zurenko GE, Munzner JB, Sweeney MT, Bammert GF, Hamel JC, Ford CW, Zhong WZ, Graber DR, Martin GE, Han F, Dolak LA, Seest EP, Ruble JC, Kamilar GM, Palmer JR, Banitt LS, Hurd AR, Barbachyn MR. Antimicrob Agents Chemother. 2008;52:2806–2812. doi: 10.1128/AAC.00247-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brandstetter H, Grams F, Glitz D, Lang A, Huber R, Bode W, Krell HW, Engh RA. J Biol Chem. 2001;276:17405–17412. doi: 10.1074/jbc.M007475200. [DOI] [PubMed] [Google Scholar]
- 33.Dunten P, Kammlott U, Crowther R, Levin W, Foley LH, Wang P, Palermo R. Protein Science. 2001;10:923–926. doi: 10.1110/ps.48401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Advanced Drug Delivery Reviews. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 35.Baell JB, Holloway GA. J Med Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 36.Ruble JC, Hurd AR, Johnson TA, Sherry DA, Barbachyn MR, Toogood PL, Bundy GL, Graber DR, Kamilar GM. J Amer Chem Soc. 2009;131:3991–3997. doi: 10.1021/ja808014h. [DOI] [PubMed] [Google Scholar]
- 37.Toellner TS. Hyperfine Interactions. 2000;125:3–28. [Google Scholar]
- 38.Sturhahn W, Toellner TS, Alp EE, Zhang X, Ando M, Yoda Y, Kikuta S, Seto M, Kimball CW, Dabrowski B. Phys Rev Lett. 1995;74:3832–3835. doi: 10.1103/PhysRevLett.74.3832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


