Abstract
The World Health Organization (WHO) classification of central nervous system (CNS) tumors was revised in 2016 with a basis on the integrated diagnosis of molecular genetics. We herein provide the guidelines for using molecular genetic tests in routine pathological practice for an accurate diagnosis and appropriate management. While astrocytomas and IDH-mutant (secondary) glioblastomas are characterized by the mutational status of IDH, TP53, and ATRX, oligodendrogliomas have a 1p/19q codeletion and mutations in IDH, CIC, FUBP1, and the promoter region of telomerase reverse transcriptase (TERTp). IDH-wildtype (primary) glioblastomas typically lack mutations in IDH, but are characterized by copy number variations of EGFR, PTEN, CDKN2A/B, PDGFRA, and NF1 as well as mutations of TERTp. High-grade pediatric gliomas differ from those of adult gliomas, consisting of mutations in H3F3A, ATRX, and DAXX, but not in IDH genes. In contrast, well-circumscribed low-grade neuroepithelial tumors in children, such as pilocytic astrocytoma, pleomorphic xanthoastrocytoma, and ganglioglioma, often have mutations or activating rearrangements in the BRAF, FGFR1, and MYB genes. Other CNS tumors, such as ependymomas, neuronal and glioneuronal tumors, embryonal tumors, meningothelial, and other mesenchymal tumors have important genetic alterations, many of which are diagnostic, prognostic, and predictive markers and therapeutic targets. Therefore, the neuropathological evaluation of brain tumors is increasingly dependent on molecular genetic tests for proper classification, prediction of biological behavior and patient management. Identifying these gene abnormalities requires cost-effective and high-throughput testing, such as next-generation sequencing. Overall, this paper reviews the global guidelines and diagnostic algorithms for molecular genetic testing of brain tumors.
Keywords: Brain neoplasms, Molecular biology, Next generation sequencing, Pathological diagnosis
The central nervous system (CNS) tumors are those of the brain and spinal cord including the meninges, pituitary gland, pineal gland, and nerves according to the 3rd edition of the International Classification of Diseases for Oncology (ICD-O-3) (http://codes.iarc.fr/). The incidence rate of all primary malignant and nonmalignant CNS tumors in the United States was 22.36 cases per 100,000 (5.67/100,000 for 0–14 years, 5.71/100,000 for 0–19 years) according to the U.S. Central Brain Tumor Registry (CBTRUS; http://www.cbtrus.org/factsheet/factsheet.html). The proportion of women with CNS tumors is higher than that of men (1.2:1). In the United States, the number of newly diagnosed primary malignant and nonmalignant CNS tumors is expected to reach 79,270 (26,070 cases of primary malignant and 53,200 cases of nonmalignant tumors) by 2017. The average annual mortality rate in the United States between 2009 and 2013 is 4.32 per 100,000 and with 73,450 deaths attributed to primary malignant brain and other CNS tumors. If we extrapolate the U.S. data with its incidence rate to the Korean population, per year we would expect about 12,500 new cases of primary CNS tumors and 2,160 deaths. According to the nationwide cancer registration (hospital based Korean Central Cancer Registry [KCCR], http://www.iacr.com.fr/index.php?option=com_comprofiler&task=userprofile&user=973&Itemid=498, http://ncc.re.kr/cancerStatsView.ncc?bbsnum=358&searchKey=total&searchValue=&pageNum=1), 10,004 patients were diagnosed with primary CNS tumors in Korea in 2010, which means that the incidence of brain tumors in Korea is lower than that of United States [1,2]. Similar to the United States, CNS tumors occurred more frequently in female (female:male, 1.59:1). The four most common tumors were meningioma (35.5%), pituitary tumors (18.7%), gliomas (15.1%), and nerve sheath tumors (10.3%). Glioblastoma (GBM) is the most common and the most malignant glioma, which comprises 34.6% of all gliomas. In children (0–19 years), sellar region tumors (pituitary adenoma and craniopharyngiomas), pilocytic astrocytomas, germ cell tumor, and embryonal tumors/medulloblastoma were the most common brain tumors [2].
The 5-year relative survival rate in the United States after the diagnosis of brain or other CNS tumors that are either primary malignant or nonmalignant is 34.7% and 90.4%, respectively. The survival rate of patients with malignant CNS tumors depends on onset age, histologic and molecular diagnosis, and the tumor grade. The overall survival rate decreases with age (0–19 years, 73.8%; 20–44 years, 61.5%; 45–54 years, 33.5%; 55–64 years, 18.5%; 65–74 years, 11.2%; 75 or older: 6.3%). Therefore, children are more likely to survive than adults after malignant CNS tumors develop. Thus, advances must be made to improve the survival rates of young patients with CNS tumors.
The completion of the human genome project led to two powerful tools: the human genome sequence and advanced molecular technologies. Over the past few decades, investigators have been constantly searching for causes of cancer. Through high-throughput studies such as the Cancer Genome Atlas (TCGA) project (https://cancergenome.nih.gov/) or the cBio-Portal project (http://www.cbioportal.org/), a large number of brain tumors have been sequenced and major genetic alterations in brain tumors have been identified [3], thus allowing for the classification of gliomas. The signaling pathways that are mainly involved in GBM are RTK/RAS/phosphoinositide 3-kinase (PI3K) signaling, p53 signaling, and Rb signaling, which were identified through analyses of common mutations and copy number variations (CNV) in gliomas [4]. In addition, according to the molecular classification by global mRNA expression and DNA methylation, GBMs can be categorized into neural, proneural, classical, and mesenchymal transcriptomic subtypes [5].
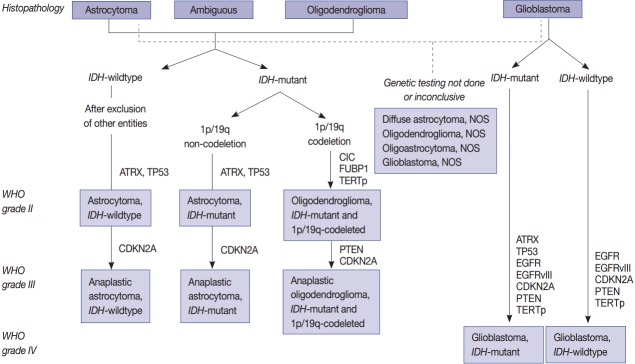
One of the most prominent molecular discoveries was made in 2008 in the genomics of diffuse gliomas. Mutations in IDH1 and IDH2 were discovered by whole exome sequencing (WES) of GBMs, thus altering the classification of gliomas [6]. IDH1 or IDH2 mutations are found in both astrocytic and oligodendroglial tumors since they act as a starting point for gliomagenesis. Therefore, the 2016 World Health Organization (WHO) classification guidelines combined astrocytic tumors and oligodendroglial tumors into one category [7-10]. Furthermore, each tumor has a typical genetic signature. For example, oligodendrogliomas are characterized by a codeletion in 1p/19q, mutations in IDH, and the promoter region of the gene encoding telomerase reverse transcriptase (TERTp). Furthermore, grade 2 and grade 3 astrocytic tumors are characterized by ATRX and TP53 mutations, while IDH-wildtype (primary) GBMs are characterized by the CNV of EGFR, PTEN, CDKN2A/B, PDGFRA, and MET genes, in addition to a lack of mutations in IDH and a codeletion in 1p/19q [11-13].
Even in tumors that are morphologically similar, those with mutations in IDH and a codeletion in 1p/19q differ in the treatment response and prognosis compared with tumors without these two molecular alterations [11,14-19]. Since only gliomas with mutations in IDH and a codeletion in 1p/19q are considered oligodendrogliomas, the so-called pediatric-type oligodendroglioma lacking these two alterations is not considered an oligodendroglioma. The molecular differences of primary IDH-wildtype and IDH-mutant (secondary) GBMs are well summarized in the paper by Cachia et al. [20]. The genetic abnormalities of the IDH-mutant GBMs are similar to grade II/III astrocytomas. In other words, they more likely have mutations in IDH, ATRX, and TP53.
Since the prognosis of each tumor depends on the mutational status of IDH1 and IDH2, astrocytic tumors are also classified according to this metric (Fig. 1) [6,8,21]. Pediatric GBMs and high-grade gliomas differ from those of adults. No genetic abnormalities in IDH1 and IDH2 are found in pediatric GBMs, but mutations are generally found in H3F3A, ATRX and DAXX (Fig. 2). The K27M in H3F3A is a major mutation found in diffuse midline gliomas [22-25]. Additionally, a C11orf95-RELA fusion was found in supratentorial ependymomas [26-34]. A V600E mutation in BRAF was found in circumscribed gliomas, such as pleomorphic xanthoastrocytoma (PXA) (66%) [35-38], gangliogliomas (25%) [35,38], and pilocytic astrocytomas (15%) [35,38]. Furthermore, a BRAF-TIAA1549 fusion was discovered in pilocytic astrocytoma (more than 70%) (Fig. 2) [39-43]. SMARCB1 (INI1) or SMARCA4 (BRG1) gene mutations or deletions were observed in atypical teratoid/rhabdoid tumors (AT/RT) [44-46], and a NAB2-STAT6 fusion was present in solitary fibrous tumor/hemangiopericytomas [47-49]. Meningiomas also have typical gene mutations according to the tumor type and grade. Sixty percent of the sporadic meningiomas have mutations in TRAF7, KLF4, AKT1, SMO, and PIK3CA [50-52]. Among them, mutations in TRAF7 and KLF4 are found in secretory-type meningiomas and mutations in TRAF7/AKT1/PIK3CA are found in meningothelial and transitional-type meningiomas [52,53]. Mutations in TRAF7 are the most common genomic aberrations and are found in 12%–15% of sporadic meningiomas, preferentially fibrous and transitional subtype, which are found concurrently with mutations in KLF4, AKT1, or PIK3CA, but are mutually exclusive with mutations in SMO and neurofibromatosis type 2 (NF2) [53]. NF2-associated meningiomas have mutations in NF2. Atypical and anaplastic meningiomas usually have mutations in TERTp or marked copy number aberrations and loss of CDKN2A/2B [53]. These findings demonstrate that tumors are genetic disorders and that certain mutations can represent different biological behaviors and result in different prognoses.
Fig. 1.
Schematic view of the classification of adult diffuse glioma according to the status of key genes.
Fig. 2.
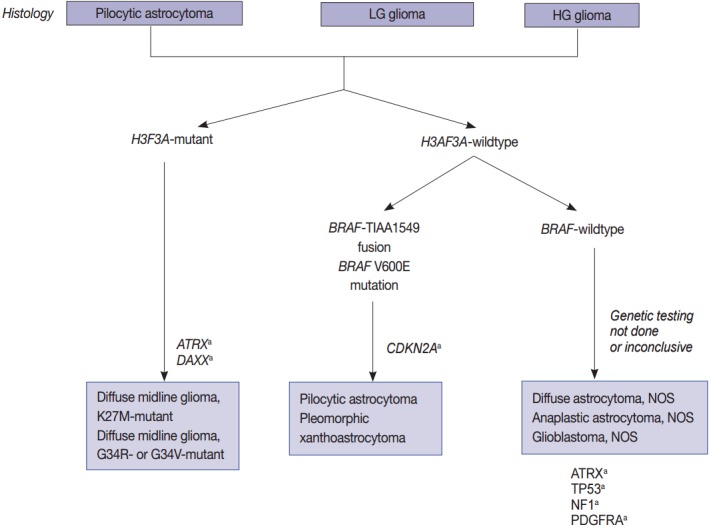
Schematic view of the classification of pediatric diffuse gliomas according to the status of key genes. LG, low grade; HG, high grade; NOS, not otherwise specified. aCharacteristic but not required for diagnosis.
Molecular testing, such as molecular profiling of brain tumors, improves diagnostic accuracy, allows for predictive prognosis, and enables target identification. At present, target therapy has become one of the most promising treatment options for many tumors. Here, we summarize and provide guidelines for the molecular testing of brain tumors. High-throughput genomic studies, such as next-generation sequencing (NGS), can be useful financially and practically by rapidly replacing time-consuming and labor-intensive conventional methods since multiple molecular genetic abnormalities must be studied in order to diagnose various brain tumors.
IMPORTANT MOLECULAR GENETICS FOUND IN BRAIN TUMORS
The indications and methods of each genetic study common to brain tumors are summarized in Tables 1–6. Integration of morphological criteria and genetic alterations is the guideline for an accurate diagnosis, risk classification, and development of therapeutic strategies.
Table 1.
| WHO grade | Altered genes | Genomic alteration (%) | |
|---|---|---|---|
| Astrocytoma | II, III | IDH mutation | 65 |
| P53 mutation | 96 | ||
| ATRX mutation | 96 | ||
| Glioblastoma | IV | EGFR amplification | 57 |
| PDGFRA amplification | 13 | ||
| EGFRvIII mutation | 20 | ||
| PTEN homozygous deletion | 25–35 | ||
| CDKN2A homozygous deletion | 61 | ||
| BRAF V600E mutation | 1–2 (epithelioid GBM) | ||
| TP53 mutation | 25–35 | ||
| Oligodendroglioma | II, III | IDH mutation | 100 |
| 1p/19q codeletion | 100 | ||
| CIC/FUBP1 mutation | 56/29 | ||
| TERTp mutation | 80–96 | ||
| Subependymal giant cell astrocytoma | I | TSC1 and TSC2 mutation | 100 |
| Pilocytic astrocytoma | I | BRAF-KIAA1549 fusion | 75 (cerebellar tumor) |
| BRAF V600E mutation | 13–15 | ||
| Pleomorphic xanthoastrocytoma | II, III | BRAF V600E mutation | 66 |
| CDKN2A homozygous deletion | 50 | ||
| Angiocentric glioma | I | MYB-QKI fusion | 100 |
| Ganglioglioma | I, III | BRAF V600E mutation | 25 |
| TSC1 and TSC2 mutation | Unknown | ||
| Craniopharyngioma, papillary type | I | BRAF V600E mutation | 100 |
| Craniopharyngioma, adamantinomatous type | I | CTNNB1 mutation | 100 |
| AT/RT | IV | SMARCB1 deletion/mutation | > 95 |
| IV | SMARCA4 mutation | < 5 | |
| Cribriform tumor | SMACB1 deletion/mutation | 100 | |
| Meningioma, fibrous type | NF2 inactivation mutation (no hot spot) | 45–5550 | |
| TRAF7/KLF4 mutation | 100 (secretory meningioma) | ||
| AKT1 (p.Glu17Lys) mutation | 2.5 | ||
| SMO (p.Trp535Leu) mutation | 5 | ||
| TERTp mutation | 10 | ||
| Langerhans cell histiocytosis | I, III | BRAF V600E mutation | 50–5754 |
WHO, World Health Organization; GBM, glioblastoma; TSC, tuberous sclerosis complex; AT/RT, atypical teratoid/rhabdoid tumor.
Table 6.
Probes and reading criteria of FISH studies used for brain tumor diagnosis
| Target probe | Control | Cut off | Indication | Biomarker | |
|---|---|---|---|---|---|
| 1p deletion | Chr1p36 | 1q25 | 1p < 0.8 and Deleted nuclei > 50% | ODG | Diagnostic, prognostic, and predictive |
| 19q deletion | Chr19q13 | 19p13 | 19q < 0.8 and Deleted nuclei > 50% | ODG, HGG | Diagnostic, prognostic, and predictive |
| BRAF gain | Chr7q34 | CEP7 | Gold signal > 3 | PA | Diagnostic, prognostic, and predictive |
| CDKN2A (9p21.3) homozygous/hemizygous deletion | Chr9p21.3 | CEP9 | HoD ≥ 10% | HGG | Diagnostic and prognostic |
| HeD ≥ 50% | |||||
| EGFR amplification | Chr711.2 | CEP7 | Ratio ≥ 2.0 | HGG | Diagnostic and prognostic |
| PTEN HoD /HeD | Chr10q23.31 | CEP10 | HoD ≥ 10% | HGG | Diagnostic and prognostic |
| HeD ≥ 50% | |||||
| RELA-C11orf95 fusion | Chr11q13.1 | CEP11 | Break apart | S-ependymoma | Diagnostic and prognostic |
| C19MC amplification | Chr19 | CEP19 | Ratio ≥ 2.0 | ODG, GBM | Diagnostic and prognostic |
| SMARCB1 | Chr22q11.23 | CEP22 | NF2 < 0.8 | AT/RT | Diagnostic and prognostic |
FISH, fluorescent in situ hybridization; Chr, chromosome; ODG, oligodendroglioma; HGG, high grade glioma; PA, pilocytic astrocytoma; HoD, homozygous deletion; HeD, hemizygous deletion; S-ependymoma, supratentorial ependymoma; GBM, glioblastoma; AT/RT, atypical teratoid/rhabdoid tumor.
Mutations in IDH1/2
The discovery of mutations affecting the enzymatic function of IDH in gliomas provided a fundamentally new insight into the biology of gliomagenesis and triggered molecular classification of gliomas by a somatic mutation. IDH catalyzes the oxidative decarboxylation of isocitrate to produce α-ketoglutarate (α-KG) and CO2, but mutant IDH1/2 has a preferential affinity for α-KG instead of isocitrate outside of the citric acid cycle, thus leading to the production and accumulation of the oncometabolite 2-hydroxyglutarate (2HG) [67-70]. Dang et al. [68] hypothesized that 2HG induces redox stress due to damage to the respiratory chain, which subsequently promotes the mutagenesis and development of gliomas. The α-KG–dependent prolyl hydroxylases modulate HIF1α levels and changes in the HIF1α downstream pathway, leading to an increase in reactive oxygen species levels and potentially contributing to the risk of cancer [71].
The glioma-CpG island methylator phenotype (G-CIMP) subset is distinctively and invariably found in the gliomas with mutant IDH, which is the proneural transcriptional group [72]. 2HG, the oncometabolite produced by mutant IDH1/2, inhibits α-KG–dependent dioxygenases including histone demethylases and the Ten-Eleven Translocation (TET) family of histone 5-methylcytosine hydroxylase, which directly induce the hypermethylated state [73]. The G-CIMP subgroup of GBMs is common in younger patients and has a longer lifespan [13,17,30,67,74-76].
All IDH1/2 mutations are common in 70%–80% of type II and III infiltrating gliomas and are found in 100% of oligoden-drogliomas and IDH-mutant GBMs [67].
TP53 mutation
TP53 is a typical tumor suppressor gene located in 17p13.1, which encodes the nuclear protein p53. The p53 protein responds to diverse cellular stresses to regulate the expression of its target genes, thereby inducing cell cycle arrest, apoptosis, senescence, DNA repair, or metabolic changes. Mutated TP53 genes and overexpressed abnormal p53 protein, which has a longer half-life than wild type p53, are associated with a variety of human cancers, including Li-Fraumeni syndrome and many hereditary gliomas. As mentioned in the introduction, the p53 signaling pathway is one of the major abrogated pathways of astrocytic tumors including GBMs. WHO class II and III astrocytic tumors show high levels of TP53 mutations (94%) and/or p53 overexpression (Tables 3, 4) [3]. However, these alterations are rare in oligodendrogliomas, well-circumscribed astrocytic tumors including pilocytic astrocytomas, pleomorphic xanthoastrocytomas, and subependymal giant cell astrocytomas, ependymomas, and embryonal tumors such as medulloblastoma, except for the SHH-type, p53-activated subgroup.
Table 3.
| Genes and molecules | Biomarker type | PA (%) | PXA (%) | DA/AA (%) | IDH- wildtype GBM (%) | IDH- mutant GBM (%) | DMG, H3 K27M (%) | ODG (%) | Epen (%) | MB (%) | AT/RT (%) | Test method |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WHO grade | I | II, III | II, III | IV | IV | IV | II/III | II/III | IV | IV | ||
| IDH1 mutation | Diagnostic and prognostic | 0 | - | > 80 | 5-6 | > 95 | 0 | 98 | 0 | 0 | 0 | IHC, sequencing |
| IDH2 mutation | Diagnostic and prognostic | 0 | - | - | 2-5 | 0 | 2 | 0 | 0 | 0 | Sequencing | |
| TP53 mutation | Diagnostic and prognostic | 0 | 6 | 94 | 25-35 | 62-65 | 45-50 | 9-44 | 0 | - | 0 | Sequencing, IHC |
| ATRX mutation | Diagnostic and prognostic | - | 86 | 0 | 90-95 | 50 | - | 0 | - | 0 | IHC, sequencing | |
| Histone K27M mutation | Diagnostic and prognostic | 0 | - | 0 | Pediatric HGG | 0 | 100 | 0 | 0 | 0 | 0 | IHC, sequencing |
| Histone G34 mutation | Diagnostic and prognostic | 0 | - | - | Uncertain | - | - | 0 | 0 | - | - | Sequencing |
| Histone K36 mutation | Diagnostic and prognostic | 0 | - | - | - | - | - | 0 | 0 | - | - | Sequencing |
| 1p/19q codeletion | Diagnostic, prognostic, and predictive | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | FISH, CGH |
| CIC mutation | Diagnostic, prognostic, and predictive | 0 | - | - | - | - | - | 49 | 0 | - | - | Sequencing, IHC |
| FUBP1 mutation | Diagnostic, prognostic, and predictive | 0 | - | - | - | - | - | 29 | 0 | - | - | Sequencing, IHC |
| EGFR amp | Prognostic | 0 | - | - | 35-45 | 4 | - | - | 0 | - | - | FISH, CGH, IHC |
| PDGFRA amp | Diagnostic | - | - | 13 | - | 50 | - | - | - | FISH, CGH, IHC | ||
| PTEN HoD | Prognostic | 0 | - | - | 25-35 | 5 | - | - | 0 | - | - | FISH, CGH, IHC |
| CDKN2A HoD | Predictive | 0 | 60 | 11 | 35-50 | - | < 5 | - | 0 | - | - | FISH, CGH, IHC |
| BRAF V600E mutation | Diagnostic and prognostic | 15 | 67 | 0 | 1-2 | 0 | 0 | 0 | 0 | 0 | 0 | Sequencing, IHC |
| BRAF-KIAA1549 fusion | Diagnostic and prognostic | 70 | - | - | - | - | - | - | - | - | - | RT-PCR, FISH (gain) |
| TERT promoter mutation | Diagnostic and prognostic | - | - | - | > 80 | 26 | - | 96 | 0 | 21 | - | Sequencing |
| C11orm-RELA fusion | Diagnostic and prognostic | 0 | - | - | - | - | - | - | <5 | - | - | RT-PCR, IHC, RNA seq |
| β-Catenin mutation | Diagnostic and prognostic | 0 | - | - | - | - | - | - | 0 | 6-9 | - | IHC, sequencing |
| MYC amp | Prognostic | 0 | - | - | - | - | - | - | - | 10–15 | - | FISH, CGH, IHC |
| MYCN amp | Prognostic | 0 | - | - | - | - | - | - | - | 10–15 | - | FISH, CGH |
| SMARCB1 mutation | Diagnostic and prognostic | 0 | - | - | - | - | - | - | 0 | 0 | 98 | IHC, sequencing |
| SMARCA4 mutation | Diagnostic and prognostic | 0 | - | - | - | - | - | - | 0 | 0 | 1-2 | IHC, sequencing |
| MGMT methylation | Predictive | - | - | 78.5 | 48.5 | 60-80 | - | 85 | 0 | 0 | 0 | MSP-PCR, pyrosequencing, IHC |
PA, pilocytic astrocytoma; PXA, pleomorphic xanthoastrocytoma; DA, diffuse astrocytoma; AA, anaplastic astrocytoma; GBM, glioblastoma; DMG, diffuse midline glioma; HGG, high grade glioma; ODG, oligodendroglioma; MB, medulloblastomas; AT/RT, atypical teratoid/ rhabdoid tumor; WHO, World Health Organization; IHC, immunohistochemistry; FISH, fluorescent in situ hybridization; CGH, comparative genomic hybridization; RT-PCR, reverse transcription polymerase chain reaction; -, not known; MSP-PCR, methylation-specific polymerase chain reaction.
Table 4.
Summary of the mutation specific antibodies and their indication in glioma diagnosis [12,36,48,81,84]
| Antibody | Positive loci | Mutated gene status | Positive result | Tumors |
|---|---|---|---|---|
| ATRX | Nucleus | Loss of function mutation | Negative | Diffuse and anaplastic astrocytoma |
| β-Catenin | Nucleus | Gain of function mutation | Focal positive | WNT type medulloblastoma, adamantinomatous craniopharyngioma |
| BRAF VE1 (BRAF V600E) | Cytoplasm | Gain of function mutation | Positive | Pleomorphic xanthoastrocytoma, ganglioglioma, pilocytic astrocytoma, epithelioid glioblastoma, papillary craniopharyngioma |
| BRG1 | Nucleus | Gain of function mutation | Negative | Atypical teratoid rhabdoid tumor |
| CIC | Nucleus | Loss of function mutation | Negative | Oligodendroglioma (47%) [15] |
| c-MET | Membrane | Overexpression | Positive | Glioblastoma, anaplastic astrocytoma |
| EGFR | Membrane | Overexpression | Positive | Glioblastoma, anaplastic astrocytoma |
| EGFRvIII | Membrane | Overexpression | Positive | Glioblastoma, anaplastic astrocytoma |
| H3 K27M | Nucleus | Gain of function mutation | Positive | Diffuse midline glioma |
| FUBP1 | Nucleus | Loss of function mutation | Negative | Oligodendroglioma (16%) [15] |
| IDH1 (H09) | Nucleus and cytoplasm | Gain of function mutation | Positive | Astrocytoma and oligodendroglioma |
| INI1 | Nucleus | Loss of function mutation | Negative | Atypical teratoid rhabdoid tumor |
| P16 | Nucleus and cytoplasm | Loss of function mutation | Negative | High grade glioma |
| P53 | Nucleus | Overexpression | Positive | Astrocytic tumors |
| PDGFRA | Membrane | Overexpression | Positive | Glioblastoma, anaplastic astrocytoma |
| PTEN | Cytoplasm | Loss of function mutation | Negative | Glioblastoma |
| RELA (NFKB3) | Cytoplasm | Gain of function mutation | Positive | Cerebral ependymoma |
| STAT6 | Nucleus | Gain of function mutation | Positive | Solitary fibrous tumor/hemangiopericytoma |
| MLH1 | Nucleus | Loss of function mutation | Negative | Gliomas |
| MSH2 | Nucleus | Loss of function mutation | Negative | Gliomas |
| PMS2 | Nucleus | Loss of function mutation | Negative | Gliomas |
| Ki67 | Nucleus | Overexpression | Positive | Brain tumors (for an ancillary test for tumor grading) |
| pHH3 | Nucleus | Overexpression | Positive | Brain tumors (for counting mitoses) |
| GFAP | Nucleus | Expression | Positive | Astrocytic tumors |
| Olig2 | Nucleus | Expression | Positive | Gliomas including astrocytomas and oligodendroglioma |
| Neither neuronal nor ependymal tumors |
ATRX is an X-linked gene of α-thalassemia and mental retardation syndrome
ATRX is located in Xq21.1, encodes a 280-kDa nuclear protein, and has been shown to be involved in a wide range of cellular functions such as DNA recombination, repair, and transcription regulation [82]. It binds strongly within the promyelocytic leukemia body of the nucleus. When ATRX interacts with DAXX, this complex functions as a histone chaperone complex for the deposition of histone variant H3.3 into heterochromatic repeats including pericentric, telomeric, and ribosomal DNA repeat sequences [83]. Functional mutations of this gene have been associated with sister chromatid clumps and defects, abnormal DNA methylation, and the maintenance of telomerase-independent telomeres, resulting in an alternative lengthening of telomeres (ALT). Decreased nuclear expression of ATRX and ATRX mutations are observed in grade 2 and 3 astrocytic tumors including IDH mutant gliomas (86%), IDH-mutant GBMs (85%), and pediatric high grade gliomas such as diffuse midline gliomas (DMGs) (Figs. 1, 2) [3].
Histone H3F3A (K27M, K36, and G34) mutations
Histone protein H3.1 and H3.2 are expressed only in the S phase and are integrated into the chromatin by chaperone CAF1 during DNA replication only, but H3.3 is expressed at all stages of the cell cycle and can be integrated into the chromatin independently of DNA replication. H3.3 is associated with active chromatin such as H3K4 trimethylation, which promotes open chromatin structure for the binding of co-factors and transcription factors to activate transcription [85]. Recurrent mutations in H3F3A, which encodes the replication-independent histone 3 variant H3.3, were first detected in the pediatric diffuse infiltrating pontine glioma (DIPG) by a comprehensive WES analysis. DIPG was later renamed as DMG and is considered a H3 K27M mutation in the new 2016 WHO classification (Fig. 2) [7,23]. Eighty percent of pontine gliomas (DIPG) and 70% of other loci DMGs contain a mutation in one allele of the H3F3A gene, which is a mutation on the histone tail (K27M, K36, G34R/G34V), an important post-translational modification factor [25].
The histone H3F3A mutant gliomas have a poor prognosis regardless of histopathological grade. K27M-H3.3 mutations occur mainy in young patients (median age, 11 years) and G34R/V-H3.3 mutations occur in older children and young adults (median age, 20 years [22]. All of the cases with G34-H3.3 mutations are in pediatric GBMs (13/13), and especially associated with mutations in ATRX and DAXX [22]. Loss of ATRX is associated with the ALT phenotype; thus, ALT often coexists with mutations in ATRX/H3F3A/TP53 [22].
Codeletion of chromosome 1p/19q and mutations in CIC and FUBP1
Simultaneous deletion of chromosome 1p/19q is a characteristic and early genetic event in oligodendroglial tumors, which is associated with better prognosis and is also a good indicator of the patient response to specific combination chemotherapy of procarbazine, lomustine, and vincristine (PCV). Recurrent mutations in CIC and FUBP1 were found in 46%–53% and 15%–24% of oligodendrogliomas, respectively, in addition to a codeletion in 1p/19q and mutations in IDH1 or IDH2 (Fig. 1) [86-88].
FUBP1 is located in 1p31.1, and mutations in FUBP1 are predicted to disrupt FUBP1 protein function (Table 1). FUBP1 acts as an RNA binding protein and alterations of its normal function can lead to tumorigenesis, which has been suggested to act either as a protooncogene or a tumor suppressor gene depending on the tumor type [36,87]. Through the c-Myc pathway, FUBP1 promotes cell migration and protects cells from apoptosis.
The CIC gene on chromosome 19q13.2 represses genes induced by RTK pathway activation [89]. Although the function of human CIC protein is not known, a recent study in cultured cells demonstrated that CIC acts together with IDH1 to regulate citrate levels in the cytoplasm. CIC mutations promote the accumulation of 2HG and reduce clonogenicity in the setting of IDH1 mutations. Loss of 19q in oligodendroglial tumors unmasks mutations in the CIC gene [87,90]. In this regard, the co-existence of IDH1 with CIC or FUBP1 mutations may partially explain the slower tumor growth and longer survival of oligodendrogliomas with mutations in IDH1 and a codeletion of 1p/19q when compared to other diffuse gliomas [90]. The overall survival rate of patients with oligodendrogliomas with CIC mutations was lower than that of patients without CIC mutations, and the FUBP1 mutation was significantly associated with unfavorable progression-free survival [15]. However, oligodendrogliomas cannot be diagnosed with CIC or FUBP1 mutations only [15].
EGFR amplification and EGFRvIII truncation mutations
EGFR, also called Erb1 or HER1, is located on chromosome 7q12 and acts as an ErbB family receptor tyrosine kinase. EGFR amplification is closely related to EGFR overexpression. EGFRvIII-positive tumor cells exhibit particularly high levels of mTOR signal and exhibit the most proliferative and aggressive phenotype similar to GBMs (Fig. 1). The EGFRvIII mutation is a 801 bp frame deletion from exons 2 to 7 of the EGFR gene, which is associated with EGFR amplification and the response to antibody therapy as well as poor prognosis [93]. In addition to EGFR amplification, EGFRvIII-positive GBM cell lines are sensitively suppressed by first-generation EGFR inhibitors (erlotinib, gefitinib, vandetanib) and second-generation EGFR inhibitors (NT113) as compared to GBM cell lines in which PTEN tumor suppressor genes are lost in vitro [94].
EGFR immunohistochemistry is usually uniform in tumors expressing high levels of the protein, however, EGFRvIII staining is heterogeneous and usually present in only a subset of tumor cells. Therefore, representative sampling of tumor tissue is important to avoid false-negative results [56,57].
Unlike EGFR point mutations, EGFR fusions with SEPT14, PSPH, or SEC61G has also been shown to play a role in GBMs, offering a unique opportunity to investigate fused oncogene dependencies [94].
Deletions in CDKN2A/2B
CDKN2A/2B is a tumor suppressor gene located on chromosome 9p21.3, because it encodes a cyclin-dependent kinase inhibitor (p16) and is a cell cycle regulator of the Rb pathway. In 60% of GBMs and 11% of low-grade gliomas including oligodendroglioma, pleomorphic xanthoastrocytoma, and pilocytic astrocytoma, the loss or inactivation of p16 protein as a result of a homozygous deletion or promoter methylation of CDKN2A/2B is observed (Fig. 1) [55]. It is also one of the more commonly altered genes in PXA and high grade gliomas in both adults and children (Fig. 2) [35,95]. After adjusting for the IDH mutational status, sex, and age, CDKN2A deletions were strongly associated with poorer overall survival in astrocytomas but not in oligodendrogliomas [18].
BRAF mutations and BRAF-KIAA1549 fusions
BRAF V600E mutations and fusions of BRAF with KIAA1549 or FAM131B are characteristics of pilocytic astrocytomas. In 2008, a tandem duplication was confirmed in 7q34, and a new fusion gene was found to be generated by a fusion between the KIAA1549 gene and BRAF genes, which was previously uncharacterized in pilocytic astrocytomas [58]. Fusions between KIAA1549 exon 15 and BRAF exon 9 and those between FAM131B exon 2 and BRAF exon 9 are derived from a 2.0 Mb tandem duplication in chromosome band 7q34, which can be detected by realtime polymerase chain reaction (PCR) or fluorescent in situ hybridization (FISH) (Fig. 2) [62].
The BRAF V600E mutation was found in two-thirds of all pleomorphic xanthoastrocytomas, one-fourth of gangliogliomas, and one-seventh of pilocytic astrocytomas [35].
In children, CDKN2A deletions and BRAF mutations are early events in low-grade gliomas (pediatric low-grade glioma [PLGG]) undergoing malignant transformation (Fig. 2) [96]. The BRAF V600E mutant PLGG has longer transformation latency periods than the BRAF wild type PLGG (median latency period, 6.65 years vs 1.59 years, respectively) [96]. As a result, all of the patients with secondary high-grade glioma (sHGGs) containing mutant BRAF were diagnosed at age 9 or older [96]. The sHGG in children showed recurrent changes of BRAF V600E mutations and CDKN2A deletions in 39% and 57%, respectively.
TERTp mutations
Human telomerase is a ribonucleoprotein that regulates the length of telomeric DNA at the ends of chromosomes; therefore, it plays an important role in cellular immortalization and oncogenesis [13]. Mutations in TERTp are molecular hallmarks of glioma, occurring in more than 80%–96% of IDH-wildtype GBMs and oligodendrogliomas, but less frequently present in grade II and III astrocytomas (38.5%) (Table 1, Fig. 1) [97]. Chan et al. (2015) [98] found that mutations in TERTp are present in 41 of 142 (28.9%) grade II gliomas and 20 of 72 (27.8%) grade III gliomas. The mutations were always found in two hotspots (chr5: 1 295 228 C > T and 1 295 250 C > T), resulting in a somatic gain-of-function mutation and an enhancement of TERTp activity [99]. A relative telomere length is strongly correlated with TERTp mutations. These two hot spot mutations are mutually exclusive in gliomas and TP53 mutations, but coincident with a 1p/19q codeletion [100].
The prognostic effect of the TERTp mutation is controversial. Some studies have suggested that TERTp mutations in low-grade gliomas are associated with shorter progression-free survival [101]. Other researchers have found that TERTp mutations are a predictor of a poorer response to temozolomide [102]. TERTp mutations are associated with good outcome in gliomas with mutant IDH, but it is also related to poor outcome in gliomas with wild type IDH [97]. In gliomas with wildtype IDH or GBMs with hypomethylated MGMT, TERTp mutations were found to predict poor prognosis [103]. Therefore, when using the TERTp mutational status as a prognostic factor, other factors such as mutations in IDH and tumor grade should be considered [101]. Primer sequences for TERTp mutation is summarized in Table 5.
Table 5.
| Gene | Full name | Gene location | Forward primer | Reverse primer | Product (bp) | Indication | Function |
|---|---|---|---|---|---|---|---|
| IDH1 | Isocitrate dehydrogenase 1 | 2q33.3 | 5´-M13-GTA AAA CGA CGG CCA GTC GGT CTT CAG AGA AGC CA-3´ | 5´-GCG GAT AAC AAT TTC ACA CAG GGC AAA ATC ACA TTA TTG C-3´ | 180–190 | Astrocytic and ODG | Affect citrate metabolism, leading to 2-HG metabolite |
| IDH2 | Isocitrate dehydrogenase 2 | 15q26.1 | 5´-GCT GCA GTG GGA CCA CTA TT-3´ | 5´-TGT GGC GTT GTA CTG CAG AG-3´ | 295–305 | Astrocytic and ODG | Same with IDH1 |
| H3F3A (K27M/K36/G34) | H3 Histone family 3A | 1q42.12 | 5´-CTG GTA AAG CAC CCA GGA AGC-3´ | 5´-CAT GGA TAG CAC ACA GGT TGG T-3´ | 330–340 | DMG | Chromatin structure, gene transcription |
| BRAF V600E | V-RAF murine sarcoma viral oncogene homolog B1 | 7q34 | 1st: 5´-GCT TGC TCT GAT AGG AAA ATG AG-3´ | 1st: 5´-GTA ACT CAG CAG CAT CTC AGG-3´ | 245–255 | Gliomas including PXA, GG, PA, epithelioid GBM | MAPK signaling |
| 2nd: 5´-TCA TAA TGC TTG CTC TGA TAG GA-3´ | 2nd: 5´-GGC CAA AAA TTT AAT CAG TGG A-3´ | 230–235 | |||||
| CTNNB1 | Catenin, beta-1 | 3p22.1 | 5´-GAT TTG ATG GAG TTG GAC ATG G-3´ | 5´-TGT TCT TGA GTG AAG GAC TGA G-3´ | 230–235 | Medulloblastoma, WNT subtype, craniopharyngioma, adamantinomatous type | The transmission of the ‘contact inhibition’ signal |
| TERTp | Telomerase reverse transcriptase | 5p15.33 | 5´-M13-GGC CGA TTC GAC CTC TCT-3´ (M13: TGT AAA ACG ACG GCC AGT) | 5´-AGC ACC TCG CGG TAG TGG-3´ (M13: GCG GAT AAC AAT TTC ACA CA) | 300–310 | Gliomas, especially ODG and GBM | Telomerase maintenance |
| MGMT | O6 mentylguanine-DNA methyltransferase | 10q26.3 | 5´-TTT CGA CGT TCG TAG GTT TTC GC-3´ | 5´-GCA CTC TTC CGA AAA CGA AAC G-3´ | 80–90 | Gliomas | Promoter methylation |
ODG, oligodendroglioma; 2-HG, 2-hydroxyglutamate; DMG, diffuse midline glioma; PXA, pleomorphic xanthoastrocytoma; GG, ganglioglioma; PA, pilocytic astrocytoma; GBM, glioblastoma; MAPK, mitogen-activated protein kinase.
C11orf95-RELA fusion
More than two-thirds of supratentorial ependymomas contain oncogenic fusions between RELA (the principle effector of canonical nuclear factor κB [NF-κB] signaling) and C11orf95 (an uncharacterized gene) (Table 2) [32,34]. Ependymomas carrying the C11orf95-RELA fusion are characterized by a nuclear accumulation of p65RelA (NFκB) indicating a pathological activation of the NFkB signaling pathway [34]. C11orf95-RELA fusions result from chromothripsis involving chromosome 11q13.1. Furthermore, the C11orf95-RELA fusion protein was found to spontaneously translocate to the nucleus to activate NF-kB target genes, and to rapidly transform neural stem cells, the cell of origin for ependymomas, to form these tumors in mice, which is a poor prognostic marker (Table 2) [34].
Table 2.
| Tumor | Molecular subgroup | Molecular feature | Prognosis |
|---|---|---|---|
| LGG | BRAFV600E | 70% of PXA, GG, DA | Good |
| KIAA-BRAF fusion; BRAF duplication | 90% of PA | Good | |
| MYB-QKI rearrangement | High proportion of AG | Good | |
| FGFR1 duplication | PA, DA, DNT | Good | |
| HGG | K27M-mutant | H3.1 and H3.3 K27 mutation, PDGFRA focal amplification, TP53 mutation, ACVR1 mutation | Poor |
| G34R/V-mutant | H3.3 G3 and TP53 mutation | Poor | |
| RTK1 amplified | PDGFRA and EGFR focal amplification, CDKN2A/2B homozygous deletion | Poor | |
| Mesenchymal | NFI mutation, PDGFRA and EGFR focal amplification, CDKN2A/2B homozygous deletion | - | |
| Ependymoma | C11orf95-RELA+ (70% of ST E) | RELA fusion transcripts | Poor |
| RELA fusion - (30% of ST E) | Possibly YAP1 fusion transcripts | Good | |
| Posterior fossa E | Group A: LAMA2 overexpression, | Poor | |
| Group B: NELL2 overexpression | Good | ||
| Spinal cord E (10% of child E) | NF2 mutation, myxopapillary histology | Good | |
| Medulloblastoma | WNT (10%) | Nuclear β-catenin positive, CTNNBI mutations in exon 3, monosomy 6 | Excellent |
| SHH (30%) | Heterogeneous molecular features depending on age of presentation; PTCH1, SMO, and SUFU mutations, GLI2 and MYCN amplification; germline TP53 mutations | Intermediate (except in infants who have a good prognosis) | |
| Group 3 (25%) | By some unknown mechanism; MYC amplification and over expression | Poor | |
| Group 4 (35%) | i(17)q; MYCN amplification | Intermediate | |
| AT/RT | SMARCB1/SHARCA4 mutation | Poor | |
| ETMR | C19MC amplification | Poor |
LGG, low grade glioma; PXA, pleomorphic xanthoastrocytoma; GG, ganglioglioma; DA, diffuse astrocytoma; PA, pilocytic astrocytoma; AG, angiocentric glioma; DNT, dysembryoplastic neuroepithelial tumor; HGG, high grade glioma; ST, supratentorial; PF, posterior fossa; E, ependymoma; WNT, wingless signaling pathway; i, isochromosome; AT/RT, atypical teratoid/rhabdoid tumor; ETMR, embryonal tumor with multilayer rosettes.
CTNNB1 mutations
Mutations in CTNNB1 on chromosome 3p21, which encodes β-catenin, promote the stabilization and nuclear translocation of itself, therefore activating the WNT signaling pathway (Table 2). The nuclear translocation of β-catenin is present in medulloblastomas, solitary fibrous tumors/hemangiopericytomas, and adamantinomatous-type craniopharyngiomas. Nuclear expression of β-catenin is always focal. Overall, these mutations can act as future therapeutic targets in nuclear β-catenin–positive tumors.
MYC and MYCN amplification
MYC is a regulator gene that encodes for transcription factors for c-myc, Mycn, and Mycl [77]. Nuclear myc protein has multiple functions such as cell cycle progression, apoptosis, and cellular transformation. MYC/MYCN amplification or overexpression is found not only in type-3 medulloblastomas, but also in other aggressive subtypes of medulloblastomas, which account for approximately 10% of medulloblastomas [78,104].
SMARCB1 and SMARCA4 mutations
AT/RT are highly malignant CNS embryonal tumors with rhabdoid morphology, where biallelic inactivation of SMARCB1 results in a loss of INI1 (BAF47) nuclear expression (Tables 2–4). The loss of BRG1 nuclear expression in AT/RT with mutations in SMARCA4 is rarely reported [105]. The tumor suppressors SMARCB1 and SMARCA4 are located on 22q11.23 and 19p13.2, respectively.
The biallelic loss of SMARCB1 and SMARCA4 can be recognized by nuclear negativity of the INI1 and BRG1 immunostaining. However, FISH and DNA sequencing can also be used to detect these genetic alterations.
WES and RNA sequencing of AT/RT revealed few somatic mutations and several deregulated signaling pathways related to the SMARCB1 deficiency [106].
C19MC amplification
Chromosomal 19 micro-ribosomal clusters (C19MC), localized at 19q13.42, are produced by chromothripsis, a phenomenon of chromosomal breakage and inaccurate recombination, resulting in the rearrangement of thousands of clustered chromosomes in a single event to a localized and restricted genomic region on one or more chromosomes [107]. C19MC amplification is a genetic feature of embryonal tumors with multilayer rosettes (ETMR), supratentorial ependymomas, and medulloepitheliomas (Table 2) [108]. This abnormality can be detected by FISH.
MGMT promoter hypermethylation
One of the most clinically important DNA methylation markers in GBMs is the promoter of MGMT, a DNA repair enzyme that can abrogate the effects of alkylating chemotherapy such as temozolomide. More than 50% of oligodendrogliomas and 30%–40% of adult high-grade gliomas (approximately 40% of IDH-wildtype GBMs) reveal MGMT methylation. MGMT promoter hypermethylation induces gene silencing and susceptibility to combined temozolomide and radiation therapy. In cases of oligodendroglioma with promoter methylation of the MGMT gene, whether or not PCV chemotherapy is advantageous remains controversial [59]. However, it is still considered to be the most accurate predictive factor for survival during PCV chemotherapy [60].
METHODS OF MOLECULAR TESTING
The molecular testing methods commonly used in pathology laboratories include immunohistochemical staining, direct sequencing, FISH, chromosomal genomic hybridization, and NGS.
Immunohistochemical studies
Some of the genetic changes can be detected by immunohistochemical studies. IDH R132H, H3 K27M, BRAF V600E, and EGFRvIII can be diagnosed using mutation-specific monoclonal antibodies (mAbs). IDH1 (H09), K27M, and BRAF VE1 antibodies have 100% sensitivity and specificity to detect gene mutations [36]. The INI-1 (SMARCB1), BRG1 (SMARCA4), p16 (CDKN2A), CIC, and FUBP1 mutations and MGMT methylation can be detected by the loss of expression. TP53 mutations are detected by the complete loss of expression or overexpression via the stabilization of the mutant protein. In addition, STAT6 (NAB2-STAT6 fusion), L1CAM (protein marker for RELA-c9ORF95 fusion), and the amplifications of EGFR, MET, and PDGFR can be analyzed by overexpression assays (Table 4) [21,24,36,55]. Mutations in several genes give rise to the overexpression or loss of proteins, depending on whether the mutation is a gain of function or a loss of function mutation, respectively. For example, p53 nuclear overexpression reflects a TP53 mutation, and p16 nuclear and cytoplasmic losses are associated with a CDKN2A homozygous deletion. Furthermore, the nuclear loss of ATRX is associated with an ATRX mutation, and MLH1, MSH2, and PMS nuclear losses are associated with methylation of these genes. Among these, all but TP53 have high correlations between immunohistochemical results and molecular genetic studies. The correlation rate of p53 between immunohistochemistry and mutation studies is low. This is because p53 immunoreactivity may have resulted from the prolongation of half-life either due to p53 mutations or the accumulation of wild type protein brought about by mechanisms other than those caused by mutations, such as a complex formation with MDM2 overexpression products [109], whereas p53 negativity can be observed by the methylation of the TP53 promoter. Therefore, overexpression of p53 protein is not always associated with mutations in conserved exons of the p53 gene.
Direct sequencing
The rapidly growing number of prognostic and predictive genetic markers in neuro-oncology has led to an increasing need for a more thorough molecular analysis of brain tumor specimens in a modern pathology setting. Although several diagnostically important mutations can be detected by immunohistochemistry, this is not the case for the full spectrum of alterations now known to be of relevance. Therefore, the mutations of IDH 1/2, BRAF, CTNNB1, H3F3A (K27M, G34), TP53, ATRX, and TERTp are usually detected by direct sequencing. The sequencing conditions, primer sequences, and PCR product sizes that are currently being used for brain tumor diagnosis are summarized in Table 5.
In order to detect mutations in CIC (missense mutations or small in-frame deletions) and FUBP1 (indel, splicing alteration, and nonsense mutations) in the functional regions such as the HMG box and CI motif, NGS is required because the mutation sites of these two genes are widely distributed along the coding regions [88].
Sanger sequencing is a method of DNA sequencing based on the selective incorporation of chain-terminating dideoxynucleotides by DNA polymerase during DNA replication in vitro, which was developed by Frederick Sanger and colleagues in 1977 [91].
Recently, one-step Sanger sequencing (combining the amplification and sequencing steps) methods such as Ampliseq and SeqSharp have been developed that allows for rapid sequencing of target genes without cloning or prior amplification.
However, covering all potentially clinically relevant genes in a routine diagnostic setting by these methods has become virtually impossible. Reliable high-throughput methods allowing for parallel analysis of multiple targets emerged as an attractive alternative for comprehensive diagnostic neuropathology. NGS provides opportunities to evaluate many genomic targets with high accuracy and sensitivity due to high sequencing coverage. However, the Sanger method is widely used for small-scale projects, and validation of NGS results, especially long serial DNA sequence reads that are greater than 500 nucleotides.
Pyrosequencing
Pyrosequencing is a method of DNA sequencing based on the “sequencing by synthesis” principle [110]. It differs from Sanger sequencing in that it relies on the detection of pyrophosphates that are released upon nucleotide incorporation rather than chain termination with dideoxynucleotides. Various gene alterations can be detected by pyrosequencing, including IDH1/IDH2 mutations, TERTp mutation and MGMT methylation [92,111-113].
NGS
More recently, high volume Sanger sequencing has been supplanted by NGS methods, especially for large-scale, automated genome analyses. NGS is a high-capacity parallel sequencing process that handles hundreds to millions of DNA fragments simultaneously [114,115]. Currently, there are second- and third-generation sequencing technologies that continue to reduce the cost of DNA sequencing and improve accuracy. The ability to multiplex several samples also leads to increased throughput and reduced cost. Soon, NGS will enter clinical practices but it is still uncertain whether it can replace the established techniques. Depending on the type of NGS panel, it can only detect single nucleotide variations and indels or embrace CNV and translocations. The performance of the NGS panel requires rigorous validations and strict quality control for its sensitivity, specificity, and accuracy. There are several NGS panels for brain tumor companion diagnosis [78,79,95,116]. Current practice and guidelines for the clinical use of a NGS-based oncology test are described in Strom’s 2016 paper [117]. A comparison of different NGS-based diagnostic tools is summarized in Lapin et al. paper (2016) [118].
FISH
FISH studies can be easily used to identify chromosomal aberrations, such as a 1p/19q codeletion, EGFR amplification or high polysomy, PTEN and CDKN2A homozygous or hemizygous deletions, C-MYC and N-MYC amplifications, BRAF fusions or copy gains, C11orf95-RELA fusions, C11orf95-YAP1 fusions, YAP1-MAMLD1 fusions, and MYB-QKI fusions. The probes and reading criteria of currently used FISH for the diagnosis and prognosis of brain tumors are summarized in Table 6.
The presence or absence of the 1p/19q codeletion can be determined by FISH, comparative genomic hybridization (CGH), or microsatellite analyses for a loss of heterozygosity (LOH) on chromosomes 1p and 19q [119,120].
When the amount of DNA is insufficient to carry out the single nucleotide polymorphism or CGH array, microsatellite analysis is performed to evaluate via PCR to determine whether there is a LOH of chromosome 1p and 19q [121]. In cases of 1p/19q codeletion, the evaluation and interpretation of FISH results are based on International Society of Pediatric Oncology (SIOP) guidelines for neuroblastoma study [80]. For each hybridization, the number of green and orange signals is assessed for a minimum of 100 nonoverlapping nuclei. An interpretation of a deletion was made when > 50% of nuclei harbored less than a single orange signal (0/2, 1/2, 0/3, 1/3, etc). Such deletions most likely correspond to a LOH (Fig. 3). However, with increasing grades of malignancy, genomic polyploidies may be encountered. These chromosomal polysomies may be balanced (e.g., 3/3, 4/4, 5/5, etc.) or imbalanced (e.g., 3/2, 4/2, 3/5, 4/5, etc.), indicating relative gains or losses. Whether an imbalance situation with relative loss of the target 1p or 19q corresponds to a hemizygous deletion in the presence of reduplication cannot be solved by FISH. In such cases, secondary assays such as PCR-based LOH or CGH studies should be used for better specificity [80].
Fig. 3.
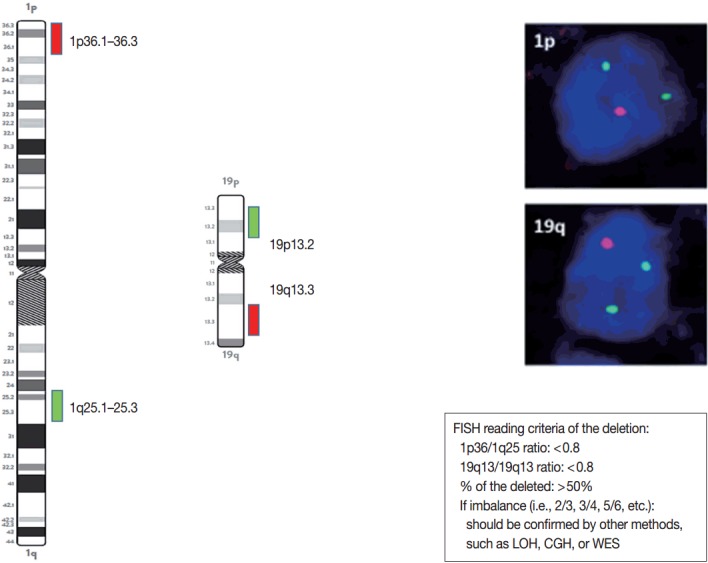
Schematic view, photographs, and reading criteria of the FISH probes for detecting 1p and 19q deletion. The FISH probes are long enough to find out the whole arm deletion. 1p/19q deleted cases show one orange signal and two green signals. The diagnostic cut-off of 1p and 10q deletion is written in the box. FISH, fluorescent in situ hybridization; LOH, loss of heterozygosity; CGH, comparative genomic hybridization; WES, whole exome sequencing.
Methylation specific PCR (O6 methyl guanine methyl transferase)
Methylation can be detected by methylation specific PCR (MSP), pyrosequencing, multiplex ligation-dependent probe assay, or BeadChip microarray [57,113,122]. In the Seoul National University Hospital, MSP is conducted using an EZ DNA methylation Kit (catalog No. D5005, Zymo Research, Irvine, CA, USA) to determine the methylation status of the MGMT promoter. The primer set is described in Table 5. The primer sequences for MGMT were as follows: methylated forward, 5′-TTT CGA CGT TCG TAG GTT TTC GC-3′; methylated reverse, 5′-GCA CTC TTC CGA AAA CGA AACG 3′; unmethylated forward, 5′-TTT GTG TTT TGA TGT TTG TAG GTT TTT GT-3′; and unmethylated reverse, 5′-AAC TCC ACA CTC TTC CAA AAA CAA AAC A-3′.
DISCUSSION
Among primary adult and pediatric CNS tumors, diffuse gliomas are the largest and most diverse group. They usually arise in the cerebral hemispheres and are defined by their widely infiltrative properties and tendency for biological progression. According to the 2016 WHO criteria, gliomas can be classified using combined histological and molecular markers as diffuse astrocytic and oligodendroglial tumors, other astrocytic tumors, ependymal tumors, or other gliomas. The most aggressive astrocytic tumors with a dismal prognosis are the GBMs with wild type IDH and DMG, histone-mutant (H3 K27, G34, and K36). Less infiltrative gliomas in children and young adults include WHO grade I pilocytic astrocytomas,WHO grade II/III pleomorphic xanthoastrocytomas, and WHO grade I subependymal giant cell astrocytomas. Other major classes of CNS tumors include neuronal and mixed glioneuronal tumors as well as embryonal tumors such as cerebellar medulloblastomas, and AT/RT, and extra-axial meningiomas.
The diagnosis of CNS tumors is historically primarily based on histopathological features. However, studies have shown that patients with morphologically identical tumors have different clinical outcomes and treatment responses due to the different molecular genetic characteristics of the tumor. Therefore, many molecular markers became deeply integrated into the diagnosis of CNS tumors and are now used to guide prognosis and treatment of patients. If molecular genetic testing is not possible, a diagnosis is made with the nonspecific term “glioma, not otherwise specified (NOS).” In this case, because the grade and the exact diagnosis of the tumor as well as the biological markers are covered, the precision diagnosis and the appropriate treatments are no longer possible. These “NOS diagnostics” must be reclassified using molecular genetic diagnostic tools.
Diffuse astrocytic and oligodendroglial tumors
Molecular genetic testing can classify diffuse astrocytic and oligodendroglial tumors (WHO grades II–III) and differentiate IDH-mutant GBMs from IDH-wildtype GBMs based on molecular signature of astrocytic and oligodendrogial tumors. Diffuse astrocytic tumors and IDH-mutant GBMs are characterized by mutations in IDH1/2, TP53, and ATRX, whereas oligodendrogliomas are characterized by mutations in IDH1/2, CIC, FUBP1, and TERTp, and a codeletion in 1p/19q (whole chromosomal arm deletion) (Fig. 1). IDH-wildtype GBMs can be identified by demonstrating a dysregulation of several critical signaling pathways and a lack of the above-mentioned genetic alterations except TERTp mutations, which are common in IDH-wildtype GBM. The main signaling pathways involved in IDH-wildtype GBMs are RTK/RAS/PI3K pathways (via amplification and mutations in EGFR [EGFRvIII], PIK3CA, RAS, NF1, and MET) and TP53 and RB1 suppressor pathways (via mutations in or loss of TP53, CDKN2A, and RB1 genes) (Fig. 1) [94].
Mutations in IDH1/2 should be studied in diffuse gliomas for the WHO classification of CNS tumors. They are favorable prognostic markers and are genomic abnormalities initially present in both astrocytic and oligodendroglial tumors. Therefore, from the revised version of 2016 fourth edition of the CNS Tumor Classification, these two tumor categories have been combined as diffuse astrocytic and oligodendroglial tumors [7]. P53 and ATRX mutations are now the hallmark of WHO grade II and grade III astrocytic tumors. TERTp mutations, such as C228T and C250T, are found in more than 90% of oligodendroglial tumors and in more than 80% of GBMs as well as about 30%–40% of lower grade gliomas. Mutations in TERTp were associated with poor survival and resistance to radiotherapy in GBMs and in lower grade gliomas (grade II and III) without IDH mutation; however, TERTp mutations were associated with a favorable prognosis in lower grade gliomas with IDH mutations [63,123].
High- and low-grade pediatric gliomas
Pediatric high-grade gliomas are unique, featuring mutations in H3F3A, ATRX, and DAXX, whereas PLGGs contain mutations or rearrangements in BRAF, FGFR1, or MYB. Furthermore, mutations in IDH are rare unless the patient is an adolescent (Fig. 2) [22,64,65]. The circumscribed low-grade gliomas such as pilocytic astrocytoma, pilomyxoid astrocytoma, ganglioglioma, and PXA often harbor the BRAF V600E or FGFR1 mutation, or gene fusions involving BRAF with KIAA1549 or FAM131B [35,62,65]. Histone H3 K27M, K36, and G34 mutations should be studied in pediatric glioma arising in the midline of CNS, i.e., the thalamus, pons, and spinal cord [124]. However, the histone G34 mutation can be present in cerebral high-grade diffuse gliomas of the adult (about 5%) and the histone K36 mutation is rarely found in pediatric midline gliomas [84].
Finally, the BRAF V600E mutation can be studied in low-grade glioneuronal tumors, such as PXA (about 60%), ganglioglioma (about 25%), pilocytic astrocytoma (about 15%), epithelioid GBM (about 50%), and papillary type of craniopharingioma (about 100%) [35,66].
Medulloblastomas and other embryonic tumors
Medulloblastomas have been recently divided into several subtypes based on specific driver mutations including WNT, SHH, group 3, and group 4 [125-127]. Mutations in CTNNB1, DDX3X or monosomy 6 in the Wnt pathway of medulloblastomas tend to lead to a much better prognosis. In contrast, MYC or MYCN/CDK6 amplification is characteristic of group 3 and 4 medulloblastomas, which are far more likely to metastasize and have a poor prognosis even with intensive therapy. Tumors with PTCH and SMO belong to the SHH class, and have a relatively intermediate prognosis between Wnt and group 3/4 tumors. CTNNB1 mutations can be present in classical-type medulloblastomas and adamantinomatous-type craniopharyngiomas [61,125,128].
SMARCB1 (INI1) or SMARCA4 (BRG1) gene mutations or deletions are essential for the diagnosis of AT/RT, which show dismal prognosis [44-46]. The molecular genetic hallmark of ETMR is chromosome 19 miRNA cluster (C19MC) amplification [129]. If certain tumors are morphologically similar to these tumors, i.e., rhabdoid feature, but molecular studies do not reveal these genetic abnormalities or molecular studies cannot be done, CNS embryonal tumor with rhabdoid feature or ETMR, NOS can be made as a pathological diagnosis [7,129]. Therefore, the molecular characterization of medulloblastomas and embryonal tumors have lasting clinical value.
Meningioma and meningeal solitary fibrous tumor/hemangiopericytoma
Meningiomas often have mutations in the NF2, AKT1, SMO, and KLF4 genes. Furthermore, meningiomas with mutant NF2 are far more likely to exhibit atypical grade II features than the other subtypes [50-52]. Recurrent mutations in KLF4, AKT1, and SMO genes are often present in NF2-negative sporadic meningiomas [50]. Clinical trials involving SMO/AKT/NF2 inhibitors are open for patients with progressive meningiomas with SMO/AKT/NF2 mutations [50,81].
Meningeal hemangiopericytomas and solitary fibrous tumors have the same genetic modification as NAB2-STAT6 gene fusion and are considered as the same tumor [48]. This gene fusion study is essential for the differential diagnosis of histologically similar tumors.
Genetic alterations as prognostic and predictive markers and therapeutic targets
Overall, the analysis of multiple molecular markers not only aids in establishing a correct morphological diagnosis, but also highlights the biological differences between morphologically similar tumors. It can also help with the clinical management of patients. For example, patients with IDH1/TERTp/CIC/FUBP1 positive lower grade gliomas (WHO grades II–III) have a significantly longer median overall survival than those with IDH1/TP53/ATRX mutations [86,130].
In addition, several molecular biomarkers showed promise in predicting responses to targeted therapies [94,131]. For example, clinical trials are now open for GBMs with EGFRvIII mutations, vemurafenib is being evaluated in BRAF V600E mutant gliomas, and clinical response has already been observed in FGFR3-TACC3–positive patients treated with an FGFR inhibitor [78]. NGS-based high-throughput molecular testing may provide potential new targets for precision medicine.
CONCLUSION
We are now in the era of precision medicine, and have slowly caught up with the rapid development of precision medicine. High-throughput assays that can test multiple genes at once are essential for pathological diagnosis in daily practice, and NGS technology has made this possible. Current practices and guidelines for the clinical use of NGS-based oncology tests are well documented in Strom’s paper [117]. However, the medical insurance system has not kept up with the speed of technical development, the advances in diagnostic modalities, and newly developed treatment options. These are obstacles that prevents the doctors from implementing the precision medicine. In order to introduce the advanced knowledge and technologies into the daily practice of pathological diagnosis and clinical fields and reduce the costs, the medical insurance system should be formulated accordingly.
Acknowledgments
This work was supported by Grant from Korean Society of Pathologist (2016).
Footnotes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Lee CH, Jung KW, Yoo H, Park S, Lee SH. Epidemiology of primary brain and central nervous system tumors in Korea. J Korean Neurosurg Soc. 2010;48:145–52. doi: 10.3340/jkns.2010.48.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jung KW, Ha J, Lee SH, Won YJ, Yoo H. An updated nationwide epidemiology of primary brain tumors in republic of Korea. Brain Tumor Res Treat. 2013;1:16–23. doi: 10.14791/btrt.2013.1.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Brat DJ, Verhaak RG, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–98. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–77. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–73. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 6.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–20. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 8.Weller M, Weber RG, Willscher E, et al. Molecular classification of diffuse cerebral WHO grade II/III gliomas using genome- and transcriptome-wide profiling improves stratification of prognostically distinct patient groups. Acta Neuropathol. 2015;129:679–93. doi: 10.1007/s00401-015-1409-0. [DOI] [PubMed] [Google Scholar]
- 9.Rajmohan KS, Sugur HS, Shwetha SD, et al. Prognostic significance of histomolecular subgroups of adult anaplastic (WHO Grade III) gliomas: applying the ‘integrated’ diagnosis approach. J Clin Pathol. 2016;69:686–94. doi: 10.1136/jclinpath-2015-203456. [DOI] [PubMed] [Google Scholar]
- 10.Malzkorn B, Reifenberger G. Practical implications of integrated glioma classification according to the World Health Organization classification of tumors of the central nervous system 2016. Curr Opin Oncol. 2016;28:494–501. doi: 10.1097/CCO.0000000000000327. [DOI] [PubMed] [Google Scholar]
- 11.Clark K, Voronovich Z, Horbinski C. How molecular testing can help (and hurt) in the workup of gliomas. Am J Clin Pathol. 2013;139:275–88. doi: 10.1309/AJCPFO8IIDNBIJ8Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reuss DE, Sahm F, Schrimpf D, et al. ATRX and IDH1-R132H immunohistochemistry with subsequent copy number analysis and IDH sequencing as a basis for an “integrated” diagnostic approach for adult astrocytoma, oligodendroglioma and glioblastoma. Acta Neuropathol. 2015;129:133–46. doi: 10.1007/s00401-014-1370-3. [DOI] [PubMed] [Google Scholar]
- 13.Aldape K, Zadeh G, Mansouri S, Reifenberger G, von Deimling A. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129:829–48. doi: 10.1007/s00401-015-1432-1. [DOI] [PubMed] [Google Scholar]
- 14.Bush NA, Chang SM, Berger MS. Current and future strategies for treatment of glioma. Neurosurg Rev. 2017;40:1–14. doi: 10.1007/s10143-016-0709-8. [DOI] [PubMed] [Google Scholar]
- 15.Chan AK, Pang JC, Chung NY, et al. Loss of CIC and FUBP1 expressions are potential markers of shorter time to recurrence in oligodendroglial tumors. Mod Pathol. 2014;27:332–42. doi: 10.1038/modpathol.2013.165. [DOI] [PubMed] [Google Scholar]
- 16.Kreth S, Thon N, Kreth FW. Epigenetics in human gliomas. Cancer Lett. 2014;342:185–92. doi: 10.1016/j.canlet.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 17.Ostrom QT, Gittleman H, Stetson L, Virk SM, Barnholtz-Sloan JS. Epidemiology of gliomas. Cancer Treat Res. 2015;163:1–14. doi: 10.1007/978-3-319-12048-5_1. [DOI] [PubMed] [Google Scholar]
- 18.Reis GF, Pekmezci M, Hansen HM, et al. CDKN2A loss is associated with shortened overall survival in lower-grade (World Health Organization Grades II-III) astrocytomas. J Neuropathol Exp Neurol. 2015;74:442–52. doi: 10.1097/NEN.0000000000000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simeonova I, Huillard E. In vivo models of brain tumors: roles of genetically engineered mouse models in understanding tumor biology and use in preclinical studies. Cell Mol Life Sci. 2014;71:4007–26. doi: 10.1007/s00018-014-1675-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cachia D, Kamiya-Matsuoka C, Mandel JJ, et al. Primary and secondary gliosarcomas: clinical, molecular and survival characteristics. J Neurooncol. 2015;125:401–10. doi: 10.1007/s11060-015-1930-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Louis DN, Perry A, Burger P, et al. International Society of Neuropathology: Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol. 2014;24:429–35. doi: 10.1111/bpa.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–31. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 23.Khuong-Quang DA, Buczkowicz P, Rakopoulos P, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012;124:439–47. doi: 10.1007/s00401-012-0998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Solomon DA, Wood MD, Tihan T, et al. Diffuse midline gliomas with histone H3-K27M mutation: a series of 47 cases assessing the spectrum of morphologic variation and associated genetic alterations. Brain Pathol. 2016;26:569–80. doi: 10.1111/bpa.12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang R, Han J, Daniels D, Huang H, Zhang Z. Detecting the H3F3A mutant allele found in high-grade pediatric glioma by real-time PCR. J Neurooncol. 2016;126:27–36. doi: 10.1007/s11060-015-1936-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nambirajan A, Malgulwar PB, Sharma MC, et al. C11orf95-RELA fusion present in a primary intracranial extra-axial ependymoma: report of a case with literature review. Neuropathology. 2016;36:490–5. doi: 10.1111/neup.12299. [DOI] [PubMed] [Google Scholar]
- 27.Wu J, Armstrong TS, Gilbert MR. Biology and management of ependymomas. Neuro Oncol. 2016;18:902–13. doi: 10.1093/neuonc/now016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Figarella-Branger D, Lechapt-Zalcman E, Tabouret E, et al. Supratentorial clear cell ependymomas with branching capillaries demonstrate characteristic clinicopathological features and pathological activation of nuclear factor-kappaB signaling. Neuro Oncol. 2016;18:919–27. doi: 10.1093/neuonc/now025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nobusawa S, Hirato J, Sugai T, et al. Atypical teratoid/rhabdoid tumor (AT/RT) arising from ependymoma: a type of AT/RT secondarily developing from other primary central nervous system tumors. J Neuropathol Exp Neurol. 2016;75:167–74. doi: 10.1093/jnen/nlv017. [DOI] [PubMed] [Google Scholar]
- 30.Olar A, Sulman EP. Molecular markers in low-grade glioma-toward tumor reclassification. Semin Radiat Oncol. 2015;25:155–63. doi: 10.1016/j.semradonc.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capper D, Reifenberger G. Classification of gliomas: current progress and perspectives. Nervenarzt. 2015;86:672–83. doi: 10.1007/s00115-014-4223-3. [DOI] [PubMed] [Google Scholar]
- 32.Cachia D, Wani K, Penas-Prado M, et al. C11orf95-RELA fusion present in a primary supratentorial ependymoma and recurrent sarcoma. Brain Tumor Pathol. 2015;32:105–11. doi: 10.1007/s10014-014-0205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nobusawa S, Hirato J, Yokoo H. Molecular genetics of ependymomas and pediatric diffuse gliomas: a short review. Brain Tumor Pathol. 2014;31:229–33. doi: 10.1007/s10014-014-0200-6. [DOI] [PubMed] [Google Scholar]
- 34.Parker M, Mohankumar KM, Punchihewa C, et al. C11orf95-RELA fusions drive oncogenic NF-kappaB signalling in ependymoma. Nature. 2014;506:451–5. doi: 10.1038/nature13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schindler G, Capper D, Meyer J, et al. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 2011;121:397–405. doi: 10.1007/s00401-011-0802-6. [DOI] [PubMed] [Google Scholar]
- 36.Tanboon J, Williams EA, Louis DN. The diagnostic use of immunohistochemical surrogates for signature molecular genetic alterations in gliomas. J Neuropathol Exp Neurol. 2016;75:4–18. doi: 10.1093/jnen/nlv009. [DOI] [PubMed] [Google Scholar]
- 37.Thomas L, Di Stefano AL, Ducray F. Predictive biomarkers in adult gliomas: the present and the future. Curr Opin Oncol. 2013;25:689–94. doi: 10.1097/CCO.0000000000000002. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Wu G, Miller CP, et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45:602–12. doi: 10.1038/ng.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korshunov A, Meyer J, Capper D, et al. Combined molecular analysis of BRAF and IDH1 distinguishes pilocytic astrocytoma from diffuse astrocytoma. Acta Neuropathol. 2009;118:401–5. doi: 10.1007/s00401-009-0550-z. [DOI] [PubMed] [Google Scholar]
- 40.Tian Y, Rich BE, Vena N, et al. Detection of KIAA1549-BRAF fusion transcripts in formalin-fixed paraffin-embedded pediatric low-grade gliomas. J Mol Diagn. 2011;13:669–77. doi: 10.1016/j.jmoldx.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cin H, Meyer C, Herr R, et al. Oncogenic FAM131B-BRAF fusion resulting from 7q34 deletion comprises an alternative mechanism of MAPK pathway activation in pilocytic astrocytoma. Acta Neuropathol. 2011;121:763–74. doi: 10.1007/s00401-011-0817-z. [DOI] [PubMed] [Google Scholar]
- 42.Collins VP, Jones DT, Giannini C. Pilocytic astrocytoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129:775–88. doi: 10.1007/s00401-015-1410-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roth JJ, Fierst TM, Waanders AJ, Yimei L, Biegel JA, Santi M. Whole chromosome 7 gain predicts higher risk of recurrence in pediatric pilocytic astrocytomas independently from KIAA1549-BRAF fusion status. J Neuropathol Exp Neurol. 2016;75:306–15. doi: 10.1093/jnen/nlw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sredni ST, Tomita T. Rhabdoid tumor predisposition syndrome. Pediatr Dev Pathol. 2015;18:49–58. doi: 10.2350/14-07-1531-MISC.1. [DOI] [PubMed] [Google Scholar]
- 45.Hasselblatt M, Gesk S, Oyen F, et al. Nonsense mutation and inactivation of SMARCA4 (BRG1) in an atypical teratoid/rhabdoid tumor showing retained SMARCB1 (INI1) expression. Am J Surg Pathol. 2011;35:933–5. doi: 10.1097/PAS.0b013e3182196a39. [DOI] [PubMed] [Google Scholar]
- 46.Fruhwald MC, Biegel JA, Bourdeaut F, Roberts CW, Chi SN. Atypical teratoid/rhabdoid tumors-current concepts, advances in biology, and potential future therapies. Neuro Oncol. 2016;18:764–78. doi: 10.1093/neuonc/nov264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diamandis P, Ferrer-Luna R, Huang RY, et al. Case report: next generation sequencing identifies a NAB2-STAT6 fusion in glioblastoma. Diagn Pathol. 2016;11:13. doi: 10.1186/s13000-016-0455-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han N, Kim H, Min SK, et al. Meningeal solitary fibrous tumors with delayed extracranial metastasis. J Pathol Transl Med. 2016;50:113–21. doi: 10.4132/jptm.2015.10.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maekawa A, Kohashi K, Yamada Y, et al. A case of intracranial solitary fibrous tumor/hemangiopericytoma with dedifferentiated component. Neuropathology. 2015;35:260–5. doi: 10.1111/neup.12181. [DOI] [PubMed] [Google Scholar]
- 50.Brastianos PK, Horowitz PM, Santagata S, et al. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet. 2013;45:285–9. doi: 10.1038/ng.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clark VE, Erson-Omay EZ, Serin A, et al. Genomic analysis of non NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339:1077–80. doi: 10.1126/science.1233009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reuss DE, Piro RM, Jones DT, et al. Secretory meningiomas are defined by combined KLF4 K409Q and TRAF7 mutations. Acta Neuropathol. 2013;125:351–8. doi: 10.1007/s00401-013-1093-x. [DOI] [PubMed] [Google Scholar]
- 53.Yuzawa S, Nishihara H, Tanaka S. Genetic landscape of meningioma. Brain Tumor Pathol. 2016;33:237–47. doi: 10.1007/s10014-016-0271-7. [DOI] [PubMed] [Google Scholar]
- 54.Mehes G, Irsai G, Bedekovics J, et al. Activating BRAF V600E mutation in aggressive pediatric Langerhans cell histiocytosis: demonstration by allele-specific PCR/direct sequencing and immunohistochemistry. Am J Surg Pathol. 2014;38:1644–8. doi: 10.1097/PAS.0000000000000304. [DOI] [PubMed] [Google Scholar]
- 55.Rodriguez FJ, Vizcaino MA, Lin MT. Recent advances on the molecular pathology of glial neoplasms in children and adults. J Mol Diagn. 2016;18:620–34. doi: 10.1016/j.jmoldx.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maire CL, Ligon KL. Molecular pathologic diagnosis of epidermal growth factor receptor. Neuro Oncol. 2014;16 Suppl 8:viii1–6. doi: 10.1093/neuonc/nou294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Masui K, Mischel PS, Reifenberger G. Molecular classification of gliomas. Handb Clin Neurol. 2016;134:97–120. doi: 10.1016/B978-0-12-802997-8.00006-2. [DOI] [PubMed] [Google Scholar]
- 58.Jones DT, Kocialkowski S, Liu L, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68:8673–7. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van den Bent MJ, Dubbink HJ, Sanson M, et al. MGMT promoter methylation is prognostic but not predictive for outcome to adjuvant PCV chemotherapy in anaplastic oligodendroglial tumors: a report from EORTC Brain Tumor Group Study 26951. J Clin Oncol. 2009;27:5881–6. doi: 10.1200/JCO.2009.24.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dubbink HJ, Atmodimedjo PN, Kros JM, et al. Molecular classification of anaplastic oligodendroglioma using next-generation sequencing: a report of the prospective randomized EORTC Brain Tumor Group 26951 phase III trial. Neuro Oncol. 2016;18:388–400. doi: 10.1093/neuonc/nov182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramaswamy V, Remke M, Bouffet E, et al. Risk stratification of childhood medulloblastoma in the molecular era: the current consensus. Acta Neuropathol. 2016;131:821–31. doi: 10.1007/s00401-016-1569-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roth JJ, Santi M, Pollock AN, et al. Chromosome band 7q34 deletions resulting in KIAA1549-BRAF and FAM131B-BRAF fusions in pediatric low-grade gliomas. Brain Pathol. 2015;25:182–92. doi: 10.1111/bpa.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang ZY, Chan AK, Ding XJ, et al. TERT promoter mutations contribute to IDH mutations in predicting differential responses to adjuvant therapies in WHO grade II and III diffuse gliomas. Oncotarget. 2015;6:24871–83. doi: 10.18632/oncotarget.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bandopadhayay P, Ramkissoon LA, Jain P, et al. MYB-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism. Nat Genet. 2016;48:273–82. doi: 10.1038/ng.3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qaddoumi I, Orisme W, Wen J, et al. Genetic alterations in uncommon low-grade neuroepithelial tumors: BRAF, FGFR1, and MYB mutations occur at high frequency and align with morphology. Acta Neuropathol. 2016;131:833–45. doi: 10.1007/s00401-016-1539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Behling F, Barrantes-Freer A, Skardelly M, et al. Frequency of BRAF V600E mutations in 969 central nervous system neoplasms. Diagn Pathol. 2016;11:55. doi: 10.1186/s13000-016-0506-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Appin CL, Brat DJ. Molecular pathways in gliomagenesis and their relevance to neuropathologic diagnosis. Adv Anat Pathol. 2015;22:50–8. doi: 10.1097/PAP.0000000000000048. [DOI] [PubMed] [Google Scholar]
- 68.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2010;465:966. doi: 10.1038/nature09132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–44. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang H, Ye D, Guan KL, Xiong Y. IDH1 and IDH2 mutations in tumorigenesis: mechanistic insights and clinical perspectives. Clin Cancer Res. 2012;18:5562–71. doi: 10.1158/1078-0432.CCR-12-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dimitrov L, Hong CS, Yang C, Zhuang Z, Heiss JD. New developments in the pathogenesis and therapeutic targeting of the IDH1 mutation in glioma. Int J Med Sci. 2015;12:201–13. doi: 10.7150/ijms.11047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–22. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Appin CL, Brat DJ. Biomarker-driven diagnosis of diffuse gliomas. Mol Aspects Med. 2015;45:87–96. doi: 10.1016/j.mam.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 74.Claus EB, Walsh KM, Wiencke JK, et al. Survival and low-grade glioma: the emergence of genetic information. Neurosurg Focus. 2015;38:E6. doi: 10.3171/2014.10.FOCUS12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cohen AL, Colman H. Glioma biology and molecular markers. Cancer Treat Res. 2015;163:15–30. doi: 10.1007/978-3-319-12048-5_2. [DOI] [PubMed] [Google Scholar]
- 76.Jha P, Pia Patric IR, Shukla S, et al. Genome-wide methylation profiling identifies an essential role of reactive oxygen species in pediatric glioblastoma multiforme and validates a methylome specific for H3 histone family 3A with absence of G-CIMP/isocitrate dehydrogenase 1 mutation. Neuro Oncol. 2014;16:1607–17. doi: 10.1093/neuonc/nou113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Swartling FJ. Myc proteins in brain tumor development and maintenance. Ups J Med Sci. 2012;117:122–31. doi: 10.3109/03009734.2012.658975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Northcott PA, Pfister SM, Jones DT. Next-generation (epi)genetic drivers of childhood brain tumours and the outlook for targeted therapies. Lancet Oncol. 2015;16:e293–302. doi: 10.1016/S1470-2045(14)71206-9. [DOI] [PubMed] [Google Scholar]
- 79.Blumenthal DT, Dvir A, Lossos A, et al. Clinical utility and treatment outcome of comprehensive genomic profiling in high grade glioma patients. J Neurooncol. 2016;130:211–9. doi: 10.1007/s11060-016-2237-3. [DOI] [PubMed] [Google Scholar]
- 80.Ambros PF, Ambros IM, SIOP Europe Neuroblastoma Pathology, Biology, and Bone Marrow Group Pathology and biology guidelines for resectable and unresectable neuroblastic tumors and bone marrow examination guidelines. Med Pediatr Oncol. 2001;37:492–504. doi: 10.1002/mpo.1242. [DOI] [PubMed] [Google Scholar]
- 81.Abedalthagafi M, Bi WL, Aizer AA, et al. Oncogenic PI3K mutations are as common as AKT1 and SMO mutations in meningioma. Neuro Oncol. 2016;18:649–55. doi: 10.1093/neuonc/nov316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Clynes D, Higgs DR, Gibbons RJ. The chromatin remodeller ATRX: a repeat offender in human disease. Trends Biochem Sci. 2013;38:461–6. doi: 10.1016/j.tibs.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 83.Gibbons RJ, McDowell TL, Raman S, et al. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat Genet. 2000;24:368–71. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- 84.Chan KM, Fang D, Gan H, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013;27:985–90. doi: 10.1101/gad.217778.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen P, Zhao J, Wang Y, et al. H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes Dev. 2013;27:2109–24. doi: 10.1101/gad.222174.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jiao Y, Killela PJ, Reitman ZJ, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:709–22. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bettegowda C, Agrawal N, Jiao Y, et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333:1453–5. doi: 10.1126/science.1210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sahm F, Koelsche C, Meyer J, et al. CIC and FUBP1 mutations in oligodendrogliomas, oligoastrocytomas and astrocytomas. Acta Neuropathol. 2012;123:853–60. doi: 10.1007/s00401-012-0993-5. [DOI] [PubMed] [Google Scholar]
- 89.Jiménez G, Shvartsman SY, Paroush Z. The Capicua repressor: a general sensor of RTK signaling in development and disease. J Cell Sci. 2012;125(Pt 6):1383–91. doi: 10.1242/jcs.092965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chittaranjan S, Chan S, Yang C, et al. Mutations in CIC and IDH1 cooperatively regulate 2-hydroxyglutarate levels and cell clonogenicity. Oncotarget. 2014;5:7960–79. doi: 10.18632/oncotarget.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–7. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Switzeny OJ, Christmann M, Renovanz M, Giese A, Sommer C, Kaina B. MGMT promoter methylation determined by HRM in comparison to MSP and pyrosequencing for predicting high-grade glioma response. Clin Epigenetics. 2016;8:49. doi: 10.1186/s13148-016-0204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tritz R, Habita C, Robbins JM, Gomez GG, Kruse CA. Catalytic nucleic acid enzymes for the study and development of therapies in the central nervous system: review article. Gene Ther Mol Biol. 2005;9A:89–106. [PMC free article] [PubMed] [Google Scholar]
- 94.Venkatesan S, Lamfers ML, Dirven CM, Leenstra S. Genetic biomarkers of drug response for small-molecule therapeutics targeting the RTK/Ras/PI3K, p53 or Rb pathway in glioblastoma. CNS Oncol. 2016;5:77–90. doi: 10.2217/cns-2015-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nikiforova MN, Wald AI, Melan MA, et al. Targeted next-generation sequencing panel (GlioSeq) provides comprehensive genetic profiling of central nervous system tumors. Neuro Oncol. 2016;18:379–87. doi: 10.1093/neuonc/nov289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mistry M, Zhukova N, Merico D, et al. BRAF mutation and CDKN2A deletion define a clinically distinct subgroup of childhood secondary high-grade glioma. J Clin Oncol. 2015;33:1015–22. doi: 10.1200/JCO.2014.58.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang P, Cai J, Yan W, et al. Classification based on mutations of TERT promoter and IDH characterizes subtypes in grade II/III gliomas. Neuro Oncol. 2016;18:1099–108. doi: 10.1093/neuonc/now021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chan AK, Yao Y, Zhang Z, et al. Combination genetic signature stratifies lower-grade gliomas better than histological grade. Oncotarget. 2015;6:20885–901. doi: 10.18632/oncotarget.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Arita H, Narita Y, Takami H, et al. TERT promoter mutations rather than methylation are the main mechanism for TERT upregulation in adult gliomas. Acta Neuropathol. 2013;126:939–41. doi: 10.1007/s00401-013-1203-9. [DOI] [PubMed] [Google Scholar]
- 100.Arita H, Narita Y, Fukushima S, et al. Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathol. 2013;126:267–76. doi: 10.1007/s00401-013-1141-6. [DOI] [PubMed] [Google Scholar]
- 101.Heidenreich B, Rachakonda PS, Hosen I, et al. TERT promoter mutations and telomere length in adult malignant gliomas and recurrences. Oncotarget. 2015;6:10617–33. doi: 10.18632/oncotarget.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen C, Han S, Meng L, Li Z, Zhang X, Wu A. TERT promoter mutations lead to high transcriptional activity under hypoxia and temozolomide treatment and predict poor prognosis in gliomas. PLoS One. 2014;9:e100297. doi: 10.1371/journal.pone.0100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Arita H, Yamasaki K, Matsushita Y, et al. A combination of TERT promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathol Commun. 2016;4:79. doi: 10.1186/s40478-016-0351-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Morrissy AS, Garzia L, Shih DJ, et al. Divergent clonal selection dominates medulloblastoma at recurrence. Nature. 2016;529:351–7. doi: 10.1038/nature16478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Masliah-Planchon J, Machet MC, Fréneaux P, et al. SMARCA4-mutated atypical teratoid/rhabdoid tumor with retained BRG1 expression. Pediatr Blood Cancer. 2016;63:568–9. doi: 10.1002/pbc.25772. [DOI] [PubMed] [Google Scholar]
- 106.Sandgren J, Holm S, Marino AM, et al. Whole exome- and mRNA sequencing of an AT/RT case reveals few somatic mutations and several deregulated signalling pathways in the context of SMARCB1 deficiency. Biomed Res Int. 2015;2015:862039. doi: 10.1155/2015/862039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rode A, Maass KK, Willmund KV, Lichter P, Ernst A. Chromothripsis in cancer cells: an update. Int J Cancer. 2016;138:2322–33. doi: 10.1002/ijc.29888. [DOI] [PubMed] [Google Scholar]
- 108.Gajjar A, Pfister SM, Taylor MD, Gilbertson RJ. Molecular insights into pediatric brain tumors have the potential to transform therapy. Clin Cancer Res. 2014;20:5630–40. doi: 10.1158/1078-0432.CCR-14-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Das A, Tan WL, Teo J, Smith DR. Glioblastoma multiforme in an Asian population: evidence for a distinct genetic pathway. J Neurooncol. 2002;60:117–25. doi: 10.1023/a:1020622415786. [DOI] [PubMed] [Google Scholar]
- 110.Zascavage RR, Shewale SJ, Planz JV. Deep-sequencing technologies and potential applications in forensic DNA testing. Forensic Sci Rev. 2013;25:79–105. [PubMed] [Google Scholar]
- 111.Cykowski MD, Allen RA, Fung KM, Harmon MA, Dunn ST. Pyrosequencing of IDH1 and IDH2 mutations in brain tumors and non-neoplastic conditions. Diagn Mol Pathol. 2012;21:214–20. doi: 10.1097/PDM.0b013e31825d802b. [DOI] [PubMed] [Google Scholar]
- 112.Quillien V, Lavenu A, Ducray F, et al. Validation of the high-performance of pyrosequencing for clinical MGMT testing on a cohort of glioblastoma patients from a prospective dedicated multicentric trial. Oncotarget. 2016;7:61916–29. doi: 10.18632/oncotarget.11322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Havik AB, Brandal P, Honne H, et al. MGMT promoter methylation in gliomas-assessment by pyrosequencing and quantitative methylation-specific PCR. J Transl Med. 2012;10:36. doi: 10.1186/1479-5876-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Worst BC, van Tilburg CM, Balasubramanian GP, et al. Next-generation personalised medicine for high-risk paediatric cancer patients: The INFORM pilot study. Eur J Cancer. 2016;65:91–101. doi: 10.1016/j.ejca.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 115.Sahm F, Schrimpf D, Jones DT, et al. Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol. 2016;131:903–10. doi: 10.1007/s00401-015-1519-8. [DOI] [PubMed] [Google Scholar]
- 116.Zacher A, Kaulich K, Stepanow S, et al. Molecular diagnostics of gliomas using next generation sequencing of a glioma-tailored gene panel. Brain Pathol. 2017;27:146–59. doi: 10.1111/bpa.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Strom SP. Current practices and guidelines for clinical next-generation sequencing oncology testing. Cancer Biol Med. 2016;13:3–11. doi: 10.28092/j.issn.2095-3941.2016.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lapin V, Mighion LC, da Silva CP, Cuperus Y, Bean LJ, Hegde MR. Regulating whole exome sequencing as a diagnostic test. Hum Genet. 2016;135:655–73. doi: 10.1007/s00439-016-1677-3. [DOI] [PubMed] [Google Scholar]
- 119.Woehrer A, Sander P, Haberler C, et al. FISH-based detection of 1p 19q codeletion in oligodendroglial tumors: procedures and protocols for neuropathological practice: a publication under the auspices of the Research Committee of the European Confederation of Neuropathological Societies (Euro-CNS) Clin Neuropathol. 2011;30:47–55. doi: 10.5414/npp30047. [DOI] [PubMed] [Google Scholar]
- 120.Jha P, Sarkar C, Pathak P, et al. Detection of allelic status of 1p and 19q by microsatellite-based PCR versus FISH: limitations and advantages in application to patient management. Diagn Mol Pathol. 2011;20:40–7. doi: 10.1097/PDM.0b013e3181e961e9. [DOI] [PubMed] [Google Scholar]
- 121.Idbaih A, Ducray F, Dehais C, et al. SNP array analysis reveals novel genomic abnormalities including copy neutral loss of heterozygosity in anaplastic oligodendrogliomas. PLoS One. 2012;7:e45950. doi: 10.1371/journal.pone.0045950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wiestler B, Capper D, Hovestadt V, et al. Assessing CpG island methylator phenotype, 1p/19q codeletion, and MGMT promoter methylation from epigenome-wide data in the biomarker cohort of the NOA-04 trial. Neuro Oncol. 2014;16:1630–8. doi: 10.1093/neuonc/nou138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gao K, Li G, Qu Y, et al. TERT promoter mutations and long telomere length predict poor survival and radiotherapy resistance in gliomas. Oncotarget. 2016;7:8712–25. doi: 10.18632/oncotarget.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gessi M, Gielen GH, Dreschmann V, Waha A, Pietsch T. High frequency of H3F3A (K27M) mutations characterizes pediatric and adult high-grade gliomas of the spinal cord. Acta Neuropathol. 2015;130:435–7. doi: 10.1007/s00401-015-1463-7. [DOI] [PubMed] [Google Scholar]
- 125.Ellison DW, Dalton J, Kocak M, et al. Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol. 2011;121:381–96. doi: 10.1007/s00401-011-0800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kool M, Korshunov A, Remke M, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123:473–84. doi: 10.1007/s00401-012-0958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Taylor MD, Northcott PA, Korshunov A, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–72. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sekine S, Shibata T, Kokubu A, et al. Craniopharyngiomas of adamantinomatous type harbor beta-catenin gene mutations. Am J Pathol. 2002;161:1997–2001. doi: 10.1016/s0002-9440(10)64477-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jakobiec FA, Kool M, Stagner AM, et al. Intraocular medulloepitheliomas and embryonal tumors with multilayered rosettes of the brain: comparative roles of LIN28A and C19MC. Am J Ophthalmol. 2015;159:1065–74. doi: 10.1016/j.ajo.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 130.Cahill DP, Louis DN, Cairncross JG. Molecular background of oligodendroglioma: 1p/19q, IDH, TERT, CIC and FUBP1. CNS Oncol. 2015;4:287–94. doi: 10.2217/cns.15.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jue TR, McDonald KL. The challenges associated with molecular targeted therapies for glioblastoma. J Neurooncol. 2016;127:427–34. doi: 10.1007/s11060-016-2080-6. [DOI] [PubMed] [Google Scholar]