Abstract
TGFβ-SMAD signaling exerts a contextual effect that suppresses malignant growth early in epithelial tumorigenesis but promotes metastasis at later stages. Longstanding challenges in resolving this functional dichotomy may uncover new strategies to treat advanced carcinomas. The Krüppel-like transcription factor KLF10 is a pivotal effector of TGFβ/SMAD signaling that mediates anti-proliferative effects of TGFβ. In this study, we show how KLF10 opposes the pro-metastatic effects of TGFβ by limiting its ability to induce epithelial-to-mesenchymal transition (EMT). KLF10 depletion accentuated induction of EMT as assessed by multiple metrics. KLF10 occupied GC-rich sequences in the promoter region of the EMT-promoting transcription factor SLUG/SNAI2, repressing its transcription by recruiting HDAC1 and licensing the removal of activating histone acetylation marks. In clinical specimens of lung adenocarcinoma, low KLF10 expression associated with decreased patient survival, consistent with a pivotal role for KLF10 in distinguishing the anti-proliferative versus pro-metastatic functions of TGFβ. Our results establish that KLF10 functions to suppress TGFβ-induced EMT, establishing a molecular basis for the dichotomy of TGFβ function during tumor progression.
Keywords: Epithelial-to-mesenchymal transition, Krüppel-like Factor 10, SNAI2, Epigenetic, Transforming Growth Factor-beta
Introduction
Non-Small Cell Lung Carcinoma (NSCLC) is the leading cause of cancer-related deaths in the United States and around the world (1). Therapeutic resistance and tumor metastasis are the most common causes of lung cancer-related death. Despite recent progress in early detection and therapeutic efficacy, most lung cancers are diagnosed at an advanced or metastatic stage and, thus, overall patient survival rates remain very poor (2).
Epithelial-to-mesenchymal transition (EMT) is an evolutionary conserved trans-differentiation process proposed to play a central role in cancer metastasis, therapeutic resistance and cancer stem-cell like features (3). EMT plays an important role during embryonic development and is indispensable for tissue and organ development in multicellular organisms (4). During EMT, epithelial cells lose apico-basolateral polarity and dissolution of adherence junctions occurs, resulting in the loss of cell-cell contact and an opening of the basement membrane barrier (5). Cells attain front-to-rear polarity, become spindle shaped, and gain migratory and invasive capacity. Upon completion of EMT, expression of epithelial markers like E-cadherin (CDH1) and Zona Occludens 1 (ZO-1) is suppressed, whereas mesenchymal markers such as N-cadherin (CDH2), Vimentin (VIM) and Fibronectin (FN1) are upregulated. Genes encoding various transcription factors including Snail (SNAI1), Slug (SNAI2), ZEB1 and ZEB2 (collectively termed as EMT-TFs) become activated following the extracellular stimuli and carry out the downstream nuclear regulation of EMT (6). EMT is proposed to play an important role in driving cancer progression by enabling tumor cells to become more invasive and migratory, which are essential features for promoting tumor metastasis (7).
One important EMT-TF is SNAI2 (also referred to as Slug), a member of the Snail family of transcriptional repressors (8). SNAI2 promotes EMT, in part, by binding to an E-box element in the promoter proximal region of the CDH1 gene (encoding E-cadherin) and repressing its transcription (9). Reduced E-cadherin expression has been shown to be associated with poorly differentiated and metastatic NSCLC. Furthermore, SNAI2 is frequently overexpressed in different types of metastatic cancers including lung adenocarcinoma (10).
EMT is controlled by various signaling pathways with one of the most potent inducers being Transforming Growth Factor-β (TGFβ). TGFβ is a pleiotropic cytokine which exerts anti-proliferative and tumor suppressive effects in normal cells and/or in early stages of cancer, but also elicits pro-metastatic functions during cancer progression (11). TGFβ signaling occurs through a heteromeric complex containing two each of the type 1 and type 2 transmembrane serine/threonine kinase receptors (TβRI and TβRII). Binding of TGFβ ligand to TβRII facilitates interaction with and phosphorylation of TβRI, which in turn phosphorylates the SMAD2 and SMAD3 proteins (also known as R-Smad). SMAD2/3 proteins then form a heteromeric complex with SMAD4 (also known as a Co-Smad) and translocate to the nucleus. Subsequently, activated SMADs regulate target genes either by directly binding to the DNA at so-called Smad binding elements (SBE) or through interactions with other transcription factors (12). Numerous studies have documented both the anti-cancer and pro-metastatic effects of TGFβ signaling, however, the mechanisms behind the switch between these two functions of TGFβ signaling is not completely understood (13).
Krüppel-Like Factor-10 (KLF10) is a zinc finger-containing transcription factor originally identified as an early response gene following TGFβ treatment of human osteoblasts and thus initially named TGFβ-Inducible Early Gene-1 (TIEG1) (14). Like other KLF family members, the KLF10 protein contains three zinc-fingers at its carboxy terminal end which allow for the regulation of gene transcription via direct binding to GC-rich DNA sequences at target genes (15). Overexpression of KLF10 was shown to mimic the anti-proliferative and apoptotic effects of TGFβ in various cell lines (16–19). Importantly, KLF10 serves as a positive feedback loop for regulating TGFβ signaling by inducing the expression of SMAD2 and inhibiting the expression of the inhibitory SMAD7 gene (20,21). KLF10 functions primarily as a transcriptional repressor and regulates target gene expression by recruiting various epigenetic co-repressors including Histone Deacetylases -1 and -2 (HDAC1 and -2) and Lysine Demethylase-5B (KDM5B) to promoter regions of target genes (22,23). Importantly, multiple studies have demonstrated a putative tumor suppressor role for KLF10 in different types of cancer (24–26).
In this study, we demonstrate that Klf10-deficient mice have a significantly increased incidence of lung tumor formation and increased tumor size following DMBA treatment compared to wild type mice. Moreover, using various cell culture-based assays as well as transcriptome- and genome-wide analyses, we demonstrate that KLF10 suppress TGFβ-induced EMT. We provide evidence that KLF10 binds to and represses the SNAI2 promoter by recruiting HDAC1. Together these findings identify KLF10 as central component of the tumor suppressor function of TGFβ.
Materials and methods
Cell culture and transfections
A549 (lung adenocarcinoma cell line) cells were kindly provided by Prof. Ekkehard Dikomey (University Hospital Eppendorf, Hamburg, Germany) and Panc1 (pancreatic cancer cell line) cells by Dr. Elisabeth Hessmann (University Medical Center, Göttingen, Germany). H1299 cells (non-small cell lung carcinoma cell line) were a kind gift from Dr. Muriel Lize (University Medical Center, Göttingen, Germany). MDA-MB-231 cells were a kind gift from Dr. Harriet Wikman (University Hospital Eppendorf, Hamburg, Germany). Cell lines used in this study were authenticated by DNA profiling using eight different and highly polymorphic short tandem repeat (STR) loci. A549 and Panc1 cells were cultured in phenol red-free Dulbecco’s Modified Eagle’s Media/F12 (DMEM/F12; Invitrogen), MDA-MB-231 cells in DMEM (Invitrogen) and H1299 cells in RPMI-1640 (Invitrogen) supplemented with 10% fetal bovine serum (Sigma) and 1X Pen/Strep (Sigma) at 37°C under 5% CO2. siRNA transfections were performed using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. SmartPoolR siRNA against KLF10 (Dharmacon) consisted of the following sequences: 5′-GAAGAACCCACCUAAAUGU-3′, 5′-GAUAAGGAGUCACAUCUGU-3′, 5′-GAAGUGAGCAAGCUAAAUG-3′, 5′-CACCAGACCUGCCCAAUGA-3′. siGENOME Non-targeting siRNA (Dharmacon; D-001206-13) was used as a negative control. For plasmid transfections, cells were transfected at ca. 70% confluency with pEGFP-C1 or pEGFP-C1-TIEG expression plasmids using Lipofectamine-2000 (Invitrogen) according to the manufacturer’s instructions. For inducing EMT, cells were treated with 5 ng/ml TGFβ (R&D systems) for 90 minutes or 72 hours.
Antibodies
The following commercially available antibodies were used for ChIP, western blotting and immunofluorescence: Vimentin (clone v9, Santa Cruz, sc-6260, 1:400 for IF), E-cadherin (clone 24E10, Cell Signaling, 3195, 1:200 for IF, 1:1000 for western blotting), ZO-1 (clone D7D12, Cell Signaling, 8193, 1:200 for IF), SNAI2 (clone C19G7, Cell Signaling, 9585, 1:200 for IF, 1:1000 for western blotting), KLF10 (clone EPR12102, Abcam, ab184182, 1:1000 for western blotting, 1μg for ChIP), H3K27ac (Diagenode, C15410196, 1μg for ChIP), H3K9ac (Diagenode, C15410004, 1μg for ChIP), HDAC1 (Diagenode, C15410053, 2μg for ChIP), HSC70 (clone B-6, Santa Cruz, sc-7298, 1:10,000 for western blotting), non-specific IgG (ab46540, 1μg for ChIP).
Migration assay
Transwell migration assays were performed to analyze the migratory potential of cells as described previously (27). Twenty-four hours after siRNA transfection cells were treated with 5 ng/ml TGFβ for 48 hours. Cells were trypsinized, counted and 2.5 × 104 cells were seeded into cell culture inserts with a pore size of 8.0 μm (BD biosciences) and TGFβ treatment was continued for another 48 hours. Migrated cells were then fixed with methanol for 10 minutes followed by staining with 0.1% (w/v) crystal violet dissolved in 10% (v/v) formaldehyde for 10 minutes.
Immunofluorescence staining
Cells were grown on coverslips in 24-well plates as described previously (27). Cells were gently washed with 1× PBS twice, fixed with 4% paraformaldehyde for 10 minutes and then washed with PBS. Permeabilization of the cells was performed using 0.1% Triton X-100 (prepared in PBS) for 10 minutes followed by two washes with PBS before blocking with PBS containing 10% FBS for 20 minutes. Cells were then incubated with the primary antibody diluted in blocking solution overnight at 4°C. The next day, cells were washed three times with PBS and then incubated for 1 hour at room temperature with secondary antibodies conjugated with Alexa-488 (A11008) or Alexa-594 (A11005) (Invitrogen, CA). Coverslips were washed twice with PBS, incubated with DAPI (Sigma, D9542, diluted at 1:15,000 in PBS) for 5 minutes at room temperature and then mounted on glass slides using mounting media (Dako, S3023). Images were captured using the AXIO Scope.A1 microscope (Zeiss) and data were analyzed using the ZEN 2 lite software.
Western blot and gene expression analysis
Cell lysates were prepared in RIPA buffer (1% (v/v) NP-40, 0.5% (w/v) sodium deoxycholate and 0.1% (w/v) SDS in 1X PBS) along with protease inhibitors (1 mM Pefabloc, 1 ng/μL Aprotinin/Leupeptin, 10 mM β-glycerophosphate and 1 mM N-ethylmaleimide) as described previously (27). Following sonication to ensure solubilization of chromatin-associated proteins, equal amounts of protein were incubated with SDS loading dye at 95 °C for 10 minutes. Samples were then separated by polyacrylamide gel electrophoresis (PAGE) and analyzed by western blotting using the indicated antibodies.
For analysis of gene expression, total RNA was isolated using QIAzol reagent (Qiagen) as per the manufacturer’s instructions. One microgram of total RNA was reverse transcribed using random nonamers (Sigma). Quantitative real-time PCR (qRT-PCR) was performed as previously described (28) using the primers listed in Supplemental Table S1. Gene expression was normalized to HNRNPK and represented as fold induction relative to the negative control siRNA transfected cells. All experiments were performed using biological triplicates.
Luciferase assay
For luciferase assays, the Dual-Luciferase Reporter Assay System (Promega) was used as previously described (29). Briefly, cells were co-transfected with a SNAI2-promoter containing luciferase promoter construct (kindly provided by S. Hüttelmaier; (30) together with a plasmid expressing Renilla luciferase (pRL-TK, Promega) and either an empty control vector (pSG5) or KLF10 expression vector. Firefly luciferase activity was normalized to Renilla luciferase activity to control for transfection efficiency. Luminescence measurements were performed using a Centro LB 960 Microplate Luminometer (Berthold Technologies).
Animal studies
Klf10 knock-out mice were described previously (31). Neonatal (2–5 day old) pups were treated (dorsally) with 0.05 ml of 0.5% DMBA (Sigma #D3254) in acetone as previously described (32). Mice were housed until four months of age at which time they were sacrificed and lungs were removed and examined under a dissecting scope for the presence of tumors. Tumor nodules were counted and measured for each mouse and group averages were determined.
Chromatin immunoprecipitation, ChIP-sequencing and data analysis
Chromatin immunoprecipitation was performed essentially as described previously (33). Cells were cross-linked with 1% formaldehyde for 15 minutes (for KLF10 ChIP) or 10 minutes (for H3K9Ac, H3K27Ac and HDAC1). Prior to library preparation, DNA samples were sonicated a second time using a Bioruptor® Pico (Diagenode) to ensure fragment sizes of approximately ≤ 300 bp. Fragmented DNA was used for library preparation using NEBNext Ultra DNA library preparation kit (New England Biolabs) according to the manufacturer’s protocol. IgG was used as a negative control. DNA was quantitated using the Qubit dsDNA HS assay and a Qubit® 2.0 Fluorimeter (Invitrogen). Three to five ng of DNA was used for library preparation. The fragment size of the libraries was analyzed using an Agilent Bioanalyzer 2100 (High Sensitivity DNA assay). Libraries were sequenced using HiSeq 2500 (Illumina) and sequence reads (single end, 50 bp) were aligned to the human reference genome (UCSC hg19) using Bowtie2 with default parameters (version 0.4) (34). Peak calling was performed using Model-based Analysis of ChIP-seq (MACS2) (version 2.1.0.20140616.0) on Galaxy Cistrome (35). The minimum FDR cutoff for peak detection (q-value) was set to 0.05. Bigwig files were generated using BamCoverage from deeptools (36). The bigwig data was visualized using Integrative Genomics Viewer (IGV, version 2.3.66) (37). Furthermore, bigwig and bed files were also used to generate aggregate profiles and relative enrichment of the ChIP regions over defined genomic locations using SitePro and CEAS tools, respectively, using the Galaxy Cistrome server. ChIP-seq and RNA-seq data have been deposited in Gene Expression Omnibus and are available under the accession number GSE90567.
RNA-sequencing and data analysis
RNA-seq was performed in triplicate for Panc1 and duplicate for A549 cells. Total RNA was isolated and RNA integrity was confirmed using an Agilent Bioanalyzer 2100. Five hundred ng total RNA was used to prepare the libraries using NEXTflex Rapid Directional RNA-Seq Kit (Bioo Scientific). Fastq files were mapped to the human transcriptome (hg19) using TopHat tool (38) on Galaxy (Bowtie2 settings with preset option set to ‘very sensitive’). BAM files were coordinate-sorted using SortSam (version 1.126.0) from Picard tools (39) on Galaxy. Subsequent files were used for read counting using the HTSeq tool (version 0.6.0) (40) and then the count files were used for measuring differential gene expression using DESeq2 (41) on R (Bioconductor version 3.2). Gene Ontology analyses were performed using DAVID bioinformatics resources (42). Significantly enriched GO categories were selected based on FDR value ≤ 0.05. Gene Set Enrichment Analysis (GSEA) (43) was performed with standard parameters (1000 permutations for gene sets, Signal2Noise metric for ranking genes) and significantly enriched pathways (c5.all gene sets) were shown.
Statistical analysis
Data are represented as mean ± SD. One-way ANOVA test was used to test the statistical significance of qRT-PCR data. 2-tailed Fisher’s exact test and 1-sided Student’s t-test were used to calculate the statistical significance of in vivo data. A p-value of ≤ 0.05 was considered statistically significant. p-values ≤ 0.05, ≤ 0.01 and ≤ 0.001 are represented by single, double and triple asterisks respectively.
Results
KLF10 is a lung cancer tumor suppressor in vivo
To investigate the role of KLF10 in lung cancer, we analyzed its expression in both normal as well as lung adenocarcinoma samples (Oncomine.org). Interestingly, KLF10 expression was significantly downregulated in tumor samples compared to the normal samples (Fig. 1A). Importantly, Kaplan-Meier analyses (44) revealed that low KLF10 expression was associated with significantly worse survival in patients with lung adenocarcinoma, demonstrating a potential tumor suppressor function of KLF10 in human cancer (Fig. 1B). In order to determine whether KLF10 indeed functions as a tumor suppressor in lung cancer, we performed an in vivo tumor incidence study in Klf10−/− mice to investigate the role for KLF10 in lung tumorigenesis. To induce lung cancer in wild type and Klf10−/− mice, neonatal littermates were treated with the chemical carcinogen 7,12-Dimethylbenz(a)anthracene (DMBA), which induces lung adenocarcinoma (45). Interestingly, we observed a significantly higher lung tumor incidence (Fig. 1D) and increased tumor size (Fig. 1E) in Klf10−/− mice as compared to wild type controls. Together these results demonstrate an inverse correlation between KLF10 expression and patient outcome, as well as lung cancer development in mice, underscoring a tumor suppressor role for KLF10.
Figure 1.
KLF10 expression is decreased in lung adenocarcinoma. A, KLF10 expression is significantly downregulated in lung adenocarcinoma. Gene expression in primary lung cancer and the corresponding normal samples were analyzed using the Oncomine database. B, Kaplan-Meier survival analysis displays that low KLF10 expression is associated with worse overall survival (OS) in lung adenocarcinoma (curves generated at kmplot.com). C, Lung tumor incidence in wild type (WT), Klf10+/− or Klf10-null mice following DMBA treatment (percentage). D, Average tumor diameter per genotype (mm). Data represent mean ± SD. n represents number of tumors per genotype.
KLF10 differentially regulates TGFβ-target genes
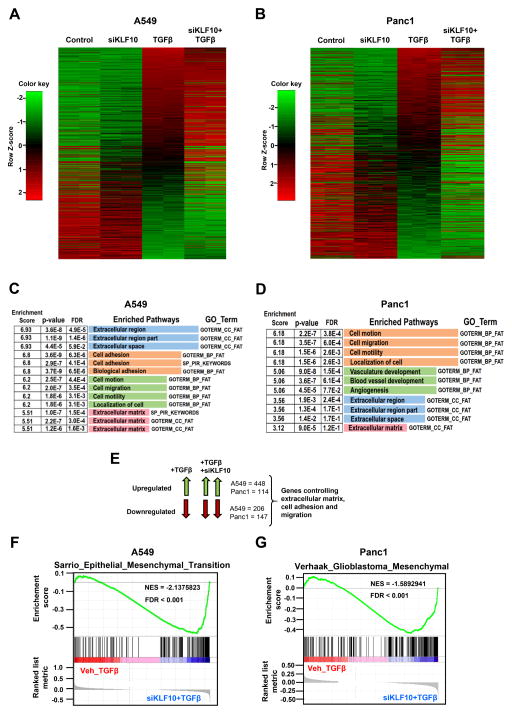
Since KLF10 has been proposed as a central regulator of TGFβ signaling, we performed transcriptome-wide analyses on TGFβ-regulated genes using RNA-seq in control and KLF10-depleted cells. We utilized two different TGFβ-responsive cell lines, A549 lung adenocarcinoma and Panc1 pancreatic ductal adenocarcinoma cells, the latter having previously been shown to display decreased proliferation and increased apoptosis upon KLF10 overexpression (16). Interestingly, heatmap analyses revealed that many TGFβ-regulated genes were positively or negatively affected in their induction following KLF10 knockdown (Fig. 2A & 2B). Further, we were interested to investigate the biological relevance of the TGFβ-regulated genes which were affected by KLF10 knockdown in TGFβ-stimulated cells. We performed Gene Ontology (GO) analyses using the DAVID (Database for Annotation, Visualization and Integrated Discovery) software. Interestingly, TGFβ-regulated genes differentially regulated in response to KLF10 depletion were associated with pathways controlling extracellular matrix, cell adhesion, cell migration and angiogenesis, pathways frequently associated with EMT (Fig. 2C & 2D). Gene Set Enrichment Analysis (GSEA) further confirmed a significant enrichment of pathways associated with EMT (Fig. 2F & 2G). Taken together these results indicate that KLF10 affects TGFβ-regulated gene sets that are associated with controlling EMT, poorly differentiated and metastatic cancer.
Figure 2.
KLF10 depletion has significant effects on TGFβ-regulated gene sets. A & B, Heatmap from RNA-seq data in A549 and Panc1 cells respectively for significantly up-(red) or down-(green) regulated genes (±1.5 log2fold, padj ≤ 0.05) upon TGFβ treatment (5 ng for 72 hours) and KLF10 knockdown. Significantly up- and down-regulated genes upon TGFβ treatment compared to the control sample were sorted from high to low. C & D, DAVID gene ontology analysis was performed on TGFβ-regulated genes which were significantly (padj ≤ 0.05) affected (±1.5 log2fold) by KLF10 knockdown. Significantly (p-value and FDR ≤ 0.05) enriched pathways from top 4 clusters (based on enrichment score) are shown. E, Schematic showing the overlapping sets of genes that were used for gene ontology analysis. F & G, Gene Set Enrichment Analysis (GSEA) was performed on RNA-Seq data from A549 and Panc1 cells. For comparison, TGFβ alone and TGFβ treatment combined with KLF10 knockdown samples were compared. EMT-related pathways were found to be significantly enriched in TGFβ-treated cells depleted with KLF10. Significance was determined based on NES (normalized enrichment score) and FDR (false discovery rate).
KLF10 occupancy is enriched at target gene promoters
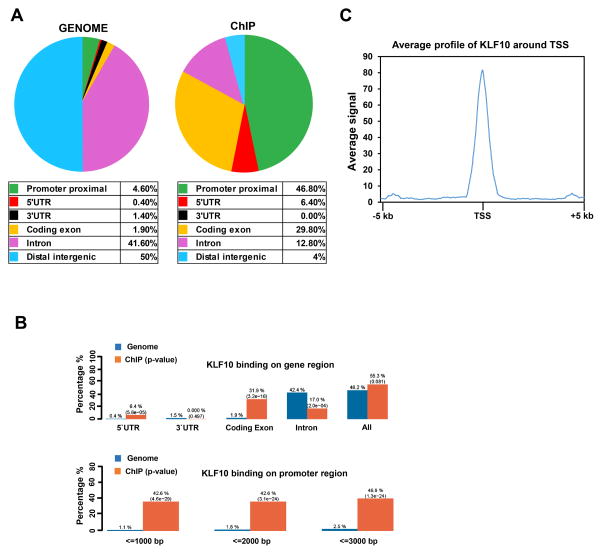
To investigate the genome-wide occupancy of KLF10, we performed chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq). We determined the enrichment of KLF10 at various functional genomic locations using the Cis-regulatory Element Annotation System (CEAS) which provides information about the enrichment of the ChIP regions at specific genomic locations. Interestingly, in comparison to the whole genome, KLF10 binding sites were localized largely to the promoter regions (46.8%) and coding exons (29.8%) of genes (Fig. 3A, 3B). Consistently, aggregate plot metagene analysis (± 5Kb around TSS) showed an enrichment of KLF10 occupancy near the transcriptional start site (TSS) of KLF10-occupied genes (Fig. 3C). Together these results revealed that KLF10 mainly occupies promoter regions of target genes.
Figure 3.
KLF10 is primarily localized to proximal promoter regions. A, B, CEAS (Cis-regulatory Element Annotation System) analysis of ChIP-seq data. Relative enrichment of specific gene regions such as promoters and gene bodies, in ChIP compared to the whole genome was obtained. Distribution of KLF10 occupancy at specific genomic locations reveals a significant enrichment at proximal promoter and 5′ regulatory regions (downstream ≤ 1000 bp; 5′ UTR) compared to the whole genome. C, Aggregate plot analysis was performed on KLF10-bound genes. Highest average signal for KLF10 enrichment was at the TSS (±5 kb) region of the KLF10-occupied genes.
KLF10 occupies the SNAI2 promoter and suppresses its expression
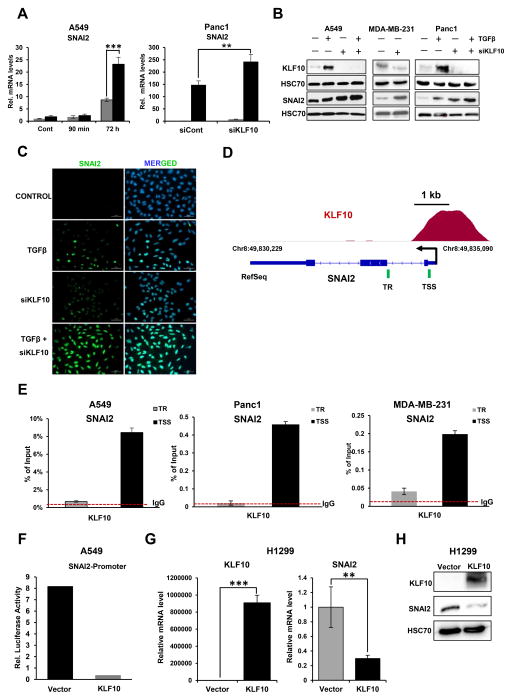
To further elucidate the mechanism underlying the role of KLF10 in controlling the TGFβ-regulated EMT-associated transcriptional program, we performed Genome Regulatory Element and Annotation Tool (GREAT) analyses to identify potential direct target genes of KLF10. By combining ChIP-seq and transcriptome data, we identified a limited number of direct target genes located within 25 kb of a KLF10 peak identified by ChIP-seq which were also differentially regulated in RNA-seq analyses (Supplementary Fig. S1A & B). Notably the EMT-TF SNAI2 (but not other EMT-TFs), as well as FosB, an AP1 transcription factor family member implicated in EMT (46), was significantly upregulated in KLF10-depleted cells treated with TGFβ. Based on this observation, we hypothesized that SNAI2 and FosB could be important targets of KLF10. Interestingly, KLF10 depletion resulted in significantly increased mRNA (Fig. 4A) and protein (Fig. 4B) levels of SNAI2 following TGFβ treatment in A549 and Panc1 cells. Consistently, we also observed increased mRNA levels of FosB in KLF10-depleted A549 cells (Supplementary Fig. S1C). Remarkably, depletion of KLF10 alone was sufficient to increase the protein level of SNAI2 in the metastatic breast cancer cell line MDA-MB-231 (Fig. 4B). Immunofluorescence staining in A549 cells confirmed these findings and showed enhanced nuclear expression of SNAI2 in KLF10-depleted cells treated with TGFβ (Fig. 4C). Consistently, analysis of our ChIP-seq data revealed significant occupancy of KLF10 at the proximal SNAI2 and FosB promoters, suggesting that KLF10 directly represses their activity (Fig. 4D and Supplementary Fig. S1D). This finding was confirmed by ChIP-qPCR assays in A549, Panc1 and MDA-MB-231 cells, which substantiated KLF10 occupancy near the TSS of SNAI2, but not on adjacent downstream site within the transcribed region (TR; Fig. 4E). Similarly, we could also confirm binding of KLF10 to the FosB gene in A549 cells (Supplementary Fig. S1E). Further, we examined the effect of TGFβ treatment on KLF10 occupancy at the SNAI2 gene and observed a significant increase (Supplementary Fig. S2).
Figure 4.
KLF10 is recruited to, and suppresses the expression of the SNAI2 gene. A, SNAI2 gene expression was analyzed by quantitative real-time (RT) PCR and shown as “relative mRNA levels” compared to the expression of an unregulated control gene (HNRNPK). Data are represented as mean ± SD. n=3. ***p ≤ 0.005, **p ≤ 0.01, *p ≤ 0.05. B, Increased SNAI2 protein levels were observed by Western blot analysis of whole cell lysate. Western blot showing the induction of KLF10 protein levels following TGFβ treatment and depletion following siRNA-mediated knockdown. HSC70 was used as a loading control. C, Immunofluorescence staining for SNAI2 in A549 cells. Marked increased expression of nuclear SNAI2 was observed in KLF10-depleted cells treated with TGFβ (5 ng/ml for 72 h) as compared to cells treated with TGFβ alone. Nuclei were stained with DAPI. Scale bar represents 50 μm. D, ChIP-seq occupancy profile of KLF10 on the SNAI2 gene. A significant peak was observed near the TSS of SNAI2. The TSS and direction of transcription are indicated by a black arrow. The green lines indicates the locations analyzed for quantitative analysis of KLF10 occupancy near the TSS and transcribed region (TR) in E. E, ChIP analysis of KLF10 occupancy on the SNAI2 gene. Immunoprecipitated DNA was compared to input and shown as percentage. IgG antibody was used as a negative control to determine the background level of binding and is shown as a red dotted line. Data are represented as mean ± SD. n=3. F, Dual luciferase reporter assay was performed in A549 cells to verify the effect of KLF10 on SNAI2 promoter activity. Cells were co-transfected with a SNAI2-promoter luciferase reporter construct and either an empty vector or a KLF10 expression vector. Renilla luciferase was used as an internal control for transfection efficiency and normalization. SNAI2 promoter activity is shown as relative firefly luciferase activity normalized to Renilla luciferase activity. G, Gene expression level of SNAI2 upon KLF10 overexpression in H1299 was analyzed by quantitative RT-PCR and shown as “relative mRNA levels” compared to the expression of an unregulated control gene (HNRNPK). Data are represented as mean ± SD. n=3. ***p ≤ 0.005, **p ≤ 0.01, *p ≤ 0.05. H, A marked reduction in SNAI2 protein levels was observed upon KLF10 overexpression in H1299 cells. HSC70 was used as a loading control.
We next further examined the potential role of KLF10 in controlling SNAI2 promoter activity using the dual luciferase reporter assay. As shown in Fig. 4F, co-transfection of a KLF10 expression vector led to a dramatic reduction in SNAI2 promoter activity as compared to vector-transfected control cells. In contrast, overexpression of KLF10 resulted in significant reduction of mRNA and protein levels of SNAI2 (Fig. 4G & 4H). Together, these results support a direct role of KLF10 in repressing the transcription of the SNAI2 gene at the promoter proximal region.
KLF10 recruits HDAC1 to repress SNAI2 transcription
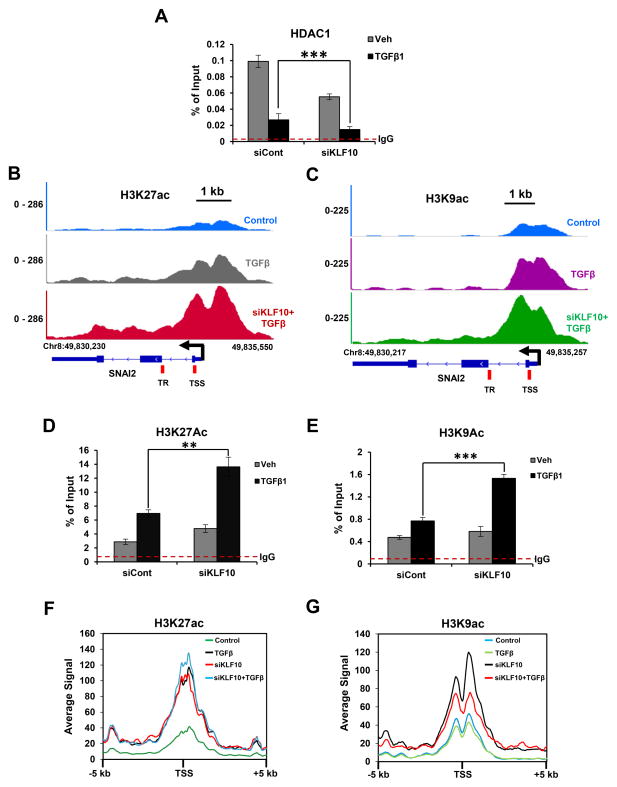
Previously, it was reported that KLF10 functions to repress transcription at least in part by interacting with co-repressors such as histone deacetylases and recruiting them to target genes (47). We therefore examined HDAC1 occupancy at the SNAI2 promoter in control or KLF10-depleted A549 cells. Importantly, consistent with the reported interaction of KLF10 with HDAC1-containing complexes, we observed significantly decreased recruitment of HDAC1 following KLF10 depletion in TGFβ-treated cells, thereby suggesting that KLF10 represses SNAI2 expression through HDAC1 recruitment (Fig. 5A). Since histone deacetylation is tightly coupled to HDAC-mediated gene repression, we performed ChIP analysis for H3K9ac and H3K27ac on the SNAI2 gene as well as ChIP-seq to examine the global effects of KLF10 on histone acetylation. These studies revealed that both acetylation marks significantly increased on SNAI2 following TGFβ treatment and were further increased upon KLF10 depletion (Fig. 5B–5E). Furthermore, aggregate plot analyses for H3K9ac and H3K27ac near (± 5Kb) KLF10-bound regions showed an increased average signal upon KLF10 depletion (Fig. 5F & 5G). Taken together, these results suggest that KLF10 recruits HDAC1 to SNAI2 and other target genes to repress transcription by removing activating histone acetylation marks.
Figure 5.
KLF10 recruits HDAC1 to repress SNAI2 gene transcription. A, ChIP analysis of HDAC1 occupancy on the SNAI2 gene. Significantly decreased recruitment of HDAC1 to the SNAI2 TSS (amplicon location indicated in B) was observed upon KLF10 depletion in TGFβ treated cells. Immunoprecipitated DNA is shown as percentage of input. IgG antibody was used as a negative control and is indicated by the red dotted line. Data are represented as mean ± SD. n=3. ***p ≤ 0.005. B & C, ChIP-seq profiles of H3K27ac and H3K9ac respectively, on the SNAI2 gene. The direction of transcription and the TSS are indicated by black arrows. Scale bar is represented in kilobases. D & E, ChIP analysis of H3K27ac and H3K9ac on the TSS of the SNAI2 gene. Immunoprecipitated DNA was compared to input and shown as percentage. IgG antibody was used as a negative control to subtract the background level and is shown as a red dotted line. Data are represented as mean ± SD. n=3. ***p ≤ 0.005. F & G, Average profile of H3K27ac and H3K9ac (5 kb) up- and down-stream of the KLF10-bound genes.
KLF10 depletion enhances TGFβ-induced EMT
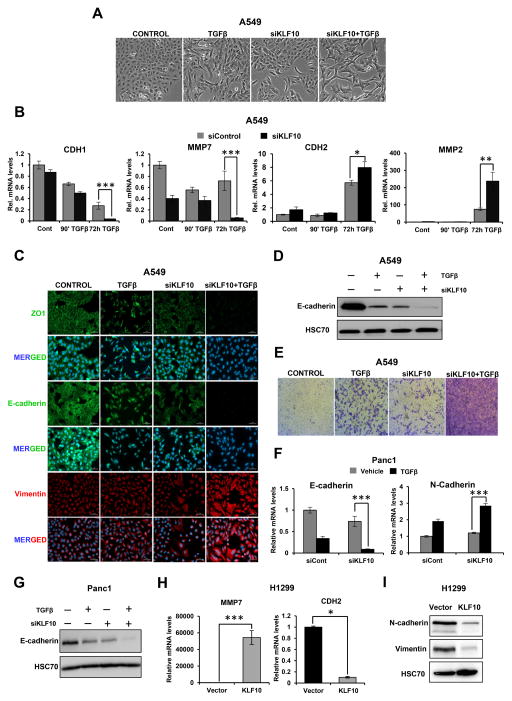
SNAI2 is one of the most important EMT-TF and emerged as a direct KLF10 target gene in our studies. Therefore, we next further investigated a role for KLF10 in TGFβ-induced EMT. During EMT, cells undergo robust changes in morphology in which they generally lose epithelial characteristics and transition to a more elongated and mesenchymal shape. Upon stimulation with TGFβ, A549 cells depleted of KLF10 acquired a mesenchymal shape which was more pronounced compared to the cells treated with TGFβ alone, suggesting EMT induction was enhanced following KLF10 depletion (Fig. 6A). To further confirm an effect on EMT, we performed gene expression analyses, western blotting and immunofluorescence staining in A549 cells to examine the effects of KLF10 perturbation on the expression of epithelial and mesenchymal markers. These analyses revealed that the mRNA levels of genes associated with an epithelial phenotype (E-cadherin and MMP7; Supplementary Fig. S3) were significantly downregulated, whereas mesenchymal markers (N-cadherin and MMP2) were significantly upregulated (Fig. 6B). Immunofluorescence staining performed in TGFβ-treated KLF10-depleted cells for classical EMT markers revealed a significant loss of epithelial markers (E-cadherin and ZO-1) and gain of the mesenchymal marker Vimentin compared to cells treated with TGFβ alone (Fig. 6C). We also observed a strong combinatory decrease of E-cadherin protein levels in KLF10-depleted cells following TGFβ treatment (Fig. 6D). Since EMT is generally associated with a more migratory phenotype, we also investigated the effect of KLF10 depletion on the migration potential of TGFβ-treated cells. Interestingly, KLF10-depleted and TGFβ-stimulated cells exhibited significantly increased migration potential in a transwell migration assay as compared to the untreated cells (Fig. 6E). A role for KLF10 in suppressing EMT was also confirmed by gene expression and western blot analyses for EMT markers in Panc1 cells (Fig. 6F & 6G). Interestingly, overexpression of KLF10 in H1299 cells resulted in a significant reduction in CDH2 mRNA levels and increased expression of MMP7 expression (Fig. 6H). Furthermore, Western blot analysis also confirmed a marked reduction in both N-cadherin and Vimentin protein levels in KLF10 overexpressing H1299 cells (Fig. 6I). Taken together, these results revealed a previously unknown function of KLF10 in suppressing TGFβ-induced EMT.
Figure 6.
KLF10 depletion enhances TGFβ-induced EMT. A, Morphological changes of A549 cells were observed following KLF10 depletion and/or treatment with TGFβ (5 ng/ml for 72 h). Phase contrast images were taken at 10× magnification. B, Gene expression levels of epithelial-(E-cadherin/CDH1 and MMP7) and mesenchymal-(N-cadherin/CDH2 and MMP2) related genes in A549 cells following transfection of control or KLF10 siRNAs and vehicle or TGFβ treatment were analyzed by quantitative RT-PCR and shown as “relative mRNA levels” normalized to an unregulated control gene (HNRNPK) and the control-transfected and vehicle-treated condition. Data are represented as mean ± SD. n=3. ***p ≤ 0.005, **p ≤ 0.01, *p ≤ 0.05. C, Immunofluorescence staining showing enhanced EMT induction upon TGFβ treatment in KLF10-depleted cells. Cells were stained for epithelial markers (ZO1 and E-cadherin) and Vimentin as a mesenchymal marker. Nuclei were stained with DAPI. Scale bar represents 50 μm. D, Western blot analysis of whole cell protein lysates showing a significant loss of E-cadherin following 72 h of TGFβ treatment in KLF10-depleted cells. HSC70 was used as a loading control. E, Transwell migration assay showing a significant increase in migration capacity of TGFβ-treated cells depleted for KLF10. Cells were stained with crystal violet. F, Gene expression analysis of epithelial and mesenchymal markers in Panc1 cells. G, Western blot analysis in Panc1 cells showing loss of E-cadherin following 72 h of TGFβ treatment in KLF10-depleted cells. HSC70 was used as a loading control. H, Gene expression levels of MMP7 and CDH2 following KLF10 overexpression in H1299 cells. I, A marked reduction in N-cadherin and Vimentin protein levels was observed upon KLF10 overexpression in H1299 cells. HSC70 was used as a loading control
Thus, these findings not only show that KLF10 functions as a tumor suppressor in an experimental lung cancer mouse model, but also provide mechanistic insight into a direct function of KLF10 in suppressing EMT by binding to the SNAI2 promoter and enhancing recruitment of HDAC1 to repress SNAI2 expression.
Discussion
Despite recent advances in cancer treatment, NSCLC remains the leading cause of cancer-related deaths worldwide, where patient mortality can largely be attributed to the development of metastatic disease.
Perturbations in the TGFβ signaling pathway are frequently associated with lung cancer metastasis (48). Under physiological circumstances, TGFβ plays an important role in maintaining tissue homeostasis through its anti-proliferative and apoptosis-inducing functions, whereby it frequently acts as a tumor suppressor. However, in the late stages of cancer development, TGFβ can promote tumor progression by exerting pro-metastatic effects (49). The intricate mechanisms by which this switch in TGFβ function occurs are not completely understood. In this study, we have demonstrated for the first time that KLF10 suppresses the pro-metastatic effects of TGFβ in lung and pancreatic cancer models. Our transcriptome-wide data demonstrates that KLF10 affects the expression of a large fraction of TGFβ-regulated genes. Interestingly, the effect of KLF10 on TGFβ-regulated genes was gene-specific, where certain genes were upregulated upon KLF10 depletion, while others were downregulated. This suggests that KLF10 may specifically promote the expression of genes involved in the tumor-suppressive activity of TGFβ while repressing genes involved in the pro-metastatic effects of TGFβ.
Despite the fact that KLF family members show remarkable similarity in their DNA-binding domains, they can have different effects on transcriptional regulation. For instance, some KLFs act predominantly as activators and others as repressors, while some can function as either activators or repressors, depending on the cellular, signaling and epigenetic context. Several KLFs were shown to act as tumor suppressors in different cancer types as well as regulate various cellular processes. For instance, both KLF10 and KLF17 (50) have been shown to promote TGFβ signaling, thus raising the possibility of functional redundancy in their functions. It remains to be elucidated to what extent the different KLFs (for example, KLF10 and KLF17) are co-expressed and regulate the same set of genes or pathways and if they can substitute for each other’s functions.
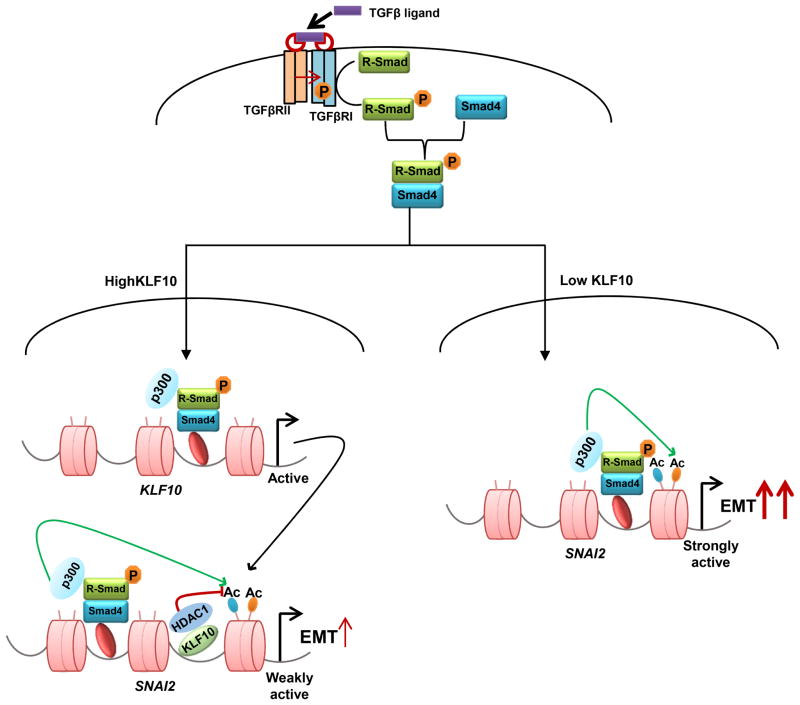
SNAI2 is one of the most important EMT-TFs and has been shown to directly repress the CDH1 gene encoding E-cadherin (51). Remarkably, KLF10 depletion resulted in significantly increased expression of the SNAI2 gene and decreased E-cadherin expression. Thus, our studies support a model in which KLF10 normally binds to the SNAI2 promoter and serves to recruit HDAC1 to remove activating histone acetylation marks, thereby decreasing SNAI2 expression and suppressing EMT (Fig. 7). Thus, we propose that KLF10 plays a central role in determining the net effect of TGFβ treatment on the transcriptional induction of SNAI2. In this model, TGFβ treatment would result in the simultaneous activation (i.e., through SMAD proteins) and repression (i.e., through KLF10) of SNAI2 expression. Consistently, we observed increased KLF10 occupancy on the SNAI2 gene following TGFβ treatment. According to this model, KLF10 would serve as a molecular rheostat to control the magnitude of TGFβ signaling and the activation of EMT. Thus, decreased KLF10 expression, which frequently occurs in cancer, may facilitate an increased induction of SNAI2 expression following activation of TGFβ/SMAD signaling, thereby increasing the potential for tumor metastasis.
Figure 7.
Proposed model for the mechanism of action of KLF10. A, In response to TGFβ ligand binding, KLF10 gene expression is induced. KLF10 protein binds to the SNAI2 gene and recruits HDAC1 to remove activating acetylation marks thereby leading to gene repression. B, In the case of low KLF10 expression and/or function, TGFβ stimulation results in uninhibited induction SNAI2 gene expression, resulting in enhanced EMT and tumor progression.
Similar to KLF10, KLF4, another member of the KLF family has been shown to inhibit EMT in hepatocellular carcinoma cells (HCC) by repressing SNAI2 gene expression (52). In this case, TGFβ inhibits the expression of KLF4, resulting in enhanced expression of the SNAI2 gene. Thus, it will be interesting to examine the interplay between KLF10 and KLF4 in the regulation of EMT by TGFβ. Another potential target of KLF10, FOSB has been shown to be involved in TGFβ-induced EMT in mammary epithelial cells (46). Interestingly, it was shown that FOSB can interact with and recruit HDAC2 to repress the E-cadherin (CDH1) gene. Thus, KLF10 likely controls TGFβ-induced EMT by both direct (e.g., SNAI2 and FOSB) and indirect effects (e.g., CDH1 and CDH2) on a number of genes.
During the course of EMT, cells undergo dynamic changes between epithelial and mesenchymal phenotypes that involve repression and activation of various genes. Regulation of gene expression is tightly coupled to the activity of transcription factors, cofactors and epigenetic regulators which modify chromatin structure in a way that favors gene activation or repression (53). A prominent example is histone acetylation, where the addition of acetyl groups to lysine residues of histone and non-histone proteins (catalyzed by histone acetyltransferases), or removal by histone deacetylases, is associated with gene activation or repression, respectively. Many EMT-TFs also function by recruiting epigenetic repressors like HDACs to target genes to induce EMT (54,55). HDACs have been frequently found to be associated with tumor development (56). Interestingly, KLF10 was shown to recruit corepressors like HDAC1 to its target genes to regulate their expression (47). In our study, we demonstrate that KLF10 recruits HDAC1 to the SNAI2 promoter and leads to decreased levels of H3K9ac and H3K27ac marks, providing a novel molecular insight behind KLF10 and its transcriptional regulation of EMT. Thus, the role of HDACs in controlling EMT appears to be multifaceted where differential interaction with either EMT-TFs or EMT-suppressing factors such as KLF10 may promote or suppress EMT in a context- and gene-dependent manner. Similarly, global inhibition of HDAC activity will likely also result in pleiotropic effects resulting from inhibition of both EMT promoting and suppressive factors, caution must be used in the interpretation of such studies.
In line with a tumor suppressor function, and consistent with our previously published data in breast cancer (24), we found that KLF10 expression in lung cancer is significantly lower compared to normal tissues. Consistently, we demonstrated that Klf10-null mice display higher lung tumor incidence compared to wild type mice, confirming a tumor suppressor function of KLF10 in vivo. Perhaps more importantly, low KLF10 mRNA levels in lung adenocarcinoma are associated with poor patient outcome, further supporting a clinical relevance for KLF10 in cancer.
In summary, we established a previously unknown function of KLF10 in regulating the pro-metastatic effects of TGFβ by suppressing TGFβ-induced EMT via SNAI2 as a direct KLF10 target gene, which is repressed in cooperation with HDAC1. Overall, loss of KLF10 expression, or inhibition of its function, may play a central role in the switch between the tumor suppressive effects of TGFβ in early stage disease and the pro-metastatic activities of this cytokine during disease progression.
Acknowledgments
Financial support,
The work presented here was supported by a NIH grant, R01 DE14036 to J.R. Hawse and M. Subramaniam, the German Academic Exchange Service (DAAD) to S. Nagarajan and the Lower Saxony Ministry for Science and Culture (grant ZN3222 to V. Ellenrieder and S.A. Johnsen).
The authors would like to thank Thomas C. Spelsberg for his excellent career mentorship, support and helpful suggestions. We thank Upasana Bedi and Zeynab Najafova for helpful discussions. We also thank Sabine Bolte and Nicole Molitor for outstanding technical support.
Footnotes
Potential conflicts of interest: The authors have no conflicts of interest
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Prudkin L, Liu DD, Ozburn NC, Sun M, Behrens C, Tang X, et al. Epithelial-to-mesenchymal transition in the development and progression of adenocarcinoma and squamous cell carcinoma of the lung. Mod Pathol. 2009;22:668–678. doi: 10.1038/modpathol.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valastyan S, Weinberg RA. Tumor Metastasis: Molecular Insights and Evolving Paradigms. Cell. 2011;147:275–92. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial–mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–42. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 5.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–28. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 7.Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Migrating cancer stem cells — an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744–9. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 8.Naber HPH, Drabsch Y, Snaar-Jagalska BE, ten Dijke P, van Laar T. Snail and Slug, key regulators of TGF-β-induced EMT, are sufficient for the induction of single-cell invasion. Biochem Biophys Res Commun. 2013;435:58–63. doi: 10.1016/j.bbrc.2013.04.037. [DOI] [PubMed] [Google Scholar]
- 9.Bolós V, Peinado H, Pérez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- 10.Shih J-Y. Transcription Repressor Slug Promotes Carcinoma Invasion and Predicts Outcome of Patients with Lung Adenocarcinoma. Clin Cancer Res. 2005;11:8070–8. doi: 10.1158/1078-0432.CCR-05-0687. [DOI] [PubMed] [Google Scholar]
- 11.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 12.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 13.Bierie B, Moses HL. Tumour microenvironment: TGFβ: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6:506–20. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- 14.Subramaniam M, Harris SA, Oursler MJ, Rasmussen K, Riggs BL, Spelsberg TC. Identification of a novel TGF-β-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic Acids Res. 1995;23:4907–4912. doi: 10.1093/nar/23.23.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chrisman HR, Tindall DJ. Identification and Characterization of a Consensus DNA Binding Element for the Zinc Finger Transcription Factor TIEG/EGR α. DNA Cell Biol. 2003;22:187–199. doi: 10.1089/104454903321655819. [DOI] [PubMed] [Google Scholar]
- 16.Tachibana I, Imoto M, Adjei PN, Gores GJ, Subramaniam M, Spelsberg TC, et al. Overexpression of the TGFbeta-regulated zinc finger encoding gene, TIEG, induces apoptosis in pancreatic epithelial cells. J Clin Invest. 1997;99:2365–74. doi: 10.1172/JCI119418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hefferan TE, Reinholz GG, Rickard DJ, Johnsen SA, Waters KM, Subramaniam M, et al. Overexpression of a Nuclear Protein, TIEG, Mimics Transforming Growth Factor-Action in Human Osteoblast Cells. J Biol Chem. 2000;275:20255–9. doi: 10.1074/jbc.C000135200. [DOI] [PubMed] [Google Scholar]
- 18.Bender H, Wang Z, Schuster N, Krieglstein K. TIEG1 facilitates transforming growth factor-β-mediated apoptosis in the oligodendroglial cell line OLI-neu. J Neurosci Res. 2004;75:344–52. doi: 10.1002/jnr.10856. [DOI] [PubMed] [Google Scholar]
- 19.Johnsen SA, Subramaniam M, Effenberger KE, Spelsberg TC. The TGFβ inducible early gene plays a central role in the anti-proliferative response to TGFβ. Signal Transduct. 2004;4:29–35. [Google Scholar]
- 20.Johnsen SA, Subramaniam M, Katagiri T, Janknecht R, Spelsberg TC. Transcriptional regulation of Smad2 is required for enhancement of TGF?/Smad signaling by TGF? inducible early gene. J Cell Biochem. 2002;87:233–41. doi: 10.1002/jcb.10299. [DOI] [PubMed] [Google Scholar]
- 21.Johnsen SA, Subramaniam M, Janknecht R, Spelsberg TC. TGFβ inducible early gene enhances TGFβ/Smad-dependent transcriptional responses. Oncogene. 2002;21:5783–90. doi: 10.1038/sj.onc.1205681. [DOI] [PubMed] [Google Scholar]
- 22.Xiong Y, Svingen PA, Sarmento OO, Smyrk TC, Dave M, Khanna S, et al. Differential coupling of KLF10 to Sin3-HDAC and PCAF regulates the inducibility of the FOXP3 gene. Am J Physiol - Regul Integr Comp Physiol. 2014;307:R608–20. doi: 10.1152/ajpregu.00085.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J, Shin S, Subramaniam M, Bruinsma E, Kim T-D, Hawse JR, et al. Histone demethylase JARID1B/KDM5B is a corepressor of TIEG1/KLF10. Biochem Biophys Res Commun. 2010;401:412–6. doi: 10.1016/j.bbrc.2010.09.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramaniam M, Hefferan Te, Tau K, Peus D, Pittelkow M, Jalal S, et al. Tissue, cell type, and breast cancer stage-specific expression of a TGF-β inducible early transcription factor gene. J Cell Biochem. 1998;68:226–36. [PubMed] [Google Scholar]
- 25.Chang VHS, Chu P-Y, Peng S-L, Mao T-L, Shan Y-S, Hsu C-F, et al. Krüppel-Like Factor 10 Expression as a Prognostic Indicator for Pancreatic Adenocarcinoma. Am J Pathol. 2012;181:423–30. doi: 10.1016/j.ajpath.2012.04.025. [DOI] [PubMed] [Google Scholar]
- 26.Song K-D, Kim D-J, Lee JE, Yun C-H, Lee WK. KLF10, transforming growth factor-β-inducible early gene 1, acts as a tumor suppressor. Biochem Biophys Res Commun. 2012;419:388–94. doi: 10.1016/j.bbrc.2012.02.032. [DOI] [PubMed] [Google Scholar]
- 27.Prenzel T, Begus-Nahrmann Y, Kramer F, Hennion M, Hsu C, Gorsler T, et al. Estrogen-Dependent Gene Transcription in Human Breast Cancer Cells Relies upon Proteasome-Dependent Monoubiquitination of Histone H2B. Cancer Res. 2011;71:5739–53. doi: 10.1158/0008-5472.CAN-11-1896. [DOI] [PubMed] [Google Scholar]
- 28.Bedi U, Scheel AH, Hennion M, Begus-Nahrmann Y, Rüschoff J, Johnsen SA. SUPT6H controls estrogen receptor activity and cellular differentiation by multiple epigenomic mechanisms. Oncogene. 2015;34:465–73. doi: 10.1038/onc.2013.558. [DOI] [PubMed] [Google Scholar]
- 29.Hossan T, Nagarajan S, Baumgart SJ, Xie W, Magallanes RT, Hernandez C, et al. Histone Chaperone SSRP1 is Essential for Wnt Signaling Pathway Activity During Osteoblast Differentiation. STEM CELLS. 2016;34:1369–76. doi: 10.1002/stem.2287. [DOI] [PubMed] [Google Scholar]
- 30.Zirkel A, Lederer M, Stöhr N, Pazaitis N, Hüttelmaier S. IGF2BP1 promotes mesenchymal cell properties and migration of tumor-derived cells by enhancing the expression of LEF1 and SNAI2 (SLUG) Nucleic Acids Res. 2013;41:6618–36. doi: 10.1093/nar/gkt410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramaniam M, Gorny G, Johnsen SA, Monroe DG, Evans GL, Fraser DG, et al. TIEG1 Null Mouse-Derived Osteoblasts Are Defective in Mineralization and in Support of Osteoclast Differentiation In Vitro. Mol Cell Biol. 2005;25:1191–9. doi: 10.1128/MCB.25.3.1191-1199.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 33.Nagarajan S, Hossan T, Alawi M, Najafova Z, Indenbirken D, Bedi U, et al. Bromodomain Protein BRD4 Is Required for Estrogen Receptor-Dependent Enhancer Activation and Gene Transcription. Cell Rep. 2014;8:460–9. doi: 10.1016/j.celrep.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based Analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–91. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Picard Tools - By Broad Institute [Internet] [cited 2016 Jan 25]. Available from: http://broadinstitute.github.io/picard/
- 40.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 43.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szász AM, Lánczky A, Nagy Á, Förster S, Hark K, Green JE, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–33. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duro de Oliveira K, Vannucci Tedardi M, Cogliati B, Zaidan Dagli ML. Higher Incidence of Lung Adenocarcinomas Induced by DMBA in Connexin 43 Heterozygous Knockout Mice. BioMed Res Int. 2013;2013:e618475. doi: 10.1155/2013/618475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pakala SB, Singh K, Reddy SDN, Ohshiro K, Li D-Q, Mishra L, et al. TGF-β1 signaling targets metastasis-associated protein 1, a new effector in epithelial cells. Oncogene. 2011;30:2230–41. doi: 10.1038/onc.2010.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin W, Chen B, Li J, Zhu H, Huang M, Gu S, et al. TIEG1 Inhibits Breast Cancer Invasion and Metastasis by Inhibition of Epidermal Growth Factor Receptor (EGFR) Transcription and the EGFR Signaling Pathway. Mol Cell Biol. 2012;32:50–63. doi: 10.1128/MCB.06152-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guasch G, Schober M, Pasolli HA, Conn EB, Polak L, Fuchs E. Loss of TGFβ Signaling Destabilizes Homeostasis and Promotes Squamous Cell Carcinomas in Stratified Epithelia. Cancer Cell. 2007;12:313–27. doi: 10.1016/j.ccr.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siegel PM, Massagué J. Cytostatic and apoptotic actions of TGF-β in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–20. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 50.Ali A, Zhang P, Liangfang Y, Wenshe S, Wang H, Lin X, et al. KLF17 empowers TGF-β/Smad signaling by targeting Smad3-dependent pathway to suppress tumor growth and metastasis during cancer progression. Cell Death Dis. 2015;6:e1681. doi: 10.1038/cddis.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hajra KM, Chen DY-S, Fearon ER. The SLUG Zinc-Finger Protein Represses E-Cadherin in Breast Cancer. Cancer Res. 2002;62:1613–8. [PubMed] [Google Scholar]
- 52.Lin Z-S, Chu H-C, Yen Y-C, Lewis BC, Chen Y-W. Krüppel-Like Factor 4, a Tumor Suppressor in Hepatocellular Carcinoma Cells Reverts Epithelial Mesenchymal Transition by Suppressing Slug Expression. PLOS ONE. 2012;7:e43593. doi: 10.1371/journal.pone.0043593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–49. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.von Burstin J, Eser S, Paul MC, Seidler B, Brandl M, Messer M, et al. E-Cadherin Regulates Metastasis of Pancreatic Cancer In Vivo and Is Suppressed by a SNAIL/HDAC1/HDAC2 Repressor Complex. Gastroenterology. 2009;137:361–371. e5. doi: 10.1053/j.gastro.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 55.Peinado H, Ballestar E, Esteller M, Cano A. Snail Mediates E-Cadherin Repression by the Recruitment of the Sin3A/Histone Deacetylase 1 (HDAC1)/HDAC2 Complex. Mol Cell Biol. 2004;24:306–19. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marks PA, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Histone deacetylases and cancer: causes and therapies. Nat Rev Cancer. 2001;1:194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]