Abstract
Tomato (Lycopersicon esculentum) Cf genes confer resistance to the fungal pathogen Cladosporium fulvum through recognition of secreted avirulence (Avr) peptides. Plant defense responses, including rapid alterations in gene expression, are immediately activated upon perception of the pathogen. Previously, we identified a collection of Avr9/Cf-9 rapidly (15 to 30 min) elicited (ACRE) genes from tobacco (Nicotiana tabacum). Many of the ACRE genes encode putative signaling components and thus may play pivotal roles in the initial development of the defense response. To assess the requirement of 42 of these genes in the hypersensitive response (HR) induced by Cf-9/Avr9 or by Cf-4/Avr4, we used virus-induced gene silencing (VIGS) in N. benthamiana. Three genes were identified that when silenced compromised the Cf-mediated HR. We further characterized one of these genes, which encodes a Ser/Thr protein kinase called Avr9/Cf-9 induced kinase 1 (ACIK1). ACIK1 mRNA was rapidly upregulated in tobacco and tomato upon elicitation by Avr9 and by wounding. Silencing of ACIK1 in tobacco resulted in a reduced HR that correlated with loss of ACIK1 transcript. Importantly, ACIK1 was found to be required for Cf-9/Avr9- and Cf-4/Avr4-mediated HRs but not for the HR or resistance mediated by other resistance/Avr systems, such as Pto/AvrPto, Rx/Potato virus X, or N/Tobacco mosaic virus. Moreover, VIGS of LeACIK1 in tomato decreased Cf-9–mediated resistance to C. fulvum, showing the importance of ACIK1 in disease resistance.
INTRODUCTION
Plant cells induce an array of defense responses upon perception of pathogen-derived molecules. General elicitors that are characteristic of whole classes of microorganisms, such as bacterial flagellin, can trigger resistance (Gómez-Gómez and Boller, 2002). However, disease resistance is often governed by a gene-for-gene interaction (Dangl and Jones, 2001), in which plants carrying a resistance (R) gene specifically recognize a pathogen carrying a corresponding avirulence (Avr) gene. R gene–mediated resistance, or race-specific resistance, is typically accompanied by rapid localized cell death at the site of infection called the hypersensitive response (HR).
Cladosporium fulvum is a biotrophic fungus that causes leaf mold in tomato (Lycopersicon esculentum). The tomato–C. fulvum interaction is useful for studying the molecular basis of race-specific resistance (Rivas and Thomas, 2002). Tomato Cf genes confer resistance to specific races of C. fulvum through recognition of Avr peptides that are secreted into the leaf apoplast during infection. For example, the R genes Cf-4 and Cf-9 from tomato confer resistance to strains of C. fulvum that express Avr4 and Avr9, respectively. Avr4 and Avr9 encode precursor proteins that are processed upon secretion to mature proteins of 86 and 28 amino acids, respectively (Van den Ackerveken et al., 1993; Joosten et al., 1994). Cf genes encode type I transmembrane glycoproteins carrying extracytoplasmic Leu-rich repeats (LRRs), a single pass membrane-spanning region, and a short cytoplasmic domain that has no similarity to known signaling domains (Thomas et al., 1998). The Cf-4 and Cf-9 proteins have identical C-terminal regions with sequence variation confined to their N-terminal LRRs. Extensive binding studies have failed to detect a direct interaction between Cf-9 and Avr9 (Luderer et al., 2001), suggesting that perception of Avr9 by Cf-9 is indirect and possibly involves a high affinity binding site that has been detected in membranes of solanaceous plants (Kooman-Gersmann et al., 1996).
Cf-4 and Cf-9 confer an Avr-dependent HR in tobacco (Hammond-Kosack et al., 1998; Thomas et al., 2000). We used Cf-9–transgenic tobacco and derived cell suspension cultures to characterize a variety of early responses to the Avr9 elicitor (Rivas and Thomas, 2002). These responses include changes in ion fluxes (Piedras et al., 1998; Blatt et al., 1999), production of active oxygen species (Piedras et al., 1998), and the activation of mitogen-activated protein kinases and a calcium-dependent protein kinase (CDPK) (Romeis et al., 1999, 2000, 2001). These events are accompanied by massive changes in gene expression (Durrant et al., 2000), which likely lead to a reprogramming of the cell toward defense. However, the molecular mechanisms by which Cf-9 activates these responses are not well understood.
Many signaling components acting downstream of R genes have been identified by forward genetic screens in tomato, barley (Hordeum vulgare), and especially in the model plant Arabidopsis thaliana (Martin et al., 2003). In tomato, Rcr-1 and Rcr-2 were found to be required for the function of Cf-9 (Hammond-Kosack et al., 1994), but the phenotypes associated with mutations in these genes did not allow molecular identification of the genes. The Rcr3 gene product is required for Cf-2 function but not the function of other Cf genes (Dixon et al., 2000). Rcr3 encodes a secreted Cys protease and likely functions upstream of Cf-2 (Krüger et al., 2002). Reverse genetics has been more successful in identifying genes required for Cf-9 function. Virus-induced gene silencing (VIGS) in Nicotiana benthamiana was used to show that silencing of the NtCDPK2 gene family causes a reduced HR mediated by Cf-4 and Cf-9 (Romeis et al., 2001). The HRs elicited by Cf-4/Avr4 and Cf-9/Avr9 were also suppressed by VIGS of SGT1 (Peart et al., 2002b). SGT1 is required for the function of SCF (Skp1-Cul1-F-box) ubiquitin ligases and other multiprotein complexes, indicating that ubiquitylation is important for Cf-mediated defense responses. VIGS of CITRX, which encodes a thioredoxin that interacts with the C-terminal cytoplasmic domain of Cf-9, causes an accelerated Cf-9/Avr9-triggered HR and causes increased resistance of tomato to C. fulvum (Rivas et al., 2004). The negative regulatory role of CITRX is important for the defense response of Cf-9 but not Cf-2.
In this study, we sought to identify additional components required for Cf-mediated defense responses. We previously used cDNA-amplified fragment length polymorphism (AFLP) analysis to identify transcripts whose expression patterns are rapidly altered during the Cf-9/Avr9-mediated defense response in tobacco (N. tabacum) cell cultures (Durrant et al., 2000). Many Avr9/Cf-9 rapidly elicited (ACRE) genes are predicted to encode regulatory proteins, including protein kinases and transcription factors. We have reannotated the ACRE genes and used VIGS to silence 42 ACRE genes. We have identified three genes that are important for generating the HRs elicited by Cf-9/Avr9 and Cf-4/Avr4. One of these genes encodes a protein kinase called ACIK1. ACIK1 was found to be an important component of the Cf-9 and Cf-4 signaling pathways but not other R gene pathways.
RESULTS
Functional Classification of ACRE Genes Using Transcript Tags
We previously identified a set of mainly active oxygen species–independent ACRE genes using cDNA-AFLP transcript profiling (Durrant et al., 2000). DNA sequences were obtained for 257 out of 290 differentially expressed cDNA-AFLP fragments. Comparison of these sequences to each other revealed considerable redundancy in the data set that was not previously recognized. Some of the AFLP fragments are nearly identical to each other. For example, AFLP fragment 2 is highly similar to AFLP fragments 123 (89% identity), 124 (95% identity), and 147 (93% identity). This is partly because tobacco is an allotetraploid originating from two distinct diploid species and has duplicate, but distinct, copies of each gene. Also, members of highly related gene family members may yield similar AFLP fragments. Because of this, only 227 out of the 257 sequenced fragments are truly distinct in the collection. We estimate that 1.6 transcript tags on average represent each full-length ACRE transcript (Durrant et al., 2000). Therefore, our collection of differentially expressed transcript tags represents ∼142 distinct genes.
We previously reported sequence similarity for 37 induced and five repressed cDNA-AFLP fragments (42 in total) (Durrant et al., 2000). We have refined the BLAST search criteria and recently searched updated genomic and EST databases for all 257 sequenced fragments. We used a two-step strategy: first, we searched nucleotide sequence databases with ACRE gene sequences, and when these revealed tomato or tobacco EST homologs, a longer sequence could then be used to search the Arabidopsis, rice (Oryza sativa), and other protein sequence databases. We have found similarities for 136 additional cDNA-AFLP fragments (178/257 in total), which represent ∼110 distinct genes (see Supplemental Table 1 online). Based on homology, transcript tags were classified into functional groups (Table 1; see Supplemental Table 1 online). Prominent amongst these groups are putative regulators of protein degradation (F-box, U-box, RING-H2, and BTB/POZ containing proteins), transcription factors (WRKY, Myb, C2H2 zinc finger, and AP2-type), and protein kinases, including receptor-like kinases. Other interesting examples include genes predicted to encode calcium binding proteins, protein phosphatases, lipases, an NDR/HIN1-like protein, AAA-type ATPases, a ligand-gated ion channel, a Cf-type R protein, a synaptobrevin-like protein, and a subunit of the Exo70 exocytosis complex.
Table 1.
Functional Classification of cDNA-AFLP Tag Sequences Altered in Cf-9/Avr9 Response
| Functional Class | Number of Tags |
|---|---|
| Metabolism | 27 |
| Transcription factors | 16 |
| Protein kinases/phosphatases | 14 |
| Protein degradation | 13 |
| Intracellular transport | 10 |
| Calcium binding proteins | 5 |
| Hormone responsive | 4 |
| Chloroplast associated | 4 |
| Pathogenesis-related genes | 3 |
| Resistance genes | 2 |
| Oxidative burst | 2 |
| Miscellaneous functions | 9 |
| Unknown function | 67 |
| No hits | 81 |
This updated bioinformatics study provides a much more complete picture of the types of genes elicited by Cf-9/Avr9 than was previously described (Durrant et al., 2000) and allowed a more informed choice of which ACRE genes to use for functional studies.
Generation of a Collection of Full-Length and Partial ACRE cDNAs
ACRE cDNA-AFLP fragments were typically in the size range of 50 to 150 bp. We wanted to obtain more sequence information for these induced transcripts to verify the homologies and then use these sequences for gene silencing. We previously isolated full-length cDNA clones for 13 ACRE genes (Table 2; Durrant et al., 2000). We obtained additional DNA sequence for a further 29 induced cDNA-AFLP fragments that were predicted to encode proteins that may play critical roles in the defense response (Table 2). Primers (primary and nested) were designed for each of these cDNA-AFLP fragments and 3′-rapid amplification of cDNA ends (RACE) was used to extend the sequence. 5′-RACE was also performed to obtain full-length cDNA sequence for eight of these ACRE genes. Individual clones were identified for which the sequence of the RACE fragments corresponded to that of the original cDNA-AFLP fragment.
Table 2.
Full-Length and Partial ACRE cDNA Sequences Used for VIGS
| ACRE Number | 3′-RACE Product (bp) | 5′-RACE Product (bp) | Full-Length cDNA Size (bp) | GenBank Accession Number | Top Match BlastX, E-Value | Description of Top Hits |
|---|---|---|---|---|---|---|
| 1 | 1166 | AF211527 | AAR37423, 8e-52 | EREBP transcription factor | ||
| 4 | 2833 | AF211528 | AAT37497, 0.0 | N resistance protein, truncated | ||
| 11 | 986 | AY775028 | AAM63226, 6e-89 | Protein kinase, S-type receptor | ||
| 14 | 476 | AY775029 | NP_567515, 4e-32 | Triacylglycerol lipase | ||
| 19 | 387 | AY775030 | AAM61056, 5e-33 | EF-hand calcium binding protein | ||
| 20 | 417 | AY775031 | NP_194458, 2e-35 | EF-hand calcium binding protein | ||
| 31 | 972 | AF211529 | Q09011, 1e-82 | EF-hand calcium binding protein | ||
| 34 | 164 | AY775032 | No hits | No hits | ||
| 36 | 805 | AY775033 | AAL11556, 8e-78 | Chloroplast nucleoid DNA binding protein, aspartyl protease | ||
| 44 | 954(+) | AY775034 | AAO48953, 0.0 | Lipoxygenase, LoxD-like | ||
| 57 | 286 | AY775035 | BAB90781, 2e-19 | Calmodulin-related | ||
| 65 | 475 | AF211539 | AAK95311, 6e-05 | Unknown | ||
| 75 | 733 | AF211540 | AAN60336, 5e-10 | Unknown | ||
| 76 | 458 | AY775036 | NP_565634, 4e-38 | NDR1/HIN1-like | ||
| 77 | 314 | AY775037 | BI209582, 3e-05 | Protein kinase | ||
| 101 | 443 | AY775038 | O23429, 3e-19 | Synaptobrevin-related | ||
| 102 | 892 | AY775039 | AAL77654, 4e-84 | AAA-type ATPase, mitochondrial | ||
| 111B | 1000 | AF211531 | AAQ88400, 1e-92 | EREBP transcription factor | ||
| 126 | 788 | 405 | 1173 | AY220477 | NP_197017, 5e-69 | WRKY transcription factor |
| 132 | 956 | AF211532 | NP_175785, 5e-42 | RING-H2 zinc finger protein | ||
| 137 | 1171 | AF211537 | NP_915467, 3e-42 | Unknown | ||
| 140 | 518 | AY775040 | NP_196452, 9e-38 | ABA-responsive protein, GRAM domain | ||
| 141 | 1593 | 1570 | 3178 | AY220478 | CAC29254, 0.0 | Ligand-gated ion channel protein |
| 146 | 927 | AF211533 | AAO42131, 1e-31 | Unknown | ||
| 150 | 362 | AY775041 | NP_172572, 9e-13 | Protein kinase | ||
| 151 | 492 | AY775042 | JQ0956, 2e-04 | Myb transcription factor | ||
| 169 | 867 | AF211534 | AAL50092, 5e-10 | Unknown | ||
| 180 | 621 | AF211538 | NP_568661, 2e-06 | Unknown | ||
| 189 | 1245 | 850 | 1879 | AY220479 | AAG21977, e-170 | F-box/LRR protein |
| 194 | 651 | AF211535 | NP_568598, 2e-33 | Unknown | ||
| 197 | 530 | AY775043 | NP_191075, 5e-26 | Exo70 exocyst complex subunit | ||
| 216 | 749 | 654 | 1351 | AY220480 | AAK16686, e-143 | Protein kinase, NAF domain |
| 231 | 1396 | AF211536 | BAB02626, e-136 | Glycosyl transferase | ||
| 236 | 671 | AY775044 | NP_177231, 8e-40 | Protein kinase | ||
| 246 | 754 | AY775045 | NP_173960, 1e-68 | α/β Hydrolase | ||
| 256 | 949 | AY775046 | S27754, 4e-93 | Protein kinase, S-type receptor | ||
| 261 | 724 | AY775047 | NP_200709, 4e-85 | Protein kinase, APK1-like | ||
| 264 | 645 | 1172 | 1744 | AY220481 | AAL07094, e-170 | Protein kinase |
| 271 | 783 | AY775048 | AAD39930, e-101 | Protein phosphatase 2A regulatory subunit | ||
| 275 | 960(+) | 541 | 1500(+) | AY220482 | AAD13301, e-174 | Cf-type resistance protein |
| 276 | 1132 | 1489 | 2444 | AY220483 | NP_174228, 0.0 | U-box/ARM repeat protein, ARC1-like |
| 284 | 806 | 873 | 1570 | AY220484 | T09640, e-127 | Protein phosphatase 2C |
Screening of ACRE Genes Required for Cf-Mediated Defense Response Using VIGS
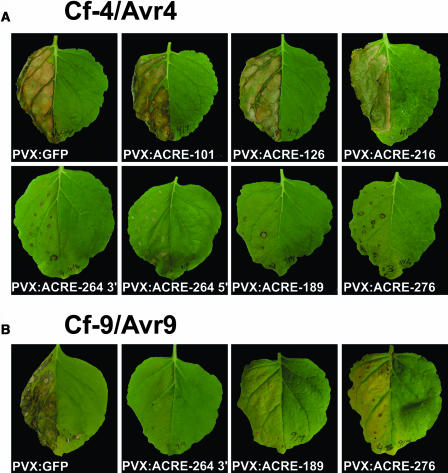
VIGS is a powerful tool for the analysis of gene function (Baulcombe, 1999). VIGS uses a viral-expressed transgene to trigger RNA interference (RNAi) against an endogenous gene. The silencing mechanism also targets closely related genes (more than ∼90% sequence identity). To examine the biological functions of ACRE genes in defense, the 42 ACRE full-length or partial cDNAs described above were subcloned into a Potato virus X (PVX) silencing vector (Lu et al., 2003a). We chose to use the extended ACRE cDNA sequences for VIGS instead of the smaller original cDNA-AFLP fragments because the use of fragments <150 bp has been shown to give less extensive and more transient silencing than larger fragments (Lu et al., 2003b). N. benthamiana plants were inoculated with PVX:ACRE gene or control PVX:GFP (green fluorescent protein) silencing constructs (Lu et al., 2003a). Three weeks after the initiation of silencing, leaf halves were infiltrated with Agrobacteria expressing Cf-4/Avr4 or Cf-9/Avr9. On PVX:GFP inoculated control plants, the HR induced by Cf-4/Avr4 and Cf-9/Avr9 occurred after 3 and 6 d, respectively (Figures 1A and 1B). Silencing of most of the ACRE genes had no effect on either Cf-4– or Cf-9–mediated HR, as shown for ACRE-101, ACRE-126, and ACRE-216 (Figure 1A). However, VIGS of three ACRE genes resulted in dramatically reduced HRs mediated by Cf-4 (Figure 1A) or by Cf-9 (Figure 1B). Two of these genes, ACRE-189 and ACRE-276, are predicted to encode proteins involved in ubiquitylation of protein substrates and will be the subject of future studies. We focus here on ACRE-264. The HR induced by Cf-4/Avr4 or Cf-9/Avr9 was abolished when either a 3′-RACE cDNA fragment of ACRE-264 or a nonoverlapping 5′-RACE cDNA fragment of ACRE-264 was used for VIGS (Figure 1). ACRE-264 is predicted to encode a protein kinase and is henceforth referred to as ACIK1 (for Avr/Cf-induced kinase 1).
Figure 1.
Identification of ACRE Genes Required for Cf-9– and Cf-4–Dependent HR.
VIGS in N. benthamiana using PVX. Seedlings were inoculated with the indicated PVX constructs to initiate silencing of the respective gene. PVX:GFP was used as a negative control.
(A) Three weeks after initiation of silencing, one leaf half was infiltrated with Agrobacteria that direct Cf-4 and Avr4 expression. Photographs of necrosis were taken 3 d after agroinfiltration.
(B) Three weeks after initiation of silencing, one leaf half was infiltrated with Agrobacteria that direct Cf-9 and Avr9 expression. Photographs of necrosis were taken 6 d after agroinfiltration.
NtACIK1 Is a Member of the Receptor-Like Cytoplasmic Kinase-VII Subfamily of Protein Kinases
The 131-bp NtACIK1 cDNA-AFLP fragment was used as a probe to screen a cDNA library established from Avr9-elicited tobacco cells (Durrant et al., 2000). Several independent cDNA clones were sequenced, and a clone containing the exact sequence of the NtACIK1 cDNA-AFLP fragment was selected for further study. We compared the sequences of the NtACIK1 cDNA clone with the 5′-RACE and 3′-RACE NtACIK1 sequences used for VIGS in N. benthamiana. The 5′-RACE sequence was 99.9% identical (1159/1160 bp) to the 5′-end of the NtACIK1 cDNA clone with the exception of an additional 12 bp of 5′-end untranslated sequence. The 3′-RACE sequence was 94.3% identical (510/541 bp) to the 3′-end of the NtACIK1 cDNA clone and had an additional 118 bp of 3′-end untranslated sequence. This indicates that the 5′-RACE cDNA fragment likely corresponds to the same gene as the NtACIK1 cDNA clone and that the 3′-RACE cDNA fragment corresponds to a highly related but distinct gene. Gel blot hybridizations were performed with tobacco genomic DNA digested with EcoRI and EcoRV under high-stringency conditions using the full-length cDNA of NtACIK1 as a probe. Two hybridizing bands were observed for N. tabacum and one band for N. sylvestris (data not shown). This indicates that there are two closely related genes of NtACIK1 in N. tabacum, each originating from its ancestral parents N. sylvestris and N. tomentosiformis.
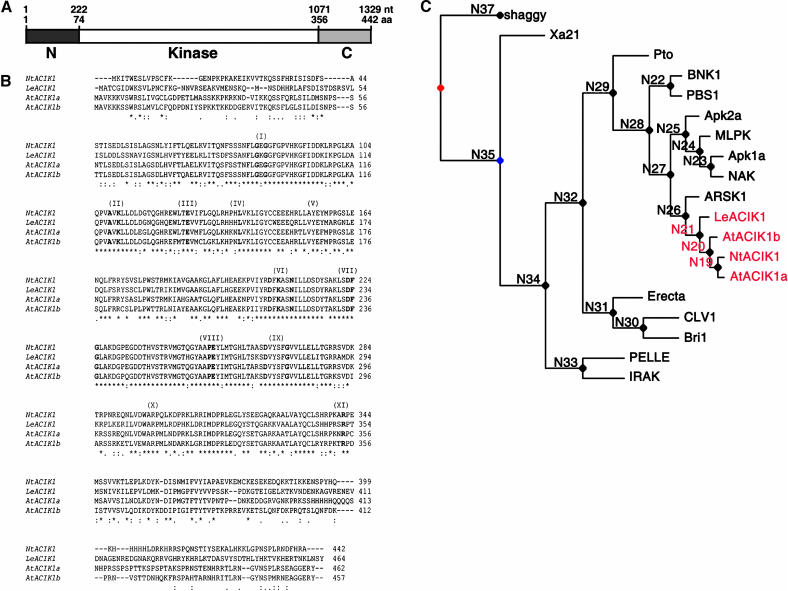
The predicted NtACIK1 protein contains 442 amino acids with a molecular mass of 50.2 kD. A Blastp search revealed that NtACIK1 has all 11 subdomains required for Ser/Thr protein kinase activity (Figure 2B). The kinase domain is flanked by two short nonkinase domains that have no obvious similarities to proteins of known function (Figure 2A). Because the protein sequence does not contain any membrane-spanning region or signal peptides, it is likely that NtACIK1 is either a cytoplasmic or nuclear kinase. Alignments of NtACIK1 with putative orthologs in tomato (LeACIK1) and Arabidopsis (AtACIK1a [At2g05940] and AtACIK1b [At5g35580]) showed an overall amino acid sequence identity of 60 to 75% (Figure 2B). The strongest similarity between the sequences was found in the kinase domains, which were 80 to 87% identical. Both the flanking N-terminal and C-terminal regions are highly divergent, sharing only 31 to 40% identity.
Figure 2.
Sequence of NtACIK1 Protein and Alignments of Homologs.
(A) Schematic representation of NtACIK1 protein.
(B) The amino acid sequences of tobacco NtACIK1, tomato LeACIK1, Arabidopsis AtACIK1a, and Arabidopsis AtACIK1b were aligned with ClustalW software (http://clustalw.genome.ad.jp/). The positions of subdomains (I to XI) characteristic of protein kinases are indicated in parenthesis above the sequences. Amino acids that are highly conserved among protein kinases are shown in bold. Identical amino acids are indicated by an asterisk, and similar residues are marked by a colon or a dot.
(C) Phylogenetic tree of NtACIK1 and related protein kinases. The tree was built using Bayesian evolutionary tree estimation (Sjölander, 1998) and is based on the full kinase domain of each protein.
Database searches revealed that the kinase domain of NtACIK1 is similar to those of receptor-like kinase domains. In Arabidopsis, ∼620 of the ∼1100 predicted protein kinases are receptor-like (Shiu and Bleecker, 2001). Many kinase sequences classified as receptor-like kinases lack an apparent signal sequence or transmembrane domain and are termed receptor-like cytoplasmic kinases. NtACIK1 and its orthologs are most related to a subgroup of 46 Arabidopsis kinases called receptor-like cytoplasmic kinase-VII (Shiu and Bleecker, 2001; Shiu et al., 2004). Members of this subgroup include NAK, APK1a, APK1b, APK2a, and ARSK1, which are a set of partially characterized protein kinases of unknown function (Hirayama and Oka, 1992; Moran and Walker, 1993; Hwang and Goodman, 1995; Ito et al., 1997). This subgroup includes PBS1, which is an Arabidopsis protein kinase required for recognition of AvrPphB from Pseudomonas syringae pv phaseolicola (Swiderski and Innes, 2001). MLPK, a membrane-anchored protein kinase involved in Brassica self-incompatibility signaling (Murase et al., 2004), is also highly related in sequence to this subgroup. The relationship between ACIK1 and other plant protein kinases was analyzed by construction of a phylogenetic tree (Figure 2C). Although several programs were used to estimate phylogenies, the tree shown was built using Bayesian evolutionary tree estimation (Sjölander, 1998). Within the clade containing ACIK1 proteins (node N28), all trees were similar; a neighbor joining tree gave bootstrap values of 73%, joining ACIK1 sequences to ARSK1 (node N26), and 82% joining to the clade containing the NAK kinase (node N27). The Bayesian evolutionary tree estimation produced the tree topology most widely recognized as being correct for the more diverse kinases. In summary, this analysis indicated that NtACIK1 and its orthologs cluster with ARSK1 (Hwang and Goodman, 1995), but in a branch distinct from NAK, MLPK, and PBS1.
NtACIK1 and LeACIK1 mRNAs Are Upregulated upon Elicitation
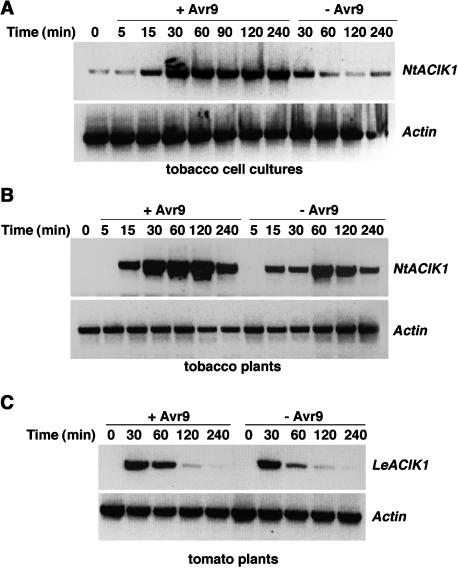
Although gene induction might not be directly linked to gene function in a particular defense response, we wanted to confirm elicitor-induced NtACIK1 gene expression. NtACIK1 transcript levels were analyzed in Cf-9-tobacco cell cultures and in Cf-9-tobacco plants over a time course of 4 h after elicitation with intracellular fluid (IF) containing Avr9 [IF(+Avr9)] or IF lacking Avr9 [IF(-Avr9)]. RT-PCR with NtACIK1-specific primers was conducted using the constitutively expressed actin gene as a control (Figure 3). A low level of NtACIK1 transcript was detected in unelicited cell cultures (Figure 3A, 0-min time point), probably because of the constant stress stimulus from shaking of the cells. However, addition of IF(+Avr9) resulted in a significant increase in NtACIK1 levels within 15 min that reached maximum levels by 30 min (Figure 3A, +Avr9). High levels of NtACIK1 mRNA were maintained over the 4-h time course. Addition of IF lacking Avr9 did not yield such an induction of NtACIK1 mRNA in cell cultures (Figure 3A, −Avr9). NtACIK1 was also rapidly induced in leaves of tobacco after infiltration of IF(+Avr9) (Figure 3B). However, control infiltrations of IF(−Avr9) also caused the induction of NtACIK1 (Figure 3B, −Avr9). Avr9 elicitation caused a stronger upregulation of NtACIK1 than the nonspecific mechanical stress caused by infiltration. This overlap in the transcriptional responses elicited by Cf-9/Avr9 and by wounding has been observed for all ACRE genes characterized to date (Durrant et al., 2000; data not shown).
Figure 3.
Expression Patterns of NtACIK1 and LeACIK1 after Elicitation by Avr9 and Wounding.
Cf-9 tobacco cell cultures (A), Cf-9 tobacco plants (B), or Cf-9 tomato plants (C) were treated with IF that contains Avr9 (+) or that does not contain Avr9 (−). At the time points indicated, leaf samples or cultured suspension cells were harvested, and total RNA was isolated and used for RT-PCR with NtACIK1 or LeACIK1 specific primers. Equal cDNA amounts were controlled by amplification of the constitutively expressed actin gene.
We also investigated the expression pattern of the putative NtACIK1 homolog from tomato, LeACIK1 (Figure 2B). The full-length cDNA sequence of a tomato homolog, LeACIK1, was identified in GenBank (AF332960). As observed for NtACIK1, LeACIK1 was strongly induced in tomato plants by both wounding [infiltration with control IF(−Avr9)] and Avr9-elicitation [infiltration with IF(+Avr9)] (Figure 3C). We therefore concluded that LeACIK1 might be the tomato homolog of NtACIK1.
LeACIK1 Is a Functional Protein Kinase
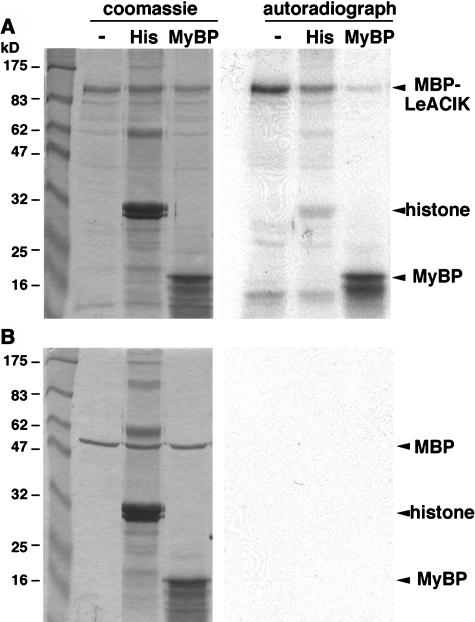
To determine whether NtACIK1 and LeACIK1 encode functional protein kinases, we performed kinase assays with purified ACIK1 fusion proteins expressed in Escherichia coli. Constructs were generated that fused maltose binding protein (MBP) to the N terminus of full-length NtACIK1 or LeACIK1. Fusion proteins were expressed and affinity purified by binding to amylose resin. Unfortunately, MBP-NtACIK1 was degraded after purification (data not shown), and so only MBP-LeACIK1 was used in the kinase assays. As a negative control, purified MBP expressed from the empty vector was used. Kinase activity was tested by adding different artificial substrates and [γ-32P]ATP to the purified proteins and assaying for the ability of MBP or MBP-LeACIK1 to phosphorylate these substrates. Autoradiography showed that MBP-LeACIK1 fusion protein autophosphorylates and phosphorylates myelin-basic protein (Figure 4A, right panel). Histone was not an efficient substrate for MBP-LeACIK1. No radioactive signals could be detected when MBP alone was used instead of the MBP-LeACIK fusion protein (Figure 4B, right panel). Thus, LeACIK1 is a functional protein kinase.
Figure 4.
LeACIK1 Is a Functional Protein Kinase.
MBP-LeACIK1 fusion protein or MBP as control were expressed in E. coli BL21 cells and affinity purified using amylose resin. Purified MBP-LeACIK1 (A) or MBP (B) were subjected to an in vitro kinase assay using histone (His) or myelin-basic protein (MyBP) as substrate. After the kinase reaction, the samples were separated on a SDS polyacrylamide gel, and the gel was stained with Coomassie blue (left panels), dried, and subjected to autoradiography (right panels).
ACIK1 Is Required for Cf-Mediated HR
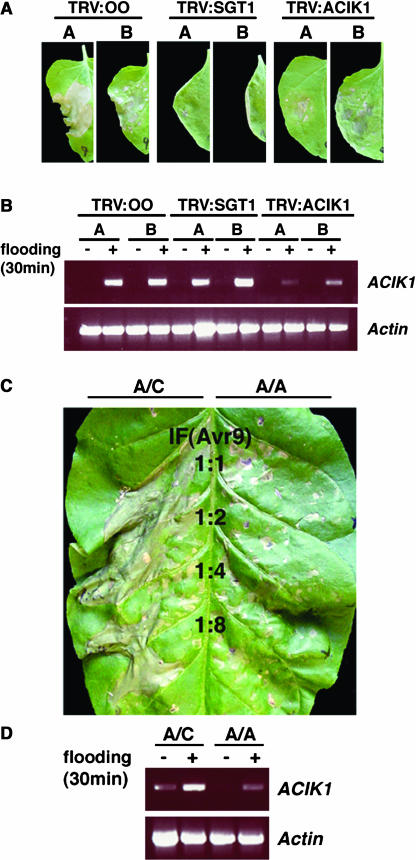
We wanted to verify the phenotype of silencing ACIK1 using a different silencing vector based on tobacco rattle virus (TRV) (Ratcliff et al., 2001). TRV-based VIGS induces milder viral symptoms than PVX-based VIGS and often gives more effective silencing but is not currently useful for high-throughput gene silencing screens. To silence NtACIK1 homologs in N. benthamiana, the 5′-RACE fragment spanning the N-terminal region of the NtACIK1 gene was cloned into a TRV silencing vector (Ratcliff et al., 2001). Wild-type N. benthamiana plants were silenced with TRV:ACIK1, TRV:SGT1, or TRV:00 (empty vector control). An HR developed in leaves of TRV:00 plants after agroinfiltration of Cf-9/Avr9 (Figure 5A, left two panels). Silencing of NbSGT1 was previously shown to eliminate the HR produced by Cf-4/Avr4 or by Cf-9/Avr9 (Peart et al., 2002b) and was used as a positive control (Figure 5A, middle two panels). Silencing of NbACIK1 resulted in a reduction of the Cf-9/Avr9-induced HR (Figure 5A, right two panels), which confirmed our initial observations using PVX-based silencing. However, we generally observed a range of phenotypes, from a complete loss of HR to a strong HR comparable with that of TRV:00 control plants. We therefore analyzed NbACIK1 transcript levels in silenced leaves from two independent experiments using RT-PCR with gene-specific primers outside of the region used for silencing. In these two experiments, 14 out of 16 leaves from TRV:00 infected plants and only two out of 16 leaves from TRV:SGT1 infected plants showed HR symptoms after Cf-9/Avr9 elicitation. In the TRV:ACIK1 silenced plants, 25 out of 40 elicited leaves showed symptoms. NbACIK1 transcript analysis of those leaves revealed that NbACIK1 mRNA accumulation after infiltration of water (flooding) was only strongly reduced in TRV:ACIK1 leaves that also showed less HR symptoms, as shown in Figure 5B for two selected leaves. All the other TRV:ACIK1 leaves had comparable NbACIK1 mRNA levels to those in plants infected with the two control constructs TRV:00 or TRV:SGT1 (Figure 5B). In conclusion, the extent of the HR strictly correlated with the amount of NbACIK1 mRNA; the more mRNA that could be detected, the less attenuated was the HR (cf. Figures 5A, right two panels, and 5B).
Figure 5.
ACIK1 Is Required for Both Cf-9– and Cf-4–Dependent HRs in N. benthamiana and Tobacco.
(A) VIGS in N. benthamiana using TRV. Seedlings were inoculated with TRV:ACIK1, TRV:SGT1, or control TRV:00. Three weeks after initiation of silencing, one leaf half was infiltrated with Agrobacteria that direct Cf-9 and Avr9 expression. Pictures of two independent leaves (A and B) were taken 3 d after agroinfiltration (top panel).
(B) To analyze NbACIK1 transcript levels, RNA was isolated from the other leaf half of each leaf shown in (A) before and 30 min after infiltration of water (flooding) to induce NbACIK1 expression. NbACIK1 transcript accumulation was analyzed by RT-PCR. Actin was used as an internal control for equal cDNA loading (bottom panel).
(C) RNAi in N. tabacum. pBIN19 constructs containing the NtACIK1/NtACIK1 hairpin (A/A) or containing a nonhairpin control NtACIK1/NtCDPK2 (A/C) were transiently expressed in Cf-9 transgenic tobacco plants. Four days after Agrobacteria were infiltrated, the leaves were challenged with various dilutions of IF(+Avr9), and the development of HR was observed for 3 d.
(D) On parallel leaves, NtACIK1 transcript accumulation was determined as described in (B).
We also investigated the effect of silencing NtACIK1 in Cf-9 transgenic tobacco plants. Because VIGS cannot be used in N. tabacum, we generated NtACIK1-RNAi hairpin constructs under the control of the 35S promoter for gene silencing in tobacco leaf tissue. As a negative control, we used a nonhairpin construct that contained one arm of NtACIK1 cDNA and one arm of NtCDPK2 cDNA (Romeis et al., 2001), which cannot form a hairpin structure for silencing. These constructs were transiently delivered into tobacco leaves by agroinfiltration. Four days after agroinfiltration, tobacco leaves were infiltrated with IF(+Avr9) and scored for development of the HR. In four out of six independent experiments, we observed strongly reduced HR symptoms in the leaf half expressing the NtACIK1-hairpin construct as compared with the control leaf half (Figure 5C). This again correlated with weaker stress-induced NtACIK1 transcript accumulation in the silenced leaf half (Figure 5D). Thus, using a second independent and nonviral silencing method, we could confirm the requirement of NtACIK1 for the Cf-9/Avr9-induced HR.
ACIK1 Is Specifically Important for the Cf-Mediated Defense Pathway
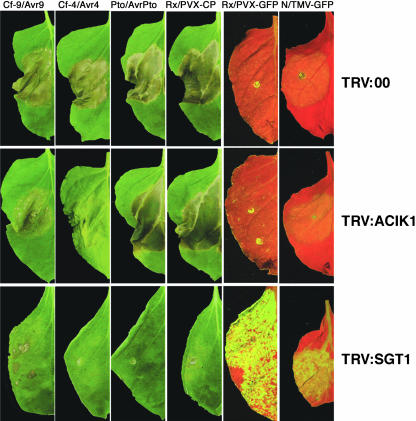
To investigate the specificity of ACIK1 as a regulator of the HR, additional R gene/Avr gene interaction systems were tested for their ability to induce an HR in ACIK1-silenced N. benthamiana plants. We tested the HR produced by Pto/AvrPto (Bogdanove and Martin, 2000) and Rx/PVX coat protein (Bendahmane et al., 1999). Pto is an R gene encoding a Ser/Thr kinase that mediates resistance to P. syringae carrying AvrPto (Martin et al., 1993). Rx encodes an R protein of the nucleotide binding (NB)-LRR class that mediates resistance to PVX (Bendahmane et al., 1999). N. benthamiana plants were silenced with TRV:00, TRV:SGT1, or TRV:ACIK1 and then infiltrated with Agrobacteria expressing the different R/Avr gene–containing constructs 3 to 4 weeks after initiation of silencing (Figure 6). Delivery of Pto/AvrPto and Rx/PVX into leaves of N. benthamiana plants silenced with TRV:00 resulted in an HR after 2 to 3 d (Figure 6, top row, columns 3 and 4). Silencing of SGT1 blocked the HR produced by these two R/Avr combinations (Figure 6, bottom row, columns 3 and 4), as previously shown (Peart et al., 2002b). However, no reduction or delay in the Pto- or Rx-mediated HR was observed in the ACIK1-silenced plants in five independent experiments each using six leaves (Figure 6, middle row, columns 3 and 4). The HR produced by Cf-9/Avr9 and Cf-4/Avr4 was compromised in plants silenced for NbACIK1 (Figure 6, middle row, columns 1 and 2), indicating that silencing of NbACIK1 was successful in these sets of experiments.
Figure 6.
ACIK1 Specifically Regulates the Cf-9– and Cf-4–Dependent HRs.
N. benthamiana leaves were silenced as described in Figure 5A and injected with Agrobacteria expressing the following R gene/Avr gene combinations: Cf-9/Avr9, Cf-4/Avr4, Pto/AvrPto, or Rx/PVX-coat protein. In addition, N. benthamiana plants carrying the N or the Rx gene were inoculated with Agrobacteria expressing either TMV-GFP or PVX-GFP, respectively. Pictures of HR were taken 3 d after Agrobacterium infection. Accumulation of TMV-GFP and PVX-GFP was monitored by GFP fluorescence 5 DAI on inoculated leaves.
To further test the specificity of ACIK1 function, N- and Rx-mediated resistance was tested in N. benthamiana plants silenced for NbACIK1. The N gene encodes an NB-LRR R protein that mediates resistance to Tobacco mosaic virus (TMV). Transgenic N. benthamiana plants expressing either the N gene (Peart et al., 2002a) or the Rx gene (Bendahmane et al., 1999) were used in these experiments. Inoculation of GFP-tagged strains of TMV (TMV-GFP) or PVX (PVX-GFP) onto N. benthamiana plants results in widespread growth of either virus that can be observed under UV light as accumulation of GFP fluorescence (Peart et al., 2002a). No green fluorescence was detected after inoculation of N-transgenic N. benthamiana with TMV-GFP or after inoculation of Rx-transgenic N. benthamiana with PVX-GFP (Figure 6, top row, columns 5 and 6; Peart et al., 2002a). In three independent experiments, each using six leaves, no effect on TMV or PVX spread was observed in NbACIK1 silenced plants because in neither case was green fluorescence detected in the inoculated leaves 5 DAI (Figure 6, middle row, columns 5 and 6). By contrast, plants that were silenced with TRV:SGT1 showed pronounced GFP fluorescence (Figure 6, bottom row, columns 5 and 6). ACIK1 therefore does not appear to play a role in resistance to TMV or PVX.
Taken together, these results indicate that ACIK1 function is likely to be specific for Cf-9– and Cf-4–mediated defense responses.
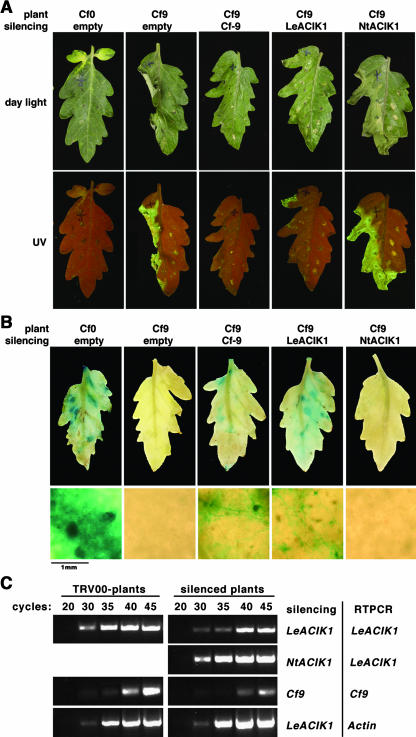
LeACIK1 Is Required for Tomato Cf-9–Mediated Resistance to C. fulvum
We used VIGS of LeACIK1 to test if ACIK1 is required not only for HR but also for resistance in tomato against C. fulvum. The last 150 bp of the LeACIK1 coding sequence were cloned into a TRV vector described by Liu et al. (2002). This region of LeACIK1 is highly gene specific. For controls, 150 bp corresponding to the C terminus of Cf-9 or the last 150 bp of the NtACIK1 coding sequence were each cloned into the TRV vector. For each construct, we used at least five leaves of three different plants. We analyzed the Cf-9/Avr9-dependent HR in tomato 3 weeks after silencing with the different constructs. Consistent with the observation in N. benthamiana, the HR induced by infiltration of IF(+Avr9) was much weaker and delayed in LeACIK1 or Cf-9 silenced plants compared with the control plants silenced with the empty vector (Figure 7A, Table 3). Furthermore, UV-fluorescing phytoalexins were detected in the control plants, whereas this symptom was much reduced or abolished in the leaves silenced for either LeACIK1 or Cf-9. In tobacco, such fluorescence is characteristic of the accumulation of the phenolic phytoalexin scopolin and its aglycone scopoletin (Costet et al., 2002) and is a marker for the HR (Gachon et al., 2004). Little or no necrosis was observed in silenced Cf0 plants or when control IF without the Avr9 peptide was used (Figure 7A, right leaf halves). Silencing with the NtACIK1 sequence did not have any effect on the development of HR after Cf-9/Avr9 elicitation. Although LeACIK1 and NtACIK1 seem to be homologous genes, the 150-bp sequences used for silencing were only 43% identical, so no cross-silencing was expected.
Figure 7.
LeACIK1 Is Required for Cf-9–Mediated Resistance to C. fulvum Expressing Avr9.
Transgenic Cf-9 or Cf0 tomato seedlings were inoculated with Agrobacteria expressing the indicated construct.
(A) Three weeks later, leaves were infiltrated with IF containing Avr9 (left leaf halves) or not containing Avr9 (right leaf halves). Pictures of HR symptoms were taken 1 d later under daylight (top panels) or UV light (bottom panels).
(B) Silenced tomato plants were infected with C. fulvum race 4 GUS. Leaves were stained with X-gluc 3 weeks after C. fulvum infection, and pictures were taken (top panels). In addition, fungal growth was monitored under the microscope (bottom panels).
(C) Tomato plants were silenced with the control vector (TRV:00) or the constructs indicated on the right. Total RNA was isolated from leaf discs, and LeACIK1, Cf-9, or actin transcript accumulation was analyzed by RT-PCR. Numbers of PCR cycles are indicated at the top. The control PCR with actin-specific primers was performed for every sample but is only shown for control and LeACIK1-silenced plants.
Table 3.
Proportion of TRV-Silenced Tomato Leaves Showing HR or C. fulvum Growth
| Silencing Construct | Tomato Genotype | HR | C. fulvum Growth |
|---|---|---|---|
| TRV:00 | Cf0 | 0/20 | 47/48 |
| TRV:00 | Cf-9 | 18/20 | 2/45 |
| TRV:Cf-9 | Cf-9 | 2/20 | 25/48 |
| TRV:LeACIK1 | Cf-9 | 5/20 | 30/48 |
| TRV:NtACIK1 | Cf-9 | 15/20 | 1/40 |
C. fulvum growth was detected by X-gluc staining.
To test the effect of silencing on Cf-9–mediated resistance, tomato plants were dip-inoculated with C. fulvum spores 3 weeks after silencing induction (Figure 7B, Table 3). The C. fulvum strain used expresses the β-glucuronidase (GUS) gene to allow easy visualization of fungal growth. After an additional 3 weeks, leaves were stained with 5-bromo-4-chloro-3-indoyl-β-glucuronide (X-gluc) to score fungal growth macroscopically and microscopically. As expected, no X-gluc staining and no fungal growth could be observed in Cf-9 tomato plants that were either untreated or silenced with the empty vector control (Figure 7B; data not shown). By contrast, Cf0 tomato plants that were untreated or silenced with the control TRV:00 had extensive fungal growth as detected by X-gluc staining (Figure 7B; data not shown). The dark blue stain was a result of fungal hyphae, growing intracellularly and emerging again through the stomata. Leaves from Cf-9 plants silenced with either TRV:LeACIK1 or TRV:Cf-9 showed significantly increased staining as compared with the empty vector controls, indicating that silencing of LeACIK1 caused a breakdown of Cf-9–mediated resistance. Fungal hyphae could be detected in LeACIK1- and Cf-9–silenced leaves under the microscope, although in both cases considerably less hyphae were observed than in infected Cf0 plants (Figure 7B, bottom panels). Also, no reproductive structures emerging from the stomata were observed, indicating that LeACIK1 and Cf-9 silencing only cause partial loss of resistance. Silencing with the NtACIK1 sequence had no effect on Cf-9–mediated resistance. These results were obtained in three independent experiments using multiple leaves from two to three silenced tomato plants. We confirmed that LeACIK1 mRNA and Cf-9 mRNA were reduced in the respective silenced leaves using RT-PCR (Figure 7C). Notably, under our conditions, silencing in tomato plants seemed to be less effective relative to N. benthamiana, and the reductions of LeACIK1 and Cf-9 mRNA levels were always partial.
DISCUSSION
Functional Genomics of ACRE Genes
Gene expression profiling using cDNA-AFLP display was previously used to identify transcripts whose expression is rapidly altered during the Avr9- and Cf-9–mediated defense response in transgenic Cf-9 tobacco (Durrant et al., 2000). In this earlier study, only a limited number of sequenced cDNA-AFLP fragments (42/257) were reported to have similarity to sequences in gene databases. By searching updated genomic and EST databases, we have provided an expanded view of the ACRE genes (178/257 fragments with hits). Many of the ACRE genes encode regulatory proteins, such as protein kinases, transcription factors, and regulators of protein degradation and are thus likely to play important roles in the Cf-9–mediated defense response. To test this, we took advantage of VIGS (Baulcombe, 1999). In a recent screen, VIGS was used to survey ∼5000 cDNAs from a normalized library for their role in Pto-mediated HR and resistance (Lu et al., 2003a). We focused here on a smaller collection of genes that were already correlated with defense because of their induced gene expression. VIGS of 42 ACRE genes implicated three genes in Cf-9/Avr9-dependent host cell death. We cannot conclude, however, that the other 39 ACRE genes are not involved in Cf-9– or Cf-4–mediated HR. Some genes may not have been sufficiently silenced to observe a phenotype. Also, because many of the ACRE genes are strongly induced, silencing may not be able to keep up with gene induction. Finally, we only screened for reduction of the HR, which is a crude assay; other aspects of the defense response may have been affected even when VIGS did not affect the HR.
In this study, we focused on the role of the protein kinase ACIK1 in the plant defense response. Future studies will focus on the roles of ACRE-189 and ACRE-276, which are predicted to be involved in protein degradation via ubiquitylation.
ACIK1 Is Induced by Avr9 and Other Stress Stimuli
As expected from the cDNA-AFLP analysis, the NtACIK1 gene and its tomato homolog LeACIK1 are transcriptionally induced upon elicitation in the Cf-9/Avr9 system. Although osmotic/mechanical stress by infiltration of water into tobacco and tomato leaves also resulted in transcript accumulation, the response to Avr9 elicitation was stronger and more prolonged. ACIK1 protein is probably present before elicitation because small amounts of ACIK1 transcript were detected before elicitation in cell cultures and also in leaf tissue after increasing PCR cycles during RT-PCR (data not shown). Upregulation of the ACIK1 transcript may be critical for maintaining ACIK1 protein levels after elicitation because activation of the kinase could result in increased protein turnover. The LeACIK1 gene was also reported to be constitutively upregulated in uninfected tomato Rio Grande plants expressing the Pto resistance gene under its native promoter (RG-PtoR) (Mysore et al., 2002). In addition, the Arabidopsis ortholog AtACIK1a (At2g05940) was induced after infection with incompatible P. syringae strains (Tao et al., 2003) and by wounding (Cheong et al., 2002), and both AtACIK1a and AtACIK1b (At5g35580) were induced in Arabidopsis cell cultures after flagellin treatment (Navarro et al., 2004). It is unknown if the ACIK1 orthologs have any regulatory roles in responses outside of Cf-mediated defense. ACIK1 function may, however, participate in the cross talk that occurs in the overlapping pathways activated by wounding and pathogen resistance as suggested for CDPKs (Ludwig et al., 2004).
So far, we cannot distinguish if the required ACIK1 function is a result of preexisting protein or protein derived from induced transcript. However, it is likely that important regulation of ACIK1 activity is achieved through posttranslational modifications that occur upon elicitation, involving changes in protein kinase activity, subcellular localization, and/or protein stability. Unfortunately, we were unsuccessful in detecting endogenous ACIK1 protein in planta using antibodies generated against NtACIK1 (data not shown). It is possible that ACIK1 protein levels are maintained at relatively low levels. We were also unable to detect elicitor-induced changes in ACIK1 protein kinase activity or protein localization using transiently expressed epitope-tagged versions of ACIK1 (data not shown).
ACIK1 Is Specifically Required for Cf-9–/Cf-4–Mediated HR and Resistance
We used two independent gene silencing techniques, VIGS in N. benthamiana and transient expression of hairpin constructs in Cf-9 tobacco, to confirm the requirement for NtACIK1 in the Cf-9/Avr9-induced HR. It has been shown, however, that the development of an HR is not necessarily required for resistance (Bendahmane et al., 1999; Sharma et al., 2003). Therefore, we also used VIGS in tomato to investigate the effect of LeACIK1 silencing on resistance to the natural pathogen C. fulvum. LeACIK1-silenced Cf-9 tomato plants were compromised in the Avr9-induced HR, consistent with the loss of HR observed in N. benthamiana and tobacco. Moreover, these plants had an increased susceptibility to a normally incompatible C. fulvum race. We did not observe any effect of ACIK1 silencing on the function of other R genes, such as Pto, Rx, or N, suggesting that ACIK1 function is specifically important for Cf-9 and Cf-4 function. Other components have been reported to be required for disease resistance, such as SGT1 (Peart et al., 2002b), Hsp90 (Lu et al., 2003b), WIPK (Ekengren et al., 2003; Sharma et al., 2003), and PAD4 (Glazebrook et al., 1996; Ekengren et al., 2003), which are each required for more than one gene-for-gene interaction. This suggests that these components are further downstream in the signaling pathways or have more general functions, as is likely for SGT1 that may be involved in assembly of SCF complexes and HSP90 as a protein chaperone. By contrast, ACIK1 appears to be specifically required for Cf-9 and Cf-4 function and is thus likely to be involved in very early steps of the signaling pathway. Although we could not detect a direct interaction between Cf-9 and LeACIK1 in yeast two-hybrid experiments (data not shown), we cannot rule out that ACIK1 is participating in a signaling complex in which Cf-9 or Cf-4 and other proteins are present.
VIGS can be effective with as little as 25 nucleotides of sequence identity between the insert in the virus vector and the target RNA (Thomas et al., 2001). Because we used 5′- or 3′-RACE ACIK1 cDNA fragments for VIGS in N. benthamiana, we cannot exclude silencing of other closely related genes. The same would apply for the gene-silencing experiments using transient expression of an NtACIK1-hairpin construct because 550 bp of coding sequence was used that included part of the region encoding the kinase domain. However, for VIGS in tomato we used only the last 150 bp of sequence corresponding to the nonconserved region at the C terminus of the protein beyond the kinase domain. Searches of the tomato EST database (The Institute for Genomic Research) revealed no significant sequence identities with this region. We therefore did not expect any cross-silencing of related kinases, and the effect of VIGS is likely to be specific for LeACIK1.
Possible Functions of ACIK1 in the Cf-9–Mediated Defense Response
ACIK1 belongs to a subfamily of protein kinases with no proven role in stress signaling. A related Arabidopsis kinase of this subfamily, ARSK1, was shown to be root specific and was transcriptionally induced after ABA or NaCl treatment (Hwang and Goodman, 1995), suggesting a putative role in signal transduction of osmotic stress. NAK-type kinases are part of a related but distinct subfamily of protein kinases, but their functions in plants are mostly unclear, and there are only a few examples described so far (Hardie, 1999). The NAK-type kinase APK1 from Arabidopsis was reported to display both Tyr and Ser/Thr kinase activity (Hirayama and Oka, 1992), but no functional data were presented. A membrane-anchored kinase, MPLK, from Brassica that is similar to APK1b was recently shown to be required for self-incompatibility signaling (Murase et al., 2004).
The kinase domains of ACIK1 orthologs cluster with receptor-like kinases (Shiu and Bleecker, 2001). Receptor-like kinases have been implicated in both race-specific and general defense responses. The Xa21 gene from rice specifies race-specific resistance against Xanthomonas oryzae (Song et al., 1995), whereas the Arabidopsis FLS2 gene is required for recognition of the general bacterial elicitor flagellin (Gómez-Gómez and Boller, 2000). The Xa21 and FLS2 proteins each consist of extracellular LRRs, a single-pass transmembrane domain, and a cytoplasmic Ser/Thr kinase domain. Because Cf-9 does not contain any apparent signaling domains, we expect that at least one additional protein provides the signaling function. In this scenario, Cf-9 and ACIK1 could be part of a receptor-like kinase complex, with Cf-9 providing the extracellular LRRs for recognition and ACIK1 providing the cytoplasmic kinase for signaling. Such an interaction between a transmembrane protein containing an extracellular LRR domain and a protein kinase has been well described. In Drosophila, the Toll protein interacts via an adaptor complex with the Pelle protein kinase (Hoffmann and Reichhart, 2002). Similarly, in mammals, Toll-like receptors such as TLR2, TLR4, or TLR5 interact via the adaptor molecule MyD88 with the protein kinase IRAK (Akira et al., 2001; Underhill and Ozinsky, 2002) to form functional receptor-like kinases.
Another possible role for LeACIK1 is the phosphorylation of negative regulators of the signaling pathway leading to defense responses. Our laboratory recently identified a thioredoxin, CITRX, as an interactor of the Cf-9 resistance protein and as a negative regulator of Cf-9–mediated signaling responses in tomato (Rivas et al., 2004). To activate the corresponding signaling pathways, negative regulators such as CITRX have to be inactivated or degraded. Proteolysis in the eukaryotic cytosol typically involves substrate ubiquitylation mediated by, for instance, the SCF complex or other E3 ligases. Polyubiquitylated proteins are then targeted for degradation via the 26S proteasome (Hochstrasser, 1995). It is known that many substrates of SCF complexes must be phosphorylated before they can bind to the SCF complex and become polyubiquitylated (Deshaies, 1999; Kumar et al., 2003). The requirement for substrate phosphorylation creates an opportunity for differential regulation of the stability of SCF substrates. Elicitation would result in an increase of ACIK1 protein kinase activity, leading to the phosphorylation and specific degradation of its substrates and induction of the corresponding signaling pathways.
A third possible role for ACIK1, which is not mutually exclusive with the first two options, is as a component of a protein kinase cascade. Two well-studied kinase families participating in phosphorylation cascades during plant defense responses are the mitogen-activated protein kinases (MAPKs) and the CDPKs. In the Cf-9/Avr9 system, Romeis et al. (1999) described the activation of the two MAPKs SIPK (salicylic acid–induced protein kinase) and WIPK (wound-induced protein kinase). It has been shown that suppression of certain MAPK activities resulted in reduced plant defense responses. In tomato, Pto-mediated resistance was compromised by silencing the genes encoding the two MAPK kinases MEK1 and MEK2 and the two MAPKs WIPK and NTF6 (Ekengren et al., 2003). In N. benthamiana, silencing of either SIPK or WIPK compromised N gene–mediated TMV resistance (Jin et al., 2003), and silencing of both SIPK and WIPK reduced the resistance to the bacterial pathogen Pseudomonas cichorii (Sharma et al., 2003). In Arabidopsis, silencing of MPK6 reduced basal and race-specific resistance to various pathogens (Menke et al., 2004). Similarly, CDPKs were shown to be required for plant defense responses. In Cf-9 transgenic tobacco plants, NtCDPK2 was activated after Avr9 elicitation, and VIGS of NtCDPK2 in N. benthamiana plants resulted in reduced Cf-9– and Cf-4–mediated HRs (Romeis et al., 2001). The tomato Pto kinase interacts with the Ser/Thr kinases Pti1 (Zhou et al., 1995), Adi2, and Adi3 (Bogdanove and Martin, 2000). This suggests that Pto is involved in phosphorylation cascades independent of the described MAPK or CDPK pathways. It is possible that ACIK1 may also take part in an as yet unidentified signaling cascade. Future work on the identification of the downstream targets and of interacting proteins will lead to a better understanding of the role of ACIK1 in the plant defense response.
METHODS
Plant Materials and Growth Conditions
Nicotiana benthamiana was germinated and grown in a glasshouse under semicontrolled conditions where temperatures ranged from ∼20 to 25°C. In the winter, supplementary lighting was used to provide a minimum 16-h daylength. Transgenic N. benthamiana plants expressing the N resistance gene (line 310A) or the Rx resistance gene (line Rx-18) were described previously (Bendahmane et al., 1999; Peart et al., 2002a). Lycopersicon esculentum cv Moneymaker was grown as above. Tobacco plants were grown in environmentally controlled growth cabinets at 24°C, with a 16-h-light/8-h-dark cycle. The Cf-9 transgenic N. tabacum cv Petite Havana line 34.1B was described previously (Hammond-Kosack et al., 1998). Suspension cultures of Cf-9 tobacco cells derived from line 34.1B were subcultured as previously described (Piedras et al., 1998).
Primers
The sequences of all primers used are listed in Supplemental Table 2 online.
Rapid Amplification of ACRE cDNA Ends
Twenty-nine partial ACRE cDNA sequences were amplified from tobacco RNA by 3′-RACE. Total RNA was isolated from suspension cells of Cf-9 tobacco 30 min after adding IF(+Avr9) (Durrant et al., 2000). First-strand cDNA was synthesized from 5 μg of total RNA using Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA) and primer AP. The 3′-ends of the ACRE cDNAs were then amplified using gene-specific primers, which contained a ClaI restriction site, and primer AUAP, which contained a SalI restriction site. Nested PCR was performed with a second gene-specific primer that contained a ClaI site and primer AUAP, except for ACREs 216, 264, and 284, where the primary PCR was sufficient for specific amplification of cDNA. 3′-RACE products were cloned into the pGEM-T Easy plasmid (Promega, Madison, WI), and sequences of independent clones were determined. Clones were identified that matched or almost matched the sequence of the original cDNA-AFLP fragment. 5′-RACE was used to obtain full-length cDNA sequence for 8 of the 29 ACRE genes of interest. The 5′-ends were determined using either the 5′-RACE system kit (Invitrogen) or the SMART RACE cDNA amplification kit (Clontech, Palo Alto, CA) according to the manufacturers' instructions. Gene-specific primers contained a ClaI site and the 5′-end primers, AUAP, or NUP-SalI contained a SalI site. 5′-RACE products were cloned into pGEM-T Easy and characterized as described above for the 3′-RACE products.
PVX-Based VIGS in N. benthamiana
The PVX vector construct for agroinoculation, pgR106, and the inoculation procedures have been described previously (Lu et al., 2003b). Thirteen ACRE cDNAs were previously isolated by screening a tobacco cDNA library (Table 1; Durrant et al., 2000). Primers ClaI-SK and M13(-20) were used to amplify the entire sequence of 12 of these cDNAs, which were in pBluescript SK−. Primers 132P4ClaI and T3 were used to amplify 900 bp of 5′-end ACRE-132 cDNA sequence. PCR products were digested with ClaI and XhoI and ligated into the ClaI and SalI sites of pgR106. Because ACRE-137 had an internal ClaI site, only 787 bp from the 3′-end of this cDNA was cloned into the silencing vector. All 29 ACRE cDNA sequences amplified by 3′-RACE and four of the ACRE cDNA sequences amplified by 5′-RACE (141, 264, 275, and 284) were isolated from the pGEM-T Easy vector by digestion with ClaI and SalI and cloned into the corresponding sites of pgR106. All plasmids were used to transform Agrobacterium tumefaciens strain GV3101 containing the helper plasmid pJICSa_RepA (Hellens et al., 2000).
For viral inoculation, second leaves of 2- to 3-week-old N. benthamiana seedlings were pierced with a toothpick that was previously touched to an Agrobacterium colony. After an additional 3 to 4 weeks, during which PVX spread systemically, the third or fourth leaves were analyzed for an HR phenotype. Individual leaves were infiltrated on one side with Agrobacterium containing binary vectors expressing Cf-9 and Avr9 (p9/456/Avr9) or expressing Cf-4 and Avr4 (p3/456/Avr4) (Thomas et al., 2000).
TRV-Based VIGS in N. benthamiana
The TRV vector construct for agroinoculation, pTV00, has been described previously (Ratcliff et al., 2001). The NtACIK1 5′-RACE cDNA fragment in the pGEM-T Easy vector was isolated and cloned into pTV00 using ClaI and SalI to create TRV:ACIK1. The empty pTV00 vector and pTV00 containing the NbSgt1.2 fragment (TRV:SGT1; Peart et al., 2002b) were used as controls. All plasmids were used to transform Agrobacterium strain GV3101 as above.
Infection of plants by agroinfiltration was done as previously described (Ratcliff et al., 2001), except that cultures containing pTV00-derived constructs were mixed with those containing pBINTRA6 in a 10:1 ratio before infiltration. The fourth and fifth leaves above the viral inoculated leaves were analyzed for ACIK1 transcript levels and for defense-related phenotypes. For elicitation of the HR, R genes and their cognate avirulence genes were coexpressed using Agrobacterium-mediated transient expression. The binary constructs containing the various R/Avr combinations have been described elsewhere: Cf-9/Avr9, Cf-4/Avr4 (Thomas et al., 2000), Pto/AvrPto (Peart et al., 2002b), and Rx/PVX-coat protein (Peart et al., 2002a). TMV-GFP and PVX-GFP inoculation of transgenic N. benthamiana plants carrying the N or the Rx gene, respectively, as well as visual detection and imaging of GFP fluorescence were performed as described by Peart et al. (2002a).
TRV-Based VIGS in Lycopersicon esculentum
A 150-bp PCR fragment spanning the last 48 amino acids of LeACIK1 was amplified from cDNA using primers sulphurLeACIKfwd and LeACIK-flrev. As controls, 150-bp fragments of Cf-9 (using primers SulphurCF9Fwd and Cf9-G3a) and NtACIK1 (using primers SulphurNtACIKFwd and NtACIK-STOP-Bgl) were also amplified by PCR. PCR products were purified and cloned into SmaI-digested pTRV-RNA2 vector (Liu et al., 2002). Silencing of tomato plants variety Moneymaker containing or not containing the Cf-9 resistance gene was performed as described by Liu et al. (2002), with the exception that cotyledons of 10-d-old seedlings were infiltrated with the Agrobacteria.
Three to four weeks after the onset of silencing, plants were treated with paclobutrazol and dip-inoculated with 5 × 106 spores of Cladosporium fulvum race 4 GUS as previously described (Balint-Kurti et al., 1994; Thomas et al., 1997). Fungal growth was scored 3 weeks later by GUS staining and analyzed by light microscopy (Axiophot; Zeiss, Jena, Germany). In parallel, leaves of silenced tomato plants were infiltrated with IF with or without Avr9 and were analyzed for development of HR.
Transient Silencing in Tobacco with RNAi
Silencing of NtACIK1 expression in Cf-9 tobacco plants by RNAi (Chuang and Meyerowitz, 2000; Smith et al., 2000) was achieved by Agrobacterium-mediated transient expression of an NtACIK1 RNAi transgene (hairpin construct) driven by the 35S promoter. For this, we made use of pSLJ1382B1 (A.A. Ludwig and T. Romeis, unpublished data), which contains an intron sequence flanked by 850 bp of NtCDPK2 sequence (Romeis et al., 2001) in sense and antisense orientations. The 189-bp intron sequence is from the potato (Solanum tuberosum) ST-LS1 gene (Vancanneyt et al., 1990), and 513 bp of the NtACIK1 cDNA sequence was amplified by PCR using primers 264RNAi5′XhoI and 264RNAi3′AscI or 264RNAi5′XbaI and 264RNAi3′FseI. The same sequence was amplified in both reactions, but each product contains different flanking sites. The PCR product containing XhoI and AscI sites was digested and used to replace the 5′ NtCDPK2 arm of pSLJ1382B1. This construct, pSLJ20234, contains one arm of NtACIK1 and one arm of NtCDPK2 and cannot form a hairpin structure when expressed, thus serving as a negative control. The NtACIK1 PCR product containing XbaI and FseI sites was then used to replace the 3′ NtCDPK2 arm of pSLJ20234 to create pSLJ20235. The EcoRI/HindIII cassettes of pSLJ20234 and pSLJ20235, which contain a 35S promoter, an intron with flanking arms, and an octopine synthase terminator, were cloned into the corresponding sites of pBIN19. Tobacco leaves were infiltrated with Agrobacterium strain GV3103 transformed with either of these binary constructs. Four days after infiltration, the infected leaves were challenged with IF(+Avr9) in different dilutions. The development of the HR was observed for 3 d.
Isolation of Full-Length NtACIK1 cDNA by Library Screening
The 131-bp cDNA-AFLP fragment for ACRE-264 was used as a probe to screen a cDNA library established from elicited tobacco cells (Durrant et al., 2000). Several independent clones were sequenced, and the clone containing the exact sequence of the ACRE-264 cDNA-AFLP fragment was selected for further study.
Phylogenetic Tree Analysis
The sequences of several protein kinase domains were identified using PFAM (Sonnhammer et al., 1998) and aligned using MAFFT (Katoh et al., 2002). Alignments were viewed and edited using the Belvu alignment editor. (Sonnhammer et al., 1998; www.cgr.ki.se/cgr/groups/sonnhammer/Belvu.html). Phylogenetic trees were estimated upon this alignment using Neighbor and Protpars from the Phylip software package (Felsenstein, 1993) using 1000 and 100 bootstrap replicates, respectively. Consensus trees were built using Consense (from the Phylip package). Bayesian evolutionary tree estimation (Sjölander, 1998) was used to build the tree shown; branching within the clade containing ACIK1 proteins (node N28) was well supported by strong bootstrap values in the other trees.
RT-PCR
Total RNA from leaves or from cultured suspension cells was isolated using the Tri Reagent method according to the manufacturer's recommendations (Sigma-Aldrich, St. Louis, MO). First-strand cDNA was synthesized from 2 μg of total RNA using Expand reverse transcriptase (Roche, Indianapolis, IN). RT-PCR was performed as described previously (Romeis et al., 2001) using the following primers: NtACIK-ATG-ClaI and NtACIK1-mutDRev, LeACIK1-flfwd and LeACIK1-RTPCRrev, or Cf9-16s and Cf9-18a. Actin was used as control for equal cDNA amounts using primers AC1 and AC2.
In Vitro Kinase Assay
The full-length cDNAs of NtACIK1 and LeACIK1 were amplified by PCR with primers NtACIK-ATG-Bgl and NtACIK-STOP-Bgl or LeACIK-ATG-Sal1 and LeACIK-STOP-Pst1, respectively. The 1326-bp NtACIK1 and the 1395-bp LeACIK1 PCR products were sequenced and inserted into either the BamHI (NtACIK1) or SalI/PstI (LeACIK1) sites of pMALc2X (New England Biolabs, Beverly, MA). pMALc2X contains the gene encoding MBP upstream of multiple cloning sites. Expression of fusion proteins in Escherichia. coli BL21 cells and protein purification using amylose resin was performed according to the manufacturer's instructions. To determine the in vitro kinase activity of MBP-LeACIK1, 200 ng of purified protein was incubated in a 30-μL reaction containing 40 mM Hepes, 2 mM DTT, 0.1 mM EGTA, 10 mM MgCl2, 10 mM MnCl2, 1 mM CaCl2, 5 μg of kinase substrate, 125 kBq [γ-32P]ATP (111 TBq/mmol; NEN Life Science Products, Boston, MA), and 50 μM ATP. Kinase substrates used were histone type III-SS (Sigma-Aldrich) and myelin-basic protein (Sigma-Aldrich). After incubation for 30 min at 30°C, the reaction was stopped by adding 15 μL of SDS loading dye. Samples were analyzed by SDS-PAGE, Coomassie Brilliant Blue stain, and autoradiography to detect incorporation of 32P into the kinase substrates.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers listed in Table 2. The accession numbers for the other sequences described in this article are as follows: NtACIK1 (AY220481), LeACIK1 (AF332960), AtACIK1a (AAL07094), and AtACIK1b (NP_198408).
Supplementary Material
Acknowledgments
We thank David Baulcombe's laboratory for the PVX and TRV vectors used for VIGS and for providing the GFP-PVX and GFP-TMV constructs as well as N- and Rx-transgenic N. benthamiana plants. We also thank Shelley Hepworth, Thorsten Nürnberger, and Frédéric Brunner for helpful comments on the manuscript. This research was supported by the Gatsby Charitable Foundation. O.R. was supported by a Human Frontiers Science Program fellowship. A.A.L. was supported by a European Community Marie Curie Fellowship (EC Grant HPMF/CT-2001-01288). L.F.-L. was supported in part by a National Science Foundation grant to K.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jonathan D.G. Jones (jonathan.jones@sainsbury-laboratory.ac.uk).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.026013.
References
- Akira, S., Takeda, K., and Kaisho, T. (2001). Toll-like receptors: Critical proteins linking innate and acquired immunity. Nat. Immunol. 2, 675–680. [DOI] [PubMed] [Google Scholar]
- Balint-Kurti, P.J., Dixon, M.S., Jones, D.A., Norcott, K.A., and Jones, J.D.G. (1994). RFLP linkage analysis of the Cf-4 and Cf-9 genes for resistance to Cladosporium fulvum in tomato. Theor. Appl. Genet. 88, 691–700. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D.C. (1999). Fast forward genetics based on virus-induced gene silencing. Curr. Opin. Plant Biol. 2, 109–113. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A., Kanyuka, K., and Baulcombe, D.C. (1999). The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatt, M.R., Grabov, A., Brearley, J., Hammond-Kosack, K.E., and Jones, J.D.G. (1999). K+ channels of Cf-9 transgenic tobacco guard cells as targets for Cladosporium fulvum Avr9 elicitor-dependent signal transduction. Plant J. 19, 453–462. [DOI] [PubMed] [Google Scholar]
- Bogdanove, A.J., and Martin, G.B. (2000). AvrPto-dependent Pto-interacting proteins and AvrPto-interacting proteins in tomato. Proc. Natl. Acad. Sci. USA 97, 8836–8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong, Y.H., Chang, H.-S., Gupta, R., Wang, X., Zhu, T., and Luan, S. (2002). Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 129, 661–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang, C.F., and Meyerowitz, E.M. (2000). Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97, 4985–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costet, L., Fritig, B., and Kauffmann, S. (2002). Scopoletin expression in elicitor-treated and tobacco mosaic virus-infected tobacco plants. Physiol. Plant. 115, 228–235. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Deshaies, R.J. (1999). SCF and cullin/RING H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Dixon, M.S., Golstein, C., Thomas, C.M., van der Biezen, E.A., and Jones, J.D.G. (2000). Genetic complexity of pathogen perception by plants: The example of Rcr3, a tomato gene required specifically by Cf-2. Proc. Natl. Acad. Sci. USA 97, 8807–8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant, W.E., Rowland, O., Piedras, P., Hammond-Kosack, K.E., and Jones, J.D.G. (2000). cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell 12, 963–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekengren, S.K., Liu, Y., Schiff, M., Dinesh-Kumar, S.P., and Martin, G.B. (2003). Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J. 36, 905–917. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (1993). PHYLIP (Phylogeny Inference Package) Version 3.5c. (Seattle: University of Washington).
- Gachon, C., Baltz, R., and Saindrenan, P. (2004). Over-expression of a scopoletin glucosyltransferase in Nicotiana tabacum leads to precocious lesion formation during the hypersensitive response to tobacco mosaic virus but does not affect virus resistance. Plant Mol. Biol. 54, 137–146. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J., Rogers, E.E., and Ausubel, F.M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143, 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez, L., and Boller, T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez, L., and Boller, T. (2002). Flagellin perception: A paradigm for innate immunity. Trends Plant Sci. 7, 251–256. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., Jones, D.A., and Jones, J.D.G. (1994). Identification of two genes required in tomato for full Cf-9-dependent resistance to Cladosporium fulvum. Plant Cell 6, 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., Tang, S., Harrison, K., and Jones, J.D.G. (1998). The tomato Cf-9 disease resistance gene functions in tobacco and potato to confer responsiveness to the fungal avirulence gene product avr 9. Plant Cell 10, 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie, D.G. (1999). Plant protein serine/threonine kinases: Classification and functions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 97–131. [DOI] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832. [DOI] [PubMed] [Google Scholar]
- Hirayama, T., and Oka, A. (1992). Novel protein kinase of Arabidopsis thaliana (APK1) that phosphorylates tyrosine, serine and threonine. Plant Mol. Biol. 20, 653–662. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M. (1995). Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr. Opin. Cell Biol. 7, 215–223. [DOI] [PubMed] [Google Scholar]
- Hoffmann, J.A., and Reichhart, J.-M. (2002). Drosophila innate immunity: An evolutionary perspective. Nat. Immunol. 3, 121–126. [DOI] [PubMed] [Google Scholar]
- Hwang, I.W., and Goodman, H.M. (1995). An Arabidopsis thaliana root-specific kinase homolog is induced by dehydration, ABA, and NaCl. Plant J. 8, 37–43. [DOI] [PubMed] [Google Scholar]
- Ito, T., Takahashi, N., Shimura, Y., and Okada, K. (1997). A serine/threonine protein kinase gene isolated by an in vivo binding procedure using the Arabidopsis floral homeotic gene product, AGAMOUS. Plant Cell Physiol. 38, 248–258. [DOI] [PubMed] [Google Scholar]
- Jin, H., Liu, Y., Yang, K.Y., Kim, C.Y., Baker, B., and Zhang, S. (2003). Function of a mitogen-activated protein kinase pathway in N gene-mediated resistance in tobacco. Plant J. 33, 719–731. [DOI] [PubMed] [Google Scholar]
- Joosten, M.H., Cozijnsen, T.J., and De Wit, P.J. (1994). Host resistance to a fungal tomato pathogen lost by a single base-pair change in an avirulence gene. Nature 367, 384–386. [DOI] [PubMed] [Google Scholar]
- Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooman-Gersmann, M., Honee, G., Bonnema, G., and De Wit, P.J. (1996). A high-affinity binding site for the AVR9 peptide elicitor of Cladosporium fulvum is present on plasma membranes of tomato and other Solanaceous plants. Plant Cell 8, 929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger, J., Thomas, C.M., Golstein, C., Dixon, M.S., Smoker, M., Tang, S., Mulde, L., and Jones, J.D.G. (2002). A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 296, 744–747. [DOI] [PubMed] [Google Scholar]
- Kumar, S.K.G., Tang, W., Ravindranath, A.K., Clark, W.A., Croze, E., and Fuchs, S.Y. (2003). SCFHOS ubiquitin ligase mediates the ligand-induced down-regulation of the interferon-a receptor. EMBO J. 22, 5480–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.L., Schiff, M., and Dinesh-Kumar, S.P. (2002). Virus-induced gene silencing in tomato. Plant J. 31, 777–786. [DOI] [PubMed] [Google Scholar]
- Lu, R., Malcuit, I., Moffett, P., Ruiz, M.T., Peart, J., Wu, A.J., Rathjen, J.P., Bendahmane, A., Day, L., and Baulcombe, D.C. (2003. a). High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22, 5690–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R., Martin-Hernandez, A.M., Peart, J.R., Malcuit, I., and Baulcombe, D.C. (2003. b). Virus-induced gene silencing in plants. Methods 30, 296–303. [DOI] [PubMed] [Google Scholar]
- Luderer, R., et al. (2001). No evidence for binding between resistance gene product Cf-9 of tomato and avirulence gene product AVR9 of Cladosporium fulvum. Mol. Plant-Microbe Interact. 14, 867–876. [DOI] [PubMed] [Google Scholar]
- Ludwig, A.A., Romeis, T., and Jones, J.D.G. (2004). CDPK-mediated signaling pathways: Specificity and cross-talk. J. Exp. Bot. 55, 181–188. [DOI] [PubMed] [Google Scholar]
- Martin, G.B., Bogdanove, A.J., and Sessa, G. (2003). Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54, 23–61. [DOI] [PubMed] [Google Scholar]
- Martin, G.B., Brommonschenkel, S.H., Chunwongse, J., Frary, A., Ganal, M.W., Spivey, R., Wu, T., Earle, E.D., and Tanksley, S.D. (1993). Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262, 1432–1436. [DOI] [PubMed] [Google Scholar]
- Menke, F.L.H., van Pelt, J.A., Pieterse, C.M.J., and Klessig, D.F. (2004). Silencing of the mitogen-activated protein kinase MPK6 compromises disease resistance in Arabidopsis. Plant Cell 16, 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran, T.V., and Walker, J.C. (1993). Molecular cloning of two novel protein kinase genes from Arabidopsis thaliana. Biochim. Biophys. Acta 1216, 9–14. [DOI] [PubMed] [Google Scholar]
- Murase, K., Shiba, H., Iwano, M., Che, F.S., Watanabe, M., Isogai, A., and Takayama, S. (2004). A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science 303, 1516–1519. [DOI] [PubMed] [Google Scholar]
- Mysore, K.S., Crasta, O.R., Tuori, R.P., Folkerts, O., Swirsky, P.B., and Martin, G.B. (2002). Comprehensive transcript profiling of Pto- and Prf-mediated host defense responses to infection with Pseudomonas syringae pv. tomato. Plant J. 32, 299–315. [DOI] [PubMed] [Google Scholar]
- Navarro, L., Zipfel, C., Rowland, O., Keller, I., Robatzek, S., Boller, T., and Jones, J.D.G. (2004). The transcriptional innate immune response to flg22: Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 135, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart, J.R., Cook, G., Feys, B.J., Parker, J.E., and Baulcombe, D.C. (2002. a). An EDS1 orthologue is required for N-mediated resistance against tobacco mosaic virus. Plant J. 29, 569–579. [DOI] [PubMed] [Google Scholar]
- Peart, J.R., et al. (2002. b). Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. USA 99, 10865–10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piedras, P., Hammond-Kosack, K.E., Harrison, K., and Jones, J.D.G. (1998). Rapid, Cf-9- and Avr9-dependent production of active oxygen species in tobacco suspension cultures. Mol. Plant-Microbe Interact. 11, 1155–1166. [Google Scholar]
- Ratcliff, F., Martin-Hernandez, A.M., and Baulcombe, D.C. (2001). Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25, 237–245. [DOI] [PubMed] [Google Scholar]
- Rivas, S., Rougon, A., Smoker, M., Schauser, L., Yoshioka, H., and Jones, J.D.G. (2004). CITRX thioredoxin is a negative regulator of cell death and defense responses that interacts with the tomato Cf-9 resistance protein. EMBO J. 23, 2156–2165. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Rivas, S., and Thomas, C.M. (2002). Recent advances in the study of tomato Cf resistance genes. Mol. Plant Pathol. 3, 277–282. [DOI] [PubMed] [Google Scholar]
- Romeis, T., Ludwig, A.A., Martin, R., and Jones, J.D.G. (2001). Calcium-dependent protein kinases play an essential role in a plant defense response. EMBO J. 20, 5556–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis, T., Piedras, P., and Jones, J.D.G. (2000). Resistance gene-dependent activation of a calcium-dependent protein kinase in the plant defense response. Plant Cell 12, 803–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis, T., Piedras, P., Zhang, S., Klessig, D.F., Hirt, H., and Jones, J.D.G. (1999). Rapid Avr9- and Cf-9 -dependent activation of MAP kinases in tobacco cell cultures and leaves: Convergence of resistance gene, elicitor, wound, and salicylate responses. Plant Cell 11, 273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, P.C., Ito, A., Shimizu, T., Terauchi, R., Kamoun, S., and Saitoh, H. (2003). Virus-induced silencing of WIPK and SIPK genes reduces resistance to a bacterial pathogen, but has no effect on the INF1-induced hypersensitive response (HR) in Nicotiana benthamiana. Mol. Genet. Genomics 269, 583–591. [DOI] [PubMed] [Google Scholar]
- Shiu, S.H., and Bleecker, A.B. (2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98, 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, S.H., Karlowski, W.M., Pan, R., Tzeng, Y.H., Mayer, K.F.X., and Li, W.H. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16, 1220–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjölander, K. (1998). Phylogenetic inference in protein superfamilies: Analysis of SH2 domains. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 165–174. [PubMed] [Google Scholar]
- Smith, N.A., Singh, S.P., Wang, M.B., Stoutjesdijk, P.A., Green, A.G., and Waterhouse, P.M. (2000). Total silencing by intron-spliced hairpin RNAs. Nature 407, 319–320. [DOI] [PubMed] [Google Scholar]
- Song, W.Y., Wang, G.L., Chen, L.L., Kim, H.S., Pi, L.Y., Holsten, T., Gardner, J., Wang, B., Zhai, W.X., Zhu, L.H., Fauquet, C., and Ronald, P. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806. [DOI] [PubMed] [Google Scholar]
- Sonnhammer, E.L.L., Eddy, S.R., Birney, E., Gateman, A., and Durbin, R. (1998). Pfam: Multiple sequence alignments and HMM. Nucleic Acids Res. 26, 320–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiderski, M.R., and Innes, R.W. (2001). The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J. 26, 101–112. [DOI] [PubMed] [Google Scholar]
- Tao, Y., Xie, Z., Chen, W., Glazebrook, J., Chang, H.-S., Han, B., Zhu, T., Zou, G., and Katagiri, F. (2003). Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, C.L., Jones, L., Baulcombe, D.C., and Maule, A.J. (2001). Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nicotiana benthamiana using potato virus X vector. Plant J. 25, 417–425. [DOI] [PubMed] [Google Scholar]
- Thomas, C.M., Dixon, M.S., Parniske, M., Golstein, C., and Jones, J.D.G. (1998). Genetic and molecular analysis of tomato Cf genes for resistance to Cladosporium fulvum. Philos. Trans. R. Soc. Lond. B Biol. Sci. 353, 1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, C.M., Jones, D.A., Parniske, M., Harrison, K., Balint-Kurti, P.J., Hatzixanthis, K., and Jones, J.D.G. (1997). Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell 9, 2209–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, C.M., Tang, S., Hammond-Kosack, K., and Jones, J.D.G. (2000). Comparison of the hypersensitive response induced by the tomato Cf-4 and Cf-9 genes in Nicotiana spp. Mol. Plant-Microbe Interact. 13, 465–469. [DOI] [PubMed] [Google Scholar]
- Underhill, D.M., and Ozinsky, A. (2002). Toll-like receptors: Key mediators of microbe detection. Curr. Opin. Immunol. 14, 103–110. [DOI] [PubMed] [Google Scholar]
- Vancanneyt, G., Schmidt, R., O'Connor-Sanchez, A., Willmitzer, L., and Rocha-Sosa, M. (1990). Construction of an intron-containing marker gene: Splicing of the intron in transgenic plants and its use in monitoring early events in Agrobacterium-mediated plant transformation. Mol. Gen. Genet. 220, 245–250. [DOI] [PubMed] [Google Scholar]
- Van den Ackerveken, G.F., Vossen, P., and De Wit, P.J. (1993). The AVR9 race-specific elicitor of Cladosporium fulvum is processed by endogenous and plant proteases. Plant Physiol. 103, 91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J.M., Loh, Y.T., Bressan, R.A., and Martin, G.B. (1995). The tomato gene Pti1 encodes a serine threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell 83, 925–935. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.