Abstract
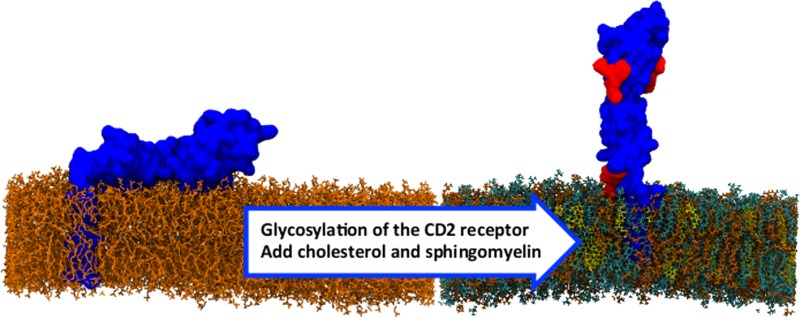
Proteins embedded in the plasma membrane mediate interactions with the cell environment and play decisive roles in many signaling events. For cell–cell recognition molecules, it is highly likely that their structures and behavior have been optimized in ways that overcome the limitations of membrane tethering. In particular, the ligand binding regions of these proteins likely need to be maximally exposed. Here we show by means of atomistic simulations of membrane-bound CD2, a small cell adhesion receptor expressed by human T-cells and natural killer cells, that the presentation of its ectodomain is highly dependent on membrane lipids and receptor glycosylation acting in apparent unison. Detailed analysis shows that the underlying mechanism is based on electrostatic interactions complemented by steric interactions between glycans in the protein and the membrane surface. The findings are significant for understanding the factors that render membrane receptors accessible for binding and signaling.
Membrane proteins and receptors direct a variety of cellular functions by, for example, being involved in cell recognition and generating signals associated with cellular communication. The fact that these molecules are tethered to membranes and interact at cellular interfaces has several important consequences.1 Principal among these is that membrane tethering limits the ability of receptors to encounter each other, and this requires the opposing surfaces to come into sufficiently close proximity to allow engagement of the binding partners driven by lateral diffusion in the membrane only. A related consideration is that cell surface proteins can vary considerably in size, which will tend to work against the interactions of pairs of smaller proteins. On the other hand, all of these proteins are tethered to the membrane either by a glycosylphosphatidylinositol (GPI) anchor, which in at least some instances is likely to be highly flexible,2 or via a relatively short (∼4–15 residue) “stalk”-like segment presumed to have extended, nonrigid structures. At present, it is unclear how the positioning of the ligand binding sites of receptors is optimized to ensure efficient ligand engagement at the cell surface.
Recent experimental work has suggested that lipids located in the extracellular leaflet of a cell membrane can influence protein behavior and activation. For example, it has been proposed that the ganglioside GM3 inhibits epidermal growth factor receptor (EGFR) autophosphorylation,3 and the activation of toll-like receptor 4 has been reported to be inhibited by another two gangliosides (GM1 and GD1A).4 These studies suggest that lipids could affect ectodomain activity, perhaps by influencing the positioning of the receptor in the membrane. Other work on the EGFR implies that protein glycosylation can also influence membrane receptor conformation and accessibility for ligand binding.5 Here, we explore the possibility that lipids and glycosylation act in concert to control the positioning of the ligand binding region of an immune receptor, CD2,6−9 using atomistic molecular dynamics simulations.
CD2 is expressed in T-cells and natural killer cells, where it functions as a cell adhesion and co-stimulatory molecule. The extracellular domain (ECD) of CD2 is relatively small (∼75 Å),10,11 which for our purposes is advantageous because it allows us to explore its full conformational space in multimicrosecond simulations. This region of CD2 consists of a membrane-distal immunoglobulin superfamily (IgSF) V-set domain supported by a C2-set domain.10,11 The positioning of the ligand binding region of CD2 at the “top” of the V-set domain suggests its optimization for ligand binding. The ECD is attached to a conventional transmembrane domain via a seven-residue, apparently extended stalk.11 An intriguing feature of CD2 is the presence of a highly conserved glycosylation site at the base of CD2 of domain 2.12 This glycosylation site is well away from the ligand binding region, and it has been shown that the ligand-binding properties of CD2 are in any case glycosylation-independent. It has therefore been proposed that the conserved site is well placed for an N-glycan positioned there to stabilize the orientation of the protein at the surface.10,12 Finally, there are suggestions that CD2 function is lipid-dependent.13,14
In general terms, N-glycosylation is one of the most common structural modifications of proteins, being involved in, for example, protein folding, structural diversification, and activation.15−17 Considering the importance of N-glycosylation and the difficulties associated with unraveling the effects of atom-scale structural modifications, the role of membrane protein glycosylation has received surprisingly little attention in the form of atomistic simulations until now; to our knowledge, only one atom-scale simulation study has explored the influence of glycosylation on membrane protein conformation and the resulting effects on presentation and ligand binding.5 The study showed glycosylation to critically determine the structural arrangement of the EGFR ectodomain and its ligand-binding domains. Further studies on peptides interacting with lipid membranes have explored, for example, the importance of glycosylation in the binding of autoantibodies with glycopeptides used as biomarkers.18
CD2 is the ideal candidate for exploring how lipids and glycosylation could act in concert to position its ectodomain, thereby enhancing its binding function. This stems from the facts that CD2 has conserved membrane-proximal glycosylation, there are data favoring the view that its function is lipid-dependent,13,14 and, above all, that it is a relatively small glycoprotein. We find that both lipid composition and glycosylation influence the orientation and dynamics of the CD2 ectodomain, but the most distinct and most pronounced effect is observed when they act in concert. The implications of these findings are discussed below.
We unraveled the importance of glycosylation (G) compared to nonglycosylation (NG) in two different lipid membranes: a single-component, liquid-disordered (Ld) fluid bilayer made of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC; referred to here as the Ld bilayer) and a bilayer with a ternary lipid mixture (DOPC, sphingomyelin (SM), and cholesterol (Chol)), which is more ordered (liquid-ordered (Lo)) and less fluid and is often taken as a physical model of putative membrane “rafts”19 (referred to here as the Lo bilayer). Introducing glycosylated (G) and nonglycosylated (NG) CD2 into these bilayers resulted in the simulation of four systems (Ld-NG, Ld-G, Lo-NG, Lo-G), which were simulated for 500 ns each, and every simulation was repeated three times to improve sampling.
Figure 1 highlights qualitatively the main conclusions of this study by means of a series of snapshots (see also movies in the Supporting Information (SI)). We find that the orientation of the ECD of CD2 can depend in a critical manner on both CD2 glycosylation and the local lipid composition of the membrane.
Figure 1.
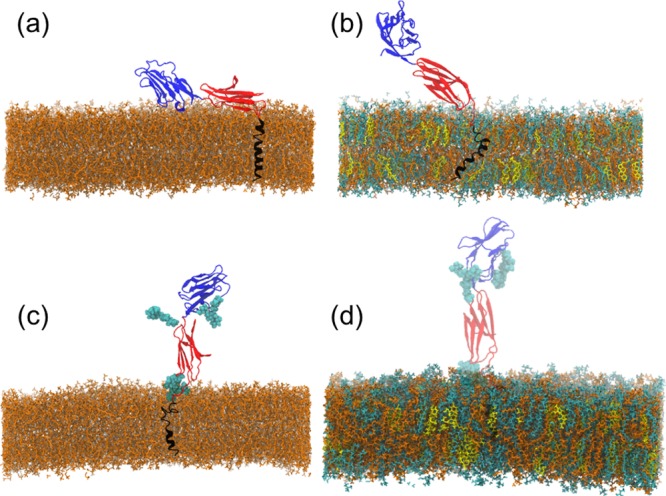
Simulation snapshots of the equilibrium configurations of nonglycosylated CD2 in (a) Ld (Ld-NG) and (b) Lo (Lo-NG) bilayers and of glycosylated CD2 in (c) Ld (Ld-G) and (d) Lo (Lo-G) bilayers. Color code: DOPC (orange), SM (cyan), Chol (yellow), CD2 domain 1 (dark blue), CD2 domain 2 (red), CD2 transmembrane helix (black), and lipid carbohydrate chains (light blue). The glycans attached to CD2 in panels (c) and (d) (Ld-G, Lo-G) are shown in light blue. For clarity, water and ions are not shown.
In the Ld bilayer without glycosylation (Figure 1a), the ECD is positioned parallel to the membrane surface. For the same Ld bilayer with glycosylation (Figure 1c), the ECD of CD2 is positioned more upright, suggesting that glycosylation plays a role in CD2 orientation. Considering CD2 in the Lo bilayer, we find that the lipid composition has an important role in CD2 ECD positioning too (Figure 1b,d). Without glycosylation in the Lo system, the ECD of CD2 fluctuates around an upright position (Figure 1b). However, the most significant change is observed when the Lo bilayer hosts glycosylated CD2, where the protein assumes a constitutively upright position (Figure 1d).
While the data shown in Figure 1 are suggestive, a more quantitative analysis of the simulation data confirmed these findings. We first calculated the tilt angle for vectors that represent the ECDs of CD2. The vectors used to characterize the domain orientations were defined as follows. We first define three points A, B, and C (Figure 2a), where point A is localized at the top of domain 1 (atom N in the LYS residue), point B is in the linker region10 between domains 1 and 2 (atom N in the GLU residue), and point C connects domain 2 to the transmembrane helix at the “top” of the membrane (atom N in ASP). Vectors RBA from B to A (in the domain 1) and RCB from C to B (in the domain 2) are then defined as vectors connecting these points (Figure 2a). The tilt angles of these vectors with respect to the membrane normal are denoted by θ1 (for domain 1) and θ2 (for domain 2), respectively.
Figure 2.
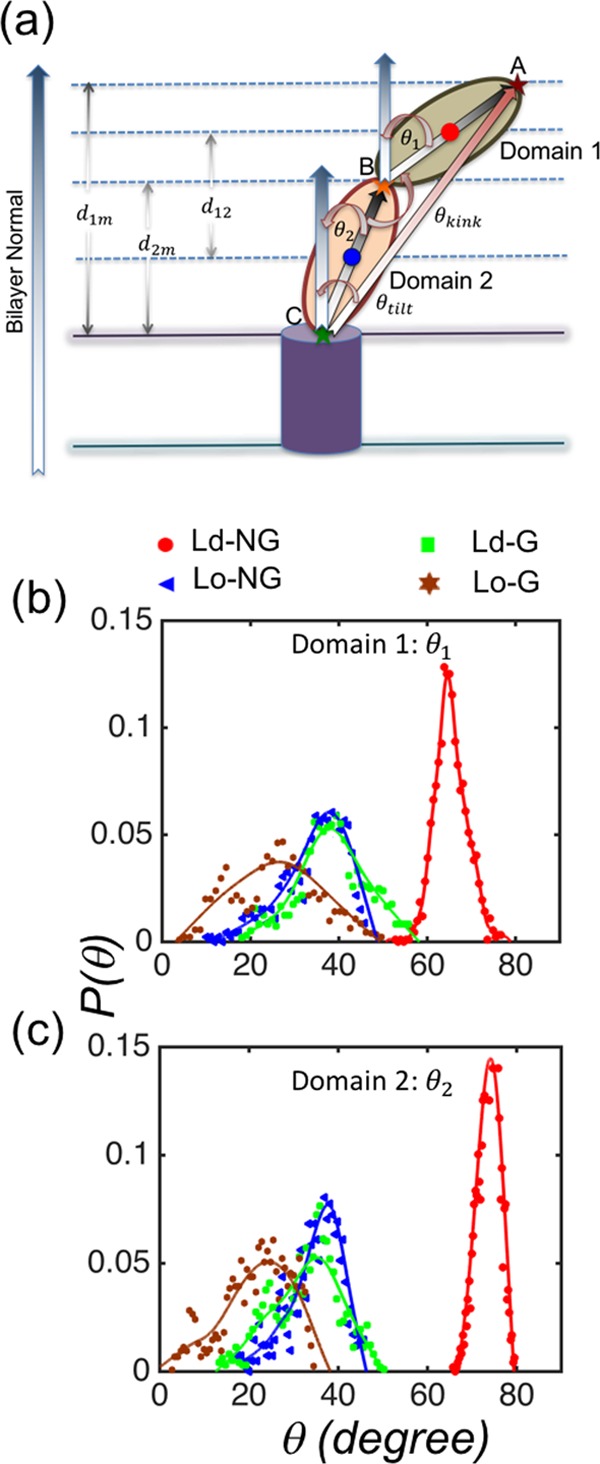
(a) Schematic diagram of CD2 embedded in a lipid membrane. The transmembrane part of CD2 is shown in violet, while the two domains in the ECD of CD2 are denoted by semidark green (domain 1) and light orange (domain 2). Points A, B, and C (see text) stand for the top of domain 1, the junction between domains 1 and 2, and the bottom of domain 2 that is connected to the transmembrane helix of CD2, respectively. (b) Probability distribution functions of the tilt angle of domain 1 with respect to the membrane normal. (c) A similar probability distribution for the tilt of domain 2. Color code: Ld-NG (red), Ld-G (green), Lo-NG (blue), Lo-G (brown).
The probability distributions P(θ1) and P(θ2) of the tilt angles are shown in Figure 2b,c, respectively. The results show that the tilt angle θ1 of domain 1 is the highest for nonglycosylated CD2 embedded in the Ld bilayer, and the smallest tilt is observed for glycosylated CD2 in the Lo bilayer. The tilts for glycosylated CD2 in the Ld bilayer and for nonglycosylated CD2 in the Lo bilayer are comparable and between the two above-mentioned extreme cases. The average values of the tilt angles are summarized in Table 1, which highlights profound differences between the four systems. A similar pattern is found for the tilt angle θ2 of domain 2 (Figure 2c). Similar conclusions are found when one analyzes the tilt angle of the entire ECD of CD2 (θtilt between RCA and the membrane normal; Figure 2a) and the kink angle between domains 1 and 2 (θkink between vectors RBA and RCB; Figure 2a); see Figure S4 in the SI.
Table 1. Results for the Average Values of the Tilt Angles (θ1, θ2) Describing the Orientation of Domains 1 and 2 of the CD2 Ectodomaina.
| system | average θ1 (deg) | average θ2 (deg) |
|---|---|---|
| Ld-NG | 65.30 ± 0.07 | 73.33 ± 0.30 |
| Ld-G | 38.40 ± 0.39 | 31.77 ± 1.11 |
| Lo-NG | 34.62 ± 0.50 | 34.98 ± 0.61 |
| Lo-G | 25.82 ± 1.00 | 21.78 ± 0.57 |
The results suggest that glycosylation and lipid composition govern the orientation of the ECD of CD2. One possible general conclusion that can be drawn is that for membrane receptors with binding surfaces or binding pockets in the extracellular region, glycosylation and the lipid environment may restrict the amount of ligand-accessible space. We therefore complemented the above analysis for ECD orientation and determined also the distances between the ECD of CD2 and the bilayer surface. To this end, the distance between the center of mass (CM) positions of domains 1 and 2 along the bilayer normal direction is defined as d12. The distances along the membrane normal between A and C and B and C are then defined as d1m and d2m, respectively (see Figure 2).
Data presented in Figure 3a show that the distance d12 is the shortest for nonglycosylated CD2 in the DOPC bilayer, and it is the longest for the glycosylated CD2 in the Lo bilayer. The distance is in-between these limits for the remaining two cases (Ld-G and Lo-NG). Consistent conclusions are found based on the distances d1m and d2m (Figure 3b,c).
Figure 3.
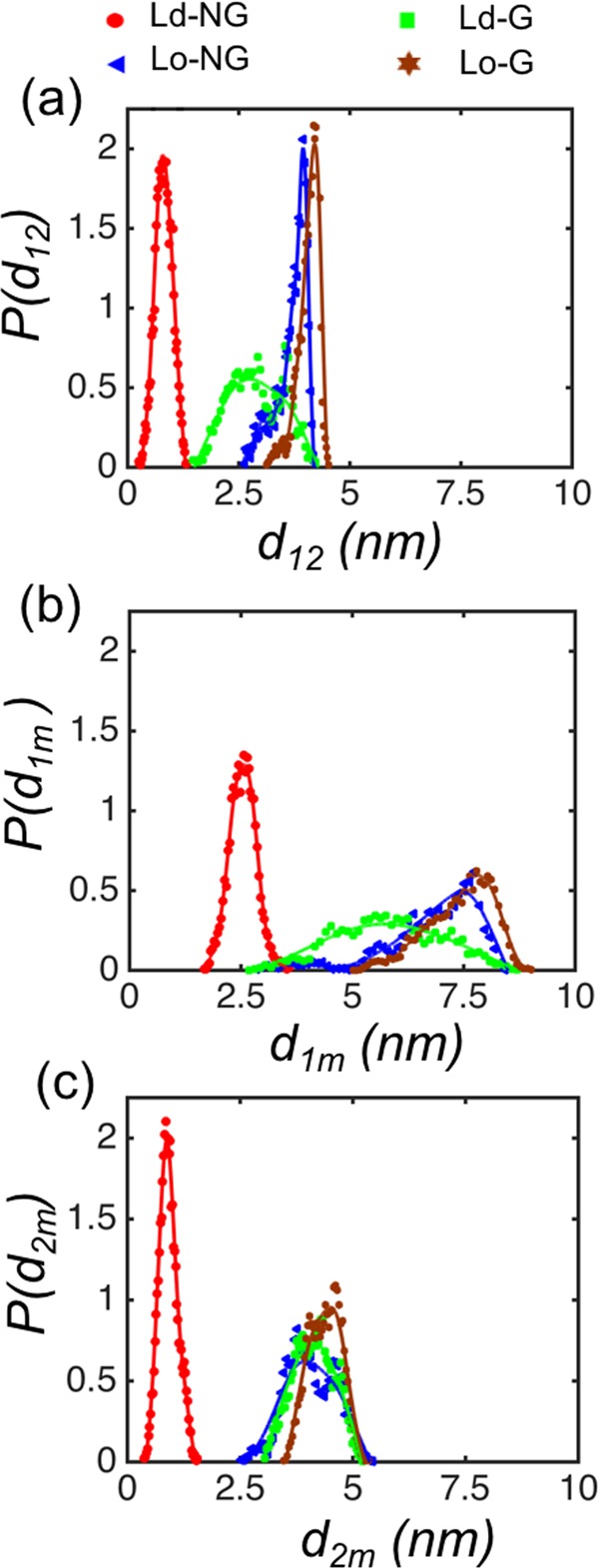
Probability distributions for distances (a) d12, (b) d1m, and (c) d2m for the different conditions studied.
Figure 3 allows us to indicate the relative importance of glycosylation and the two lipid compositions for the orientation of the ECD. First, Figure 3 provides compelling evidence that the ECD generally stands most upright when CD2 (either glycosylated or nonglycosylated) is in the Lo bilayer. Second, in both Ld and Lo bilayers, the glycosylated form of CD2 stands more upright than the nonglycosylated protein. This feature is quite weak in the Lo systems but strong in the Ld membranes.
The largest effect on the positioning of the ECD of CD2 appears to be the Lo lipid composition in the bilayer, compared to the Ld bilayer. Glycosylation also influences ECD positioning, but this effect is somewhat less pronounced compared to the change induced when going from the Ld to the Lo lipid environment. In order to understand what governs the orientation of the CD2 ectodomain, and in particular why the ectodomain of nonglycosylated CD2 collapses in the Ld bilayer, we next explored the orientation of the CD2 transmembrane domain and its dependence on glycosylation and membrane lipid content. Figure 4 shows that the orientation of the transmembrane domain is distinctly different in the Ld and Lo bilayers, irrespective of the presence of glycosylation. In the case of the Ld environment, where the membranes are ∼0.4 nm thicker than those in the Lo systems, the transmembrane region is more “upright” on average (Figure 4). Further analysis showed that the orientation of the transmembrane domain does not correlate with the position of the ectodomain (Figure S5).
Figure 4.
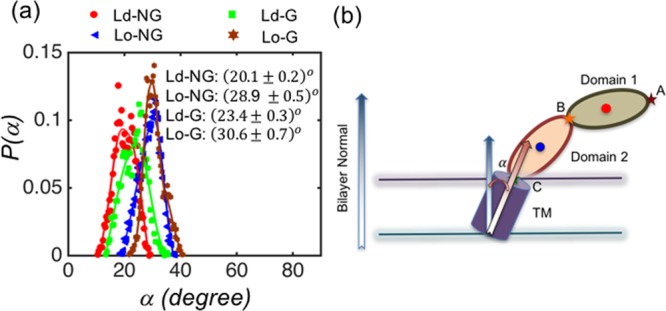
(a) Probability distributions for the orientation of the transmembrane region of CD2 characterized by α, which is the angle between the principal axis of the CD2 transmembrane domain and the membrane normal. Average values of α together with error bars are listed in the inset. (b) Schematic illustration of the angle α.
We next turned our attention to lipid–protein interactions. We identified the amino acids on nonglycosylated CD2 that interacted with the lipid membrane environment in the Ld system. These were positively charged lysines and arginines forming a patch, present in both human and rat CD2, and suggesting electrostatic interactions to play a role in CD2 tilting. We therefore performed additional simulations wherein Lys125, Lys132, Lys185, Lys183, Lys206, Lys134, Lys175, Lys166, Arg170, and Arg129 were deprotonated to a neutral form (Figure S10). Figure 5 depicts the significance of deprotonation. Whereas the ECD of intact nonglycosylated CD2 associates directly with the membrane surface of the Ld bilayer, deprotonated nonglycosylated CD2 is positioned upright in a largely similar manner to glycosylated CD2 in the Lo bilayer. These data suggest that the main cause of the CD2 ectodomain’s tilt is the electrostatic attractive interaction between the charged residues and the strongly polar lipid head groups (of DOPC) in the bilayer. When weakly polar Chol replaces strongly polar DOPC, as in a change from the Ld to the Lo system, or when CD2 is deprotonated, the tilt is reduced (Figures 2b,c and 5). In addition, the glycans bound to CD2 and, in particular, the glycosylation in the juxtamembrane region of CD2 prevent collapse of the ectodomain by both shielding the charged residues and holding the ECD upright through steric effects.
Figure 5.
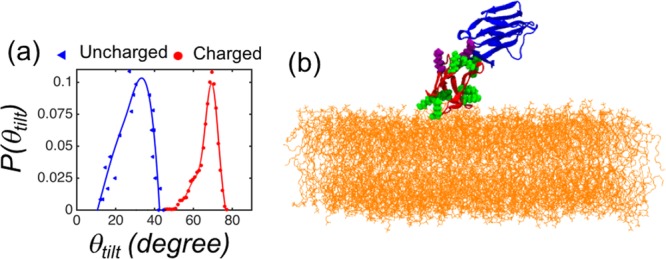
(a) Distribution of the tilt angle θtilt in the Ld-NG system (blue line) when a set of 10 positively charged amino acids of CD2 were deprotonated (see text). The tilt θtilt is the angle between the membrane normal and a vector from the point C to A (see Figure 2a). The corresponding data for the intact Ld-NG system without deprotonation are shown for comparison (red line). (b) Snapshot of the equilibrium configuration for the deprotonated Ld-NG system. The neutralized lysines are shown in green and the neutralized arginines in violet.
The above-described conclusions are supported by analysis of the total electrostatic energy that results from electrostatic interactions between CD2 and all of the membrane lipids (see Figure S8). Here, the basis is the idea that the larger the (attractive) electrostatic interaction, the more tilted the ECD. Accordingly, we observe that the electrostatic energy is the highest in the Ld-NG system, followed by (in decreasing order) Ld-G, Lo-NG, and Lo-G. The correlation with the results describing the ECD orientation in Figure 2 is evident. Further, we find that the total electrostatic interaction is lowest in the deprotonated system, where the ECD of CD2 stands the most upright.
As discussed above, the positively charged lysines and arginines form a patch that is present in both human and rat CD2. As a matter of fact, a patch of charged residues is present in this region even beyond these species. We carried out sequence alignment for CD2 for all mammal species for which the sequence was deposited in UNIPROT (see Figure S11). Regarding the charged residues, lysines 125 and 132 were found to be highly conserved, and lysines 185, 175, and 166 as well as arginine 170 were also frequently conserved. As for glycosylation, positions 89 and 141 were also found to be highly conserved.
Our results suggest that positioning of the ECD in CD2 is affected by protein glycosylation and the lipid composition of the membrane environment that hosts the protein. On the basis of analysis for CD2, the most decisive factor is the lipid content (Figure 2). We found that when CD2 is transferred from a bilayer that is characterized by a DOPC-rich Ld membrane environment to a Chol- and SM-rich bilayer with a more ordered Lo membrane environment, the orientation of the ECD undergoes significant reorganization; in the Ld bilayer, the ECD associates with the membrane, while in the Lo bilayer, the ECD stands upright. Glycosylation had a similar effect, but its significance was less pronounced versus that of lipid-induced effects. Importantly, the largest effects were observed when lipids and glycosylation acted in unison.
Lipids are proposed to affect membrane protein conformation in a number of ways. These include Chol-induced changes in membrane order, ordering of water, ion binding and ion-mediated interactions, lipid–protein interactions, or changes in the pressure profile inside of a membrane.20−24 We tested many of the potential contributions in our simulations (e.g., leaving out Chol, considering differing water contents, and assessing the importance of lipid–protein interactions), but each of these was found to be insignificant (see the SI). Instead, the results point to a conclusion that the lipid-induced tilt of the membrane protein ectodomain was driven by electrostatic interactions. The results in Figures 5 and S8 demonstrate this convincingly. Our simulations indicate that the significant tilt of the CD2 ectodomain largely disappeared when the positively charged amino acids (two arginines and eight lysines), which interact with the headgroup of DOPC, were deprotonated (Figure 5). Further, when we considered the total electrostatic interaction between CD2 and the lipid membrane, we found a direct correlation between the electrostatic interaction and the ECD tilt (Figures S8 and 2).
The effect on CD2 orientation of glycosylation appears to be due to steric effects; glycans in the ECD of CD2 act as a steric barrier, preventing association with the membrane surface. Similar effects were recently observed for the EGFR,5 wherein loss of glycosylation led to strong interactions between the protein and the membrane surface, resulting in a reduction in the accessibility of the ligand-binding site. In the present work, the most important glycosylation is that linked to Asn141 and Asn150 located close to the membrane surface (Figure 1, Figure S9). The influence of these glycans was enhanced in the Lo membrane environment, presumably due to mechanical effects such as Chol-induced increased order and reduced elasticity. However, the glycosylation at Asn150 is likely also to act as a shield, preventing electrostatic interactions mediated by lysine and arginine residues,25 which in CD2 form a patch of positive electrostatic potential (see Figures S9 and S10).
In conclusion, our results show that lipids and glycosylation act in concert in modulating the orientation of the CD2 ECD. These effects are likely to have a crucial role in ensuring that CD2 binds efficiently to its ligands, enhancing T-cell interactions and signaling. In more general terms, given the abundance of glycosylation among membrane proteins, our results suggest their orientation and presentation to be quite broadly influenced by the concerted interplay of lipids and glycosylation.
Methods
The crystal structures of the SH2 and SH3 domains of CD2 were obtained from the PDB database (id: 1HNF),26 and the transmembrane helical part of CD2 was built using the VMD package.27 Hence, we simulated residues 25–235 of CD2. For numbering of the residues, we used the UNIPROT scheme. As for glycosylation, N-linked glycans display an extraordinary diversity.28 However, because all N-linked glycans share the common pentasaccharide core (GlcNAc2Man3; see Figure S1), this primary sugar core is here attached to the N-terminus of residues 89, 141, and 150 (Figure S9) using the protocol described elsewhere.5 The present study shows that even this primary sugar core that is an appropriate choice for simulation purposes to demonstrate the effects of glycosylation has a major influence on CD2 orientation and presentation. The effects of glycosylation can be expected to strengthen with longer and more specific glycans.
The multicomponent Lo membrane used in our simulations was comprised of 35.3 mol % SM (SM d16:1/16:0), 43.0 mol % DOPC, and 28.7 mol % Chol. As a control system in the Ld phase, we used a single-component DOPC bilayer. The number of lipids ranged between 508 and 982, and the number of water molecules was between ∼85 000 and ∼220 000. Details of the systems’ compositions and dimensions are given in Table S1.
All simulations were performed with the GROMACS 4 package.29 The refined OPLS all-atom force field was used for SM and DOPC,30−33 and the standard OPLS all-atom force field was employed for the protein and the sugars together with the OPLS-compatible TIP3P water model.34 The Nosé–Hoover thermostat35 and the Parrinello–Rahman barostat36 were used to maintain the temperature and pressure at 310 K and 1 atm. A semi-isotropic pressure coupling with a compressibility of 4.5 × 10–5 bar–1 was used in the NpT ensemble. Long-range electrostatic interactions were incorporated through the Particle Mesh Ewald (PME) method with a cutoff of 1 nm (between real and reciprocal space descriptions), and the same cutoff was also used for Lennard-Jones interactions.37 For each of the systems, we carried out three independent 500 ns simulations. The first 300 ns of the 500 ns trajectories was considered as an equilibration period based on convergence of the ECD orientation distributions (see the SI), and the last 200 ns was used for analysis unless mentioned otherwise. The results of the three independent simulations were used as the basis for error analysis. The results of the three simulations, for each of the systems considered, were found to be consistent with each other.
Acknowledgments
We wish to thank the Academy of Finland (Center of Excellence in Biomembrane Research (T.R., I.V.)) and the European Research Council (Advanced Grant project CROWDED-PRO-LIPIDS) (I.V.) for financial support. We also thank the Medical Research Council (MRC, Grant Number MC_UU_12010/unit Programmes G0902418 and MC_UU_12025) and MRC/BBSRC/EPSRC (Grant Number MR/K01577X/1) for generous funding of J.B.d.l.S., S.D., and C.E., the Wellcome Trust for funding to S.D., and funding by the Marie Curie Career Integration Grant to J.B.d.l.S. For computational resources, we wish to thank the CSC – IT Center for Science (Espoo, Finland).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jpclett.6b02824.
Additional results and a discussion of their interpretation, including time dependence plots, probability distributions, electrostatic energy plots and surfaces, and the sequence alignment for CD2 (PDF)
Figure S11 (PDF)
Simulation of the Ld-NG system (MPG)
Simulation of the Lo-NG system (MPG)
Movie of the Ld-G simulation (MPG)
Movie of the Lo-G system (MPG)
The authors declare no competing financial interest.
Supplementary Material
References
- Davis S. J.; Ikemizu S.; Evans E. J.; Fugger L.; Bakker T. R.; van der Merwe P. A. The Nature of Molecular Recognition by T cells. Nat. Immunol. 2003, 4, 217–224. 10.1038/ni0303-217. [DOI] [PubMed] [Google Scholar]
- Zuegg J.; Gready J. E. Molecular Dynamics Simulation of Human Prion Protein Including Both N-linked Oligosaccharides and the GPI anchor. Glycobiology 2000, 10, 959–974. 10.1093/glycob/10.10.959. [DOI] [PubMed] [Google Scholar]
- Coskun Ü.; Grzybek M.; Drechsel D.; Simons K. Regulation of Human EGF Receptor by Lipids. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 9044–9048. 10.1073/pnas.1105666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaeva S.; Bayunova L.; Sokolova T.; Vlasova Y.; Bachteeva V.; Avrova N.; Parnova R. GM1 and GD1a Gangliosides Modulate Toxic and Inflammatory Effects of E. coli Lipopolysaccharide by Preventing TLR4 Translocation into Lipid Rafts. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2015, 1851, 239–247. 10.1016/j.bbalip.2014.12.004. [DOI] [PubMed] [Google Scholar]
- Kaszuba K.; Grzybek M.; Orlowski A.; Danne R.; Rog T.; Simons K.; Coskun U.; Vattulainen I. N-glycosylation as Determinant of Epidermal Growth Factor Receptor Conformation in Membranes. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 4334–4339. 10.1073/pnas.1503262112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg J. S. The Biology of CD2: Adhesion, Transmembrane Signal, and Regulatory Receptor of Immunity. J. Surg. Res. 1993, 54, 258–267. 10.1006/jsre.1993.1041. [DOI] [PubMed] [Google Scholar]
- Driscoll P. C.; Cyster J. G.; Campbell I. D.; Williams A. F. Structure of Domain 1 of Rat T Lymphocyte CD2 Antigen. Nature 1991, 353, 762–765. 10.1038/353762a0. [DOI] [PubMed] [Google Scholar]
- Rogers C. J.; Brissette-Storkus C. S.; Chambers W. H.; Cameron J. L. Acute Stress Impairs NK Cell Adhesion and Cytotoxicity through CD2, but Not LFA-1. J. Neuroimmunol. 1999, 99, 230–241. 10.1016/S0165-5728(99)00125-3. [DOI] [PubMed] [Google Scholar]
- Yang J. J.; Ye Y.; Carroll A.; Yang W.; Lee H. W. Structural Biology of the Cell Adhesion Protein CD2: Alternatively Folded States and Structure-Function Relation. Curr. Protein Pept. Sci. 2001, 2, 1–17. 10.2174/1389203013381251. [DOI] [PubMed] [Google Scholar]
- Jones E. Y.; Davis S. J.; Williams A. F.; Harlos K.; Stuart D. I. Crystal Structure at 2.8 Å Resolution of a Soluble Form of the Cell Adhesion Molecule CD2. Nature 1992, 360, 232–239. 10.1038/360232a0. [DOI] [PubMed] [Google Scholar]
- Bodian D. L.; Jones E. Y.; Harlos K.; Stuart D. I.; Davis S. J. Crystal Structure of the Extracellular Region of the Human Cell Adhesion Molecule CD2 at 2.5 Å Resolution. Structure 1994, 2, 755–766. 10.1016/S0969-2126(94)00076-X. [DOI] [PubMed] [Google Scholar]
- Tavernor A. S.; Kydd J. H.; Bodian D. L.; Jones E. Y.; Stuart D. I.; Davis S. J.; Butcher G. W. Expression Cloning of an Equine T-Lymphocyte Glycoprotein CD2 cDNA. Structure-Based Analysis of Conserved Sequence Elements. Eur. J. Biochem. 1994, 219, 969–976. 10.1111/j.1432-1033.1994.tb18579.x. [DOI] [PubMed] [Google Scholar]
- Nunes R. J.; Castro M. A. A.; Gonçalves C. M.; Bamberger M.; Pereira C. F.; Bismuth G.; Carmo A. M. Protein Interactions between CD2 and Lck Are Required for the Lipid Raft Distribution of CD21. J. Immunol. 2008, 180, 988–997. 10.4049/jimmunol.180.2.988. [DOI] [PubMed] [Google Scholar]
- Walzel H.; Blach M.; Hirabayashi J.; Kasai K.; Brock K. Involvement of CD2 and CD3 in Galectin-1 Induced Signaling in Human Jurkat T-cells. Glycobiology 2000, 10, 131–140. 10.1093/glycob/10.2.131. [DOI] [PubMed] [Google Scholar]
- Moremen K. W.; Tiemeyer M.; Nairn A. V. Vertebrate Protein Glycosylation: Diversity, Synthesis and Function. Nat. Rev. Mol. Cell Biol. 2012, 13, 448–462. 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A.; Aebi M. Intracellular Functions of N-linked Glycans. Science 2001, 291, 2364–2369. 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- Meyer B.; Möller H.. Glycopeptides and Glycoproteins. In Conformation of Glycopeptide and Glycoproteins; Wittmann V, Ed.; Springer-Verlag: Berlin, Heidelberg, Germany, 2006; pp 187–251. [Google Scholar]
- Bruno A.; Scrima M.; Novellino E.; D’Errico G.; D’Ursi A. M.; Limongelli V. The Glycan Role in the Glycopeptide Immunogenicity Revealed by Atomistic Simulations and Spectroscopic Measurements on the Multiple Sclerosis Biomarker CSF114(Glc). Sci. Rep. 2015, 5, 9200. 10.1038/srep09200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D.; Simons K. Lipid Rafts as a Membrane-Organizing Principle. Science 2010, 327, 46–50. 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- Lingwood D.; Binnington B.; Róg T.; Vattulainen I.; Grzybek M.; Coskun U.; Lingwood C. A.; Simons K. Cholesterol Modulates Glycolipid Conformation and Receptor Activity. Nat. Chem. Biol. 2011, 7, 260–262. 10.1038/nchembio.551. [DOI] [PubMed] [Google Scholar]
- Magarkar A.; Dhawan V.; Kallinteri P.; Viitala T.; Elmowafy M.; Róg T.; Bunker A. Cholesterol Level Affects Surface Charge of Lipid Membranes in Saline Solution. Sci. Rep. 2014, 4, 5005. 10.1038/srep05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesnar E.; Subczynski W. K.; Pasenkiewicz-Gierula M. Saturation with Cholesterol Increases Vertical Order and Smoothes the Surface of the Phosphatidylcholine Bilayer: A Molecular Simulation Study. Biochim. Biophys. Acta, Biomembr. 2012, 1818, 520–529. 10.1016/j.bbamem.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rog T.; Vattulainen I. Cholesterol, Sphingolipids, and Glycolipids: What Do We Know About Their Role in Raft-Like Membranes. Chem. Phys. Lipids 2014, 184, 82–104. 10.1016/j.chemphyslip.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Ollila O. H. S.; Risselada H. J.; Louhivuori M.; Lindahl E.; Vattulainen I.; Marrink S. J. 3D Pressure Field in Lipid Membranes and Membrane Complexes. Phys. Rev. Lett. 2009, 102, 078101. 10.1103/PhysRevLett.102.078101. [DOI] [PubMed] [Google Scholar]
- Sun D.; Forsman J.; Woodward C. E. Evaluating Force Fields for the Computational Prediction of Ionized Arginine and Lysine Side-Chains Partitioning into Lipid Bilayers and Octanol. J. Chem. Theory Comput. 2015, 11, 1775–1791. 10.1021/ct501063a. [DOI] [PubMed] [Google Scholar]
- Bodian D. L.; Jones E. Y.; Harlos K.; Stuart D. I.; Davis S. J. Crystal Structure of the Extracellular Region of the Human Cell Adhesion Molecule CD2 at 2.5 Å Resolution. Structure 1994, 2, 755–766. 10.1016/S0969-2126(94)00076-X. [DOI] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. VMD: Visual Molecular Dynamics. J. Mol. Graphics 1996, 14, 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Stick R. V.; Williams S. J.. Carbohydrates: The Essential Molecules of Life; Elsevier: Amsterdam, The Netherlands, 2009. [Google Scholar]
- Hess B.; Kutzner C.; van der Spoel D.; Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Maciejewski A.; Pasenkiewicz-Gierula M.; Cramariuc O.; Vattulainen I.; Róg T. Refined OPLS–AA Force Field for Saturated Phosphatidylcholine Bilayers at Full Hydration. J. Phys. Chem. B 2014, 118, 4571–4581. 10.1021/jp5016627. [DOI] [PubMed] [Google Scholar]
- Kulig W.; Pasenkiewicz-Gierula M.; Róg T. Cis and Trans Unsaturated Phosphatidylcholines Bilayers: Molecular Dynamics Simulation Study. Chem. Phys. Lipids 2016, 195, 12–20. 10.1016/j.chemphyslip.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Kulig W.; Pasenkiewicz-Gierula M.; Róg T. Topologies, Structures and Parameter Files for Lipid Simulations in GROMACS with the OPLS-AA Force Field: DPPC, POPC, DOPC, PEPC, and Cholesterol. Data in Brief 2015, 5, 333–336. 10.1016/j.dib.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Róg T.; Orłowski A.; Llorente A.; Skotland T.; Sylvänne T.; Kauhanen D.; Ekroos K.; Sandvig K.; Vattulainen I. Package of GROMACS Input Files for Molecular Dynamics Simulations of Mixed, Asymmetric Bilayers Including Molecular Topologies, Equilibrated Structures, and Force Field for Lipids Compatible with OPLS-AA Parameters. Data in Brief 2016, 7, 1171–1174. 10.1016/j.dib.2016.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- Hoover W. G. Canonical Dynamics: Equilibrium Phase-Space Distributions. Phys. Rev. A: At., Mol., Opt. Phys. 1985, 31, 1695–1697. 10.1103/PhysRevA.31.1695. [DOI] [PubMed] [Google Scholar]
- Parrinello M.; Rahman A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J. Appl. Phys. 1981, 52, 7182–7190. 10.1063/1.328693. [DOI] [Google Scholar]
- Essmann U.; Perera L.; Berkowitz M. L.; Darden T.; Lee H.; Pedersen L. G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593. 10.1063/1.470117. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


