Abstract
Geminivirus replication enhancer (REn) proteins dramatically increase the accumulation of viral DNA species by an unknown mechanism. In this study, we present evidence implicating SlNAC1, a new member of the NAC domain protein family from tomato (Solanum lycopersicum), in Tomato leaf curl virus (TLCV) REn function. We isolated SlNAC1 using yeast (Saccharomyces cerevisiae) two-hybrid technology and TLCV REn as bait, and confirmed the interaction between these proteins in vitro. TLCV induces SlNAC1 expression specifically in infected cells, and this upregulation requires REn. In a transient TLCV replication system, overexpression of SlNAC1 resulted in a substantial increase in viral DNA accumulation. SlNAC1 colocalized with REn to the nucleus and activated transcription of a reporter gene in yeast, suggesting that in healthy cells it functions as a transcription factor. Together, these results imply that SlNAC1 plays an important role in the process by which REn enhances TLCV replication.
INTRODUCTION
Geminiviruses are a large and diverse family of plant-infecting pathogens segregated into four genera based on genome structure, insect vectors, and host range (van Regenmortel et al., 2000). They possess small, single-stranded DNA genomes comprising one or two components of 2.6 to 2.8 kb. Of the proteins they encode, only the replication-associated protein (Rep) is essential for virus replication. Rep, the product of the C1 (also designated AC1, L1, or AL1) gene, specifically recognizes and binds the viral origin (Fontes et al., 1994; Behjatnia et al., 1998), and nicks and religates viral DNA to initiate and terminate rolling-circle replication (Laufs et al., 1995). However, it does not have a DNA polymerase function. Therefore, geminiviruses are heavily dependent on host factors to amplify their genome, but many infect differentiated cells that have exited the cell division cycle and cannot support DNA replication (Nagar et al., 1995; Lucy et al., 1996; Sudarshana et al., 1998; Morra and Petty, 2000). As a consequence, an early step in the geminivirus infection process is induction of the required replication machinery.
Reprogramming of the plant cell to facilitate geminivirus replication appears to rely heavily on an interaction between Rep and retinoblastoma (Rb; Xie et al., 1995; Collin et al., 1996; Grafi et al., 1996; Ach et al., 1997). In animal cells, Rb modulates the activity of E2F transcription factors, which are involved in the transcriptional regulation of genes expressed at the G1/S boundary of the cell division cycle (Harbour and Dean, 2000). Mammalian DNA oncoviruses encode proteins that interact with Rb and consequently disrupt the Rb control pathway (Chellappan et al., 1992; Zamanian and La Thangue, 1992). The idea that geminiviral Rep proteins may act in an analogous manner was first suggested from the observation that induction of proliferating cell nuclear antigen (PCNA) occurred in transgenic plants expressing Rep from Tomato golden mosaic virus (TGMV; Nagar et al., 1995). PCNA is an accessory factor for DNA polymerase δ that is normally present only in S-phase cells (Daidoji et al., 1992). Two lines of evidence imply that induction of PCNA is mediated by the Rep–Rb interaction. First, analysis of Rep mutants revealed that the ability of Rep to activate PCNA expression is linked tightly to its capacity to interact with Rb (Kong et al., 2000). Second, induction of PCNA occurs at the transcriptional level, and the PCNA promoter is under E2F negative control (Egelkrout et al., 2001).
It is becoming increasingly evident that other host factors are involved in geminivirus replication. Xie et al. (1999) identified two wheat (Triticum aestivum) proteins, GRAB1 and GRAB2, which interact with Wheat dwarf virus (WDV) RepA. Overexpression of these proteins in cultured cells inhibited WDV DNA replication, suggesting that RepA disrupts a GRAB-mediated response that represses viral infection. GRAB1 and GRAB2 are both members of the recently identified NAC family of genes found in many plant species but, so far, not in other eukaryotes. NAC proteins share a common structure consisting of a conserved N-terminal region (the NAC domain) and a highly variable C terminus. The name is derived from the three type members, NO APICAL MERISTEM (NAM) from petunia (Petunia hybrida; Souer et al., 1996) and the ATAF and CUP-SHAPED COTYLEDON (CUC) genes from Arabidopsis thaliana (Aida et al., 1997). Since the identification of these genes, many more NACs have been found; Ooka et al. (2003) studied the rice (Oryza sativa) and Arabidopsis genomes and found 75 and 105 predicted NAC proteins in each species, respectively. NACs possess roles as diverse as pattern formation in embryos (Souer et al., 1996), flower development (Sablowski and Meyerowitz, 1998), leaf senescence (John et al., 1997), and auxin-dependent lateral foot formation (Xie et al., 2000). In addition to these developmental roles, they have also been implicated in plant defense responses (Collinge and Boller, 2001).
Geminiviral replication enhancer (REn) proteins (also designated C3, AC3, L3, or AL3) are able to increase viral DNA accumulation (Elmer et al., 1988; Sunter et al., 1990) and enhance infectivity and symptom expression (Hormuzdi and Bisaro, 1995). Although little molecular information regarding this process is available, replication accessory factors encoded by mammalian oncoviruses often interact with host proteins to generate a cellular environment suited to DNA replication (Jansen-Durr, 1996). Consistent with this idea, REn was recently shown to bind Rb (Settlage et al., 2001), implying that, like Rep, this protein is involved in disruption of cell cycle controls. In this study, we screened a tomato (Solanum lycopersicum) library for proteins that interact with the REn protein from the Australian Tomato leaf curl virus (TLCV) to determine whether other host factors are involved in REn function. A new member of the NAC domain family, which we have named SlNAC1 (for S. lycopersicum NAC1), was shown to interact with REn in yeast (Saccharomyces cerevisiae) and in vitro. Here, we present evidence implicating SlNAC1 in REn-mediated enhancement of viral DNA accumulation.
RESULTS
Identification of a NAC Domain Protein That Interacts with REn
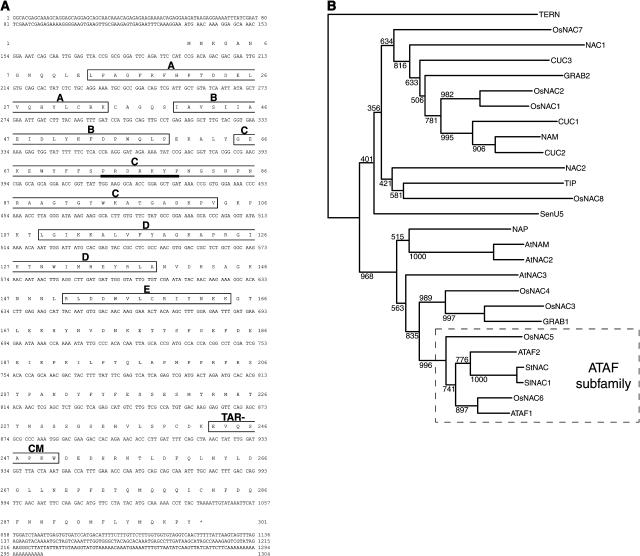
To identify host proteins interacting with the TLCV REn protein, a yeast two-hybrid screen of a tomato cDNA library fused to the B42 activation domain (AD)–encoding sequence (Zhou et al., 1995) was performed using REn fused to the LexA DNA-binding domain (BD) as bait. A total of 2 × 106 transformants were assayed for Leu prototrophic growth and green fluorescent protein (GFP) activity. Of these transformants, one was able to activate both reporter genes. Plasmid DNA was rescued, verified by retransformation into yeast with the bait, and the sequence of the insert determined. The plasmid contained a cDNA of 1304 bp encoding a predicted full-length translation product of 301 amino acids. Nucleotides 52 to 597 of the cDNA are identical to an EST generated from tomato carpel tissue (GenBank accession number AI486942). A BLAST query of the protein sequence revealed that the N-terminal 169 amino acid residues contain the five conserved blocks of homology that comprise the NAC domain (Figure 1A, boxed). Based on this defining characteristic, we named the protein SlNAC1 for S. lycopersicum NAC1 (GenBank accession number AY498713).
Figure 1.
Nucleotide Sequence of SlNAC1 and Alignment of Its Putative Translation Product with Other NAC Domain Proteins.
(A) Nucleotide and amino acid sequences of SlNAC1. The five subdomains (A to E) comprising the NAC domain are shown in empty boxes. A putative nuclear localization signal is indicated by a bold line under the sequence PRDRKYP. The TAR-CM of the ATAF subgroup is also boxed.
(B) The predicted amino acid sequence of SlNAC1 (Figure 1A) and known NAC family proteins were subjected to phylogenetic analysis. Multiple sequence alignment of the proteins was conducted using ClustalX (Thompson et al., 1997), and phylogenetic analysis was performed by the neighbor-joining method (Saitou and Nei, 1987). A bootstrap analysis of 1000 resampling replicates was conducted with ClustalX. The rooted phylogenetic tree was displayed using the NJPlot program included with ClustalX. The gene names and references for other NACs are as follows: A. thaliana, ATAF1 and ATAF2 (Aida et al., 1997), AtNAC2 (Takada et al., 2001), AtNAC3 (Takada et al., 2001), AtNAM (Duval et al., 2002), CUC1 (Takada et al., 2001), CUC2 (Takada et al., 2001), CUC3 (Vroemen et al., 2003), NAC1 (Xie et al., 2000), NAC2, NAP (Sablowski and Meyerowitz, 1998), and TIP (Ren et al., 2000); rice, OsNAC1 to OsNAC8 (Kikuchi et al., 2000); petunia, NAM (Souer et al., 1996); tomato, SenU5 (John et al., 1997); potato, StNAC (Collinge and Boller, 2001); and wheat, GRAB1 and GRAB2 (Xie et al., 1999).
A recent phylogenetic analysis of the NAC domains from known NAC family proteins and putative Arabidopsis and rice NACs separated them into 18 subgroups (Ooka et al., 2003). We compared the NAC domains from SlNAC1 and other known NAC family proteins. According to dendograms obtained by the neighbor-joining method (Figure 1B) and the maximum-parsimony method (data not shown), SlNAC1 falls into the so-called ATAF subgroup. The C-terminal region of NAC proteins, termed the transcriptional activation region (TAR), is highly divergent, but Ooka et al. (2003) found 13 common motifs (CMs) in 12 of the 18 subgroups. Members of the ATAF subgroup contain the sequence EVQS[E/x]PK[W/l], which is also present in SlNAC1 (Figure 1A, boxed and labeled TAR-CM). This supports our classification of SlNAC1 into this subgroup. Analysis of the primary sequence of SlNAC1 using PSORT II (http://bioweb.pasteur.fr/seqanal/interfaces/psort2.html) identified a putative classical (SV40 large T antigen–type) nuclear localization signal in subdomain C from amino acids 74 to 80 (Figure 1A, underlined). This sequence, PRDRKYP, was conserved amongst 12 NACs in a study performed by Kikuchi et al. (2000), suggesting that it may be functional in vivo.
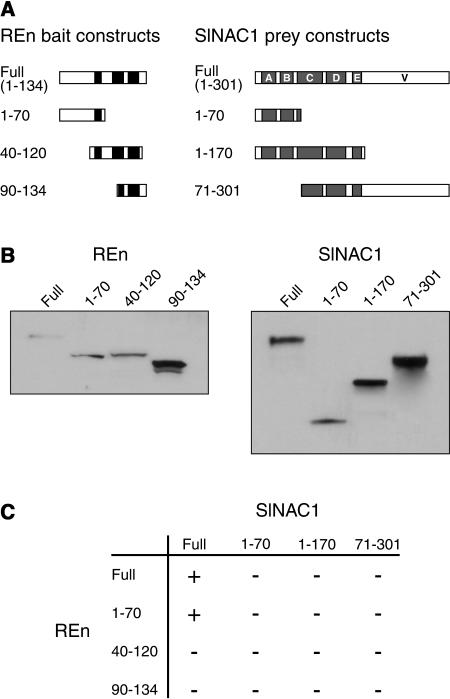
To map the domains responsible for the interaction between REn and SlNAC1, truncations of the genes encoding both proteins were made and cloned into pLexA and pB42AD to create fusions with the LexA DNA BD and B42 AD, respectively. The secondary structure of REn, predicted using PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/), contains three α-helices found between amino acids 56 to 65, 79 to 95, and 101 to 116. Because α-helices are frequently important in protein–protein interactions, we generated three truncations of REn based on the location of these putative structures (Figure 2A). REn 1-70 contains only the first helix, REn 40-120 contains all three helices, whereas REn 90-134 contains the third helix and a part of the second. Three truncations of SlNAC1 were made based on the location of NAC subdomains (Figure 2A): SlNAC1 1-70 contains subdomains A, B, and a small part of C; SlNAC1 1-170 contains all of the five subdomains that make up the NAC domain; and SlNAC1 71-301 contains subdomains D and E and the majority of C, and all of the variable C terminus.
Figure 2.
Deletion Analysis of REn and SlNAC1 to Identify Regions Required for Interaction between the Two Proteins.
(A) Diagrammatic representation of REn (bait) and SlNAC1 proteins (prey) tested for interaction. The REn proteins were expressed as LexA DNA BD fusions, and the SlNAC1 proteins were expressed as B42 AD fusions. The positions of three putative α-helices in REn are indicated by closed boxes. In SlNAC1, the positions of the NAC subdomains are shown in shaded boxes (A to E), whereas the variable C terminus is denoted V.
(B) Immunoblot analysis of yeast cells demonstrating that noninteracting REn-LexA fusions and SlNAC1-B42 fusions are expressed at levels similar to those of interacting fusion proteins. Total protein from yeast cultures containing different REn and SlNAC1 fusion proteins was extracted, fractionated on 4 to 20% SDS-polyacrylamide gels, and immunoblotted with anti-LexA (to detect REn-LexA fusions) or anti-hemagglutinin (HA) (to detect SlNAC1-B42 fusions).
(C) The N-terminal region of REn is important for SlNAC1 binding. Interaction was indicated by the ability of cells transformed with bait, prey, and displayREPORTER plasmids to grow on medium lacking Leu. As an additional indicator of interaction, colonies were monitored for GFP expression by visualization under UV light.
Each of the REn and SlNAC1 truncations as well as the full-length proteins were coexpressed in yeast and their interaction assayed by Leu prototrophic growth and GFP expression. REn 1-70 was able to interact with full-length SlNAC1 (Figure 2C), whereas the other two REn truncations could not. This suggests that the first putative α-helix of REn may be involved in SlNAC1 binding. None of the three truncations of SlNAC1 were able to interact with REn in yeast. This may indicate the involvement of a larger proportion of SlNAC1 in the interaction or reflect structural constraints imposed on the functional REn-interacting domain. Immunoblot analysis of yeast cells demonstrated that noninteracting REn and SlNAC1 truncations were expressed at levels similar to those of interacting proteins (Figure 2B), confirming that negative results were not as a result of an absence of protein.
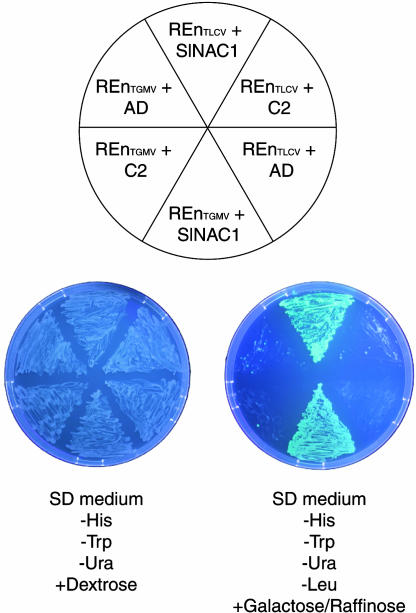
We examined the general significance of SlNAC1 binding to TLCV REn by testing whether SlNAC1 could also interact with REn encoded by TGMV. TGMV is a bipartite begomovirus encoding a REn protein that is 54.2% identical in sequence (65.6% similar) to TLCV REn. In the same yeast two-hybrid assay, TGMV REn also interacted with SlNAC1, as shown in Figure 3. Neither REn protein interacted with the control protein TLCV C2 (also designated AC2, L2, AL2, or TrAP) nor with the AD alone. A test of Tomato yellow leaf curl Sardinia virus (TYLCSV) REn–SlNAC1 interaction was not possible because TYLCSV REn exhibited weak autoactivation activity in our yeast system (data not shown).
Figure 3.
SlNAC1 Interacts with Both TLCV and TGMV REn.
Yeast two-hybrid assays testing the ability of SlNAC1 to interact with REn of TLCV (REnTLCV) and TGMV (REnTGMV). Yeast coexpressing proteins as indicated (top) were grown on SD – His – Trp – uracil (Ura) medium (bottom left), and interaction was tested by Leu prototrophy and GFP expression on an inductive carbon source (galactose and raffinose; bottom right). REn proteins were fused to the LexA DNA BD, whereas SlNAC1 was fused to the B42 AD. Negative controls included REnTLCV and REnTGMV coexpressed with TLCV C2 fused to the AD, or coexpressed with AD alone.
SlNAC1 Acts as a Transcriptional Activator in Yeast
There is considerable evidence to suggest that NAC domain proteins function as transcription factors. First, several NACs, including ATAF1 and ATAF2 (Souer et al., 1996), AtNAM (Duval et al., 2002), NAC1 (Xie et al., 2000), TIP (Ren et al., 2000), and a group of Brassica napus NACs (Hegedus et al., 2003), are able to activate transcription of a reporter gene in yeast, an activity mediated by the divergent C-terminal sequences. Second, AtNAM and NAC1 bind a specific DNA sequence found in the 35S promoter of Cauliflower mosaic virus (CaMV; Xie et al., 2000; Duval et al., 2002). Third, overexpression of NAC1 in Arabidopsis caused upregulation of the auxin-responsive genes AIR3 and DBP (Xie et al., 2000), whereas CUC1 activated the expression of genes involved in the development of the shoot apical meristem (Hibara et al., 2003). Finally, a nuclear localization pattern has been observed for NAC1 (Xie et al., 2000).
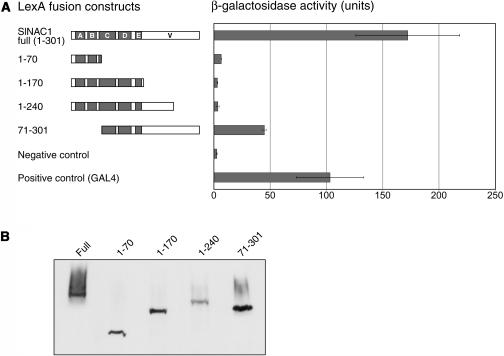
Based on these data, we tested for the presence of an AD in SlNAC1 using yeast as an assay system. A SlNAC1 fusion to the LexA DNA BD was coexpressed with pSH18-34, which contains eight LexA operators that direct transcription of the lacZ gene (Golemis et al., 1994). Cells were assayed for β-galactosidase activity using a liquid culture assay (see Methods). As predicted, the LexA:SlNAC1 fusion was able to activate expression of the reporter gene, and its transactivation activity was at least as strong as the positive control, a LexA fusion to the GAL4 AD (Figure 4A). Four truncations of the SlNAC1 gene were fused to LexA to determine the domains required for transcriptional activation (Figure 4A). This deletion analysis revealed that the variable C-terminal region (amino acids 71 to 301) could activate transcription of lacZ but more weakly than full-length SlNAC1. None of the N-terminal fragments (1-70, 1-170, and 1-240) were able to promote expression of lacZ. These data indicate that SlNAC1 has a transcriptional AD that is active in yeast and is located near its C terminus. Immunoblotting confirmed that all LexA:SlNAC1 fusion proteins were produced at similar levels in yeast (Figure 4B).
Figure 4.
The Divergent C-Terminal Region of SlNAC1 Is Able to Activate Transcription in Yeast.
(A) Regions of SlNAC1 able to activate transcription in yeast. The LexA:SlNAC1 fusion proteins are represented diagrammatically at left, with the positions of the NAC subdomains shown in shaded boxes (A to E) and the variable C terminus denoted V. The ability of LexA:SlNAC1 fusion proteins to activate transcription in yeast is shown at right. Activities were assayed by measuring β-galactosidase activity in total protein extracts from cells containing pLexA-SlNAC1 plasmids and pSH18-34, which contains a lacZ reporter gene downstream of the LexA recognition site. Positive control corresponds to yeast containing pSH18-34 and expressing a LexA fusion with the GAL4 AD. Negative control corresponds to yeast containing pSH18-34 and expressing LexA. Error bars indicate the standard deviation for each sample.
(B) Immunoblot analysis of yeast cells demonstrating that nontransactivating LexA:SlNAC1 fusions are expressed at levels similar to those of transactivating fusion proteins. Total protein from yeast cultures containing fusion proteins was extracted, fractionated on SDS-polyacrylamide gels, and immunoblotted with anti-LexA.
In Vitro Binding of SlNAC1 to TLCV REn
The specificity of the REn/SlNAC1 protein interaction was tested using an in vitro pull down assay. A 6×His-REn fusion protein was expressed in Escherichia coli, purified to homogeneity, and mixed with crude soluble protein extracted from E. coli cells induced to express a SlNAC1-calmodulin binding peptide (CBP) fusion protein containing a FLAG epitope (CBP-SlNAC1). The mixture was incubated with nickel-nitrilotriacetic acid agarose (Ni-NTA), washed extensively to remove unbound protein, resuspended in loading buffer, electrophoresed, and transferred to polyvinylidene fluoride (PVDF) membrane. The presence of CBP-SlNAC1 and 6×His-REn was determined by immunoblotting using antibodies directed against FLAG and polyHis, respectively.
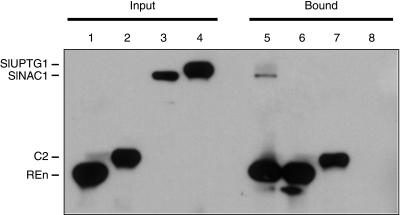
Bound CBP-SlNAC1 was detectable when incubated with 6×His-REn (Figure 5, lane 5). To determine the specificity of CBP-SlNAC1 binding, it was added to Ni-NTA resin alone (lane 8) or in combination with purified 6×His-C2, another TLCV-encoded protein (lane 7). In both of these reactions, CBP-SlNAC1 was not detected in the bound fraction, indicating that it was interacting specifically with 6×His-REn. To determine if 6×His-REn was specifically pulling down CBP-SlNAC1, we mixed it with total soluble protein extracted from cells induced to express CBP-SlUPTG1, a control CBP-tagged protein (lane 6). No CBP-SlUPTG1 was detectable in the bound fraction, indicating that 6×His-REn does not indiscriminately bind abundant proteins in a mixture. SlUPTG1 is a tomato homolog of potato (Solanum tuberosum) UDP-glucose:protein transglucosylase identified in another of our yeast two-hybrid screens (accession number AY622990). All reactions were performed at least twice with consistent results.
Figure 5.
REn Interacts with SlNAC1 in Vitro.
Purified 6×His-tagged proteins were mixed with crude CBP-tagged protein mixtures, incubated with Ni-NTA, and washed extensively to remove any unbound protein. Bound protein was resuspended in loading buffer, resolved by SDS-PAGE, and analyzed by immunoblotting using anti-polyHis and anti-FLAG (CBP-tagged proteins also contain a FLAG epitope) antibodies. Reactions were as follows: 6×His-REn and CBP-SlNAC1 (lane 5), 6×His-REn and CBP-SlUPTG1 (lane 6), 6×His-C2 and CBP-SlNAC1 (lane 7), and CBP-SlNAC1 alone (lane 8). Protein inputs for each reaction are shown: 6×His-REn (lane 1), 6×His-C2 (lane 2), CBP-SlNAC1 (lane 3), and CBP-SlUPTG1 (lane 4).
REn and SlNAC1 Are Targeted to the Nucleus
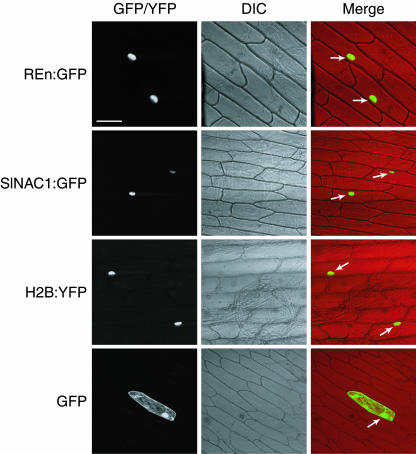
To investigate the potential role of SlNAC1 in REn function in vivo and to further verify the putative interaction between these proteins, we examined the subcellular localization of SlNAC1 and REn in plant cells. Each of the open reading frames (ORFs) was fused to GFP downstream of the CaMV 35S promoter. The fusion proteins (REn:GFP and SlNAC1:GFP) were transiently expressed in onion (Allium cepa) epidermal cells after biolistic delivery of vector DNA and analyzed by confocal microscopy.
Free GFP was distributed in both the cytoplasm and the nucleus of bombarded cells (Figure 6, bottom). By contrast, both REn:GFP and SlNAC1:GFP localized exclusively to nuclei (top and second rows), which were clearly visible as dense ovoid structures when cells were viewed with differential interference optics (middle column). Further verifying this result, the distribution pattern of REn:GFP and SlNAC1:GFP matched that of the Arabidopsis HISTONE 2B:yellow fluorescent protein (H2B:YFP) fusion protein (third row), a control for nuclear localization (Boisnard-Lorig et al., 2001). NAC1 from Arabidopsis (Xie et al., 2000) and CmNACP from pumpkin (Cucurbita maxima; Ruiz-Medrano et al., 1999) were also found to be nuclear proteins, implying that this is a general characteristic of NAC proteins and supporting the idea that they function as transcription factors. More importantly, however, the localization of REn and SlNAC1 to the nucleus suggests that an opportunity exists for binding between these proteins in TLCV-infected plants.
Figure 6.
REn and SlNAC1 Localize to the Nucleus of Onion Cells.
REn:GFP (top row) and SlNAC1:GFP (second row), as well as GFP alone (bottom row), were expressed in onion epidermal cells using the CaMV 35S promoter after biolistic delivery of vector DNA. A positive control for nuclear localization, H2B:YFP, is also shown (third row). Cells were analyzed for GFP and YFP fluorescence (left column) by confocal microscopy. Differential interference contrast (DIC) images and merge images are shown in the middle and right columns, respectively. Nuclei in merge images are indicated by arrows. Bar = 100 μm.
TLCV Infection Induces the Expression of SlNAC1
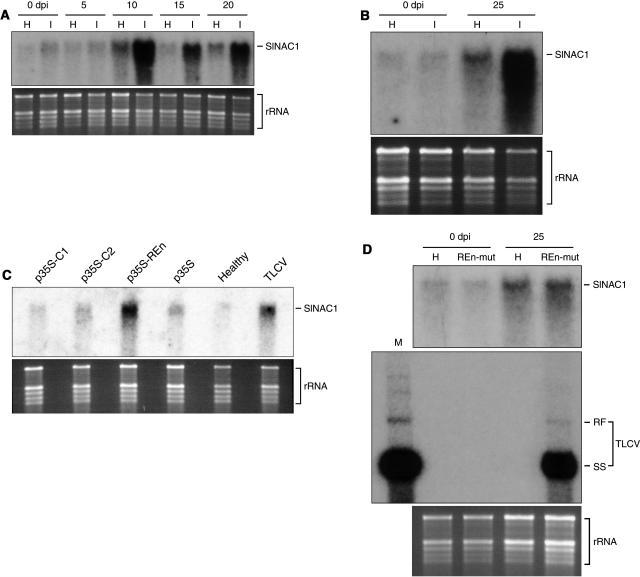
To analyze the endogenous expression of SlNAC1, we performed an RNA gel blot analysis of total RNA preparations from tomato leaf tissue. SlNAC1 mRNA of the predicted size (∼1300 nucleotides) was detectable at low levels in healthy tomato leaves (data not shown). To test whether SlNAC1 transcription might be regulated by TLCV infection, total RNA from new, emerging leaves of infected and healthy plants sampled at various time points postinoculation was analyzed (Figure 7A). SlNAC1 expression was strongly induced in infected plants at 10 d postinoculation (dpi) and maintained to at least 20 dpi, a result observed in three independent experiments. Infection with TYLCSV caused a similar increase in the levels of SlNAC1 transcript (Figure 7B), suggesting that induction of this gene is a general response to geminivirus infection. Some fluctuation in the level of SlNAC1 gene expression in healthy plants over the course of these experiments was also observed, although this was minimal compared with the induction caused by geminiviral infection.
Figure 7.
SlNAC1 Is Induced by TLCV Infection.
(A) TLCV infection results in an upregulation of SlNAC1 gene expression. RNA gel blot showing the expression of SlNAC1 in healthy (H) or TLCV-infected (I) tomato plants. Tissue samples were obtained at 0, 5, 10, 15, and 20 dpi.
(B) TYLCSV infection results in an upregulation of SlNAC1 gene expression. RNA gel blot showing the expression of SlNAC1 in healthy or TYLCSV-infected tomato plants. Tissue samples were obtained 0 and 25 dpi.
(C) Transient expression of REn is sufficient to induce SlNAC1 gene expression. Tomato leaves were infiltrated with A. tumefaciens cells containing a replication-competent TLCV 1.1mer, p35S, or p35S expressing the TLCV genes C1, C2, and REn. RNA was extracted from tissues 5 d postinfiltration and SlNAC1 expression analyzed by RNA gel blotting.
(D) A TLCV REn mutant cannot induce SlNAC1 gene expression. RNA gel blot showing the expression of SlNAC1 in healthy plants or plants infected with a TLCV REn mutant (REn-mut) at 0 and 25 dpi (top). The presence of replicating TLCV REn mutant was confirmed by DNA gel blotting (middle). In this blot, we also ran an extract obtained from plants infected with wild-type virus (left, designated M); the ratio of REn mutant:wild-type virus total nucleic acid extracts is 20:1. TLCV DNA species are marked RF (supercoiled double-stranded replicative form) and SS (single stranded).
We asked whether REn, given its physical interaction with SlNAC1, plays a role in the regulation of SlNAC1 gene expression. Tomato leaf tissue was infiltrated with Agrobacterium tumefaciens cells harboring a REn expression construct (p35S-REn), and changes in SlNAC1 transcript accumulation were analyzed (Figure 7C). Expression of REn induced SlNAC1 gene expression to levels similar to that observed when tissue was infiltrated with cells containing a replicating TLCV construct. By contrast, tissue that was infiltrated with A. tumefaciens containing an empty expression vector or vectors designed to express two other TLCV-encoded genes, C1 and C2, contained levels of SlNAC1 transcript similar to untreated tissue. These results suggested that induction of SlNAC1 in response to TLCV infection is mediated by REn, and also demonstrated that SlNAC1 is not induced nonspecifically in response to A. tumefaciens infection or wounding associated with the infiltration procedure. In a subsequent experiment, a TLCV derivative containing a mutation in the C3 gene that prevents translation of the REn protein (Rigden et al., 1996) was tested for its effect on SlNAC1 expression. The level of SlNAC1 transcript in tomato plants agroinoculated with the REn mutant was comparable to healthy controls 25 dpi (Figure 7D, top). The presence of replicating REn mutant virus was confirmed by DNA gel blotting the same total nucleic acid samples and hybridizing with a TLCV-specific probe (Figure 7D, middle). Together, our results strongly imply that REn alone is responsible for induction of SlNAC1. It must be noted that, as expected, the amount of viral DNA in extracts obtained from REn mutant–infected plants (middle, right lane) was much lower than equivalent samples from plants infected with wild-type virus (middle, lane M; the ratio of REn mutant:wild-type total nucleic acid extracts is 20:1). Thus, this experiment does not rule out the possibility that the absence of SlNAC1 induction in REn mutant–infected plants is because of reduced viral load.
In all RNA gel-blot analyses, indistinguishable results were obtained when membranes were hybridized with probes synthesized from the full-length SlNAC1 gene or from only the divergent 3′ sequence (data not shown), indicating that variation in the expression of other putative NAC genes in response to TLCV infection or to transient REn expression was insignificant.
TLCV Replication Is Tissue Specific, and SlNAC1 Induction Occurs Only in TLCV-Infected Cells
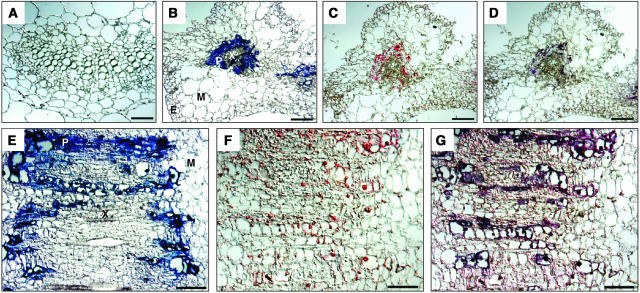
SlNAC1 upregulation may be a systemic stress response, or alternatively TLCV may act to specifically induce expression of this gene in infected cells. To distinguish between these possibilities, we performed in situ hybridization experiments to analyze the specific regions of SlNAC1 mRNA accumulation compared with sites of TLCV infection. Hybridization of tomato tissue with TLCV and SlNAC1 probes produced only very weak chromogenic signals (data not shown). Because TLCV-derived nucleic acid accumulates to much higher levels in Nicotiana benthamiana, leaf tissue derived from this host was analyzed. A single-stranded RNA (ssRNA) complementary to the TLCV V2 gene produced a strong signal that was observed mainly in phloem cells but also in some xylem parenchyma and bundle sheath cells (Figures 8B and 8E). This indicates that TLCV is limited to vascular tissue, a characteristic also reported for Abutilon mosaic virus, Squash leaf curl virus, and Tomato yellow leaf curl virus from the Dominican Republic (Horns and Jeske, 1991; Sanderfoot and Lazarowitz, 1996; Rojas et al., 2001). No signal was obtained when healthy N. benthamiana leaf tissue was hybridized with a probe complementary to the divergent 3′ SlNAC1 sequence, which should not detect unrelated N. benthamiana NAC proteins (Figure 8A). However, in TLCV-infected sections, a SlNAC1 homolog was detected in some phloem cells (Figures 8C and 8F). To test whether induction of this gene was occurring only in cells infected with TLCV, dual-color in situ hybridizations were performed (Jowett, 2001). Hybridization of the TLCV probe to sections exhibiting a SlNAC1 signal produced a distinctive purple chromogenic output (Figures 8D and 8G). This color is produced by the masking of the red SlNAC1 signal by the blue viral signal, and confirms that almost every cell that accumulated substantial amounts of SlNAC1 mRNA also contained TLCV. Thus, induction of a N. benthamiana SlNAC1 homolog in response to TLCV infection is not a systemic response but rather is restricted to cells infected with the virus.
Figure 8.
Induction of SlNAC1 by TLCV Occurs Only in Infected Cells.
Tissue sections derived from mock-inoculated (A) and TLCV-infected ([B] to [G]) leaves of N. benthamiana were hybridized with either fluorescein-labeled ssRNA probe complementary to SlNAC1 ([A], [C], [D], [F], and [G]) or DIG-labeled ssRNA probe complementary to TLCV ([A], [B], [D], [E], and [G]). (A) to (D) are cross sections and (E) to (G) are longitudinal sections taken from the main leaf vein. Bar = 100 μm. Cell types present are indicated: E, epidermal; M, mesophyll; P, phloem; and X, xylem.
The Expression Level of SlNAC1 Is a Determinant of Geminiviral Replication
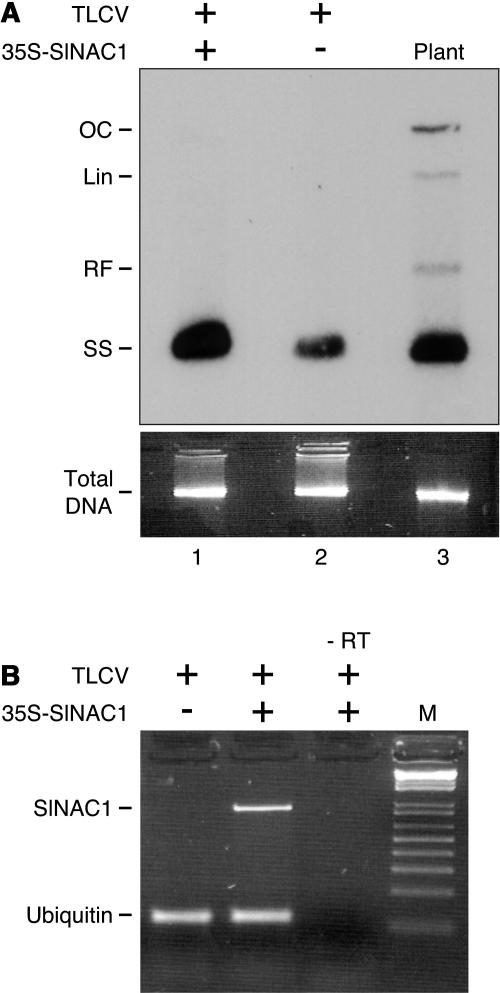
To investigate the possible function of SlNAC1 in relation to TLCV infection, the effect of constitutive, high-level expression of this gene on TLCV replication was analyzed. A transient TLCV replication system, based on Agrobacterium-mediated delivery of an infectious TLCV construct into N. benthamiana leaf strips (Dry et al., 1997), was used in this study. When A. tumefaciens cells harboring the infectious TLCV construct were combined with A. tumefaciens containing an SlNAC1 expression construct, TLCV ssDNA accumulated to a level equivalent to that observed in TLCV-infected plants (Figure 9A, cf. lanes 1 and 3). The level of TLCV ssDNA produced in the presence of 35S-driven SlNAC1 expression was considerably more than that observed in our control treatment, in which TLCV was combined with an empty expression construct (lane 2). This response was observed in four independent experiments in which all treatments were performed in duplicate. The level of SlNAC1 expression in all leaf strip samples was concurrently analyzed by semiquantitative real-time PCR, which confirmed that the enhancement of TLCV ssDNA accumulation was associated with SlNAC1 expression by p35S-SlNAC1 (Figure 9B). Together, these results suggest that SlNAC1 is involved in TLCV replication in planta.
Figure 9.
SlNAC1 Expression Enhances TLCV ssDNA Accumulation.
(A) Expression of SlNAC1 enhances TLCV ssDNA accumulation in a transient replication assay. A. tumefaciens cells harboring Bin19-TLCV1.1 were combined with A. tumefaciens cells containing either an empty expression construct (lane 2) or p35S-SlNAC1 (lane 1) and cocultivated for 48 h with leaf strips from N. benthamiana plants. DNA was extracted from tissue samples 3 d later and replication of TLCV analyzed by DNA gel blotting. Lane 3 (Plant) is a sample extracted from TLCV-infected N. benthamiana used as a marker for TLCV DNA forms, marked OC (open circular double stranded), Lin (linear double stranded), RF (supercoiled double-stranded replicative form), and SS (single stranded). OC, Lin and RF DNA forms were observed in extracts from N. benthamiana leaf strips after longer exposures.
(B) Analysis of SlNAC1 expression by p35S-SlNAC1 in N. benthamiana leaf strips by semiquantitative RT-PCR. Total RNA was prepared from leaf strips treated with TLCV plus an empty expression construct or TLCV plus p35S-SlNAC1. Ubiquitin mRNA served as an internal control. RT reaction mix without reverse transcriptase was used as a negative control (marked –RT). M, size markers.
DISCUSSION
Because of their limited coding capacities, geminiviruses depend on host factors to amplify their genomes. In quiescent cells that have exited the cell division cycle and cannot support DNA replication, these pathogens must therefore induce the required replicational machinery. To achieve this, they encode proteins that increase the expression level of growth-promoting genes and/or alter the function of cell-cycle regulatory proteins, often by physically interacting with host factors. For example, the geminiviral Rep protein upregulates expression of PCNA, possibly by interfering with Rb/E2F-mediated transcriptional repression of the PCNA gene through its interaction with Rb (Egelkrout et al., 2001). Rep also binds histone H3 (Kong and Hanley-Bowdoin, 2002), suggesting that it may act to alleviate repression of virus replication and transcription processes induced by the packaging of geminiviral double-stranded DNA species into minichromosomes (Abouzid et al., 1988; Pilartz and Jeske, 1992). The other viral protein required for high levels of viral DNA accumulation, REn, is involved in several protein–protein interactions. It binds to Rep and may increase the affinity of this protein for the viral origin of replication (Fontes et al., 1994; Settlage et al., 1996; Gladfelter et al., 1997), an activity proposed to enhance viral replication (Hanley-Bowdoin et al., 1999). REn also interacts with the host Rb and PCNA proteins, suggesting that its role in replication is multifaceted (Settlage et al., 2001; Castillo et al., 2003). Consistent with this idea, we report here that a new tomato protein of the NAC domain family, SlNAC1, is induced by and interacts with REn and appears to be involved in viral replication.
The Role of SlNAC1 in TLCV Infection
In a transient replication system, expression of SlNAC1 considerably enhanced the accumulation of TLCV ssDNA (Figure 9A), suggesting that this gene may facilitate TLCV replication. It is not logical to consider that tomato plants would retain a gene that promotes disease, and, therefore, SlNAC1 must perform some essential cellular function. However, our results suggest that geminiviruses, through the action of REn proteins, have hijacked the innate role of SlNAC1. Several mechanisms can be envisioned to explain this result. One is that SlNAC1 acts indirectly in TLCV replication as a positive regulator of cellular genes required during viral infection. For example, it may activate transcription of genes required for S-phase functions that are normally absent in differentiated cells, a strategy analogous to the putative release of E2F transcription factors when geminiviral Rep proteins bind Rb. This explanation does not appear to correlate with the proposed function of other NAC proteins in meristem development and plant senescence pathways, where these factors contribute to a decision of cells to leave the proliferative state and take a certain differentiation pathway. For example, NAM is thought to interfere with cell division around the developing shoot apical meristem (Souer et al., 1996) to drive flower development. Further, the observation that GRAB proteins from wheat interfere with replication of WDV led Xie et al. (1999) to speculate that these NACs play a role in the pathway leading to cell differentiation. However, the family of genes encoding NAC domains is very large and members appear to possess highly diverse functions. Thus, it is reasonable to speculate that some NACs could upregulate genes involved in processes advantageous to geminivirus replication, such as DNA replication, transcription, or the G1/S transition of the cell cycle. Supporting this idea, NAC1 from Arabidopsis is involved in the initiation of lateral root development (Xie et al., 2000), whereas CUC1 promotes adventitious SAM formation by maintaining epidermal cells in an undifferentiated state in transgenic Arabidopsis (Takada et al., 2001; Hibara et al., 2003).
Another possibility is that SlNAC1 functions directly in geminiviral replication. There are numerous examples in which host transcription factors play an important and direct role in activating the DNA replication of mammalian oncoviruses by binding the viral origin of replication and increasing the initiation frequency (Li et al., 1998, and references cited therein). Alternatively, REn may recruit SlNAC1 into a DNA replication complex, where it could promote amplification of the viral genome. This idea is supported by the observation that REn interacts with PCNA (Castillo et al., 2003), a host factor that acts as a sliding clamp and modulates the interaction of other proteins, including polymerases, with DNA (reviewed in Hingorani and O'Donnell, 2000).
A third possible scenario is that SlNAC1 could positively modulate transcription of viral genes. Although the geminiviral C2 protein is responsible for activating virion-sense gene expression (Sunter and Bisaro, 1992; Sunter et al., 1994; Dry et al., 2000), expression of the complementary-sense genes is probably controlled by host factors. SlNAC1 mediates expression of a reporter gene in yeast (Figure 4), suggesting that it may function endogenously as a transcription factor and, therefore, positively modulate cis-acting promoter elements in the geminiviral genome.
Mechanism of TLCV-Mediated SlNAC1 Induction
SlNAC1 gene expression was upregulated in response to TLCV infection (Figures 7 and 8). Two lines of evidence presented in this study support the idea that this induction is mediated by REn. First, transient delivery of a REn expression construct resulted in increased accumulation of SlNAC1 mRNA, whereas control constructs were unable to engender this response (Figure 7C). Second, a TLCV REn mutant was unable to upregulate SlNAC1 despite accumulating to moderate levels in infected tissue (Figure 7D). Several mechanisms by which SlNAC1 is induced can be envisioned. First, REn could act directly as a transactivator of SlNAC1 gene expression. Analysis of the peptide sequence of TGMV REn revealed that its acidic N terminus resembles some transcriptional ADs (Hanley-Bowdoin et al., 1999). Second, SlNAC1 induction may be a side effect of the presence of REn in a plant cell. It is doubtful that it occurs via the putative REn–Rb or REn–PCNA interactions because Rep, which also binds these host factors, was unable to stimulate SlNAC1 expression. However, SlNAC1 induction may occur because REn is impinging on other cellular processes, possibly through an as yet uncharacterized protein interaction. This explanation is supported by the observation that REn and Rep produced highly disparate phenotypic effects when transiently expressed in host plants (Selth et al., 2004). Finally, although at this time we have no evidence to suggest that induction of SlNAC1 relies on the REn–SlNAC1 protein interaction, this possibility cannot be discounted. For example, by sequestering SlNAC1 through physical interaction, REn may relieve a negative feedback mechanism by which SlNAC1 inhibits transcription of its gene. Such a function is not unprecedented: AtWRKY6, a member of the large WRKY family of plant-specific transcriptional regulators, is able to suppress its own promoter activity while positively influencing the expression of genes involved in senescence and pathogen defense (Robatzek and Somssich, 2002).
It was originally proposed that NAC proteins could be divided into three subfamilies (Kikuchi et al., 2000). More recently, Ooka et al. (2003) performed a more comprehensive phylogenetic analysis of known NACs and putative Arabidopsis and rice NACs and identified 18 subfamilies. Members of the so-called ATAF subfamily, identified in both studies, appear to share a conserved role in the response to stress. Genes belonging to this group are induced by wounding (Collinge and Boller, 2001), fungal infection (Collinge and Boller, 2001; Hegedus et al., 2003), bacterial infection (Mysore et al., 2002), insect damage (Hegedus et al., 2003), and cold shock (Hegedus et al., 2003). SlNAC1, which also belongs to the ATAF subfamily (Figure 1B), is induced by TLCV (this study) and Pseudomonas syringae (Mysore et al., 2002) infection, suggesting that it may play a general role in stress responses. However, four lines of evidence support the idea that stimulation of SlNAC1 gene expression by TLCV is a specific response and that SlNAC1 plays an active role in TLCV infection. First, neither A. tumefaciens infection nor wounding associated with the agroinfiltration procedure induced SlNAC1. Second, induction of SlNAC1 by TLCV is restricted to infected cells and appears to be mediated by the REn protein. Third, SlNAC1 interacts with the TLCV-encoded REn protein. Finally, overexpression of SlNAC1 enhances the accumulation of TLCV DNA species in a transient replication system.
NACs Are Involved in Other Viral Infections
Xie et al. (1999) found an interaction between the WDV RepA protein and two wheat NACs, GRAB1 and GRAB2. The N terminus (amino acids 1 to 208) of TLCV Rep shares 39.5% sequence identity with full-length WDV RepA. However, we were unable to detect binding between bacterially expressed TLCV Rep and SlNAC1 in vitro (data not shown). Another apparent difference between the WDV–GRAB and TLCV–SlNAC1 interactions is that, although expression of both GRAB genes was shown to interfere with WDV DNA replication in cultured wheat cells, SlNAC1 expression enhanced TLCV ssDNA accumulation in a transient replication assay. The distinct roles of GRABs and SlNAC1 in geminivirus infection may again reflect the functional diversity that exists between members of the NAC domain family. Supporting the idea of NACs possessing diverse roles in viral pathogenesis, the NAC domain–containing Arabidopsis TIP protein is involved in the Turnip crinkle virus resistance response pathway by interacting with the Turnip crinkle virus coat protein (Ren et al., 2000). Alternatively, it could denote different DNA replication strategies used by the highly divergent dicot-infecting TLCV and monocot-infecting WDV. It would be useful to examine the effect WDV infection has on the expression level of GRAB1 and GRAB2 to see whether, in contrast with the situation with TLCV and SlNAC1, the virus downregulates these detrimental genes.
REn has previously been reported to physically interact with itself, Rep, Rb, and PCNA. Thus, it probably plays several roles in geminiviral infection, including the establishment of a cellular environment competent for DNA replication and an involvement in initiation of viral DNA replication. Despite its apparent multifunctionality, our results strongly imply that the mechanism by which REn increases viral ssDNA accumulation involves its interaction with SlNAC1. We do not yet know at which stage REn/SlNAC1 binding is involved in TLCV replication: it may have a direct role in this process or could simply be required to induce SlNAC1, which in turn acts to facilitate TLCV replication. However, the data presented here suggest that tomato plants silenced for SlNAC1 expression, if not impaired in other functions, may exhibit tolerance to TLCV infection. We are currently attempting to generate transgenic tomato plants stably expressing a SlNAC1-hairpin construct to test this resistance strategy.
METHODS
Oligonucleotides Used in This Study
Oligonucleotide sequences shown in Table 1 were synthesized by Geneworks (Adelaide, Australia). Bold letters in the oligonucleotide sequences indicate added restriction sites used for cloning.
Table 1.
Oligonucleotide Primers Used in This Study
| Name | Sequence |
|---|---|
| P1 | 5′-TTGAATTCGATTCACGCACAGGGGAACC-3′a |
| P2 | 5′-GGGGCTCGAGTTAATAAAAATTAAATTTTA-3′ |
| P3 | 5′-AAGCCTCGAGTCATGTGAAGTCCAGGAA-3′ |
| P4 | 5′-GGGAATTCAACTACAACCACGAC-3′ |
| P5 | 5′-AAGCCTCGAGTCATGAGTCTAGTACATT-3′ |
| P6 | 5′-GGGGAATTCAAGTATTTAGATAGT-3′ |
| P7 | 5′-GGGAATTCAACAAAGGAGCAAACGGA-3′ |
| P8 | 5′-AAGCCTCGAGTCAATACCACTCTTTTTC-3′ |
| P9 | 5′-AAGCCTCGAGTCAATGCTTCTCAAGTGT-3′ |
| P10 | 5′-GGGGAATTCTTTTTCTCACCAAGG-3′ |
| P11 | 5′-TTGCGGCCGCTTAGTAAGGTTTTTGCAT-3′ |
| P12 | 5′-AAAAGCGGCCGCTCAACATGGCGACAAGAC-3′ |
| P13 | 5′-TTTGGATCCGAT TCACGCACAGGGGA-3′ |
| P14 | 5′-GGGGGGGGGAAGCTTTTAATAAAAATTAAATTT-3′ |
| P15 | 5′-TTGGATCCCAGAATTCATCACCC-3′ |
| P16 | 5′-GGGGAAGCTTTTAAATACCCTCAAG-3′ |
| P17 | 5′-TTTGGATCCAACAAAGGAGCAAACGGA-3′ |
| P18 | 5′-TTGCGGCCGCTTAGTAAGGTTTTTGCAT-3′ |
| P19 | 5′-TTGAATTCGCAGCAGCAACACCA-3′ |
| P20 | 5′-GGCTCGAGCTACTTTTTAGTCTT-3′ |
| P21 | 5′-GGGGGTCTAGATTAATAAAAATTAAATTTTA-3′ |
| P22 | 5′-GGGTCTAGATTAGTAAGGTTTTTGCAT-3′ |
| P23 | 5′-TCCCCCGGGATGACTAGACCAAAGTCATTCCGTATAAATGCTAA-3′ |
| P24 | 5′-CGCGGATCCTCAATTCTCTTCCTCCGGATGG-3′ |
| P25 | 5′-GGGGATCCATGTTCAGAATTCATCACCCTCAAC-3′ |
| P26 | 5′-GGGGATCCTTAAATACCCTCAAGAAACG-3′ |
| P27 | 5′-CGGGGTACCATGGATTCACGCACAGGGGAACC-3′ |
| P28 | 5′-TGCTCTAGAGTTAATAAAAATTAAATTTTATATCATGAT-3′ |
| P29 | 5′-AATCTAGAGAATTCAGTACCGCCTCGCCAACG-3′ |
| P30 | 5′-GGGGATCCGGTACCTTAGTAAGGTTTTTGCAT-3′ |
| P31 | 5′-TTGGATCCAGCAAGCGACCAGCAGAT-3′ |
| P32 | 5′-GGGGGGAAGCTTTTAATTCTGAATCGAATC-3′ |
| P33 | 5′-CGGGGTACCATGAACAAAGGAGCAAACGGAAATCAG-3′ |
| P34 | 5′-TGCTCTAGATTAGTAAGGTTTTTGCATGTATAGG-3′ |
| P35 | 5′-CGGCATGCTTAACACATGCA-3′ |
| P36 | 5′-AGCCGTTTCCAGCTGTTGTTC-3′ |
Sequences shown in boldface correspond to specific restriction enzyme sites.
Yeast Two-Hybrid Screen
Yeast (Saccharomyces cerevisiae) strain displayYEAST-L (MATα, trp1, his3, ura3, leu2∷2 LexAop-LEU2; Display Systems Biotech, Vista, CA), containing a Leu biosynthesis gene downstream of the DNA recognition sequence for the LexA DNA BD, was used in two-hybrid screening. The REn ORF was amplified by PCR using P1 and P2. The product was digested with EcoRI and XhoI and ligated into similarly digested pLexA (HIS3 marker) to generate pLexA-Ren, which expresses a fusion of REn to the LexA DNA BD. Yeast was first transformed with displayREPORTER (URA3 marker), a vector containing the GFP ORF downstream of the DNA recognition sequence for the LexA DNA BD. Cells were then sequentially transformed with pLexA-REn and with a pJG4-5 (TRP1 marker) tomato (Solanum lycopersicum) Rio Grande cDNA library (Zhou et al., 1995). Colonies were selected on agar plates lacking uracil, His, Trp, and Leu but containing galactose and raffinose to induce the GAL1 promoter driving expression of the tomato cDNAs fused to the B42 AD. Large colonies appearing within 5 d and exhibiting GFP expression were spread on plates lacking uracil, His, and Trp, and then transferred back to plates selecting for activation of the LEU2 gene to remove false positives. Cells were then grown in media lacking Trp to select for pJG4-5 and yeast plasmid DNA purified using an RPM yeast plasmid isolation kit (Q-Biogene, Carlsbad, CA). Escherichia coli KC8 was transformed with purified pJG4-5 plasmid DNA because this strain is trp1− and its defect can be complemented by the TRP1 gene present in pJG4-5. To further eliminate false positives, plasmid DNA purified from KC8 was transformed back into displayYEAST-L containing displayREPORTER and pLexA-REn and the activation of GFP and LEU2 reassessed.
To analyze the domains of the proteins responsible for their interaction, truncations of the REn and SlNAC1 genes were cloned into pLexA and pJG4-5, respectively. The fragments amplified were as follows: REn encoding amino acids 1 to 70 (P1 and P3), REn 40-120 (P4 and P5), REn 90-134 (P6 and P2), SlNAC1 1-70 (P7 and P8), SlNAC1 1-170 (P7 and P9), and SlNAC1 71-301 (P10 and P11). Products were digested with EcoRI/XhoI (REn 1-70, REn 40-120, REn 90-134, SlNAC1 1-70, and SlNAC1 1-170) or EcoRI/NotI (SlNAC1 71-301) and ligated into similarly digested pLexA or pJG4-5.
The SlNAC1 truncation sequences described above were transferred into pLexA to delineate the protein's putative transcriptional AD in yeast. Two other SlNAC1 sequences were cloned into EcoRI/NotI-digested pLexA for this yeast one-hybrid study: full-length SlNAC1 (amplified using primers P7 and P11) and a fragment encoding amino acids 1 to 240 (P7 and P12).
To measure the ability of different LexA-SlNAC1 fusion proteins to act as transcriptional activators, a reporter plasmid that contains eight LexA operators that direct transcription of the lacZ gene was used (pSH18-34; Golemis et al., 1994). Quantitative β-galactosidase assays from liquid cultures were performed according to the Yeast Protocols Handbook 2001 (Clontech, Palo Alto, CA) using o-nitrophenyl-β-d-galactopyranoside as substrate. A Microplate Reader 450 (Bio-Rad, Hercules, CA) was used to measure accumulation of the o-nitrophenol product. One unit of β-galactosidase is defined as the amount of activity hydrolyzing 1 nmol o-nitrophenyl-β-d-galactopyranoside per minute per cell. The assay was performed twice using three independent transformants for each construct. The positive control plasmid used in this study, pSH17-4, expresses a LexA fusion to the GAL4 AD (Golemis et al., 1994).
To monitor fusion protein production in yeast, total protein (0.3 mL of yeast culture equivalent) was extracted and size fractionated on 4% to 20% Tris-Gly-SDS polyacrylamide gels (Gradipore, Frenchs Forest, Australia). Electrophoresed protein samples were transferred to Immobilon P PVDF membrane (Millipore, Billerica, MA) and blocked with 5% (w/v) nonfat dry milk before incubation with either rabbit anti-LexA polyclonal antibody (Invitrogen, Carlsbad, CA) to detect LexA-fusion proteins or anti-HA monoclonal antibody (Sigma-Aldrich, St. Louis, MO) to detect B42-fusion proteins. Donkey anti-Rabbit (Sigma) or Goat anti-Mouse IgG-horseradish peroxidase conjugate (Promega, Madison, WI) was used as the secondary antibody and detected using SuperSignal West Pico chemiluminescent substrate (Pierce Biotechnology, Rockford, IL).
Production of Recombinant Proteins and in Vitro Binding Experiments
Production of 6×His-tagged REn and C2 proteins was achieved using the pQE30 vector (Qiagen, Clifton Hill, Australia). The coding region of REn was amplified using oligonucleotides P13 and P14, digested with BamHI/HindIII, and ligated into similarly digested pQE30 to generate pQE30-REn. pQE30-C2 was constructed in the same way, using oligonucletides P15 and P16 to amplify the C2 gene.
A 6×His-REn recombinant protein was purified using a protocol developed by Behjatnia et al. (1998) for the preparation of 6×His-Rep protein, with minor modifications. Briefly, E. coli M15 cells were transformed with pQE30-REn, grown to an OD of 0.9, and induced with 1 mM isopropyl-β-d-galactopyranoside (IPTG) for 3 h at room temperature. Cells were harvested, resuspended in Ni-NTA binding buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 10 mM β-mercaptoethanol, and 1% Tween-20), and lysed by 1 mg/mL lysozyme, freeze-thawing, and sonication. Crude soluble protein was retrieved by centrifugation and 6×His-REn protein purified using Ni-NTA agarose (Qiagen). For the production of 6×His-C2, cells were transformed with pQE30-C2, grown to an OD of 0.7, and induced with 1 mM IPTG for 3 h at 37°C. Total soluble protein was extracted using sarkosyl by the method of Frangioni and Neel (1993) and dialysed against Ni-NTA binding buffer before purification of the recombinant protein.
Production of CBP-tagged SlNAC1 and SlUPTG1 proteins containing a FLAG epitope was achieved using the pCAL-n-FLAG vector (Stratagene, La Jolla, CA). The coding region of SlNAC1 was amplified by PCR using the oligonucleotides P17 and P18, digested with BamHI, and ligated into BamHI/SmaI-digested pCAL-n-FLAG vector to generate pCAL-SlNAC1. The SlUPTG1 ORF was amplified using the oligonucleotides P19 and P20, digested with EcoRI/XhoI, and ligated into similarly digested pCAL-n-FLAG to yield pCAL-SlUPTG1.
E. coli B834-pLysS cells were transformed with pCAL-SlNAC1 and pCAL-SlUPTG1, grown to an OD of 0.7, and induced with 0.5 mM IPTG at 37°C for 3 h. Crude soluble protein was extracted using sarkosyl as described by Frangioni and Neel (1993).
Binding experiments were performed by adding 50 ng of a purified 6×His-tagged protein and 200 ng of total soluble protein extracted from cells induced to express the CBP-tagged protein of interest to 10 μL of Ni-NTA agarose in 300 μL of binding buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0) in an Eppendorf tube. Tubes were then mixed gently on a rotating platform at 4°C for 40 min. The resin was washed three times by brief centrifugation and resuspension in 400 μL of binding buffer, resuspended in 50 μL of sample loading buffer (0.5 M Tris-HCl, 10% glycerol, 2% SDS, 5% β-mercaptoethanol, and 1% bromophenol blue), and incubated at 94°C for 10 min. Aliquots (10 μL) of eluate from the pelleted beads were size fractionated on 4 to 20% Tris-Gly-SDS polyacrylamide gels. Electrophoresed protein samples were transferred to Immobilon P PVDF membrane and blocked with 5% (w/v) nonfat dry milk before incubation with mouse anti-polyHis and anti-FLAG monoclonal antibodies (Sigma). Goat anti-Mouse IgG-horseradish peroxidase conjugate was used as the secondary antibody and detected using SuperSignal West Pico chemiluminescent substrate.
Analysis of GFP Fusion Proteins by Microprojectile Bombardment
A variant of the shuttle vector pART7 (Gleave, 1992) termed pART7-C′gfp, which contains the full-length GFP ORF (lacking the stop codon) upstream of the multiple cloning site (T. Franks, unpublished data), was used to transiently express REn:GFP and SlNAC1:GFP fusion proteins in onion (Allium cepa) tissue. Full-length REn was amplified using primers P1 and P21, and the SlNAC1 ORF was amplified using primers P7 and P22. After restriction enzyme digestion with EcoRI and XbaI, fragments were ligated into similarly digested pART7-C′gfp to generate C-terminal fusions with GFP. Also used in this experiment were pART7-ATG:GFP, which expresses free GFP (T. Franks, unpublished data), and pBI121-H2B:YFP, which expresses Arabidopsis H2B fused to the GFP yellow variant YFP (Boisnard-Lorig et al., 2001).
Onion epidermal strips on agar containing MS salt mixture (Invitrogen) were bombarded with each of the vectors. For four shots, 400 μg of gold particles in 100 μL of ethanol were vortexed for 2 min, spun down for 10 s in a microfuge, drained, washed twice with sterile water, and resuspended in 25 μL of 40% glycerol. While gently vortexing, 4 μL of the plasmid solutions (400 ng/μL), 10 μL of cold 0.1 M spermidine, and 25 μL of 2.5 M CaCl2 were added dropwise and the resulting mixture incubated on ice for 10 min. The particles were spun down, washed with 70% ethanol, resuspended in 24 μL of cold 100% ethanol, and 6-μL aliquots were placed onto sterile filter holders. After bombardment, tissue was stored in the dark for 48 h and GFP/YFP expression visualized using a Bio-Rad Radiance 2100 confocal laser scanning microscope system. The excitation wavelength used for both GFP and YFP analysis was 488 nm.
Analysis of SlNAC1 Gene Expression
Three-week-old tomato plants were inoculated with TLCV, TYLCSV, or the TLCV REn mutant (Rigden et al., 1996) using Agrobacterium tumefaciens (Grimsley et al., 1987). Total nucleic acid was extracted at various time points from new, emerging leaves and subjected to RNA gel blot analysis as described (Selth et al., 2004). To detect replication of the REn mutant, the same samples were analyzed by DNA gel blotting as described (Dry et al., 1993).
The binary vector pART27 was used to express individual TLCV genes to analyze their effect on SlNAC1 mRNA production. Primers to amplify C1 (P23 and P24), C2 (P25 and P26), and C3 (P27 and P28) were designed. The PCR products were digested with SmaI/HindIII (C1), BamHI (C2), and KpnI/XbaI (C3) and ligated into similarly digested pART7. Fragments containing the CaMV 35S promoter upstream of the TLCV gene were liberated from these plasmids by digestion with NotI and ligated into pART27 previously cut with NotI and dephosphorylated. The resultant vectors were designated p35S-C1, p35S-C2, and p35S-C3. Young tomato leaves were infiltrated with A. tumefaciens C58 cells containing the p35S constructs as described (Selth et al., 2004). At 5 d postinfiltration, SlNAC1 expression was analyzed by RNA gel blot analysis.
In Situ Hybridizations
Templates for the generation of ribonucleic probes were constructed as follows. A fragment comprising nucleotides 401 to 906 of SlNAC1 was amplified using primers P29 and P30 and ligated into pGEM-T-Easy (Promega). The full-length TLCV V2 ORF was amplified using primers P31 and P32 and ligated into pGEM-T-Easy. Plasmids were linearized with NdeI (SlNAC1) or SalI (V2) and transcribed with T7 RNA polymerase. RNA probes labeled with fluorescein-12-UTP (for SlNAC1 probes) and digoxigenin (DIG)-11-dUTP (for TLCV probes) were prepared using fluorescein or DIG RNA labeling mix, respectively (Roche Diagnostics, Castle Hill, Australia).
Plant material was collected from TLCV-infected plants 3 weeks postinoculation. Preparation of tissue sections and hybridization of DIG- and fluorescein-labeled probes was performed as described by Guerin et al. (2000). Probes were detected using Fast Red (Roche Diagnostics) or Western Blue substrates (Promega). For dual-color in situ hybridizations, probes were applied simultaneously and detected sequentially (Jowett, 2001).
Analysis of TLCV DNA Replication
The vector pART27 (Gleave, 1992) was used to transiently overexpress SlNAC1 in Nicotiana benthamiana leaf strips. The entire SlNAC1 ORF was amplified by PCR using primers P33 and P34, digested with KpnI and XbaI, and ligated into KpnI/XbaI-digested pART7. A DNA fragment containing the CaMV 35S promoter and the SlNAC1 ORF was released by NotI digestion and ligated into similarly digested pART27 to generate p35S-SlNAC1. A. tumefaciens strain C58 was transformed separately with p35S-SlNAC1, empty pART27, and a Bin19 construct containing a TLCV 1.1mer (Bin19-TLCV1.1; Rigden et al., 1996). Cultures were grown at 28°C for 48 h and used in leaf strip transient replication assays as described (Dry et al., 1997). A. tumefaciens containing Bin19-TLCV1.1 was cocultivated with leaf strips in combination with A. tumefaciens harboring empty pART27 or p35S-SlNAC1 at a ratio of 1:2. Viral replication in agroinoculated tissues was analyzed by DNA gel blotting as described (Dry et al., 1993).
Quantitation of SlNAC1 mRNA Expression by Semiquantitative Reverse Transcription–PCR
Total RNA from N. benthamiana leaf strips was prepared using an RNeasy plant mini kit (Qiagen), which includes a treatment with RNase-free DNase. Semiquantitative reverse transcription (RT)–PCR was performed using a SuperScript one-step RT-PCR kit (Invitrogen) and 80 ng of RNA as template. The SlNAC1 primers (P33 and P34) were used at a final concentration of 0.2 μM. The internal control, ubiquitin, was amplified with primers P35 and P36 (Jin et al., 2002) used at a final concentration of 0.05 μM. RT reaction mix without reverse transcriptase served as a negative control. After the linear phase of DNA amplification (26 cycles), the PCR products were examined by electrophoresis in a 2.0% agarose gel.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY498713 (SlNAC1) and AY622990 (SlUPTG1).
Acknowledgments
We thank Mandy Walker, Ian Dry, Ming-Bo Wang, and Masumi Robertson for helpful discussions and careful reading of the manuscript. We are indebted to Anna Koltunow and Susan Johnson for insight regarding the in situ experiments, Jamus Stonor for excellent technical assistance, Ghafar Sarvestani for assistance with the confocal scanning laser microscope, Jim Haseloff for supplying pBI121-H2B:GFP, and Gregory Martin for the gift of the tomato cDNA library. S.D. was supported by Australian Research Council Grant A09802106.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: M. Ali Rezaian (ali.rezaian@csiro.au).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.027235.
References
- Abouzid, A.M., Frischmuth, T., and Jeske, H. (1988). A putative replicative form of the Abutilon mosaic virus (gemini group) in a chromatin-like structure. Mol. Gen. Genet. 212, 252–258. [Google Scholar]
- Ach, R.A., Durfee, T., Miller, A.B., Taranto, P., Hanley-Bowdoin, L., Zambryski, P.C., and Gruissem, W. (1997). RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol. Cell. Biol. 17, 5077–5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behjatnia, S.A.A., Dry, I.B., and Rezaian, M.A. (1998). Identification of the replication-associated protein binding domain within the intergenic region of tomato leaf curl geminivirus. Nucleic Acids Res. 26, 925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisnard-Lorig, C., Colon-Carmona, A., Bauch, M., Hodge, S., Doerner, P., Bancharel, E., Dumas, C., Haseloff, J., and Berger, F. (2001). Dynamic analyses of the expression of the HISTONE∷YFP fusion protein in arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13, 495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo, A., Collinet, D., Deret, S., Kashoggi, A., and Bejerano, E.R. (2003). Dual interaction of plant PCNA with geminivirus replication accessory protein (REn) and viral replication protein (Rep). Virology 312, 381–394. [DOI] [PubMed] [Google Scholar]
- Chellappan, S., Kraus, V.B., Kroger, B., Munger, K., Howley, P.M., Phelps, W.C., and Nevins, J.R. (1992). Adenovirus E1A, simian virus 40 tumour antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc. Natl. Acad. Sci. USA 89, 4549–4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin, S., Fernandez-Lobato, M., Gooding, P.S., Mullineaux, P.M., and Fenoll, C. (1996). The two nonstructural proteins from wheat dwarf virus involved in viral gene expression and replication are retinoblastoma-binding proteins. Virology 219, 324–329. [DOI] [PubMed] [Google Scholar]
- Collinge, M., and Boller, T. (2001). Differential induction of two potato genes, Stprx2 and StNAC, in response to infection by Phytophthora infestans and to wounding. Plant Mol. Biol. 46, 521–529. [DOI] [PubMed] [Google Scholar]
- Daidoji, H., Takasaki, Y., and Nakane, P.K. (1992). Proliferating cell nuclear antigen (PCNA/cyclin) in plant proliferating cells: Immunohistochemical and quantitative analysis using autoantibody and murine monoclonal antibodies to PCNA. Cell Biochem. Funct. 10, 123–132. [DOI] [PubMed] [Google Scholar]
- Dry, I.B., Krake, L.R., Mullineaux, P.M., and Rezaian, M.A. (2000). Regulation of tomato leaf curl viral gene expression in host tissues. Mol. Plant Microbe Interact. 13, 529–537. [DOI] [PubMed] [Google Scholar]
- Dry, I.B., Krake, L.R., Rigden, J.E., and Rezaian, M.A. (1997). A novel subviral agent associated with a geminivirus: The first report of a DNA satellite. Proc. Natl. Acad. Sci. USA 94, 7088–7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dry, I.B., Rigden, J.E., Krake, L.R., Mullineaux, P.M., and Rezaian, M.A. (1993). Nucleotide sequence and genome organization of tomato leaf curl geminivirus. J. Gen. Virol. 74, 147–151. [DOI] [PubMed] [Google Scholar]
- Duval, M., Hsieh, T.-F., Kin, S.Y., and Thomas, T.L. (2002). Molecular characterization of AtNAM: A member of the Arabidopsis NAC domain superfamily. Plant Mol. Biol. 50, 237–248. [DOI] [PubMed] [Google Scholar]
- Egelkrout, E.M., Robertson, D., and Hanley-Bowdoin, L. (2001). Proliferating cell nuclear antigen transcription is repressed through an E2F consensus element and activated by geminivirus infection in mature leaves. Plant Cell 13, 1437–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmer, J.S., Brand, L., Sunter, G., Gardiner, W.E., Bisaro, D.M., and Rogers, S.G. (1988). Genetic analysis of the tomato golden mosaic virus. II. The product of the AL1 coding sequence is required for replication. Nucleic Acids Res. 16, 7043–7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes, E.P.B., Eagle, P.A., Sipe, P.S., Luckow, V.A., and Hanley-Bowdoin, L. (1994). Interaction between a geminivirus replication protein and origin DNA is essential for viral replication. J. Biol. Chem. 269, 8459–8465. [PubMed] [Google Scholar]
- Frangioni, J.V., and Neel, B.G. (1993). Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 210, 179–187. [DOI] [PubMed] [Google Scholar]
- Gladfelter, H.J., Eagle, P.A., Fontes, E.P.B., Batts, L.A., and Hanley-Bowdoin, L. (1997). Two domains of the AL1 protein mediate geminivirus origin recognition. Virology 239, 186–197. [DOI] [PubMed] [Google Scholar]
- Gleave, A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20, 1203–1207. [DOI] [PubMed] [Google Scholar]
- Golemis, E.A., Gyuris, J., and Brent, R. (1994). Interaction trap/two-hybrid systems to identify interacting proteins. In Current Protocols in Molecular Biology, F.M. Ausubel and K. Struhl, eds (New York: John Wiley & Sons), pp. 13.14.1–13.14.17.
- Grafi, G., Burnett, R.J., Helentjaris, T., Larkins, B.A., Decaprio, J.A., Sellers, W.R., and Kaelin, W.G. (1996). A maize cDNA encoding a member of the retinoblastoma protein family: Involvement in endoreduplication. Proc. Natl. Acad. Sci. USA 93, 8962–8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsley, N., Hohn, T., Davies, J.W., and Hohn, B. (1987). Agrobacterium-mediated delivery of infectious maize streak virus into maize plants. Nature 325, 177–179. [Google Scholar]
- Guerin, J., Rossel, J.B., Robert, S., Tsuchiya, T., and Koltunow, A. (2000). A DEFICIENS homologue is down-regulated during apomictic initiation in ovules of Hieracium. Planta 210, 914–920. [DOI] [PubMed] [Google Scholar]
- Hanley-Bowdoin, L., Settlage, S.B., Orozco, B.M., Nagar, S., and Robertson, D. (1999). Geminiviruses: Models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Plant Sci. 18, 71–106. [PubMed] [Google Scholar]
- Harbour, J.W., and Dean, D.C. (2000). The Rb/E2F pathway: Expanding roles and emerging paradigms. Genes Dev. 14, 2393–2409. [DOI] [PubMed] [Google Scholar]
- Hegedus, D., Yu, M., Baldwin, D., Gruber, M., Sharpe, A., Parkin, I., Whitwill, S., and Lydiate, D. (2003). Molecular characterization of Brassica napus NAC domain transcriptional activators in response to biotic and abiotic stress. Plant Mol. Biol. 53, 383–397. [DOI] [PubMed] [Google Scholar]
- Hibara, K., Takada, S., and Tasaka, M. (2003). CUC1 gene activates the expression of SAM-related genes to induce adventitious shoot formation. Plant J. 36, 687–696. [DOI] [PubMed] [Google Scholar]
- Hingorani, M.M., and O'Donnell, M. (2000). Sliding clamps: A tail(ored) fit. Curr. Biol. 10, R25–R29. [DOI] [PubMed] [Google Scholar]
- Hormuzdi, S.G., and Bisaro, D.M. (1995). Genetic analysis of beet curl top virus: Examination of the roles of L2 and L3 genes in viral pathogenesis. Virology 206, 1044–1054. [DOI] [PubMed] [Google Scholar]
- Horns, T., and Jeske, H. (1991). Localization of abutilon mosaic virus (AbMV) DNA within leaf tissue by in situ hybridization. Virology 181, 580–588. [DOI] [PubMed] [Google Scholar]
- Jansen-Durr, P. (1996). How viral oncogenes make the cell cycle. Trends Genet. 12, 270–275. [DOI] [PubMed] [Google Scholar]
- Jin, H., Axtell, M.J., Dahlbeck, D., Ekwenna, O., Zhang, S., Staskawicz, B., and Baker, B. (2002). NPK1, an MEKK1-like mitogen-activated protein kinase kinase kinase, regulates innate immunity and development in plants. Dev. Cell 3, 291–297. [DOI] [PubMed] [Google Scholar]
- John, I., Hackett, R., Cooper, W., Drake, R., Farrell, A., and Grierson, D. (1997). Cloning and characterization of tomato leaf senescence-related cDNAs. Plant Mol. Biol. 33, 641–651. [DOI] [PubMed] [Google Scholar]
- Jowett, T. (2001). Double in situ hybridization techniques in zebrafish. Methods 23, 345–358. [DOI] [PubMed] [Google Scholar]
- Kikuchi, K., Ueguchi-Tanaka, M., Yoshida, K.T., Nagato, Y., Matsusoka, M., and Hirano, H.-Y. (2000). Molecular analysis of the NAC gene family in rice. Mol. Gen. Genet. 262, 1047–1051. [DOI] [PubMed] [Google Scholar]
- Kong, L.J., and Hanley-Bowdoin, L. (2002). A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection. Plant Cell 14, 1817–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, L.J., Orozco, B.M., Roe, J.L., Nagar, S., Ou, S., Feiler, H.S., Durfee, T., Miller, A.B., Gruissem, W., Robertson, D., and Hanley-Bowdoin, L. (2000). A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 19, 3485–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs, J., Traut, W., Heyraud, F., Matzeit, V., Rogers, S., Schell, J., and Gronenborn, B. (1995). In vitro cleavage and joining at the viral origin of replication by the replication initiator protein of tomato yellow leaf curl virus. Proc. Natl. Acad. Sci. USA 92, 3879–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R., Yu, D.S., Tanaka, M., Zheng, L., Berger, S.L., and Stillman, B. (1998). Activation of chromosomal DNA replication in Saccharomyces cerevisiae by acidic transcriptional activation domains. Mol. Cell. Biol. 18, 1296–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucy, A.P., Boulton, M.I., Davies, J.W., and Maule, A.J. (1996). Tissue specificity of Zea mays infection by maize streak virus. Mol. Plant Microbe Interact. 9, 22–31. [Google Scholar]
- Morra, M.R., and Petty, I.T.D. (2000). Tissue specificity of geminivirus infection is genetically determined. Plant Cell 12, 2250–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysore, K.S., Crasta, O.R., Tuori, R.P., Folkerts, O., Swirsky, P.B., and Martin, G.B. (2002). Comprehensive transcript profiling of Pto- and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J. 32, 299–315. [DOI] [PubMed] [Google Scholar]
- Nagar, S., Pedersen, T.J., Carrick, K.M., Hanley-Bowdoin, L., and Robertson, D. (1995). A geminivirus induces expression of a host DNA synthesis protein in terminally differentiated plant cells. Plant Cell 7, 705–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooka, H., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10, 239–247. [DOI] [PubMed] [Google Scholar]
- Pilartz, M., and Jeske, H. (1992). Abutilon mosaic geminivirus double-stranded DNA is packed into minichromosomes. Virology 189, 800–802. [DOI] [PubMed] [Google Scholar]
- Ren, T., Qu, F., and Morris, T.J. (2000). HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus. Plant Cell 12, 1917–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigden, J.E., Dry, I.B., Krake, L.R., and Rezaian, M.A. (1996). Plant virus DNA replication processes in Agrobacterium: Insight into the origins of geminiviruses? Proc. Natl. Acad. Sci. USA 93, 10280–10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek, S., and Somssich, I.E. (2002). Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 16, 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas, M.R., Jiang, H., Salati, R., Xoconostle-Cazares, B., Sudarshana, M.R., Lucas, W.J., and Gilbertson, R.L. (2001). Functional analysis of proteins involved in movement of the monopartite begomovirus, Tomato yellow leaf curl virus. Virology 291, 110–125. [DOI] [PubMed] [Google Scholar]
- Ruiz-Medrano, R., Xoconostle-Cazares, B., and Lucas, W.J. (1999). Phloem long-distance transport of CmNACP mRNA: Implications for supracellular regulation in plants. Development 126, 4405–4419. [DOI] [PubMed] [Google Scholar]
- Sablowski, R.W.M., and Meyerowitz, E.M. (1998). A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92, 93–103. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Sanderfoot, A.A., and Lazarowitz, S.G. (1996). Getting it together in plant virus movement: Cooperative interactions between bipartite geminivirus movement proteins. Trends Cell Biol. 6, 353–358. [DOI] [PubMed] [Google Scholar]
- Selth, L.A., Randles, J.W., and Rezaian, M.A. (2004). Host responses to transient expression of individual genes encoded by Tomato leaf curl virus. Mol. Plant Microbe Interact. 17, 27–33. [DOI] [PubMed] [Google Scholar]
- Settlage, S.B., Miller, A.B., Gruissem, W., and Hanley-Bowdoin, L. (2001). Dual interaction of a geminivirus replication accessory factor with a viral replication protein and a plant cell cycle regulator. Virology 279, 570–576. [DOI] [PubMed] [Google Scholar]
- Settlage, S.B., Miller, A.B., and Hanley-Bowdoin, L. (1996). Interactions between geminivirus replication proteins. J. Virol. 70, 6790–6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souer, E., van Houwelingen, A., Kloos, D., Mol, J., and Koes, R. (1996). The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85, 159–170. [DOI] [PubMed] [Google Scholar]
- Sudarshana, M.R., Wang, H.L., Lucas, W.J., and Gilbertson, R.L. (1998). Dynamics of bean dwarf mosaic geminivirus cell-to-cell and long-distance movement in Phaseolus vulgaris revealed, using the green fluorescent protein. Mol. Plant Microbe Interact. 11, 277–291. [Google Scholar]
- Sunter, G., and Bisaro, D.M. (1992). Transactivation of geminivirus AR1 and BR1 gene expression by the viral AL2 gene product occurs at the level of transcription. Plant Cell 4, 1321–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunter, G., Hartitz, M.D., Hormuzdi, S.G., Brough, C.L., and Bisaro, D.M. (1990). Genetic analysis of tomato golden mosaic virus: ORF AL2 is required for coat protein accumulation while ORF AL3 is necessary for efficient DNA replication. Virology 179, 69–77. [DOI] [PubMed] [Google Scholar]
- Sunter, G., Stenger, D.C., and Bisaro, D.M. (1994). Heterologous complementation by geminivirus AL2 and AL3 genes. Virology 203, 203–210. [DOI] [PubMed] [Google Scholar]
- Takada, S., Hibara, K., Ishada, T., and Tasaka, M. (2001). The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128, 1127–1135. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Regenmortel, M.H., Mayo, M.A., Fauquet, C.M., and Maniloff, J. (2000). Virus nomenclature: Consensus versus chaos. Arch. Virol. 145, 2227–2232. [DOI] [PubMed] [Google Scholar]
- Vroemen, C.W., Mordhorst, A.P., Albrecht, C., Kwaaitaal, M.A., and de Vries, S.C. (2003). The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15, 1563–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Q., Frugis, G., Colgan, D., and Chua, N.-H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14, 3024–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Q., Sanz-Burgos, A.P., Guo, H., Garcia, J.A., and Gutierrez, C. (1999). GRAB proteins, novel members of the NAC domain family, isolated by their interaction with a geminivirus protein. Plant Mol. Biol. 39, 647–656. [DOI] [PubMed] [Google Scholar]
- Xie, Q., Suarez-Lopez, P., and Gutierrez, C. (1995). Identification and analysis of a retinoblastoma binding motif in the replication protein of a plant DNA virus: Requirement for efficient viral DNA replication. EMBO J. 14, 4073–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian, M., and La Thangue, N.B. (1992). Adenovirus E1A prevents the retinoblastoma gene product from repressing the activity of a cellular transcription factor. EMBO J. 11, 2603–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Loh, Y.-T., Bressan, R.A., and Martin, G.B. (1995). The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell 83, 925–935. [DOI] [PubMed] [Google Scholar]