Abstract
Group II introns are ribozymes that catalyze a splicing reaction with the same chemical steps as spliceosome-mediated splicing. Many group II introns have lost the capacity to self-splice while acquiring compensatory interactions with host-derived protein cofactors. Degenerate group II introns are particularly abundant in the organellar genomes of plants, where their requirement for nuclear-encoded splicing factors provides a means for the integration of nuclear and organellar functions. We present a biochemical analysis of the interactions between a nuclear-encoded group II splicing factor and its chloroplast intron target. The maize (Zea mays) protein Chloroplast RNA Splicing 1 (CRS1) is required specifically for the splicing of the group II intron in the chloroplast atpF gene and belongs to a plant-specific protein family defined by a recently recognized RNA binding domain, the CRM domain. We show that CRS1's specificity for the atpF intron in vivo can be explained by CRS1's intrinsic RNA binding properties. CRS1 binds in vitro with high affinity and specificity to atpF intron RNA and does so through the recognition of elements in intron domains I and IV. These binding sites are not conserved in other group II introns, accounting for CRS1's intron specificity. In the absence of CRS1, the atpF intron has little uniform tertiary structure even at elevated [Mg2+]. CRS1 binding reorganizes the RNA, such that intron elements expected to be at the catalytic core become less accessible to solvent. We conclude that CRS1 promotes the folding of its group II intron target through tight and specific interactions with two peripheral intron segments.
INTRODUCTION
Group II introns are large catalytic RNAs that are found in bacteria and in the organellar genomes of fungi, algae, and plants (Lambowitz et al., 1999; Bonen and Vogel, 2001; Dai and Zimmerly, 2002, 2003). Group II introns are characterized by small regions of sequence similarity and a conserved secondary structure that is typically depicted as six helical domains emanating from a central core (Michel et al., 1989; Michel and Ferat, 1995; Qin and Pyle, 1998; Bonen and Vogel, 2001). Biochemical analyses of model group II introns have assigned specific roles in splicing to several of these domains (Qin and Pyle, 1998) and have provided evidence suggesting an evolutionary relationship between group II introns and the spliceosome (Valadkhan and Manley, 2001; Villa et al., 2002).
Group II introns are particularly abundant and diverse in the organellar genomes of plants (Bonen and Vogel, 2001); for example, angiosperm mitochondria and chloroplasts each house ∼20 group II introns (reviewed in Barkan, 2004). However, none of these introns have been reported to self-splice in vitro. In some instances, this recalcitrance to self-splicing is reflected by the clear absence of important structural elements (Bonen and Vogel, 2001), but many chloroplast introns appear to contain the essential group II intron features (Michel et al., 1989), and these must be compromised in more subtle ways. This loss of self-splicing activity has been accompanied by the evolutionary acquisition of protein partners that provide compensatory functions.
An ancient class of intron–protein partners involves a conserved maturase protein encoded within the intron itself (reviewed in Lambowitz et al., 1999). Many extant group II introns lack maturase open reading frames (ORFs), but all appear to be derived from maturase-encoding ancestors (Toor et al., 2001). Plant chloroplasts harbor a single maturase-like ORF, called MatK, which is encoded in the trnK intron. MatK has been implicated in splicing through phylogenetic arguments (Mohr et al., 1993; Ems et al., 1995) and because an unidentified chloroplast-encoded protein is required for the splicing of chloroplast introns in subgroup IIA (Jenkins et al., 1997; Vogel et al., 1997, 1999). However, a role for MatK in splicing has yet to be confirmed.
More recently acquired protein cofactors for group II introns were recruited from the host genome. For example, genetic analyses have shown that the splicing of at least 10 of the 17 group II introns in maize (Zea mays) chloroplasts requires nuclear-encoded proteins that are unrelated to maturases (Jenkins et al., 1997; Jenkins and Barkan, 2001; Till et al., 2001; Ostheimer et al., 2003). The participation of nuclear-encoded proteins in chloroplast splicing provides a means to control the biogenesis of the photosynthetic apparatus; indeed, the splicing of maize chloroplast introns is developmentally regulated (Barkan, 1989). Genetic screens in the green alga Chlamydomonas reinhardtii demonstrated that the splicing of the two group II introns in its chloroplast genome, both of which are transcribed in pieces and spliced in trans, is also dependent upon a complex set of nuclear-encoded proteins (Goldschmidt-Clermont et al., 1990; Perron et al., 1999, 2004; Rivier et al., 2001).
The nature of the biochemical interactions between group II introns and their protein partners is just starting to be explored. The best characterized example involves the Lactococcus lactis LtrA maturase and its host intron, LtrB. The LtrB intron will self-splice in the presence of elevated [Mg2+], but LtrA promotes LtrB splicing at physiological [Mg2+] (Saldanha et al., 1999). LtrA binds tightly to the LtrB intron, primarily through a binding site in intron domain IV (Wank et al., 1999). This interaction nucleates the binding of LtrA to low-affinity sites in core intron elements, and together, these interactions are thought to promote tertiary interactions within the intron that are important for RNA catalysis (Matsuura et al., 2001; Noah and Lambowitz, 2003; Rambo and Doudna, 2004).
By contrast, very little is known about the interactions between host-encoded protein cofactors and group II introns. Nuclear-encoded group II intron splicing factors in maize and C. reinhardtii chloroplasts have been shown to be associated with their genetically defined intron targets in vivo (Rivier et al., 2001; Till et al., 2001; Ostheimer et al., 2003), but no reports have addressed how those proteins recognize intron RNA or how those interactions promote splicing. Given the diversity of host-encoded group II splicing factors, it is likely that their analysis will reveal mechanisms of protein-assisted splicing that differ from those in the maturase system.
In this study, we explore the biochemical interactions between a host-encoded splicing factor, Chloroplast RNA Splicing 1 (CRS1), and its group II intron target. CRS1 is a nuclear-encoded protein in maize that is required solely for the splicing of the chloroplast atpF intron (Jenkins et al., 1997; Vogel et al., 1999), which is a member of the subgroup IIA intron class (Michel et al., 1989). CRS1 is related to two other group II intron splicing factors in maize, CRS2 Associated Factor 1 (CAF1) and CAF2, both of which form complexes with a peptidyl-tRNA hydrolase-like protein called CRS2 (Jenkins and Barkan, 2001) and facilitate the splicing of chloroplast subgroup IIB introns (Jenkins et al., 1997; Ostheimer et al., 2003). The similarity among CRS1, CAF1, and CAF2 is limited to repeated 10-kD domains that are related to a conserved protein in prokaryotes (Till et al., 2001; Ostheimer et al., 2003). These domains were dubbed chloroplast RNA splicing and ribosome maturation (CRM) domains (Ostheimer et al., 2003); the crystal structure of the Escherichia coli representative of this family provided evidence that CRM domains are previously unrecognized RNA binding domains (Ostheimer et al., 2002).
CRS1 has three CRM domains and is bound specifically to the atpF intron RNA in vivo (Till et al., 2001; Ostheimer et al., 2003). Here, we show that purified recombinant CRS1 dimerizes and binds with high affinity and specificity to the atpF intron in vitro. Sites of interaction between CRS1 and the intron were localized to two intron segments that are predicted to be on the periphery of the folded intron, in intron domains I and IV. These binding sites are not conserved in other group II introns and thus can account for CRS1's specificity for the atpF intron. We show further that CRS1 binding causes a shift in the structure of the intron population, such that the average structure becomes more compact and more closely resembles that expected of a catalytically active intron. These results suggest that the requirement for CRS1 is attributable to its participation in the productive folding of its group II intron substrate.
RESULTS
Recombinant CRS1 Is a Dimer
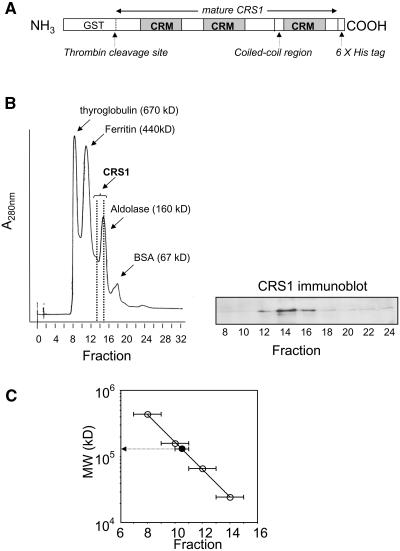
The organization of domains and functional motifs in CRS1 is shown in Figure 1A. CRS1 has three CRM domains (Till et al., 2001; Ostheimer et al., 2002, 2003). Immediately preceding its third CRM domain is an ∼50–amino acid segment that is predicted to participate in coiled-coil interactions. To determine whether CRS1 can form homomultimers, a recombinant CRS1 fusion protein with an NH2-terminal glutathione S-transferase (GST) tag and a COOH-terminal 6xHis tag (Figure 1A) was expressed in E. coli, purified by consecutive nickel and glutathione affinity chromatography, and eluted from the glutathione resin by incubation with thrombin to release CRS1 from the GST moiety. This recombinant CRS1 protein has a calculated molecular mass of 77 kD but eluted from a Superdex-200 gel filtration column with globular proteins of ∼170 kD (Figure 1B). CRS1's rate of sedimentation through a glycerol gradient suggested a size of ∼150 kD, slightly smaller than the aldolase marker (160 kD) (Figure 1C). The agreement of these two assays, which fractionate proteins based on different physical parameters, strongly suggests that the bulk of the recombinant CRS1 is dimeric (molecular mass =154 kD) at the concentration analyzed (∼2 μM), although we cannot exclude the possibility that a small proportion is trimeric or tetrameric. It seems likely that dimerization is mediated by CRS1's coiled-coil domain.
Figure 1.
Recombinant CRS1 Is Dimeric.
Diagram of recombinant CRS1 fusion protein (A). CRS1 lacking its predicted N-terminal chloroplast targeting sequence (mature CRS1, starting at residue 40 of the primary translation product) was expressed as a fusion protein with an N-terminal GST tag and a C-terminal 6xHis tag. The three CRM domains and the predicted coiled-coil region are indicated. CRS1 was released from GST by thrombin cleavage and fractionated on a Superdex-200 gel-filtration column (B) or by glycerol gradient sedimentation (C). Data points in (C) represent the peak fraction for each protein; bars show the adjacent fractions in which each protein was also detected. Size standards (open circles in [C]) were chymotrypsinogen A (25 kD), BSA (67 kD), aldolase (160 kD), ferritin (440 kD), and thyroglobulin (670 kD). CRS1 was detected by immunoblot analysis ([B]; data not shown).
CRS1 Binds atpF Intron RNA with High Affinity and Specificity in Vitro
Among the 17 group II introns in the maize chloroplast genome, only the atpF intron fails to splice in crs1 mutants (Jenkins et al., 1997; Vogel et al., 1999). Furthermore, CRS1 is associated specifically with the atpF intron in vivo (Till et al., 2001; Ostheimer et al., 2003). To determine whether CRS1 has intrinsic RNA binding activities that can account for these interactions, the intron binding properties of purified recombinant CRS1 dimers were studied with filter binding assays (Wong and Lohman, 1993; Weeks and Cech, 1995). Binding conditions were initially optimized by varying pH, salt, and temperature; optimal binding was observed at pH 7 to 7.5, 75 to 150 mM K+, 10 mM Mg2+, at temperatures between 25 and 30°C (data not shown). Standard binding conditions of 150 mM KOAc, 10 mM MgCl2, pH 7.0, and 25°C were used for subsequent experiments (unless otherwise stated) because these mimic physiological conditions. CRS1's RNA binding activity dropped precipitously when the protein was preheated to 50°C (data not shown), as expected for an activity that requires a native protein conformation.
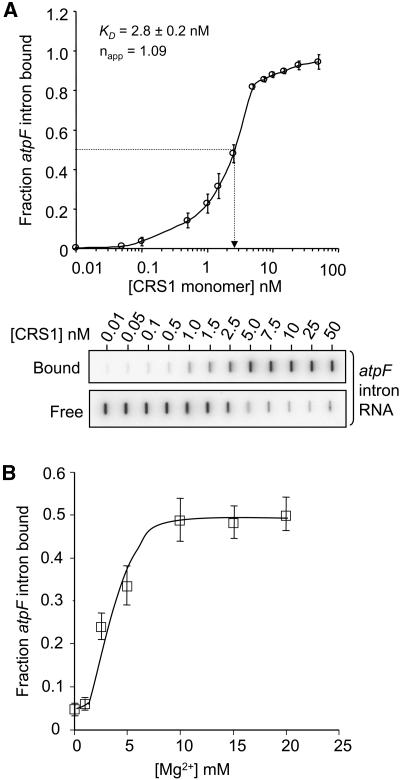
To estimate CRS1's equilibrium binding constant for atpF intron RNA, the formation of RNA/protein complexes was measured when increasing amounts of CRS1 were incubated with trace amounts of radiolabeled atpF intron (Figure 2A). Under the standard binding conditions, CRS1 bound the atpF intron RNA with a Kd ≈ 3 nM (CRS1 monomer concentration); the apparent Kd varied from ∼0.5 to ∼3 nM between different protein preparations but was highly reproducible within each preparation. The apparent Hill coefficient was ∼1, suggesting noncooperative binding. CRS1's affinity for intron increased dramatically as [Mg2+] was increased from 0 to 10 mM, suggesting that CRS1 recognizes a Mg2+-dependent RNA structure (Figure 2B).
Figure 2.
Equilibrium Binding Constant and Mg2+ Dependence of the CRS1–Intron Interaction.
(A) Equilibrium binding reactions contained 25 pM 32P-labeled atpF intron RNA and the indicated concentrations of CRS1. Binding reactions were filtered through stacked nitrocellulose and nylon membranes; protein-RNA complexes were retained on the nitrocellulose membrane (bound); RNA that passed through the nitrocellulose was retained on the nylon membrane underlay (free). Raw slot blot data are shown from a representative experiment. The Hill coefficient (napp) was determined by calculating the slope of a Hill plot, where the y axis is the log(bound RNA/unbound RNA) and the x axis is log[CRS1]. The Kd value and each data point are the mean ± sd of three experiments performed with the same CRS1 preparation.
(B) Mg2+ dependence of the CRS1/intron interaction. CRS1 (2.5 nM) was incubated with 32P-labeled atpF intron RNA (25 pM) in 150 mM KOAc, with increasing [MgCl2]. Protein-RNA complexes were quantified by nitrocellulose retention as above. Each data point is the mean ± sd of four experiments.
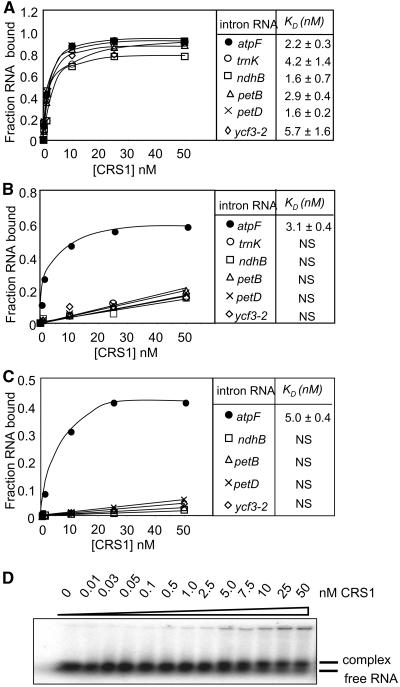
In the absence of nonspecific competitor and in the standard binding salts (150 mM K+, 10 mM Mg2+), CRS1 bound with similar affinities to atpF intron and to five other chloroplast group II introns (Figure 3A). However, the addition of 50 μg/mL of tRNA (Figure 3B) or the addition of K+ to 330 mM (Figure 3C) revealed CRS1's specificity for the atpF intron. The abundance of other RNAs in vivo and the fact that CRS1 is not in excess of its atpF intron substrate in vivo (Till et al., 2001) presumably serve to minimize CRS1's interactions with noncognate intron RNAs in the chloroplast. These results show that the in vivo specificity of CRS1 for the atpF intron reflects activities that are intrinsic to CRS1, rather than depending upon other molecules. The presence of competitor reduced the maximal amount of atpF intron bound from 100 to ∼50% (Figure 3), suggesting that the specific binding sites are inaccessible in ∼50% of the intron molecules, presumably because of intron misfolding. Gel mobility shift assays gave similar results (Figure 3D) but were not used routinely because of the difficulty of resolving the unbound intron RNA substrate (831 nucleotides) from the RNA/protein complex.
Figure 3.
CRS1 Binds with Specificity to the atpF Intron in Vitro.
(A) to (C) Equilibrium filter binding assays were performed with the indicated chloroplast group II intron RNA (25 pM) and increasing concentrations of CRS1 in the standard binding buffer (150 mM K+, 10 mM Mg2+ salt concentrations) (A), in the standard buffer containing 50 μg/mL E. coli tRNA (B), or in the standard buffer supplemented with KOAc to bring the K+ concentration to 330 mM (C). The atpF and trnK introns are in subgroup IIA, whereas the ndhB, petB, petD, and ycf3-2 introns are in subgroup IIB (Michel et al., 1989). The data points are the average of at least three experiments. The Kd values are the means ± sd of three to five experiments. NS, nonspecific binding.
(D) Gel-mobility shift assay showing homogeneity of the CRS1-intron complex formed in vitro. Binding reactions performed under the conditions used in (C) were electrophoresed through a 1.5% agarose gel under native conditions. Bands representing the CRS1/intron complex and free intron RNA are indicated. Because of the large size of the RNA substrate, CRS1 binding causes only a small shift in mobility.
Stoichiometry of the Intron/CRS1 Complex
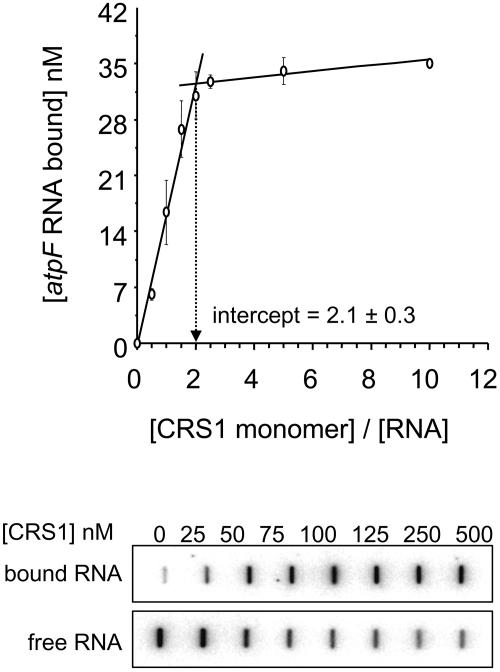
To determine the protein:RNA stoichiometry in the CRS1/intron complex, binding assays were performed with 50 nM atpF intron RNA (a concentration well above the Kd for the CRS1/intron interaction) and increasing amounts of CRS1 (Figure 4). The inflection point occurs at a [CRS1 monomer]:[intron RNA] ratio of ∼2, suggesting that two molecules of CRS1 interact with a single molecule of atpF intron RNA. Taken together with the fact that recombinant CRS1 dimerizes (Figure 1), these results suggest that a single CRS1 dimer binds to each atpF intron RNA molecule. However, some ambiguity is introduced by the fact that ∼30% of the intron RNA molecules remained unbound at the point where the curve plateaus, presumably because of the presence of misfolded isomers with inaccessible CRS1 binding sites. Thus, the effective concentration of intron RNA in these reactions is somewhat lower than its total concentration, and we cannot rule out the possibility that two CRS1 dimers bind to the atpF intron.
Figure 4.
Stoichiometry of the CRS1/Intron Complex.
Filter binding assays were performed in the standard buffer supplemented with 50 μg/mL E. coli tRNA, with increasing amounts of CRS1 (0 to 500 nM monomer concentration) and a fixed amount of atpF intron RNA (50 nM). Raw slot blot data from a representative experiment are shown below. The top panel shows RNA retained on the nitrocellulose membrane (bound); the bottom panel shows RNA that passed through the nitrocellulose and that was retained on the nylon membrane underlay (free).
CRS1 Binds to Nonconserved Elements in Intron Domains I and IV
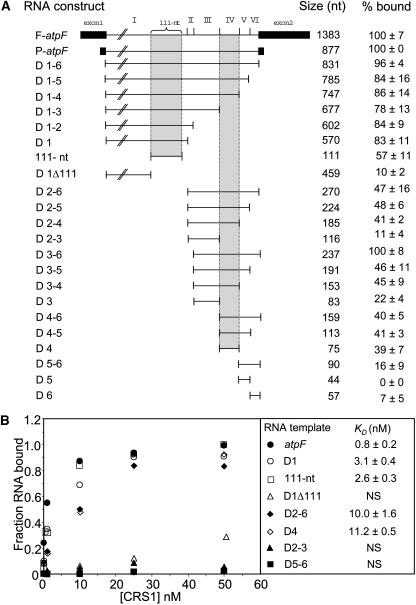
To identify the sites of CRS1 interaction in the 831-nucleotide atpF intron, the affinity of CRS1 for different intron fragments was determined with filter binding assays (Figure 5A). Assays used CRS1 at a concentration of 2.5 nM and included E. coli tRNA (50 μg/mL) to reduce nonspecific interactions. CRS1 bound with the highest affinity to intron fragments that included either domain IV or a 111-nucleotide accessory loop near the 3′ end of domain I that is inserted between subdomains Ii and Iii (see Figure 7) and that is unique to chloroplast atpF introns (Michel et al., 1989). The presence of exon sequences did not significantly influence CRS1 binding (cf. D1-6, F-atpF, and P-atpF in Figure 5A).
Figure 5.
Binding of CRS1 to atpF Intron Fragments.
(A) Equilibrium filter binding assays were performed with CRS1 (2.5 nM monomer concentration) and radiolabeled RNAs (25 pM) containing the indicated atpF intron sequences. Reactions used the standard binding buffer with 50 μg/mL E. coli tRNA added as nonspecific competitor. Results were normalized to the fraction of P-atpF RNA retained on the filter (∼50% of P-atpF RNA was bound at this protein concentration) and are an average of at least five assays ± sd. The junctions between the six intron domains are diagrammed at the top.
(B) Equilibrium Kd for selected intron RNA fragments. Binding reactions were performed in the standard buffer with the addition of 50 μg/mL tRNA competitor, and complex formation was assayed by filter binding. Data points are the average of at least three experiments involving a single CRS1 preparation. The fraction of RNA bound is normalized to that observed at saturation for the intact intron (∼70% of the input RNA). Calculated Kd values are the mean ± sd of at least three experiments. The Kd values obtained with different CRS1 preparations varied between ∼0.7 and 2 nM, but the results within each preparation were highly reproducible, as were the relative affinities for the different intron fragments.
Figure 7.
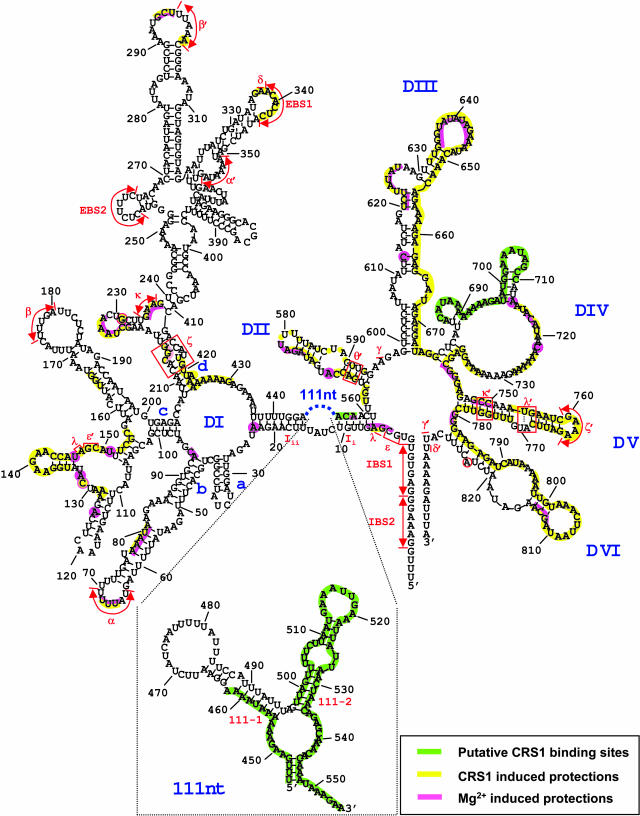
Secondary Structure Model of the Maize atpF Intron and Summary of Hydroxyl-Radical Protection Data.
The sequence shown includes 16 nucleotides of the 5′ exon and 11 nucleotides of the 3′ exon, with residue numbering beginning at the first intron nucleotide. The structure of this intron, which belongs to subgroup IIA, was modeled based on a previous structural model of chloroplast atpF introns (Michel et al., 1989), more recent data on conserved structural features of subgroup IIA introns (Toor et al., 2001), and the secondary structure prediction algorithms MFOLD (Zuker, 2003) and Alifold (Hofacker et al., 2002). Nucleotides predicted to be involved in the canonical subgroup IIA tertiary interactions IBS1–EBS1, IBS2–EBS2, α–α′, β–β′, γ–γ′, δ–δ′, ε–ε′, ζ–ζ′, κ–κ′, and λ–λ′ are indicated. A putative θ′ sequence is marked at the base of domain II, but an unambiguous θ element in domain I could not be identified. Similarly, η−η′ interaction sites are not marked because their positions are ambiguous. The sequence and predicted secondary structure of the 111-nucleotide insertion at the base of domain I are shown in the inset. High-resolution data were not obtained for several regions (the 3′ half of domain II, the 5′ half of domain III, and the 3′ region of domain VI) (see Figure 6 legend); lack of shading in these regions indicates lack of data, rather than lack of protection.
Equilibrium binding assays using a subset of these RNAs (Figure 5B) gave analogous results. CRS1 bound to the 111-nucleotide region of domain I and to intact domain I with similar affinities (Kd ∼ 3 nM); deletion of the 111-nucleotide region from domain I (D1Δ111) resulted in low-affinity nonspecific binding. Therefore, the 111-nucleotide region contains all of the sequences responsible for CRS1's high-affinity interaction with domain I. CRS1's affinity for RNA containing domains II through VI (D2-6) was similar to that for domain IV alone (Kd ≈ 10 nM); furthermore, domains II to III (D2-3) and domains V to VI (D5-6) lacked significant affinity for CRS1. Thus, CRS1's interaction with domain IV is largely responsible for CRS1's binding to intron sequences downstream of domain I.
Hydroxyl-Radical Footprinting of CRS1–Intron Complexes
To understand the mechanism of protein-assisted splicing, it is necessary to identify intron residues that are bound by CRS1 and to assess how CRS1 binding impacts intron conformation. CRS1/intron complexes assembled in vitro were analyzed by hydroxyl-radical footprinting to gain insight into both issues (Figure 6). Hydroxyl-radicals cleave solvent-exposed regions of the RNA backbone in a manner that is independent of RNA secondary structure (Powers and Noller, 1995). CRS1 could protect residues from cleavage either directly, by masking its binding sites on the RNA, or indirectly, by inducing a conformational change in the RNA that reduces their accessibility to solvent.
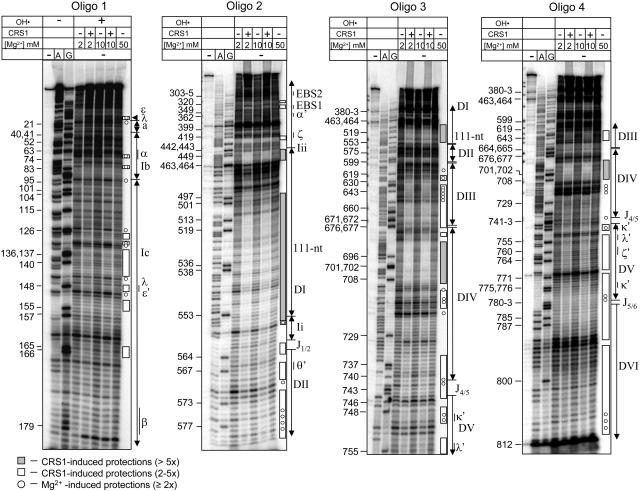
Figure 6.
Hydroxyl-Radical Footprinting of CRS1/atpF Intron Complexes.
atpF intron RNA (40 nM) was incubated in the presence or absence of CRS1 (100 nM) at the indicated Mg2+ concentration and then subjected to hydroxyl-radical cleavage (OH•). Cleavage sites were mapped by primer extension using one of four 5′-labeled oligonucleotides (oligos 1 through 4). The first three lanes show primer extension reactions with untreated intron RNA template and all four deoxyribonucleotides (first lane) or that included dideoxyribonucleotides that terminate extension at either A or G residues (second and third lanes) to provide sequencing ladders. Nucleotide residue numbers marked to the left and intron structural features marked to the right correspond with those shown on the intron map in Figure 7. The results with no Mg2+ and 2 mM Mg2+ in the absence of CRS1 were found to be similar in preliminary experiments; only the 2 mM Mg2+ data are shown. The data shown do not provide high-resolution information for several regions, including subdomain d of domain I, the 3′ half of domain II, the 5′ half of domain III, and the 3′ region of DVI (nucleotides 814 to 831). Subdomain d of domain I contains several sites of important tertiary interactions and was examined at high resolution using a primer corresponding to residues 404 to 380 (data not shown); the indicated protections in this region reflect this supplementary data.
In vitro–transcribed atpF intron RNA (40 nM) was incubated in the absence or presence of CRS1 (100 nM monomer concentration) at several Mg2+ concentrations and in the presence of 330 mM K+ to reduce nonspecific interactions; the CRS1/intron complex formed under similar conditions appears homogeneous in a gel mobility shift assay, although the resolution of that assay was rather limited (Figure 3D). The complex was subjected to hydroxyl-radical cleavage, and the positions of backbone cleavage were determined by primer extension using a series of oligonucleotide primers that together surveyed the entire intron. Figure 6 shows representative footprinting data, with the positions of landmark structural features in the intron indicated to the right, and intron residue numbers indicated to the left. Intron features and nucleotide positions are shown in the context of a secondary structure model in Figure 7.
CRS1 induced two types of protection: (1) widespread protection of relatively small magnitude suggested that CRS1 promotes a global reorganization of the intron such that it adopts a more compact and/or uniform structure, and (2) discrete regions of stronger protection suggested points of protein-RNA proximity or particularly efficient internalization to the intron core upon protein binding. These effects were distinguished by considering the following: (1) CRS1's affinity for various intron fragments (Figure 5), (2) published results that have identified tertiary interactions and intron segments in the solvent-inaccessible core of catalytically active group II introns (Matsuura et al., 2001; Swisher et al., 2001; Noah and Lambowitz, 2003), and (3) comparison with results obtained in the absence of CRS1 but in the presence of 50 mM Mg2+, which induces native structures in some self-splicing group II introns (Matsuura et al., 2001; Swisher et al., 2001; Noah and Lambowitz, 2003; Su et al., 2003). The results of the footprinting experiments are summarized on the secondary structure model of the atpF intron in Figure 7 and are discussed below.
CRS1 Binding Sites in Domains I and IV
The highest degree of protein-induced protection (between fivefold and 20-fold, indicated by shaded bars in Figure 6) was observed in three regions: two segments of the 111-nucleotide accessory loop of domain I (nucleotides 445 to 462 and 498 to 557) and the 5′ region of domain IV (nucleotides 684 to 710). These protections are likely to result primarily from direct protection by CRS1 because they lie within intron fragments that bind with highest affinity to CRS1 (Figure 5), their protection is not induced by 50 mM Mg2+ (Figure 6, last lanes), and they are predicted to be on the periphery of the folded intron based on studies with self-splicing group II introns (Qin and Pyle, 1998; Costa et al., 2000; Gordon and Piccirilli, 2001; Matsuura et al., 2001; Swisher et al., 2001; Pyle, 2002; Fedorova et al., 2003). The interactions between CRS1 and its putative binding sites in domains I and IV are explored in more detail below.
The hydroxyl-radical footprinting data suggest the possibility of an additional region of CRS1 contact involving residues ∼662 to 674 in domain III. The magnitude of protection by CRS1 in this region is only slightly less than that observed within the domains I and IV binding sites, and protection was not induced by elevated Mg2+. On the other hand, the RNA binding data involving intron fragments did not provide evidence for a high-affinity binding site in domain III (Figure 5). This region may therefore contain a lower-affinity secondary binding site for CRS1, or it may become protected in the presence of CRS1 because of CRS1-induced structural changes in the RNA.
Evidence for CRS1-Induced Intron Folding
Numerous intron segments that did not show significant affinity for CRS1 in filter binding assays (Figure 5) were protected severalfold in the presence of CRS1 (Figure 6, open bars). These protections were clustered in regions anticipated to reside in the introns' catalytic core and/or that have been shown to be internalized in self-splicing group II introns; they include most of domains V and VI, much of domains II and III, the linker between domains I and II (J1/2), and several segments of domain I. The magnitude of these protections and the sites of protection in domain I correlated with the results from Mg2+-induced folding of an ai5γ group II intron derivative (Swisher et al., 2001): EBS1, λ−ɛ/λ−ɛ′, and ζ were protected in both experiments, whereas EBS2 and β were protected in neither (Figure 6, with higher resolution data not shown). These results suggest that CRS1 binding promotes the formation or stability of a compact intron conformation that resembles that expected for the active intron.
A striking difference between these results and those described for self-splicing group II introns is the relatively small effect of elevated [Mg2+]: 50 mM Mg2+ induced only limited protection of the atpF intron from hydroxyl-radicals (residues designated with circles in Figure 6). Furthermore, the Mg2+-induced protection was much less pronounced in scope and magnitude than that induced by CRS1 (e.g., compare circles to open bars in D2, D5, and D6). Thus, the atpF intron is much more compromised in its self-directed folding capability than the ai5γ and L1.LtrB introns, for which elevated Mg2+ promotes a catalytically active, compact conformation (Matsuura et al., 2001; Swisher et al., 2001; Noah and Lambowitz, 2003; Su et al., 2003). In light of these findings, it is not surprising that the maize atpF intron does not self-splice in vitro and that it did not splice when expressed in E. coli (data not shown). We have also been unable to detect CRS1-promoted splicing in vitro or in E. coli (data not shown), suggesting that additional factors are required to arrive at the catalytically active intron conformation.
Analysis of CRS1 Binding Sites in Domains I and IV
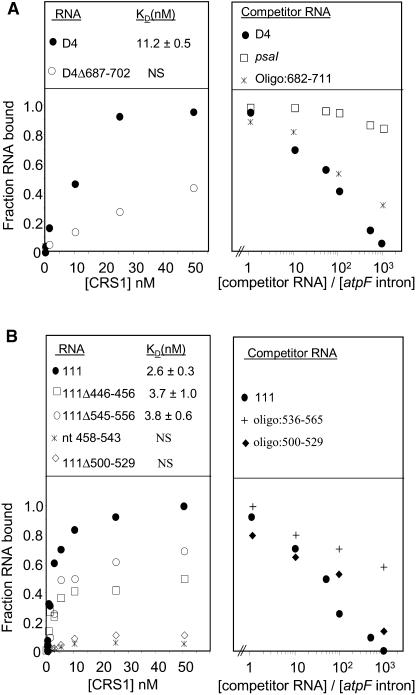
A series of experiments was performed to define the intron sequences that are essential for a high affinity interaction with CRS1. Deletion of residues 687 to 702 in domain IV (D4Δ687-702), which map to the center of the domain IV segment that is most heavily protected from hydroxyl-radicals by CRS1, led to a dramatic reduction in CRS1 affinity (Figure 8A, left panel). In addition, an oligonucleotide representing nucleotides 682 to 711 competed effectively with the intact intron for binding to CRS1 (Figure 8A, right panel). Taken together with the footprinting data, these results provide strong evidence that CRS1 recognizes nucleotides within the 5′ region of domain IV (see green shading in Figure 7).
Figure 8.
Detailed Analysis of CRS1 Binding Sites in Domains I and IV.
Binding reactions were done in the standard binding salts supplemented with 50 μg/mL E. coli tRNA. The data points are the average of at least three experiments, and calculated Kd values are the mean ± sd of at least three experiments.
(A) Analysis of the CRS1 binding site in domain IV. The left panel shows the results of equilibrium filter binding assays with intact domain IV RNA (D4) and a deletion derivative lacking residues 687 to 702. The fraction of RNA bound is normalized to that observed at saturation for intact domain IV (∼80% of the input RNA). The right panel shows the results of competition binding assays involving radiolabeled atpF intron and increasing amounts of the indicated nonradioactive competitor RNAs. The molar ratio of competitor RNA to the radiolabeled intron is indicated.
(B) Analysis of the CRS1 binding site in domain I. The left panel shows the results of equilibrium filter binding assays with the intact 111-nucleotide region and several deletion derivatives. The fraction of RNA bound is normalized to that observed at saturation for the intact 111-nucleotide region (∼80% of the input RNA). The right panel shows the results of competition binding assays involving radiolabeled atpF intron and the indicated molar excess of competitor RNA.
Analogous experiments were performed to dissect CRS1's interactions with the 111-nucleotide region of domain I. The CRS1 footprint in this region included two segments that are likely to be clustered in the folded RNA (see Figures 6 and 7). Deletion of residues 500 to 529 eliminated specific binding to CRS1 (Figure 8B, left panel), and an oligonucleotide containing only nucleotides 500 to 529 competed effectively for binding to the atpF intron (Figure 8B, right panel). These results suggest that CRS1 recognizes residues in the 500- to 529-nucleotide region.
Additional data showed that interactions in this region are complex and that they may be influenced by the formation of an RNA structure involving the 3′ and 5′ extremes of the footprinted region. Deletion of residues at the termini of the region (446 to 456 or 545 to 556) had a relatively small effect on CRS1 binding (in comparison with deletion of residues 500 to 529) (Figure 8B, left panel), and an oligonucleotide containing only the 3′ terminal residues 536 to 565 was a relatively poor competitor for CRS1 binding (Figure 8B, right panel), supporting the idea that CRS1 binding does not require nucleotides at the termini of the footprinted region. Strikingly, however, the binding curves for fragments 111Δ446-456 and 111Δ545-556 (Figure 8B, left panel) plateaued with a relatively small fraction of RNA molecules bound, indicating a high fraction of RNA molecules that were incompetent for CRS1 binding. Thus, the 5′ and 3′ borders of the footprinted region in domain I may contribute to CRS1 recognition by promoting the formation of a specific three-dimensional structure. The involvement of RNA structure in this CRS1 binding site is further supported by the fact that binding to the 111-nucleotide region is Mg2+ dependent (data not shown).
Whereas deletion of either the 5′ or 3′ border of the footprinted region had a relatively small effect on CRS1 binding, deletion of both regions (Figure 8B, left panel, nucleotides 458 to 543) eliminated detectable interaction with CRS1. This is puzzling in light of the fact that an oligonucleotide containing only residues 500 to 529 competed effectively for binding to the intact intron (Figure 8B, right panel) and again suggests that alternative RNA structures influence the accessibility of the CRS1 interaction site. One potential scenario involves the unpaired nucleotides 505 to 508 (5′-UUCU-3′) in the 111-2 stem (see Figure 7, inset). These nucleotides are complementary to the sequence 5′-AGAA-3′, which is found at both ends of the footprinted region (nucleotides 450 to 453 and 552 to 555) in a predicted single-stranded context. The results are consistent with the possibility that a pseudoknot involving nucleotides 505 to 508 and either of these two complementary sequences guides the correct folding of the 111-nucleotide loop, such that nucleotides ∼500 to 530 are accessible for CRS1 binding. However, a more detailed analysis will be required to fully define the interactions in this region.
In summary, the footprinting, equilibrium binding, and competition experiments yielded a consistent picture in which CRS1 binds nucleotides ∼500 to 530 in the 111-nucleotide accessory loop of domain I and nucleotides ∼680 to 710 in domain IV. Binding in these regions can account for CRS1's affinity for the intact intron, but we cannot eliminate the possibility that CRS1 interacts with lower affinity at additional sites.
DISCUSSION
Self-splicing group II introns have given rise to a diverse collection of degenerate introns that have lost the capacity to self-splice (Bonen and Vogel, 2001) and that have presumably acquired compensatory interactions with host proteins. Several nuclear-encoded proteins required for the splicing of group II introns in chloroplasts have been identified through genetic screens, but little is known about the biochemical properties of these proteins or the nature of their interactions with intron RNA. This study presents a biochemical analysis of the interactions between a host-encoded group II intron splicing factor and its cognate group II intron. The nuclear-encoded protein CRS1 is required specifically for the splicing of the group II intron in the maize chloroplast atpF gene (Jenkins et al., 1997) and is tightly associated with that intron in vivo (Till et al., 2001; Ostheimer et al., 2003). The results presented here show that the in vivo specificity of CRS1 for the atpF intron can be explained by CRS1's intrinsic RNA binding properties. Purified recombinant CRS1 is a dimer and binds in vitro to atpF intron RNA with high affinity and specificity under conditions that mimic those in the chloroplast stroma (150 mM K+, 10 mM Mg2+, pH 7.0, 50 μg/mL E. coli tRNA, 25°C) (Horlitz and Klaff, 2000; Shaul, 2002). We showed further that in the absence of CRS1, the atpF intron has little uniform tertiary structure even in the presence of the high Mg2+ concentrations that induce the folding of model autocatalytic group II introns (Matsuura et al., 2001; Swisher et al., 2001; Noah and Lambowitz, 2003). CRS1 binding induced a change in intron conformation at physiological Mg2+ concentrations, such that the intron population was dominated by molecules with large solvent-inaccessible regions involving intron elements found at the core of catalytically active group II introns (Matsuura et al., 2001; Swisher et al., 2001). These results lead to a picture in which CRS1 promotes the correct folding of its group II intron target through tight and specific interactions with two peripheral intron segments.
CRS1 Binds to Nonconserved Elements in Domains I and IV
Hydroxyl-radical footprinting experiments identified intron segments that are protected from solvent as a result of CRS1 binding. Sites that were protected by virtue of their proximity to CRS1 were distinguished from those protected by CRS1-induced RNA structure by performing binding assays with intron fragments and deletion derivatives. The results showed that CRS1 binds with high affinity to two intron segments. The highest affinity interaction maps within a 111-nucleotide subdomain at the base of domain I; the second site maps to the 5′ half of domain IV, adjacent to the conserved domain IV stem (Figure 7). In other group II introns, corresponding features are either nonexistent (the domain I-111 nucleotide subdomain) or not conserved in sequence (the domain IV site), so these results account for CRS1's specificity for the atpF intron. The CRS1 binding sites are likely to lie on the periphery of the folded intron, given their lack of conservation and their lack of proximity to intron elements implicated in catalysis.
CRS1 contains three putative RNA binding domains, called CRM domains (see Figure 1) (Till et al., 2001; Ostheimer et al., 2002, 2003). We have found that CRS1's third CRM domain expressed in isolation binds with a Kd of ∼40 nM to the atpF intron, providing direct evidence that CRM domains can bind RNA (O. Ostersetzer and A. Barkan, unpublished data). Data shown here support the idea that a single CRS1 dimer binds each intron (although the possibility that a second dimer binds cannot be eliminated), so at least six CRM domains are potentially involved in intron recognition. Our results do not address how these domains work together for RNA recognition: it is possible that one CRS1 monomer in each dimer recognizes each of the two binding sites, that the pair of CRS1 monomers cooperate for binding at each site, or that one CRS1 dimer binds to each of the two sites. CRM domains are derived from a conserved prokaryotic protein (Till et al., 2001; Ostheimer et al., 2002), and they have been incorporated into a family of single-domain and multidomain proteins specifically in the plant lineage. In Arabidopsis thaliana, this family has 16 members, including predicted orthologs of the maize chloroplast group II splicing factors CRS1, CAF1, and CAF2 (Ostheimer et al., 2003). CRS1, CAF1, and CAF2 are the only proteins in this family that have been studied, but it seems likely that they all interact with RNA. With the CRS1 binding sites in the atpF intron now defined, the CRS1/atpF intron system is poised to serve as a model to understand the principles underlying RNA recognition by CRM domains.
Predicted orthologs of both CRS1 and the chloroplast atpF intron are found in both monocots and dicots. The atpF intron sequences corresponding to the CRS1 binding sites are very highly conserved among other graminacious monocots but diverge considerably in the dicots (see Supplemental Figure 1 online). Dicot atpF introns do have an insertion that corresponds in position to the domain I:111-nucleotide subdomain in maize, but the putative CRS1 binding site (residues ∼498 to 525) is poorly conserved, and the 3′ end of the subdomain (nucleotides corresponding to residues 526 to 555) is missing entirely. CRS1's binding site in domain IV is somewhat more conserved but still shows substantial variation between monocots and dicots (see Supplemental Figure 1 online). Assuming that CRS1 plays a similar role in the splicing of the atpF intron in monocots and dicots, the CRS1 orthologs provide a series of natural protein variants with modified RNA specificities that will be useful for understanding how CRS1 recognizes its binding sites and how RNA binding proteins coevolve with their RNA targets.
The CRS1 binding site in domain IV maps to the analogous position as the primary binding site for group II intron maturases in their host introns (Wank et al., 1999; Huang et al., 2003; Rambo and Doudna, 2004). Thus, the domain IV binding site may have retained its function as a protein-interaction site during the evolution of the ORF-less atpF intron from a maturase-encoding ancestor (Toor et al., 2001). This region of domain IV appears to be particularly malleable because it is also the site of insertion of maturase ORFs and of the discontinuities in fragmented group II introns that are spliced in trans (Bonen and Vogel, 2001).
It is possible that insertions at the base of domain I provide another feature that is commonly exploited for protein binding in group II introns. Although the atpF intron is so far unique in having a subdomain positioned on the 3′ side of the domain I stem, an insertion on the opposite strand of the domain I stem (at residue ∼13) is relatively common (Toor et al., 2001). It was hypothesized that such an insertion in the GroEL intron in Azotobacter vinelandii might serve as a protein-interaction site (Adamidi et al., 2003); the fact that CRS1 binds to sequences protruding from the opposite strand of the same stem lends credence to this idea. Furthermore, the L. lactis LtrA maturase binds with high affinity to domain I of its host intron, although the precise position of this binding site has not been defined (Rambo and Doudna, 2004). The fact that both the host-encoded protein CRS1 and the intron-encoded LtrA maturase interact with their cognate introns primarily through domains I and IV despite the independent evolutionary origins of the proteins suggests that domains I and IV are particularly amenable to the acquisition of protein-interaction sites through which proteins can guide intron folding without disrupting essential intron elements. It will be interesting to learn whether the analogous regions of domains I and IV serve as binding sites for host factors in other group II introns.
There is strong evidence that CRS1 is not sufficient to promote the splicing of the atpF intron. In chloroplast extract, CRS1 is found together with the atpF intron RNA in a complex of ∼700 kD (Till et al., 2001; Ostheimer et al., 2003). The complex that survives extract preparation includes little, if any, exon RNA because of endogenous RNase activity (Till et al., 2001). Only ∼420 kD of the ∼700-kD complex can be accounted for by the combined molecular masses of the atpF intron and a CRS1 dimer. Furthermore, neither incubation of recombinant CRS1with unspliced atpF RNA nor their coexpression in E. coli resulted in detectable splicing (data not shown). Indeed, the splicing of the atpF intron in vivo is known to require chloroplast ribosomes or a chloroplast translation product (Jenkins et al., 1997), so MatK, the sole maturase-like molecule encoded in the maize chloroplast genome, is a good candidate for an additional splicing factor. If MatK does prove to be involved in the splicing of the atpF intron, it will be interesting to learn where it binds the intron, given that CRS1 occupies the site analogous to the primary binding site for characterized maturases in microorganisms (Wank et al., 1999; Huang et al., 2003).
Impact of CRS1 on Intron Conformation
Our results suggest that the atpF intron is highly compromised in its capacity for self-directed folding and that CRS1 binding helps to compensate for this deficiency. We have been unable to detect any self-splicing activity in the atpF intron despite exploration of a wide variety of salt, temperature, and intron-folding conditions. This correlates well with the hydroxyl-radical protection data, which provided little evidence for uniform tertiary structure when the atpF intron was incubated under the conditions of high [Mg2+] that promote the folding of the ai5γ and LtrB introns. The addition of CRS1 at physiological salt concentrations caused the internalization of intron elements that have been shown to be protected from solvent in catalytically active model group II introns. CRS1's binding to the intact intron, to isolated domain IV, and to the isolated domain I:111-nucleotide region was dependent upon the presence of Mg2+ (Figure 2B; data not shown), suggesting that CRS1 recognizes Mg2+-dependent RNA structures within each of its binding sites.
The LtrA maturase/LtrB intron interaction is the only other instance in which the biochemical interactions between a group II intron and a protein cofactor have been explored. In both cases, protein binding induces the formation or stabilization of tertiary interactions in the intron. However, the impact of CRS1 on the conformation of the atpF intron contrasts in both magnitude and scope with the impact of the LtrA maturase on the LtrB intron. First, LtrA's function in intron folding can largely be replaced by high [Mg2+] (Matsuura et al., 2001), whereas CRS1's function cannot. Second, the addition of CRS1 at physiological [Mg2+] results in the protection of large segments of intron from solvent, whereas the impact of LtrA is more limited. LtrA belongs to the family of group II intron maturases, which often retain dual functions in splicing and intron mobility (Lambowitz et al., 1999). When both the splicing and mobility functions of a maturase are under selection, the coevolution of intron/maturase partners may be more constrained than the coevolution of introns with host-derived splicing factors, where intron-degeneration may be more easily compensated by interactions with proteins of diverse origin. The degenerate group II introns that are typical in the organelles of plants (Bonen and Vogel, 2001) can be anticipated to interact with a diverse set of host proteins whose study is likely to reveal new mechanisms for protein-facilitated RNA catalysis.
METHODS
Expression and Purification of Recombinant CRS1
A DNA fragment encoding mature CRS1 (i.e., lacking the predicted chloroplast targeting sequence) was generated by PCR from a CRS1 cDNA template (Till et al., 2001) using the primers 5′-CCACATGTTAGCCGAGGCAACTCCCAA-3′ and 5′-AAGCGGCCGCGTGGGTTCGATACCTGATTT-3′. This PCR fragment was digested with AflIII and NotI and subcloned into the NcoI and NotI sites of pET28b (Novagen, Madison,WI), such that sequences encoding a 6xHis tag were fused in frame to the 3′ end of the CRS1 ORF. This modified CRS1 ORF was then PCR amplified with primers 5′-GGTGGATCCATGGCCGAGGCAACTCCTTCC-3′ and 5′-GGTTCTAGAGGGCTTTGTTAGCAGCCG-3′, digested with XbaI, the ends filled in by incubation with deoxynucleotide triphosphate (dNTP) and T4 DNA polymerase, digested with BamHI, and ligated into the pGEX-2TK vector (Amersham-Pharmacia, Uppsala, Sweden) that had been digested with SmaI and BamHI. The resulting plasmid was named pGEX-CRS1-His and encodes mature CRS1 with GST fused to its N terminus and a 6xHis tag fused to its C terminus (Figure 1A). The DNA sequence of this and all other clones and PCR products used in this work was confirmed before use.
Based on analysis of CRS1 expression from pGEX-CRS1-His in a variety of Escherichia coli host cells and under various growth and induction conditions, the following conditions for the recovery of soluble recombinant CRS1 were chosen: pGEX-CRS1-His was transformed into E. coli BL21 Star (DE3) cells (Invitrogen, Carlsbad, CA), grown at 37°C to OD600 ∼ 0.8, and induced with 0.3 mM isopropylthio-β-galactoside for 16 h at 20°C. Cells were lysed in a cold French press without prior freezing of the cell pellet, in lysis buffer (10 mM Tris-HCl, pH 8, 50 mM NaH2PO4, 750 mM NaCl, 5 mM imidazole, and 5 mM β-mercaptoethanol) that was adjusted to pH 8.0 and supplemented with 2 μg/mL of leupeptin, 1 μg/mL of pepstatin, 4 μg/mL of aprotinin, 1 mM phenylmethylsulfonyl fluoride, and 75 units/mL of DNase I. The GST-CRS1 fusion protein was purified from the soluble lysate by nickel-affinity chromatography (nickel-nitrilotriacetic acid agarose beads; Qiagen, Valencia, CA). After washing the nickel beads with lysis buffer (lacking protease inhibitors and DNase), protein was eluted in the same buffer supplemented with 250 mM imidazole, diluted 1:20 in PBS, and immediately added to glutathione-agarose affinity resin (Sigma-Aldrich, St. Louis, MO). After a 30-min incubation at room temperature, the beads were washed with PBS, followed by a wash with high salt buffer (50 mM Tris-HCl, 750 mM NaCl, 0.1% Triton X-100, and 5 mM β-mercaptoethanol). The high-salt wash reduced protein aggregation and removed traces of E. coli nucleic acids, as indicated by the results of gel-filtration chromatography and the ratio of absorbance at 260/280 nm, respectively. After washing the glutathione beads briefly in PBS, CRS1 was released from the GST moiety by on-column cleavage with thrombin protease (Sigma-Aldrich; 1.25 units/mL in PBS) for 15 min at 25°C. Thrombin activity in the eluate was terminated by the addition of one-quarter volume of Benzamidine-Sepharose beads (Amersham-Pharmacia). The released CRS1 was separated from the benzamidine beads by centrifugation at 15,000g for 5 min and was further purified by gel-filtration chromatography on Superdex-200 resin (Amersham-Pharmacia) in 50 mM Hepes-KOH, pH 7.0, 250 mM NaCl, 5 mM β-mercaptoethanol, and 1% glycerol. The resulting mature CRS1 (mCRS1) was dialyzed against a buffer containing 50 mM Hepes-KOH, pH 7.0, 500 mM KCl, 5 mM β-mercaptoethanol, and 50% glycerol and stored at −20°C. This procedure resulted in a highly purified mCRS1-dimer preparation at a yield of 15 to 50 μg protein/liter of culture that retained its intron binding activity for at least 3 weeks.
Glycerol Gradient Sedimentation
Glycerol gradient sedimentation was performed as previously described (Weeks and Cech, 1995). Purified mCRS1 (100 μL of a 0.2-μg/μL solution, equivalent to a concentration of 2.5 μM) was mixed with size standards (chymotrypsinogen A, 25 kD; BSA, 67 kD; aldolase, 158 kD; ferritin, 440 kD) and applied to a 5-mL linear 10 to 50% glycerol gradient in 50 mM Tris-HCl, pH 7, 250 mM NaCl, and 5 mM β-mercaptoethanol, pH 7.0. Gradients were centrifuged at 4°C for 15 h at 50,000 rpm in a Beckman SW55Ti rotor (Fullerton, CA). Fractions of 250 μL were removed from the top of the tube, and an aliquot of each fraction was analyzed by SDS-PAGE and immunoblot analysis with anti-CRS1 antibodies.
RNA Substrates for Binding Studies
RNAs used for binding studies are described in Table 1. With the exception of the three synthetic oligonucleotides indicated, all RNAs were generated by in vitro transcription from PCR fragments in which a T7 promoter was incorporated into the 5′ PCR primer. In vitro transcription was performed with T7 RNA polymerase (Promega, Madison, WI) with 0.5 mM each of ATP, GTP, and CTP and 0.05 mM UTP in the presence of 20 μCi [α32P]UTP (3000 Ci/mmol). Transcripts were gel purified after electrophoresis in 4% polyacrylamide gels containing 7 M urea, eluted from the gel slice by incubation in 0.3 M NaOAc, 2 mM EDTA, and 0.1% SDS for 1 h at 60°C, and subjected to phenol-chloroform extraction and ethanol precipitation. RNAs were resuspended in deionized water and stored at −20°C for up to 1 week. All solutions used with RNA were autoclaved and then filtered through 0.2-μm nitrocellulose syringe filters to remove trace proteins.
Table 1.
RNAs Used for in Vitro Binding Studies
| Transcript Name | atpF Intron Residues Included (Intron Residues Numbered as in Figure 7) |
|---|---|
| F-atpF | atpF pre-mRNA, including the complete protein-coding exon sequences and intron |
| P-atpF | Complete atpF intron flanked by 22 nucleotides of exon 1 and 24 nucleotides of exon 2 |
| D1–6 | Intron nucleotides 1–831 |
| D1–5 | Nucleotides 1–785 |
| D1–4 | Nucleotides 1–747 |
| D1–3 | Nucleotides 1–677 |
| D1–2 | Nucleotides 1–602 |
| D1 | Nucleotides 1–570 |
| D1Δ111a | Joined nucleotides 1–445 with 557–570 |
| 111 nucleotide | Nucleotides 446–556 |
| 111Δ446-456 | Nucleotides 457–556 |
| 111Δ500-529a | Joined nucleotides 446–499; 530–556 |
| 111Δ545-556 | Nucleotides 446–544 |
| Nucleotides 458–543 | Nucleotides 458–543 |
| D2–6 | Nucleotides 562–831 |
| D2–5 | Nucleotides 562–785 |
| D2–4 | Nucleotides 562–747 |
| D2–3 | Nucleotides 562–677 |
| D2 | Nucleotides 562–602 |
| D3–6 | Nucleotides 595–831 |
| D3–5 | Nucleotides 595–785 |
| D3–4 | Nucleotides 595–747 |
| D3 | Nucleotides 595–677 |
| D4–6 | Nucleotides 673–831 |
| D4–5 | Nucleotides 673–785 |
| D4 | Nucleotides 673–747 |
| D4Δ687-702a | Joined nucleotides 673–686; 703–747 |
| D5–6 | Nucleotides 742–831 |
| D5 | Nucleotides 742–785 |
| D6 | Nucleotides 775–831 |
| Oligo 500–529 | Nucleotides 500–529 (synthetic oligonucleotide) |
| Oligo 536–565 | Nucleotides 536–565 (synthetic oligonucleotide) |
| Oligo 682–711 | Nucleotides 682–711 (synthetic oligonucleotide) |
Deletions obtained by PCR-mediated mutagenesis.
RNA Binding Assays
For standard equilibrium binding assays [32P]-labeled RNAs (25 pM or as indicated) were denatured by heating to 95°C for 2 min in 10 mM Tris-HCl, pH 7.0, and 1 mM EDTA and transferred to 55°C. An equal volume of 2× binding buffer (100 mM Tris-HCl, pH 7.0, 300 mM KOAc, 20 mM MgCl2, 20 μg/mL BSA, and 10 mM DTT) prewarmed to 55°C was added, and the mixture was transferred to 25°C for 5 min. rRNAsin was added to 0.5 units/μL, and the folded RNA was aliquoted into tubes for individual binding reactions (20 μL total volume) containing CRS1 (0 to 100 nM). Binding reactions were incubated at 25°C for 15 min; pilot experiments established that a 15-min incubation was sufficient to reach equilibrium. Samples were applied by vacuum filtration to stacked nitrocellulose (Nitrobind; 0.22 μM; Fisher Scientific, Portland, OR) and nylon membranes (Magna Nylon; 0.44 μM; Fisher) in a slot blot manifold (Wong and Lohman, 1993; Weeks and Cech, 1995). Slots were washed once by vacuum filtration with 100 μL of wash buffer (50 mM Tris-HCl, pH 7.0, 150 mM KOAc, and 10 mM MgCl2; or alternative salts to match those in the binding buffer). The membranes were dried and analyzed with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). For competition assays, the radiolabeled and unlabeled competitor RNAs were added simultaneously in molar ratios varying between 1/1 and 1/106.
For the gel mobility shift assay, binding reactions were performed as described above and were then chilled on ice. Glycerol and bromophenol blue were added to 10 and 0.025%, respectively, and samples were loaded onto a refrigerated 1.5% agarose gel prepared in 25 mM Tris-HCl, pH 7.0, 50 mM KOAc, and 5 mM MgCl2. Electrophoresis was performed in the same buffer at 50 V for ∼3 h at 4°C with buffer recycling. Gels were dried and imaged with a Storm PhorphorImager (Molecular Dynamics). Data were quantified with ImageQuant software (version 5.1; Molecular Dynamics).
Hydroxyl-Radical Footprinting
Hydroxyl-radical footprinting assays were performed largely as described by Powers and Noller (1995). Before each assay, purified recombinant CRS1 was dialyzed against 50 mM Hepes-KOH and 0.5 M KOAc, pH 7.0. Nonradioactive atpF intron RNA was gel purified and folded according to the procedure above in a buffer lacking DTT. Binding reactions (20 μL) contained 40 nM RNA, 100 nM mCRS1 (monomer concentration), 50 mM Hepes-KOH, pH 7.0, 150 mM KOAc, and the specified concentrations of MgCl2 and were incubated at 25°C for 10 min. The reactions were then transferred to ice for 30 min. Production of hydroxyl-radicals was then induced by pipetting 1 μL each of freshly prepared solutions containing 50 mM ammonium Fe (II) sulfate, 100 mM EDTA, 0.25 M ascorbic acid, and 2.5% H2O2 onto the walls of the tubes. Tubes were centrifuged briefly to mix the reagents and incubated on ice for 10 min. The reactions were terminated by the addition of thiourea to 20 mM and E. coli tRNA (Sigma-Aldrich) to 10 μg/mL, followed by phenol extraction and ethanol precipitation in the presence of 0.3 M NaOAc, pH 6.5. Cleavage sites were mapped by primer extension using primers complementary to the following intron residues: oligo 1, nucleotides 190 to 219; oligo 2, nucleotides 585 to 602; oligo 3, nucleotides 768 to 785; oligo 4, nucleotides 832 to 855. An additional oligonucleotide, complementary to EBS1 (5′-TTCTAGTAAATAGTTCAAACTATT-3′) was used to acquire higher resolution data for portions of domain I (data not shown). The primers were combined with the RNA in 100 mM Tris-HCl, pH 9.0, and 100 mM KCl, heated to 95°C and cooled slowly to 48°C to allow annealing. The extension reactions were performed in 50 mM Tris-HCl, pH 9.0, 50 mM KCl, 5 mM MgCl2, 0.25 mM dNTP, 0.5 units/μL rRNasin, and 0.25 units/mL AMV-reverse transcriptase (Promega) for 90 min at 48°C. A partial sequencing ladder was generated with the untreated RNA, in primer extension reactions including either ddTTP (0.13 mM) or ddCTP (0.065 mM), with the corresponding dNTP at 0.25 mM. Primer extension reactions were stopped by the addition of two volumes of 90% formamide, 0.5× TBE, and 25 mM EDTA, heated to 80°C for 3 min, resolved by electrophoresis in 5% polyacrylamide gels containing 7 M urea, dried and imaged with a Storm PhosphorImager. Data were quantified with ImageQuant software (version 5.1).
Supplementary Material
Acknowledgments
We thank Andy Berglund for synthesizing RNA oligonucleotides used in this work and Ken Prehoda, Clarke Conant, and Andy Berglund for useful discussions. This work was supported by grants to A.B. from the National Science Foundation (MCB0314597) and the USDA (2003-35318-13578) and by a Binational Agricultural Research and Development postdoctoral fellowship (FI-320-2001) to O.O.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Alice Barkan (abarkan@molbio.uoregon.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.027516.
References
- Adamidi, C., Fedorova, O., and Pyle, A.M. (2003). A group II intron inserted into a bacterial heat-shock operon shows autocatalytic activity and unusual thermostability. Biochemistry 42, 3409–3418. [DOI] [PubMed] [Google Scholar]
- Barkan, A. (1989). Tissue-dependent plastid RNA splicing in maize: Transcripts from four plastid genes are predominantly unspliced in leaf meristems and roots. Plant Cell 1, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan, A. (2004). Intron splicing in plant organelles. In Molecular Biology and Biotechnology of Plant Organelles, H. Daniell and C. Chase, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 281–308.
- Bonen, L., and Vogel, J. (2001). The ins and outs of group II introns. Trends Genet. 17, 322–331. [DOI] [PubMed] [Google Scholar]
- Costa, M., Michel, F., and Westhof, E. (2000). A three-dimensional perspective on exon binding by a group II self-splicing intron. EMBO J. 19, 5007–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, L., and Zimmerly, S. (2002). Compilation and analysis of group II intron insertions in bacterial genomes: Evidence for retroelement behavior. Nucleic Acids Res. 30, 1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, L., and Zimmerly, S. (2003). ORF-less and reverse-transcriptase-encoding group II introns in achaebacteria, with a pattern of homing into related group II intron ORFs. RNA 9, 14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ems, S.C., Morden, C.W., Dixon, C.K., Wolfe, K.H., dePamphilis, C.W., and Palmer, J.D. (1995). Transcription, splicing and editing of plastid RNAs in the nonphotosynthetic plant Epifagus virginiana. Plant Mol. Biol. 29, 721–733. [DOI] [PubMed] [Google Scholar]
- Fedorova, O., Mitros, T., and Pyle, A. (2003). Domains 2 and 3 interact to form critical elements of the group II intron active site. J. Mol. Biol. 330, 197–209. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont, M., Girard-Bascou, J., Choquet, Y., and Rochaix, J.-D. (1990). Trans-splicing mutants of Chlamydomonas reinhardtii. Mol. Gen. Genet. 223, 417–425. [DOI] [PubMed] [Google Scholar]
- Gordon, P., and Piccirilli, J. (2001). Metal ion coordination by the AGC triad in domain 5 contributes to group II intron catalysis. Nat. Struct. Biol. 8, 893–898. [DOI] [PubMed] [Google Scholar]
- Hofacker, I.L., Fekete, M., and Stadler, P.F. (2002). Secondary structure prediction for aligned RNA sequences. J. Mol. Biol. 319, 1059–1066. [DOI] [PubMed] [Google Scholar]
- Horlitz, M., and Klaff, P. (2000). Gene-specific trans-regulatory functions of magnesium for chloroplast mRNA stability in higher plants. J. Biol. Chem. 275, 35638–35645. [DOI] [PubMed] [Google Scholar]
- Huang, H.-R., Chao, M., Armstrong, B., Wang, Y., Lambowitz, A., and Perlman, P. (2003). The DIVa maturase binding site in the yeast group II intron aI2 is essential for intron homing but not for in vivo splicing. Mol. Cell. Biol. 23, 8809–8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins, B., and Barkan, A. (2001). Recruitment of a peptidyl-tRNA hydrolase as a facilitator of group II intron splicing in chloroplasts. EMBO J. 20, 872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins, B., Kulhanek, D., and Barkan, A. (1997). Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell 9, 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz, A., Caprara, M., Zimmerly, S., and Perlman, P. (1999). Group I and group II ribozymes as RNPs: Clues to the past and guides to the future. In The RNA World, R. Gesteland, T. Cech, and J. Atkins, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 451–485.
- Matsuura, M., Noah, J., and Lambowitz, A. (2001). Mechanism of maturase-promoted group II intron splicing. EMBO J. 20, 7259–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, F., and Ferat, J.-L. (1995). Structure and actvities of group II introns. Annu. Rev. Biochem. 64, 435–461. [DOI] [PubMed] [Google Scholar]
- Michel, F., Umesono, K., and Ozeki, H. (1989). Comparative and functional anatomy of group II catalytic introns: A review. Gene 82, 5–30. [DOI] [PubMed] [Google Scholar]
- Mohr, G., Perlman, P., and Lambowitz, A. (1993). Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Res. 21, 4991–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noah, J., and Lambowitz, A. (2003). Effects of maturase binding and Mg2+ concentration on group II intron RNA folding investigated by UV cross-linking. Biochemistry 42, 12466–12480. [DOI] [PubMed] [Google Scholar]
- Ostheimer, G., Barkan, A., and Matthews, B. (2002). Crystal structure of E. coli YhbY: A representative of a novel class of RNA binding proteins. Structure 10, 1593–1601. [DOI] [PubMed] [Google Scholar]
- Ostheimer, G., Williams-Carrier, R., Belcher, S., Osborne, E., Gierke, J., and Barkan, A. (2003). Group II intron splicing factors derived by diversification of an ancient RNA binding module. EMBO J. 22, 3919–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron, K., Goldschmidt-Clermont, M., and Rochaix, J.-D. (1999). A factor related to pseudouridine synthases is required for chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J. 18, 6481–6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron, K., Goldschmidt-Clermont, M., and Rochaix, J.D. (2004). A multiprotein complex involved in chloroplast group II intron splicing. RNA 10, 704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers, T., and Noller, H. (1995). Hydroxyl radical footprinting of ribosomal proteins on 16S rRNA. RNA 1, 194–209. [PMC free article] [PubMed] [Google Scholar]
- Pyle, A. (2002). Metal ions in the structure and function of RNA. J. Biol. Inorg. Chem. 7, 679–690. [DOI] [PubMed] [Google Scholar]
- Qin, P.Z., and Pyle, A.M. (1998). The architectural organization and mechanistic function of group II intron structural elements. Curr. Opin. Struct. Biol. 8, 301–308. [DOI] [PubMed] [Google Scholar]
- Rambo, R.P., and Doudna, J.A. (2004). Assembly of an active group II intron-maturase complex by protein dimerization. Biochemistry 43, 6486–6497. [DOI] [PubMed] [Google Scholar]
- Rivier, C., Goldschmidt-Clermont, M., and Rochaix, J. (2001). Identification of an RNA-protein complex involved in chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J. 20, 1765–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha, R., Chen, B., Wank, H., Matsuura, M., Edwards, J., and Lambowitz, A. (1999). RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry 38, 9069–9083. [DOI] [PubMed] [Google Scholar]
- Shaul, O. (2002). Magnesium transport and function in plants: The tip of the iceberg. Biometals 15, 309–323. [DOI] [PubMed] [Google Scholar]
- Su, L.J., Brenowitz, M., and Pyle, A.M. (2003). An alternative route for the folding of large RNAs: Apparent two-state folding by a group II intron ribozyme. J. Mol. Biol. 334, 639–652. [DOI] [PubMed] [Google Scholar]
- Swisher, J., Duarte, C., Su, L., and Pyle, A. (2001). Visualizing the solvent-inaccessible core of a group II intron ribozyme. EMBO J. 20, 2051–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till, B., Schmitz-Linneweber, C., Williams-Carrier, R., and Barkan, A. (2001). CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA 7, 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor, N., Hausner, G., and Zimmerly, S. (2001). Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA 7, 1142–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadkhan, S., and Manley, J. (2001). Splicing-related catalysis by protein-free snRNAs. Nature 413, 701–707. [DOI] [PubMed] [Google Scholar]
- Villa, T., Pleiss, J., and Guthrie, C. (2002). Spliceosomal snRNAs: Mg2+-dependent chemistry at the catalytic core? Cell 109, 149–152. [DOI] [PubMed] [Google Scholar]
- Vogel, J., Boerner, T., and Hess, W. (1999). Comparative analysis of splicing of the complete set of chloroplast group II introns in three higher plant mutants. Nucleic Acids Res. 27, 3866–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J., Hubschmann, T., Borner, T., and Hess, W. (1997). Splicing and intron-internal RNA editing of trnK-matK transcripts in barley plastids: Support for MatK as an essential splice factor. J. Mol. Biol. 270, 179–187. [DOI] [PubMed] [Google Scholar]
- Wank, H., SanFilippo, J., Singh, R., Matsuura, M., and Lambowitz, A. (1999). A reverse transcriptase/maturase promotes splicing by binding at its own coding segment in a group II intron RNA. Mol. Cell 4, 239–250. [DOI] [PubMed] [Google Scholar]
- Weeks, K.M., and Cech, T.R. (1995). Efficient protein-facilitated splicing of the yeast mitochondrial bI5 intron. Biochemistry 34, 7728–7738. [DOI] [PubMed] [Google Scholar]
- Wong, I., and Lohman, T. (1993). A double-filter method for nitrocellulose-filter binding: Application to protein-nucleic acid interactions. Proc. Natl. Acad. Sci. USA 90, 5428–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.