Abstract
Background
Eukaryotic whole genome sequences are accumulating at an impressive rate. Effective methods for comparing multiple whole eukaryotic genomes on a large scale are needed. Most attempted solutions involve the production of large scale alignments, and many of these require a high stringency pre-screen for putative orthologs in order to reduce the effective size of the dataset and provide a reasonably high but unknown fraction of correctly aligned homologous sites for comparison. As an alternative, highly efficient methods that do not require the pre-alignment of operationally defined orthologs are also being explored.
Results
A non-alignment method based on the Singular Value Decomposition (SVD) was used to compare the predicted protein complement of nine whole eukaryotic genomes ranging from yeast to man. This analysis resulted in the simultaneous identification and definition of a large number of well conserved motifs and gene families, and produced a species tree supporting one of two conflicting hypotheses of metazoan relationships.
Conclusions
Our SVD-based analysis of the entire protein complement of nine whole eukaryotic genomes suggests that highly conserved motifs and gene families can be identified and effectively compared in a single coherent definition space for the easy extraction of gene and species trees. While this occurs without the explicit definition of orthologs or homologous sites, the analysis can provide a basis for these definitions.
Background
Several methods have been developed for the detailed global comparison of multiple whole genomes and the production of global phylogenies. Most of these methods require the prior identification and selection of a reasonably small subset of putative orthologs within which individual homologous sites are identified with some degree of confidence using alignment [1-7]. Frequently, detailed alignment information is subdivided and compressed into a smaller number of complex characters (such as gene content or gene order), which are then used for quantitative comparison [[4,5]; see [6] for review], but the more or less direct use of large scale sequence alignments have also been attempted [7].
Though generally less developed, many non-alignment methods, considered initially by Blaisdell [8], are currently being explored for a similar purpose [[9-15]; see [17] for review]. Rarely do such methods simultaneously provide 1) detailed and unbiased comparisons of a high fraction of biomolecular sequences within full genome datasets, and 2) globally consistent gene and species trees based on this exhaustive comparison. We have recently developed an SVD-based phylogenetic method that provides accurate comparisons of a high fraction of sequences within whole genomes without the prior identification of orthologs or homologous sites [13]. This method has been successfully applied to a number of diverse genome datasets, including mitochondrial genomes, bacterial genomes, and viral genomes [13-15]. Here we apply this method to a diverse set of nine complete eukaryotic nuclear genomes, resulting in the production of a species tree based on the automatic identification and simultaneous comparison of over 400 conserved amino acid motifs and gene families.
Results and discussion
Proteome data sets and sequence conversion
The nine eukaryotic genomes compared in this analysis are listed in Table 1. The protein sets obtained from NCBI for the malaria parasite (Pfal) and the budding yeast (Scer) each contributed only 3% of the 175,559 total proteins in the dataset, while the proteins for Frub provided nearly 21% of the total. Only the Frub proteins were obtained from the Ensemble Genome Browser [16], since protein predictions for this organism were not available from NCBI. Differences in methods used to predict proteins by these two organizations might be responsible for the large difference in the number of proteins predicted for comparable vertebrate genomes (>37,000 for Frub, but only 21–25,000 for all other vertebrates). These differences could, in principle, drastically effect the gene and species trees derived from a global comparison of all proteins. However, the position of Frub in the final species tree suggests that these effects were relatively minor (see below). We have noted previously that even drastic genome size differences can be accommodated by our method [14].
Table 1.
Genes and Genomes Compared
| Organism | SVD "top 5" | Genome Total |
| Hsap | 996 (23%) | 25,319 (14%) |
| Mmus | 881 (20%) | 25,371 (14%) |
| Rnov | 670 (15%) | 21,204 (12%) |
| Frub | 536 (12%) | 37,439 (21%) |
| Agam | 573 (13%) | 16,091 (9%) |
| Dmel | 443 (10%) | 18,107 (10%) |
| Cele | 135 (3%) | 21,124 (12%) |
| Scer | 113 (3%) | 5,855 (3%) |
| Pfal | 23 (1%) | 5,049 (3%) |
| 4370 (100%) | 175559 (100%) |
SVD-derived vector definitions for motifs and gene families
All the proteins in the dataset were recoded as overlapping tetrapeptide frequency vectors and the resulting data matrix was decomposed by the SVD. A total of 437 singular triplets were obtained as output. The "protein" vectors provided in the "right" factor matrix are known to provide reduced dimensional definitions for all proteins in the dataset as linear combinations of the orthogonal "right" singular vectors (rsv's). Conversely, the right singular vectors themselves frequently represent "ideal" versions of proteins defining a given gene family [13,14] Protein vectors having the strongest projections on a given rsv are therefore likely to represent members of a given gene family. In this analysis, the proteins with the five strongest projections (referred to as the "top 5") for each rsv were used to identify and summarize a number of gene families. The total number of proteins from each species that appear in the "top 5" for all 437 right singular vectors are listed in Table 1. Although the fraction of "top 5" proteins identified by the SVD roughly parallels the fraction of total proteins from each species, the mammalian proteins tend to dominate the analysis.
Each right singular vector can potentially define two distinct gene families. In this case, the highest positive elemental values within a vector identify proteins associated with one protein family, while the highest negative values identify proteins associated with an anti-correlated family (i.e. proteins that rarely share the same tetrapeptides). Frequently, however, strong family definitions are provided for just one protein family. In this case, the anti-correlated proteins are seen to be derived from a mixture of two or more families. Since the choice of sign is arbitrary, strong family definitions are equally likely to be provided by either the positive or the negative values within a vector. Family definitions provided by positive vector values are denoted below using the simple vector index (e.g. 277 = the 277th singular vector). Those provided by negative vector values are followed by an "a" (e.g. 277a). Its worth noting at this point that protein family definitions provided by the SVD necessarily account for not only "what is there" (tetrapeptides that form the motifs that define the family), but also "what is not there" (tetrapeptides excluded by that family of proteins, but likely to form anti-correlated motifs within other families of proteins).
Protein family definitions provided by right singular vectors
An abbreviated list of 58 protein families identified within the 437 SVD-derived singular triplets are provided in Table 2. For each listed singular triplet, the gi# of an example protein chosen from among the "top 5" values within the right singular vector is provided, along with its corresponding Name and a Protein Description provided within the NCBI annotation for that protein. In general, proteins described by the more dominant singular triplets were selected for presentation from the complete list of 437 triplets. However, some were chosen due to their historical utility for evolutionary comparisons (ribosomal proteins) and/or their tendency to be accompanied by strongly correlated peptide motifs (last column of Table 2). Relatively few families appear in the table due to the fact that some vectors strongly describe only one family rather than two, some vectors describe only families from species that lack annotation or are poorly annotated at NCBI (i.e. Frub proteins, Agam proteins, etc.), some vectors describe protein families listed by NCBI merely as "unknown" or "conserved unknown", some vectors describe proteins with weakly conserved motifs, and some vectors describe distinct subfamilies of proteins. In the latter case, multiple right singular vectors are apparently required in combination to describe some of the more diverse families of proteins. Included in Table 2 is the number of singular vectors that include the chosen example protein within its "top 5". When multiple vectors are involved in defining multiple related subfamilies, the most "dominant" vector (the one on which the example protein casts its strongest projection) is listed in the first column. Thus, some proteins are seen to have multiple subfamily affiliations. The multiple vectors observed per family effectively subdivide the 58 families into 179 distinct subfamilies. For instance, Table 2 includes a set of 18 ribosomal protein families described by a total of 65 singular vectors (highlighted in bold). Ribosomal proteins are frequently well conserved, effectively aligned, and commonly used for estimating evolutionary relationships. Their presence within our list of dominant singular vectors suggests their utility for establishing evolutionary relationships even in the absence of explicit alignments and explicit a priori assignments of orthology.
Table 2.
A selected list of protein family/motifs identified by SVD-derived singular triplets (st's). In this summary table, unique example proteins (rsv-gi#) were chosen from the 5 to 40 "top five" proteins identified as members of a given family by as many as 8 distinct right singular vectors. As examples, six individual ras proteins representing six broad categories of ras (highlighted in italics) are defined by a total of 13 right singular vectors, and 18 ribosomal proteins (highlighted in bold) are defined by a total of 65 right singular vectors. The lengths of continuous copep strings identified from the corresponding left singular vectors and their specificities (E-values) as revealed by pairwise BLAST are also provided.
| triplet | # | rsv-gi# | Name | Protein Description | lsv copep string (E-value) |
| 421a | 1 | 11415030 | HIST1H4J | H4 histone family, member E | 62 aa's (1e-54) |
| 417a | 2 | 21166389 | HIST1H2BC | H2B histone family, member L | 75 aa's (4e-67) |
| 413a | 1 | 31560385 | Rpl21 | ribosomal protein L21 | 60 aa's (2e-55) |
| 408 | 1 | 4501885 | ACTB | beta actin; beta cytoskeletal actin | 42 aa's (9e-38) |
| 405 | 1 | 4506661 | Rpl7a | ribosomal protein 7a | 79 aa's (3e-62) |
| 392a | 1 | 5174735 | TUBB2 | tubulin, beta, 2 | 45 aa's (7e-41) |
| 389a | 2 | 13569962 | RAB1B | RAB1B, RAS oncogene family; small GTP-binding | 14 aa's (2e-11) |
| 389 | 3 | 6677781 | Rpl29 | ribosomal protein L29 | 77 aa's (3e-60) |
| 387 | 3 | 31981690 | Hspa8 | heat shock 70kD protein 8 | 40 aa's (2e-35) |
| 385a | 1 | 11024714 | UBB | ubiquitin B precursor; polyubiquitin B | 77 aa's (2e-68) |
| 378a | 5 | 26051216 | CAMK2B | calmodulin-dependent protein kinase IIB isoform 7 | 14 aa's (2e-10) |
| 373a | 2 | 4502201 | ARF1 | ADP-ribosylation factor 1 | 86 aa's (1e-41) |
| 371a | 3 | 6679439 | Ppia | peptidylprolyl isomerase A; cyclophilin A | 55 aa's (2e-48) |
| 368a | 5 | 25150942 | Tcb-1 | transposable element tcb1 transposase (1O615) | 88 aa's (7e-74) |
| 363 | 3 | 33149310 | UBE2D3 | ubiquitin-conjugating enzyme E2D 3 isoform 1 | 138 aa's (7e-91) |
| 354 | 3 | 4502549 | CALM2 | calmodulin 2; phosphorylase kinase delta | 40 aa's (1e-19) |
| 352a | 4 | 17105394 | RPL23A | ribosomal protein L23a | 44 aa's (3e-33) |
| 350a | 4 | 9845511 | RAC1 | ras-related C3 botox sub 1 isoform Rac1, rho | 15 aa's (2e-12) |
| 347a | 3 | 51873060 | Eef1a1 | eukaryotic translation elongation factor 1 alpha 1 | 24 aa's (4e-19) |
| 345 | 2 | 27679110 | Rpl17 | ribosomal protein L17 (L23) | 92 aa's (2e-89) |
| 341a | 5 | 31980772 | Ppp1cc | protein phosphatase 1, catalytic, gamma isoform | 20 aa's (5e-17) |
| 337 | 5 | 24648716 | mod(mdg4) | modifier of mdg4 | 32 aa's (2e-29) |
| 334 | 5 | 24653107 | Galpha49B | G protein alpha49B | 19 aa's (9e-18) |
| 333a | 3 | 4506633 | RPL31 | ribosomal protein L31 | 78 aa's (8e-74) |
| 329a | 2 | 34878793 | Pcdha13 | protocadherin alpha 13 | 17 aa's (8e-14) |
| 327 | 3 | 32307119 | PPP2R2B | Serine/threonine protein phosphatase 2A, neuronal | 23 aa's (7e-20) |
| 324 | 1 | 31982919 | ZNF430 | zinc finger protein 430 | 18 aa's (3e-11) |
| 322a | 3 | 34871376 | LOC287293 | similar to high mobility group 1 protein | 15 aa's (9e-13) |
| 321a | 3 | 4504445 | HNRPA1 | heterogeneous nuclear ribonucleoprotein A1 | 23 aa's (2e-18) |
| 320a | 2 | 25141298 | kin-1 | cyclic AMP-dependent catalytic subunit (kin-1) | 66 aa's (4e-62) |
| 316a | 5 | 22094075 | Slc25a5 | solute carrier family 25; adenine nucleotide | 27 aa's (7e-22) |
| 308a | 3 | 9845502 | LAMR1 | laminin receptor 1 (67kD, ribosomal protein SA) | 68 aa's (1e-60) |
| 304 | 3 | 6978809 | Eno1 | enolase 1, alpha | 32 aa's (3e-27) |
| 301 | 4 | 27676004 | LOC365206 | similar to ribosomal protein L9 | 139 aa's (1e-13) |
| 295 | 2 | 31083250 | PPP2R5C | Ser/threo protein phosphatase 2A, 56 kD regulator, | 16 aa's (6e-12) |
| 292 | 4 | 31560517 | Rpl27a | ribosomal protein L27a | 58 aa's (7e-56) |
| 291 | 2 | 15011936 | RPS26 | ribosomal protein S26 | 77 aa's (7e-64) |
| 288 | 1 | 22129671 | Olfr493 | olfactory receptor MOR204–35 | 12 aa's (3e-08) |
| 287 | 2 | 38076430 | LOC193565 | similar to T-cell receptor alpha chain | 16 aa's (2e-12) |
| 285a | 3 | 6754140 | H2-Q7 | histocompatibility 2, Q region locus 7 | 19 aa's (5e-16) |
| 280a | 5 | 16418339 | Rpl10 | ribosomal protein 10 | 27 aa's (4e-23) |
| 277a | 1 | 15718763 | KRAS2 | cellular c-Ki-ras2 proto-oncogene | 9 aa's (2e-06) |
| 277 | 2 | 27689505 | Rab5c | similar to Rab5c protein | 17 aa's (4e-13) |
| 276 | 4 | 24580529 | M(2)21AB | Minute (2) 21AB CG2674-PA | 25 aa's (5e-20) |
| 272 | 1 | 25742772 | Kcna2 | potassium voltage-gated channel, shaker-related, | 12 aa's (1e-09) |
| 270 | 4 | 33186863 | Rpl13 | ribosomal protein L13 | 11 aa's (3e-09) |
| 266 | 4 | 4506697 | RPS20 | ribosomal protein S20 | 54 aa's (2e-49) |
| 256 | 3 | 4506597 | RPL12 | ribosomal protein L12 | 34 aa's (8e-30) |
| 253a | 6 | 15809016 | MRLC2 | myosin regulatory light chain MRCL2 | 19 aa's (7e-16) |
| 247 | 3 | 31981515 | Rpl7 | ribosomal protein L7 | 10 aa's (4e-08) |
| 240a | 5 | 24639734 | Dlc | dynein light chain ATPase | 22 aa's (4e-21) |
| 237a | 4 | 34865959 | gpdh | similar to glyceraldehyde-3-phosphate | 16 aa's (7e-13) |
| 236a | 2 | 10835049 | ARHA | Aplysia ras-related homolog 12; oncogene RHO | 9 aa's (9e-07) |
| 230 | 6 | 15431293 | RPL15 | ribosomal protein L15 | 11 aa's (6e-09) |
| 224 | 5 | 13592069 | Rps10 | ribosomal protein S10 | 81 aa's (1e-78) |
| 197a | 2 | 14249144 | Rab11b | RAB11B, member RAS oncogene family | 15 aa's (4e-12) |
| 190a | 6 | 4506621 | RPL26 | ribosomal protein L26 | 16 aa's (8e-14) |
| 183a | 5 | 14277700 | RPS12 | ribosomal protein S12 | 13 aa's (1e-10) |
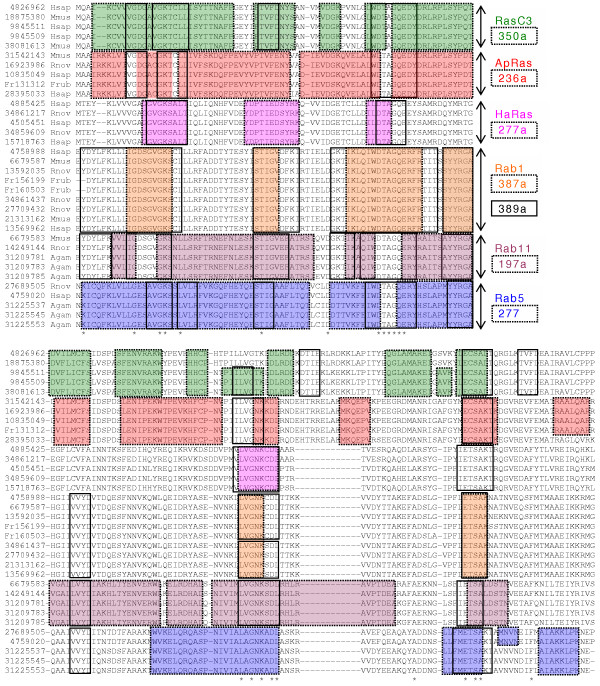
The diverse families of ras proteins present within the eukaryotic data set provide good examples of the ability of SVD-derived singular triplets to identify and describe both superfamilies and subfamilies of proteins. The ras proteins are well described by at least 13 vectors, including the 6 dominant vectors highlighted in italics in Table 2. All the "top 5" members of the protein families identified by these 6 vectors are listed in Table 3. Vector 197a summarizes the brain-associated ras11 subfamily (Rab11), vector 236a summarizes the Aplysia-related ras subfamily (ApRas), vectors 277 and 277a summarize the brain-associated ras 5 subfamily (Rab5) and the complex Ha/K/Nras subfamily (HaRas) respectively, vector 350a summarizes the ras-related C3 botulinum toxin substrate 1 subfamily (Rac1), and vector 387a summarizes the brain-associated ras1B subfamily (Rab1B). The most dominant ras vector, 389a, appears to describe a more generalized version of the Rab1 subfamily, since this vector includes both Rab1A and Rab1B proteins within the "top five". In addition, as explained below, this vector also summarizes a high fraction of the entire set of 34 ras sequences within all subfamilies.
Table 3.
Comparison of seven ras family clusters provided by right singular vectors with KOG and Homologen clusters. Only proteins having one of the five strongest projections ("top five") for a given singular vector are used in the comparison. Few genomes have KOG members specifically identified by NCBI, however, most or all of the "top 5" proteins for a given rsv would likely be identified as members of the same KOG family. For 197a (Rab11), the KOG # provided in parentheses is that of the closely related human protein.
| rsv# | gi# | projection | organism | GeneName | kog# | hg# |
| 197a | 6679583 | 0.06900 | Mmus | Rab11b | (0087) | 3109 |
| (Rab11) | 14249144 | 0.06892 | Rnov | Rab11b | na | 3109 |
| 31209781 | 0.06827 | Agam | na | na | 3109 | |
| 31209783 | 0.06827 | Agam | na | na | 3109 | |
| 31209785 | 0.06826 | Agam | na | na | 3109 | |
| 236a | 31542143 | 0.05883 | Mmus | Arha | na | 1257 |
| (ApRas) | 16923986 | 0.05883 | Rnov | Arha2 | na | 1257 |
| 10835049 | 0.05873 | Hsap | RHOA | 0393 | 1257 | |
| 28395033 | 0.05610 | Hsap | ARHC | 0393 | 22408 | |
| en131312 | 0.05412 | Frub | na | na | na | |
| 277 | 27689505 | 0.07229 | Rnov | Rab5c | na | 20961 |
| (Rab5) | 4759020 | 0.07214 | Hsap | RAB5C | 0092 | 20961 |
| 31225537 | 0.07022 | Agam | na | na | 20961 | |
| 31225545 | 0.07022 | Agam | na | na | 20961 | |
| 31225553 | 0.07022 | Agam | na | na | 20961 | |
| 277a | 15718763 | 0.04278 | Hsap | KRAS2 | 0395 | 2159 |
| (HaRas) | 4885425 | 0.04243 | Hsap | HRAS | 0395 | 3907 |
| 34861217 | 0.04243 | Rnov | Hras1 | na | 3907 | |
| 4505451 | 0.04176 | Hsap | NRAS | 0395 | 20564 | |
| 34859609 | 0.04165 | Rnov | Nras | na | 20564 | |
| 350a | 9845511 | 0.07403 | Hsap | RAC1 | 0393 | 23126 |
| (RasC3) | 38081613 | 0.07403 | Mmus | Rac1 | na | 23126 |
| 9845509 | 0.06942 | Hsap | RAC1b | 0393 | 23126 | |
| 4826962 | 0.06820 | Hsap | RAC3 | 0393 | 3705 | |
| 18875380 | 0.06820 | Mmus | Rac3 | na | 3705 | |
| 387a | 34861437 | 0.03486 | Rnov | Rab1B | na | 23689 |
| (Rab1) | 21313162 | 0.03413 | Mmus | Rab1B | na | 23689 |
| 13569962 | 0.03400 | Hsap | RAB1B | 0084 | 23689 | |
| 27709432 | 0.03400 | Rnov | Rab1B-like | na | 27733 | |
| en156199 | 0.03396 | Frub | na | na | na | |
| 389a | 4758988 | 0.04851 | Hsap | RAB1A | 0084 | 3067 |
| (Rab/Ras) | 6679587 | 0.04851 | Mmus | Rab1A | na | 3067 |
| 13569962 | 0.04840 | Hsap | RAB1B | 0084 | 23689 | |
| en160503 | 0.04824 | Frub | na | na | na | |
| 13592035 | 0.04811 | Rnov | Rab1A | na | 3067 |
For comparison, KOG and Homologen memberships are also listed, when available, for each of the "top 5" proteins listed in Table 3. Table 4 provides a similar comparison for a set of four arbitrarily selected protein families unrelated to ras or to each other (potassium channel, enolase, solute carrier protein, and ADP-ribosylation factor). Since most of the genomes used in our study have not yet been included within the KOG classification scheme, only fly and human proteins have official KOG affiliations. However, we expect with high likelihood that most if not all of the top 5 proteins listed in Tables 3 and 4 would also be members of the particular KOG family listed for each vector. Given this, there would be a good correspondence in Tables 3 and 4 between KOG family members and the proteins identified by singular vectors. In contrast, the Homologen resource appears to provide a more selective classification method, dividing the KOG protein families into two or more subfamilies within which members are more likely to represent specific orthologs.
Table 4.
Comparison of four unrelated protein clusters provided by right singular vectors with KOG and Homologen clusters. Descriptions for each of these clusters are provided in Table 2. Only proteins having one of the five strongest projections ("top five") for a given singular vector are used in the comparison.
| rsv# | gi# | projection | organism | GeneName | kog# | hg# |
| 272a | en165011 | 0.06928 | Frub | na | na | na |
| (Kcna) | 25742772 | 0.06865 | Rnov | Kcna2 | na | 21034 |
| 4826782 | 0.06834 | Hsap | Kcna2 | 1545 | 21034 | |
| 31543024 | 0.06821 | Mmus | Kcna2 | na | 21034 | |
| 27465523 | 0.06632 | Rnov | Kcna1 | na | 183 | |
| 304 | 12963491 | 0.101507 | Mmus | Eno1 | na | 1093 |
| (Eno) | 6978809 | 0.101252 | Rnov | Eno1 | na | 1093 |
| 4503571 | 0.097337 | Hsap | Eno1 | 2670 | 1093 | |
| 51770896 | 0.092899 | Mmus | Eno1 | na | 1093 | |
| en150208 | 0.091209 | Frub | na | na | na | |
| 316a | 32189350 | 0.11376 | Rnov | Slc25a5 | na | 37448 |
| (Slc25) | 22094075 | 0.11343 | Mmus | Slc25a5 | na | 37448 |
| 4502099 | 0.11202 | Hsap | Slc25a5 | 0749 | 37448 | |
| en159404 | 0.1034 | Frub | na | na | na | |
| 20863388 | 0.10117 | Mmus | Slc25a4 | na | 36058 | |
| 373a | 4502201 | 0.12887 | Hsap | Arf1 | 0070 | 1253 |
| (Arf) | 6680716 | 0.12887 | Mmus | Arf1 | na | 1253 |
| 11968098 | 0.12887 | Rnov | Arf1 | na | 1253 | |
| 24668762 | 0.12856 | Dmel | Arf79F | 0070 | 1253 | |
| 24668773 | 0.12856 | Dmel | Arf79F | 0070 | 1253 |
Conserved motif definitions provided by left singular vectors
Members of any particular ras subfamily represented by a given right singular vector share a uniquely conserved set of correlated tetrapeptides we have previously referred to as a "copep motif". These motifs are explicitly described by the corresponding "left" singular vectors (lsv's) comprising a given singular triplet. The lsv's describe these copep motifs as linear combinations of the 160,000 possible tetrapeptides. Those with high positive values identify peptides found with high probability in the conserved motif of a given subfamily, while those with a high negative value identify peptides excluded with high probability. Therefore, like the rsv's, the lsv's frequently describe two distinct anti-correlated entities (in this case motifs rather than protein families) using either positive or negative values within the vector. Using essentially the same procedure described above for any given rsv, the tetrapeptides having the largest positive or largest negative projections on any given lsv were identified in order to provide a focused summary of the motifs described by that vector. For motif extraction, however, an arbitrary cut-off value (absolute value > 0.025) was used to identify dominant tetrapeptides.
In most cases, it is possible to cluster the resulting short list of dominant tetrapeptides into several uninterrupted copep strings formed by tetrapeptides that overlap in 3 of 4 consecutive amino acid positions. Using this procedure, one long copep string was identified for each of the singular triplets listed in Table 2. The length of the identified long copep string and its corresponding E-value (resulting from pairwise BLAST) are provided as a summary in the last column. The precise amino acid sequences of the long copep strings identified for all listed vectors are provided in a supplementary table [see Additional file 1]. The E-values listed provide a measure of the specificity with which each corresponding protein is identified by the copep string extracted from a given lsv. Its important to note that the long copep string provides only an approximate summary of the lsv from which it is extracted, yet the small E-values clearly indicate that the vast majority of the proteins identified in Table 2 are very specifically recognized by their corresponding copep string.
Figure 1 provides a more detailed demonstration of how correlated peptide motifs and their associated gene families are simultaneously identified and described by SVD-derived singular vectors. In order to allow a clear comparison of SVD-derived motifs with alignment-derived motifs, the dominant tetrapeptides were superimposed over matching regions of a standard ClustalX alignment of the 34 ras proteins identified in the "top five" of the corresponding right singular vectors listed in Table 3. In this example, the dominant tetrapeptides extracted from the six selected left singular vectors are demarcated within (shaded/colored) boxes. Many of the dominant tetrapeptides are seen to form extended strings of overlapping peptides that correspond well to conserved contiguous regions within particular subsets of the ras proteins. For example, vectors 350a and 236a identify and provide distinct descriptions for motifs within the Ras-related botulinum toxin C3 substrate proteins (RasC3) and the Aplysia-related ras proteins (ApRas), respectively. The two most dominant left singular vectors of Figure 1 (389a and 387a) describe motifs within overlapping subsets of the nine Rab1 proteins. In addition, the most dominant left singular vector (389a) appears to describe a highly conserved motif within the entire set of 34 ras proteins reasonably well (solid clear boxes). This vector conspicuously identifies dominant tetrapeptides that span the two regions of the alignment in which unbroken strings of two or more invariant amino acids (asterisks) are present. These two regions are known to be required for ras GTPase activity [18]. It is notable that although these 34 ras proteins have only one stretch with more than two globally conserved consecutive amino acids (DTAGQE), vector 389a is capable of describing large regions of all 34 proteins by recognizing the latent similarity of multiple equivalent tetrapeptides. For example, this single vector recognizes KSAL, KSCL, and KTCL (residues 18–21 of the alignment) as dominant tetrapeptides that occupy equivalent positions within four of the six subtypes of ras proteins (Figure 1). Vector 389a also provides a reasonably strong summary of the large number of other ras proteins present within the genomes of these organisms, but not included in Figure 1 (not shown). In general, the most dominant singular vectors appear to identify highly conserved peptides present in a high fraction of individual members of a protein family or superfamily, while the less dominant vectors appear to describe conserved tetrapeptides present within a restricted set of proteins comprising a subfamily.
Figure 1.
Ras families and sub-families defined by singular vectors (labeled at right). For comparison, dominant peptide strings identified by SVD (boxes) are shown within a Clustal-X alignment. The aligned region corresponds to the first 181aa's of the 192aa Human Rac3 protein. Protein sequences are labeled by gi# (or ensemble# for Frup). Asterisks (*) indicate globally conserved residues. Subfamily motifs associated with negative vector values are denoted with an "a" suffix (e.g. 350a).
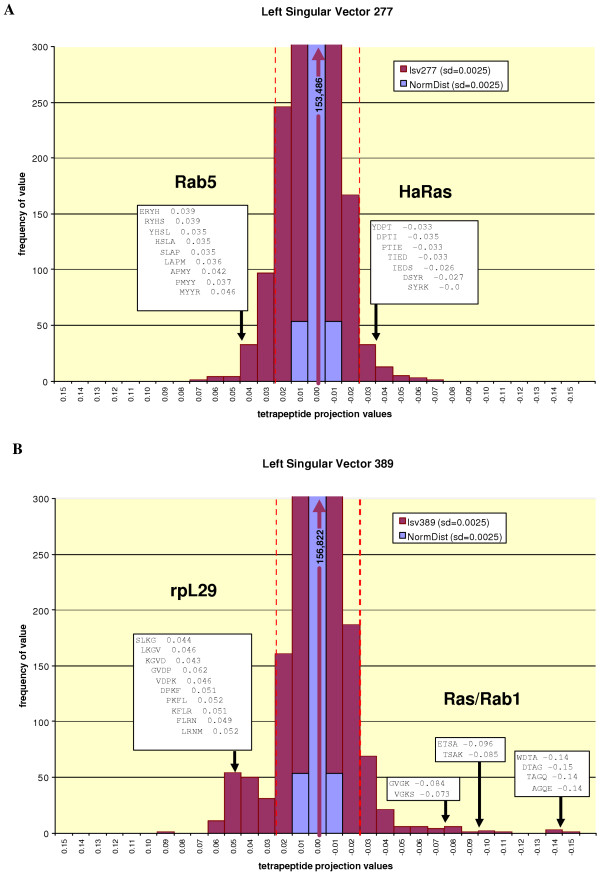
Instead of simply providing restricted motif summaries using the most dominant elements of the left singular vectors, we have also attempted to examine entire vectors in order to gain a better understanding of the motifs (and associated protein families) they describe. A reasonably efficient method for depicting left singular vectors is presented in Figure 2, using vectors 389 and 277 as examples. Both vectors are shown as frequency distributions (purple bars) that summarize the approximate magnitudes of the projections provided by all 160,000 tetrapeptides on the vector in question. These distributions are compared to a normal distribution having the same standard deviation (blue bars). In both examples, a significant fraction of tetrapeptides have high or low values in considerable excess of that expected from a normal distribution. Many of these also exceed the arbitrary cut-off value of 0.025 (dashed lines) used to extract the dominant tetrapeptides that serve to summarize the corresponding motifs. Parts of the Rab5 and HaRas motifs extracted from vector 277 are shown in Figure 2A as overlapping dominant tetrapeptides with associated projection values. Similar motifs extracted from vector 389 are shown in Figure 2B. In the latter case, a motif from the large subunit ribosomal protein rpL29 represents the "anti-motif"of the Ras/Rab proteins described by the extreme vector elements of opposite sign.
Figure 2.
Left singular vectors depicted as tetrapeptide projection value frequency distributions. Distributions for singular vectors 277 (A) and 389 (B) are shown in purple, normal distributions having the same standard deviation are shown in blue. For both distributions, the vast majority of values fall between 0.015 and -0.015. Dashed lines mark the cut-off values used to extract dominant tetrapeptides summarizing correlated peptide (copep) motifs. Selected strings of overlapping tetrapeptides describing parts of these motifs are shown boxed above the approximate regions in the distribution in which they appear.
Species vectors for the production of species phylogenies
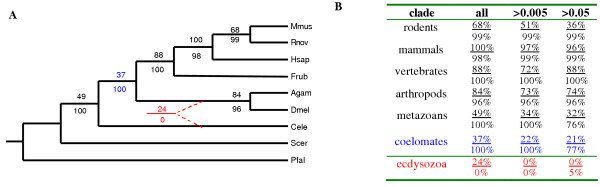
The detailed comparative information contained within the hundreds of singular vectors and their corresponding motifs and gene families was subsequently used to build a species phylogeny by summing all the SVD-derived right protein vectors separately for each organism and then comparing the relative orientation of the resulting species vectors [13]. Figure 3A shows the SVD-based topology obtained for the nine eukaryotes compared in this study. This tree supports a coelomate rather than ecdysozoan lineage. Two distinct re-sampling methods were used to estimate branch statistics for this tree. The top value of each pair of support values for each branch shown in Figure 3A was generated using a traditional bootstrap procedure [19]. In this case, 100 random sets of 437 re-sampled singular vectors were made and used to construct 100 species trees. Alternatively, a novel "successive, delete one" jackknife procedure [14] was used to generate the bottom value shown for each branch. In this case, the least dominant singular vector was removed successively (down to 10 vectors) to generate 427 ordered sets of singular vectors, and a new tree was estimated following each removal. Although bootstrap support values for the branches grouping arthropods with vertebrates (37%) and worms with other metazoa (49%) are relatively weak, support values for these branches are strong (100%) using the modified jackknife procedure. All other branches are strongly supported by both procedures. The branch separating Cele from the coelomates is of special interest, since the weak bootstrap support observed (37%) might suggest a significant affinity between Cele and the arthropods consistent with the "ecdysozoan" model (Figure 3A – alternative branching pattern shown in red). Bootstrap support for the alternative ecdysozoan cluster, however, was only 24%.
Figure 3.
SVD-based proteome phylogeny (A) of nine eukaryotes with percentage branch support: top – bootstrap; bottom – novel jackknife. An unsupported alternative phylogeny containing the "ecdysozoan" lineage is indicated by the dashed red branches. Percentage branch support values for the various clades of the tree are also provided to the left (B) for trees built using all proteins, as well as trees built after poorly described proteins are removed using either of two alternative vector magnitude inclusion values (>0.005, >0.05).
Use of the "successive, delete-one" jackknife procedure as a species tree branch statistic is justified by the fact that SVD provides singular triplets in order of their "dominance" in explaining the data set [20]. Mathematical dominance provides an objective measure of importance that can be utilized to weight characters. Since the modified jackknife procedure used here deletes the least dominant singular vectors one at a time in order, the more dominant singular vectors (i.e. conserved motifs/families) are automatically weighted more heavily within the consensus tree. Hence, one can argue that our novel jackknife procedure provides stronger support for the derived phylogeny because the most dominant singular vectors generally contain stronger information about gene and species relationships.
Poorly described proteins and species tree quality
While our SVD-based analysis technically considers all proteins present within all nine genomes of the data set, it is likely that accurate vector definitions are provided for only a small fraction of these proteins. Theoretically, the 437 singular triplets could effectively describe as many as 2 × 437 = 874 protein families. However, many of these vectors appear to best describe particular subfamilies of larger groups of closely related proteins. Thus, the 58 protein families listed in Table 2 are each represented by anywhere from 1 to 8 triplets. Although, as mentioned earlier, some protein families lacking clear functional annotation were omitted from this table, it still serves to provide a conservative lower estimate of the number of well-described protein families provided by the SVD. Assuming the number of identifiable protein families in our nine genome data set significantly exceeds the 58 to 179 protein families unambiguously demarcated and subdivided in our analysis, then hundreds or perhaps thousand of the poorly described proteins included in our species vector sums might be contributing a high fraction of "noise" to the definition of species.
In an attempt to increase the fraction of well described proteins used to define species, proteins having poor projections on all 437 right singular vectors were ignored during the summation process. Arbitrary vector magnitude cut-off values of 0.005 or 0.05 were applied to reduce the number of poorly described proteins used to build species trees. Even though the highest and most stringent cut-off value removed the majority of proteins during summation, both new species trees had identical topologies to that of the tree shown in Figure 3A in which all proteins were included. Bootstrap and modified jackknife branch support values for these tree are shown in Figure 3B along with those derived from the inclusive analysis. The removal of only a small fraction of poorly described proteins (cut-off = 0.005, about 103 proteins removed) resulted in 22% bootstrap and 100% modified jackknife support for the coelomate lineage, but 0% support for the ecdysozoan lineage. Removal of a much higher fraction of poorly described proteins (cut-off = .05, about 105 proteins removed) produced an equivalent result. Hence, poorly described proteins contribute little to the support that our analysis provides for the coelomate model.
Conclusions
As demonstrated above, an SVD-based analysis of multiple genomes automatically interprets proteins from input genomes as potential members of a limited list of hierarchically defined protein families and subfamilies. Each subfamily is defined in detail by one or more singular vectors as linear combinations of a large number of peptides (160,000 tetrapeptides, in this case). Potentially, a large number of proteomes lacking annotation can be directly interpreted using this method, assuming a sufficient number of annotated proteomes are included in the analysis. Although most of the genomes used in the present analysis were already accompanied by detailed protein annotations, formal annotations of the Frub and Agam proteins were not readily available. Nevertheless, our SVD-based analysis was able to provide precise protein motif descriptions and subfamily affiliations, not only for the six Frub or Agam proteins shown in Figure 1, but also for any of the hundreds of other Frub or Agam proteins exhibiting strong vector projections on any of the 437 derived singular vectors (see "SVD top five" of Table 1).
Our method bears partial resemblance to a recently described graph-theoretic method for rapidly clustering massive datasets of whole genome protein sequence [22]. In this case, the protein definitions generated were not used to derive gene or species trees, but to provide for a comprehensive clustering of all proteins into families having one or more members. The nodes of their graphs, like the vectors from the right matrix in our analysis, represent proteins, while the edges between nodes in their graphs, like the angles between vectors in our analysis, contain the distance information used to compare proteins. However, the distance information in their analysis was obtained ultimately from exhaustive pairwise BLAST alignments. In contrast, our distance information was derived without alignment, by reference to the 437 most dominant SVD-derived orthonormal left singular vectors. These vectors provide "motif models" expressed as particular linear combinations of the 160,000 possible tetrapeptides. The projections of these motif models on a given protein vector serve to quantitatively define the protein. Since no more than 874 motif models would be provided by our truncated SVD, our method would be less effective than other methods for providing comprehensive family designations for all proteins in a dataset [22,23]. However, a high fraction of these protein families are found to contain only one or a few members [22]. Singletons and small families would generally provide unimportant contributions to relative species definitions, since the majority of species would lack a homolog for comparison. Hence small or poorly conserved protein families, presumably represented by the weaker singular triplets in a complete SVD, are profitably ignored in our analysis.
Although our descriptive analysis of singular triplets (e.g. Table 2, Figure 1) suggests that the protein vectors in our high dimensional definition space can be effectively clustered, we have not applied any specific clustering algorithm. Hence no explicit clustering of proteins, equivalent to the identification of orthologs or homologs, is required. Nevertheless, the application of a clustering algorithm to our vector based symmetric protein distances is clearly feasible and results in accurate clustering for a high fraction of proteins. In fact, the accuracy with which proteins are clustered into known families via Neighbor Joining was used previously to establish optimal dimensionality for a well characterized data set [13]. In addition, unlike other methods, our method provides a straightforward vector addition mechanism for converting relative protein definitions into relative species definitions for the production of species phylogenies.
Alternative non-alignment methods exist for comparing sequences [reviewed in [17]]. Some of these methods may prove to be scalable and adaptable to the problem of whole genome phylogeny. For example, a comprehensive bacterial phylogeny was recently derived using species vectors that include a set of background corrected pentapeptide or hexapeptide (K-tuple) frequency values [12]. Although apparently effective for producing global species phylogenies, this method fails to provide quantitatively comparable protein definitions or interpretable predictions for conserved motifs. While many phylogenetically informative pentapeptides and hexapeptides are likely derived from homologs or orthologs, no mechanism exists for extracting, summarizing, and interpreting this information in terms of motif and gene family definitions. This high stringency method provides a low false positive rate (strong connections between probable orthologous peptides), but comes at the expense of a high false negative rate (little or no recognition of other homologous regions within proteins). For organisms exhibiting a significant level of horizontal gene transfer [24-26], models for motifs and protein families may be crucial tools for identifying "borrowed" genes and assessing their impact on phylogenetic hypotheses.
Our SVD-based species tree supports the traditional "coelomate" model of animal phylogeny. Other large-scale, genome level analyses also tend to support this model [27,28]. The alternative "ecdysozoan" model is supported by comparative analyses of rRNA and analyses that include morphological characters [28,29]. Although genome-scale analyses should perhaps carry considerable weight due to the higher fraction of "total information" used as input, the separation of "signal" from "noise" represents a serious hurdle for these methods. Our method represents a uniquely independent solution that provides a noise-reduced simultaneous global comparison of all proteins within multiple genomes without the need for alignments and without the prior application of operational definitions of orthology. As such, it provides a global perspective on gene and species relationships that is based on a much larger subset of information than that normally used. Since it is a non-alignment method, it provides a fundamentally different kind of analysis, and to the extent that the resulting species phylogenies agree with those provided by other analyses that depend upon highly filtered subsets of aligned orthologs or close homologs, we may derive an additional degree of confidence in these relationships. However, the balanced comparison of a large number of additional whole genome sequences from a variety of animals will likely be required in order to produce an unambiguous and universally accepted animal phylogeny.
Methods
Datasets
Complete reference protein sequences for nine whole eukaryotic genomes ranging from yeast to man were compiled into a single dataset (Figure 1a). Curated protein sequence files were obtained from NCBI dated as follows: human (Hsap) 10/10/03, mouse (Mmus) 10/31/03, rat (Rnov) 9/23/03, mosquito (Agam) 10/24/03, fly (Dmel) 10/24/03, worm (Cele) 11/12/03, malaria (Pfal) 10/17/02, and yeast (Scer) 11/12/03. Pufferfish (Frub) protein sequences dated 8/26/02 were obtained from the DOE Joint Genome Institute.
Peptide frequencies and SVD
Each protein sequence in the dataset was recoded as a high dimensional vector containing raw frequencies for each of the 160,000 possible tetrapeptides. Previous work has established that although tripeptides work well for estimating similarities between highly divergent proteins contained within small sets of viral genomes [15], tetrapeptides work better for larger data sets derived from vertebrate mitochondrial genomes or whole bacterial genomes [13,14]. Although pentapeptides also worked well with the mitochondrial datasets (unpublished), our computational capacity precluded the use of pentapeptides (3.2 million patterns) and larger data sets, like the one used here. Following a log-entropy transformation [21], the singular value decomposition of the resulting data matrix was computed. The log-entropy transformation tends to down-weight evenly distributed high frequency peptides that are likely sources of homoplasy. After 1500 Lanczos iterations (residual errors less than 10-6), three output matrices were obtained, consisting of 437 singular triplets (left and right singular vectors and their corresponding singular value). Each left singular vector produced by the SVD defines one or two conserved motifs within the dataset as particular linear combinations of tetrapeptides [13,14]. Similarly, each of the right singular vectors defines one or two conserved gene families (or subfamilies) as particular linear combinations of proteins. Each gene family identified by a given right singular vector contains motifs described by the corresponding left singular vector. Two distinct motif/families are frequently identified per triplet, since each triplet describes both a correlated motif/family (positive values) and an anti-correlated motif/family (negative values).
Vector based motif and protein family models
"Dominant" vector elements (absolute values in excess of 0.025) were extracted from the left singular vectors and summarized using the C++ program "Copepx" [14]. These values were associated with the most "dominant" (i.e. highly conserved) tetrapeptides found within the motifs described by a given left vector. In addition, the "top five" positive and "top five" negative elements were extracted from the right singular vectors and summarized using the C++ program "Coprotx". These values represent the most dominant members of the gene families described by a given right vector.
Species trees and branch support
Distance matrices were derived by summing all the SVD-derived right protein vectors for a given organism and then comparing the relative orientation of the resulting species vectors using the program Cosdist [13,14]. Species trees were subsequently derived from distance matrices using Phylip-Neighbor [30]. Two distinct resampling methods were used to provide branch support: a traditional bootstrap procedure [19], and a modified jackknife procedure. For the bootstrap, 100 random sets of 437 resampled singular vectors were made and used to construct 100 species trees. For the "successive, delete one" jackknife procedure [14], the least dominant singular vector was removed successively (down to 10 vectors) to generate 427 ordered sets of singular vectors, and a new tree was estimated following each removal.
List of abbreviations used
Homo Sapiens (Hsap), Mus musculus (Mmus), Rattus Norvegicus (Rnov), Anopheles gambiae (Agam), Drosophila melanogaster (Dmel), Caenorhabditis elegans (Cele), Plasmodium falciparum (Pfal), and Saccharomyces cerevisiae (Scer), Fugu rubripes (Frub), correlated peptide (copep), correlated protein (coprot), right singular vector (rsv), left singular vector (lsv).
Authors' contributions
GS conceived the study, gathered the input data, provided primary interpretation of the output, and drafted the manuscript. MB wrote and adapted software, performed computational analysis on the input data, and provided manuscript modifications. All authors read and approved the final manuscript.
Supplementary Material
Copep Motifs. Long copep strings identified within the left singular vectors of a given s-triplet.
Acknowledgments
Acknowledgements
This work was supported in part by a Computer and Information Technology Resources challenge grant from the University of Tennessee, Knoxville.
Contributor Information
Gary W Stuart, Email: G-Stuart@indstate.edu.
Michael W Berry, Email: berry@cs.utk.edu.
References
- House CH, Fitz-Gibbon ST. Using homolog groups to create a whole-genomic tree of free-living organisms: an update. J Mol Evol. 2002;54:539–547. doi: 10.1007/s00239-001-0054-5. [DOI] [PubMed] [Google Scholar]
- Lerat E, Daubin V, Moran NA. From Gene Trees to Organismal Phylogeny in Prokaryotes: The Case of the gamma-Proteobacteria. PLoS Biol. 2003;1:E19. doi: 10.1371/journal.pbio.0000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Rogozin IB, Grishin NV, Koonin EV. Genome trees and the tree of life. Trends Genet. 2002;18:472–479. doi: 10.1016/S0168-9525(02)02744-0. [DOI] [PubMed] [Google Scholar]
- Lin J, Gerstein M. Whole-genome trees based on the occurrence of folds and orthologs: implications for comparing genomes on different levels. Genome Res. 2000;10:808–818. doi: 10.1101/gr.10.6.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA. Genome phylogeny based on gene content. Nat Genet. 1999;21:108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- Sawa G, Dicks J, Roberts IN. Current approaches to whole genome phylogenetic analysis. Brief Bioinform. 2003;4:63–74. doi: 10.1186/1471-2105-4-63. [DOI] [PubMed] [Google Scholar]
- Wolf YI, Rogozin IB, Koonin EV. Coelomata and not Ecdysozoa: evidence from genome-wide phylogenetic analysis. Genome Res. 2004;14:29–36. doi: 10.1101/gr.1347404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaisdell BE. A measure of the similarity of sets of sequences not requiring sequence alignment. Proc Natl Acad Sci U S A. 1986;83:5155–5159. doi: 10.1073/pnas.83.14.5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu KH, Qi J, Yu ZG, Anh V. Origin and phylogeny of chloroplasts revealed by a simple correlation analysis of complete genomes. Mol Biol Evol. 2004;21:200–206. doi: 10.1093/molbev/msh002. [DOI] [PubMed] [Google Scholar]
- Li M, Badger JH, Chen X, Kwong S, Kearney P, Zhang H. An information-based sequence distance and its application to whole mitochondrial genome phylogeny. Bioinformatics. 2001;17:149–154. doi: 10.1093/bioinformatics/17.2.149. [DOI] [PubMed] [Google Scholar]
- Pe'er I, Felder CE, Man O, Silman I, Sussman JL, Beckmann JS. Proteomic signatures: amino acid and oligopeptide compositions differentiate among phyla. Proteins. 2004;54:20–40. doi: 10.1002/prot.10559. [DOI] [PubMed] [Google Scholar]
- Qi J, Wang B, Hao BI. Whole proteome prokaryote phylogeny without sequence alignment: a K-string composition approach. J Mol Evol. 2004;58:1–11. doi: 10.1007/s00239-003-2493-7. [DOI] [PubMed] [Google Scholar]
- Stuart GW, Moffett K, Leader JJ. A comprehensive vertebrate phylogeny using vector representations of protein sequences from whole genomes. Mol Biol Evol. 2002;19:554–562. doi: 10.1093/oxfordjournals.molbev.a004111. [DOI] [PubMed] [Google Scholar]
- Stuart GW, Berry MW. A comprehensive whole genome bacterial phylogeny using correlated peptide motifs defined in a high dimensional vector space. J Bioinfo Comp Biol. 2003;19:475–494. doi: 10.1142/S0219720003000265. [DOI] [PubMed] [Google Scholar]
- Stuart GW, Moffet K, Bozarth R. A whole genome perspective on the phylogeny of the plant virus family Tombusviridae. Arch Virol. 2004;149:1595–1610. doi: 10.1007/s00705-004-0298-7. [DOI] [PubMed] [Google Scholar]
- Ensemble Genome Browser http://www.ensembl.org
- Vinga S, Almeida J. Alignment free sequence comparison – a review. Bioinformatics. 2002;4:513–523. doi: 10.1093/bioinformatics/btg005. [DOI] [PubMed] [Google Scholar]
- Macaluso M, Russo G, Cinti C, Bazan V, Gebbia N, Russo A. Ras family genes: an interesting link between cell cycle and cancer. J Cell Physiol. 2002;192:125–130. doi: 10.1002/jcp.10109. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence-limits on phylogenies – an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Berry MW, Browne M. Understanding Search Engines: Mathematical Modeling and Text Retrieval. SIAM, Philadelphia; 1999. [Google Scholar]
- Frakes WB, Baeza-Yates R. Information Retrieval: Data Structures and Algorithms. Prentice Hall, Englewood Cliffs, NJ; 1992. [Google Scholar]
- Enright AJ, Kunin V, Ouzounis CA. Protein families and TRIBES in genome sequence space. Nucleic Acids Res. 2003;31:4632–4638. doi: 10.1093/nar/gkg495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Fedorova ND, Jackson JD, Jacobs AR, Krylov DM, Makarova KS, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5:R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubin V, Moran NA, Ochman H. Phylogenetics and the cohesion of bacterial genomes. Science. 2003;301:829–832. doi: 10.1126/science.1086568. [DOI] [PubMed] [Google Scholar]
- Philippe H, Douady CJ. Horizontal gene transfer and phylogenetics. Curr Opin Microbiol. 2003;6:498–505. doi: 10.1016/j.mib.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Raymond J, Zhaxybayeva O, Gogarten JP, Blankenship RE. Evolution of photosynthetic prokaryotes: a maximum-likelihood mapping approach. Philos Trans R Soc Lond B Biol Sci. 2003;358:223–230. doi: 10.1098/rstb.2002.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JE, Ikeo K, Gojobori T, Hedges SB. The evolutionary position of nematodes. BMC Evol Biol. 2002;2:7. doi: 10.1186/1471-2148-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguinaldo AM, Turbeville JM, Linford LS, Rivera MC, Garey JR, Raff RA, Lake JA. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature. 1997;387:489–493. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- Mallatt J, Winchell CJ. Testing the new animal phylogeny: first use of combined large-subunit and small-subunit rRNA gene sequences to classify the protostomes. Mol Biol Evol. 2002;19:289–301. doi: 10.1093/oxfordjournals.molbev.a004082. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6a3. Department of Genome Sciences, University of Washington, Seattle. 2002.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Copep Motifs. Long copep strings identified within the left singular vectors of a given s-triplet.