Abstract
Entamoeba histolytica infection remains a public health concern in developing countries. Early diagnosis of amoebiasis can avoid disease complications, thus this study was aimed at developing a test that can rapidly detect the parasite antigens in stool samples. Rabbits were individually immunized with recombinant pyruvate phosphate dikinase (rPPDK) and E. histolytica excretory-secretory antigens to produce polyclonal antibodies. A rapid dipstick test was produced using anti-rPPDK PAb lined on the dipstick as capture reagent and anti-EhESA PAb conjugated to colloidal gold as the detector reagent. Using E. histolytica-spiked in stool sample of a healthy individual, the detection limit of the dipstick test was found to be 1000 cells ml−1. Meanwhile when rPPDK was spiked in the stool sample, the minimum concentration detected by the dipstick test was 0.1 μg ml−1. The performances of the dipstick, commercial Techlab E. histolytica II enzyme-linked immunosorbent assays (ELISA) and real-time PCR were compared using 70 stool samples from patients infected with Entamoeba species (n = 45) and other intestinal pathogens (n = 25). When compared to real-time PCR, the diagnostic sensitivity of the dipstick for detection of E. histolytica was 65.4% (n = 17/26); while the diagnostic specificity when tested with stool samples containing other intestinal pathogens was 92% (23/25). In contrast, Techlab E. histolytica II ELISA detected 19.2% (5/26) of the E. histolytica-positive samples as compared to real-time PCR. The lateral flow dipstick test produced in this study enabled rapid detection of E. histolytica, thus it showed good potential to be further developed into a diagnostic tool for intestinal amoebiasis.
Keywords: Entamoeba histolytica, stool antigen detection, pyruvate phosphate dikinase, excretory-secretory antigens, lateral flow rapid test
Introduction
Amoebiasis is caused by Entamoeba histolytica and is one of the most common parasitic infections worldwide. It was estimated that about 50 million cases of symptomatic amoebiasis and amoebic liver abscess (ALA) and up to 100,000 deaths occur annually [1]; however improvements in socio-economic conditions and health facilities in many parts of the world may necessitate re-evaluation of the prevalence. The diagnosis of intestinal E. histolytica infection has traditionally relied upon microscopic examination of fresh or fixed stool specimens [2]. However, it is often misleading due to morphological similarities between E. histolytica and the non-pathogenic species such as E. dispar, E. moshkovskii and E. bangladeshi [3,4]. It is important to correctly diagnose amoebiasis patients to reduce the morbidity and mortality, and to minimize unnecessary treatment of individuals who harbored non-pathogenic species in their stool samples. Isoenzyme analysis of E. histolytica culture has been used to differentiate E. histolytica from other non-pathogenic species, however, this method is not widely available and not practical for routine diagnosis [2,5].
Several newer diagnostic tests such as enzyme-linked immunosorbent assays (ELISAs), rapid immunochromatographic assays and DNA based methods have been developed to detect amoebic antigens in stool [6−10]. The available antigen detection assays vary in their sensitivities and specificities, and many cannot reliably distinguish between E. histolytica and E. dispar [11]. PCR-based assays have been reported to demonstrate excellent diagnostic sensitivity and specificity when compared to microscopy in the diagnosis of amoebiasis [2,3,12]. In other evaluation studies, similar diagnostic sensitivity and specificity were reported for PCR and ELISA [6,13]. Nevertheless, PCR-based assays are not widely employed and remain impractical in many developing and underdeveloped countries [2,4,14].
Therefore a simple, rapid, sensitive and specific antigen detection test that can be transported at room temperature is needed for diagnosis of intestinal amoebiasis. Towards achieving this aim, the present study was aimed at developing a lateral flow dipstick test for the detection of E. histolytica antigen in stool sample.
Materials and methods
Stool samples
A total of 70 stool samples were used, which previously had been examined by microscopy. They were from the laboratories of the co-authors: (1) Department of Microbiology and Parasitology, School of Medical Sciences, USM (n = 23) and; (2) Department of Parasitology, Faculty of Medicine, University of Malaya (n = 47). The use of the stool samples were approved by the Human Research Ethics Committees of the respective institutions. Most of the samples were formed stools, and stored either in 2.5% potassium dichromate at 4 °C (n = 36), or frozen at −20 °C (n = 34). From the 70 samples, microscopic examination showed that 45 were positive for Entamoeba spp. with single infection (n = 18) and Entamoeba spp. with multiple infection (n = 27), the latter comprised co-infections with Ascaris lumbricoides (n = 6), Giardia lamblia (n = 2), Trichuris trichiura (n = 5), A. lumbricoides and T. trichiura (n = 6), G. lamblia and T. trichiura (n = 6), G. lamblia and A. lumbricoides (n = 1), G. lamblia, A. lumbricoides and T. trichiura (n = 1). In addition, 25 stool samples were positive with other intestinal pathogens: Necator americanus (n = 3), Ancylostoma spp. (n = 3), Strongyloides stercoralis (n = 1), Clostridium spp. (n = 1), Salmonella spp. (n = 4), Shigella spp. (n = 4), T. trichiura (n = 1), Campylobacter spp. (n = 1), enteropathogenic E. coli (EPEC) (n = 1), rotavirus (n = 1), adenovirus (n = 1), Aeromonas hydrophilia (n = 1); and multiple infections of A. hydrophilia and Shigella flexnari (n = 1), T. trichiura and Giardia spp. (n = 1), A. lumbricoides, Entamoeba coli and T. trichiura (n = 1).
Detection of E. histolytica and E. dispar in stool samples by real-time polymerase chain reaction
During genomic extraction, InhibitEX tablet (Qiagen, Hilden, Germany) was added to absorb DNA-damaging substances and PCR inhibitors in the stool sample. Real-time PCR was performed according to our previous protocol [15]. Each amplification reaction was performed in a total volume of 25 μl with 12.5 μl HotStarTaq Master Mix (Qiagen), 5 mg ml−1 MgCl2 (Fermentas, MA), 0.1 mg ml−1 bovine serum albumin (BSA) [Sigma, MO], 10 μM each Ehd-239F and Ehd-88R primers, 0.25 μM E. histolytica-specific MGB-Taqman probe/0.25 μM E. dispar-specific MGB-Taqman probe and 2.5 μl of DNA templates. The BSA was added to the master mix to reduce PCR inhibition and to improve the specificity of PCR. Table 1 shows the sequences of primers and probes used to detect E. histolytica and E. dispar in this study. The amplification parameters were as follows: 95 °C for 15 min, followed by 40 cycles of 95 °C for 9 seconds and 60 °C for 1 min. Amplification detection and data analysis were performed using the Applied Biosystems 7500/7500 Fast Real-Time PCR System (Applied Biosystems, CA). Fluorescence was measured during the annealing step of each cycle. For each PCR run, two types of control reactions were included i.e. two positive controls namely E. histolytica genomic DNA extracted from trophozoites cultured in TYI-S-33 media (supplemented with 12.5% bovine serum) and E. dispar plasmid DNA; and a negative control comprising PCR mixture without DNA template i.e. non-template control. The latter ruled out the possibility of contamination being as a cause of false positive results.
Table 1.
Primers and probes for the DNA detection of E. histolytica and E. dispar.
| Primers and Probes ID | Sequences (5′-3′) | Target |
|---|---|---|
| Ehd-239F | 5′ATTGTCGTGGCATCCTAACTCA 3′ | E. histolytica |
| Ehd-88R | 5′GCGGACGGCTCATTATAACA 3′ | |
| Histolytica 96T | VIC 5 TCATTGAATGAATTGGCCATTT 3 NFQ | |
| Ehd-239F | 5′ATTGTCGTGGCATCCTAACTCA 3′ | E. dispar |
| Ehd-88R | 5′GCGGACGGCTCATTATAACA 3′ | |
| E. dispar | FAM 5′TTACTTACAATAAATTGGCCACTTTG3′ MGB |
Detection of E. histolytica in stool samples by antigen detection test
The Techlab E. histolytica II ELISA antigen detection test (Techlab, VA) was used to detect E. histolytica in the stool samples. The test detects the amoebic Gal/GalNAc-specific adherence lectin and was performed according to the manufacturer’s instructions.
Production and purification of polyclonal antibodies
Recombinant PPDK (rPPDK) protein was expressed and purified according to our previous report [16]. Meanwhile E. histolytica excretory-secretory antigens (EhESA) was produced using the method we have described earlier [17]. New Zealand white rabbits (Oryctolagus cuniculus; female, 2.8–3.0 kg, 11–13 weeks old) were used for immunizations, with electro-eluted rPPDK, or E. histolytica ESA. On the first day of the immunization, 1 mg ml−1 of each antigen was mixed with Freund’s complete adjuvant (Sigma, MO). Subsequent immunizations with the similar dosages of the antigens were each mixed with incomplete Freund’s adjuvant (Sigma), and performed on the 21st and 42nd days. On the 60th day, the rabbits were bled by cardiac puncture and the serum samples were collected. The rPPDK and EhESA-antisera were stored in small aliquots at −20 °C. The use of rabbits in this study was approved by the Animal Research Ethics Committee at USM (ref. no: USM/Animal Ethics Approval/2012/(84)(456)). Purified polyclonal IgGs to rPPDK and EhESA were produced using Melon IgG Spin Purification Kit (Thermo Scientific, MA) according to the manufacturer’s instructions.
SDS-PAGE and Western blot
EhESA and rPPDK were separately resolved on SDS-PAGE gel. The protein was then transferred onto a nitrocellulose membrane (Bio-Rad, CA) and blocked with TBS (150 mM NaCl, 50 mM Tris–HCl, pH 7.5) containing 0.05% Tween 20 (TBS-T) and 5% skim milk at room temperature for 1 h. Subsequently, the membrane was incubated with 1:5,000 dilution of purified rabbit antiserum (anti-rPPDK IgG or anti-EhESA IgG) overnight, followed by three times of TBS-T wash. The blot was then incubated with 1:10,000 dilution of goat anti-rabbit IgG conjugated to horseradish peroxidase (Thermo Scientific) at room temperature for 1 h. The nitrocellulose membrane was developed using enhanced chemiluminescence (ECL) substrate system (Thermo Scientific) and the image captured on X-ray film (Thermo Scientific).
SDS-PAGE and Western blot were also performed on E. histolytica lysate (40 μg per lane), using anti-rPPDK IgG as primary antibody (1:5,000) and the same secondary antibody as above. For all Western blots, a nitrocellulose membrane strip incubated with pre-immune rabbit anti-serum as primary antibody was used as control.
Preparation of the lateral flow dipstick test
Hi-flow Plus 90 membrane card (301 mm length) with nitrocellulose membrane flow rate of 90 ± 23 sec/4 cms (Millipore, MA) was used to make the lateral flow rapid dipstick test. Diagnostic sensitivity and specificity of the rapid test was determined using dipstick which had two lines i.e. test (bottom) and control (top) lines. The test line comprised anti-rPPDK IgG and the control line comprised polyclonal goat anti-rabbit IgG (Thermo Scientific) at 0.2 mg ml−1. The lines were jetted linearly onto the membrane card (0.1 μl per mm) using IsoFlow™ Dispenser (Imagene Technology, CA). After the lined reagents have dried, blocking solution (Roche Diagnostics, Mannheim, Germany) was pipetted on the nitrocellulose membrane section of the card. After overnight drying, the top sticker portion of the card was removed and an absorbent pad was attached in its place, while ensuring a 2 mm overlap with the nitrocellulose membrane. The membrane card with the attached absorbent pad was cut into 5 mm strips using Index Cutter-I (A-Point Technologies, NJ), then stored in a dry cabinet at the room temperature.
For optimization experiments and to determine the limit of detection, a dipstick dot test was used. This was made by first cutting the membrane card with attached absorbent pad into strips, then 1 μl of purified rabbit anti-rPPDK IgG was dotted, one dot per dipstick. After drying, the dipstick was blocked, then dried again. Another component of the dipstick test was the anti-EhESA IgG conjugated to colloidal gold (anti-EhESA IgG–gold), this was prepared as described in our previous report [18].
Test procedure
For sample preparation, 0.15 to 0.20 g of solid stool or 400 μl of liquid stool sample was suspended in 400 μl phosphate-buffered saline (PBS, pH 7.2) with 0.05% Tween 20. The mixture was mixed well and briefly centrifuged at 6000×g for 1 min. Five μl of anti-EhESA IgG-gold with the optical density (OD) of 4 was added to 20 μl of the diluted stool in a well of a microtiter plate. The dipstick was then dipped into the well. After the line (s) was well-developed, the dipstick was transferred into the adjacent well containing 40 μl chase buffer to wash the excess colloidal gold-conjugated IgG. The result was interpreted within 15 min. The sample that showed two red-coloured lines (i.e. test and control lines) on the dipstick was determined as positive, while a sample that showed only the control line was determined as negative. For the dot dipstick test, appearance of a red dot showed a positive result, and no dot was read as a negative result.
Optimization of the lateral flow dipstick test and evaluation of the diagnostic sensitivity and specificity
The concentration of the polyclonal antibody (anti-rPPDK IgG) on the test line and OD of anti-EhESA IgG-gold were optimized. Various antibody concentrations on the test line (1.0, 1.5, 2.0 and 2.5 mg ml−1) were tested with an initial anti-ESA IgG-gold at OD4. Then using the optimum concentration of the antibody on the test line, various ODs of anti-EhESA IgG-gold ranging from OD3 to OD8 were tested.
To determine the detection limit of the test, trophozoites from a culture of E. histolytica HM1: IMSS were washed with PBS. Next, serial ten-fold dilutions of the trophozoites (10 to 10,000 cells ml−1) were prepared and separately spiked into stool sample of a healthy individual. The mixture of trophozoites and stool sample was sonicated for 1 min, then tested with the dipstick test. The limit of detection of the dipstick test was also assessed by spiking rPPDK (0.001 to 100 μg ml−1) into the stool sample.
The diagnostic sensitivity of the dipstick test was evaluated using stool samples which were positive for E. histolytica by real-time PCR, using the following formula: (number of dipstick-positive samples / number of real-time PCR-positive samples) × 100. Meanwhile diagnostic specificity was evaluated using stool samples which were positive for other intestinal pathogens, using the following formula: (number of dipstick-negative samples / number of samples containing other intestinal pathogens) × 100. Agreements between various tests were quantified using Cohen kappa indices (κ).
Results
Analysis of stool samples by real-time PCR and Techlab E. histolytica II ELISA
From the 45 samples which were microscopy-positive for Entamoeba spp., real-time PCR detected E. histolytica in 26 (57.8%) samples and E. dispar in 16 (35.5%) samples with median Ct of 34.1 (28.4 < Ct < 35.3) and 34.6 (32.8 < Ct < 35.1), respectively. Standard curves for DNA copy numbers of plasmids harboring E. histolytica and E. dispar sequences showed R2 = 0.95; 88% efficiency and R2 = 0.98; 88% efficiency, respectively. Based on the standard curves, the limit of detections (LoD) of both E. histolytica and E. dispar exhibited 0.3 DNA copy number with Ct values of 36.40 and 36.32, respectively. The Ct values of all stool samples positive for E. histolytica and E. dispar were less than 36. Sample was confirmed as negative via real-time PCR when the Ct value was above than LoD or when no amplification curve was obtained. There were 19 and 29 samples which were microscopy-positive for Entamoeba spp., but negative for E. histolytica and E. dispar, respectively by the real-time PCR. Among the samples positive for Entamoeba spp. (n = 34) by real-time PCR, E. histolytica single infection was most dominant (52.9%; 18/34), followed by E. dispar (23.5%; 8/34) and E. histolytica and E. dispar mixed infections (23.5%; 8/34).
As shown in Table 2, out of 45 samples which were microscopy-positive for Entamoeba spp., the Techlab E. histolytica II ELISA was positive for only 5 samples. All the five stool samples were also positive by the real-time PCR. As compared to real-time PCR, the diagnostic sensitivity of the Techlab E. histolytica II ELISA was 19.2% (5/26) (Table 3). The 25 stool samples of other intestinal pathogens gave negative results by both real-time PCR and Techlab E. histolytica II ELISA.
Table 2.
Detection of E. histolytica by Techlab E. histolytica II ELISA from microscopy-positive samples.
| Microscopy-positive | Techlab E. histolytica II ELISA |
|
|---|---|---|
| Positive | Negative | |
| Entamoeba spp. | 5 | 40 |
| Other intestinal pathogens* | 0 | 25 |
| Total | 5 | 65 |
N. americanus, Ancylostoma spp., S. stercoralis, Clostridium spp., Salmonella spp., Shigella spp., T. trichiura, Campylobacter spp., enteropathogenic E. coli (EPEC), Rotavirus, Adenovirus, A. hydrophilia.
Table 3.
Diagnostic sensitivity of Techlab E. histolytica II ELISA when compared to real-time PCR.
| Techlab E. histolytica II ELISA | Real-time PCR for detection of E. histolytica |
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 5 | 0 | 5 |
| Negative | 21 | 19 | 40 |
| Total | 26 | 19 | 45 |
Production of polyclonal antibodies
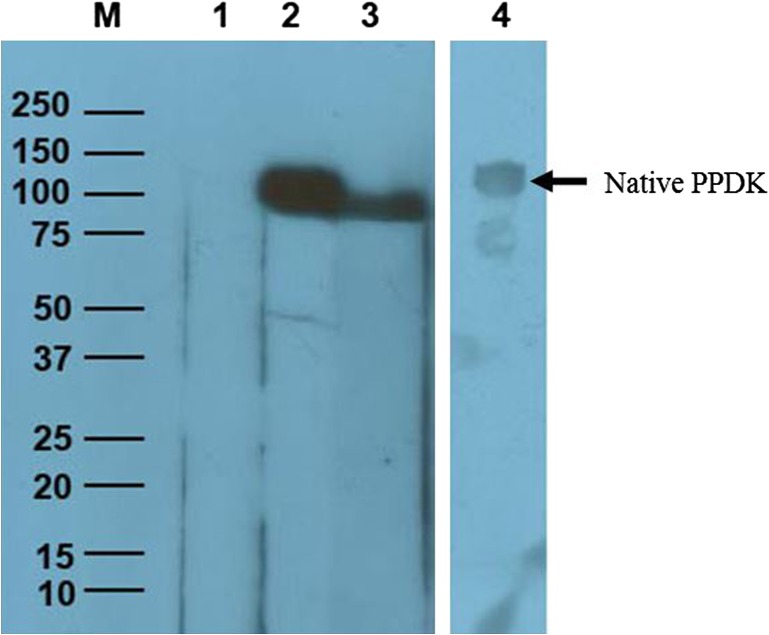
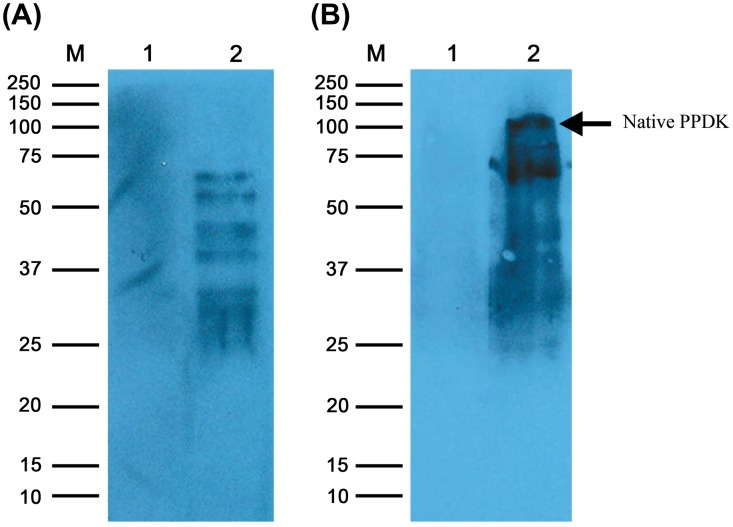
Both anti-PPDK IgG and anti-EhESA IgG antiserum showed a titre of 2,048,000. From the Western blot (Figure 1), the purified rPPDK was recognized by both anti-rPPDK IgG and the anti-EhESA IgG antibodies as a single band with approximate molecular mass of 98 kDa. Furthermore, native PPDK in ESA was recognized by anti-PPDK IgG antibody (Figure 1, lane 4). Figure 2 shows that anti-rPPDK polyclonal antibody recognized multiple proteins in the E. histolytica lysate. It also shows that the band corresponding to the molecular weight of PPDK (110 kDa) was seen in the Western blot when a higher amount of protein was loaded into the well.
Figure 1.
Western blot of E. histolytica rPPDK and native PPDK using anti-rPPDK IgG and anti-EhESA IgG polyclonal antibodies.
Notes: Lane M: Precision Plus ProteinTM Unstained Standard (Bio-Rad, CA) is indicated in kilodaltons (kDa). Lane 1: rPPDK protein probed with pre-immune rabbit anti-serum. Lanes 2 and 3: rPPDK protein (~98 kDa) probed with anti-rPPDK IgG and anti-EhESA IgG antibodies, respectively; Lane 4: E. histolytica ESA probed with anti-rPPDK IgG antibody. Arrow shows native PPDK in ESA at ~110 kDa.
Figure 2.
Western blot of E. histolytica lysate using anti-rPPDK IgG.
Notes: Lane M: Precision Plus ProteinTM Unstained Standard (Bio-Rad, CA) is indicated in kilodaltons (kDa). Panels A and B are Western blots performed with SDS-PAGE gels loaded with 20 and 90 μg E. histolytica lysate protein per well, respectively; Lane 1: E. histolytica lysate protein probed with pre-immune rabbit serum. Lane 2: E. histolytica lysate protein probed with anti-rPPDK IgG. Arrow shows native PPDK at ~110 kDa.
Evaluation of the diagnostic sensitivity and specificity of the lateral flow dipstick test
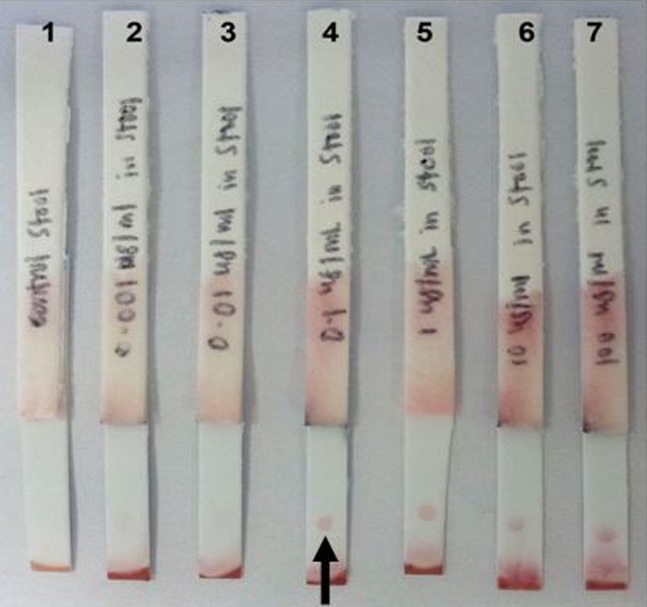
The optimum concentration of anti-rPPDK IgG on the test line and OD of anti-EhESA IgG gold were determined to be 2 mg ml−1 and OD4, respectively. The detection limit of the dipstick test was 1,000 cells ml−1 (Figure 3) and 0.1 μg ml−1 of rPPDK (Figure 4). Figure 5 shows examples of the reactivities of the dipstick test with patients stool samples. The diagnostic sensitivity of the dipstick test for the detection of E. histolytica was 65.4% (17/26) as compared to real-time PCR (Table 4). Eighteen samples were negative by both tests, whereas one sample was positive using the dipstick test but negative using real-time PCR. Meanwhile, nine samples were negative by the dipstick test but positive by the real-time PCR. A Cohen’s kappa value of 0.57 (95% CI, 0.35 to 0.79; p < 0.005) was obtained, which indicated a moderate agreement between the two assays with the calculative relative agreement of 77.8%.
Figure 3.
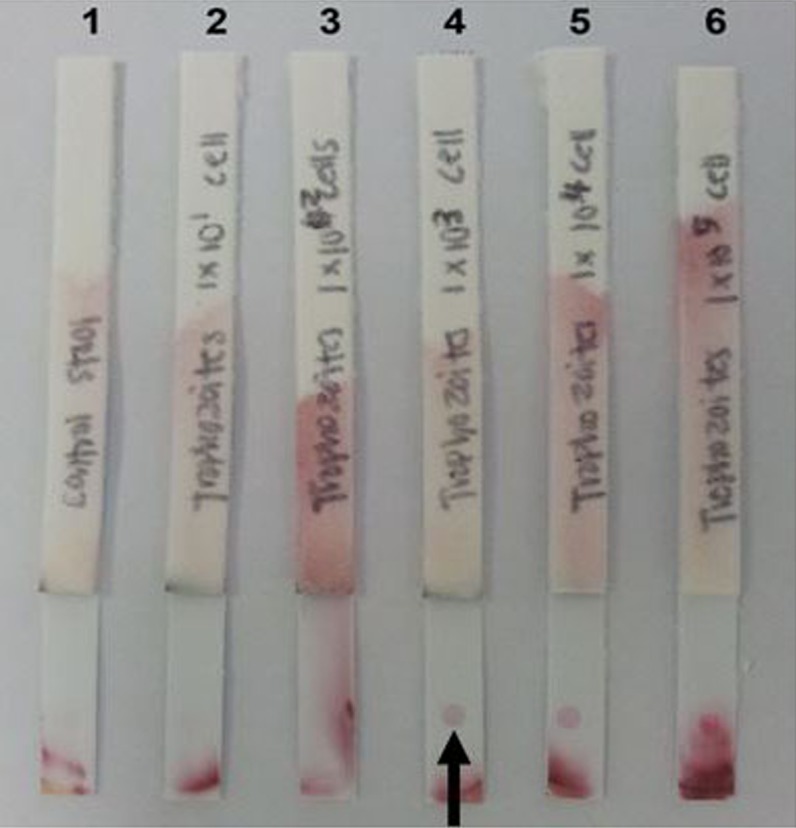
Analytical sensitivity of the lateral flow dipstick test in detecting antigen from lysed E. histolytica trophozoites spiked in a stool sample of healthy individual.
Notes: Arrow indicates the position of the visible dot. Lane 1: dipstick tested with non-spiked stool; Lanes 2–6: dipsticks tested with stool spiked with 10, 100, 1,000, 10,000 and 100,000 cells ml−1 of trophozoites, respectively. The limit of detection of the dipstick was determined to be 1,000 cells ml−1.
Figure 4.
Analytical sensitivity of lateral flow dipstick test strip in detecting rPPDK spiked in a stool sample of healthy individual. Arrow indicates the position of the visible dot.
Notes: Lane 1: dipstick tested with non-spiked stool sample; Lanes 2–7: dipsticks tested with stool spiked with 0.001, 0.01, 0.1, 1, 10 and 100 μg ml-1 of rPPDK, respectively. The limit of detection of the dipstick was determined to be 0.1 μg ml−1 rPPDK.
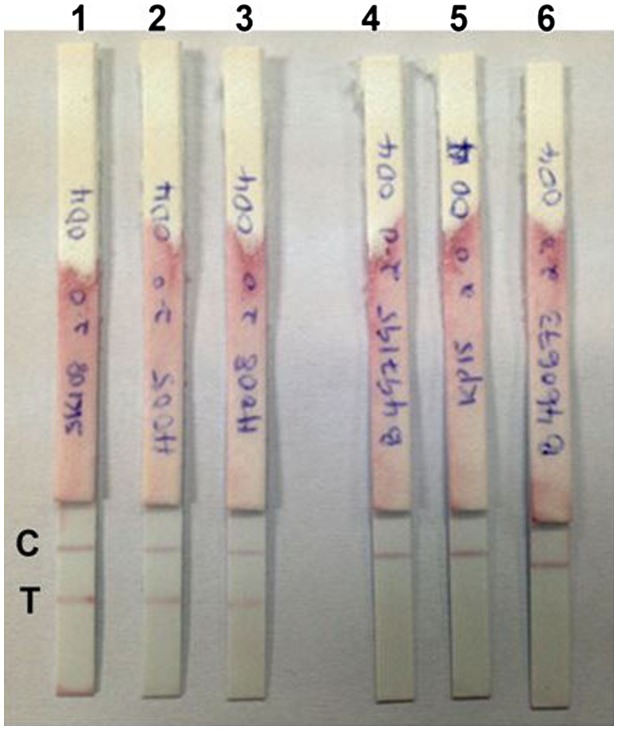
Figure 5.
Representative lateral flow dipstick tests using positive and negative stool samples.
Notes: Lanes 1–3: positive result when tested with three different stool samples with E. histolytica; Lanes 4–6: negative results when tested with stool samples containing other pathogens (Adenovirus, N. americanus and Salmonella spp., respectively). C: control line; T: test line.
Table 4.
Diagnostic sensitivity of the lateral flow dipstick test as compared to real-time PCR.
| Lateral flow dipstick test | Real-time PCR for detection of E. histolytica |
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 17 | 1 | 18 |
| Negative | 9 | 18 | 27 |
| Total | 26 | 19 | 45 |
Among stools with E. histolytica single infections, the dipstick detected 13 positive samples (72.2%). Meanwhile, the dipstick detected 5 of 8 (62.5%) samples with mixed E. histolytica and E. dispar. Only one of 8 samples (12.5%) which contained E. dispar (single ‘infection’) was positive by the dipstick test. All stool samples which were negative by real-time PCR (n = 11, microscopic positive) were also negative by the dipstick.
The diagnostic sensitivity of the dipstick test was 80% (4/5) when compared to the Techlab E. histolytica II ELISA. Twenty six samples tested negative by both dipstick and Techlab E. histolytica II ELISA, whereas 14 samples were positive using the dipstick test but negative with the ELISA. Only one sample was negative by the dipstick test but positive by the ELISA. The relative agreement was calculated to be 66.7% while Cohen’s kappa value of 0.21 (95% CI, −0.02 to 0.44; p > 0.005), this indicated a fair agreement between the lateral flow dipstick test and the ELISA. The diagnostic specificity of the dipstick test was 92% (23/25) when tested with samples containing other intestinal pathogens, the two false positive samples were from enteropathogenic E. coli (EPEC) and Shigella spp.
Discussion
Microscopy remains the routine method for laboratory diagnosis of intestinal amoebiasis despite its inability to discriminate among E. histolytica, E. dispar, E. moshkovskii and E. bangladeshi [2,4]. The outcome of microscopic examination depends on sample storage condition, duration of sample processing, personnel skill and parasite density [19]. More advanced techniques such as PCR, real-time PCR, and isoenzyme characterization can differentiate among the Entamoeba species. However these methods may take hours or days to produce results and require well-trained personnel, sophisticated laboratory equipment and expensive reagents, thus are not practical for most diagnostic laboratories [8,20]. Therefore, commercially available antigen-detection assays currently represent the most practical method for the identification of E. histolytica in stool samples [21].
Goñi [19] reported that an immunochromatographic dip strip test (RIDA® QUICK Cryptosporidium/Giardia/Entamoeba Combi; R-biopharm AG, Germany) showed 62.5% diagnostic sensitivity and 96.1% specificity for the detection of E. histolytica/E. dispar. Triage parasite panel (BioSite Diagnostics, CA) was reported to show 96 to 100% diagnostic sensitivity and 99.1 to 100% specificity when compared to microscopy in detecting E. histolytica/E. dispar [10,22,23]. Meanwhile ProSpecT ELISA (Thermo Fisher, MA), an FDA-approved test, showed diagnostic sensitivity and specificity of 78 and 99%, respectively for detection of Entamoeba species as compared to microscopy [24]. In another study by Gatti [25], the reported sensitivity and specificity of ProSpecT ELISA in identifying E. histolytica/E. dispar were 54.5 and 94%, respectively as compared to culture and zymodeme. However, since the above tests cannot differentiate among E. histolytica and E. dispar, they are not methods of choice for a diagnostic laboratory [26].
Merlin Optimum S (Merlin Diagnostika, Bornheim, Germany), Entamoeba CELISA PATH (Cellabs Pty Ltd., Australia) and Techlab E. histolytica II are ELISAs that specifically detect E. histolytica. Merlin Optimum S detects serine-rich protein of E. histolytica, but showed only 4.2% sensitivity as compared to the combined results of two other coproantigens ELISAs [27]. The latter two tests utilize monoclonal antibody to Gal/GalNAc-specific adherence lectin. Entamoeba CELISA PATH was reported to detect 28% of PCR-positive samples [14]. Meanwhile, Techlab E. histolytica II ELISA was reported to be more sensitive than the combination of culture and microscopy, however it was 79% sensitive and 96% specific when compared to real-time PCR [28].
Recently, a rapid test version of the Techlab E. histolytica II ELISA has been commercialized, known as E. histolytica Quik Chek (Techlab). It is a flow-through test consisting of a membrane device containing a strip lined with two antibodies. The control line binds the conjugate, and the second line contains monoclonal antibody against E. histolytica lectin which binds to the antigen (in stool)-conjugate complex. A study by Korpe [29] reported that the E. histolytica Quik Chek assay exhibited 100% sensitivity and specificity compared to the E. histolytica II ELISA. In another study, the E. histolytica Quik Chek assay exhibited 97% sensitivity and 100% specificity as compared to the ProSpecT ELISA [11].
In this study, we described the development and preliminary evaluation of a lateral flow dipstick test for rapid detection of E. histolytica stool sample. This assay used two different polyclonal antibodies as capture and detector reagents, it is easy to perform and can produce result in less than 15 min. In contrast to microscopy, the dipstick test can detect disintegrated or degraded parasites [30]. One of the polyclonal antibodies was against rPPDK, its native form is a 110 kDa protein in E. histolytica ESA. Western blot analysis using patients serum samples revealed the high diagnostic value of native and recombinant forms of PPDK for detection of ALA [16,17]. Since PPDK is a component of E. histolytica ESA, it is conceivable that it may be also useful as a target for detection of this parasite in stool sample. Therefore, in the present study, polyclonal antibodies against rPPDK and EhESA were raised and used to develop a lateral flow dipstick test, followed by preliminary evaluation using stool samples from patients with intestinal amoebiasis.
Out of 70 stool samples, 45 (64.3%) were positive for Entamoeba by microscopy, 26 (37.1%) by real-time PCR, 5 (7.1%) by Techlab E. histolytica II ELISA and 18 (25.7%) by the dipstick. Among the 45 microscopy-positive stool samples, the dipstick detected 18 (40%) as positive. Meanwhile among the 26 samples positive by real-time PCR, 65.4% (17/26) tested positive by the dipstick. The Ct values of all stool samples positive for either Entamoeba species were less than 36, which is below the LoD, thus demonstrating true positive results. The stool samples that were detected positive by the real-time PCR but negative by the dipstick were probably samples with very low number of parasites. On the other hand, among 8 samples positive for E. dispar (negative for E. histolytica) by real-time PCR, one positive sample was detected by dipstick. This false positive sample gave a Ct value of 34.75 which is comparable with the Ct values of the other 7 stool samples, thus the false positivity cannot be attributed to a higher density of E. dispar in the stool sample. There were also 11 microscopy-positive samples which were negative by both real-time PCR and the dipstick; these were probably samples which contained morphologically similar but non-pathogenic amoeba such as E. moshkovskii, E. bangladeshi, E. coli, E. polecki and E. hartmanni [5,31,32].
In comparison, Techlab E. histolytica II ELISA detected 5 (11.1%) samples among the 45 microscope positive samples. Similarly, among the PCR positive samples, the ELISA showed lower sensitivity (5/26 or 19.2%). This finding is comparable to those by Gatti [25], who reported that the Techlab E. histolytica II ELISA showed low diagnostic sensitivity (14.3%) when compared to culture and zymodeme identification. Another study conducted in Australia also reported that the ELISA did not identify any of the E. histolytica samples which were positive by PCR. In addition, cross-reactivity was observed for three specimens, one of which was positive for both E. dispar and E. moshkovskii while the other two samples contained E. moshkovskii [14]. There are also other studies that showed Techlab E. histolytica II had lower sensitivity (55/95, 57.9%) when compared to microscopy [9] and to real-time PCR (10/14, 71.4%) [33].
The results of Techlab E. histolytica II ELISA in this study and others (as described above) were different from the results obtained in countries where E. histolytica is highly endemic whereby high sensitivities (95–100%) were documented [3,6,13]. It is notable that the samples collected by Haque [6] were fresh and unpreserved diarrheic stool as recommended by the ELISA kit manufacturer; while in the present study, the stools were either frozen or preserved in 2.5% potassium dichromate, thus this may contribute to the low sensitivity of the ELISA observed in this study. The commercial ELISA recognizes antigens from the trophozoite stage, which are generally found in diarrheic stool samples during an acute amoebic infection and not present in formed stool [25]. In addition, the antigen level in the stool samples may be below the ELISA detection limit [7].
The present study demonstrated that the detection limit of our dipstick with stool sample was 1,000 trophozoites/ml or 0.1 μg ml−1 of rPPDK. This detection limit is comparable to the Triage parasite panel that required >1,000 trophozoites per ml for an unequivocal positive signal [33]. The detection limit of the dipstick seemed to be better than that reported for Techlab E. histolytica II ELISA which required lysate from 10,000 parasites for a positive reaction [14]. Entamoeba CELISA PATH kit was also shown to be more sensitive than Techlab E. histolytica II ELISA, the former was able to detect approximately 1,000 trophozoites per well [14]. Another study conducted by Pillai and Kain [27] found the ProSpecT ELISA detected E. histolytica/E. dispar antigen at 250 trophozoites per ml. However, Triage parasite and ProSpecT ELISA were unable to distinguish between pathogenic E. histolytica and the non-pathogenic and prevalent E. dispar. The dipstick developed in this study showed 92% (23/25) diagnostic specificity when tested with stool samples from patients with other intestinal pathogens. This is comparable with Techlab E. histolytica II ELISA, E. histolytica Quik Chek, ProSpecT ELISA, Triage parasite panel which demonstrated specificities ranging from 93 to 100% [6,22,25,29,33]. Nevertheless, cross-reactivity was detected with one of four samples of Shigella spp. and one sample from enteropathogenic E. coli (EPEC). There is low similarity (17.3%) between PPDK amino acid sequences of E. histolytica and Shigella flexnari (Assession No.:CDX06869.1), and the protein is not present in enteropathogenic E. coli (EPEC). Thus, the false positive results may be due to non-specific binding. Many more stool samples from Shigella spp. and EPEC need to be tested to determine whether these were true cases of cross-reactivity.
Although there is almost a 100% similarity between the PPDK protein sequences of E. histolytica and E. dispar (Accession: XP_657332.1 and XP_001736561.1 respectively), the dipstick developed in the present study showed good specificity against E. dispar. This could be due to a combination of reasons. A previous report on proteomic analysis of lysates of the two Entamoeba species showed 141 spots expressed at a substantially (> 5- fold) higher level in E. histolytica HM-1:IMSS than in E. dispar [34]. Thus similar observation may be expected with ESA components of the two Entamoeba species. Since ESA of E. histolytica was used to produce the anti-EhESA polyclonal antibody, the latter can be expected to be dominated by antibodies to ESA proteins which are highly expressed by E. histolytica as compared to E. dispar. Thus it is likely that E. histolytica proteins in the stool samples bind much better to anti-EhESA, as compared to the binding of E. dispar proteins to the same antibody. In performing the test, first the stool solution (containing Entamoeba proteins) was mixed with gold conjugated anti-EhESA (anti-EhESA-gold). The parasite proteins in the stool bind to the antibody mixture in the anti-EhESA-gold, thus creating antigen-antibody complexes. As alluded above, we expect that E. histolytica antigens will form much more of the complexes than E. dispar antigens. The antigen-antibody complexes (including complexes formed with native PPDK) were then allowed to bind to the lined anti-rPPDK polyclonal antibody on the dipstick, thus giving rise to the appearance of a positive test line. In the present study, multiple Western blot bands were observed when E. histolytica lysate was probed with anti-rPPDK polyclonal antibody, this showed that other Entamoeba antigens (besides native PPDK) bind/cross-bind to the lined anti-rPPDK antibody. Besides, the protein band corresponding to the molecular mass of PPDK (110 kDa) was only seen when a high amount of lysate protein was loaded. When the dipstick was dipped in a stool sample containing E. dispar, the amount of antigen complexed with anti-EhESA-gold was probably very much less (than with a stool containing E. histolytica), since the gold-conjugated antibody came from a different Entamoeba species. This in turn will lead to relatively smaller number of the complexes bound to the lined anti-rPPDK, resulting in the appearance of a negative dipstick result. Nevertheless, in future studies, it would good to test the dipsticks with stool samples spiked with cultured trophozoites of E. dispar, E. moshkovskii and E. bangladeshi to confirm its specificity against these morphologically similar species.
The results of the present study showed proof-of-concept of a lateral flow dipstick test for detection of E. histolytica using anti-rPPDK and anti-EhESA polyclonal antibodies. Although the diagnostic sensitivity of the dipstick was not very high, it was notably much higher than Techlab E. histolytica II ELISA. Further work will be needed to increase the diagnostic sensitivity of the dipstick. One way this may be achieved is by developing a flow-through (instead of a lateral flow) assay since this test format allows a greater sample volume to be used.
Disclosure statement
Rahmah Noordin, Nurulhasanah Othman and Zeehaida Mohamed are named as inventors in a related patent application made by Universiti Sains Malaysia, entitled “Diagnosis of Entamoeba histolytica”, which has been granted in South Africa (2014/00469), and pending in Malaysia (PI 2011003202) and several other countries.
Funding
This work was supported by Malaysian Ministry of Higher Education (MOHE) FRGS [grant number 203/CIPPM /6711241].
Acknowledgements
We would like to thank Zunulhisham Suhaimi and Nor Dyana Zakaria for their technical contributions in this study.
References
- [1].World Health Organization (WHO) WHO/PAHO/UNESCO report. A consultation with experts on amoebiasis. Epidemiol Bull. 1997;18:13–14. [PubMed] [Google Scholar]
- [2].Fotedar R, Stark D, Beebe N, et al. A laboratory diagnostic techniques for Entamoeba species. Clin Microbiol Rev. 2007;20(3):511–532. 10.1128/CMR.00004-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Haque R, Faruque ASG, Hahn P, et al. Entamoeba histolytica and Entamoeba dispar infection in children in Bangladesh. J Infect Dis. 1997;175(3):734–736. 10.1093/infdis/175.3.734 [DOI] [PubMed] [Google Scholar]
- [4].Tanyuksel M, Petri WA Jr. Laboratory diagnosis of amebiasis. Clin Microbiol Rev. 2003;16(4):713–729. 10.1128/CMR.16.4.713-729.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Petri WA. Jr, Haque R, Lyerly D, et al. 2000. Estimating the impact of amoebiasis on health. Parasitol Today. 2000; 16(8):320–321. 10.1016/S0169-4758(00)01730-0 [DOI] [PubMed] [Google Scholar]
- [6].Haque R, Neville LM, Hahn P, et al. Rapid diagnosis of Entamoeba infection by using Entamoeba and Entamoeba histolytica stool antigen detection kits. J Clin Microbiol. 1995;33(10):2558–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mirelman D, Nuchamowitz Y, Stolarsky T. Comparison of use of enzyme-linked immunosorbent assay-based kits and PCR amplification of rRNA genes for simultaneous detection of Entamoeba histolytica and E. dispar. J Clin Microbiol. 1997;35(9):2405–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Haque R, Ali IK, Akther S, et al. Comparison of PCR, isoenzyme analysis, and antigen detection for diagnosis of Entamoeba histolytica infection. J Clin Microbiol. 1998;36(2):449–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gonin P, Trudel L. Detection and differentiation of Entamoeba histolytica and Entamoeba dispar isolates in clinical samples by PCR and enzyme-linked immunosorbent assay. J Clin Microbiol. 2003;41(1):237–241. 10.1128/JCM.41.1.237-241.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sharp SE, Suarez CA, Duran Y, et al. Evaluation of the triage micro parasite panel for detection of Giardia lamblia, Entamoeba histolytica/Entamoeba dispar, and Cryptosporidium parvum in patient stool specimens. J Clin Microbiol. 2001;39(1):332–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Verkerke HP, Hanbury B, Siddique A, et al. Multisite clinical evaluation of a rapid test for Entamoeba histolytica in stool. J Clin Microbiol. 2015;53(2):493–497. 10.1128/JCM.02836-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Stark D, Fotedar R, van Hal S, et al. Prevalence of enteric protozoa in HIV-positive and HIV-negative men who have sex with men from Sydney. Am J Trop Med Hyg. 2007;76(3):549–552. [PubMed] [Google Scholar]
- [13].Solaymani-Mohammadi S, Rezaian M, Babaei Z, et al. Comparison of a stool antigen detection kit and PCR for diagnosis of Entamoeba histolytica and Entamoeba dispar infections in asymptomatic cyst passers in Iran. J Clin Microbiol. 2006;44(6):2258–2261. 10.1128/JCM.00530-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Stark D, Hal SV, Butcher A, et al. Comparison of stool antigen detection kits to PCR for diagnosis of amebiasis. J Clin Microbiol. 2008;46(5):1678–1681. 10.1128/JCM.02261-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Othman N, Mohamed Z, Verweij JJ, et al. Application of real-time polymerase chain reaction in detection of Entamoeba histolytica in pus aspirates of amoebic liver abscess patients. Foodborne Pathog Dis. 2010;7(6):637–641. 10.1089/fpd.2009.0427 [DOI] [PubMed] [Google Scholar]
- [16].Saidin S, Yunus MH, Zakaria ND, et al. Production of recombinant Entamoeba histolytica pyruvate phosphate dikinase and its application in a lateral flow dipstick test for amoebic liver abscess. BMC Infect Dis. 2014;14(1):182–190. 10.1186/1471-2334-14-182 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [17].Wong WK, Tan ZN, Othman N, et al. Analysis of Entamoeba histolytica excretory-secretory antigen and identification of a new potential diagnostic marker. Clin Vaccine Immunol. 2011;18(11):1913–1917. 10.1128/CVI.05356-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Makhsin SR, Razak KA, Noordin R, et al. The effects of size and synthesis methods of gold nanoparticle-conjugated MαHIgG4 for use in an immunochromatographic strip test to detect Brugian filariasis. Nanotechnology. 2012;23(49):495719. 10.1088/0957-4484/23/49/495719 [DOI] [PubMed] [Google Scholar]
- [19].Goñi P, Martín B, Villacampa M, et al. Evaluation of an immunochromatographic dip strip test for simultaneous detection of Cryptosporidium spp, Giardia duodenalis, and Entamoeba histolytica antigens in human faecal samples. Eur J Clin Microbiol Infect Dis. 2012;31(8):2077–2082. [DOI] [PubMed] [Google Scholar]
- [20].Blessmann J, Buss H, Tonun P. Real-time PCR for detection and differentiation of Entamoeba histolytica and Entamoeba dispar in fecal samples. J Clin Microbiol. 2002;40(12):4413–4417. 10.1128/JCM.40.12.4413-4417.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Buss S, Kabir M, Petri WR Jr, et al. Comparison of two immunoassays for detection of Entamoeba histolytica. J Clin Microbiol. 2008;46(8):2778–2779. 10.1128/JCM.00652-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Garcia LS, Shimizu RY, Bernard CN. Detection of Giardia lamblia, Entamoeba histolytica/Entamoeba dispar and Cryptosporidium parvum antigens in human fecal specimens using the triage parasite panel enzyme immunoassay. J Clin Microbiol. 2002;38(9):3337–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Swierczewski B, Odundo E, Ndonye J, et al. Comparison of the triage micro parasite panel and microscopy for the detection of Entamoeba histolytica/Entamoeba dispar, Giardia lamblia, and Cryptosporidium parvum in stool samples collected in Kenya. J Trop Med. 2012;1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ong SJ, Cheng MY, Liu KH, et al. Use of the ProSpecT microplate enzyme immunoassay for the detection of pathogenic and non-pathogenic Entamoeba histolytica in faecal specimens. Trans R Soc Trop Med Hyg. 1996;90(3):248–249. [DOI] [PubMed] [Google Scholar]
- [25].Gatti S, Swierczynski G, Robinson F, et al. Amebic infections due to the Entamoeba histolytica-Entamoeba dispar complex: a study of the incidence in a remote rural area of Ecuador. Am. J Trop Med Hyg. 2002;67(1):123–127. [DOI] [PubMed] [Google Scholar]
- [26].Tengku SA, Norhayati M. Public health and clinical importance of amoebiasis in Malaysia: a review. Trop Biomed. 2011;28(2):194–194. [PubMed] [Google Scholar]
- [27].Pillai DR, Kain KC. Immunochromatographic strip-based detection of Entamoeba histolytica-E. dispar and Giardia lamblia coproantigen. J Clin Microbiol. 1999;37(9):3017–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Roy S, Kabir M, Mondal D, et al. Real-time-PCR assay for diagnosis of Entamoeba histolytica infection. Clin Microbiol. 2005;43(5):2168–2172. 10.1128/JCM.43.5.2168-2172.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Korpe PS, Stott BR, Farida N, et al. Evaluation of a rapid point-of-care fecal antigen detection test for Entamoeba histolytica. Am J Trop Med Hyg. 2012;86(6):980–981. 10.4269/ajtmh.2012.11-0661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bhaskar S, Singh S, Sharma M. A single-step immunochromatographic test for the detection of Entamoeba histolytica antigen in stool samples. J Immunol Methods. 1996;196(2):193–198. 10.1016/0022-1759(96)00125-1 [DOI] [PubMed] [Google Scholar]
- [31].Hamzah Z, Petmitr S, Mungthin M, et al. Differential detection of Entamoeba histolytica, Entamoeba dispar and Entamoeba moshkovskii by a single-round PCR assay. J Clin Microbiol. 2006;44(9):3196–3200. 10.1128/JCM.00778-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ngui R, Angal L, Fakhrurrazi SA, et al. Differentiating Entamoeba histolytica, Entamoeba dispar and Entamoeba moshkovskii using nested polymerase chain reaction (PCR) in rural communities in Malaysia. Parasit Vectors. 2012;5(1):187–193. 10.1186/1756-3305-5-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Visser LG, Verweij JJ, Van Esbroeck M, et al. Diagnostic methods for differentiation of Entamoeba histolytica and Entamoeba dispar in carriers: performance and clinical implications in a non-endemic setting. Int J Med Microbiol. 2006;296(6):397–403. 10.1016/j.ijmm.2006.03.001 [DOI] [PubMed] [Google Scholar]
- [34].Davis PH, Chen M, Zhang X, et al. Proteomic Comparison of Entamoeba histolytica and Entamoeba dispar and the Role of E. histolytica Alcohol Dehydrogenase 3 in Virulence. PLoS Negl Trop Dis. 2009;3(4):e415. 10.1371/journal.pntd.0000415 [DOI] [PMC free article] [PubMed] [Google Scholar]