See Mercado and Hetz (doi:10.1093/brain/awx107) for a scientific commentary on this article.
Signalling through the PERK/eIF2α-P branch of the Unfolded Protein Response is increased in many neurodegenerative diseases. Halliday et al. identify two safe compounds – one licensed – that act on this pathway and are neuroprotective in mice with neurodegeneration. These drugs can now be repurposed in clinical trials for the treatment of dementia.
Keywords: neurodegeneration, drug repurposing, therapeutics, dementia
Abstract
See Mercado and Hetz (doi:10.1093/brain/awx107) for a scientific commentary on this article.
Signalling through the PERK/eIF2α-P branch of the unfolded protein response plays a critical role in controlling protein synthesis rates in cells. This pathway is overactivated in brains of patients with Alzheimer’s disease and related disorders and has recently emerged as a promising therapeutic target for these currently untreatable conditions. Thus, in mouse models of neurodegenerative disease, prolonged overactivation of PERK/eIF2α-P signalling causes sustained attenuation of protein synthesis, leading to memory impairment and neuronal loss. Re-establishing translation rates by inhibition of eIF2α-P activity, genetically or pharmacologically, restores memory and prevents neurodegeneration and extends survival. However, the experimental compounds used preclinically are unsuitable for use in humans, due to associated toxicity or poor pharmacokinetic properties. To discover compounds that have anti-eIF2α-P activity suitable for clinical use, we performed phenotypic screens on a NINDS small molecule library of 1040 drugs. We identified two compounds, trazodone hydrochloride and dibenzoylmethane, which reversed eIF2α-P-mediated translational attenuation in vitro and in vivo. Both drugs were markedly neuroprotective in two mouse models of neurodegeneration, using clinically relevant doses over a prolonged period of time, without systemic toxicity. Thus, in prion-diseased mice, both trazodone and dibenzoylmethane treatment restored memory deficits, abrogated development of neurological signs, prevented neurodegeneration and significantly prolonged survival. In tauopathy-frontotemporal dementia mice, both drugs were neuroprotective, rescued memory deficits and reduced hippocampal atrophy. Further, trazodone reduced p-tau burden. These compounds therefore represent potential new disease-modifying treatments for dementia. Trazodone in particular, a licensed drug, should now be tested in clinical trials in patients.
Introduction
Overactivation of the unfolded protein response (UPR) has emerged as a major pathogenic mechanism across the spectrum of neurodegenerative diseases, for which no disease-modifying treatments currently exist (Hetz et al., 2013; Smith and Mallucci, 2016). Dysregulation of the pancreatic endoplasmic reticulum kinase (PERK) branch of the UPR is particularly prominent: high levels of activated PERK (PERK-P) and its downstream target, the phosphorylated alpha subunit of eukaryotic initiation factor 2 (eIF2α) are seen in brains of patients with Alzheimer’s and Parkinson’s diseases, progressive supranuclear palsy and frontotemporal dementia (FTD) (Hoozemans et al., 2007, 2009; Stutzbach et al., 2013), and also in the rare prion disorders. In these disorders, PERK-P and eIF2α-P accumulation are temporally and spatially associated with the deposition of disease-specific misfolded proteins—notably phosphorylated tau in Alzheimer’s disease and progressive supranuclear palsy (Nijholt et al., 2012; Stutzbach et al., 2013).
eIF2α-P controls global protein synthesis rates by inhibiting translation at the level of initiation (Sonenberg and Hinnebusch, 2009). Phosphorylation of eIF2α is controlled by activation of PERK, and also by the eIF2α kinases of the closely related integrated stress response (ISR) (Ron and Walter, 2007). In the brain, eIF2α represents a hub for controlling rates of protein synthesis essential for learning and memory formation and for maintaining neuronal integrity in health and disease. Sustained overactivation of PERK/eIF2α-P signalling causes chronic translational attenuation leading to synapse loss and neurodegeneration in prion-diseased and FTD mice (Moreno et al., 2012; Radford et al., 2015). This is prevented by genetic and pharmacological interventions that reduce or inhibit eIF2α-P signalling, restoring translation rates (Moreno et al., 2012, 2013; Halliday et al., 2015; Radford et al., 2015). Similar interventions restore memory in various mouse models of Alzheimer’s disease (Ma et al., 2013; Devi and Ohno, 2014) and boost memory in wild-type mice (Sidrauski et al., 2013). This occurs downstream and irrespective of the disease-specific misfolded protein/s involved, with broad relevance for potential treatment of a range of neurodegenerative diseases. However, preclinical pharmacological approaches have not yet yielded compounds suitable for translation to the clinic. Despite being markedly neuroprotective, the selective PERK inhibitor, GSK2606414, is toxic to the pancreas due to the extent of UPR inhibition and on-target effects of PERK inhibition (Moreno et al., 2013; Halliday et al., 2015). In contrast, the small molecule ISRIB (Sidrauski et al., 2013), which acts downstream of eIF2α-P (Sekine et al., 2015; Sidrauski et al., 2015), is neuroprotective without pancreatic toxicity, due to an inherent limitation in its potency for UPR/ISR inhibition (Halliday et al., 2015). ISRIB, however, is highly insoluble and unsuitable for use in humans in its current form. Thus, despite the promise of the mechanistic target, safe and effective drugs acting on the pathway do not exist.
The development of such therapeutics is the focus of many drug discovery initiatives. However, this is in an inherently time-consuming and costly endeavour. In contrast, drug repurposing can bypass much of this process, if existing compounds with the desired activity can be found amongst the current pharmacopeia. Thus, in an effort to identify safe drugs with anti-eIF2α-P therapeutic activity, but lacking associated toxicity and/or adverse pharmacokinetic properties, we carried out phenotypic screens using the NINDS (National Institute for Neurological Disorders and Stroke) Custom Collection 2 Library of 1040 compounds containing ∼75% FDA-approved drugs (often already licensed for use in patients). Our aim was to identify compounds that can reverse UPR/ISR activation induced by UPR stressors such as tunicamycin or thapsigargin in cells/model systems. This would allow detection of compounds with pharmacological activity on the UPR/ISR, which could be tested for therapeutic efficacy in vivo.
Our screens yielded two compounds with anti-eIF2α-P activity that we took forward to test in mouse models of neurodegeneration. Biochemically, trazodone hydrochloride, a licensed anti-depressant, and dibenzoylmethane (DBM) restored protein synthesis rates in prion-diseased and tauopathy FTD mice with established disease. Clinically, both drugs prevented the emergence of clinical signs in most treated prion-diseased mice and restored memory in FTD mice; pathologically, both drugs markedly reduced neuronal loss and hippocampal atrophy in both models. There was no toxicity, in particular to the pancreas. Given the prevalence of UPR activation in Alzheimer’s disease, other tauopathies and related disorders, we propose that these compounds could be rapidly repositioned for clinical trials to determine the efficacy of this therapeutic approach for the treatment of dementia.
Materials and methods
Cell culture
CHO-KI CHOP::luciferase cells (gift of David Ron) and Chinese hamster ovary (CHO) cells were cultured in Dulbecco’s modified Eagle medium (DMEM)/F12(Ham) (Gibco) supplemented with 10% foetal calf serum, 2 mM l-glutamine and 1× penicillin/streptomycin. HEK293 cells (Invitrogen), and mouse neuroblastoma N2A cells were cultured in DMEM (Gibco) supplemented with 10% foetal calf serum, 2 mM l-glutamine and 1× penicillin/streptomycin. All cells were maintained at 37°C with 5% CO2. Cell lines were chosen due to known robust UPR responses, and regularly checked for mycoplasma contamination.
CHOP::luciferase assay
CHO-KI cells stably transfected with a CHOP::luciferase reporter (Harding et al., 2005) were plated at a density of 105 per well in a 24-well plate and left to grow overnight. Cells were treated for 6 h with 5 μg/ml tunicamycin or vehicle only [100% dimethylsulphoxide (DMSO)] and then extracted using the Steady-Glo® luciferase assay system (Promega) before being quantified using the GloMax® 96 microplate luminometer (Promega). The hits from the Caenorhabditis elegans screen (Supplementary Table 1) were incubated with 5 μg/ml tunicamycin for 6 h at 20 μM before assaying as above. Drugs that reduced CHOP::luciferase expression by ∼50% (in a similar manner to ISRIB) (Halliday et al., 2015) are predicted to be efficacious without toxicity.
Immunoblotting
Protein samples were isolated from hippocampi or cells using RIPA lysis buffer (150 mM NaCl, 1% Triton™ X-100, 0.5% sodium deoxycholate, 0.1% SDS and 50 mM Tris pH8.0) supplemented with PhosSTOP™ and protease inhibitors (Roche). Protein levels were determined by resolving 20 μg of protein on SDS-PAGE gels, transferred onto nitrocellulose or PVDF membranes and incubated with primary antibodies for eIF2α-P (1:1000; Cell Signaling 3597s), eIF2α (1:1000; Cell Signaling 2103s), ATF4 (CREB-2, 1:1000; Santa Cruz sc200), ATF6 (1:1000; Genetek 70B1413), GSK3β (1:2000, Cell Signaling, 9832), pSer9-GSK3β (1:1000, Cell Signaling, 9322), total tau (tau-5, 1:2000; Invitrogen ANB0042), p-tau (AT100, 1:2000; Thermo Fisher Scientific MN1060). Horseradish peroxidase-conjugated secondary antibodies (1:5000; Dako) were applied and protein visualized using enhanced chemiluminescence (GE Healthcare) and quantitated using ImageJ. Antibodies against GAPDH (1:5000; Santa Cruz sc32233), β-actin (1:5000; Abcam ab8227) and β-tubulin (1:5000; Millipore MAB1637) were used to determine loading. To detect PrPSc (prion protein) homogenized samples were digested with 50 μg/ml of proteinase K (PK) at 37°C for 1 h prior to electrophoresis. Membranes were then probed with ICSM-35 (1:10 000; D-GEN 0130-03501) and goat anti-mouse (1:10 000; Dako).
XBP1 splicing assay
Total RNA was extracted from CHO-KI cells with the mirVana™ RNA/miRNA isolation kit (Ambion Inc.). RNA samples were reverse-transcribed with ImProm-II™ reverse transcriptase (Promega) by priming with oligo(dT). XBP1 (x-box binding protein 1) mRNA was amplified with primers flanking the 26 base pair intron (5’-GGAGTGGAGTAAGGCTGGTG and 5’-CCAGAATGCCCAAAAGGATA) with Phusion® High-Fidelity Taq Polymerase (New England Biolabs). Polymerase chain reaction (PCR) products were resolved on 3% agarose gels. Mouse neuroblastoma cells (N2A) were treated with tunicamycin (5 μg/ml) for 8 h and used as a positive control for XBP1 splicing (Quaglio et al., 2011).
Quantification of XBP1 splicing by quantitative PCR
HEK293 cells were treated with tunicamycin (5 μg/ml) and either trazodone, DBM (both 20 μM) or DMSO for 6 h. Total RNA was extracted with the mirVana™ RNA/miRNA isolation kit (Ambion Inc.). RNA samples were reverse-transcribed with ImProm-II™ reverse transcriptase (Promega) by priming with oligo(dT). Quantitative PCR was carried out at 95°C for an initial 3 min followed by 35 cycles of denaturation at 95°C for 10 s, annealing at 65°C for 15 s and extension at 72°C for 30 s using SYBR® Green supermix and StepOnePlus™ thermocycler (Applied Biosystems). Spliced XBP1 was detected using primers: forward 5'TGCTGAGTCCGCAGCAGGTG3' and reverse 5'GCTGGCAGGCTCTGGGGAAG3' and compared to the β-actin reference gene (forward 5’CCGATCCACACGGAGTACTTG3’ and reverse 5’GGCACCCAGCACAATGAAG3’).
Puromycin labelling and immunoblot analysis
The effects of endoplasmic reticulum stress on puromycinylated protein levels were determined as previously described (Halliday et al., 2015). In brief, 106 HEK293 cells were plated in 6-well plates. Two days later, culture media was changed to fresh media, and cells were treated with vehicle (DMSO) or thapsigargin in the presence or absence of the indicated concentration of inhibitors for 2.5 h. For puromycin labelling, 10 μg/ml puromycin was added during the last 10 min before harvest. Cells were lysed with passive lysis buffer (Sigma) supplemented with protease inhibitor cocktail (Roche). After centrifugation at 13 000 rpm for 20 min, supernatants were mixed with SDS-PAGE sample buffer. To detect puromycinylated protein 20 μg of total protein, respectively, was subjected to 12% SDS-PAGE and transferred onto PVDF membrane. Immunoblot detection was conducted using primary antibodies for puromycinylated protein (1:5000; Proteintech). Scanned images were quantified using ImageJ software.
Measurement of ternary complex activity
To obtain pRLSV40 ATF4 5’UTR constructs, the human ATF4 5’UTR (the upstream untranslated region) was amplified using the following primer pair: Forward 5’CTGgctagcGCCCTTTTTCTACTTTGCCCG3’; Reverse 5’CTGCTCAGGgctagcATTTCGGTCATGTTG 3’. Upstream 5’UTR ATG codons were removed by site-directed mutagenesis to obtain pRLSV40 ATF4 5’UTR mutant using the following primer pairs:
ATF4 mutant external F 5’gctagcGCCCTTTTTCTACTTTGCCCGCCCACAGAGGTAG3’;
ATF4 mutant external R 5’gctagCATTTCGGTCATGTTGCGGTGCTTTG3’;
ATF4 mutant internal F 5’GTCCACGGCCACCAGGGCGTATTAGGGGCAG3’;
ATF4 mutant internal R 5’CTGCCCCTAATACGCCCTGGTGGCCGTGGAC3’;
ATF4 mutant internal F2 5’CAGCGGCTTAAGCCAGGGCGCTTCTCACGG3’;
ATF4 mutant internal R2 5’CCGTGAGAAGCGCCCTGGCTTAAGCCGCTG3’.
The PCR product was inserted into the NheI site of the pRLSV40 construct (Meijer et al., 2013) upstream of the Renilla luciferase coding region, creating pATF4 with the intact ATF4 5’UTR, and pATF4mu with the upstream open reading frames removed from the ATF4 5’UTR. The internal control firefly luciferase-encoding plasmid pGL3 was purchased from Promega and used to normalize luciferase expression.
CHO cells were grown in 12-well plates and transfected with 200 ng reporter plasmid and 30 ng control plasmid per well using Lipofectamine® 3000 and the manufacturer’s instructions. Twenty-four hours later the cells were stressed with 1 μM thapsigargin and treated with either GSK2606414 (5 μM), ISRIB (1 μM), trazodone (20 μM) or DBM (20 μM). Cells were lysed after the indicated incubation time using Passive Lysis Buffer (Promega), and 10 µl of lysate were assayed using the Dual-Luciferase Reporter Assay System (Promega) on a GloMax® 96 Microplate Luminometer (Promega). For DNA transfections, relative luciferase activity was calculated as a ratio of Renilla luciferase (Rluc) to firefly luciferase (Fluc).
Protein synthesis rates in ex vivo slices
Protein synthesis rates were calculated by measuring 35S-methionine incorporation into proteins in acute hippocampal slices, as described (Moreno et al., 2012, 2013). In brief, hippocampal slices were prepared with a tissue chopper (McIlwain) and dissected in an oxygenated cold (2–5°C) sucrose artificial CSF containing 26 mM NaHCO3, 2.5 mM KCl, 4 mM MgCl2, 0.1 mM CaCl2 and 250 mM sucrose. Slices were allowed to recover in normal artificial CSF buffer while being oxygenated at 37°C for 1 h in 95% O2/5% CO2, and then incubated with 5.7 mBq of 35S-methionine label for 1 h. Samples were washed and then homogenized in 1× passive lysis buffer (Promega), and proteins were precipitated with 25% trichloroacetic acid (TCA) (Sigma). TCA lysates were then placed on Whatman filters, washed with 70% industrial methylated spirits and acetone, and then placed into scintillation cocktail buffer. Incorporation of radiolabel was measured by scintillation counting (WinSpectral, Wallac).
Histology
Paraffin-embedded brain and pancreas were sectioned at 5 μm and stained with haematoxylin and eosin or NeuN antibody (1:200; Millipore) for neuronal counts as described (Moreno et al., 2012, 2013). All images were taken on using AxioVision 4.8 software (Zeiss) and counted using Volocity imaging system. CA1 pyramidal neuron counts were determined using three serial sections from five separate mice (Moreno et al., 2012, 2013). All neuronal counting was performed with the investigator being blind to the sample group being analysed. Immunohistochemistry for pSer202/Thr205 tau was performed using AT8 (1:100; Thermo Scientific).
Mice
Wild-type FVB mice were obtained from Charles River, UK. rTg4510 tauP301L+ and tauP301L− mice were generated from crossing FVB-Tg(tetO-MAPT*P301L)#Kha/JlwsJ (015815; The Jackson Laboratory) with B6.Cg-Tg(Camk2a-tTA)1Mmay/DboJ (007004; The Jackson Laboratory).
Prion infection of mice
Tg37+/− mice were inoculated with 1% brain homogenate of Chandler/RML prions aged 3–4 weeks, as described (Mallucci et al., 2003). Animals were culled when they developed clinical signs of prion disease or lost 20% of body weight from the start of the study. Control mice received 1% normal brain homogenate.
Pharmacological treatment of mice
Mice were intraperitoneally injected once daily with 40 mg/kg trazodone hydrochloride or vehicle (sterile saline), or fed powdered diet 5LF2 containing 0.5% dibenzoylmethane ad libitum. Treatment was from 7 weeks post-infection until terminal clinical sign appeared in tg37+/− mice, or from 4 months until 8 months in rTg4510 mice. Sample sizes are based on our previous papers (Moreno et al., 2012, 2013; Mallucci et al., 2002, 2003), 12–15 mice are used per group as this gives adequate statistical power to detect changes in longevity and behaviour. Mice were randomly assigned a treatment by cage number, and no mice were excluded from the analysis. Experimenters were blind to the treatment group of the mice when clinical signs were being assessed.
Detection of compounds by LC-MS/MS
Wild-type FVB mice for these experiments were obtained from Charles River. Blood and brain tissue were collected 2 or 8 h after dosing from mice treated with one intraperitoneal dose of 40 mg/kg trazodone hydrochloride or vehicle. Blood and brain tissue was also collected from mice given free access to food mixture containing 0.5% dibenzoylmethane for 24 h before testing. Blood plasma (up to 0.2 ml, exact volume measured) was diluted with water to 0.2 ml and extracted with 0.4 ml of chloroform:methanol 2:1(v/v). After vortex mixing (10 min) and centrifugation (10 000g, 10 min), the lower layer was dried with vacuum centrifugation and reconstituted in 50 μl of methanol. Brain tissue (one complete half, ∼0.25 g weighed exactly) was homogenized in 0.5 ml of chloroform: methanol 2:1 (v/v) and further processed exactly as the plasma samples. Quantitative analysis (using external standards) was performed by tandem liquid chromatography-mass spectrometry (LC-MS/MS) using a 4000 QTRAP mass spectrometer (Applied Biosystems) equipped with a turbo ion source and LC series 10 AD VP (Shimadzu). The mobile phase was a water/acetonitrile gradient modified with 0.1% formic acid using an Agilent 2 Poroshell 120 SB-C18 2.1 × 50 mm (2.7 μm particle size) column, which was maintained at 40°C. LC-MS/MS multiple reaction monitoring used precursors and product ions of mass/charge ratio (m/z) 372 and 148 for trazodone and m/z 225 and 105 for dibenzoylmethane in positive electrospray ionization mode. Data analysis was carried out with Analyst 1.4.1 in the quantitative mode.
Novel object recognition memory test
Novel object recognition memory test was carried out as described (Moreno et al., 2013). Briefly, mice were tested in a black cylindrical arena (69-cm diameter) mounted with a 100 light-emitting diode cluster infrared light source and a high-resolution day/night video camera (Sony). Mice were acclimatized to the arena 5 days before testing. During the learning phase, two identical objects were placed 15 cm from the sides of the arena. Each mouse was placed in the arena for two blocks of 10 min for exploration of the objects with an intertrial interval of 10 min. Two hours later, one of the objects was exchanged for a new one, and the mouse was replaced in the arena for 5 min (test phase). The amount of time spent exploring all objects was tracked and measured for each animal with EthoVision software (Tracksys Ltd). All objects and the arena were cleansed thoroughly between trials to ensure the absence of olfactory cues.
Burrowing
Briefly, mice were placed in a large cage with a Perspex tube full of food pellets, as described (Moreno et al., 2013). The natural tendency of rodents is to displace (burrow) the food pellets. The percentage of burrowing activity is calculated from the difference in the weight of pellets in the tube before and after 2 h.
Statistical analyses
Statistical analyses were performed using Prism V6 software. Data were analysed using one-way ANOVA and Tukey’s post hoc test for multiple variables. For Kaplan-Meier analysis, Mantel-Cox test was used. All data in bar charts shows mean ± standard error of the mean (SEM).
Experimental design
All animal work conformed to the ARRIVE guidelines, UK Home Office regulations and institutional guidelines. Mice were randomly assigned treatment groups by cage number. Experimenter was blind to group allocation during the experiments and when assessing clinical signs. For behavioural testing no formal randomization was needed or used.
Results
Screening approach of small compound library identifies potential PERK branch UPR inhibitors
First, in a novel primary screen, we tested the ability of all 1040 compounds of the NINDS Custom Collection 2 in a phenotypic screen in C. elegans. C. elegans have many experimental advantages as a model organism, many of which pertain to drug screening. In contrast to cell lines, they contain many cell types (in particular, a functioning nervous system) and have easily observable phenotypes, increasing the likelihood of detection of representative and translatable effects of drugs on signalling pathways. For this screen, we tested the ability of the NINDS compounds to prevent tunicamycin-induced developmental delay in C. elegans (Richardson et al., 2011) (Supplementary material and Supplementary Fig. 1). Briefly, C. elegans develop through four larval stages (L1–L4) before reaching adulthood, with a generational time of 3 days. When C. elegans are exposed to tunicamycin—a nucleoside antibiotic that inhibits N-linked glycosylation and induces UPR activation—from hatching, the development of the majority of worms stalls between the L2 and L3 larval stages. We considered any compound in the screen that overcame the developmental delay as a potential UPR inhibitor suitable for further investigation. Twenty compounds (Supplementary Table 1, cropped) of the 1040 screened (Supplementary Table 1, full) overcame the tunicamycin-induced developmental delay in the nematodes.
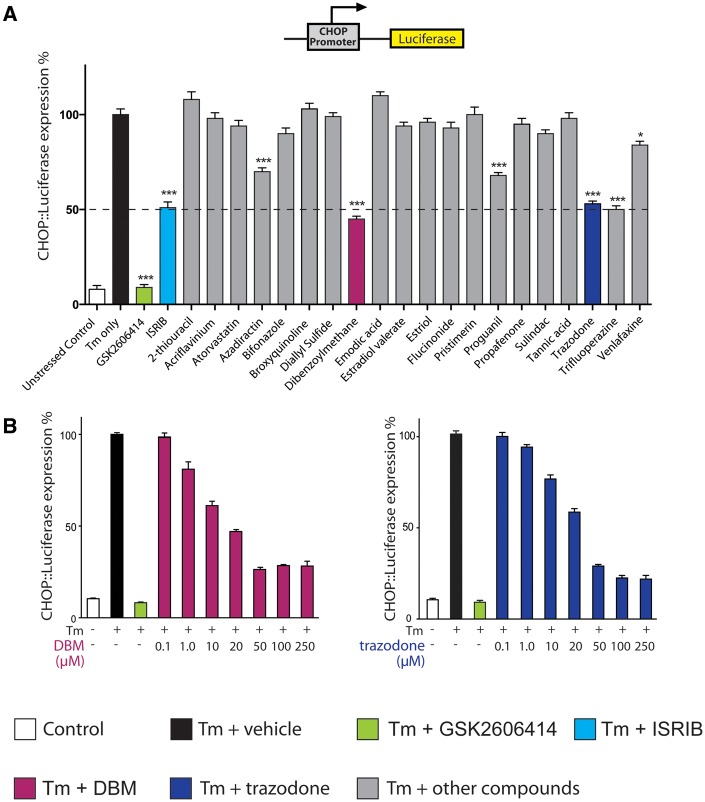
To further delineate the mechanisms of action of the 20 ‘hit’ compounds, we next used a system in which signal transduction could be further investigated. Tunicamycin is a generic UPR stressor, activating all three UPR branches (Oslowski and Urano, 2011). We used a mammalian cell model as a secondary screening system to identify compounds acting through PERK branch mediated eIF2α-P signalling. eIF2α phosphorylation leads to general translational repression, while simultaneously stimulating the selective translation of a number of specific mRNAs, including activating transcription factor 4 (ATF4). ATF4, in turn induces expression of CHOP. CHOP expression is thus a downstream effect of eIF2α phosphorylation. To detect activation of the PERK/eIF2α pathway, we used a CHO cell line containing a reporter construct encoding the DDIT3 (CHOP) promoter and 5’ UTR region driving a firefly luciferase transgene (Fig. 1A). Exposure to UPR stressors induces robust luciferase expression in CHO CHOP::luciferase cells (Harding et al., 2005). Thus, we screened the 20 hit compounds from our primary screen for degree of inhibition of luciferase response to tunicamycin in CHOP::luciferase cells as a measure of inhibition of eIF2α-P signalling. We were interested in compounds with partial UPR inhibitory activity (as occurs with ISRIB) (Halliday et al., 2015), as we previously determined that this allows sufficient restoration of translation rates to protect the brain, without compromising secretory tissue (Halliday et al., 2015). Five drugs met the criteria for a ‘hit’ in this screen. This was defined as a partial reduction in luciferase signal induction in tunicamycin-stressed cells, similar to ∼50% inhibition induced by ISRIB. In contrast, GSK2606414 produced almost 100% inhibition of luciferase signal induction by tunicamycin, consistent with previous findings (Fig. 1A; Halliday et al., 2015).
Figure 1.
A screening approach uncovers two partial inhibitors of the UPR. (A) Luciferase expression in CHOP::luciferase cells treated with tunicamycin (Tm) (3 μg/ml) and compounds from primary screen (grey bars) (Supplementary Table 1), ISRIB (turquoise bar), GSK2606414 (green bar) or tunicamycin alone (black bar). ‘Hits’, including DBM (magenta bar) and trazodone (navy bar), repress luciferase expression to similar extent to ISRIB (dotted line). All drugs at 20 μM, except ISRIB, 1 μM; n = 3 and all experiments performed in triplicate. (B) DBM and trazodone inhibit luciferase expression in a dose-dependent manner. Concentrations of tunicamycin and GSK2606414 and n as in A.
We selected two of these five, trazodone hydrochloride and DBM, for further testing because of their suitability for treating patients in eventual translational studies. Thus, trazodone, a licensed antidepressant (whose primary action is as a serotonin antagonist and reuptake inhibitor, and which has mild sedative effects due to a degree of histamine receptor activity), is safely used in Alzheimer’s disease for management of agitation and insomnia, albeit usually in relatively advanced disease (McCleery et al., 2014). Further, in humans, trazodone is rapidly and almost completely absorbed orally, freely crosses the blood–brain barrier and has a biological half-life of ∼10 h. DBM is a naturally occurring structural analogue of curcumin, with widely reported anti-cancer properties (Khor et al., 2009), which has no known toxicity. Two of the other three hits in this screen were rejected because of lack of suitability for eventual translational studies in humans: trifluorperazine is an anti-psychotic contraindicated in the elderly (due to increased risk of death when used to treat behavioural/psychological problems caused by dementia in older people); azadirectin is a pesticide found in neem oil and is poorly brain penetrant. The third hit, proguanil, is an anti-malarial that is safe in humans; however, it is toxic to mice, precluding our carrying out the relevant preclinical experiments. We then tested the effects of increasing concentrations of trazodone and DBM in CHOP::luciferase cells. Both drugs showed a dose-response effect in inhibiting luciferase expression in the reporter cell line reaching a plateau at 50 μM concentration, consistent with an intrinsic limitation of inhibitory activity (Fig. 1B), as reported for ISRIB (Halliday et al., 2015).
Trazodone and dibenzoylmethane inhibit UPR-induced eIF2α-P signalling and restore protein synthesis rates in vitro
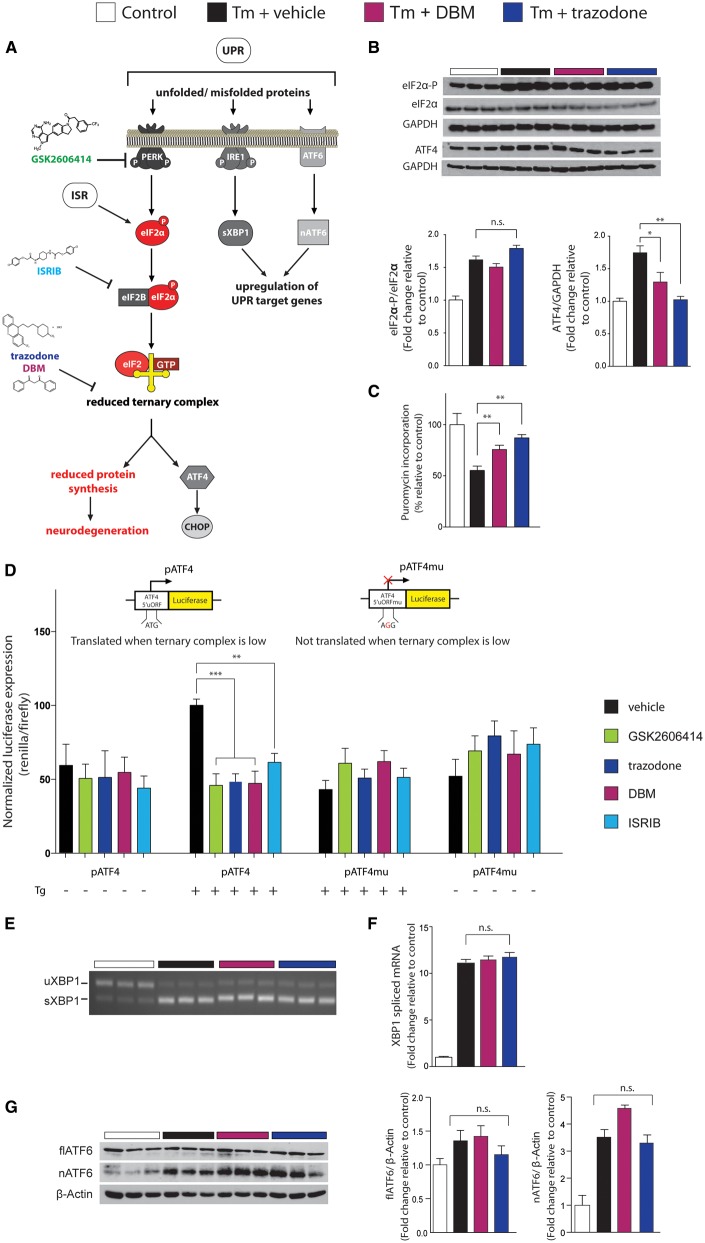
We then tested both compounds to identify their site of action in the pathway (Fig. 2A). Both trazodone and DBM (20 μM) reduced ATF4 levels, but not eIF2α-P levels in tunicamycin stressed cells (Fig. 2B), placing their site of action downstream of eIF2α-P, like ISRIB. Consistent with inhibition of eIF2α-P activity, DBM and trazodone partially restored protein synthesis rates in tunicamycin-treated cells (Fig. 2C). Due to the apparent similarities between trazodone and DBM and the mechanism of action of ISRIB (which stabilizes eIF2B, facilitating ternary complex formation) (Sekine et al., 2015; Sidrauski et al., 2015), we next determined if the compounds exerted a similar effect on eIF2B and ternary complex levels as ISRIB. Under normal circumstances, eIF2α-P reduces ternary complex availability for initiation of translation, reducing protein synthesis rates. However, selected mRNAs, including ATF4, are able to overcome this block on translation due to the structure of their upstream translated region (5’UTR) that contains multiple upstream open reading frames (Vattem and Wek, 2004) and are translated more efficiently when eIF2α-P levels are high. We used a luciferase reporter construct downstream of the ATF4 5’UTR to transfect cells. The ATF4 5’UTR directly responds to reduced levels of ternary complex, resulting in activation of luciferase when eIF2α-P levels are high, as on UPR induction. Following activation of the UPR by thapsigargin treatment in transfected CHO cells, both trazodone and dibenzoylmethane, like ISRIB, prevented the wild-type ATF4 5’UTR activation normally associated with endoplasmic reticulum stress (Fig. 2D). This effect is lost when the ATF4 5’UTR with the upstream open reading frames removed by a single nucleotide substitution of the upstream ATG start codons is expressed and treated in the same manner. Thus, these compounds prevent eIF2α-P from reducing ternary complex levels, allowing translation to proceed when eIF2α-P levels are high.
Figure 2.
Trazodone and DBM inhibit UPR-induced eIF2α-P signalling in vitro. (A) Schematic of the UPR, showing site of action of compounds modulating PERK branch dysregulation. (B) Western blots showing DBM and trazodone reduce ATF4 levels without affecting eIF2α-P levels; bar graphs on right show quantitation. Repeated in triplicate. (C) DBM and trazodone partially restore protein synthesis rates after thapsigargin (1 μM) stress in HEK293 cells, assessed by puromycin incorporation into nascent proteins quantified from western blots. Repeated in triplicate. (D) Trazodone and DBM reduce luciferase expression under control of the ATF4 5’UTR (pATF4), but have no effect when upstream open reading frames in the 5’UTR are removed (pATF4mu), demonstrating an ability to increase ternary complex levels. Levels of pATF4 and pATF4mu normalized to firefly luciferase expressed by the pGL3 plasmid in CHO cells stressed with 1 μM thapsigargin for 6 h. n = 3, repeated in triplicate. (E) XBP1 splicing is unchanged by DBM or trazodone after 3 h tunicamycin stress, determined by RT-PCR, or after 6 h stress and measured by quantitative PCR (F). (G) Full-length ATF6 (flATF6) cleavage to nuclear fragment (nATG6) is not affected by DBM or trazodone. Repeated in triplicate. All bar charts show means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, n.s. = non-significant. One-way ANOVA and Tukey’s post hoc test.
We next investigated if trazodone and DBM affect eIF2B dimerization in the same way as ISRIB (Sidrauski et al., 2015). We observed that ISRIB caused a shift in both eIF2Bδ and eIF2Bϵ subunit distribution in sucrose gradients consistent with increased dimerization of eIF2B, in agreement with previously published work (Sidrauski et al., 2015). However, neither trazodone nor DBM caused dimerization of eIF2B (Supplementary Fig. 2). Thus, the data show that while these drugs act at a similar level to ISRIB to uncouple eIF2α-P-mediated reduction of ternary complex, they are functioning via distinct mechanisms.
The compounds did not, however, act on the other UPR branches, having no effect on tunicamycin-induced XBP1 splicing, as detected by PCR (Fig. 2E), or measured semiquantitatively by quantitative PCR (Fig. 2F), or cleavage of ATF6 (Fig. 2G). Thus, these data, together with the evidence from our primary and secondary screens, support activity for both trazodone and DBM in restoring translation rates in vitro under conditions of UPR stress. Further, the evidence shows that both compounds show partial inhibition of UPR-mediated translational repression (Fig. 1A) and have an intrinsic limitation in this activity (Fig. 1B), as previously seen with ISRIB (Halliday et al., 2015). This is important, because this is known to be a safe level of UPR/ISR reversal in vivo, in contrast to the toxicity produced by complete inhibition of this signalling, as occurs with the PERK inhibitor, GSK2606414 (Moreno et al., 2013). Also, like ISRIB, it appears that the site of action of both trazodone and DBM in inhibiting this activity is downstream of eIF2α-P, at the level of ternary complex (Fig. 2D). Given these favourable in vitro features, both compounds were put forward for testing in preclinical mouse models of neurodegenerative disease in which ISRIB and/or GSK2606414 have been shown to be neuroprotective (despite pancreatic toxicity of the latter compound) (Moreno et al., 2013; Halliday et al., 2015; Radford et al., 2015).
Trazodone and dibenzoylmethane penetrate the blood–brain barrier
We next tested the therapeutic effects of trazodone and DBM in vivo. For clinical relevance, we used a daily dose of 40 mg/kg of trazodone delivered intraperitoneally in mice, equivalent to 194 mg/day in humans. Patients usually receive 150–375 mg trazodone per day. We used a standard conversion formula for dose translation to calculate the corresponding mouse dose (Reagan-Shaw et al., 2008). DBM oral suspension was provided in food as a 0.5% mixture available ad libitum, as described (Khor et al., 2009). Pharmacokinetic data pertaining to plasma levels and brain penetration were investigated by quantitative LC-MS/MS measurements in brain and plasma samples in wild-type mice. Trazodone levels were measured at 2 and 8 h post-administration; DBM levels were measured after 24 h, to allow for sufficient consumption of food containing the compound. The data confirmed both drugs penetrated the blood–brain barrier with mean brain:plasma ratios consistent with efficacious dosing (Supplementary Table 2).
Trazodone and dibenzoylmethane prevent neurological signs of prion disease in prion-infected mice
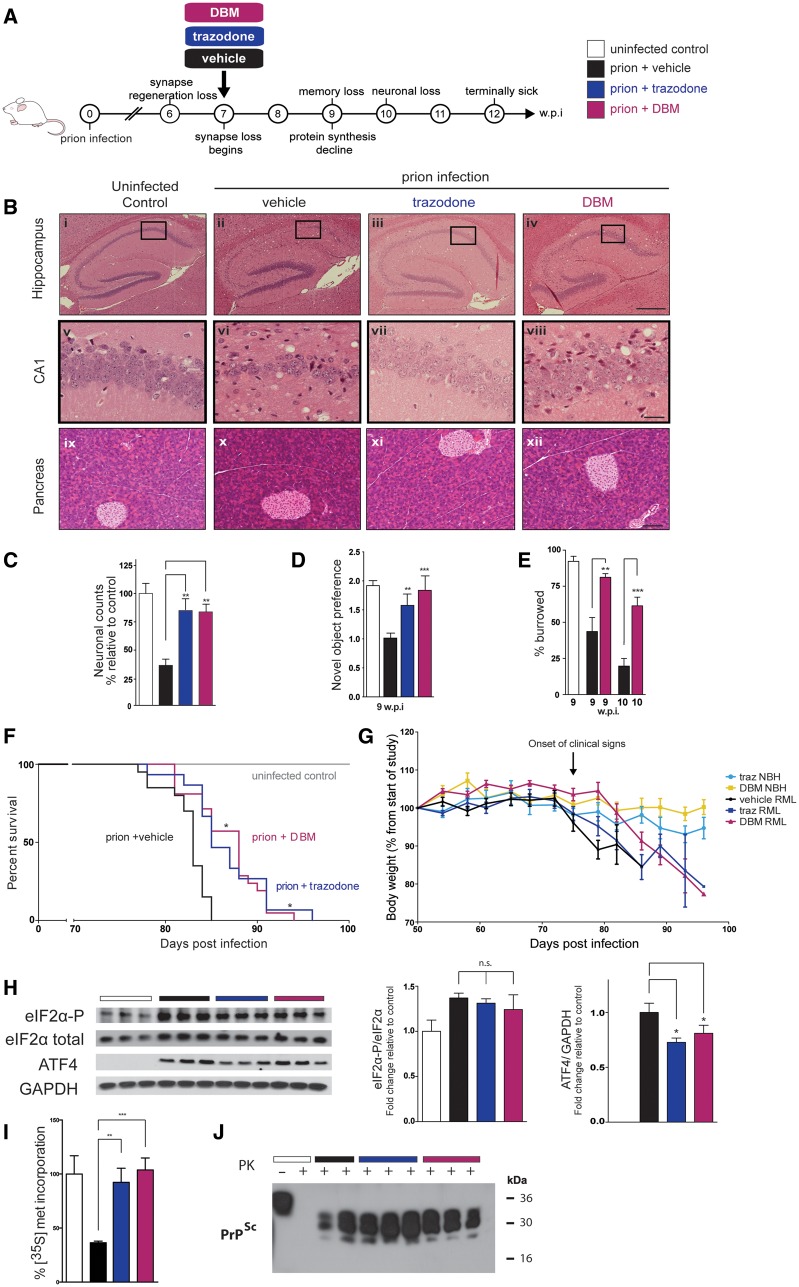
We first tested the drugs in mice with prion disease, for comparison of their therapeutic efficacy with that of the experimental compounds GSK2606414 and ISRIB (Moreno et al., 2013; Halliday et al., 2015; Radford et al., 2015). We used hemizygous tg37+/− mice (see below), as in our previous investigations (Mallucci et al., 2002, 2003; Moreno et al., 2012, 2013; Halliday et al., 2015; Radford et al., 2015) so that we could directly compare the efficacy of trazodone and DBM with previous genetic and pharmacological approaches. These mice overexpress PrP at ∼3-fold wild-type levels and therefore have a relatively rapid prion incubation time, succumbing to intracerebral Rocky Mountain laboratory (RML) prion inoculation at ∼12 weeks post-infection (∼84 days) (Mallucci et al., 2002). Misfolded PrP is detectable by 5–6 weeks post-infection, regenerative capacity is impaired at 6 weeks post-infection (Peretti et al., 2015) and overt synapse loss occurs from 7 weeks post-infection in RML-infected tg37+/− mice (Moreno et al., 2012). Dysregulated PERK signalling leads to eIF2α-P-mediated translational repression from ∼9 weeks post-infection, when cognitive deficits and memory loss are detected. Irreversible neurodegeneration occurs at 10 weeks post-infection, with signs of clinical neurological disease evident by 12 weeks post-infection (Moreno et al., 2012) (Fig. 3A). Restoring translation rates either upstream of eIF2α-P by inhibiting PERK with GSK2606414 (Moreno et al., 2013), or downstream using ISRIB (Halliday et al., 2015), has previously been shown to prevent clinical disease and neurodegeneration in prion-infected mice, independent of PrP levels. As in our previous studies (Moreno et al., 2013; Halliday et al., 2015), RML-infected tg37+/− mice were treated daily with trazodone, DBM or vehicle from 7 weeks post-infection, a time point when early degenerative changes—specifically synapse loss and spongiosis—are established, and which are considered equivalent to early stage symptoms of cognitive impairment in humans (Moreno et al., 2012). We used tg37+/− mice, as opposed to wild-type, in these as in our previous experiments, as we have previously shown UPR activation occurs in both strains, with identical effects on protein synthesis rates and clinical signs and neuronal loss, differing only in time course of disease (Moreno et al., 2012). In line with local animal husbandry guidelines, we use the mice with shorter disease course.
Figure 3.
Trazodone and DBM are neuroprotective in prion disease. (A) Schematic of prion disease course in tg37+/− mice. (B) Representative images (chosen from n = 10–12) of haematoxylin and eosin stained hippocampal and pancreatic sections, from uninfected controls and prion-infected mice treated with vehicle, trazodone and DBM. Both drugs are markedly neuroprotective (vii, viii compared to vi) and do not harm the pancreas (xi and xii). Scale bars = 400 μm (i–iv), 50 μm (v–viii) 200 μm (ix–xii). (C) Neuronal counts of CA1 region (n = 5 mice for each condition, with three slices counted from each animal). (D) Both drugs prevent loss of object recognition memory and (E) DBM prevents decline in burrowing behaviour at 9 and 10 weeks post-infection (n = 15 for each group) (F) Kaplan-Meier plot shows significantly increased survival with DBM (n = 21) and trazodone (n = 15) compared to vehicle (n = 20) treatment, *P < 0.05, Mantel–Cox analysis used. (G) Weights loss occurs in prion infected mice but not uninfected control, regardless of drug dosing. Normal brain homogenate, treated with trazodone (light blue) or DBM (yellow) n = 5. Prion infected mice treated with vehicle (black, n = 20) trazodone (dark blue, n = 15) or DBM (magenta n = 21). (H) Western blots show high levels of eIF2α-P in prion-disease brains are unaffected by treatment with DBM and trazodone, but ATF4 levels are significantly reduced; bar graphs on right show quantitation relative to loading controls (n = 3; repeated in triplicate). (I) DBM and trazodone restore global protein synthesis rates measured by 35S-met incorporation into hippocampal slices at 10 weeks post-infection (n = 6 per treatment group). (J) Levels of misfolded PrP, PrPSc, detected after PK digestion from brains of terminally sick mice is unaffected by either drug. All bar charts show means ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001, n.s. = non-significant, one-way ANOVA with Tukey’s post hoc test.
All mice were observed for effects of treatment with trazodone or DBM compared to vehicle on their clinical progression. Clinically confirmed prion disease in mice (which is terminal) is diagnosed by the appearance of a range of clinical signs indicative of neurological disease. These include the appearance of at least one of several possible early indicator signs (which are on their own non-specific) in combination with at least one of several possible confirmatory signs (Table 1). Confirmatory signs occur later in disease, when neuronal loss is advanced, and at least one is required for diagnosis. (Some, for example, impaired righting reflex occur more commonly than others). All 20 prion-infected mice treated with vehicle developed one or more confirmatory clinical signs and succumbed to prion infection in 83 ± 2 days (Table 1), consistent with previous data (Mallucci et al., 2002, 2003; Moreno et al., 2012, 2013; Halliday et al., 2015; Radford et al., 2015). Remarkably, both trazodone and DBM treatment prevented the appearance of confirmatory neurological signs diagnostic of clinical prion disease in majority of treated animals. Twelve of 15 (80%) prion-infected mice treated with trazodone and 16/21 mice (71%) treated with DBM, did not show diagnostic neurological signs of prion disease at the point of culling. While all animals developed non-specific early signs, most of these were not sustained and only 3/15 mice (trazodone) and 6/21 mice (DBM) progressed to develop prion confirmatory neurological signs.
Table 1.
Trazodone and DBM prevent clinical signs of neurological prion disease in most mice
| Clinical signs used in diagnosis of neurologically definite prion disease | Vehicle | Trazodone | DBM |
|---|---|---|---|
| Early indicator signs (not diagnostic of clinical prion disease in absence of confirmatory signs) | |||
|  Hind limb clasping | 13/20 | 3/15 | 9/21 |
|  Unsustained hunched posture | 1/20 | 2/15 | 7/21 |
|  Mild loss of coordination | 20/20 | 14/15 | 17/21 |
| Confirmatory signs (two confirmatory signs or one in combination with early indicator signs for diagnosis of clinical prion disease) | |||
|  Impairment of righting reflex | 18/20 | 2/15 | 5/21 |
|  Dragging of limbs | 1/20 | 0/15 | 0/21 |
|  Sustained hunched posture | 0/20 | 1/15 | 1/21 |
|  Abnormal breathing | 1/20 | 0/15 | 0/21 |
| Animals with clinical prion disease (at point of culling), n (time to diagnosis, days ± SEM) | 20/20 (83 ± 2) | 3/15 (83 ± 4) | 6/21 (84 ± 4) |
| Animals not manifesting neurological signs of prion disease (at point of culling), n | 0/20 | 12/15 | 15/21 |
Prion-infected mice were treated daily with trazodone, DBM or vehicle from 7 weeks post-infection, by which stage early neurological prion disease is established and significant synapse loss occurs (Moreno et al., 2012). All mice were assessed for appearance of one or more of both early indicator and confirmatory signs of neurological prion disease (‘scrapie’). One confirmatory sign and >2 early indicator signs or two confirmatory signs are required for the diagnosis of clinically definite prion disease. All vehicle-treated animals (n = 20) had confirmatory signs of terminal prion disease by 83 ± 2 days. Only 3/15 trazodone-treated and 6/21 DBM-treated mice developed neurologically definite clinical prion disease (83 ± 4 and 84 ± 4 days, respectively). The remaining mice showed no confirmatory neurological clinical signs. Other than the occurrence of one or more non-specific early indicator signs, which were mostly transient, they were clinically well.
Thus, both trazodone and DBM prevented development of neurological disease in the majority of treated animals: those animals developing confirmatory clinical signs and succumbing to prion infection may have received lower doses of drug, particularly in the DBM group, where drug levels were dependent on food consumption and varied between mice.
Trazodone and dibenzoylmethane prevent neurodegeneration and rescue behavioural deficits in prion-infected mice without pancreatic toxicity
Consistent with clinical efficacy of trazodone and DBM in preventing clinical signs of neurodegeneration, both drugs were markedly neuroprotective at the histological level in prion-infected mice. Thus, both drugs substantially reduced neuronal loss in the hippocampus in those mice in which they prevented neurological signs of prion disease. In contrast, vehicle-treated mice had extensive atrophy of CA1-3 hippocampal cells at the time of sacrifice, when they also had confirmed clinical diagnosis of prion disease. This is markedly different to the protective effects of both trazodone and DBM treatment on hippocampal morphology and integrity [Fig. 3B(ii–iv) and (vi–viii)]. Indeed, neuronal counts from CA1 regions confirmed significant protective effects of both trazodone and DBM (Fig. 3C). Critically, neither drug produced any pancreatic toxicity of either endocrine or exocrine tissue [Fig. 3B(ix–xii), Supplementary Fig. 3A and B]. Both compounds rescued the loss of object recognition memory at 9 weeks post-infection (Fig. 3D), and DBM also prevented loss of burrowing behaviour characteristic of prion infection at 9 and 10 weeks post-infection (Moreno et al., 2012, 2013) (Fig. 3E). (Trazodone treatment precluded testing for burrowing activity due to effects of sedation immediately after dosing, reducing spontaneous activity. Sedation was not sustained, but nonetheless it did not permit testing within the relevant period).
Trazodone and dibenzoylmethane significantly increase survival in prion-infected mice
Importantly, both drugs significantly increased lifespan in the 12/15 trazodone-treated and 15/21 DBM-treated animals that did not show neurological signs of prion infection (P = 0.0103) (Fig. 3F). However, the increase in lifespan was less prolonged than would have been expected, given both the lack of diagnostic signs of prion disease seen clinically (Table 1) and the extent of neuroprotection seen histologically (Fig. 3B). This occurred as, in accordance with UK Home Office regulations, these mice had to be culled because of weight loss of 20% of their original body weight, despite being devoid of neurological signs of prion disease and being otherwise in every way clinically well (Fig. 3G). This very likely masked probable much longer-lasting clinical efficacy of treatment with trazodone or DBM. Importantly, weight loss was not an effect of treatment with trazodone or DBM in itself. Control mice, inoculated with normal brain homogenate and treated with the drugs over the same time period, did not lose weight (Fig. 3G). Further, the rTg4510 tauopathy mice did not lose weight with trazodone or DBM treatment (Fig. 4G), even though treatment lasted for several months before the animals were sacrificed for analysis.
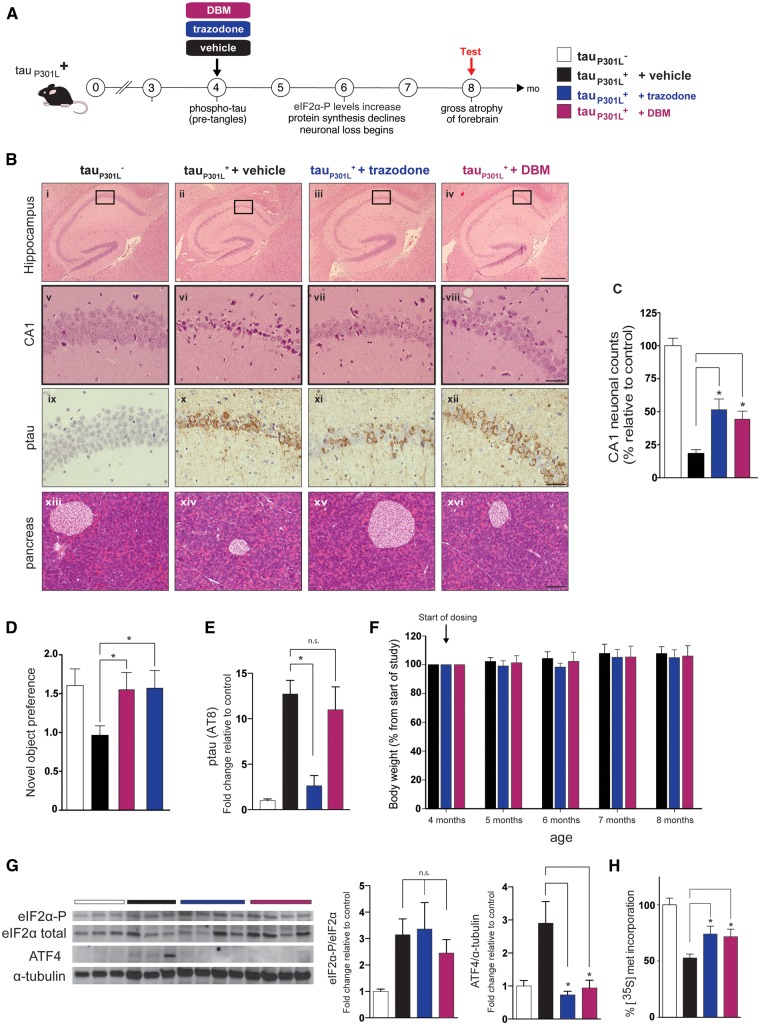
Figure 4.
Trazodone and DBM are neuroprotective in rTg4510 model of the tauopathy FTD. (A) Schematic of disease progression in rTg4510 tauP301L+ mice. (B) Representative images (chosen from n = 10–12) of hippocampal sections from 8-month-old mice stained with haematoxylin and eosin (i–viii) and pSer202/Thr205-tau (ix–xii) from tauP301L− control mice and tauP301L+ mice treated with vehicle, trazodone or DBM. Scale bars = 400 μm (i–iv), 50 μm (v–xii). (C) Neuronal counts of CA1 region at 8 months (n = 5 for each condition, three consecutive slices). (D) Both drugs prevent loss of object recognition memory (n = 12 for each group). (E) Quantification of p-tau levels measured by the AT100 antibody. Trazodone but not DBM reduces levels. n = 3 mice per group. (F) Trazodone and DBM do not cause weight loss in rTG4510 mice during the 4-month dosing period, n = 10 per group. (G) Protein levels of ATF4 are reduced after treatment with trazodone or DBM compared to vehicle; however, eIF2α-P and eIF2α levels do not change. Repeated in triplicate. (H) DBM and trazodone partially restore global protein synthesis rates measured by 35S-met incorporation into hippocampal slices at 8 months (n = 4 per treatment group). All bar charts show mean ± SEM. *P < 0.05, n.s. = non-significant, using one-way ANOVA and Tukey’s post hoc analysis.
Thus, it appears that loss of body mass is a feature of systemic prion infection (albeit not associated with other signs of ill-health), which we have previously also described with ISRIB treatment (Halliday et al., 2015). With both trazodone and DBM, as with ISRIB, this feature of prion infection is dissociated from the neuroprotective effects of treatment.
Trazodone and dibenzoylmethane restore protein synthesis downstream of eIF2α-P in prion-diseased mice
The site of action of both compounds in vivo is downstream of eIF2α-P, in agreement with our in vitro findings. Thus, elevated eIF2α-P levels in prion-diseased hippocampi were unchanged by treatment with trazodone and DBM, while ATF4 levels were reduced (Fig. 3H), replicating our observations in vitro (Fig. 2B and D). Consistent with this, both drugs increased protein synthesis rates in ex vivo hippocampal slices compared to ∼60% reduction in protein synthesis rates seen in the brains of vehicle-treated animals (Fig. 3I). DBM and trazodone treatment had no effect on the levels of misfolded PrP levels (PrPSc) (Fig. 3J), consistent with the fact that neuroprotection is likely due to effects on eIF2α-P signalling, not on prion replication.
Trazodone and dibenzoylmethane are neuroprotective in the rTg4510 model of frontotemporal dementia
PERK branch UPR activation is seen in human tauopathies, including Alzheimer’s disease and progressive supranuclear palsy (Hoozemans et al., 2009; Stutzbach et al., 2013). We thus tested trazodone and DBM in a mouse model of tauopathy. rTg4510 mice overexpress the human tau mutation P301L associated with FTD, with onset of tau pathology from 3 months of age (Santacruz et al., 2005). Raised eIF2α-P and ATF4 levels and a decline in protein synthesis rates, associated with the onset of neurodegeneration, occur at between 5–6 months of age, becoming severe—with marked forebrain atrophy—by 8 months of age (Fig. 4A). Treatment of tauP301L+ mice with PERK inhibitor GSK2606414 has been shown to be neuroprotective (Radford et al., 2015). Thus, tauP301L+ mice received trazodone or DBM daily from 4 months and were examined at 8 months, when neuronal loss is advanced in the CA1 region of untreated (vehicle-treated) mice compared to transgene negative tauP301L− mice (Fig. 4B), with shrinkage of whole hippocampus in tauP301L+ mice (Fig. 4B) (Ramsden et al., 2005; Santacruz et al., 2005). Trazodone and DBM reduced hippocampal atrophy in rTg4510 mice compared to vehicle treatment, where this was pronounced [Fig. 4B, cf. (ii), (iii) and (iv), and Fig. 4C]. They were notably neuroprotective in the hippocampus, significantly reducing loss of CA1 pyramidal neurons compared to tauP301L+ mice treated with vehicle [Fig. 4B, cf. (vi), (vii) and (viii), and Fig. 4C]. As in the prion-infected mice, treatment was not toxic to the pancreas [Fig. 4B(xiii–xvi), Supplementary Fig. 3C and D]. Both compounds rescued the loss of object recognition memory at 5 months. (Fig. 4D). Trazodone also produced a reduction in p-tau staining at 8 months compared to vehicle- or DBM-treated mice [Fig. 4B, cf. (xi) to (x) or (xii), and Fig. 4E]. This is likely due to inhibition of the tau kinase GSK3β by phosphorylation at Ser9 induced by trazodone (Supplementary Fig. 4) (Radford et al., 2015). The life-span of rTg4510 mice is long (Ramsden et al., 2005), so survival studies were not possible as an outcome of treatment, but the effects on hippocampal volume and neuronal integrity and on the general condition of the mice was marked with both drugs. Of note, both drugs were well tolerated for the duration of the experiment—several months treatment. Importantly, drug treatment did not have any effect on weight loss (Fig. 4F).
As in prion-infected mice, both compounds reduced ATF4 levels in the brains of rTg4510 without lowering eIF2α-P levels (Fig. 4G), supporting a site of action downstream of eIF2α-P, as seen in vitro (Fig. 2B and D). Trazodone and DBM also partially restored protein synthesis rates in tauopathy-treated mice (Fig. 4H), again consistent with inhibition of signalling at this level.
Discussion
There is a tremendous unmet clinical need for effective pharmacological interventions against protein-misfolding neurodegenerative disorders such as Alzheimer’s and Parkinson’s diseases. The PERK branch of the UPR, in particular, has emerged as a promising new therapeutic target in these disorders, due to its overactivation in human Alzheimer’s disease brain tissue and mechanistic implications from disease models (Hetz et al., 2013; Halliday and Mallucci, 2014). Thus, in preclinical models, many lines of evidence support increased signalling via eIF2α-P—and the ensuing attenuation of protein synthesis rates—as contributing to cognitive impairment and disease progression (Moreno et al., 2012, 2013; Ma et al., 2013; Devi and Ohno, 2014; Kim et al., 2014; Halliday et al., 2015; Radford et al., 2015). Moreover, two compounds, the PERK inhibitor GSK2606414 (Moreno et al., 2013) and the ISR inhibitor ISRIB (Halliday et al., 2015) have been shown to interfere with the translational inhibitory effects of increased eIF2α phosphorylation (acting upstream and downstream of eIF2α-P, respectively). Both of these compounds have been shown to have significant neuroprotective effects in mouse models of neurodegeneration, reducing disease progression (Moreno et al., 2013; Halliday et al., 2015; Radford et al., 2015). GSK2606414 also improves neuronal phenotypes in fly models of disease (Kim et al., 2014; Celardo et al., 2016). Further, ISRIB boosts cognition in wild-type mice (Sidrauski et al., 2013). However, neither of these substances is suitable for translation to treatment of human disease, due to associated toxicity (GSK2606414) or insolubility (ISRIB) (Moreno et al., 2013; Halliday et al., 2015). There is therefore an urgent need to find safe compounds with similar therapeutic effects to test the potential efficacy of this approach in human disease. This need drove us to develop new screens to find potential repurposable drugs for translation to the clinic as new disease-modifying treatments for dementia.
The development of experimental models amenable to live animal compound screening is an attractive approach for discovering effective pharmacological therapies. Drug screens are often a balance between choosing the most biologically relevant readout, and maintaining a high-enough throughput to remain useful. We chose the nematode worm C. elegans for our primary in vivo screen due to the worms’ quick generational time, the ability to breed hundreds of developmentally synchronized animals for testing and easily observable phenotypes. Importantly, the UPR is conserved across metazoa, and C. elegans contains orthologues of all the major UPR genes. Tunicamycin induces developmental arrest in the worms at stages L2/L3. This likely occurs as they normally greatly increase in size between the L2 and L3 stages, requiring large amounts of new protein synthesis, which is attenuated by tunicamycin (Supplementary Fig. 1). Due to the large difference in size between worms that were stalled at the L2 stage and older worms that had overcome the developmental block, compounds that exerted any beneficial effects were easily observed (Supplementary Table 1). We used a NINDS Custom Collection 2 drug library including mostly FDA-approved drugs, many of which were known to be brain-penetrant and neurologically active in humans. Twenty of the 1040 NINDS compounds overcame the developmental delay in tunicamycin-stressed worms and were therefore potential UPR inhibitors (Supplementary Table 1). However, this screen did not determine whether the drugs were direct inhibitors of the UPR. The upregulation of chaperones, or the drugs themselves acting as chemical chaperones, could similarly facilitate normal development in the worms. We therefore screened the 20 hits in mammalian CHO CHOP::luciferase cells to test for inhibition of PERK-mediated UPR signalling (Fig. 1A and B). Five of the 20 compounds repressed CHOP::luciferase signalling induced by tunicamycin. Of these, trazodone, a licensed anti-depressant safe to use in the elderly, with excellent oral pharmacokinetic profile and excellent safety profile and DBM, a curcumin analogue with notable anti-cancer properties in preclinical models, were the best candidates to take forward.
Trazodone is an antidepressant in the serotonin antagonist and reuptake inhibitor class, which has additional anxiolytic and hypnotic effects. It has been shown to reduce the behavioural and psychological symptoms of dementia (BPSD) in Alzheimer’s disease (Lopez-Pousa et al., 2008) and in FTD (Lebert et al., 2004), but no studies have looked at the progression of neurodegeneration with trazodone treatment. Although its pharmacological actions in humans are not fully understood, it is thought to have more than one mechanism of therapeutic action, making it a multifunctional drug. It is the first antidepressant with a dual mechanism of action involving inhibition of the serotonin transporter (SERT) and antagonism of the serotonin type 2 (5-HT2) receptor, producing its antidepressant effect by blocking SERT, and increasing serotonin concentrations in the brain. Trazodone exerts antagonistic properties against α1- and α2-adrenergic receptors and histamine H1 receptors, with minimal anticholinergic effects (Monti et al., 1986; Stahl, 2009). Trazodone has previously shown benefit in models of Huntington’s disease, where it improved mitochondrial respiratory complex activity (Kumar et al., 2011) and Morris water maze performance (Kumar et al., 2010)
DBM is a minor constituent of liquorice that has been found to have antineoplastic effects, with efficacy against prostate and mammary tumours (Huang et al., 1998; Khor et al., 2009). Carcinogen detoxification has been proposed as a possible mechanism of action as DBM has been reported to potently induce phase 2 hepatic detoxification enzymes (Dinkova-Kostova and Talalay, 1999). DBM has also been reported to induce the Nrf2 survival pathway (Thimmulappa et al., 2008), which is activated downstream of UPR activation (He et al., 2001). DBM has also been shown to upregulate GRP78/BiP (Frazier et al., 2004). DBM derivatives have been shown to induce protection from necrotic cell death (Hegedus et al., 2013) and protect dopaminergic neurons against both oxidative stress and endoplasmic reticulum stress (Takano et al., 2007). It is unclear if DBM itself shares these effects with its derivatives.
In this study, we found that both trazodone and DBM inhibited the effects of UPR activation and eIF2α phosphorylation, reversing translational attenuation and lowering levels of ATF4 and CHOP in mammalian cells (Figs 1A and 2B). This occurred without lowering eIF2α-P levels in vitro (Fig. 2B), and in vivo (Figs 3H and 4G), as has been described for ISRIB (Halliday et al., 2015; Sekine et al., 2015; Sidrauski et al., 2015). Both trazodone and DBM prevented eIF2α-P from lowering ternary complex levels, due to their ability to reduce ATF4 5’UTR regulation of luciferase (Fig. 2D), but this was via an independent mechanism than that of ISRIB (Supplementary Fig. 2).
Based on their ability to reverse UPR activation, trazodone and DBM were predicted to be potential therapeutic candidates for neurodegenerative disorders. Indeed, both compounds were found to be substantially neuroprotective in two different mouse models of neurodegeneration, prion disease and the rTg4510 tauopathy model of FTD. Both drugs showed beneficial neuroprotective effects similar to those of the experimental compounds GSK2606414 and ISRIB. Importantly, in both models, the drugs were first administered at a stage of early but established disease, equivalent to early symptomatic human disease—from 7 weeks post-infection in prion-disease mice and from 4 months of age in tauopathy mice. Treatment with each drug prevented neuronal loss in the hippocampus (Fig. 3B and C) and confirmatory neurological clinical signs (Table 1) in the majority of prion-infected mice, also restoring memory and preventing behavioural decline associated with prion infection (Fig. 3D and E). In tauopathy mice, both drugs were strongly protective against the marked hippocampal neuronal loss and forebrain atrophy that is a feature of this mouse model (Fig. 4B and C). These mice were also devoid of clinical signs of neurodegeneration. There was also significant increase in lifespan of prion-infected mice treated with trazodone or DBM (Fig. 3F), although this was more modest than was expected considering the neuroprotection observed, due to coincidental weight loss requiring early sacrifice of otherwise entirely healthy animals (Fig. 3G). This weight loss was not caused by trazodone or DBM (Figs 3G and 4F), which, critically, were also devoid of pancreatic toxicity [Figs 3B(ix–xii), 4B(xiii–xvi) and Supplementary Fig. 2), likely because both compounds only partially reverse eIF2α-P-mediated translational attenuation (Fig. 1A and B), which is associated with lack of toxicity to secretory tissue (Halliday et al., 2015). Thus, neither trazodone nor DBM suffer from the shortcomings of GSK2606414 and ISRIB: they are non-toxic to the pancreas and have favourable pharmacokinetic properties. Critically, however, they share the benefits of GSK2606414 and ISRIB, being markedly neuroprotective.
Interestingly, another unanticipated and previously unreported action of trazodone was to lower phosphorylated (p)-tau levels in rTg4510 mice (Fig. 4B, E and Supplementary Fig. 4). P-tau is associated with Alzheimer’s disease pathology and with the tauopathies FTD and progressive supranuclear palsy, and is in itself an intense focus for Alzheimer’s therapeutics. This additional effect of trazodone could be considered a desirable bonus in the treatment of these disorders. UPR activation is known to induce tau phosphorylation via activation of GSK3β (Nijholt et al., 2013) and treatment with the PERK inhibitor GSK2606414 also lowers tau phosphorylation (van der Harg et al., 2014; Radford et al., 2015). Trazodone was also able to inhibit GSK3β (Supplementary Fig. 4). However, the similar degree of neuroprotection afforded by trazodone and DBM (Table 1, Figs 3B, C, 4B and C) suggests that reduction of the stress response downstream of eIF2α-P and partial restoration of protein synthesis is the primary driver of neuroprotection in both tauopathy and prion-diseased mice. This is irrespective of sustained high levels of misfolded protein, both prion (Fig. 3J) and p-tau (in DBM treated mice; Fig. 4B and E), further demonstrating the central role the UPR plays in neurodegeneration.
In conclusion, we selected trazodone and DBM as potential therapeutic agents in neurodegenerative disease, based on their consistent ability to inhibit UPR/ISR-induced translational repression, rather than target disease-specific misfolded proteins, in organisms ranging from nematodes, through mammalian cell models, to different mouse models of neurodegeneration. The two drugs were markedly neuroprotective in both prion-diseased and FTD mice at clinically relevant doses over a sustained treatment period. These drugs therefore represent an important step forward in the pursuit of disease-modifying treatments for Alzheimer’s and related disorders. Trazodone in particular, is already licensed for use in elderly patients. These drugs should now be tested in clinical trials in the treatment of dementia.
Supplementary Material
Acknowledgements
We thank: MRC Technology for gift of NINDS library and D. Ron (University of Cambridge) for gift of CHOP::luciferase cells, J. Edwards (MRC Toxicology Unit) and CRF staff at the University of Leicester for technical assistance, D. Ron (Cambridge) for discussions of data.
Glossary
Abbreviations
- ATF
activating transcription factor
- CHO
Chinese hamster ovary
- CHOP
C/EBP homologous protein
- DBM
dibenzoylmethane
- eIF2α
eukaryotic initiation factor 2α
- FTD
frontotemporal dementia
- ISR
integrated stress response
- PERK
pancreatic endoplasmic reticulum kinase
- PrP
prion protein
- UPR
unfolded protein response
Funding
This work was funded by the Medical Research Council, UK (MRC 5TR50) and by a grant to GRM from the Alzheimer’s Society & Alzheimer’s Drug Discovery Foundation (RG78185). GRM holds an ERC Consolidator award.
Supplementary material
Supplementary material is available at Brain online.
References
- Celardo I, Costa AC, Lehmann S, Jones C, Wood N, Mencacci NEet al. Mitofusin-mediated ER stress triggers neurodegeneration in pink1/parkin models of Parkinson’s disease. Cell Death Dis 2016; 7: e2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi L, Ohno M. PERK mediates eIF2 alpha phosphorylation responsible for BACE1 elevation, CREB dysfunction and neurodegeneration in a mouse model of Alzheimer’s disease. Neurobiol Aging 2014; 35: 2272–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Talalay P. Relation of structure of curcumin analogs to their potencies as inducers of Phase 2 detoxification enzymes. Carcinogenesis 1999; 20: 911–14. [DOI] [PubMed] [Google Scholar]
- Frazier MC, Jackson KM, Jankowska-Stephens E, Anderson MG, Harris WB. Proteomic analysis of proteins altered by dibenzoylmethane in human prostatic cancer LNCaP cells. Proteomics 2004; 4: 2814–21. [DOI] [PubMed] [Google Scholar]
- Halliday M, Mallucci GR. Modulating the unfolded protein response to prevent neurodegeneration and enhance memory. Neuropathol Appl Neurobiol 2015; 41: 414–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday M, Radford H, Sekine Y, Moreno J, Verity N, le Quesne Jet al. Partial restoration of protein synthesis rates by the small molecule ISRIB prevents neurodegeneration without pancreatic toxicity. Cell Death Dis 2015; 6: e1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Khersonsky S, Marciniak S, Scheuner D, Kaufman RJet al. Bioactive small molecules reveal antagonism between the integrated stress response and sterol-regulated gene expression. Cell Metab 2005; 2: 361–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He CH, Gong P, Hu B, Stewart D, Choi ME, Choi AMet al. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation. J Biol Chem 2001; 276: 20858–65. [DOI] [PubMed] [Google Scholar]
- Hegedus C, Lakatos P, Kiss-Szikszai A, Patonay T, Gergely S, Gregus Aet al. Cytoprotective dibenzoylmethane derivatives protect cells from oxidative stress-induced necrotic cell death. Pharmacol Res 2013; 72: 25–34. [DOI] [PubMed] [Google Scholar]
- Hetz C, Chevet E, Harding HP. Targeting the unfolded protein response in disease. Nat Rev Drug Discov 2013; 12: 703–19. [DOI] [PubMed] [Google Scholar]
- Hoozemans JJ, van Haastert ES, Eikelenboom P, de Vos RA, Rozemuller JM, Scheper W. Activation of the unfolded protein response in Parkinson’s disease. Biochem Biophys Res Commun 2007; 354: 707–11. [DOI] [PubMed] [Google Scholar]
- Hoozemans JJ, van Haastert ES, Nijholt DA, Rozemuller AJ, Eikelenboom P, Scheper W. The unfolded protein response is activated in pretangle neurons in Alzheimer’s disease hippocampus. Am J Pathol 2009; 174: 1241–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MT, Lou YR, Xie JG, Ma W, Lu YP, Yen Pet al. Effect of dietary curcumin and dibenzoylmethane on formation of 7,12-dimethylbenz[a]anthracene-induced mammary tumors and lymphomas/leukemias in Sencar mice. Carcinogenesis 1998; 19: 1697–700. [DOI] [PubMed] [Google Scholar]
- Khor TO, Yu S, Barve A, Hao X, Hong JL, Lin Wet al. Dietary feeding of dibenzoylmethane inhibits prostate cancer in transgenic adenocarcinoma of the mouse prostate model. Cancer Res 2009; 69: 7096–102. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Raphael AR, LaDow ES, McGurk L, Weber RA, Trojanowski JQet al. Therapeutic modulation of eIF2alpha phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nature Genet 2014; 46: 152–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Kalonia H, Kumar A. Nitric oxide mechanism in the protective effect of antidepressants against 3-nitropropionic acid-induced cognitive deficit, glutathione and mitochondrial alterations in animal model of Huntington’s disease. Behav Pharmacol 2010; 21: 217–30. [DOI] [PubMed] [Google Scholar]
- Kumar P, Kalonia H, Kumar A. Novel protective mechanisms of antidepressants against 3-nitropropionic acid induced Huntington’s-like symptoms: a comparative study. J Psychopharmacol 2011; 25: 1399–411. [DOI] [PubMed] [Google Scholar]
- Lebert F, Stekke W, Hasenbroekx C, Pasquier F. Frontotemporal dementia: a randomised, controlled trial with trazodone. Dement Geriatr Cogn Disord 2004; 17: 355–9. [DOI] [PubMed] [Google Scholar]
- Lopez-Pousa S, Garre-Olmo J, Vilalta-Franch J, Turon-Estrada A, Pericot-Nierga I. Trazodone for Alzheimer’s disease: a naturalistic follow-up study. Arch Gerontol Geriatr 2008; 47: 207–15. [DOI] [PubMed] [Google Scholar]
- Ma T, Trinh MA, Wexler AJ, Bourbon C, Gatti E, Pierre Pet al. Suppression of eIF2alpha kinases alleviates Alzheimer’s disease-related plasticity and memory deficits. Nat Neurosci 2013; 16: 1299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallucci G, Dickinson A, Linehan J, Klohn PC, Brandner S, Collinge J. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science 2003; 302: 871–4. [DOI] [PubMed] [Google Scholar]
- Mallucci GR, Ratte S, Asante EA, Linehan J, Gowland I, Jefferys JGet al. Post-natal knockout of prion protein alters hippocampal CA1 properties, but does not result in neurodegeneration. EMBO J 2002; 21: 202–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleery J, Cohen DA, Sharpley AL. Pharmacotherapies for sleep disturbances in Alzheimer’s disease. Cochrane Database Syst Rev 2014; 3: CD009178. [DOI] [PubMed] [Google Scholar]
- Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SWet al. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science 2013; 340: 82–5. [DOI] [PubMed] [Google Scholar]
- Monti JM, Pellejero T, Jantos H. Effects of H1- and H2-histamine receptor agonists and antagonists on sleep and wakefulness in the rat. J Neural Transm 1986; 66: 1–11. [DOI] [PubMed] [Google Scholar]
- Moreno JA, Halliday M, Molloy C, Radford H, Verity N, Axten JMet al. Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Sci Transl Med 2013; 5: 206ra138. [DOI] [PubMed] [Google Scholar]
- Moreno JA, Radford H, Peretti D, Steinert JR, Verity N, Martin MGet al. Sustained translational repression by eIF2alpha-P mediates prion neurodegeneration. Nature 2012; 485: 507–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijholt DA, Nolle A, van Haastert ES, Edelijn H, Toonen RF, Hoozemans JJet al. Unfolded protein response activates glycogen synthase kinase-3 via selective lysosomal degradation. Neurobiol Aging 2013; 34: 1759–71. [DOI] [PubMed] [Google Scholar]
- Nijholt DA, van Haastert ES, Rozemuller AJ, Scheper W, Hoozemans JJ. The unfolded protein response is associated with early tau pathology in the hippocampus of tauopathies. J Pathol 2012; 226: 693–702. [DOI] [PubMed] [Google Scholar]
- Oslowski CM, Urano F. Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol 2011; 490: 71–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretti D, Bastide A, Radford H, Verity N, Molloy C, Martin MGet al. RBM3 mediates structural plasticity and protective effects of cooling in neurodegeneration. Nature 2015; 518: 236–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaglio E, Restelli E, Garofoli A, Dossena S, De Luigi A, Tagliavacca Let al. Expression of mutant or cytosolic PrP in transgenic mice and cells is not associated with endoplasmic reticulum stress or proteasome dysfunction. PLoS One 2011; 6: e19339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford H, Moreno JA, Verity N, Halliday M, Mallucci GR. PERK inhibition prevents tau-mediated neurodegeneration in a mouse model of frontotemporal dementia. Acta Neuropathol 2015; 130: 633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsden M, Kotilinek L, Forster C, Paulson J, McGowan E, SantaCruz Ket al. Age-dependent neurofibrillary tangle formation, neuron loss, and memory impairment in a mouse model of human tauopathy (P301L). J Neurosci 2005; 25: 10637–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reagan-Shaw S, Nihal M, Ahmad N. Dose translation from animal to human studies revisited. FASEB J 2008; 22: 659–61. [DOI] [PubMed] [Google Scholar]
- Richardson CE, Kinkel S, Kim DH. Physiological IRE-1-XBP-1 and PEK-1 signaling in Caenorhabditis elegans larval development and immunity. PLoS Genet 2011; 7: e1002391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 2007; 8: 519–29. [DOI] [PubMed] [Google Scholar]
- Santacruz K, Lewis J, Spires T, Paulson J, Kotilinek L, Ingelsson Met al. Tau suppression in a neurodegenerative mouse model improves memory function. Science 2005; 309: 476–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine Y, Zyryanova A, Crespillo-Casado A, Fischer PM, Harding HP, Ron D. Mutations in a translation initiation factor identify the target of a memory-enhancing compound. Science 2015; 348: 1027–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Acosta-Alvear D, Khoutorsky A, Vedantham P, Hearn BR, Li Het al. Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife 2013; 2: e00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Tsai JC, Kampmann M, Hearn BR, Vedantham P, Jaishankar Pet al. Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. eLife 2015; 4: e07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HL, Mallucci GR. The unfolded protein response: mechanisms and therapy of neurodegeneration. Brain 2016; 139 (Pt 8): 2113–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 2009; 136: 731–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl SM. Mechanism of action of trazodone: a multifunctional drug. CNS Spectr 2009; 14: 536–46. [DOI] [PubMed] [Google Scholar]
- Stutzbach LD, Xie SX, Naj AC, Albin R, Gilman S, Group PSPGSet al. The unfolded protein response is activated in disease-affected brain regions in progressive supranuclear palsy and Alzheimer’s disease. Acta Neuropathol Commun 2013; 1: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano K, Kitao Y, Tabata Y, Miura H, Sato K, Takuma Ket al. A dibenzoylmethane derivative protects dopaminergic neurons against both oxidative stress and endoplasmic reticulum stress. Am J Physiol Cell Physiol 2007; 293: C1884–94. [DOI] [PubMed] [Google Scholar]
- Thimmulappa RK, Rangasamy T, Alam J, Biswal S. Dibenzoylmethane activates Nrf2-dependent detoxification pathway and inhibits benzo(a)pyrene induced DNA adducts in lungs. Med Chem 2008; 4: 473–81. [DOI] [PubMed] [Google Scholar]
- van der Harg JM, Nolle A, Zwart R, Boerema AS, van Haastert ES, Strijkstra AMet al. The unfolded protein response mediates reversible tau phosphorylation induced by metabolic stress. Cell Death Dis 2014; 5: e1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A 2004; 101: 11269–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.