Abstract
Dry eye disease is the most prevalent pathological condition in aging eyes. One potential therapeutic strategy is the transplantation of lacrimal glands, generated in vitro from pluripotent stem cells such as human embryonic stem cells, into patients. One of the preceding requirements is a method to differentiate human embryonic stem cells into lacrimal gland epithelium cells. As the first step for this approach, this study aims to identify a set of transcription factors whose overexpression can promote the differentiation of human embryonic stem cells into lacrimal gland epithelium-like cells. We performed microarray analyses of lacrimal glands and lacrimal glands-related organs obtained from mouse embryos and adults, and identified transcription factors enriched in lacrimal gland epithelium cells. We then transfected synthetic messenger RNAs encoding human orthologues of these transcription factors into human embryonic stem cells and examined whether the human embryonic stem cells differentiate into lacrimal gland epithelium-like cells by assessing cell morphology and marker gene expression. The microarray analysis of lacrimal glands tissues identified 16 transcription factors that were enriched in lacrimal gland epithelium cells. We focused on three of the transcription factors, because they are expressed in other glands such as salivary glands and are also known to be involved in the development of lacrimal glands. We tested the overexpression of various combinations of the three transcription factors and PAX6, which is an indispensable gene for lacrimal glands development, in human embryonic stem cells. Combining PAX6, SIX1, and FOXC1 caused significant changes in morphology, i.e., elongated cell shape and increased expression (both RNAs and proteins) of epithelial markers such as cytokeratin15, branching morphogenesis markers such as BARX2, and lacrimal glands markers such as aquaporin5 and lactoferrin. We identified a set of transcription factors enriched in lacrimal gland epithelium cells and demonstrated that the simultaneous overexpression of these transcription factors can differentiate human embryonic stem cells into lacrimal gland epithelium-like cells. This study suggests the possibility of lacrimal glands regeneration from human pluripotent stem cells.
Regenerative medicine: lacrimal gland epithelium in vitro
One possible approach to treat dry eye diseases is to transplant lacrimal glands generated in vitro from human embryonic stem cells into patients. As a first step, we developed a novel method to generate lacrimal gland epithelium-like cells. As a model, we first studied the gene expression patterns of mouse embryonic lacrimal gland and identified key transcription factors involved in the process. Subsequently, we introduced four transcription factors in the form of synthetic mRNAs into human embryonic stem cells and successfully generated lacrimal gland epithelium-like cells, which showed elongated cell shape and increased expression of markers for epithelia, branching morphogenesis, and lacrimal glands. This study suggests the possibility of treating dry eye diseases through regeneration of lacrimal gland from human pluripotent stem cells.
Introduction
The lacrimal gland (LG) is a crucial organ for protecting the ocular surface epithelium through secreting aqueous fluid in tears and maintaining a healthy local microenvironment.1, 2 The LG develops from initial buds of the embryonic ocular mucosal epithelium, which subsequently forms an LG bud consisting of lacrimal gland epithelium (LGE) and lacrimal gland mesenchyme (LGM). The LGE differentiates into mature secretory glands with 3D structures consisting of acini, ducts, and myoepithelial cells through reciprocal epithelial and mesenchymal interactions during organogenesis.1 This branching morphogenesis, which is a common developmental process in secretory organs such as salivary glands, is governed by various signaling pathways in the LGE.3, 4 It has been shown that the expression of Pax6 in LGE is required for LG development,5 and Barx2 plays an essential role in the progress of branching morphogenesis in LGE cells to form a 3D secretory gland structure.6 The aqueous fluid secreted from the acini of the LG through water channels such as aquaporin5 (AQP5) contains electrolytes, water and various kinds of proteins,including lactoferrin (LTF), peptides, glycoproteins for wetness, lubrication, and antibiotic effects on the ocular surface.1, 7, 8
A chronic shortage of tears secreted from LGs leads to dry eye disease (DED), which is one of the most prevalent eye diseases that causes epithelial damage to the ocular surface.9, 10 Aging is known to be a predisposing factor for DED, which results in ocular discomfort, loss of vision, and a decrease in quality of life.11, 12 The current clinical therapy is an artificial tear solution, which is mainly constituted of water. For severe DED, autologous serum eye drops are used to supply tear protein alternatives.13–15
Recently, a novel method for functional LG regeneration has been proposed to cure DED in a mouse model by transplanting bioengineered LG using LGE cells and LGM cells obtained from mice on embryonic day 16.5 (E16.5).16, 17 E16.5 LGE cells are the progenitors of mature epithelial cells and retain the ability to reconstitute functional LG precursors ref. 16 and to differentiate into complex, mature LG structures such as acini and ducts.18, 19 To apply this mouse model to potential human therapy, one possible approach is to differentiate human pluripotent stem cells into human LGE-like cells (equivalent to mouse E16.5 LGE cells) and LGM-like cells (equivalent to mouse E16.5 LGM cells) and to reconstitute human LG for therapeutic transplantation. Considering anticipated difficulty in generating a complex mature organ—LG in vitro—this stepwise strategy may be justified. As the first step toward this approach, we have aimed at generating human LGE-like cells from human pluripotent stem cells.
Recent works have established that a set of transcription factors (TFs) regulates downstream gene expression and forms a network of TFs, which defines the identity of cells.20, 21 Direct conversion of cell lineages from stem cells or fibroblasts to desired cell lineages using overexpression of a specific set of TFs has been demonstrated for various cell types such as neural cells, myocardial cells, and hepatocytes.22–27 Therefore, our goal is to identify a set of TFs that promotes the differentiation of human pluripotent stem cells into LGE-like cells by using the corresponding mouse tissues, i.e., E16.5 LGE.
Here, we have compared gene expression patterns of mouse E16.5 LGE to other closely related tissues, determined TFs that are specifically expressed in E16.5 LGE, and identified three TFs—PAX6, FOXC1, and SIX1—for their potent effects on the differentiation of human embryonic stem cells (hESCs) into LGE-like cells. This study has thus provided insights into the mechanism of LG development and suggested a possibility for LG regeneration from human stem cells for future therapy of DED.
Results
Identification of TFs expressed specifically in mouse E16.5 LGE
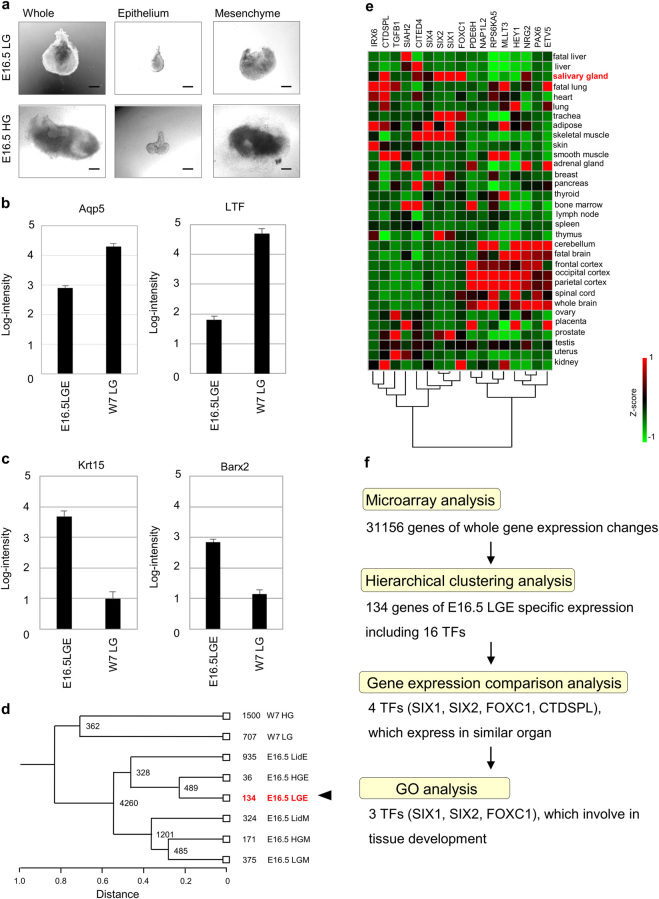
To identify candidate TFs that are highly and specifically expressed in mouse E16.5 LGE, we first carried out gene expression profiling of the E16.5 LGE and other closely related tissues by using DNA microarrays with 31,156 gene probes. The tissues examined here included LGE, LGM, harderian gland epithelium (HGE), harderian gland mesenchyme (HGM), eyelid conjunctiva epithelium (LidE), eyelid conjunctiva mesenchyme (LidM)—all from E16.5 mouse embryos. For comparison, LG and HG obtained from a 7-week-old adult mouse (W7) were also used. The separation of epithelial and mesenchymal tissues in E16.5 tissues was carefully carried out manually under the dissection microscope (Fig. 1a and Supplementary Fig. S1a).28 Their proper separation was clearly noted by principal component analysis (PCA) of the transcriptome data obtained by gene expression profiling of each tissue sample: E16.5 Mesenchyme (LGM, HGM, LidM); E16.5 Epithelium (LGE, HGE, LidE); and W7 adult glands (LG, HG) (Supplementary Fig. S1b). The surgical technique to separate epithelial and mesenchymal tissues from whole gland cannot be applied to W7 LG because it has developed epithelial structure in the gland. The transcriptome data were also consistent with previously known gene expression patterns in epithelium: the expression of aquaporin5 (AQP5), a water channel in acinar cells, and LTF, a protein secreted from acinar cells, was higher in W7 LG than in E16.5 LGE (Fig. 1b); the expression of Krt15 and Barx2 was higher in E16.5 LGE than in W7 LG (Fig. 1c). Therefore, we referred the expression data from mature gland to investigate gene expression changes in epithelium. Consistent with the PCA results, hierarchical clustering analysis also showed that the transcriptome of the E16.5 LGE was similar to that of the E16.5 HGE and, to some extent, to that of LidE (Fig. 1d). On the other hand, the transcriptome of the W7 HG was most different from that of the E16.5 LGE (Fig. 1d). Of 134 genes expressed highly in E16.5 LGE, 16 genes were TFs based on gene ontology (GO) (Supplementary Table S1). For further analyses, we added PAX6 to the TF list, as it has been reported that PAX6 is indispensable for LG development.5 Indeed, Pax6 was highly expressed in E16.5 LGE (Supplementary Fig. S1c). To further narrow down the TF list, we used previously published transcriptome data of various organs ref. 21 and examined the expression of the 17 TFs (Fig. 1e). Because the transcriptome of LG was not available in this data set, we used the transcriptome of the salivary glands as a representative secretory gland. Among the 17 TFs, four TFs—Foxc1, Six1, Six2, and Ctdspl—were highly expressed in the salivary gland (Fig. 1e). The GO annotation analysis further narrowed down the list to three TFs: SIX1, SIX2, and FOXC1 (Fig. 1f, Supplementary Fig. S1d). After adding PAX6—a TF known to be indispensable for LG development to the list—we decided to further investigate PAX6, SIX1, SIX2, and FOXC1 as primary candidate TFs for generating LGE-like cells from human pluripotent stem cells.
Fig. 1.
Identification of tissue specific TFs for LGE. a Phase-contrast microscopic images of E16.5 mouse lacrimal gland (upper) and harderian gland (lower). Images of whole (left), separated epithelium (center), and separated mesenchyme (right) are shown. Scale bar 100 µm. b The gene expression comparison of mature LGE markers in the microarray analysis between E16.5 LGE and W7 LG. E16.5 embryonic day 16.5, LGE lacrimal gland epithelium, W7 LG 7-week-old mouse lacrimal gland. c The gene expression comparison of LGE markers in the microarray analysis between E16.5 LGE and W7 LG. E16.5 embryonic day 16.5, LGE lacrimal gland epithelium, W7 LG 7-week-old mouse lacrimal gland. d Hierarchical clustering analysis. The number of specific genes is shown. W7 LG 7-week-old mouse lacrimal gland, W7 HG 7-week-old mouse harderian gland, E16.5 embryonic day 16.5, LGE lacrimal gland epithelium, LGM lacrimal gland mesenchyme, HGE harderian gland epithelium, HGM harderian gland mesenchyme, LidE eyelid conjunctiva epithelium, LidM eyelid conjunctive mesenchyme. e Gene expression profiles of 16 TFs among 134 genes and PAX6 analyzed by web database tool. Foxc1, Six1, Six2, and Ctdspl express highly also in salivary glands, which are secretory glands similar to the lacrimal glands. Heat map (Green to red) represents a Z-score among organs. f Flowchart of an approach to identify premature lacrimal gland epithelium specific transcription factors
Establishment of culture and transfection conditions for generating LGE-like cells
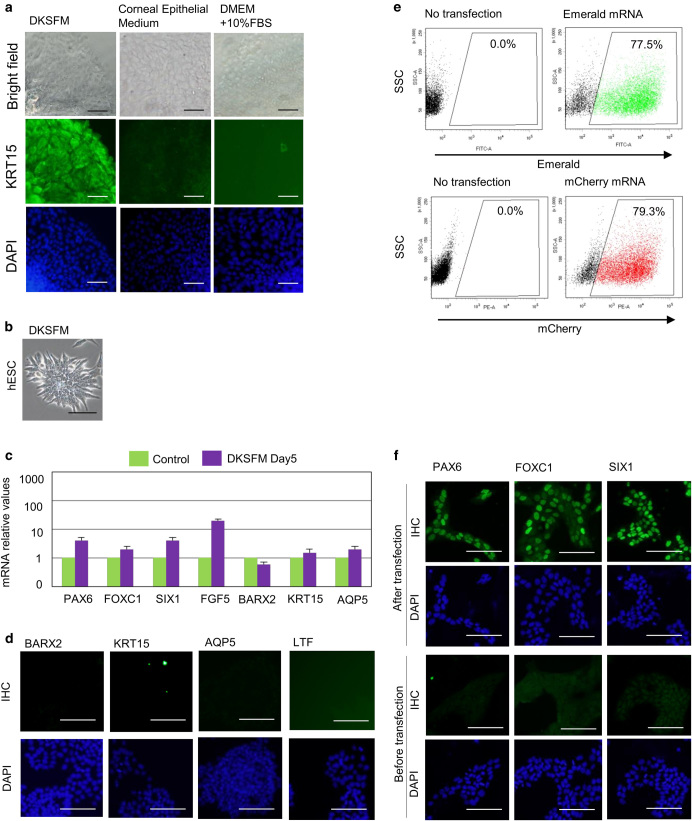
To find cell culture conditions suitable for LGE-like cells, we tested three types of cell culture media: DKSFM with CT, which has been reported as a suitable culture medium for primary mouse LGE cells;29 basal media with serum; and corneal epithelial media.30 Mouse E16.5 LGE cells survived in the DKSFM condition, but could not survive for more than five days (i.e., detached from the plate) in the basal media with serum and the corneal epithelial media. We examined the expression of KRT15 in cultured E16.5 LGE in these media. Consistent with the previous study,31 the presence of KRT15 was detected in the E16.5 LGE cultured in the DKSFM condition at culture Day 5, but was not detected in the other conditions (Fig. 2a). These results prompted us to test whether hESCs can be cultured in the DKSFM condition. We found that the DKSFM condition not only sustained the hESC culture, but also changed some hESCs into elongated morphology (Fig 2b). However, these morphological changes were not accompanied by the significant expression changes of LGE markers in either mRNA levels (Fig. 2c) or protein levels (Fig. 2d). These results suggest that the DKSFM condition alone cannot differentiate hESCs into LGE-like cells, but can support the culture. (Table 1)
Fig. 2.
Establishment of a culture condition for the overexpression of transcription factors. a Immunohistological analysis of mouse E16.5 LGE in DKSFM (left), corneal epithelial medium (center), and DMEM with serum (right). Scale bar 100 µm. b Phase-contrast images of hESCs in DKSFM at day 5. Scale bar 100 µm. c Relative mRNA expression profiles in hESCs after culture in DKSFM and the control cells that also grown for the same length of time (5 days) in basal media as the experimental cells. Error bars represent mean ± standard deviation (SD) of three samples. d Immunohistochemical analysis of the cultured hESCs in DKSFM with antibodies against BARX2, KRT15, AQP5, LTF at Day 5. Scale bar 100 µm. e The analysis of expression rate of GFP and mCherry proteins by modified mRNA transfection into hESCs using FACS analysis. f PAX6, FOXC1, and SIX1 expression in cells 8 h after transfection with the synthetic modified mRNAs
Table 1.
Candidates of specific transcription factors for mouse LGE identified by microarray analysis
| Gene symbol | Description | Fold-change |
|---|---|---|
| Six1 | Sine oculis-related homeobox 1 homolog | 26.977 |
| Six2 | Sine oculis-related homeobox 2 homolog | 13.273 |
| Foxc1 | Forkhead box C1 | 9.616 |
As a method to deliver TFs into cells, we used synthetic mRNAs prepared by in vitro transcription (Supplementary Fig. S2a) based on our previous experience with mouse ESCs.21 After optimizing the procedure as described in the methods section, we used the method to transfect hESCs with synthetic mRNAs four times during the first two days (Supplementary Fig. S2b). Two days after transfection with synthetic mRNAs of green fluorescent protein (GFP) and mCherry as controls, we observed fluorescence of both GFP and mCherry by fluorescent microscopy (Supplementary Fig. S2c). Cell sorting analysis revealed that 77.5% of hESCs transfected with GFP mRNAs were positive for GFP fluorescence and 79.3% of hESC transfected with mCherry mRNAs were positive for mCherry fluorescence (Fig. 2e). The same transfection procedure was successfully used to express TFs (Fig. 2f).
Overexpression of PAX6 and FOXC1 in hESCs induced LGE marker expression
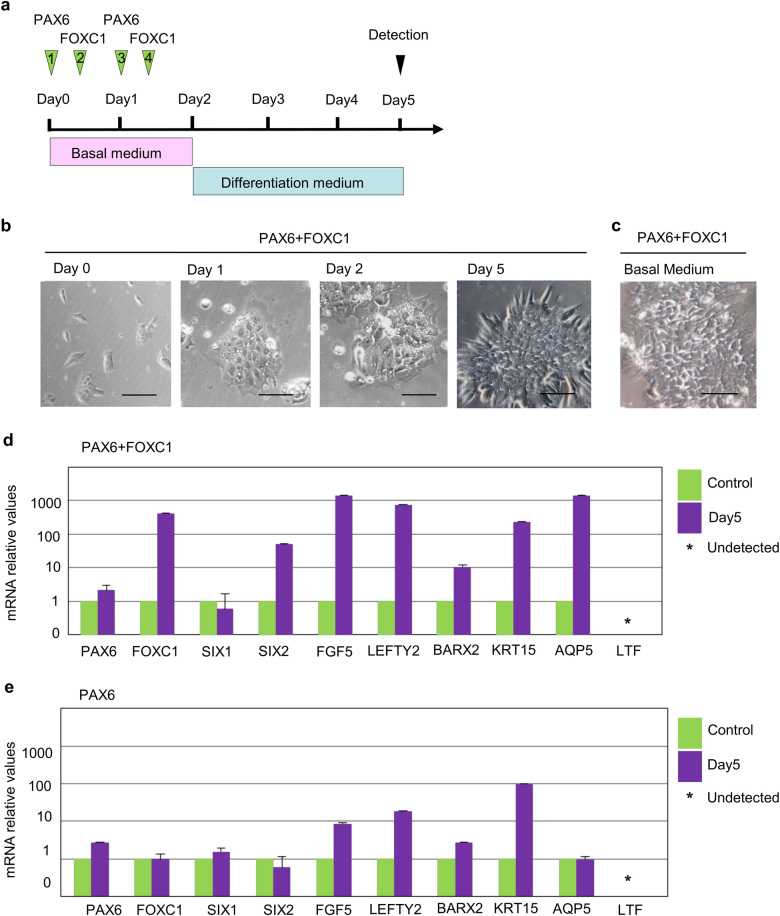
Next, we investigated whether the overexpression of candidate TFs promotes the differentiation of hESCs into LGE-like cells. We tested each—or their combinations of the synthetic mRNAs for PAX6, FOXC1, SIX1, and SIX2—and found that by Day 5 the morphology of hESCs transfected with PAX6 and FOXC1 changed into an elongated cell shape, which was especially conspicuous in the periphery of cell colonies (Fig. 3a, b). These morphological changes were not observed if transfected hESCs were kept in the standard hESC culture condition, which made these cells to grow to a confluency (Fig. 3c). It is known that BARX2 is involved in the branching of the ductal tree of LG during organogenesis in embryos and is, therefore, essential for the development of the LGs.6 Therefore, we analyzed the mRNA expression of LG-related factors including BARX2 and LGE markers, such as cytokeratin 15 (KRT15),32 AQP5, and LTF. Real-time reverse transcription-polymerase chain reaction (RT-PCR) analysis showed that co-transfection of PAX6 and FOXC1 induced the expression of endogenous FOXC1 gene and LG marker genes on Day 5, compared to the expression of these genes in control cells that grown for the same length of time (5 days) as the experimental cells (Fig. 3d). PAX6 alone and SIX2 alone or together with FOXC1 induced incomplete expression of some marker genes (Fig. 3e, Supplementary Figs. S2d, S2e, S3a, S3b, S3e). Other combinations of two TFs (PAX6+SIX1, FOXC1+SIX1, FOXC1+SIX2, PAX6+SIX2) did not induce significant changes in marker expression (Table 2, Supplementary Fig. S3a). These findings indicate that PAX6 and FOXC1 together may induce LGE-like cell differentiation.
Fig. 3.
The effect of PAX6 and FOXC1 overexpression in hESCs. a Schematic representation of time course of mRNA transfection of PAX6 and FOXC1. b Phase-contrast images of hESCs transfected with PAX6 and FOXC1 mRNAs. Scale bar 100 µm. c A phase-contrast image of PAX6 and FOXC1 transfected hESCs at Day 5 in basal medium. Scale bar 100 µm. d Relative mRNA expression levels of representative ectodermal markers (FGF5, LEFTY2), branching morphogenesis marker (BARX2), and lacrimal gland epithelial markers (KRT15, AQP5, and LTF) after transfecting PAX6 and FOXC1 mRNAs into hESCs. Quantitative PCR analyses of total RNA extracts on day 5, or the control cells also grown for the same length of time (5 days) as the experimental cells. Error bars represent mean ± SD of three samples. e Relative mRNA expression profiles of the developmental markers including lacrimal gland epithelial markers after PAX6 mRNA induction in hESCs on Day 5, or the control cells also grown for the same length of time (5 days) as the experimental cells. Error bars represent mean ± SD of three samples
Table 2.
Morphological changes of cells transfected with each transcription factor and their combinations in hESCs
| Combination of TF | Morphological change (Day 5) | mRNA expression of PAX6, BARX2, KRT15, AQP5, and LTF (Day 5) |
|---|---|---|
| None | Spindle | No |
| PAX6 | Spindle | No |
| FOXC1 | Smooth | No |
| SIX1 | Smooth | No |
| SIX2 | Round | BARX2a |
| PAX6 + FOXC1 | Elongate | BARX2, KRT15, AQP5 |
| PAX6 + SIX1 | Smooth/spindle | No |
| PAX6 + SIX2 | Round/spindle | No |
| FOXC1 + SIX1 | Spindle | No |
| FOXC1 + SIX2 | Elongate | BARX2, AQP5a |
| PAX6 + FOXC1 + SIX1 | Elongate | PAX6, BARX2, KRT15, AQP5, LTF |
| PAX6 + FOXC1 + SIX2 | Spindle/elongate | BARX2a |
aThe details of partial expression of lacrimal gland-related markers are shown in Supplementary information
SIX1 accelerated the expression of LGE markers
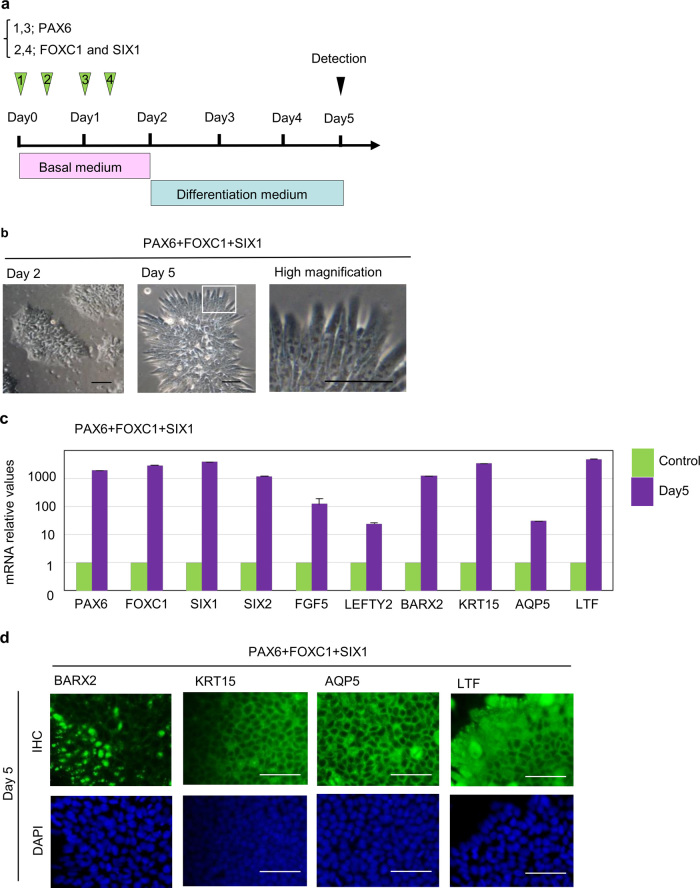
Next, we tested combinations of three TFs among candidate TFs (PAX6+FOXC1+SIX1 or PAX6+FOXC1+SIX2). On day 5 after transfection with a combination of PAX6, FOXC1, and SIX1, we observed significant morphological changes in differentiated cells, compared to those transfected with a combination of only two TFs, PAX6 and FOXC1 (Fig. 4a, b). RT-PCR analysis revealed that the expression of LG-related markers including endogenous TFs such as PAX6, FOXC1, SIX1, and SIX2, branching morphogenesis factor BARX2, AQP5, and LTF further increased by Day 5 by a combination of PAX6, FOXC1, and SIX1 compared to the expression of these genes in the control cells that grown for the same length of time (5 days) as the experimental cells (Fig. 4c). The expression of endogenous SIX2 gene increased by the overexpression of PAX6, FOXC1, and SIX1. Furthermore, immunohistochemical analysis revealed that these differentiated cells expressed LG-related markers such as AQP5 and LTF (Fig. 4d). By contrast, a combination of PAX6, FOXC1, and SIX2 could not induce differentiation by Day 5 (Table 2, Supplementary Fig. S3c, S3d). These results suggest that SIX2 is a downstream gene for SIX1 and may not be sufficient for LGE-like cell differentiation even together with PAX6 and FOXC1. Taken together, these findings indicate that only a combination of PAX6, FOXC1, and SIX1 among the TF combinations examined in this study promoted the differentiation of hESCs toward LGE-like cells.
Fig. 4.
The effect of overexpression of the combination of PAX6, FOXC1, and SIX1. a Schematic representation of mRNA transfection procedure for PAX6, FOXC1, and SIX1. b Phase-contrast images of hESCs transfected with a combination of three TFs: PAX6, FOXC1, and SIX1 at Day 2 (left) and Day 5 (center), and an enlarged image of the boxed area in the center panel (right). Scale bar 100 µm. c The expression profiles of representative markers after PAX6, FOXC1, and SIX1 induction in hESCs. Quantitative PCR analysis of total RNA extracts at Day 5, or the control cells also grown for the same length of time (5 days) as the experimental cells. Error bars represent mean ± SD of three samples. d Immunohistochemical analysis of differentiated cells with antibodies against BARX2, KRT15, AQP5, and LTF at day 5. The control cells also grown for the same length of time (5 days) as the experimental cells. Scale bar 100 µm
Discussion
In this study, we have analyzed the gene expression profiles of E16.5 LGE and other developmentally related organs, and identified three TFs—PAX6, FOXC1, and SIX1—for their involvement in LGE development. We have further demonstrated that the overexpression of these TFs in hESCs in the form of synthetic mRNAs can rapidly induce differentiation of hESCs into LGE-like cells. Overall, our study thus demonstrates a possible method for the induction of LGE cells from human pluripotent stem cells.
All organs, including LG, are organized into a complex 3D structure, which is gradually formed during embryonic development.33 The LGE cells, which exist only in embryonic stages, have the ability to differentiate into various cells in the mature branching structure of LGs including acini and duct.1, 16 Recently, LG organ regeneration has been demonstrated using LGE cells to treat DED in mouse models.17 These results suggest a possible therapeutic strategy for the human dry eye condition by differentiating human pluripotent stem cells into LGE cells and transplanting them to patients. TFs identified in this study have successfully induced not only AQP5, LTF, and LGE cell markers such as KRT15, but also branching morphogenesis markers, which are usually expressed only in the branching period during organogenesis in embryos.6, 34 These results suggest that the TFs are involved in branching development of LGs, and the differentiated cells have similar characteristics with the LGE cells. Further optimization of the differentiation conditions by adding various growth factors and by extending culture duration may help to elucidate the mechanism of maturation in LG epithelial cells.
In developmental biology, it has been well established that multiple TFs generally work in a cascade-like manner and development proceeds in a stepwise manner.21, 35–37 Accordingly, we expected stepwise differentiation from hESCs to the primary ectoderm, to ocular surface ectoderm, and then to LGE, each of which can be mediated by specific TFs. Forced expression of lineage-specific TFs has demonstrated direct conversion of stem cells or fibroblasts into other desired cell lineages such as neurons, myocardial cells, and hepatocytes.25, 26, 38–40 In the case of LG regeneration, we focused on LGE, not mature acinar cells, because the LGE can develop into a functional 3D secretory structure by transplantation in vivo.16 In this study, we have shown that only simultaneous overexpression of three TFs, but not a single TF, directly differentiates hESCs into LGE-like cells in ectodermal differentiation media, DKSFM. The development of LGs including the branching process has been reported as a result of reciprocal epithelial and mesenchymal interactions, which involve various genes and molecules such as PAX6, SIX1, FOXC1, BARX2, and growth factors.5, 6, 41–43 Our results suggest that the three TFs play an important role in the signaling network for expression of LG-related markers during LG development. To our knowledge, this is the first demonstration of the induction of LGE-like cells from human cells.
Technologies including genome engineering or viral transduction have previously been reported to overexpress TFs in cells.5, 21, 44, 45 Because these DNA-based methods have the potential to cause insertional mutagenesis, there is a pressing need for safe and efficient methodology without genome editing to redirect cell fate, particularly for clinical application.46 Synthetic mRNA-based technology has recently been proposed as a novel strategy that enables highly efficient reprogramming or differentiation of pluripotent stem cells toward desired cell lineages.47 The transient and non-mutagenic features of a RNA-based protein expression system may be particularly beneficial for clinical applications.45 In our optimized protocol, the synthetic mRNAs encoding three TFs were divided into two groups, each of which was introduced to cells separately within a day for two consecutive days. PAX6 was introduced as the first group, because PAX6 is a well-known factor indispensable for LG development.5 The other procedure we tried did not work well: for example, when hESCs were transfected with three TFs at the same time, we failed to induce exogenous gene expression, most likely due to cell toxicity. Further studies are necessary to realize an efficient transfection procedure with less toxicity for stable expression of more kinds of TFs. Our results may encourage the strategy of rapid differentiation of the pluripotent stem cells to LGE-like cells without compromising genomic integrity.
In conclusion, the current study has revealed the gene expression profiles of LG tissues, and identified specific combination of TFs for differentiation of hESCs into LGE-like cells in vitro by mRNA-based technology. These findings will be the first step for organogenesis for future functional bioengineered organ replacement therapy for DED.
Methods
Ethical statement
The Ethics Committee of Keio University approved all experimental protocols (approval in October 2012). All experiments handling human cells and tissues were carried out in accordance with the Tenets of the Declaration of Helsinki. C57BL/6 mice were purchased from CLEA Japan Inc. (Shizuoka, Japan). The hESC line (SEES3) was obtained from the National Center for Child Health and Development.48 The care and handling of the animals were performed in accordance with NIH guidelines.
Sample preparation for microarray analysis
Adult LG and harderian glands (HG) were obtained from 7-week-old (W7) mice. Embryonic LG, HG, and eyelid were obtained from embryonic day 16.5 (E16.5) mice and treated with 50 U/ml of dispase (BD, Franklin Lakes, NJ, USA) for 1.5 min at room temperature as previously described.16, 49 Then, epithelial cells (LGE, HGE, and LidE) and mesenchymal cells (LGM, HGM, and LidM) were separated by microsurgery using needles.
Real-time reverse transcription polymerase chain reaction (RT-PCR)
Total ribonucleic acid (RNA) was extracted from cells using an RNeasy Mini Kit (QIAGEN, Hilden, Germany). The expression levels of the messenger RNAs (mRNAs) in each RNA sample were determined using the Thermal Cycler Dice Real Time System (Takara Bio Inc., Otsu, Japan). RT-PCR was performed using a One Step SYBR PrimeScript PLUS RT-PCR Kit (Takara Bio Inc.). The conditions were as follows: initial hold at 42 °C for 5 min, incubation at 95 °C for 10 s, and then 50 cycles at 95 °C for 5 s and 60 °C for 31 s. The expression of mRNA was assessed by evaluating threshold cycle (CT) values. The control cells also grown for the same length of time (5 days) as the experimental cells. The CT values were normalized by the expression levels of glyceraldehyde-3-phosphate dehydrogenase, and the relative amount of mRNA specific to each of the target genes was calculated (n = 3 in each experiment).
Microarray data analysis
Total RNA was extracted with RNeasy Mini Kit (QIAGEN). The microarray was performed using Agilent 028005, Sure Print G3, Mouse GE 8×60k, 1 color arrays. Microarray data was analyzed in duplicate in adult LG and triplicate in other samples from independent samples using the NIH NIA array analysis tool ref. 31 (http://lgsun.grc.nia.nih.gov/ANOVA/) and Genome Network Platform Viewer from the National Institute of Genetics in Japan (http://genomenetwork.nig.ac.jp/). The analyses were done for 31,156 probes with the error variance model: threshold z-value to remove outliers = 8, proportion of highest variance values = 0.01, desirable degrees of freedom for Bayesian error model = 10, FDR threshold = 0.05, 0 of number of permutations for building an empirical F-distribution. Differential gene expression was defined using the statistics/threshold combination. Hierarchical clustering was used for analysis of specific gene expression with minimum fold change as 10.
Cell culture and differentiation
Human ESC lines (SEES3, passage No. 17) were maintained on feeder-free, laminin-coated dishes in StemFit AK-03 medium (Ajinomoto, Tokyo, Japan). Defined Keratinocyte Serum Free Medium (DKSFM; Life Technologies, Carlsbad, CA, USA) and corneal epithelial media ref. 30 were used for differentiation. The following factors were added to DKSFM: human recombinant EGF (10 ng/mL; Peprotech, Rocky Hill, NJ, USA) and cholera toxin (CT) (100 μg/mL; Funakoshi Co., Tokyo, Japan) as previously reported.29 Differentiated cells were examined by immunostaining.
In vitro synthesis of modified mRNAs and transfection procedure
In vitro mRNA synthesis and transfections were performed according to previous protocols.46 In brief, T7 promoter and polyA tail were added by polymerase chain reaction (PCR) using KAPA taq kit (Kapabiosystems, London, UK). RNA was transcribed from the template using MEGAscript T7 kit (Ambion, Carlsbad, CA, USA), with ARCA cap analog (New England Biolabs, Ipswich, MA, USA); ATP; GTP; 5-Methyl-CTP (TriLink, San Diego, CA, USA); and pseudo-UTP (TriLink). Synthesized RNAs were purified with the MEGAclear kit (Ambion). RNA transfections were performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. To increase the viability of transfected cells, B18R interferon inhibitor (eBioscience, San Diego, CA, USA) was supplemented to the culture medium. One day before transfection, 30,000 hESCs were seeded on a culture plate, and 1 µg/well of each synthetic modified mRNA was induced. The hESCs were subjected to four consecutive transfections with TF-encoding RNAs, or GFP or mCherry mRNAs as controls with Lipofectamine 2000 (Life Technologies) in StemFit AK-03 with B18R (eBioscience) for the first two days of differentiation. After two consecutive days of transfection, the culture medium was changed to DKSFM with 100 μg/mL CT and 10 ng/mL EGF.
Immunohistochemistry
Immunostaining was carried out using whole cells fixed on the dish as described previously.21 In short, cells were fixed with 4% paraformaldehyde (pH 7.0) in phosphate buffered saline (PBS) for 20 min at room temperature. After two rinses with PBS, cells were incubated with 0.1% Triton X-100 in PBS for 15 min at room temperature and then washed three times with PBS for 5 min each. Cells were then incubated with 10% bovine serum albumin in PBS for 30 min at room temperature followed by primary antibody incubation for 16 h at 4 °C. Primary antibodies used were as follows: rabbit anti-PAX6 (1:200; Abcam, Cambridge, UK); rabbit anti-FOXC1 (1:200; Abcam); rabbit anti-SIX1 (1:200; Sigma-Aldrich, St. Louis, MO, USA); rabbit anti-cytokeratin15 (1:200; Proteintech, Chicago, IL, USA); rabbit anti-AQP5 (1:200; Abcam); rabbit anti-Lactoferrin (1:500; Abcam); and goat anti-BARX2 (1:500; Santa Cruz Biotechnology, Dallas, TX, USA). Secondary antibody reactions were carried out by an incubation with corresponding species-specific Alexa Fluor-488-conjugated antibodies (1:500; Life Technologies) for one hour at room temperature in the dark. After four washes with PBS for 5 min each, specimens were nuclear-stained with DAPI (Life Technologies) and observed with an IX73 inverted microscope (Olympus, Tokyo, Japan). The control cells also grown for the same length of time (5 days) as the experimental cells.
Electronic supplementary material
Acknowledgements
We thank Keio-Med Open Access Facility, Keio University; Developmental Genomics and Aging Section, Laboratory of Genetics, National Institute on Aging, National Institute of Health, USA; and National Institute of Genetics in Japan for web analysis of microarray data. Gene expression profiles used in this study were deposited in NCBI’s Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/, accession number GSE71867). We thank Akihiro Ko and Vanessa Harrington for critical reading of the manuscript. This work was in part supported by the Keio University Medical Science Fund—The Mitsunada Sakaguchi Laboratory, the CREST program from the Japan Science and Technology Agency (JST), and the Research Center Network for Realization of Regenerative Medicine, Japan Agency for Medical Research and Development (AMED). This research was supported by a Grant-in-Aid for Research Activity Start-up to M.H. and in part by Grant-in-Aid for Scientific Research (KAKENHI), Japan Society for the Promotion of Science (JSPS), to S.K. and M.K. M.H. received a Research Grant for Life Sciences and Medicine from Sakaguchi Memorial Keio University Medical Science Fund. This research was also supported in part by CREST and Strategic International Collaborative Research Program (SICORP), Japan Science and Technology Agency, Research Center Network for Realization of Regenerative Medicine, Japan Agency for Medical Research and Development, and the Keio University Medical Science Fund—The Sakaguchi Laboratory.
Author contributions
M.K., K.T., T.K., S.K., and M.H. designed the research plan; M.H., T.A., N.N., A.S., and M.S. performed the experiments; M.H., S.G., T.A., Y.N., S.S., and S.K. developed new assay methods and discussed the results; M.H. and T.A. analyzed the data; and M.H., S.K., and M.K. wrote the paper.
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information accompanies the paper on the npj Aging and Mechanisms of Disease website (doi:10.1038/s41514-016-0001-8).
References
- 1.Schechter JE, Warren DW, Mircheff AK. A lacrimal gland is a lacrimal gland, but rodent's and rabbit’s are not human. Ocul. Surf. 2010;8:111–134. doi: 10.1016/S1542-0124(12)70222-7. [DOI] [PubMed] [Google Scholar]
- 2.Mishima S. Some physiological aspects of the precorneal tear film. Arch. Ophthalmol. 1965;73:233–241. doi: 10.1001/archopht.1965.00970030235017. [DOI] [PubMed] [Google Scholar]
- 3.Sakai T. Epithelial branching morphogenesis of salivary gland: exploration of new functional regulators. J. Med. Invest. 2009;56:234–238. doi: 10.2152/jmi.56.234. [DOI] [PubMed] [Google Scholar]
- 4.Sakai T, Larsen M, Yamada KM. Fibronectin requirement in branching morphogenesis. Nature. 2003;423:876–881. doi: 10.1038/nature01712. [DOI] [PubMed] [Google Scholar]
- 5.Makarenkova HP, et al. FGF10 is an inducer and Pax6 a competence factor for lacrimal gland development. Development. 2000;127:2563–2572. doi: 10.1242/dev.127.12.2563. [DOI] [PubMed] [Google Scholar]
- 6.Tsau C, et al. Barx2 and Fgf10 regulate ocular glands branching morphogenesis by controlling extracellular matrix remodeling. Development. 2011;138:3307–3317. doi: 10.1242/dev.066241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohashi Y, Dogru M, Tsubota K. Laboratory findings in tear fluid analysis. Clin. Chim. Acta. 2006;369:17–28. doi: 10.1016/j.cca.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 8.Seal DV, et al. Bacteriology and tear protein profiles of the dry eye. Br. J. Ophthalmol. 1986;70:122–125. doi: 10.1136/bjo.70.2.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemp MA. Tear film: new concepts and implications for the management of the dry eye. Trans. New Orleans Acad. Ophthalmol. 1987;35:53–64. [PubMed] [Google Scholar]
- 10.Cui X, et al. Assessment of corneal epithelial thickness in dry eye patients. Optom. Vis. Sci. 2014;91:1446–1454. doi: 10.1097/OPX.0000000000000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schein OD, Munoz B, Tielsch JM, Bandeen-Roche K, West S. Prevalence of dry eye among the elderly. Am. J. Ophthalmol. 1997;124:723–728. doi: 10.1016/S0002-9394(14)71688-5. [DOI] [PubMed] [Google Scholar]
- 12.Friedman NJ. Impact of dry eye disease and treatment on quality of life. Curr. Opin. Ophthalmol. 2010;21:310–316. doi: 10.1097/ICU.0b013e32833a8c15. [DOI] [PubMed] [Google Scholar]
- 13.Tsubota K, Satake Y, Shimazaki J. Treatment of severe dry eye. Lancet. 1996;348:123. doi: 10.1016/S0140-6736(96)24028-0. [DOI] [PubMed] [Google Scholar]
- 14.Shimmura S, et al. Albumin as a tear supplement in the treatment of severe dry eye. Br. J. Ophthalmol. 2003;87:1279–1283. doi: 10.1136/bjo.87.10.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kojima T, et al. The effect of autologous serum eyedrops in the treatment of severe dry eye disease: a prospective randomized case–control study. Am. J. Ophthalmol. 2005;139:242–246. doi: 10.1016/j.ajo.2004.08.040. [DOI] [PubMed] [Google Scholar]
- 16.Hirayama M, et al. Functional lacrimal gland regeneration by transplantation of a bioengineered organ germ. Nat. Commun. 2013;4:2497. doi: 10.1038/ncomms3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirayama M, Tsubota K, Tsuji T. Bioengineered lacrimal gland organ regeneration in vivo. J. Funct. Biomater. 2015;6:634–649. doi: 10.3390/jfb6030634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirayama M., Kawakita T., Tsubota K. & Shimmura S. Challenges and strategies for regenerating the lacrimal gland. Ocul. Surf.14, 135–143 (2015). [DOI] [PubMed]
- 19.Hirayama M, Oshima M, Tsuji T. Development and prospects of organ replacement regenerative therapy. Cornea. 2013;32:S13–S21. doi: 10.1097/ICO.0b013e3182a18e6c. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 21.Yamamizu K, et al. Identification of transcription factors for lineage-specific ESC differentiation. Stem Cell Rep. 2013;1:545–559. doi: 10.1016/j.stemcr.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-X. [DOI] [PubMed] [Google Scholar]
- 23.Pang ZP, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Son EY, et al. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ieda M, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 28.Payne AP. The harderian gland: a tercentennial review. J. Anat. 1994;185:1–49. [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi S, et al. Characterization of cultivated murine lacrimal gland epithelial cells. Mol. Vis. 2012;18:1271–1277. [PMC free article] [PubMed] [Google Scholar]
- 30.Miyashita H, et al. Long-term maintenance of limbal epithelial progenitor cells using rho kinase inhibitor and keratinocyte growth factor. Stem Cells Transl. Med. 2013;2:758–765. doi: 10.5966/sctm.2012-0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharov AA, Dudekula DB, Ko MS. A web-based tool for principal component and significance analysis of microarray data. Bioinformatics. 2005;21:2548–2549. doi: 10.1093/bioinformatics/bti343. [DOI] [PubMed] [Google Scholar]
- 32.Hirayama M, Liu Y, Kawakita T, Shimmura S, Tsubota K. Cytokeratin expression in mouse lacrimal gland germ epithelium. Exp. Eye Res. 2015;146:54–59. doi: 10.1016/j.exer.2015.11.020. [DOI] [PubMed] [Google Scholar]
- 33.Pispa J, Thesleff I. Mechanisms of ectodermal organogenesis. Dev. Biol. 2003;262:195–205. doi: 10.1016/S0012-1606(03)00325-7. [DOI] [PubMed] [Google Scholar]
- 34.Hirayama M. Proof of a concept for bioengineered organ replacement to restore lacrimal gland function. Nippon Ganka Gakkai Zasshi. 2015;119:799–806. [PubMed] [Google Scholar]
- 35.Snykers S, De Kock J, Rogiers V, Vanhaecke T. In vitro differentiation of embryonic and adult stem cells into hepatocytes: state of the art. Stem Cells. 2009;27:577–605. doi: 10.1634/stemcells.2008-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 37.Zaret KS, Grompe M. Generation and regeneration of cells of the liver and pancreas. Science. 2008;322:1490–1494. doi: 10.1126/science.1161431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caiazzo M, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 39.Efe JA, et al. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat. Cell. Biol. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 40.Huang P, et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 41.Dean C, Ito M, Makarenkova HP, Faber SC, Lang RA. Bmp7 regulates branching morphogenesis of the lacrimal gland by promoting mesenchymal proliferation and condensation. Development. 2004;131:4155–4165. doi: 10.1242/dev.01285. [DOI] [PubMed] [Google Scholar]
- 42.Mattiske D, Sommer P, Kidson SH, Hogan BL. The role of the forkhead transcription factor, Foxc1, in the development of the mouse lacrimal gland. Dev. Dyn. 2006;235:1074–1080. doi: 10.1002/dvdy.20702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Z, et al. FGF signaling activates a Sox9-Sox10 pathway for the formation and branching morphogenesis of mouse ocular glands. Development. 2014;141:2691–2701. doi: 10.1242/dev.108944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 45.Li M, Sancho-Martinez I, Izpisua Belmonte JC. Cell fate conversion by mRNA. Stem Cell Res. Ther. 2011;2:5. doi: 10.1186/scrt46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mandal PK, Rossi DJ. Reprogramming human fibroblasts to pluripotency using modified mRNA. Nat. Protoc. 2013;8:568–582. doi: 10.1038/nprot.2013.019. [DOI] [PubMed] [Google Scholar]
- 47.Warren L, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akutsu H, et al. Xenogeneic-free defined conditions for derivation and expansion of human embryonic stem cells with mesenchymal stem cells. Regen. Ther. 2015;1:18–29. doi: 10.1016/j.reth.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ogawa M, et al. Functional salivary gland regeneration by transplantation of a bioengineered organ germ. Nat. Commun. 2013;4:2498. doi: 10.1038/ncomms3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.