Preface
The information encoded in DNA is influenced by the presence of non-canonical nucleotides, the most frequent of which are ribonucleotides. In this review we discuss recent discoveries about ribonucleotide incorporation into DNA during replication by the three major eukaryotic replicases, DNA polymerases α, δ and ε. The presence of ribonucleotides in DNA causes short deletion mutations and may result in the generation of DNA single- and double-strand breaks, leading to genomic instability. We describe how these ribonucleotides are removed from DNA by ribonucleotide excision repair and by topoisomerase 1. We discuss the biological consequences and the physiological roles of ribonucleotides in DNA, and consider how deficiencies in their removal from DNA may be important in the etiology of disease.
Introduction
Cellular organisms store genetic information in DNA rather than RNA partly because DNA is more stable: DNA lacks of the reactive 2′-hydroxyl (OH) group found on the ribose sugar in RNA (FIG. 1a), which can attack the sugar-phosphate backbone of nucleic acids to generate strand breaks that can have genotoxic consequences. The presence of this 2′-OH group makes polynucleotides very sensitive to cleavage in the presence of alkali, providing a convenient method of screening for the presence of ribonucleotides within genomic DNA. As an important way of preventing genome instability and mutations, DNA polymerases select incoming nucleotides with both the correct base and sugar moieties, preferring deoxyribose rather than ribose for incorporation into the growing DNA chain. However, as for any enzymatic process, selection errors do occur, such that ribonucleotides are occasionally stably incorporated into DNA. As others have beautifully reviewed, ribonucleotide incorporation into DNA can also occur during initiation of DNA synthesis by RNA primases1 and primase-polymerases2, during specialized DNA repair and translesion synthesis reactions3,4 (Table 1), and when R-loops form during transcription5–7.
Figure 1. Ribonucleotide incorporation into DNA by polymerases.
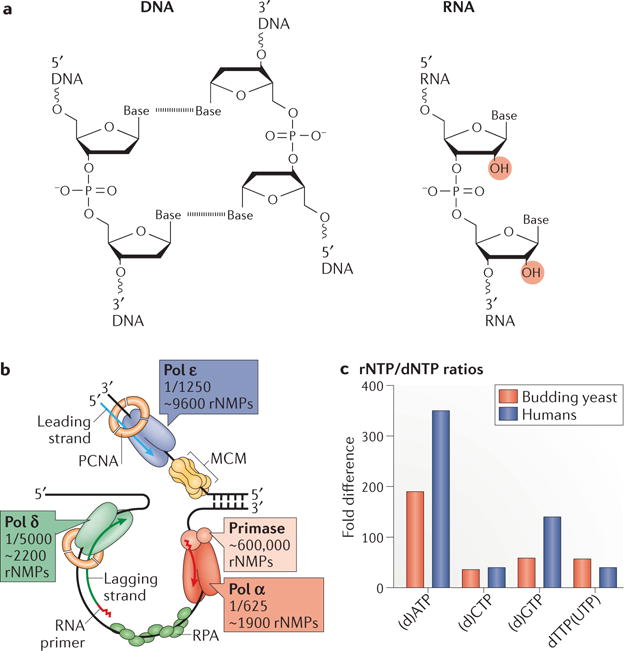
a | A comparison of the chemical structures of DNA and RNA. The dashed line between bases indicates hydrogen bonding in the double stranded DNA structure. The presence of a 2′-OH group on the ribose ring (highlighted in red) is the distinguishing feature that renders RNA less stable than DNA. b | A model of the division of labor at the budding yeast replication fork. RNA primase initiates synthesis of both strands by laying down a short RNA primer which is extended by the major leading strand polymerase, Polymerase ε (Pol ε), in a continuous fashion on the leading strand. On the lagging strand, the polymerase activity of Pol α takes over for a short stretch which is then extended by Pol δ to synthesize Okazaki fragments which are subsequently processed and joined during Okazaki fragment maturation. Both in vitro and in vivo studies have demonstrated the propensity of DNA polymerases to incorporate a significant number of ribonucleotides during synthesis. The fraction represents one ribonucleotide incorporated per x number of deoxyribonucleotides. The estimates of the total number of ribonucleotides (as ribonucleoside monophosphates (rNMPs)) incorporated into the 12 million base pair S. cerevisiae genome are indicated for leading and lagging strands. PCNA, proliferating cell nuclear antigen, a sliding clamp that binds to Pols δ and ε and enhances their processivity; RPA, replication protein A, single-strand DNA binding protein; MCM, minichromosome maintenance protein that serves as the eukaryotic replicative helicase complex. c | The nucleotide pool imbalances present in asynchronous cycling S. cerevisiae and human cells. Displayed is the ribonucleotide (rNTP) to deoxyribonucleotide (dNTP) ratio for each.
Table 1.
Ribonucleotide incorporation, processing and removal in eukaryotes
| Feature | Saccharomyces cerevisiae | Schizosaccharomyces pombe | Mus musculus | Homo sapiens |
|---|---|---|---|---|
| Incorporation | ||||
| Replicative polymerases | Pols α, δ and ε16,33,35–39,50,66,133 | Pols δ and ε52,53 | Pol*44,45 | Pols δ and ε33,34,67,117 |
| Repair polymerases | Pol ζ122 Pol η127 |
Pol4123 | TdT145 | Pol μ124 Pol β46,125,126 Pol ι121 Pol λ128 TdT124 |
| Mitochondrial DNA polymerase | Pol γ38,39 | Unknown | Pol γ146,147 | Pol γ48,147 |
| Replicase Extension | Pol ε37 Pol δ98 |
Unknown | Unknown | Pol ε34 |
| Replicase Bypass | Pols α, δ and ε97,98,100 | Unknown | Unknown | Pol ε34
Pol δ33 |
| Removal | ||||
| Replicase 3′ exonucleolytic proofreading | Pol ε (weak)37
Pol δ (very weak)33 |
Unknown | Unknown | Pol ε (weak)34
Pol δ (very weak)33 |
| RER | Yes35–39,43,50,59,66,69,91,133,134 | Yes52,53 | Yes44,45 | Yes67,117 |
| MMR | No91 Yes (for mismatched ribonucleotides)148 |
Unknown | Unknown | Unknown |
| NER | Unknown | Unknown | Unknown | No65 |
| Top1 | Yes35,66,69,92,93 | Unknown | Unknown | Unknown |
The replicative DNA polymerase has not yet been identified.
‘Yes’ indicates that the pathway has been identified. ‘No’ indicates that the pathway is not involved in ribonucleotide removal in the indicated organism. ‘Unknown’ indicates that measurements have not been performed or the specific process has not yet been identified. Pol, polymerase; TdT, Terminal deoxynucleotidyl transferase; RER, ribonucleotide excision repair; NER, nucleotide excision repair; Top1, topoisomerase 1.
This review considers another possibility, namely ribonucleotides incorporated during DNA replication by the major eukaryotic nuclear DNA replicases, DNA polymerases: Polymerase (Pol) alpha (α), delta (δ) and epsilon (ε). Given that all are proficient but imperfect at excluding ribonucleotides during synthesis8,9, we first consider how and with what efficiency these replicases prevent ribonucleotide incorporation. We then consider how ribonucleotides incorporated during eukaryotic nuclear DNA replication are removed by ribonucleotide excision repair, and how, in its absence, by topoisomerase 1. We further discuss the impact of ribonucleotides on the physical and chemical properties of DNA and how this affects DNA stability and repair. Finally, we discuss the consequences of the presence of ribonucleotides in DNA, linking the failure of their removal to human diseases. We also consider physiological roles that ribonucleotides serve when incorporated into the genome.
Ribonucleotide incorporation into DNA
Replication of the nuclear genome begins with synthesis of short RNA primers by the RNA primase component of Pol α (reviewed in1). RNA primers comprised of approximately 10 nucleotides initiate leading strand replication at origins of replication and lagging strand replication at each Okazaki fragment. Because Okazaki fragments are about 200 base pairs long, priming of the 12,000,000 base pair Saccharomyces cerevisiae nuclear genome may require the incorporation of as many as 600,000 ribonucleotides, and the mammalian nuclear genomes may require 250 times that. These numbers far exceed any other source of genomic ribonucleotides (FIG. 1b). Because they are efficiently removed during Okazaki fragment maturation (see BOX 1), their presence in the genome is usually transient. Eukaryotic RNA primases also incorporate ribonucleotides into DNA during restart of stalled replication forks (reviewed in10) and during initiation of translesion DNA synthesis by a human protein that has both primase and polymerase activities, PrimPol2,11–14. In addition, a reverse transcriptase of the human immunodeficiency virus type 1 efficiently incorporates ribonucleotides during proviral DNA synthesis in non-dividing macrophages (at a frequency of 1 in every 146 nucleotides)15. Though replicative DNA polymerases are highly selective, their massive overall contribution to DNA synthesis makes even infrequent incorporation a significant contribution to the overall genomic ribonucleotide landscape, as discussed below.
Box 1. Okazaki fragment maturation.
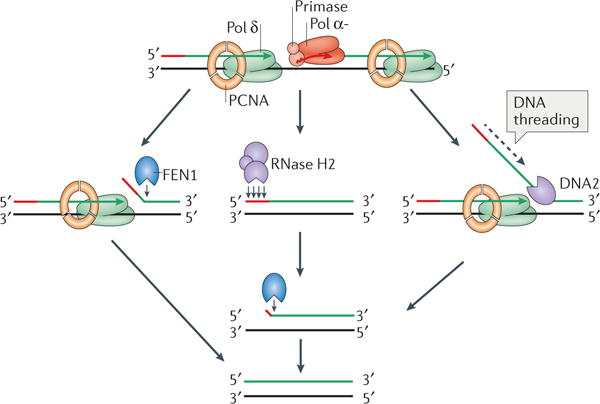
A major source of ribonucleotides incorporated into DNA, particularly in the lagging strand, are RNA primers, which are synthesized by the primase activity of Polymerase α (Pol α) (Figure 1b). In order to maintain fidelity of the lagging strand, these ribonucleotides need to be efficiently removed. This occurs through the process of Okazaki fragment maturation, which includes several pathways (reviewed in55,138–140).
During Okazaki fragment synthesis, a flap of varying lengths that includes the Pol α-synthesized RNA-DNA primer is generated when Pol δ reaches the 5′ end of the downstream Okazaki fragment and continues synthesis. This flap must be removed in order for DNA ligase 1 to join lagging strand DNA fragments to produce a continuous nascent lagging strand. One method of flap removal involves Pol δ-dependent strand displacement synthesis coupled with flap cleavage by FEN1 (flap endonuclease 1; left panel)141. An alternative mechanism involves multiple RNase H2 (or RNase H1) incisions that can occur up to the final ribonucleotide of an RNA primer, at which point FEN1 is capable of removing the final ribonucleotide (middle panel)142. Finally, RNA primer removal can also be achieved during a long flap removal pathway involving the DNA2 helicase/nuclease143 (and reviewed in144), a pathway that would be predicted to remove both ribonucleotides and errors incorporated into DNA by the proofreading-deficient Pol α enzyme (right panel). After binding the flap base, DNA threading occurs when the free 5′ end of the single-strand DNA flap is passaged through the DNA2 active site to position it for cleavage.
Nucleotide pool imbalances
The propensity of a replicative DNA polymerase to incorporate deoxyribonucleotides rather than ribonucleotides is influenced by the great excess of ribonucleotides over deoxyribonucleotides within a cell. In budding yeast, this excess is between 30 and 200-fold, depending on the base attached to the sugar16 (FIG. 1c). Similar nucleotide pool imbalances exist in mammalian cells17. Interestingly, nucleotide pool imbalances are also evident in non-cycling post-mitotic neurons where deoxyribonucleotide concentrations are very low18. The importance of nucleotide pool imbalances to replication fidelity and genome stability has been well-documented in yeast19–26. Mutations in the Rnr1 subunit of the Ribonucleotide Reductase (RNR) enzyme affect deoxyribonucleotide concentrations, causing mutations that depend on the nature and the degree of the change. Excess deoxyribonucleotides result in both misinsertions and mismatch extension at the expense of proofreading, while decreased deoxyribonucleotides result in deletions due to strand slippage. In addition, specific nucleotide pool imbalances cause a growth defect and cell cycle checkpoint activation in S. cerevisiae20, and nucleotide pools may also be critical in human cells, where such imbalances could contribute to the development or acceleration of cancer25 and/or neurodegeneration. This may be particularly relevant in tumour cells harboring mutations that impair DNA mismatch repair (MMR) and/or proofreading by Pol ε or δ27–32, in which mutagenesis is further enhanced by nucleotide pool imbalances24,25. For example, yeast cells harboring the colon cancer-associated Polδ-R696W mutant have extraordinarily high mutation rates due to both reduced nucleotide selectivity and expansion of deoxyribonucleotide pools25.
Ribonucleotide incorporation by DNA polymerases
DNA polymerase discrimination against ribonucleotide incorporation has been studied for many years and shown to vary widely among DNA polymerases in different families8,9 (Table 1). Ribonucleotide incorporation by budding yeast Pols α, δ and ε was measured using endogenous deoxyribonucleotide and ribonucleotide concentrations estimated from actively growing yeast cells16 (FIG. 1b), revealing that more than 13,000 ribonucleotides may be incorporated into the yeast genome during each round of replication. The human Pol ε and Pol δ enzymes have similar ribonucleotide incorporation propensities33,34, predicting that up to three million ribonucleotides may be incorporated during replication of the human genome. Biochemical experiments16,33,35–37 reveal ribonucleotide incorporation preferences, with rCMP and rGMP incorporated more frequently than rAMP and rUMP38,39.
Results from these biochemical analyses formed the foundation for in vivo studies of ribonucleotide incorporation into DNA by the major replicases. Ribonucleotide detection in S. cerevisiae genomic DNA was made possible by deleting RNH201, the gene encoding the catalytic subunit of RNase H2. RNase H2 is a conserved heterotrimeric enzyme that efficiently cleaves both single ribonucleotides and stretches of consecutive ribonucleotides within duplex DNA (reviewed in40). Earlier work41,42 suggested that ribonucleotides may be removed from DNA through the combined action of RNase H2 and the flap endonuclease, Fen1 (see BOX 1). Consistent with this, genomic DNA isolated from an RNH201 mutant (rnh201Δ) budding yeast strain is sensitive to alkali treatment36, with fragment sizes suggesting a ribonucleotide density of approximately one in every 6,500 deoxyribonucleotides43. Similar densities have also been detected in nuclear DNA isolated from RNase H2-deficient fission yeast and mouse cells, suggesting that more than 1,000,000 ribonucleotides may be incorporated into DNA during each round of vertebrate DNA replication44,45.
Biochemical and structural work on the B-family DNA polymerases has provided important mechanistic insights into how these enzymes choose the nucleotide containing the correct sugar for incorporation. A conserved ‘steric gate’ tyrosine in the polymerase active site prevents ribonucleotide incorporation, by clashing with the 2′-OH of the incoming ribonucleotide8,9,46–49. Replacement of this tyrosine with an alanine in Pol α (Y869A), or mutation of an adjacent conserved hydrophobic residue (Pol ε M644G, Pol δ L612M or L612G and Pol α L868M), yields yeast replicases that are less efficient at ribonucleotide discrimination35,36,38,50. DNA isolated from strains expressing these mutant replicases is more alkali-sensitive35,36,38,39,43,50, and the strand-preferences for ribonucleotide incorporation by these replicases strongly supports the model for the division of labor among polymerases at the replication fork that was originally inferred from studies of polymerase error signatures51. Thus, the M644G variant of Pol ε inserts ribonucleotides primarily in the nascent leading strand, and the L868M or Y869A Pol α variants and the L612M or L612G Pol δ variants insert ribonucleotides primarily in the nascent lagging strand. These same strand-specific ribonucleotide incorporation profiles are also observed in fission yeast52,53, suggesting evolutionary conservation. The altered ribonucleotide incorporation propensities of these variant alleles have allowed the use of ribonucleotides as a biomarker for polymerase activity and provided detailed information regarding such aspects of DNA replication enzymology as the positions of replication origins, origin timing and strength and replication termination zones (reviewed in54). Furthermore, use of DNA polymerase variants has also enabled mutational and phenotypic analyses of the biological consequences of ribonucleotides in DNA (discussed below).
Removal of ribonucleotides from DNA
Their abundance and potential to generate multiple forms of genome instability (discussed in detail below) predicts that ribonucleotides incorporated by Pols α, δ and ε must be efficiently removed from the nuclear genome. Recent studies demonstrate that this is achieved through several pathways and with varying efficiencies.
Exonucleolytic proofreading
Immediately following their insertion into DNA, ribonucleotides can be proofread, albeit weakly, by the 3′–5′ exonuclease activities of DNA polymerases δ and ε33,34,37,55. For yeast and human Pol ε, this proofreading is not nearly as efficient as proofreading of base-base mismatches. For example, biochemical analyses and mutation rate measurements demonstrate that proofreading-proficient yeast Pol ε excises about one third of the ribonucleotides37 and at least 92% of base-base mismatches incorporated by the polymerase56, suggesting that proofreading comprises only a relatively minor pathway of ribonucleotide removal34,37. This is also true for proofreading-proficient yeast and human Pol δ, which exhibit little ribonucleotide proofreading ability33. Although it has recently been suggested that Pol ε proofreads ribonucleotides incorporated by Pol δ in trans57, the in vivo relevance of this process is questionable given poor ability of Pol ε to proofread ribonucleotides in cis or base-base mismatches in trans58.
Ribonucleotide excision repair
Ribonucleotides are efficiently removed from DNA by ribonucleotide excision repair (RER; FIG. 2a; Table 1). RER is initiated by RNase H2-mediated recognition of ribonucleotides incorporated into DNA, followed by incision of the DNA backbone 5′ to the ribonucleotide36,42. Studies in vitro showed that this cleavage is followed by strand displacement synthesis of the incised DNA strand by Pol δ (or less efficiently by Pol ε), flap cleavage by flap endonuclease 1, Fen1 (or by the exonuclease Exo1 in the absence of Fen1), and finally the DNA is sealed by DNA ligase 1 to restore the DNA to its “all-deoxy”, intact form59. The PIP box (PCNA-interacting motif) in the C-terminus of the RNase H2B subunit (S. cerevisiae Rnh202) binds proliferating cell nuclear antigen (PCNA) to localize RNase H2 to the replication fork60,61, potentially coupling RER with replication. Consistent with these results, RNase H2 is recruited into DNA replication and repair foci in human cells in both a PCNA-dependent and –independent manner62.
Figure 2. Ribonucleotide removal during ribonucleotide excision repair (RER) or Topoisomerase 1 (Top1)-processing.
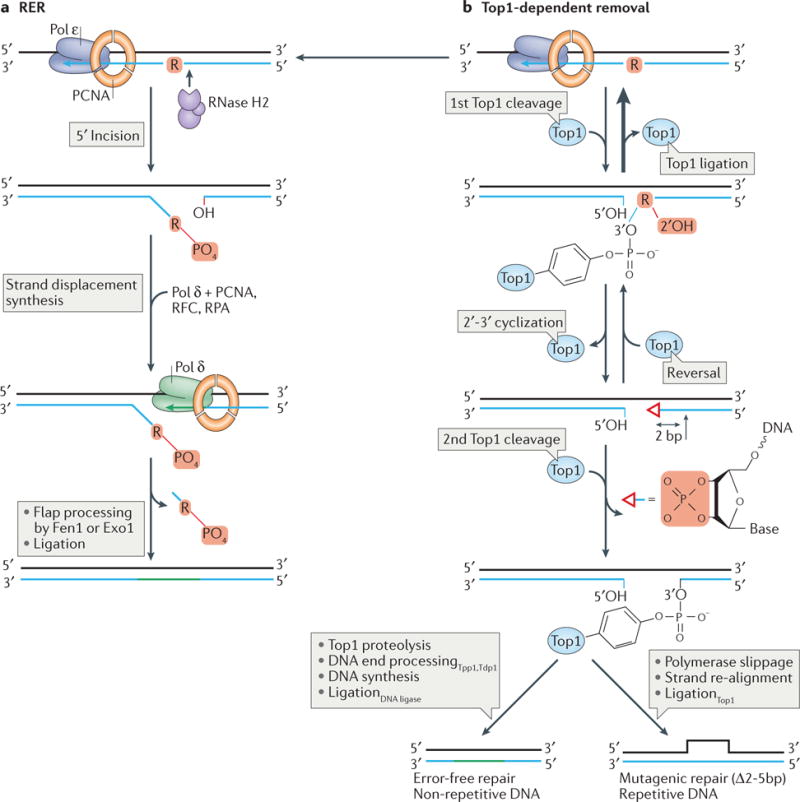
a | RER is initiated when RNase H2 incises 5′ to an embedded ribonucleotide. This is followed by a nick-translation reaction during which the nicked strand is displaced, allowing Pol δ (or Pol ε) in complex with proliferating cell nuclear antigen (PCNA) to bind and fill the gap. Replication factor C (RFC) is an ATP-dependent PCNA clamp-loader and RPA is a single-strand DNA binding protein. The flap generated following strand displacement is then nucleolytically-processed by the flap structure-specific endonuclease 1 (Fen1) or exonuclease 1 (Exo1). Although it is not depicted here, RER is important for removal of ribonucleotides incorporated during both leading and lagging strand DNA synthesis. b | In addition to RNase H2, Top1 is also able to cleave at genomic ribonucleotides. Top1 can incise the DNA on the 3′ side of a ribonucleotide, creating a nick that is typically reversed by Top1-mediated ligation. This reversal generates the original ribonucleotide-containing substrate and allows for another repair opportunity via RER (panel a). Alternatively, as occurs in cells that are RER-deficient, nucleophilic attack by the 2′-OH group on the ribose generates a 2′-3′-cyclic-PO4 (depicted by the open red triangle). This cyclic product can either be reversed through the activity of Top1 or efficiently removed following a second, this time irreversible Top1 cleavage reaction two-nucleotides upstream (5′) of the first cleavage site to generate a small gap. When the ribonucleotide is located in non-repetitive DNA, repair can proceed in an error-free fashion through the action of Top1-proteolysis and DNA end-processing enzymes (tyrosyl-DNA phosphodiesterase 1 (Tdp1), which hydrolyses the DNA-tyrosyl bond, and three prime phosphatase 1 (Tpp1), which processes the 3′ end) together with DNA polymerase(s) and DNA ligase to ultimately seal the nick. When the ribonucleotide is incorporated into a region containing repetitive DNA, mutagenic repair can occur to create a deletion of 2–5 base pairs (Δ2–5bp) in size, depending on the size of the repeat unit. This occurs as a result of re-alignment of the DNA strands followed by Top1-mediated ligation across the gap to generate a deletion of one of the repeat units. Not depicted here is the ability of Exo1 nuclease and the Srs2 helicase to enlarge this gap to prevent deletion mutations.
Nucleotide excision repair (NER), a multistep DNA repair process that excises bulky ultraviolet- (UV) or chemical-induced lesions, has been implicated to act as a backup pathway for ribonucleotide removal in RER-deficient bacterial cells63,64. However, ribonucleotides in DNA are a poor substrate for the human NER system in vitro and ribonucleotides are not efficiently removed by NER in human cells65. It remains to be determined whether NER is involved in repair of genomic ribonucleotides in other eukaryotic systems.
In line with the importance of RNase H2 in removing ribonucleotides from DNA, deletion of the genes encoding RNase H2 subunits causes the accumulation of alkali-sensitive sites (unrepaired ribonucleotides) in genomic DNA35,36,43,50,52,66. When RNase H2 loss is combined with the Pol ε mutator allele with decreased ribonucleotide discrimination (Pol ε M644G discussed above), the resulting pol2-M644G rnh201Δ budding yeast strain contains a large number of alkali-sensitive sites in nascent DNA and displays phenotypes indicative of genome instability. These phenotypes include slow growth, replication stress and elevated spontaneous mutagenesis dominated by 2–5 bp deletions in repetitive sequences (discussed below). Although dispensable for viability in yeast, RNase H2 is essential in mice, and its loss results in embryonic lethality after activation of a p53-dependant DNA damage response.44,45. Genomes of RNase H2-deficient mouse cells contain more than 1,000,000 unrepaired ribonucleotides and display genome instability, including increased levels of micronuclei, γH2AX foci, and chromosomal translocations. In accordance with observations in yeasts and mice, knockdown of RNase H2 expression in human cells causes genomic ribonucleotide accumulation and endogenous replication stress67.
RNase H2 is important for removal of both single ribonucleotides and more extensive RNA-DNA hybrids40. In order to identify its primary substrates, a separation-of-function (SOF) mutant of S. cerevisiae RNase H2 (Rnh201-P45D-Y219A) was created which abolishes its activity on single ribonucleotides while retaining its ability to cleave more extensive RNA-DNA hybrids68. Spontaneous mutation rate and specificity measurements from a strain expressing the Rnh201-SOF mutant firmly established that unrepaired single ribonucleotides, and not consecutive ribonucleotides, initiate the 2–5 bp deletion mutagenesis observed in an rnh201Δ strain. A subset of transcription-associated R-loops are processed uniquely by RNase H268. Thus, use of an RNase H2-SOF mutant in mammalian cells should provide important insights into the nature of the ribonucleotide-containing substrates that trigger genome instability and replication stress in higher eukaryotes.
Topoisomerase 1-mediated ribonucleotide removal
In the absence of RER, ribonucleotides can be removed through the activity of Topoisomerase 1 (Top1) (FIG. 2b, Table 1 and see below69,70. Most of these incision events occur at non-repetitive DNA sequences, and ultimately result in accurate ribonucleotide removal and replacement with a deoxyribonucleotide (FIG. 2b and66). However, when Top1-incision occurs in a repetitive DNA sequence, this can lead to the generation of short, 2–5 base pair (bp) deletions (FIG. 2b and see below). Larger-scale genome rearrangements have also recently been demonstrated to be Top1-dependent in a pol2-M644G rnh201Δ yeast strain71. These recombination-based events are initiated by formation of a DNA double-strand break (DSB), suggesting that DSBs are generated following Top1 cleavage at ribonucleotides. Because Top1 is critical for maintaining genome stability during transcription72, Top1-mediated ribonucleotide removal may be mechanistically linked to transcription. This is supported by the observation that Top1-dependent deletions in repeat sequences are driven by high levels of transcription73,74, some of which are also ribonucleotide-dependent75.
Studies in vivo using yeast variants of Pols ε (M644G), δ (L612M) and α (L868M) reveal a strand asymmetry that applies to Top1-initiated cleavage at ribonucleotides and mutagenesis. Specifically, ribonucleotides incorporated into nascent leading strand DNA by Pol ε are subject to Top1-initiated removal, mutagenesis and genome instability, while those incorporated into nascent lagging strand DNA by Pols α and δ are not35,66,76. Three hypotheses for these differences are currently being investigated. These include: the possibility of lower ribonucleotide density on the nascent lagging strand, alterative ribonucleotide-removal mechanisms on the nascent lagging strand, or the presence of torsional stress in the nascent leading strand that requires Top1 activity35,77. The replication strand asymmetry of Top1-dependent events is lost under high transcription conditions76. In this case, ribonucleotides present in the non-transcribed DNA strand are specifically subject to Top1-incision76. The mechanistic basis for this asymmetry between the non-transcribed DNA strand and the transcribed DNA strand for Top1-cleavage remains to be determined.
Consequences of ribonucleotides in DNA
Unrepaired ribonucleotides cause structural and chemical perturbations of nuclear DNA that are associated with several types of genome instability (recently reviewed in78), including single strand breaks (SSBs), DSBs, spontaneous mutagenesis, replication stress, cell cycle checkpoint activation, aberrant recombination and the formation of protein-DNA crosslinks.
Structural implications of genomic ribonucleotides
The structural implications of single ribonucleotides in duplex DNA have been investigated using a number of approaches, including biochemistry, nuclear magnetic resonance (NMR), X-ray crystallography, molecular dynamic simulations and atomic force microscopy (AFM)79–84. A single ribonucleotide alters helical parameters, causing a localized transition from B-form to A-form79,81,84, and changing the elastic properties of duplex DNA85, both of which can impact protein-DNA interactions. Chemically, RNA is 100,000 times more susceptible than DNA to spontaneous hydrolysis under physiological conditions (FIG. 3a;86). Additionally, the presence of ribonucleotides can affect Hoogsteen base pairing, in which a unique base pairing geometry is formed that changes the structural properties of DNA. The formation of these alternative base pairs in DNA may have functional importance as it expands the structural repertoire of duplex DNA, potentially influencing protein-DNA interactions and processes that include replication, repair and recombination (87 and references therein). Chromatin assembly and reassembly as well as nucleosome positioning may also be affected by ribonucleotides, as nucleosome binding to DNA in vitro is reduced when ribonucleotides are embedded in DNA88,89. Altogether, the structural perturbations caused by ribonucleotides incorporated into DNA have the potential to impact multiple important fundamental biological processes, including DNA replication, transcription, DNA repair, recombination and chromosome segregation.
Figure 3. Consequences of unrepaired ribonucleotides in DNA.
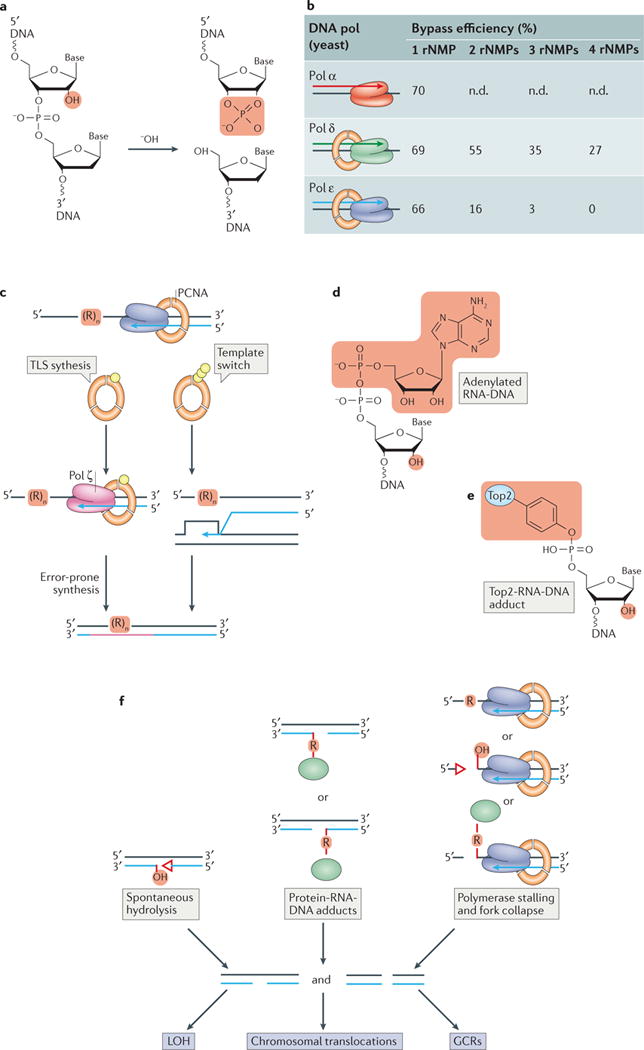
a | Hydrolysis at a ribonucleotide (occurring spontaneously or induced by alkali treatment) generates a ‘dirty’ DNA single strand break containing unligatable 2′–3′-cyclic-PO4 and 5′-OH termini. Not shown is the other intact DNA strand in the duplex molecule. b | DNA polymerases can bypass ribonucleotides in template DNA with varying proficiencies. This is dependent on the polymerase and the number of consecutive ribonucleotides present in the DNA as well as the identity of the base and the sequence context (not included in the table); n.d., not determined. c | In the absence of RNase H activity, the presence of ribonucleotides in template DNA causes DNA polymerase stalling and activates post-replication DNA repair (PRR) through ubiquitylation (yellow spheres) of proliferating cell nuclear antigen (PCNA). Through poly-ubiquitylation of PCNA ribonucleotide bypass can be achieved by a template switch mechanism. This is a form of DNA damage-tolerance in which a switch in template strands occurs during replication in order to bypass DNA damage and ensure completion of replication (right panel). Monoubiquitylation of PCNA initiates translesion DNA synthesis (TLS, left panel). This involves the recruitment of TLS polymerases such as Polymerase ζ (Pol ζ), which can bypass the lesion. However, Pol ζ lacks proofreading activity and has relatively low fidelity, causing this synthesis to be error-prone and thus mutagenic. d and e | Formation of compound DNA lesions is promoted by unrepaired genomic ribonucleotides. d | An abortive ligation lesion, adenylated RNA–DNA (5′-AMP-RNA-DNA), is formed following failure of DNA ligase to seal an RNase H2-generated nick. Highlighted in red are the adenylate group (top) and the ribose 2′-OH group (bottom). e | An irreversible topoisomerase 2 (Top2)–RNA–DNA adduct is formed when Top2 incises 5′ to a ribonucleotide. Highlighted in red are the covalently-linked Top2 adduct (top) and the ribose 2′-OH group (bottom). f | Failure of RNase H2-dependent repair can lead to spontaneous DNA hydrolysis (left), formation of protein–RNA–DNA adducts (green ellipses in the middle; see also panel e) or DNA (or RNA) polymerase stalling and fork collapse when encountering a ribonucleotide or ribonucleotide-provoked lesion (right). These events can trigger the formation of DNA single- (SSBs) and double-strand breaks (DSBs) that promote recombination and various genome rearrangements observed in yeast and mouse cells deficient in RNase H2. LOH, loss of heterozygosity; GCRs, gross chromosomal rearrangements.
Short deletion mutagenesis
Failure of RNase H2-dependent repair causes a strongly elevated rate of 2–5 base pair (bp) deletions in tandem repeat sequences36,69,90,91. These events are not corrected by DNA mismatch repair (MMR)91, and are further elevated in a yeast strain (pol2-M644G) that incorporates ribonucleotides more frequently than a wild-type enzyme36. These deletions depend on the ribonuclease activity of Top169,70 and the generation of these short deletions was proposed to involve the introduction of two consecutive incisions by Top1 adjacent to a ribonucleotide75, as was recently demonstrated in vitro92,93. To prevent these short deletions, the Srs2 helicase and the Exo1 5′–3′ exonuclease are important for processing 5′-OH DNA ends, which are produced by Top1 cleavage at ribonucleotides94. Otherwise, the 2′-OH group on the ribose sugar can execute a nucleophilic attack on the 3′-phosphotyrosine linkage with Top1, releasing Top1 and leaving an un-ligatable nick with 5′-OH and cyclic 2′–3′ phosphate (PO4) ends (FIG. 2b). A second Top1 cleavage event located 2 base pairs upstream of the 2′–3′ PO4 will result in a small DNA gap. After formation of this gap, proteolysis of Top1 followed by hydrolysis of the DNA-tyrosyl bond by tyrosyl-DNA phosphodiesterase 1 (Tdp1) and 3′-end processing by the three prime phosphatase 1 (Tpp1) have been demonstrated to occur in an in vitro system. Both Tdp1 and Tpp1 are enzymes involved in DNA strand break repair, and their combined action creates a substrate that is suitable for gap repair synthesis by Pol δ and ligation by DNA ligase to seal the nick and maintain fidelity92.
However, these repair mechanisms are insufficient to prevent mutagenesis when the ribonucleotide is present in a repetitive DNA sequence. In this case, sequence slippage-realignment can occur following the initial Top1-cleavage. This involves extrusion of the non-cleaved DNA strand and DNA strand misalignment, where unpaired nucleotides are stabilized by adjacent correct base pairs to then facilitate Top1-mediated ligation across the gap (FIG. 2b), leading to the loss of one repeat unit. This mutagenic consequence occurs at a high rate in the absence of RER, particularly on the nascent leading strand35 or, under conditions of high transcription, on the non-transcribed DNA strand76. The strand-asymmetric determinants of Top1-dependent genome instability may be related to the mechanism of Top1 recruitment to DNA, which may be mediated by interactions with the Cdc45–Mcm2-7–GINS (CMG) complex during leading strand replication95 and interaction with the C-terminus of Pol II during transcription96.
Ribonucleotide bypass by Pols α, δ and ε
Following incorporation of a ribonucleotide into DNA during synthesis, failure of RNase H2-dependent removal leaves ribonucleotides in what will be template DNA for the next round of replication. Therefore, the ability of a DNA polymerase to bypass a ribonucleotide present in the template strand is critical to the preservation of genome integrity. DNA Pols α, δ and ε can bypass template ribonucleotides33,97,98 (Table 1) with varying efficiencies depending on the replicase, ribonucleotide base identity (adenine, guanine, cytosine or uracil), the local sequence, and the number of consecutive ribonucleotides present (FIG. 3b). During RNA-templated DNA repair (RTDR), which can occur during repair of a chromosomal DSB in yeast using transcript RNA as a template, Pols α and δ can bypass a stretch of as many as four consecutive ribonucleotides, though with poor efficiency99. Subsequent biochemical experiments using nucleotide concentrations measured in yeast cells16 demonstrated that Pols α, δ and ε can also bypass a single template ribonucleotide with varying efficiencies97 (FIG. 3b). For both yeast and human Pols δ and ε, bypass capability decreases as the number of template ribonucleotides increases33,34. This is particularly evident for Pol ε, where bypass efficiency decreases from 66% to 0% as the number of ribonucleotides is increased from one to four (FIG. 3b;98). Structural analysis of the model B-family DNA polymerase of bacteriophage RB69, a homolog of Pol α, Pol δ and Pol ε, indicates that the presence of multiple ribonucleotides in template DNA increases DNA polymerase termination probability through structural perturbations that inhibit catalysis. In contrast to the major replicases, the specialized translesion (TLS) polymerase Pol zeta (Pol ζ), can readily bypass ribonucleotide-containing DNA100, and efficiently copies DNA templates containing four consecutive ribonucleotides. Such a specialized activity may be particularly advantageous during RTDR of DSBs99 or in response to replication fork stalling at unresolved stretches of genomic ribonucleotides in RNase H1/RNase H2 double mutant yeast100 (see below).
Replication stress and post-replication repair
As discussed above, replicative polymerases are not proficient at bypassing ribonucleotides that have been incorporated into DNA. As a result, ribonucleotides can impede replication fork progression33,34,98, leading to replication stress. Another source of replication stress may involve polymerase stalling at DNA breaks generated by either spontaneous hydrolysis (FIG. 3a) or Top1 cleavage at ribonucleotides, especially when considering the unligatable 2′–3′-cyclic-PO4 and 5′-OH DNA ends (FIG. 2b) that are produced. Paused DNA polymerase complexes and strand breaks may interfere with transcription, DNA segregation and DNA replication in the next round of synthesis. Consistent with these possibilities, impaired cell cycle progression and checkpoint activation are caused by unrepaired ribonucleotides in both yeast66,100 and human cells67.
Activation of post-replication DNA repair (PRR) pathways is an important means of bypassing replication stalling and ribonucleotide-induced DNA damage in RNase H2-defective cells. PRR pathways that include template switching and TLS are activated in budding yeast cells lacking both RNase H2 and RNase H1 activities100 (FIG. 3c). These repair pathways allow bypass of DNA lesions and involve ubiquitylation of the sliding clamp, proliferating cell nuclear antigen (PCNA)101. TLS is triggered by monoubiquitylation of PCNA. This initiates mutagenic DNA synthesis by enhancing the interaction between the error-prone DNA polymerases, such as Pol ζ and this modified form of PCNA. This interaction facilitates recruitment of DNA damage-tolerant repair polymerases to the lesion site, followed by error-prone DNA synthesis through the lesion (FIG. 3c). Alternatively, polyubiquitylation of PCNA activates error-free template switching, a homology-directed form of DNA repair during which the newly synthesized sister chromatid is used as the template for synthesis when a primer terminus is stalled at a lesion. PRR activation has been observed in RNase H2-depleted human cells and in cells isolated from patients suffering from Aicardi Goutières Syndrome67, a human inflammatory disorder in which RNase H2 function is often impaired.
Ribonucleotide-triggered abortive DNA ligation
As outlined above, RER serves as the primary mechanism of ribonucleotide removal from DNA (FIG. 2a). Despite its important function, this process can, in some cases, also contribute to genome instability. Namely, if following RNase H2 cleavage DNA ligase attempts to seal the generated nick prior to the next step in RER, abortive ligation will be triggered. The product of this failed ligation reaction is a compound RNA-DNA lesion containing a bulky adenylate group (5′-AMP) linked to a ribonucleotide (5′-AMP-RNA-DNA; FIG. 3d) that requires the hydrolase, Aprataxin (Aptx), for reversal102. Aptx-dependent reversal involves RNA–DNA binding and conformational changes. This repair mechanism may be particularly important when the ribonucleotide:deoxyribonucleotide ratio is very high, such as in post-mitotic neurons in the brain. The human neurodegenerative disorder, Ataxia with Oculomotor Apraxia 1 (AOA1), is characterized by mutations in APTX103, and AOA1 pathogenesis may involve the accumulation of RNA–DNA damage (see also below)102.
Protein–RNA–DNA adducts
Because of structural distortions caused by a ribonucleotide in DNA, the presence of RNA-DNA substrates in a cell may impact a number of protein-nucleic acid interactions. One example of this is the ability of a ribonucleotide in a DNA template to promote the production of a cleavage complex of RNA–DNA with Topoisomerase 2 (Top2; FIG. 3e;104,105) or Topoisomerase 1 (Top1)92, which, as discussed above, plays a role in ribonucleotide removal from DNA (Figure 2b). Like 5′-AMP-RNA-DNA lesions102, a protein-RNA-DNA crosslink is a compound DNA lesion whose toxicity is greater than the initial damage (a single ribonucleotide in DNA). Both adenylated and Top2-linked RNA-DNA lesions distort DNA, may lead to DNA breaks, and are a potential source of chromosomal rearrangements and other types of genome instability106.
Genomic rearrangements
Multiple lines of evidence suggest that failure of ribonucleotide repair results in nuclear DNA rearrangements. For example, deleting the genes encoding RNase H2 subunits in budding yeast causes an increased rate of spontaneous recombination94,107. These recombination events may be initiated by spontaneous DNA hydrolysis, SSBs and /or DSBs that occur at sites of unrepaired single ribonucleotides as well as more extensive RNA/DNA hybrids. Furthermore, polymerase blockage at ribonucleotides, or at the associated products, may promote the collapse of replication forks or transcription machinery, subsequently leading to break-induced replication. This form of recombination-dependent DNA replication can occur during repair of a DSB to promote survival but may result in complex chromosomal genome rearrangements108. Gross chromosomal rearrangements, presumably resulting from DSB formation, are also elevated in RNase H2-deficient yeast strains109,110, as are gene conversion events94,107,111. RNase H2-defective diploid yeast have elevated rates of loss-of-heterozygosity (LOH), resulting from mitotic inter-homolog recombination and non-allelic homologous recombination (NAHR), causing chromosomal translocations and copy number variation71,112. Evidence for these LOH and NAHR events being triggered by failure of RER of ribonucleotides incorporated during replication comes from the observation that the rates of these events are elevated in the Pol ε mutator variant (pol2-M644G) that has an increased propensity for ribonucleotide incorporation during synthesis compared to a wild type strain71.
Together, these data support a role for ribonucleotide removal in prevention and tolerance of RNA-DNA damage, the failure of which can lead to large-scale genome instability (FIG. 3f). Use of the RNase H2-SOF mutant (discussed above) and/or a yeast strain deficient in RNase H1 activity will provide important mechanistic insights into the ribonucleotide-containing lesions that initiate such events. Importantly, there is mounting evidence for the deleterious impact of the presence of ribonucleotides in DNA on genomic stability also in mammals, exemplified by the presence of large-scale chromosomal rearrangements and micronuclei formation in RNase H2null mouse embryonic fibroblasts44.
Genomic ribonucleotides and disease
Evidence of the importance of proper ribonucleotide processing and removal to human health is rapidly emerging. The first example of a connection between genomic ribonucleotides and human disease was provided with the discovery that mutations in the genes that encode any of the three subunits of the heterotrimeric RNase H2 enzyme (RNASEH2A, RNASEH2B and RNASEH2C in humans) cause Aicardi Goutières Syndrome (AGS)113. This rare, autosomal recessive neurodegenerative disorder causes symptoms that mimic congenital viral infection (reviewed in114). AGS patients display increased type 1 interferon (IFN) activation. Although it is currently unknown how RNase H2-deficiency activates IFN, this innate immune response may be related to unprocessed nucleic acid species or DSBs triggered by unrepaired ribonucleotides in DNA (FIG. 4a). Reduced RNase H2 function may be the result of AGS mutations in RNase H2 that perturb enzymatic activity, decrease protein expression, affect localization to replication and repair sites and/or alter complex assembly and stability62,115,116.
Figure 4. The connections between genomic ribonucleotides and human disease.
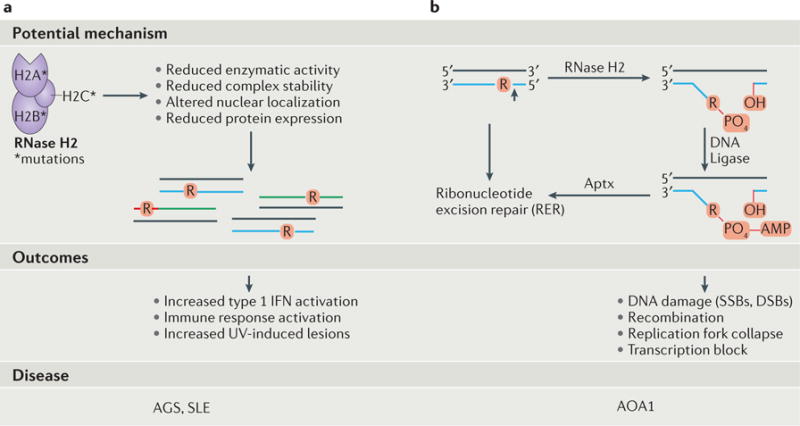
A schematic diagram illustrating the potential mechanisms, outcomes, and diseases associated with failure of ribonucleotide removal or processing. a | Mutations in RNase H2 are associated with Aicardi Goutières Syndrome (AGS) and Systemic Lupus Erythematosus (SLE). Patients suffering from AGS display increased type 1 interferon (IFN) activation, an immune response potentially related to the accumulation of aberrant ribonucleotide-containing nucleic acid species upon reduction of RNase H2-dependent repair. This reduced RNase H2 function may be related to the fact that these mutations in RNase H2 perturb enzymatic activity, decrease protein expression, affect localization to replication and repair sites and alter complex assembly and stability62,115,116. Photosensitivity and skin disease are common in SLE patients, and the demonstration of an increase in ultraviolet (UV)-induced lesions in both AGS and SLE patient cells117 raises the possibility that unrepaired genomic ribonucleotides renders DNA more susceptible to UV-induced DNA damage. b | Mutations in Aprataxin (Aptx) are associated with the neurological disease, Ataxia with Oculomotor Apraxia 1 (AOA1). Following RNase H2 incision at a ribonucleotide in DNA to initiate ribonucleotide excision repair (RER), premature engagement of this RNA-DNA substrate by DNA ligase triggers abortive ligation and transfer of an adenylate (AMP) group from DNA ligase to the RNA-DNA junction to generate a compound 5′-AMP-RNA-DNA lesion102. Failure of removal of this bulky adenylate group by Aprataxin causes accumulation of adenylated RNA-DNA and may lead to the formation of DNA breaks, affect processes such as replication and transcription, and contribute to AOA1.
Mutations in the genes encoding RNase H2 subunits also cause the related autoimmune disorder, Systemic Lupus Erythematosus (SLE)117. Similar to AGS associated phenotypes, SLE patients show the activation of type 1 IFN and the accumulation of antibodies against nucleic acids and proteins. These phenotypes may also be associated with defective RNase H2 removal of genomic ribonucleotides (FIG. 4a)117. SLE patients are photosensitive and prone to skin disease, suggesting that unrepaired genomic ribonucleotides render DNA more susceptible to UV-induced DNA damage. This is supported by the observation that both AGS and SLE patient cells exposed to UV light harbor increased numbers of cyclobutane pyrimidine dimers (CPDs), typical UV-induced lesions117. The shared immune response activation in AGS and SLE patients thus implicates ribonucleotide removal in prevention of human autoimmune diseases.
Another human disorder related to mutation of a ribonucleotide-processing enzyme is the neurological disease, AOA1 (reviewed in118) (Figure 4b). Accumulation of adenylated RNA– DNA may cause the formation of DNA breaks (both SSBs and DSBs), impair processes such as replication and transcription, and has been associated with AOA1 phenotypes. Consistent with this, biochemical and structural experiments demonstrate that AOA1-associated mutations in APTX impair the ability of this protein to resolve RNA–DNA junctions102. In fact, one mutation, K197Q, is located in the cleft responsible for RNA–DNA substrate interaction, and thus distorts the substrate-binding pocket, thereby significantly impairing deadenylation activity of APTX on a 5′-AMP-RNA-DNA substrate. Recent evidence demonstrating that 5′-AMP-DNA adducts form in mitochondrial DNA and persist in APTX-deficient cells raises the possibility that 5′-AMP-RNA-DNA lesions may also arise in mitochondrial and not only nuclear DNA119.
Physiological roles of genomic ribonucleotides
In addition to the negative consequences of genomic ribonucleotides described above, the abundance and non-random distribution of ribonucleotides in the eukaryotic nuclear genome suggests that they may also play physiological roles in specific cellular contexts120. Although this review is focused on ribonucleotides incorporated during replication, they are also incorporated by DNA repair polymerases46,121–128 (Table 1). Additionally, a residual di-ribonucleotide left by the RNA primase associated with Pol α acts as a developmental signal for mating-type switching in S. pombe129–132. Thus there may be a delicate balance between ribonucleotide incorporation and removal, as their presence may potentially impact physiological processes and features such as chromatin regulation, transcription, DNA segregation, DNA fragile site stability, G4 quadruplex formation, telomere biology, ribosomal biology, stem cell pluripotency and meiotically-programmed DSBs.
The locations of ribonucleotides were mapped genome-wide in budding and fission yeast strains lacking RNase H2 by sequencing genomic DNA fragmented at unrepaired ribonucleotides by either alkaline hydrolysis or RNase H2 cleavage38,39,53,133,134. Using replicase mutants that facilitate increased ribonucleotide incorporation into DNA, the positions of strand-specific ribonucleotide were utilized as robust biomarkers of polymerase activity. Several hypotheses about replication mechanisms were tested in this way. One discovery was that in yeast strains containing wild-type polymerases, ribonucleotides are preferentially found in the newly synthesized leading strand of nuclear DNA, raising the possibility of the existence of important signalling roles for ribonucleotides in nascent leading strand DNA50,135. One example of such a role comes from genetic and biochemical studies of MMR, a critical process used to correct replication errors that include single-base errors (base substitutions, insertions or deletions). MMR involves mismatch recognition, mismatch removal by excision, and correct re-synthesis of the DNA followed by ligation to complete repair (reviewed in136) (Figure 5). MMR is initiated by binding of the MutSα heterodimer to the mismatch. ATP activates MutSα, inducing a conformational change that allows it to interact with the MutLα heterodimer. During MMR of mismatches in nascent leading strand DNA, the entry point for MutLα may be provided by RNase H2. RNase H2 nicks 5′ to the ribonucleotide, which has been introduced during DNA synthesis by Pol ε, thereby providing a mechanism to discriminate between the newly synthesized and the template DNA strands. Once loaded, MutLα can incise the nascent strand through activation by PCNA (which can be delivered to the mismatch by RNase H2), allowing removal of the mismatch followed by DNA synthesis by Pol δ or ε (FIG. 5). This mechanism allows for efficient repair of single base mismatches introduced into nascent leading DNA strand by providing a clear discrimination between the template and newly synthesized strands50,135. Notably, the mechanism of MMR initiation differs in the lagging strand, as here frequent DNA termini prior to Okazaki fragment maturation may potentially act as entry points for MutLα. Pol ε harbors a strictly conserved methionine residue in its active site adjacent to the steric gate tyrosine (amino acid 644 in S. cerevisiae), whereas Pols α and δ bear a conserved leucine50. Replacement of this methionine- with leucine decreases the propensity for ribonucleotide incorporation36, suggesting that Pol ε has evolved to incorporate ribonucleotides into the nascent leading strand, thus enabling it to contribute to strand-discrimination during MMR.
Figure 5. Ribonucleotides act as a strand-discrimination signal during DNA mismatch repair (MMR).
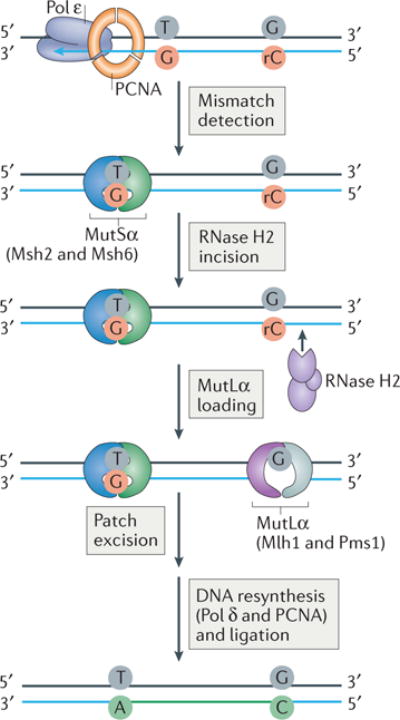
A model depicting how ribonucleotides incorporated into DNA by Polymerase ε (Pol ε) during leading strand synthesis may provide a signal for MMR in yeast. MMR involves mismatch recognition by MutSα, a heterodimer of Msh2 and Msh6, which are homologs of the bacterial MutS protein required for MMR. This recognition is followed by mismatch removal by MutLα (comprised of a heterodimer of Mlh1 and (yeast) Pms1), which are homologs of bacterial MutL.An entry point for MutLα is provided by RNase H2-mediated incision at a ribonucleotide incorporated into DNA by the replicative polymerase (Pol ε) during DNA replication to create a nick. Following MutLα loading at the incision site, excision of the mismatch and correct re-synthesis of the DNA by Pol δ or ε is followed by ligation to complete repair. Thus, the incorporated ribonucleotide serves to distinguish the newly synthesized mismatch-containing strand from the template strand.
Conclusions
The study of the causes and consequences of ribonucleotide incorporation into DNA is a rapidly emerging field. Considering their abundance, non-random distribution and biological importance, we expect that further elucidation of the effects of embedded ribonucleotides on genome stability will continue to be informative. For example, it is possible that the presence of a ribonucleotide in DNA influences its susceptibility to endogenous or exogenous DNA damage. This is supported by the observation that the expression levels of multiple DNA repair genes are increased in a yeast strain lacking RNase H2 activity137. The fact that ribonucleotides can be processed by pathways that include RER and Top1-mediated removal suggests that in addition to identification of other repair mechanisms, it will be beneficial to gain insight into the parameters that govern how and when an embedded ribonucleotide is removed from DNA as a way of elucidating the biological significance of such processing. For example, do tissue-specificities or differences related to mitotic versus quiescent cells exist with respect to ribonucleotide incorporation and removal? Furthermore, it will be important to identify additional proteins, which are involved in nucleic acid transactions and impacted by the presence of ribonucleotides in DNA, as this will provide further understanding of mechanisms linking genomic ribonucleotides to human disease, thereby offering potential avenues for therapeutic strategies. As genome-wide approaches38,39,53,133,134 become feasible in mouse and human cells, identification of ribonucleotide distribution over the mammalian genomic landscape through mapping studies may also provide insight into clinical aspects of human disorders stemming from mutations in genes important for preventing ribonucleotide-dependent genome instability, such as partial loss-of-function alleles of RNASEH2 or APTX. Further, the use of ribonucleotides as a robust biomarker of DNA polymerase activity across the nuclear genome will continue to yield information regarding replication enzymology under both unchallenged and stressed conditions.
Online summary.
Ribonucleotides are incorporated into DNA during replication.
Ribonucleotides can be removed during ribonucleotide excision repair or topoisomerase-1-initiated processing.
Failure of ribonucleotide removal is associated with genome instability in the form of mutagenesis, replication stress, DNA breaks, and chromosomal rearrangements.
Human diseases that include autoimmune disorders and neurodegenerative disease may be associated with failure to process genomic ribonucleotides
Genomic ribonucleotides act as a strand-discrimination signal during DNA mismatch repair and may have other physiological roles.
Acknowledgments
We thank Dmitry Gordenin and Scott Williams for critical reading of the manuscript. The laboratory is supported by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences. The authors apologize to colleagues whose primary research articles are not cited due to space limitations.
Glossary
- DNA polymerases
Enzymes that synthesize chains of deoxyribonucleotides
- Translesion synthesis
DNA synthesis that occurs across a template DNA lesion using a specialized DNA polymerase
- R-loop
A three-stranded structure that includes both an RNA-DNA hybrid and single DNA strand. R-loops are formed during transcription when the nascent mRNA hybridizes with the complementary template DNA strand and if not removed, can threaten genome stability or regulate gene expression
- Topoisomerase 1
A type 1B topoisomerase that relieves both positive and negative DNA supercoils by generating a reversible single strand DNA nick in duplex DNA. It acts during both DNA replication and transcription
- Okazaki fragment
A short DNA fragment synthesized by DNA polymerases α and δ with a length determined by nucleosome periodicity. Following processing to remove the RNA primer synthesized by Pol α to initiate synthesis, these segments are joined to form the continuous lagging strand by DNA ligase I
- Proofreading
The 3′–5′ exonuclease activity possessed by some DNA polymerases that facilitates removal of DNA mismatches before synthesis continues
- RNase H2
A heterotrimeric enzyme that cleaves the RNA portion of a DNA/RNA hybrid, hydrolyzing both single and multiple consecutive ribonucleotides in DNA
- B-family DNA polymerase
One of a family of related DNA polymerases found among all domains of cellular life. The family includes the highly accurate eukaryotic and archaeal replicases and the polymerases from phages T4 and RB69. Some possess a 3′–5′ exonucleolytic proofreading activity
- Strand displacement synthesis
An activity possessed by some DNA polymerases that involves displacement of the downstream DNA to produce a flap
- Proliferating Cell Nuclear Antigen (PCNA)
A homotrimeric sliding clamp complex that plays an essential role at the replication fork through recruitment of many enzymes required for DNA replication and repair. Post-translational modifications of PCNA initiate specific DNA repair processes critical for maintaining genome stability
- Micronuclei
Aberrant nuclear structures located within the cytoplasm that are comprised of chromosomal fragments that were not properly incorporated into a daughter nucleus during cell division. Micronuclei are formed in cells undergoing DNA damage
- γH2AX foci
A variant of histone H2A, H2AX, that is phosphorylated at serine 139 by checkpoint kinases during DNA damage response activation. γH2AX is modified over a large region of chromatin in the vicinity of a DSB and forms nuclear foci that serve as biomarkers for DSBs
- Replication stress
A cascade of responses that result from difficulties during DNA synthesis, such as polymerase stalling at a DNA lesion in the template strand. Replication stress may slow replisome progression and sensitizes cells to exogenous replication stress-inducing agents
- Mismatch Repair (MMR)
A post-replication repair process used to remove base-base mismatches and deletions that result from DNA synthesis errors. MMR occurs in a strand-specific manner and involves error recognition, excision and gap re-synthesis
- CMG complex
The Cdc45/Mcm2-7/GINS complex is the helicase that unwinds DNA at eukaryotic replication forks. It is physically associated with the leading strand DNA polymerase, Pol ε
- Post-replication repair
A process that occurs at sites of DNA damage to facilitate the completion of replication using error-free (Mms2-dependent template switching) or error-prone (pol zeta-dependent translesion synthesis) DNA damage-tolerance pathways
- RNA-DNA damage
Genome instability caused by the presence of ribonucleotides in DNA. RNA-DNA damage is associated with mutagenesis, chromosomal rearrangements, replication stress and DNA breaks
- Gene conversion events
A DNA repair event that involves the transfer of genetic information from a donor sequence to a homologous recipient such that the two become identical
- Loss-of-heterozygosity
A chromosomal rearrangement in diploid cells that occurs when the template for DNA synthesis during homologous recombination repair is a homologous chromosome. This may result in loss of a functional allele and occurs frequently in tumor cells
- Mitotic inter-homolog recombination
Mitotic crossover recombination that occurs between homologous chromosomes during the repair of a DNA lesion
- Non-allelic homologous recombination (NAHR)
A form of homologous recombination that occurs between similar sequences to cause chromosomal translocations and copy number variation
- DNA fragile site
A specific genomic locus susceptible to spontaneous DNA breaks that may result in chromosomal rearrangements and contribute to human disease
- G4 quadruplex
A higher-order DNA structural motif comprised of 4 strands of guanine-rich sequences that form a stable planar stacked structure at specific genomic locations, including telomeres and gene promoters, where they appear to play important positive and negative biological roles
Biographies
Jessica S. Williams is a staff scientist in the Laboratory of Genome Integrity and Structural Biology at the National Institute of Environmental Health Sciences in Research Triangle Park, North Carolina. Her current research focus is on the causes and biological consequences of incorporation of ribonucleotides into DNA. She obtained her Ph.D. from the University of Alberta, Canada and worked as a postdoctoral fellow at the Scripps Research Institute, California, before joining NIEHS.
Scott A. Lujan is a bioinformatics contractor in the Laboratory of Genome Integrity and Structural Biology at the National Institute of Environmental Health Sciences in Research Triangle Park, North Carolina. His current research focus is on the development and application of bioinformatics approaches to address biological questions related to DNA replication and repair. He obtained his Ph.D. from the University of North Carolina at Chapel Hill, before joining NIEHS as a postdoctoral fellow in the Replication Fidelity Group.
Thomas A. Kunkel is an NIH Distinguished Investigator in the Laboratory of Genome Integrity and Structural Biology at the National Institute of Environmental Health Sciences in Research Triangle Park, North Carolina. Research in his group is focused on the determinants of DNA replication fidelity using biochemical, structural, genetic and bioinformatics approaches. He obtained his Ph.D. from the University of Cincinnati and was a postdoctoral fellow at the University of Washington before joining NIEHS as a principal investigator.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Kuchta RD, Stengel G. Mechanism and evolution of DNA primases. Biochim Biophys Acta. 2010;1804:1180–1189. doi: 10.1016/j.bbapap.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez-Jimenez MI, et al. Alternative solutions and new scenarios for translesion DNA synthesis by human PrimPol. DNA Repair (Amst) 2015;29:127–138. doi: 10.1016/j.dnarep.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Goodman MF, Woodgate R. Translesion DNA polymerases. Cold Spring Harb Perspect Biol. 2013;5:a010363. doi: 10.1101/cshperspect.a010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boiteux S, Jinks-Robertson S. DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae. Genetics. 2013;193:1025–1064. doi: 10.1534/genetics.112.145219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sollier J, Cimprich KA. Breaking bad: R-loops and genome integrity. Trends Cell Biol. 2015;25:514–522. doi: 10.1016/j.tcb.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santos-Pereira JM, Aguilera A. R loops: new modulators of genome dynamics and function. Nature reviews. Genetics. 2015;16:583–597. doi: 10.1038/nrg3961. [DOI] [PubMed] [Google Scholar]
- 7.Jinks-Robertson S, Bhagwat AS. Transcription-associated mutagenesis. Annual review of genetics. 2014;48:341–359. doi: 10.1146/annurev-genet-120213-092015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown JA, Suo Z. Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry. 2011;50:1135–1142. doi: 10.1021/bi101915z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joyce CM. Choosing the right sugar: how polymerases select a nucleotide substrate. Proc Natl Acad Sci U S A. 1997;94:1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeeles JT, Poli J, Marians KJ, Pasero P. Rescuing stalled or damaged replication forks. Cold Spring Harb Perspect Biol. 2013;5:a012815. doi: 10.1101/cshperspect.a012815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan L, et al. hPrimpol1/CCDC111 is a human DNA primase-polymerase required for the maintenance of genome integrity. EMBO reports. 2013;14:1104–1112. doi: 10.1038/embor.2013.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Gomez S, et al. PrimPol, an archaic primase/polymerase operating in human cells. Mol Cell. 2013;52:541–553. doi: 10.1016/j.molcel.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bianchi J, et al. PrimPol Bypasses UV Photoproducts during Eukaryotic Chromosomal DNA Replication. Mol Cell. 2013;52:566–573. doi: 10.1016/j.molcel.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mouron S, et al. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nature structural & molecular biology. 2013;20:1383–1389. doi: 10.1038/nsmb.2719. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy EM, Amie SM, Bambara RA, Kim B. Frequent incorporation of ribonucleotides during HIV-1 reverse transcription and their attenuated repair in macrophages. J Biol Chem. 2012;287:14280–14288. doi: 10.1074/jbc.M112.348482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nick McElhinny SA, et al. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci U S A. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 18.Ferraro P, Franzolin E, Pontarin G, Reichard P, Bianchi V. Quantitation of cellular deoxynucleoside triphosphates. Nucleic Acids Res. 2010;38:e85. doi: 10.1093/nar/gkp1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar D, Viberg J, Nilsson AK, Chabes A. Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res. 2010;38:3975–3983. doi: 10.1093/nar/gkq128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar D, et al. Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res. 2011;39:1360–1371. doi: 10.1093/nar/gkq829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chabes A, et al. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003;112:391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 22.Buckland RJ, et al. Increased and imbalanced dNTP pools symmetrically promote both leading and lagging strand replication infidelity. PLoS Genet. 2014;10:e1004846. doi: 10.1371/journal.pgen.1004846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davidson MB, et al. Endogenous DNA replication stress results in expansion of dNTP pools and a mutator phenotype. EMBO J. 2012;31:895–907. doi: 10.1038/emboj.2011.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams LN, et al. dNTP pool levels modulate mutator phenotypes of error-prone DNA polymerase epsilon variants. Proc Natl Acad Sci U S A. 2015;112:E2457–2466. doi: 10.1073/pnas.1422948112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mertz TM, Sharma S, Chabes A, Shcherbakova PV. Colon cancer-associated mutator DNA polymerase delta variant causes expansion of dNTP pools increasing its own infidelity. Proc Natl Acad Sci U S A. 2015;112:E2467–2476. doi: 10.1073/pnas.1422934112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watt DL, Buckland RJ, Lujan SA, Kunkel TA, Chabes A. Genome-wide analysis of the specificity and mechanisms of replication infidelity driven by imbalanced dNTP pools. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shinbrot E, et al. Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome research. 2014;24:1740–1750. doi: 10.1101/gr.174789.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Church DN, et al. DNA polymerase epsilon and delta exonuclease domain mutations in endometrial cancer. Human molecular genetics. 2013;22:2820–2828. doi: 10.1093/hmg/ddt131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshida R, et al. Concurrent genetic alterations in DNA polymerase proofreading and mismatch repair in human colorectal cancer. European journal of human genetics : EJHG. 2011;19:320–325. doi: 10.1038/ejhg.2010.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palles C, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45:136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kandoth C, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clausen AR, Zhang S, Burgers PM, Lee MY, Kunkel TA. Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase delta. DNA Repair (Amst) 2013;12:121–127. doi: 10.1016/j.dnarep.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goksenin AY, et al. Human DNA polymerase epsilon is able to efficiently extend from multiple consecutive ribonucleotides. J Biol Chem. 2012;287:42675–42684. doi: 10.1074/jbc.M112.422733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams JS, et al. Evidence that processing of ribonucleotides in DNA by topoisomerase 1 is leading-strand specific. Nature structural & molecular biology. 2015;22:291–297. doi: 10.1038/nsmb.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nick McElhinny SA, et al. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. Show that ribonucleotides are incorporated into yeast genomic DNA, and together with reference 42, that misincorporated ribonucleotides can be removed by RNase H2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams JS, et al. Proofreading of ribonucleotides inserted into DNA by yeast DNA polymerase epsilon. DNA Repair (Amst) 2012;11:649–656. doi: 10.1016/j.dnarep.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clausen AR, et al. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nature structural & molecular biology. 2015;22:185–191. doi: 10.1038/nsmb.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koh KD, Balachander S, Hesselberth JR, Storici F. Ribose-seq: global mapping of ribonucleotides embedded in genomic DNA. Nature methods. 2015;12:251–257. doi: 10.1038/nmeth.3259. 253 p following 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eder PS, Walder RY, Walder JA. Substrate specificity of human RNase H1 and its role in excision repair of ribose residues misincorporated in DNA. Biochimie. 1993;75:123–126. doi: 10.1016/0300-9084(93)90033-o. [DOI] [PubMed] [Google Scholar]
- 42.Rydberg B, Game J. Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc Natl Acad Sci U S A. 2002;99:16654–16659. doi: 10.1073/pnas.262591699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lujan SA, et al. Mismatch repair balances leading and lagging strand DNA replication fidelity. PLoS Genet. 2012;8:e1003016. doi: 10.1371/journal.pgen.1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reijns MA, et al. Enzymatic removal of ribonucleotides from DNA is essential for Mammalian genome integrity and development. Cell. 2012;149:1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hiller B, et al. Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity. J Exp Med. 2012;209:1419–1426. doi: 10.1084/jem.20120876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cavanaugh NA, et al. Molecular insights into DNA polymerase deterrents for ribonucleotide insertion. J Biol Chem. 2011;286:31650–31660. doi: 10.1074/jbc.M111.253401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang W, Wu EY, Hellinga HW, Beese LS. Structural factors that determine selectivity of a high fidelity DNA polymerase for deoxy-, dideoxy-, and ribonucleotides. J Biol Chem. 2012;287:28215–28226. doi: 10.1074/jbc.M112.366609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kasiviswanathan R, Copeland WC. Ribonucleotide discrimination and reverse transcription by the human mitochondrial DNA polymerase. J Biol Chem. 2011;286:31490–31500. doi: 10.1074/jbc.M111.252460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeLucia AM, Grindley ND, Joyce CM. An error-prone family Y DNA polymerase (DinB homolog from Sulfolobus solfataricus) uses a ‘steric gate’ residue for discrimination against ribonucleotides. Nucleic Acids Res. 2003;31:4129–4137. doi: 10.1093/nar/gkg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lujan SA, Williams JS, Clausen AR, Clark AB, Kunkel TA. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Mol Cell. 2013;50:437–443. doi: 10.1016/j.molcel.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008;18:521–527. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miyabe I, Kunkel TA, Carr AM. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet. 2011;7:e1002407. doi: 10.1371/journal.pgen.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Daigaku Y, et al. A global profile of replicative polymerase usage. Nature structural & molecular biology. 2015;22:192–198. doi: 10.1038/nsmb.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jinks-Robertson S, Klein HL. Ribonucleotides in DNA: hidden in plain sight. Nature structural & molecular biology. 2015;22:176–178. doi: 10.1038/nsmb.2981. [DOI] [PubMed] [Google Scholar]
- 55.Williams JS, Kunkel TA. Ribonucleotides in DNA: origins, repair and consequences. DNA Repair (Amst) 2014;19:27–37. doi: 10.1016/j.dnarep.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shcherbakova PV, et al. Unique error signature of the four-subunit yeast DNA polymerase epsilon. J Biol Chem. 2003;278:43770–43780. doi: 10.1074/jbc.M306893200. [DOI] [PubMed] [Google Scholar]
- 57.Johnson RE, Klassen R, Prakash L, Prakash S. A Major Role of DNA Polymerase delta in Replication of Both the Leading and Lagging DNA Strands. Mol Cell. 2015;59:163–175. doi: 10.1016/j.molcel.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Flood CL, et al. Replicative DNA polymerase delta but not epsilon proofreads errors in Cis and in Trans. PLoS Genet. 2015;11:e1005049. doi: 10.1371/journal.pgen.1005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sparks JL, et al. RNase H2-Initiated Ribonucleotide Excision Repair. Molecular Cell. 2012;47:980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chon H, et al. Contributions of the two accessory subunits, RNASEH2B and RNASEH2C, to the activity and properties of the human RNase H2 complex. Nucleic Acids Res. 2009;37:96–110. doi: 10.1093/nar/gkn913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bubeck D, et al. PCNA directs type 2 RNase H activity on DNA replication and repair substrates. Nucleic Acids Res. 2011;39:3652–3666. doi: 10.1093/nar/gkq980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kind B, et al. Altered spatio-temporal dynamics of RNase H2 complex assembly at replication and repair sites in Aicardi-Goutieres syndrome. Human molecular genetics. 2014;23:5950–5960. doi: 10.1093/hmg/ddu319. [DOI] [PubMed] [Google Scholar]
- 63.Cai Y, Geacintov NE, Broyde S. Ribonucleotides as nucleotide excision repair substrates. DNA Repair (Amst) 2014;13:55–60. doi: 10.1016/j.dnarep.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vaisman A, et al. Removal of misincorporated ribonucleotides from prokaryotic genomes: an unexpected role for nucleotide excision repair. PLoS Genet. 2013;9:e1003878. doi: 10.1371/journal.pgen.1003878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lindsey-Boltz LA, Kemp MG, Hu J, Sancar A. Analysis of Ribonucleotide Removal from DNA by Human Nucleotide Excision Repair. J Biol Chem. 2015;290:29801–29807. doi: 10.1074/jbc.M115.695254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams JS, et al. Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol Cell. 2013;49:1010–1015. doi: 10.1016/j.molcel.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pizzi S, et al. Reduction of hRNase H2 activity in Aicardi-Goutieres syndrome cells leads to replication stress and genome instability. Human molecular genetics. 2015;24:649–658. doi: 10.1093/hmg/ddu485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chon H et al. RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic Acids Res. 2013;41:3130–3143. doi: 10.1093/nar/gkt027. Demonstrate that the functions of RNase H2 can be unlinked and establish that short deletion mutagenesis caused by unrepaired genomic ribonucleotides is due to failure of removal of single ribonucleotides from DNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim N, et al. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. Show that the short deletion mutagenesis associated with loss of RNase H2 activity in yeast is caused by topoisomerase 1 cleavage at a ribonucleotide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sekiguchi J, Shuman S. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol Cell. 1997;1:89–97. doi: 10.1016/s1097-2765(00)80010-6. [DOI] [PubMed] [Google Scholar]
- 71.Conover HN, et al. Stimulation of Chromosomal Rearrangements by Ribonucleotides. Genetics. 2015;201:951–961. doi: 10.1534/genetics.115.181149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ashour ME, Atteya R, El-Khamisy SF. Topoisomerase-mediated chromosomal break repair: an emerging player in many games. Nature reviews. Cancer. 2015;15:137–151. doi: 10.1038/nrc3892. [DOI] [PubMed] [Google Scholar]
- 73.Lippert MJ, et al. Role for topoisomerase 1 in transcription-associated mutagenesis in yeast. Proc Natl Acad Sci U S A. 2011;108:698–703. doi: 10.1073/pnas.1012363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Takahashi T, Burguiere-Slezak G, Van der Kemp PA, Boiteux S. Topoisomerase 1 provokes the formation of short deletions in repeated sequences upon high transcription in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2011;108:692–697. doi: 10.1073/pnas.1012582108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cho JE, Kim N, Li YC, Jinks-Robertson S. Two distinct mechanisms of Topoisomerase 1-dependent mutagenesis in yeast. DNA Repair (Amst) 2013;12:205–211. doi: 10.1016/j.dnarep.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cho JE, Kim N, Jinks-Robertson S. Topoisomerase 1-dependent deletions initiated by incision at ribonucleotides are biased to the non-transcribed strand of a highly activated reporter. Nucleic Acids Res. 2015;43:9306–9313. doi: 10.1093/nar/gkv824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu H, Droge P. Replication-induced supercoiling: a neglected DNA transaction regulator? Trends in biochemical sciences. 2014;39:219–220. doi: 10.1016/j.tibs.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 78.Caldecott KW. Molecular biology. Ribose–an internal threat to DNA. Science. 2014;343:260–261. doi: 10.1126/science.1248234. [DOI] [PubMed] [Google Scholar]
- 79.DeRose EF, Perera L, Murray MS, Kunkel TA, London RE. Solution structure of the Dickerson DNA dodecamer containing a single ribonucleotide. Biochemistry. 2012;51:2407–2416. doi: 10.1021/bi201710q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koh KD, Chiu HC, Riedo E, Storici F. Measuring the Elasticity of Ribonucleotide(s)-Containing DNA Molecules Using AFM. Methods Mol Biol. 2015;1297:43–57. doi: 10.1007/978-1-4939-2562-9_3. [DOI] [PubMed] [Google Scholar]
- 81.Jaishree TN, van der Marel GA, van Boom JH, Wang AH. Structural influence of RNA incorporation in DNA: quantitative nuclear magnetic resonance refinement of d(CG)r(CG)d(CG) and d(CG)r(C)d(TAGCG) Biochemistry. 1993;32:4903–4911. doi: 10.1021/bi00069a027. [DOI] [PubMed] [Google Scholar]
- 82.Egli M, Usman N, Rich A. Conformational influence of the ribose 2′-hydroxyl group: crystal structures of DNA-RNA chimeric duplexes. Biochemistry. 1993;32:3221–3237. [PubMed] [Google Scholar]
- 83.Ban C, Ramakrishnan B, Sundaralingam M. A single 2’-hydroxyl group converts B-DNA to A-DNA – Crystal structure of the DNA-RNA chimeric decamer duplex d(CCGGC)R(CCGG) with a novel intermolecular G-center-dot-C base-paired quadruplet. J Mol Biol. 1994;236:275–285. doi: 10.1006/jmbi.1994.1134. [DOI] [PubMed] [Google Scholar]
- 84.Rychlik MP, et al. Crystal structures of RNase H2 in complex with nucleic acid reveal the mechanism of RNA-DNA junction recognition and cleavage. Mol Cell. 2010;40:658–670. doi: 10.1016/j.molcel.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chiu HC, et al. RNA intrusions change DNA elastic properties and structure. Nanoscale. 2014;6:10009–10017. doi: 10.1039/c4nr01794c. [DOI] [PubMed] [Google Scholar]
- 86.Li Y, Breaker RR. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. Journal of the American Chemical Society. 1999;121:5326–5372. [Google Scholar]
- 87.Alvey HS, Gottardo FL, Nikolova EN, Al-Hashimi HM. Widespread transient Hoogsteen base pairs in canonical duplex DNA with variable energetics. Nature communications. 2014;5:4786. doi: 10.1038/ncomms5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dunn K, Griffith JD. The presence of RNA in a double helix inhibits its interaction with histone protein. Nucleic Acids Res. 1980;8:555–566. doi: 10.1093/nar/8.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hovatter KR, Martinson HG. Ribonucleotide-induced helical alteration in DNA prevents nucleosome formation. Proc Natl Acad Sci U S A. 1987;84:1162–1166. doi: 10.1073/pnas.84.5.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen JZ, Qiu J, Shen B, Holmquist GP. Mutational spectrum analysis of RNase H(35) deficient Saccharomyces cerevisiae using fluorescence-based directed termination PCR. Nucleic Acids Res. 2000;28:3649–3656. doi: 10.1093/nar/28.18.3649. Demonstrate that the spontanous mutation spectrum of an RNase H2-deficient budding yeast strain is dominated by a 4 base pair deletion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Clark AB, Lujan SA, Kissling GE, Kunkel TA. Mismatch repair-independent tandem repeat sequence instability resulting from ribonucleotide incorporation by DNA polymerase epsilon. DNA Repair (Amst) 2011;10:476–482. doi: 10.1016/j.dnarep.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sparks JL, Burgers PM. Error-free and mutagenic processing of topoisomerase 1-provoked damage at genomic ribonucleotides. EMBO J. 2015;34:1259–1269. doi: 10.15252/embj.201490868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang SY, Ghosh S, Pommier Y. Topoisomerase I alone is sufficient to produce short DNA deletions and can also reverse nicks at ribonucleotide sites. J Biol Chem. 2015;290:14068–14076. doi: 10.1074/jbc.M115.653345. Together with reference 92, show that topoisomerase 1-processing of genomic ribonucleotides can be either error-free or mutagenic and biochemically define these pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Potenski CJ, Niu H, Sung P, Klein HL. Avoidance of ribonucleotide-induced mutations by RNase H2 and Srs2-Exo1 mechanisms. Nature. 2014;511:251–254. doi: 10.1038/nature13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gambus A, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nature cell biology. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 96.Wu J, Phatnani HP, Hsieh TS, Greenleaf AL. The phosphoCTD-interacting domain of Topoisomerase I. Biochemical and biophysical research communications. 2010;397:117–119. doi: 10.1016/j.bbrc.2010.05.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Watt DL, Johansson E, Burgers PM, Kunkel TA. Replication of ribonucleotide-containing DNA templates by yeast replicative polymerases. DNA Repair (Amst) 2011;10:897–902. doi: 10.1016/j.dnarep.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Clausen AR, Murray MS, Passer AR, Pedersen LC, Kunkel TA. Structure-function analysis of ribonucleotide bypass by B family DNA replicases. Proc Natl Acad Sci U S A. 2013;110:16802–16807. doi: 10.1073/pnas.1309119110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Storici F, Bebenek K, Kunkel TA, Gordenin DA, Resnick MA. RNA-templated DNA repair. Nature. 2007;447:338–341. doi: 10.1038/nature05720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lazzaro F, et al. RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol Cell. 2012;45:99–110. doi: 10.1016/j.molcel.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ulrich HD. Timing and spacing of ubiquitin-dependent DNA damage bypass. FEBS Lett. 2011;585:2861–2867. doi: 10.1016/j.febslet.2011.05.028. [DOI] [PubMed] [Google Scholar]
- 102.Tumbale P, Williams JS, Schellenberg MJ, Kunkel TA, Williams RS. Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity. Nature. 2014;506:111–115. doi: 10.1038/nature12824. Demonstrates that abortive DNA ligation occurs at RNase H2 incision sites in DNA to generate an adenylated 5' end that requires the Aprataxin deadenylase for repair. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Moreira MC, et al. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat Genet. 2001;29:189–193. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- 104.Gao R, et al. Proteolytic degradation of topoisomerase II (Top2) enables the processing of Top2.DNA and Top2.RNA covalent complexes by tyrosyl-DNA-phosphodiesterase 2 (TDP2) J Biol Chem. 2014;289:17960–17969. doi: 10.1074/jbc.M114.565374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Andres SN, Schellenberg MJ, Wallace BD, Tumbale P, Williams RS. Recognition and repair of chemically heterogeneous structures at DNA ends. Environmental and molecular mutagenesis. 2015;56:1–21. doi: 10.1002/em.21892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wallace BD, Williams RS. Ribonucleotide triggered DNA damage and RNA-DNA damage responses. RNA biology. 2014;11:1340–1346. doi: 10.4161/15476286.2014.992283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Aguilera A, Klein HL. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics. 1988;119:779–790. doi: 10.1093/genetics/119.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Anand RP, Lovett ST, Haber JE. Break-induced DNA replication. Cold Spring Harb Perspect Biol. 2013;5:a010397. doi: 10.1101/cshperspect.a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wahba L, Amon JD, Koshland D, Vuica-Ross M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell. 2011;44:978–988. doi: 10.1016/j.molcel.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Allen-Soltero S, Martinez SL, Putnam CD, Kolodner RD. A saccharomyces cerevisiae RNase H2 interaction network functions to suppress genome instability. Mol Cell Biol. 2014;34:1521–1534. doi: 10.1128/MCB.00960-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ii M, Ii T, Mironova LI, Brill SJ. Epistasis analysis between homologous recombination genes in Saccharomyces cerevisiae identifies multiple repair pathways for Sgs1, Mus81-Mms4 and RNase H2. Mutation research. 2011;714:33–43. doi: 10.1016/j.mrfmmm.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.O’Connell K, Jinks-Robertson S, Petes TD. Elevated Genome-Wide Instability in Yeast Mutants Lacking RNase H Activity. Genetics. 2015;201:963–975. doi: 10.1534/genetics.115.182725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Crow YJ, et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat Genet. 2006;38:910–916. doi: 10.1038/ng1842. Reports a genetic association between mutations in RNase H2 and the rare neuroinflammatory disorder, AGS. [DOI] [PubMed] [Google Scholar]
- 114.Reijns MA, Jackson AP. Ribonuclease H2 in health and disease. Biochemical Society transactions. 2014;42:717–725. doi: 10.1042/BST20140079. [DOI] [PubMed] [Google Scholar]
- 115.Reijns MA, et al. The structure of the human RNase H2 complex defines key interaction interfaces relevant to enzyme function and human disease. J Biol Chem. 2011;286:10530–10539. doi: 10.1074/jbc.M110.177394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Figiel M, et al. The structural and biochemical characterization of human RNase H2 complex reveals the molecular basis for substrate recognition and Aicardi-Goutieres syndrome defects. J Biol Chem. 2011;286:10540–10550. doi: 10.1074/jbc.M110.181974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gunther C, et al. Defective removal of ribonucleotides from DNA promotes systemic autoimmunity. The Journal of clinical investigation. 2015;125:413–424. doi: 10.1172/JCI78001. Reports a genetic association between mutations in RNase H2 and the autoimmune disease, systemic lupus erythematosus, and links defective ribonucleotide removal to enhanced photosensitivity and immune signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schellenberg MJ, Tumbale PP, Williams RS. Molecular underpinnings of Aprataxin RNA/DNA deadenylase function and dysfunction in neurological disease. Progress in biophysics and molecular biology. 2015;117:157–165. doi: 10.1016/j.pbiomolbio.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Akbari M, Sykora P, Bohr VA. Slow mitochondrial repair of 5'-AMP renders mtDNA susceptible to damage in APTX deficient cells. Scientific reports. 2015;5:12876. doi: 10.1038/srep12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Potenski CJ, Klein HL. How the misincorporation of ribonucleotides into genomic DNA can be both harmful and helpful to cells. Nucleic Acids Res. 2014;42:10226–10234. doi: 10.1093/nar/gku773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Donigan KA, McLenigan MP, Yang W, Goodman MF, Woodgate R. The steric gate of DNA polymerase iota regulates ribonucleotide incorporation and deoxyribonucleotide fidelity. J Biol Chem. 2014;289:9136–9145. doi: 10.1074/jbc.M113.545442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Makarova AV, Nick McElhinny SA, Watts BE, Kunkel TA, Burgers PM. Ribonucleotide incorporation by yeast DNA polymerase zeta. DNA Repair (Amst) 2014;18:63–67. doi: 10.1016/j.dnarep.2014.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sastre-Moreno G, Sanchez A, Esteban V, Blanco L. ATP insertion opposite 8-oxo-deoxyguanosine by Pol4 mediates error-free tolerance in Schizosaccharomyces pombe. Nucleic Acids Res. 2014;42:9821–9837. doi: 10.1093/nar/gku711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nick McElhinny SA, Ramsden DA. Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol Cell Biol. 2003;23:2309–2315. doi: 10.1128/MCB.23.7.2309-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cilli P, Minoprio A, Bossa C, Bignami M, Mazzei F. Formation and Repair of Mismatches Containing Ribonucleotides and Oxidized Bases at Repeated DNA Sequences. J Biol Chem. 2015;290:26259–26269. doi: 10.1074/jbc.M115.679209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bergoglio V, Ferrari E, Hubscher U, Cazaux C, Hoffmann JS. DNA polymerase beta can incorporate ribonucleotides during DNA synthesis of undamaged and CPD-damaged DNA. J Mol Biol. 2003;331:1017–1023. doi: 10.1016/s0022-2836(03)00837-4. [DOI] [PubMed] [Google Scholar]
- 127.Donigan KA, et al. Unlocking the steric gate of DNA polymerase eta leads to increased genomic instability in Saccharomyces cerevisiae. DNA Repair (Amst) 2015;35:1–12. doi: 10.1016/j.dnarep.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gosavi RA, Moon AF, Kunkel TA, Pedersen LC, Bebenek K. The catalytic cycle for ribonucleotide incorporation by human DNA Pol lambda. Nucleic Acids Res. 2012;40:7518–7527. doi: 10.1093/nar/gks413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vengrova S, Dalgaard JZ. RNase-sensitive DNA modification(s) initiates S. pombe mating-type switching. Genes Dev. 2004;18:794–804. doi: 10.1101/gad.289404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vengrova S, Dalgaard JZ. The wild-type Schizosaccharomyces pombe mat1 imprint consists of two ribonucleotides. EMBO reports. 2006;7:59–65. doi: 10.1038/sj.embor.7400576. Introduces the concept of genomic ribonucleotides as providing an important developmental signal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sayrac S, Vengrova S, Godfrey EL, Dalgaard JZ. Identification of a novel type of spacer element required for imprinting in fission yeast. PLoS Genet. 2011;7:e1001328. doi: 10.1371/journal.pgen.1001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dalgaard JZ. Causes and consequences of ribonucleotide incorporation into nuclear DNA. Trends Genet. 2012;28:592–597. doi: 10.1016/j.tig.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 133.Reijns MA, et al. Lagging-strand replication shapes the mutational landscape of the genome. Nature. 2015;518:502–506. doi: 10.1038/nature14183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ding J, Taylor MS, Jackson AP, Reijns MA. Genome-wide mapping of embedded ribonucleotides and other noncanonical nucleotides using emRiboSeq and EndoSeq. Nature protocols. 2015;10:1433–1444. doi: 10.1038/nprot.2015.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ghodgaonkar MM, et al. Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair. Mol Cell. 2013;50:323–332. doi: 10.1016/j.molcel.2013.03.019. Together with reference 50, demonstrate the ability of ribonucleotides to act as a strand-discrimination signal during MMR of replication errors introduced during leading strand DNA synthesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kunkel TA, Erie DA. Eukaryotic Mismatch Repair in Relation to DNA Replication. Annual review of genetics. 2015;49:291–313. doi: 10.1146/annurev-genet-112414-054722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Arana ME, et al. Transcriptional responses to loss of RNase H2 in Saccharomyces cerevisiae. DNA Repair (Amst) 2012;11:933–941. doi: 10.1016/j.dnarep.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem. 2009;284:4041–4045. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zheng L, Shen B. Okazaki fragment maturation: nucleases take centre stage. J Mol Cell Biol. 2011;3:23–30. doi: 10.1093/jmcb/mjq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Balakrishnan L, Bambara RA. Okazaki fragment metabolism. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Garg P, Stith CM, Sabouri N, Johansson E, Burgers PM. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18:2764–2773. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Qiu J, Qian Y, Frank P, Wintersberger U, Shen B. Saccharomyces cerevisiae RNase H(35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol Cell Biol. 1999;19:8361–8371. doi: 10.1128/mcb.19.12.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Levikova M, Cejka P. The Saccharomyces cerevisiae Dna2 can function as a sole nuclease in the processing of Okazaki fragments in DNA replication. Nucleic Acids Res. 2015;43:7888–7897. doi: 10.1093/nar/gkv710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kang YH, Lee CH, Seo YS. Dna2 on the road to Okazaki fragment processing and genome stability in eukaryotes. Critical reviews in biochemistry and molecular biology. 2010;45:71–96. doi: 10.3109/10409230903578593. [DOI] [PubMed] [Google Scholar]
- 145.Boule JB, Rougeon F, Papanicolaou C. Terminal deoxynucleotidyl transferase indiscriminately incorporates ribonucleotides and deoxyribonucleotides. J Biol Chem. 2001;276:31388–31393. doi: 10.1074/jbc.M105272200. [DOI] [PubMed] [Google Scholar]
- 146.Yang MY, et al. Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell. 2002;111:495–505. doi: 10.1016/s0092-8674(02)01075-9. [DOI] [PubMed] [Google Scholar]
- 147.Grossman LI, Watson R, Vinograd J. The presence of ribonucleotides in mature closed-circular mitochondrial DNA. Proc Natl Acad Sci U S A. 1973;70:3339–3343. doi: 10.1073/pnas.70.12.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shen Y, Koh KD, Weiss B, Storici F. Mispaired rNMPs in DNA are mutagenic and are targets of mismatch repair and RNases H. Nature structural & molecular biology. 2012;19:98–104. doi: 10.1038/nsmb.2176. [DOI] [PubMed] [Google Scholar]


