Abstract
Purpose
Implantation of the mammalian embryo in the uterus is preceded by escape from the zona pellucida. In some species, hatching from the zona occurs preferentially from one or the other poles of the embryo. The situation for the bovine embryo, in which hatching precedes attachment to the uterus by more than a week, is unclear. The purpose was to describe whether hatching of the bovine embryo from the zona pellucida occurs preferentially from the embryonic or abembryonic pole.
Methods
Bovine blastocysts undergoing hatching were examined by light microscopy (n = 84) and epifluorescence imaging using antibodies for markers of epiblast, hypoblast, and trophectoderm (TE) (n = 26). The location of hatching was classified as being at the embryonic pole, if hatching occurred ipsilateral to the inner cell mass (ICM), or abembryonic, if hatching occurred contralateral to the ICM.
Results
A total of 55% of blastocysts exited the zona pellucida through an opening at the embryonic pole. In these cases, 68% of the cells emerging through the zona pellucida were derived from the ICM. The remainder of blastocysts hatched from an opening either contralateral or to the side of the ICM. In these cases, 87% of hatched cells were TE.
Conclusion
For the bovine embryo, there is nearly equal probability of hatching from the embryonic or abembryonic poles. Given that the surface area of the zona pellucida in contact with the TE overlying the ICM is less than for the remainder of the blastocyst, there is some preference for hatching through the embryonic pole. Thus, the bovine embryo is distinct from the mouse and human, where hatching occurs preferentially at the abembryonic pole.
Keywords: Bovine, Embryo, Blastocysts, Hatching, Embryonic pole, Abembryonic pole
Introduction
Hatching from the zona pellucida is a prerequisite for the preimplantation embryo to attach to the uterus and initiate placentation. Failure of the process could conceivably lead to pregnancy loss, as indicated by some studies where assisted hatching improved clinical pregnancy rate in women [1] and cattle [2]. Three mechanisms are known to be involved in the process of blastocyst hatching: mechanical forces exerted on the zona pellucida by blastocyst expansion, [3–5], weakening of the zona pellucida by enzymatic degradation [6–8], and penetration of the zona pellucida by projections of trophectodermal cells [9, 10]. The relative importance of these mechanisms varies between species. In the hamster, for example, the blastocoelic cavity shrinks in size prior to hatching [10]. There is also variation between species in the nature of the proteinases implicated in dissolution of the zona pellucida including a trypsin-like enzyme in the mouse [11, 12], cathepsins in the hamster [13] and a urokinase-type plasminogen activator in cattle [8, 14].
There is evidence that the blastocyst preferentially hatches from the abembryonic pole (i.e., opposite the inner cell mass (ICM) and involving mural trophectoderm (TE)) regardless of whether attachment of the blastocyst to the endometrium occurs at the abembryonic (guinea pig, hamster, mouse) or embryonic pole (human). Hatching is more frequent from the abembryonic pole in mice [12] and humans [15]. Also, trophectodermal projections predominate in this part of the TE for guinea pig [16], hamster [9] and human, [15], and trypsin-like proteinase is limited to mural TE in mice [12].
All of the species mentioned in the previous paragraph undergo implantation in the uterine endometrium shortly after hatching. Whether species that undergo a prolonged period of time after hatching before attachment to the endometrium are similarly polarized with respect to the site of hatching has not been established. The cow is one such species. While hatching occurs about 7 to 10 days after fertilization [17], the first attachments between TE and endometrium do not occur for about another 10 days, at day 20 of gestation [18]. In the only study conducted to date, it was found that 48% of bovine embryos hatched through an opening in the zona pellucida near the embryonic pole while the remainder hatched from either the TE near the side of the ICM (embryonic mural TE; 36%) or from the abembryonic polar TE (16%) [19]. Here, we reexamined the question of the location of hatching through the zona pellucida in the bovine using a combination of light microscopy and epifluorescence microscopy of embryos labeled with various markers of cell lineage. Among the markers used were CDX2 and YAP1, both markers of TE [20, 21], NANOG, which is specific to epiblast cells of the ICM [22] and GATA6, which is most abundant in cells of the hypoblast [22].
Materials and methods
In vitro production of embryos
Production of embryos was performed as described earlier [23], using sperm and oocytes from a mixture of animals of various breeds including Bos taurus, Bos indicus, and admixture of the two genetic groups. Embryos were cultured in groups of 30 in 50 μl oil-covered microdrops of a serum-free culture medium, synthetic oviduct fluid-bovine embryo 2 (SOF-BE2) [24] at 38.5 °C in a humidified environment consisting of 5% (v/v) O2, 5% (v/v) CO2, and the balance nitrogen.
A total of 110 hatching blastocysts at days 7 or 8 after insemination were collected for analysis. Each of these blastocysts had an ICM that could be clearly identified using a digital inverted microscope (Evos® FL, Thermo Fisher Scientific, Waltham, MA, USA). A subset of these blastocysts (n = 26) were also subjected to analysis by immunofluorescence.
Immunolocalization of cells labeled with epiblast, hypoblast, and TE markers
A set of hatching blastocysts were labeled using Hoescht 33342 (to label all nuclei) and a combination of two antibodies against GATA6 and CDX2 (n = 4), CDX2 and YAP1 (n = 2), or GATA6 and NANOG (n = 8). In addition, another set of hatching blastocysts were labeled using Hoescht 33342 and either CDX2 (n = 8), β-catenin (n = 3), or non-phospho (active) β-catenin (n = 1). Primary antibodies used were mouse monoclonal antibody against CDX2 (Biogenex, Fremont, CA, USA); rabbit polyclonal antibody against human GATA6 (Santa Cruz Biotechnology, Dallas, TX, USA); rabbit monoclonal anti-YAP1 (Cell Signaling Technology, Danver, MA, USA); rabbit polyclonal anti-β-catenin (Abcam, Cambridge, MA, USA); rabbit monoclonal anti non-phospho (active) β-catenin (Cell Signaling Technology)l and mouse polyclonal antibody against human NANOG (eBioscience, San Diego, CA, USA). The secondary antibodies were fluorescein isothiocyanate (FITC) conjugated goat polyclonal anti-mouse IgG (Abcam) and Alexa Fluor 555 conjugated goat polyclonal anti-rabbit IgG (ThermoFisher). All antibodies were used at 1 μg/ml except for mouse monoclonal antibody against CDX2 (Biogenex, Fremont, CA, USA), which was used at the working concentration provided by the manufacturer. Non-specific binding was evaluated by substituting IgG for the primary antibody.
All steps for immunolocalization proceeded at room temperature unless otherwise stated. Briefly, blastocysts were collected, washed three times in Dulbecco’s phosphate buffered saline (DPBS) containing 0.1% (w/v) polyvinylpyrrolidone (PVP) (DPBS/PVP), fixed for 15 min in 4% (w/v) paraformaldehyde diluted in DPBS/PVP, washed three times in PBS/PVP, incubated for 30 min in permeabilization buffer [DPBS/PVP containing 0.25% (v/v) Triton X-100], and then incubated for 1 h in blocking buffer [5% (w/v) bovine serum albumin (BSA) in DPBS]. Blastocysts were then incubated overnight with the first primary antibody at 4 °C, washed three times in washing buffer [DPBS containing 0.1% (w/v) BSA and 0.1% (v/v) Tween-20], and for 1 h in secondary antibody. The immunolabeling procedure was then repeated with a second primary antibody for those blastocysts labeled with two primary antibodies. Following labeling with antibodies, blastocysts were washed three times in washing buffer, incubated with 1 μg/ml Hoescht 33342 in DPBS/PVP for 15 min to label nuclei, washed once in DPBS/PVP and mounted on glass slides in 5–10 μl of SlowFade Gold antifade reagent (ThermoFisher Scientific). Blastocysts were visualized at ×40 objective using a Zeiss Axioplan 2 epifluorescence microscope (Zeiss, Göttingen, Germany) and Zeiss filter sets 02 [4′,6-diamidino-2-phenylindole (DAPI)], 03 (FITC), and 04 (rhodamine). Digital images were acquired using AxioVision software (Zeiss) and a high-resolution black and white Zeiss AxioCam MRm digital camera. ImageJ V. 1.48 (National Institutes of Health, Bethesda, MD, USA) was used to visualize images, count the number of cells, and measure the embryo diameter and length of the hatching opening.
One hatching blastocyst was subjected to confocal microscopy after immunolocalization of GATA6 and NANOG. The blastocyst was examined on a spinning disk confocal scanner mounted on an Olympus DSU-IX81 inverted fluorescent microscope. Images were captured with a ×40 objective using 4′,6-diamidino-2-phenylindole (DAPI), FITC and red fluorescent protein (RFP) filter sets. Digital images were taken using an attached Hamamatsu C4742-80-12AG monochrome CCD camera. SlideBook 6 Reader (Intelligent Imaging Innovations, Inc., Denver, CO, USA) was used to visualize images and count total number of cells.
Identification of cell types and embryonic poles
Hatching embryos were separated in two categories based on the location of the hatching opening: (1) embryonic pole, if hatching occurred ipsilateral to the ICM or (2) abembryonic pole, if hatching occurred from the opposite end to the ICM. Because of occasional difficulties in assigning exact location of the initial site of penetration of the zona pellucida, the abembryonic group included blastocysts in which hatching occurred from the lateral side of the embryo. The orientation of the hatching site was determined by locating the ICM by light microscopy, and for embryos that were immunolabeled, by examining the cell type present in the hatched portion of the blastocyst. Nuclei that were either NANOG+, bright GATA6+, YAP1−, or CDX2− were considered to be ICM. Nuclei that were CDX2+ cells, dim GATA6+, or YAP1+ were considered TE. Immunoreactive β-catenin was detected on the membrane of all cells but was more intense for cells of the ICM. Accordingly, cells with bright β-catenin labeling were considered ICM and cells with less intense labeling were considered TE.
Statistical analysis
Data were analyzed using the SAS v 9.4 software package (SAS Institute Inc., Cary, NC, USA); embryo was considered the experimental unit. The frequency procedure (Proc FREQ) was used to calculate the proportion of blastocysts that hatched from the embryonic and abembryonic pole. Differences between the two types of embryos in terms of proportion of ICM and TE cells in the hatched portion of the embryo was determined by analysis of variance using the generalized linear models procedure (Proc GLM) of SAS. Data shown are least-squares means ± SEM.
Results
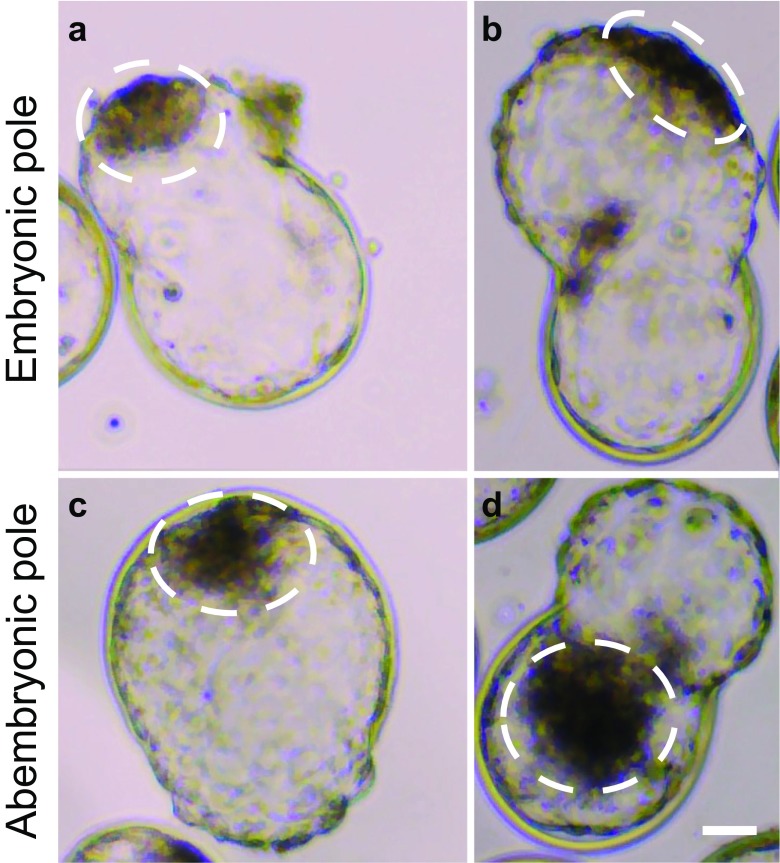
Examples of blastocysts hatching from the embryonic and abembryonic poles as determined by light microscopy are shown in Fig. 1. A total of 55% (60/110) of blastocysts hatched through the embryonic pole and 45% (50/110) through the abembryonic pole. Of these 50 embryos, 31 hatched from the lateral TE (i.e., to the side of the ICM) and 19 from the contralateral TE (i.e., opposite from the ICM). Note that, in many cases, blastocysts were examined when hatching was extensive and classification as to lateral vs contralateral locations is tentative. There was no difference in frequency between day 7 (55% embryonic pole vs 45% abembryonic pole) and day 8 (54% embryonic pole vs 46% abembryonic pole) (Table 1).
Fig. 1.
Representative images of embryos hatching through the embryonic or abembryonic pole. a, b Embryos escaping the zona pellucida through the embryonic pole. c, d Embryos hatching through the abembryonic pole completely opposite to the ICM (c, d). The area encircled with the dotted line represents the inner cell mass. Scale bar = 50 μM
Table 1.
Percent and frequency of embryos hatching from the embryonic or abembryonic pole at days 7 and 8
| Hatching pole related to the inner cell mass (ICM) | Embryonic | Abembryonic | |
|---|---|---|---|
| Adjacent to the ICM | Lateral to the ICM | Opposite to the ICM | |
| Day 7 | 55% (32/58) | 28% (16/58) | 17% (10/58) |
| Day 8 | 54% (28/52) | 29% (15/52) | 17% (9/52) |
A total of 110 embryos were evaluated after bright field and epifluorescence microscopy imaging; 58 at day 7 and 52 at day 8
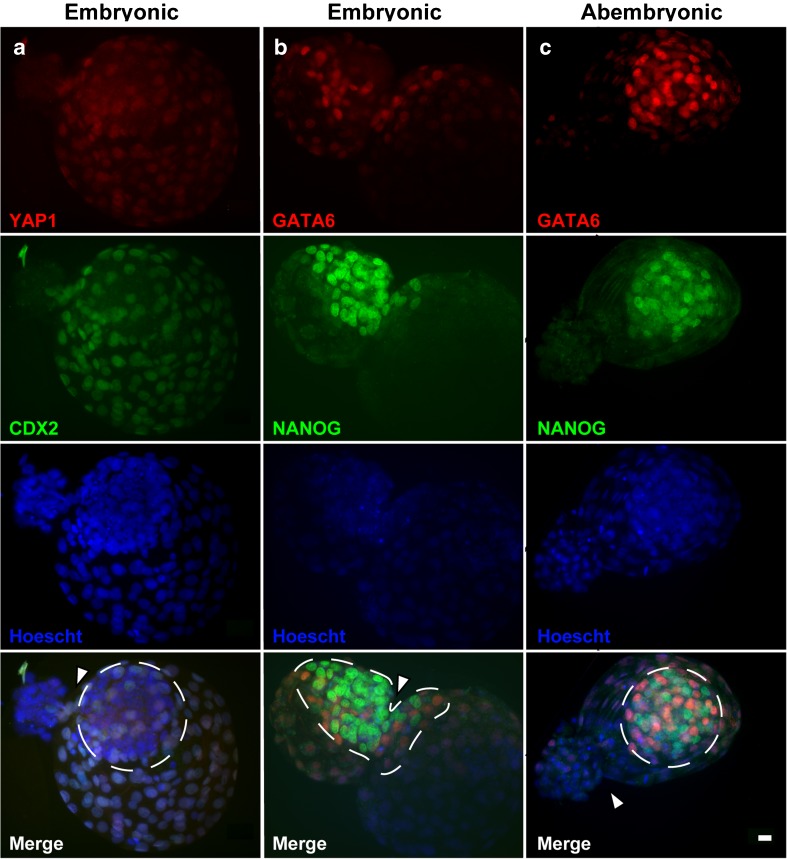
Use of immunofluorescence to examine the cells that had passed through the zona pellucida demonstrated how the site of hatching affects the composition of the hatched portion of the blastocyst. For blastocysts hatching through the embryonic pole, the hatched portion of the blastocyst contained cells of TE and/or ICM origin. For example, the embryo in Fig. 2a, none of the cells in the hatched area expressed the TE markers CDX2 or YAP1. For the blastocyst in Fig. 2b, the hatched area contained numerous cells that were positive for the ICM marker NANOG but also cells negative for ICM markers. In contrast, when hatching was through the abembryonic pole, all or most cells in the hatched region were TE. For example, in Fig. 2c, the hatched region was devoid of NANOG+ and bright GATA6+ cells. Overall, the proportion of cells in the hatched portion of the blastocyst that was ICM was higher (P < 0.0001) for blastocysts experiencing hatching through the embryonic pole than for blastocysts hatching through the abembryonic pole (68.3 vs 13.0%) (Table 2). In addition, 49.3% of the cells of the ICM were in the hatched portion of the blastocyst for those hatching through the embryonic pole vs 8.1% for those hatching through the abembryonic pole (P < 0.0001; Table 2).
Fig. 2.
Examples of immunolocalization of inner cell mass (ICM) and trophectoderm (TE) in blastocyst experiencing hatching through the embryonic pole (a, b) and abembryonic pole (c). Immunofluorescence was evaluated using epifluorescence microscopy. a Blastocyst labeled with anti-YAP1 (red) and anti-CDX2 (green). b, c Blastocysts labeled with anti-GATA6 (red) and anti-NANOG (green). Nuclei were labeled with Hoescht 33342 (blue). For each blastocyst, immunofluorescence is shown separately for the red, green, and blue channels. a ICM cells were identified as those were the nuclei were YAP1− and CDX2− while TE cells had nuclei that were YAP1+ and CDX2+. b, c Cells of the ICM that are epiblast are those with nuclei that are NANOG+; cells of the ICM that are hypoblast are those with nuclei that have bright GATA6+. Cells of the TE are those with nuclei that are NANOG− and have dim GATA6+. b Fifty percent of the ICM cells are in the hatched area and 50% remain inside. c The hatched area devoid of NANOG+ and bright GATA6+ nuclei. The area encircled with the dotted line represents the ICM. The white arrowheads indicate the opening through which the embryo is exiting. Scale bar = 20 μM
Table 2.
Proportion of hatched cells that were inner cell mass (ICM) and trophectoderm (TE) as affected by hatching pole
| Hatching pole | Embryonic | Abembryonic |
|---|---|---|
| Percent of hatched cells that were ICM | 68.3 ± 5.6*** | 13.0 ± 10.2 |
| Percent of ICM cells that hatched | 49.3 ± 5.2*** | 8.1 ± 9.6 |
A total of 26 embryos (7 at day 7 and 19 at day 8) were evaluated by epifluorescence microscopy
***Values with asterisks indicate significant difference between groups (P < 0.001)
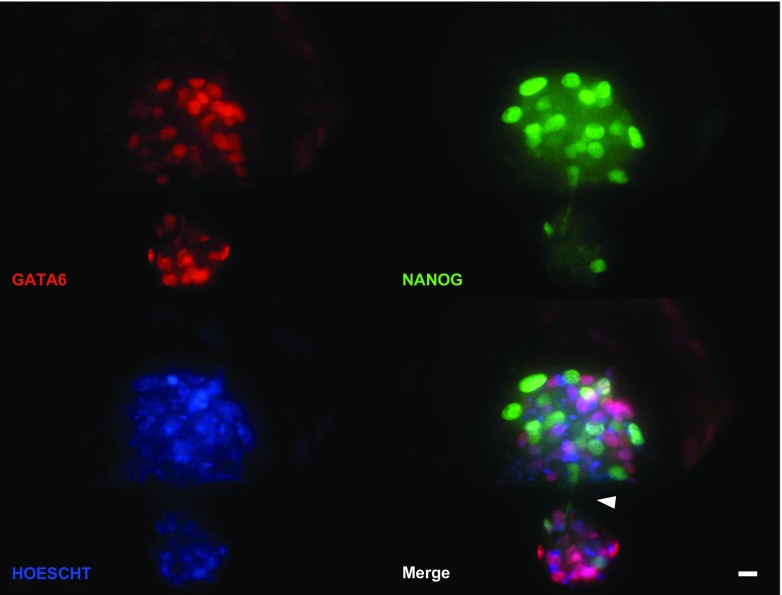
One hatching blastocyst labeled with antibodies to NANOG and GATA6 was examined by confocal microscopy (Fig. 3). This embryo was hatching through the embryonic pole. Sections taken through the plane of focus where the zona pellucida had been penetrated by cells of the blastocyst show clearly that the ICM has been stretched across the opening in zona pellucida with the hatching portion outside the zona pellucida, the larger inner portion still within the zona, and with two NANOG+ cells on either side of the opening—one that is passing through the zona and another that appears to be following behind the first cell.
Fig. 3.
Analysis of a blastocyst hatching through the embryonic pole using confocal microscopy. The blastocyst was labeled using antibodies against NANOG (green) and GATA6 (red). Nuclei were labeled with Hoescht 33342 (blue). Cells of the inner cell mass that are epiblast are NANOG+ and GATA6− while hypoblast cells are NANOG− and GATA6+. The white arrowhead points to the hatching opening. Note the pair of NANOG+ epiblast cells (pointed by the arrowhead) exiting the zona pellucida. Scale bar = 20 μM
Discussion
Present results confirm earlier results using light microscopy [19] that the in vitro developed bovine blastocyst can hatch from either the embryonic or abembryonic pole, with about 50% of blastocysts experiencing hatching through the embryonic pole. Given that less than 50% of the surface area of the zona pellucida is adjacent to the ICM, a preference for hatching through the embryonic pole is indicated. Results also confirm earlier results [19] that, when hatching does not occur through the embryonic pole, it is more likely to commence in a region of the TE lateral to the ICM than directly opposite to it. The present results extend earlier findings [19] by demonstrating how the site of hatching affects the composition of the hatched portion of the blastocyst. When hatching is from the embryonic pole, a majority of the cells in the hatched region of the blastocyst are ICM whereas TE cells predominate in the hatched region of blastocysts hatching from the abembryonic pole. Thus, the nature of the first physical contact of the cells of the embryo with the female reproductive tract is different for blastocysts hatching from the embryonic vs abembryonic pole. In the cow, hatching takes place in the uterus [17] and it remains to be seen whether the endometrium responds differently to a blastocyst hatching from the embryonal vs abembryonal poles. This is a possibility because gene expression varies between ICM and TE [25] and recent experiments in cattle indicate that the cleavage-stage embryo can interact with the oviduct to change gene expression [26, 27].
The cow is distinct from other species studied because abembryonal hatching predominates in the mouse [12], human [15], guinea pig [16], and hamster [28]. The reason for the difference is not known. Except for the cow, all of the above-named species attach to the endometrium soon after hatching whereas the bovine blastocyst resides in the uterus for 10 or more days after hatching before attaching to the endometrium. Perhaps orientation of hatching is less critical in species where the embryo spends a prolonged period free of permanent attachment to the endometrium. Examination of the location of hatching in species that share this characteristic with the cow (sheep, pig, and horse) could provide illumination on this point.
It also remains to be determined why some bovine blastocysts hatch from one location whereas others hatch from another location. In species in which hatching is biased towards the abembryonal pole, trophectodermal projections and proteinase activity is localized to this region [12, 15, 16, 28]. One possibility is that trophectodermal projections or proteinase activity develops more uniformly in the bovine blastocyst and that the site of hatching depends on physical characteristics of the zona pellucida. Alternatively the specific location of trophectodermal projections or proteinase activity could vary between embryos.
Acknowledgements
Verónica Negrón-Pérez was supported by a McKnight Doctoral Fellowship from the Florida Education Fund, Inc. The authors thank owners and employees of Central Beef Packing Co. (Center Hill, FL), Adena Meat Products L.P. (Fort McCoy, FL), and Florida Beef Inc. (Zolfo Springs, FL) for providing ovaries; William Rembert and Eddie Cummings for ovary collection; and Doug Smith and the McKnight Brain Institute Cell Tissue and Analysis Core of the University of Florida for assistance with imaging and processing of confocal microscopy figures.
Compliance with ethical standards
Funding
This study was funded by USDA-NIFA AFRI Grant No. 2011-67015-30688 and the L.E. “Red” Larson Endowment.
Conflict of interest
The authors declare that they have no conflict of interest. This article does not contain any studies with human participants or animals performed by any of the authors.
Grant support
USDA-NIFA AFRI Grant No. 2011-67015-30688.
References
- 1.Carney S-KK, Das S, Blake D, Farquhar C, Seif MWMW, Nelson L. Assisted hatching on assisted conception (in vitro fertilisation (IVF) and intracytoplasmic sperm injection (ICSI)) Cochrane Database Syst Rev. 2012;12:CD001894. doi: 10.1002/14651858.CD001894.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taniyama A, Watanabe Y, Nishino Y, Inoue T. Assisted hatching of poor-quality bovine embryos increases pregnancy. J Reprod Dev. 2011;57:543–546. doi: 10.1262/jrd.10-096T. [DOI] [PubMed] [Google Scholar]
- 3.Cole RJ. Cinemicrographic observations on the trophoblast and zona pellucida of the mouse blastocyst. J Embryol Exp Morphol. 1967;17:481–490. [PubMed] [Google Scholar]
- 4.Massip A, Mulnard J. Time-lapse cinematographic analysis of hatching of normal and frozen-thawed cow blastocysts. J Reprod Fertil. 1980;58:475–478. doi: 10.1530/jrf.0.0580475. [DOI] [PubMed] [Google Scholar]
- 5.Massip A, Mulnard J, Vanderzwalmen P, Hanzen C, Ectors F. The behaviour of cow blastocyst in vitro: cinematographic and morphometric analysis. J Anat. 1982;134:399–405. [PMC free article] [PubMed] [Google Scholar]
- 6.Sawada H, Yamazaki K, Hoshi M. Trypsin-like hatching protease from mouse embryos: evidence for the presence in culture medium and its enzymatic properties. J Exp Zool. 1990;254:83–87. doi: 10.1002/jez.1402540112. [DOI] [PubMed] [Google Scholar]
- 7.Mishra A, Seshagiri PB. Evidence for the involvement of a species-specific embryonic protease in zona escape of hamster blastocysts. Mol Hum Reprod. 2000;6:1005–1012. doi: 10.1093/molehr/6.11.1005. [DOI] [PubMed] [Google Scholar]
- 8.Berg DA, Menino AR., Jr Bovine embryos produce urokinase-type plasmogen activator. Mol Reprod Dev. 1992;31:14–19. doi: 10.1002/mrd.1080310104. [DOI] [PubMed] [Google Scholar]
- 9.Gonzales DS, Jones JM, Pinyopummintr T, Carnevale EM, Ginther OJ, Shapiro SS, Bavister BD. Trophectoderm projections: a potential means for locomotion, attachment and implantation of bovine, equine and human blastocysts. Hum Reprod. 1996;11:2739–2745. doi: 10.1093/oxfordjournals.humrep.a019201. [DOI] [PubMed] [Google Scholar]
- 10.Seshagiri PB, Sen Roy S, Sireesha G, Rao RP. Cellular and molecular regulation of mammalian blastocyst hatching. J Reprod Immunol. 2009;83:79–84. doi: 10.1016/j.jri.2009.06.264. [DOI] [PubMed] [Google Scholar]
- 11.O’Sullivan CM, Rancourt SL, Liu SY, Rancourt DE. A novel murine tryptase involved in blastocyst hatching and outgrowth. Reproduction. 2001;122:61–71. doi: 10.1530/rep.0.1220061. [DOI] [PubMed] [Google Scholar]
- 12.Perona RM, Wassarman PM. Mouse blastocysts hatch in vitro by using a trypsin-like proteinase associated with cells of mural trophectoderm. Dev Biol. 1986;114:42–52. doi: 10.1016/0012-1606(86)90382-9. [DOI] [PubMed] [Google Scholar]
- 13.Sireesha GV, Mason RW, Hassanein M, Tonack S, Navarrete Santos A, Fischer B, Seshagiri PB. Role of cathepsins in blastocyst hatching in the golden hamster. Mol Hum Reprod. 2008;14:337–346. doi: 10.1093/molehr/gan026. [DOI] [PubMed] [Google Scholar]
- 14.Coates AA, Menino AR. Effects of blastocoelic expansion and plasminogen activator activity on hatching and zona pellucida solubility in bovine embryos in vitro. J Anim Sci. 1994;72:2936–2942. doi: 10.2527/1994.72112936x. [DOI] [PubMed] [Google Scholar]
- 15.Sathananthan H, Menezes J, Gunasheela S. Mechanics of human blastocyst hatching in vitro. Reprod BioMed Online. 2003;7:228–234. doi: 10.1016/S1472-6483(10)61757-9. [DOI] [PubMed] [Google Scholar]
- 16.Spee GF. Beitrag zur entwickelungsgeschichte der fruheren stadien des meerschweinchens bis zur vollendung der keimblase. Arch Anat Physiol. 1883;7:44–60. [Google Scholar]
- 17.Betteridge KJ, Flechon JE. The anatomy and physiology of pre- attachement bovine embryos. Theriogenology. 1988;29:155–187. doi: 10.1016/0093-691X(88)90038-6. [DOI] [Google Scholar]
- 18.King GJ, Atkinson BA, Robertson HA. Development of the intercaruncular areas during early gestation and establishment of the bovine placenta. J Reprod Fert. 1981;61:469–474. doi: 10.1530/jrf.0.0610469. [DOI] [PubMed] [Google Scholar]
- 19.Niimura S, Ogata T, Okimura A, Sato T, Uchiyama Y, Seta T, Nakagawa H, Nakagawa K, Tamura Y. Time-lapse videomicrographic observations of blastocyst hatching in cattle. J Reprod Dev. 2010;56:649–654. doi: 10.1262/jrd.10-069H. [DOI] [PubMed] [Google Scholar]
- 20.Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- 21.Chen L, Wang D, Wu Z, Ma L, Daley GQ. Molecular basis of the first cell fate determination in mouse embryogenesis. Cell Res. 2010;20:982–993. doi: 10.1038/cr.2010.106. [DOI] [PubMed] [Google Scholar]
- 22.Denicol AC, Block J, Kelley DE, Pohler KG, Dobbs KB, Mortensen CJ, Ortega MS, Hansen PJ. The WNT signaling antagonist Dickkopf-1 directs lineage commitment and promotes survival of the preimplantation embryo. FASEB J. 2014:1–12. [DOI] [PMC free article] [PubMed]
- 23.Ortega MS, Wohlgemuth S, Tribulo P, Siqueira LGB, Null DJ, Cole JB, Da Silva M V., Hansen PJ. A single nucleotide polymorphism in COQ9 affects mitochondrial and ovarian function and fertility in Holstein cows. Biol Reprod 2017; 0:1–12. [DOI] [PubMed]
- 24.Kannampuzha Francis J, Tribulo P, Hansen PJ. Actions of activin A, connective tissue growth factor, hepatocyte growth factor and teratocarcinoma—derived growth factor 1 on the development of the bovine preimplantation embryo. Reprod Fertil Dev 2017:1–13 (in press). [DOI] [PubMed]
- 25.Nagatomo H, Kagawa S, Kishi Y, Takuma T, Sada A, Yamanaka K-I, Abe Y, Wada Y, Takahashi M, Kono T, Kawahara M. Transcriptional wiring for establishing cell lineage specification at the blastocyst stage in cattle. Biol Reprod. 2013;88:158. doi: 10.1095/biolreprod.113.108993. [DOI] [PubMed] [Google Scholar]
- 26.Lonergan P, Forde N. Maternal-embryo interaction leading up to the initiation of implantation of pregnancy in cattle. Animal. 2014;8(Suppl 1):64–69. doi: 10.1017/S1751731114000470. [DOI] [PubMed] [Google Scholar]
- 27.Gómez E, Muñoz M. Multiple-embryo transfer for studying very early maternal-embryo interactions in cattle. Reproduction. 2015;150:R35–R43. doi: 10.1530/REP-14-0465. [DOI] [PubMed] [Google Scholar]
- 28.Gonzales DS, Bavister BD. Zona pellucida escape by hamster blastocysts in vitro is delayed and morphologically different compared with zona escape in vivo. Biol Reprod. 1995;52:470–480. doi: 10.1095/biolreprod52.2.470. [DOI] [PubMed] [Google Scholar]