The genetic landscape of amyotrophic lateral sclerosis (ALS) is poorly understood. By examining known ALS-associated genes in a large cohort, Morgan, Shatunov et al. report an increase in mutations within the untranslated prime regions of the genes and a greater than expected number of patients with multiple potentially pathogenic variants.
Keywords: amyotrophic lateral sclerosis, neurodegeneration, complex trait, polygenic inheritance, association study
Abstract
Amyotrophic lateral sclerosis is a progressive neurodegenerative disease of motor neurons. About 25 genes have been verified as relevant to the disease process, with rare and common variation implicated. We used next generation sequencing and repeat sizing to comprehensively assay genetic variation in a panel of known amyotrophic lateral sclerosis genes in 1126 patient samples and 613 controls. About 10% of patients were predicted to carry a pathological expansion of the C9orf72 gene. We found an increased burden of rare variants in patients within the untranslated regions of known disease-causing genes, driven by SOD1, TARDBP, FUS, VCP, OPTN and UBQLN2. We found 11 patients (1%) carried more than one pathogenic variant (P = 0.001) consistent with an oligogenic basis of amyotrophic lateral sclerosis. These findings show that the genetic architecture of amyotrophic lateral sclerosis is complex and that variation in the regulatory regions of associated genes may be important in disease pathogenesis.
Introduction
Amyotrophic lateral sclerosis (ALS) is a fatal, neurodegenerative disorder with a life expectancy of 3–5 years from symptom onset. Presentation is with weakness of voluntary muscles, representing degeneration of the upper and lower motor neurons. The incidence of ALS is approximately 2 per 100 000 person–years with a slightly higher proportion of males (Logroscino et al., 2010). About 15% have concomitant frontotemporal dementia (FTD) and up to 50% may have subtle cognitive impairment (Abrahams et al., 2014).
The complexity of the molecular mechanisms implicated in ALS is paralleled by multifaceted genetics that are still not fully understood despite extensive research (Al-Chalabi and Hardiman, 2013; Morgan and Orrell, 2016). To address this issue, there has been a move towards next-generation sequencing (NGS) as a high-throughput, relatively inexpensive tool to uncover the genetic architecture of ALS and other heterogeneous neurological diseases, including Charcot–Marie–Tooth disease, dementia, ataxia and Parkinson’s disease (Johnson et al., 2010; Mencacci et al., 2014; Cady et al., 2015; Cirulli et al., 2015; van Rheenen et al., 2016).
About 5–10% of ALS is considered familial (familial ALS), which may be an underestimate depending on the definition used (Byrne et al., 2012), and there is extensive evidence that the distinction between familial and sporadic ALS is not clear-cut (Al-Chalabi and Lewis, 2011). Over 100 genes have been implicated in ALS to varying degrees (Abel et al., 2012; http://alsod.iop.kcl.ac.uk), with ∼25 of these having been replicated in subsequent studies. The four most important ALS genes by frequency are SOD1, TARDBP/TDP-43, C9orf72 and FUS. Variants in these genes are more likely to be of large effect, and carrying the genotype greatly increases the probability of ALS; in other words, these variants show moderate to high penetrance. Gene variants of low penetrance are also of interest, even though they only increase risk a little for any individual, as the overall variance in genetic risk explained is high, and such genes contribute to our understanding of the pathway to ALS. In at least some cases, ALS is oligogenic, with affected individuals carrying more than one rare variant implicated in ALS (van Blitterswijk et al., 2012; Cady et al., 2015). If a significant number of cases in ALS are indeed caused by more than one risk variant, this has implications for genetic counselling and treatment.
We therefore aimed to explore the genetics of ALS in a cohort of 1126 patients using a specific ALS-gene panel for NGS.
Materials and methods
Patients
A total of 1126 cases and 613 controls of European ancestry were used as part of this study. This was composed of 131 individuals with familial ALS (64 female, 67 male) and 995 with sporadic ALS (428 female, 567 male). The average age of onset was 56 years for familial ALS (range 24–85) and 61 for sporadic ALS (range 25–88). Control samples were composed of 232 females and 381 males. These clinical data are presented in Supplementary Table 1. Averaged across loci, 13 cases and 15 controls failed to sequence across some of the target genome. Three hundred and seventeen patients did not have complete C9orf72 data because of insufficient DNA. Patient samples were obtained predominantly from the UK National DNA Bank for Motor Neuron Disease (MND) Research (Smith et al., 2015) as well as from University College London and Partners (UCLP) MND clinics. A small subset (n = 95) of these samples overlap with our previous proof-of-principle publication and were included in this study to increase the power to detect mutation burden (Morgan et al., 2015). Control samples were either sequenced using MiSeq technology (see below), whole-exome sequencing or both. They were selected based on their ethnicity, age (over 60 years old) and whether they were free from any neurological disorder. Additionally, subjects were excluded from the study if they had a first-degree relative with a neurological disorder including Alzheimer’s disease, ALS, ataxia, autism, bipolar disorder, cerebrovascular disease, dementia, dystonia, Parkinson’s disease and schizophrenia.
Next-generation sequencing
An ALS-specific gene panel was designed for use on the Illumina MiSeq platform by means of the Illumina TruSeq Custom Amplicon Assay. This uses PCR amplicon-based target enrichment, and screens for variants across 23 genes that were selected based on their association with ALS, so that all the chief causal genes were included as well as several risk factors and a selection of variants with an uncertain relationship to ALS, either through a lack of evidence or through the gene more commonly causing a related disease. The panel included exons and flanking regions for ALS2, ANG, CHMP2B, DAO, DCTN1, FIG4, FUS, NEFH, OPTN, PFN1, PON1, PON2, PON3, PRPH, SETX, SOD1, SQSTM1, TARDBP, TREM2, UBQLN2, VAPB, VCP and VEGFA. The following genes were also extensively covered in both 5’ and 3’ untranslated regions (UTRs): three were selected because they are regarded as major ALS genes (SOD1, TARDBP, FUS) and three because the design of the assay made sequencing through the untranslated region simple to achieve (OPTN, VCP, UBQLN2; Supplementary Table 2). This study was initiated before the discovery of the ALS genes C21orf2, CHCHD10, MATR3, NEK1, TBK1 and TUBA4A.
Bioinformatic analysis
The raw FASTQ files from the Illumina MiSeq were aligned to the human reference genome build 19 (GRCh37) using Novoalign v3 hg19 (for transcripts see Supplementary Table 3), and variants were called using SAMtools v0.1.18 and GATK v3.3. Low quality variants were filtered as described previously (Morgan et al., 2015). Annotation was performed using ANNOVAR Nov2014 (Wang et al., 2010) and Variant Effect Predictor (VEP v84) tools (McLaren et al., 2010) and compared against the ExAC database of genetic variants (http://exac.broadinstitute.org/), 1000 Genomes (www.1000genomes.org), ESP6500 (evs.gs.washington.edu), UK10K (http://www.uk10k.org/) and cg69 (www.completegenomics.com/public-data/69-Genomes) to remove common variants [minor allele frequency (MAF) > 1% of the European-derived population]. Any locus with significant missingness between cases and controls was excluded using PLINK v1.09 (Purcell et al., 2007). An in-house coverage analysis software (CovCheck) determined that 92% of the desired regions were covered by at least 10 reads. The data were independently analysed by two separate groups to ensure different methods produced matching results, and every locus was visually inspected to guarantee high-quality data. Variants called with 10–20 reads were flagged to be visually inspected to remove false positives. A selection of these were checked using Sanger sequencing to ensure calling was correct.
Repeat expansions
A major drawback of NGS is its inability to reliably assay variation represented by repeats such as those in C9orf72 and ATXN2. Therefore, repeat-primed PCR was used to detect the expansion mutation in C9orf72 and standard fragment length analysis for the microsatellite repeat in ATXN2. The primers and methods used for C9orf72 and ATXN2 repeat detection have been previously described (Pulst et al., 1996; DeJesus-Hernandez et al., 2011).
Statistical analysis
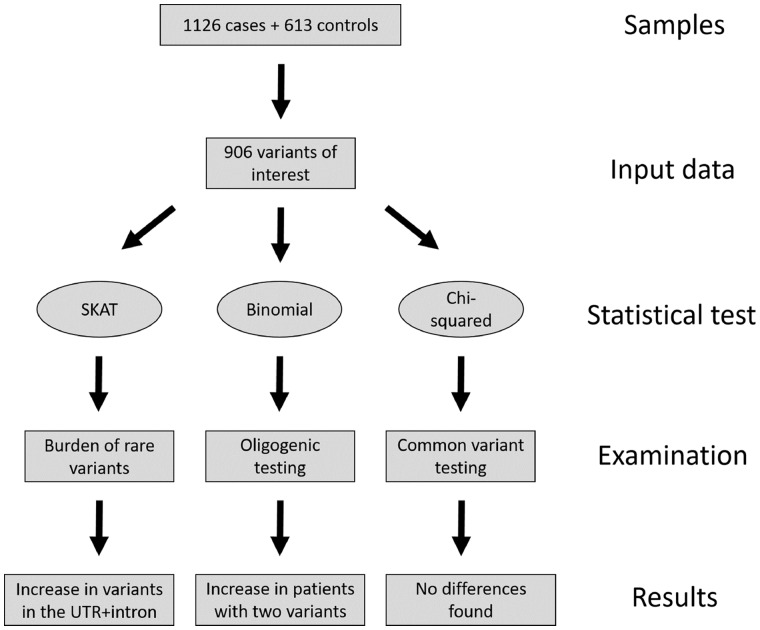
Because rare variation is too infrequent to test statistically in a sample of this size, we used a region-based test comparing the rare-variant burden in cases and controls in the form of the SNP (single nucleotide polymorphism)-set sequence kernel association test (SKAT v1.1.2; Wu et al., 2011; Fig. 1). We included intronic, exonic, synonymous, coding, and known causal variants only if they were novel or with a MAF < 0.01 and were of high-quality with no bias in the proportion of missing data between cases and controls. Comparing controls post-filtering, which had been examined using whole-exome and MiSeq-targeted sequencing, revealed differences only in the calling of indels of two or more nucleotides and not in any of the SNPs. We therefore used whole-exome sequenced samples as controls for SNP data only and did not include indels of two or more nucleotides in the analysis. The genes C9orf72, ATXN2, PON1–3 and VEGFA were not included in SKAT analysis due to the nature of their association with the disease (repeat expansions or common variation), but were instead analysed separately as described below. Sex was used as a covariate. A total of five tests were carried out; adjusted P-values are reported using the formula:
| (1) |
Figure 1.
Simplified flowchart describing the steps taken from sample to result in this study.
Where P is the critical P-value obtained in the test, n is the number of tests completed and B is the Bonferroni corrected P-value.
To examine a potential oligogenic basis of ALS, variants were analysed using a binomial test in R v3.2.3 as described previously (van Blitterswijk et al., 2012; Fig. 1). Heterozygous and homozygous variants were treated as a single event and tested using the binomial distribution by the formula:
| (2) |
Where f(x) is the probability of observing more than one rare variant in a single individual, n is the total number of patients, x the number of people carrying more than one variant, and p is the expected frequency of rare variation derived from the averaged frequency of rare variation observed in cases and controls.
We performed this test twice, first on reported ALS-variants only and then on variation found in C9orf72, SOD1, TARDBP, FUS, ANG, ALS2, VCP, OPTN, NEFH and UBQLN2 where we included all rare (MAF < 0.01), exonic, coding variation in these genes (excluding C9orf72 where only repeat expansions were included). These 10 genes were selected based on those previously tested by van Blitterswijk et al. (2012) in addition to those we believed to be of importance to the disease. Reported P-values were corrected for two tests.
For variants in the genes PON1–3 and VEGFA, association with ALS is with common variation, and so these loci were selected out of the data and filtered for variants present in dbSNP (Sherry et al., 2001; Fig. 1). Chi-squared SNP-based association tests in cases versus controls were performed using PLINK for these 20 loci and corrected for each iteration.
Results
We performed NGS on 1126 cases and 613 controls that identified, on average, 31 variants per individual passing quality control filters. Findings are presented in Table 1.
Table 1.
Previously published variants in ALS or other diseases that have been found in the current study in either patients or controls
| Chromo- some | Base pair | Gene | 1000 g | UK10K | ExAC | dbSNP137 | PhyloP | SIFT | PolyPhen2 | LRT | Exon | Nucleotide change | Amino acid change | Patients, n | Controls, n |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 11076931 | TARDBP | . | 0.00052 | 2.11 × 10−4 | rs80356715 | C | B | B | D | 3 | C269T | A90V | 2 | 1 |
| 1 | 11082325 | TARDBP | . | . | 8.13 × 10−6 | rs80356719 | C | B | B | D | 6 | G859A | G287S | 2 | 0 |
| 1 | 11082428 | TARDBP | . | . | . | . | C | D | B | D | 6 | C962T | A321V | 1 | 0 |
| 1 | 11082475 | TARDBP | . | . | . | rs80356730 | C | B | D | D | 6 | A1009G | M337V | 1 | 0 |
| 1 | 11082509 | TARDBP | . | . | . | . | C | D | P | D | 6 | G1043T | G348V | 1 | 0 |
| 1 | 11082588 | TARDBP | . | . | . | . | N | NA | NA | D | 6 | T1122G | Y374X | 1 | 0 |
| 1 | 11082598 | TARDBP | . | . | . | . | C | D | B | D | 6 | A1132G | N378D | 2 | 0 |
| 2 | 74588717 | DCTN1 | 0.0005 | 0.0047 | 3.11 × 10−3 | rs72466496 | C | D | B | N | 32 | C3746T | T1249I | 0 | 5 |
| 2 | 74594023 | DCTN1 | . | 0.00078 | 1.95 × 10−4 | rs121909344 | C | D | D | N | 21 | C2353T | R785W | 1 | 1 |
| 2 | 202626437 | ALS2 | 0.02 | 0.026 | 0.025 | rs3219154 | N | B | B | N | 4 | A280G | I94V | 0 | 33 |
| 3 | 87289899 | CHMP2B | . | 0.00052 | 1.22 × 10−4 | rs63750818 | N | B | B | N | 2 | A85G | I29V | 2 | 2 |
| 3 | 87294943 | CHMP2B | . | 0.00026 | 4.07 × 10−5 | rs200792883 | C | D | D | D | 3 | G206A | R69Q | 1 | 0 |
| 3 | 87294985 | CHMP2B | . | . | . | . | C | B | D | D | 3 | C248T | T83I | 0 | 1 |
| 5 | 179251013 | SQSTM1 | . | . | 2.20 × 10−4 | rs145056421 | N | B | B | N | 3 | G457A | V153I | 0 | 1 |
| 5 | 179252184 | SQSTM1 | 0.0032 | 0.0018 | 2.42 × 10−3 | rs11548633 | C | B | D | D | 5 | A712G | K238E | 11 | 4 |
| 5 | 179263445 | SQSTM1 | 0.0046 | 0.0021 | 8.78 × 10−4 | rs104893941 | C | D | P | N | 8 | C1175T | P392L | 5 | 2 |
| 6 | 41129105 | TREM2 | 0.03 | 0.0023 | 0.012 | rs2234253 | C | D | D | D | 2 | C287A | T96K | 0 | 1 |
| 6 | 41129207 | TREM2 | 0.01 | 0.0088 | 8.25 × 10−3 | rs143332484 | N | B | B | N | 2 | G185A | R62H | 0 | 10 |
| 6 | 41129252 | TREM2 | 0.0027 | 0.0021 | 2.07 × 10−3 | rs75932628 | C | B | D | N | 2 | G140A | R47H | 7 | 4 |
| 6 | 110036336 | FIG4 | 0.0009 | 0.0031 | 9.68 × 10−4 | rs121908287 | C | D | D | D | 2 | T122C | I41T | 3 | 5 |
| 9 | 35066777 | VCP | . | . | 8.95 × 10−5 | . | C | B | B | D | 4 | A340G | I114V | 1 | 0 |
| 9 | 35068298 | VCP | 0.0009 | 0.00052 | 5.37 × 10−4 | rs140913250 | C | B | B | D | 2 | A79G | I27V | 0 | 1 |
| 9 | 135140020 | SETX | 0.0027 | 0.0078 | 3.40 × 10−3 | rs151117904 | N | D | B | N | 26 | T7640C | I2547T | 12 | 8 |
| 9 | 135204004 | SETX | 0.0018 | . | 5.77 × 10−4 | rs149546633 | N | D | B | N | 10 | A2981G | D994G | 0 | 1 |
| 9 | 135204010 | SETX | 0.01 | 0.013 | 0.015 | rs61742937 | N | D | B | N | 10 | A2975G | K992R | 0 | 16 |
| 9 | 135224757 | SETX | 0.01 | 0.0075 | 9.04 × 10−3 | rs79740039 | N | D | B | N | 3 | G59A | R20H | 0 | 13 |
| 10 | 13152400 | OPTN | 0.07 | 0.033 | 0.045 | rs11258194 | N | B | B | N | 5 | T293A | M98K | 55 | 33 |
| 10 | 13178802 | OPTN | . | . | . | . | C | D | D | N | 16 | A1670G | K557R | 0 | 1 |
| 12 | 49689009 | PRPH | 0.02 | 0.0068 | 0.014 | rs57451017 | C | D | P | N | 1 | G26A | R9Q | 0 | 12 |
| 12 | 49690798 | PRPH | 0.03 | 0.016 | 0.022 | rs62636520 | C | B | B | N | 4 | G829A | A277T | 0 | 20 |
| 14 | 21161845 | ANG | 0.0005 | 0.0031 | 1.42 × 10−3 | rs121909536 | N | D | B | U | 2 | A122T | K41I | 0 | 2 |
| 14 | 21161973 | ANG | 0.01 | . | 1.55 × 10−3 | rs17560 | N | B | B | U | 2 | A250G | K84E | 1 | 0 |
| 16 | 31201719 | FUS | 0.0009 | . | 6.51 × 10−5 | rs186547381 | C | D | D | D | 12 | C1292T | P431L | 1 | 0 |
| 16 | 31202410 | FUS | . | . | . | . | C | D | D | D | 14 | G1520A | G507D | 1 | 0 |
| 16 | 31202739 | FUS | . | . | . | rs121909670 | N | D | D | D | 15 | C1561T | R521C | 1 | 0 |
| 16 | 31202740 | FUS | . | . | . | rs121909671 | N | D | D | D | 15 | G1562A | R521H | 2 | 0 |
| 16 | 31202740 | FUS | . | . | . | . | N | D | D | D | 15 | G1562T | R521L | 1 | 0 |
| 17 | 4849268 | PFN1 | 0.0005 | . | 4.15 × 10−4 | rs140547520 | C | D | D | D | 3 | A350G | E117G | 1 | 1 |
| 20 | 57014075 | VAPB | 0.0009 | 0.0026 | 1.34 × 10−3 | rs146459055 | C | B | B | D | 4 | T390G | D130E | 0 | 1 |
| 20 | 57016076 | VAPB | 0.0018 | 0.0013 | 1.35 × 10−3 | rs143144050 | C | B | P | D | 5 | G510A | M170I | 7 | 1 |
| 20 | 57016117 | VAPB | . | 0.0005 | . | rs145483046 | C | B | B | N | 5 | G551A | R184Q | 1 | 0 |
| 21 | 33032107 | SOD1 | . | . | . | . | N | D | B | N | 1 | C25G | L9V | 1 | 0 |
| 21 | 33038821 | SOD1 | . | . | . | . | C | NA | D | D | 3 | G229T | D77Y | 3 | 0 |
| 21 | 33039603 | SOD1 | 0.0009 | 0.00078 | 1.11 × 10−3 | rs80265967 | N | B | B | N | 4 | A272C | D91A | 3 | 0 |
| 21 | 33039636 | SOD1 | . | . | . | . | C | NA | D | D | 4 | A305G | D102G | 1 | 0 |
| 21 | 33039650 | SOD1 | . | . | . | . | C | NA | D | D | 4 | C319T | L107F | 1 | 0 |
| 21 | 33039666 | SOD1 | . | . | . | . | N | NA | NA | D | 4 | G335A | C112Y | 1 | 0 |
| 21 | 33039672 | SOD1 | . | 0.00026 | . | rs121912441 | C | NA | D | D | 4 | T341C | I114T | 5 | 0 |
| 21 | 33040829 | SOD1 | . | . | . | . | C | NA | NA | D | 5 | A403G | S135G | 1 | 0 |
| 22 | 29885016 | NEFH | 0.04 | 0.09 | 0.069 | rs59371099 | C | B | NA | D | 4 | G1387A | E463K | 173 | 109 |
| 22 | 29885473 | NEFH | 0.17 | 0.23 | 0.182 | rs5763269 | C | D | D | D | 4 | C1844T | P615L | 414 | 205 |
| 22 | 29885997 | NEFH | . | . | 8.78 × 10−4 | rs59551486 | . | . | . | . | 4 | 2368_2370 delAAG | K790del | 1 | 2 |
| 22 | 29886043 | NEFH | 0.12 | 0.17 | 0.153 | rs165602 | C | D | NA | N | 4 | A2414C | E805A | 309 | 152 |
| X | 56591796 | UBQLN2 | . | . | . | . | C | B | NA | N | 1 | C1490A | P497H | 1 | 0 |
The references for these variants are found in Supplementary Table 3.
1000g = 1000 Genomes project; prediction scores: C = conserved; D = damaging; B = benign; N = not conserved; NA = not applicable.
Variant interpretation
One of the major difficulties in NGS data is how to interpret the pathogenicity of variants, especially those that are novel or extremely rare. We found 906 alterations that were defined as potentially pathogenic, of which 225 were exonic and 55 were previously published as associated with ALS (48 variants) or another disease. However, some of these variants were also found in the control cohort (Table 1). We therefore defined variants as potentially pathogenic for ALS if they were published previously in more than one study and not found in control cohorts. Under this interpretation, 109 of 1125 cases were predicted to carry a pathological expansion of the C9orf72 gene, 19 cases with a likely causal SOD1 mutation, 14 within TARDBP and nine with a FUS variant.
Burden of rare variants
SKAT analysis showed an increased number of rare variants in cases compared to controls (P = 0.003). As this may be solely due to known pathogenic variants, those previously reported in ALS were excluded, regardless of putative pathogenicity, and SKAT was repeated. The result was still significant (P = 0.01). The burden lay primarily in the UTR and intronic regions rather than exons. We therefore tested introns and UTRs (P = 0.04) independently of exons [P = 0.1 (synonymous) and P = 0.1 (non-synonymous)]. Additionally, there were more rare variants in UTRs than in introns in patients but we did not have the statistical power to investigate this in detail (see Supplementary Table 5 for full results).
Oligogenic analysis
A binomial test restricted to exonic variants previously published in ALS did not show an excess of patients with two mutations (P = 0.4; see Supplementary Table 4 for variant references). Unrestricted testing of exons and adjacent regions for C9orf72, SOD1, TARDBP, FUS, ANG, ALS2, VCP, OPTN, NEFH and UBQLN2 showed 11 patients with more than one mutation, significantly higher than expected by chance based on the mutation rates in cases and controls (P = 0.001; see Supplementary Table 6 for variant and patient information).
Common variation in amyotrophic lateral sclerosis
The genes PON1–3 and VEGFA have both been reported as potential risk factors for ALS. We selected 20 loci of common variation within these genes to analyse but did not find any significant differences in SNP frequencies between controls and cases and, in fact, some reported important SNPs were present at a higher rate in controls than in cases (Supplementary Table 7). The frequencies in our cohort were higher than those observed in the ExAC database.
Discussion
We analysed 24 ALS genes in 1736 subjects. We detected 55 variants previously published in ALS or other diseases and 845 rare variants of uncertain significance, with a higher burden of variants in cases, an excess in the untranslated regions and introns, and oligogenic inheritance in 1% of patients. A limitation of this study is its focus on a specific set of ALS genes rather than being a truly unbiased survey of the exome or genome, but this has allowed us to test specific hypotheses. Cases with likely pathogenic SOD1, TARDBP and FUS mutations are more likely to report a family history while those with a C9orf72 expansion are more commonly sporadic and bulbar onset.
Untranslated region analysis in amyotrophic lateral sclerosis
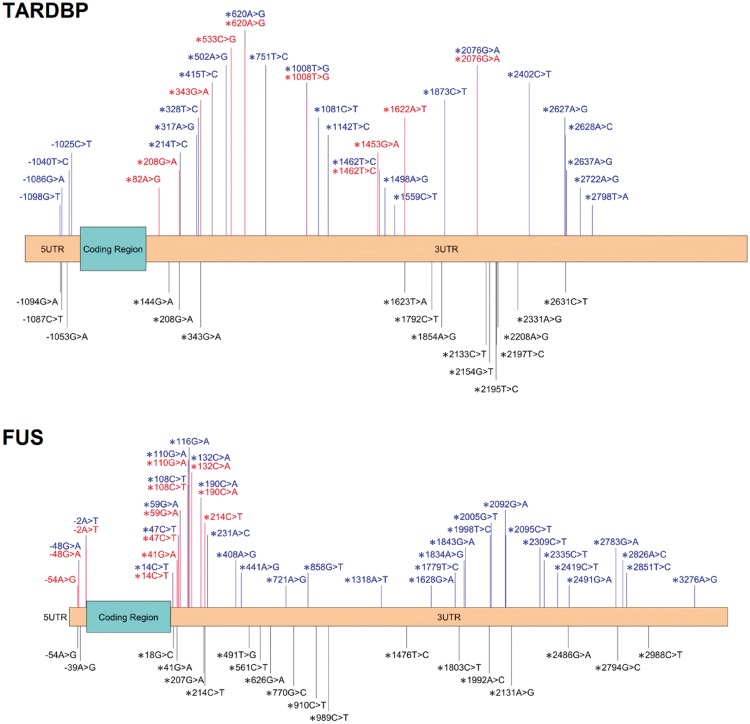
The UTRs of genes are often ignored in genetic studies of disease, partly because of the difficulty in interpreting findings. We included UTRs to address this lack of knowledge targeting SOD1, TARDBP, FUS, OPTN, VCP and UBQLN2, as well as partial coverage in the remaining genes. SKAT analysis revealed a significant excess of rare variants in patients in the UTRs and introns, and inspection of the data suggested that the UTRs contained most of this burden. Previous studies have also suggested this effect (Fig. 1). An Italian study of 420 people with ALS and 480 controls found non-coding mutations in the 3’UTR of FUS in patients, with four unique rare variants in five individuals and no rare variants in controls (Sabatelli et al., 2013). Three variants were studied further in primary fibroblast cultures (c.*59G > A, c.*108C > T and c.*110G > A). The UTR variants and a known pathogenic exonic FUS variant all cause a mislocalization of the FUS protein, an effect not seen in the other patients or controls. Similarly, the c.*48G > A variant, found in two subjects with a rapidly progressive form of ALS, increase FUS expression dramatically (Dini Modigliani et al., 2014); overexpression of wild-type FUS causes an ALS-like syndrome in mice (Mitchell et al., 2013). The 3’UTR of FUS is known to be involved in a feedback loop for its own expression via the alternative splicing of exon 7 (Zhou et al., 2013). On the other hand, FUS-knockout mice exhibit abnormalities but not ALS (Kino et al., 2015), and the ExAC database shows no loss of function FUS variants in any of the 58 787 individuals sequenced despite a statistical expectation of there being 28.6. This number is calculated using the mutation rate across the whole genome to produce a per gene probability of each type of mutation. Factoring in coverage metrics, an expected number of variants, in this case 28.6, can be obtained for the number of individuals sequenced (Lek et al., 2015). These studies combined suggest that tight control of FUS expression is necessary in humans (Dini Modigliani et al., 2014).
There are similar findings for TARDBP: a 3’ UTR variant c.*2076G > A found in two affected members of a family with ALS and FTD (Gitcho et al., 2009), doubled TARDBP RNA expression and was not present in 982 controls. Like FUS, TARDBP also regulates its own expression through the 3’UTR (Ayala et al., 2011), although this is achieved through RNA instability rather than splicing. Again, as with FUS, there are no loss-of-function variants in TARDBP in the ExAC database when 11.8 are expected.
These findings suggest variants in the UTRs of FUS and TARDBP have a role in ALS pathogenesis (Fig. 2).
Figure 2.
Schematic representation of the UTRs of TARDBP and FUS and the variants found in this study within cases (blue) and controls (black) in these regions. Variants within the 5’UTR are preceded with a minus while those contained in the 3’UTR are headed by an asterisk. This includes variants previously published in ALS marked in red (Supplementary Table 8).
Oligogenic basis of amyotrophic lateral sclerosis
We implemented a binominal test based on the probability distribution of mutations in both cases and controls to identify a greater than chance observation of combined pathogenic mutations. There was no excess of oligogenic ALS when the analysis was restricted to previously reported ALS variants, but previous studies have been unrestricted, examining all rare variants in targeted genes (van Blitterswijk et al., 2012). Performing this analysis, 1% of our patients had two mutations, significantly higher than expected based on known mutation rates. Most had C9orf72 repeat expansion combined with another mutation (e.g. VCP R155H or TARDBP A321V; Supplementary Table 6). A single control also had two mutations, P372R in ALS2 and A90V in TARDBP. ALS2 pathogenicity has only been observed in homozygotes, and this individual was heterozygous. Furthermore, the TARDBP variant has been previously identified in controls and has unclear status, although it is associated with abnormal localization and aggregation of TARDBP (Guerreiro et al., 2008; Winton et al., 2008).
What constitutes a pathogenic combination of mutations is debatable as some variants are of uncertain significance (Richards et al., 2015), and the combination of a pathogenic variant with one of uncertain significance has been considered oligogenic inheritance by some (Nakamura et al., 2016). Similarly, variation in ANG or NEFH is generally considered a weak contributor to ALS risk, and missense variants in SPG11 are often benign unless resulting in loss of function. We find oligogenic inheritance even when these genes are excluded from the analysis. Notably, one of our controls harboured a loss-of-function mutation in SPG11. Allowing a looser definition of oligogenic inheritance, oligogenic ALS is reported in ∼1.6% of cases (4% in familial and 1.3% in sporadic ALS) (Kenna et al., 2013). Oligogenic ALS involving the convergence of two ALS families has also been observed, with the proband carrying a pathogenic TARDBP variant and C9orf72 repeat expansion (Chiò et al., 2012). This appears to be associated with a more severe phenotype and an earlier age of onset. In another study of 391 cases, 3.8% had more than one mutation and an earlier age of onset by 10 years (Cady et al., 2015).
Common variation
Several previous studies have examined common variations in VEGFA and PON1–3, which are inconsistently associated with ALS (Lambrechts et al., 2003; Wills et al., 2009). We characterized 20 loci in patients and controls to find no relationship with ALS, but this may be due to low call rates in patients for these particular genes (Supplementary Table 7). For example, rs7493 and rs12026 were associated with controls, but only 95 cases had adequate data. Furthermore, 14 of the 20 tested loci were at higher frequencies in our controls than in the ExAC and 1000 Genomes databases, with six being considerably higher. This highlights the importance of collecting adequate controls for each study rather than solely relying on public data.
The sole known pathogenic DAO variant R199W has been reported in two studies and correlates with survival (Mitchell et al., 2010; Cirulli et al., 2015). We identified two subjects with sporadic ALS who harboured a different variant at the same codon: R199Q, and another alteration two amino acids away: Q201R. Both of these DAO mutants are predicted to be damaging by PhyloP, SIFT, PolyPhen and LRT, and were not found in our control samples.
Our large-scale sequencing study in ALS has identified a number of rare variations, many novel, and shown that the UTR of TARDBP and FUS are potentially important in the pathogenesis of ALS. We have also provided further support for oligogenic inheritance of ALS in a proportion of cases.
Supplementary Material
Acknowledgements
Samples used in this research were in part obtained from the UK National DNA Bank for MND Research, funded by the MND Association and the Wellcome Trust. We thank people with MND and their families for their participation in this project. We acknowledge sample management undertaken by Biobanking Solutions funded by the Medical Research Council at the Centre for Integrated Genomic Medical Research, University of Manchester.
Glossary
Abbreviations
- ALS
amyotrophic lateral sclerosis
- MND
motor neuron disease
- NGS
next-generation sequencing
- SKAT
sequence kernel association test
- SNP
single nucleotide polymorphism
- UTR
untranslated region
Funding
This project was funded by the MND Association and the Wellcome Trust. This is an EU Joint Programme - Neurodegenerative Disease Research (JPND) project. The project is supported through the following funding organisations under the aegis of JPND - www.jpnd.eu (United Kingdom, Medical Research Council (MR/L501529/1) and Economic and Social Research Council (ES/L008238/1)). C.E.S. and A.A.C. receive salary support from the National Institute for Health Research (NIHR) Dementia Biomedical Research Unit at South London and Maudsley NHS Foundation Trust and King’s College London. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health. The work leading up to this publication was funded by the European Community’s Health Seventh Framework Programme (FP7/2007–2013; grant agreement number 259867) and Horizon 2020 Programme (H2020-PHC-2014-two-stage; grant agreement number 633413).
Supplementary material
Supplementary material is available at Brain online.
References
- Abel O, Powell JF, Andersen PM, Al-Chalabi A. ALSoD: a user-friendly online bioinformatics tool for amyotrophic lateral sclerosis genetics. Hum Mutat 2012; 33: 1345–51. [DOI] [PubMed] [Google Scholar]
- Abrahams S, Newton J, Niven E, Foley J, Bak TH. Screening for cognition and behaviour changes in ALS. Amyotroph Lateral Scler Front Degener 2014; 15: 9–14. [DOI] [PubMed] [Google Scholar]
- Al-Chalabi A, Hardiman O. The epidemiology of ALS: a conspiracy of genes, environment and time. Nat Rev Neurol 2013; 9: 617–28. [DOI] [PubMed] [Google Scholar]
- Al-Chalabi A, Lewis CM. Modelling the effects of penetrance and family size on rates of sporadic and familial disease. Hum Hered 2011; 71: 281–8. [DOI] [PubMed] [Google Scholar]
- Ayala YM, De Conti L, Avendaño-Vázquez SE, Dhir A, Romano M, D’Ambrogio A et al. . TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J 2011; 30: 277–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne S, Elamin M, Bede P, Hardiman O. Absence of consensus in diagnostic criteria for familial neurodegenerative diseases. J Neurol Neurosurg Psychiatry 2012; 83: 365–67. [DOI] [PubMed] [Google Scholar]
- Cady J, Allred P, Bali T, Pestronk A, Goate A, Miller TM et al. . Amyotrophic lateral sclerosis onset is influenced by the burden of rare variants in known amyotrophic lateral sclerosis genes. Ann Neurol 2015; 77: 100–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A, Restagno G, Brunetti M, Ossola I, Calvo A, Canosa A et al. . ALS/FTD phenotype in two Sardinian families carrying both C9ORF72 and TARDBP mutations. J Neurol Neurosurg Psychiatry 2012; 83: 730–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Lasseigne BN, Petrovski S, Sapp PC, Dion PA, Leblond CS et al. . Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science 2015; 347: 1436–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ et al. . Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011; 72: 245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dini Modigliani S, Morlando M, Errichelli L, Sabatelli M, Bozzoni I. An ALS-associated mutation in the FUS 3′-UTR disrupts a microRNA–FUS regulatory circuitry. Nat Commun 2014; 5: 4335. [DOI] [PubMed] [Google Scholar]
- Gitcho MA, Bigio EH, Mishra M, Johnson N, Weintraub S, Mesulam M et al. . TARDBP 3’-UTR variant in autopsy-confirmed frontotemporal lobar degeneration with TDP-43 proteinopathy. Acta Neuropathol 2009; 118: 633–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro RJ, Schymick JC, Crews C, Singleton A, Hardy J, Traynor BJ. TDP-43 is not a common cause of sporadic amyotrophic lateral sclerosis. PLoS One 2008; 3: e2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ et al. . Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron 2010; 68: 857–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna KP, McLaughlin RL, Byrne S, Elamin M, Heverin M, Kenny EM et al. . Delineating the genetic heterogeneity of ALS using targeted high-throughput sequencing. J Med Genet 2013; 50: 776–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino Y, Washizu C, Kurosawa M, Yamada M, Miyazaki H, Akagi T et al. . FUS/TLS deficiency causes behavioral and pathological abnormalities distinct from amyotrophic lateral sclerosis [Internet]. Acta Neuropathol Commun 2015; 3 [cited 2016 Jun 18] Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4408580/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts D, Storkebaum E, Morimoto M, Del-Favero J, Desmet F, Marklund SL et al. . VEGF is a modifier of amyotrophic lateral sclerosis in mice and humans and protects motoneurons against ischemic death. Nat Genet 2003; 34: 383–94. [DOI] [PubMed] [Google Scholar]
- Lek M, Karczewski K, Minikel E, Samocha K, Banks E, Fennell T et al. . Analysis of protein-coding genetic variation in 60,706 humans. bioRxiv 2015: 30338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logroscino G, Traynor BJ, Hardiman O, Chiò A, Mitchell D, Swingler RJ et al. . Incidence of amyotrophic lateral sclerosis in Europe. J Neurol Neurosurg Psychiatry 2010; 81: 385–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP effect predictor. Bioinformatics 2010; 26: 2069–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencacci NE, Isaias IU, Reich MM, Ganos C, Plagnol V, Polke JM et al. . Parkinson’s disease in GTP cyclohydrolase 1 mutation carriers. Brain J Neurol 2014; 137: 2480–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J, Paul P, Chen H-J, Morris A, Payling M, Falchi M et al. . Familial amyotrophic lateral sclerosis is associated with a mutation in D-amino acid oxidase. Proc Natl Acad Sci USA 2010; 107: 7556–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JC, McGoldrick P, Vance C, Hortobagyi T, Sreedharan J, Rogelj B et al. . Overexpression of human wild-type FUS causes progressive motor neuron degeneration in an age- and dose-dependent fashion. Acta Neuropathol 2013; 125: 273–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan S, Orrell RW. Pathogenesis of amyotrophic lateral sclerosis. Br Med Bull 2016; 119: 87–98. [DOI] [PubMed] [Google Scholar]
- Morgan S, Shoai M, Fratta P, Sidle K, Orrell R, Sweeney MG et al. . Investigation of next-generation sequencing technologies as a diagnostic tool for amyotrophic lateral sclerosis. Neurobiol. Aging 2015; 36: 1600.e5–8. [DOI] [PubMed] [Google Scholar]
- Nakamura R, Sone J, Atsuta N, Tohnai G, Watanabe H, Yokoi D et al. . Next-generation sequencing of 28 ALS-related genes in a Japanese ALS cohort. Neurobiol Aging 2016; 39: 219.e1–e8. [DOI] [PubMed] [Google Scholar]
- Pulst SM, Nechiporuk A, Nechiporuk T, Gispert S, Chen XN, Lopes-Cendes I et al. . Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat Genet 1996; 14: 269–76. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D et al. . PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J et al. . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatelli M, Moncada A, Conte A, Lattante S, Marangi G, Luigetti M et al. . Mutations in the 3’ untranslated region of FUS causing FUS overexpression are associated with amyotrophic lateral sclerosis. Hum Mol Genet 2013; 22: 4748–55. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM et al. . dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001; 29: 308–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L, Cupid BC, Dickie BGM, Al-Chalabi A, Morrison KE, Shaw CE et al. . Establishing the UK DNA Bank for motor neuron disease (MND). BMC Genet 2015; 16: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blitterswijk M, van Es MA, Hennekam EAM, Dooijes D, van Rheenen W, Medic J et al. . Evidence for an oligogenic basis of amyotrophic lateral sclerosis. Hum Mol Genet 2012; 21: 3776–84. [DOI] [PubMed] [Google Scholar]
- van Rheenen W, Shatunov A, Dekker AM, McLaughlin RL, Diekstra FP, Pulit SL et al. . Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat Genet 2016; 48: 1043–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills A-M, Cronin S, Slowik A, Kasperaviciute D, Es MAV, Morahan JM et al. . A large-scale international meta-analysis of paraoxonase gene polymorphisms in sporadic ALS. Neurology 2009; 73: 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winton MJ, Van Deerlin VM, Kwong LK, Yuan W, Wood EM, Yu C-E et al. . A90V TDP-43 variant results in the aberrant localization of TDP-43 in vitro. FEBS Lett 2008; 582: 2252–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet 2011; 89: 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liu S, Liu G, Oztürk A, Hicks GG. ALS-associated FUS mutations result in compromised FUS alternative splicing and autoregulation. PLoS Genet 2013; 9: e1003895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.