Abstract
Long-chain acyl-CoA synthetases (ACSLs) convert fatty acids to fatty acyl-CoAs to regulate various physiologic processes. We characterized the ACSL isoforms in a cell line of homogeneous rat beta cells (INS-1 832/13 cells) and human pancreatic islets. ACSL4 and ACSL3 proteins were present in the beta cells and human and rat pancreatic islets and concentrated in insulin secretory granules and less in mitochondria and negligible in other intracellular organelles. ACSL1 and ACSL6 proteins were not seen in INS-1 832/13 cells or pancreatic islets. ACSL5 protein was seen only in INS-1 832/13 cells. With shRNA-mediated gene silencing we developed stable ACSL knockdown cell lines from INS-1 832/13 cells. Glucose-stimulated insulin release was inhibited ~ 50% with ACSL4 and ACSL3 knockdown and unaffected in cell lines with knockdown of ACSL5, ACLS6 and ACSL1. Lentivirus shRNA-mediated gene silencing of ACSL4 and ACSL3 in human pancreatic islets inhibited glucose-stimulated insulin release. ACSL4 and ACSL3 knockdown cells showed inhibition of ACSL enzyme activity more with arachidonate than with palmitate as a substrate, consistent with their preference for unsaturated fatty acids as substrates. ACSL4 knockdown changed the patterns of fatty acids in phosphatidylserines and phosphatidylethanolamines. The results show the involvement of ACLS4 and ACLS3 in insulin secretion.
Keywords: Human pancreatic islets, Insulin secretion, Insulin secretory vesicles, Acyl-CoA synthetases, Gene silencing, Phosphatidylserine, Phosphatidylethanolamine
INTRODUCTION
There is extensive evidence to suggest that rapid lipid modifications directly participate in signaling and supportive pathways for glucose-stimulated insulin secretion in the pancreatic beta cell. We previously noticed that glucose stimulation of INS-1 832/13 insulinoma cells acutely increased by about 20% the level of many lipids with C14–C24 fatty acid side chains, including phospholipids and triglycerides [1]. Others [2–6] and we [1] have observed that glucose carbon is rapidly incorporated into lipids in pancreatic islets and insulin cell lines indicating de novo lipid synthesis from glucose carbon occurs over a time course that coincides with insulin secretion. The enzyme patterns in pancreatic islets and pancreatic beta cell lines also support the idea that pancreatic beta cells are a lipogenic tissue. Pancreatic beta cells in human pancreatic islets and the insulin cell line INS-1 832/13 contain very high levels of enzymes needed for lipid synthesis including pyruvate carboxylase [7, 8], fatty acid synthase [1, 8] and acetyl-CoA carboxylase [1]. Acetyl-CoA carboxylase catalyzes the formation of malonyl-CoA that cells use for fatty acid synthesis as well as possibly, in the case of the beta cell, for signaling purposes [9–11]. Of the two isoforms of acetyl-CoA carboxylase (ACC1 or ACC2)1 the one that is present in pancreatic islets of humans and rats, as well as the INS-1 832/13 insulinoma cell line, is ACC1 which is the isoform found in lipogenic tissues [1]. The knockdown of pyruvate carboxylase or fatty acid synthase in the INS-1 832/13 cell line [7, 12] lowers numerous phospholipids and inhibits glucose-induced insulin release establishing the importance of these enzymes in insulin secretion.
Modifications of beta cell lipids include alterations that affect fluidity of the plasma membrane and the membrane of the insulin secretory granules. Changes in levels of phospholipids and increases in unsaturation of fatty acid side chains in phospholipids increase the fluidity and fusion of intracellular membranes. Phosphatidylserine is fivefold higher in the insulin secretory granules compared to the whole beta cell [13]. The negatively charged phosphatidylserine in the insulin secretory granule membrane facilitates its fusion with the plasma membrane by its interaction with the positively charged domains of SNARE proteins in the plasma membrane [14]. The action of P4 ATPases (flippases) that rapidly transport phosphatidylserine across the membrane of insulin secretory granules also enhance the coupling of the insulin granule membrane with the plasma membrane thus promoting the extrusion of the insulin from the insulin secretory granules into the circulation [14]. P4 ATPases are highly concentrated in insulin secretory granules of beta cells and knockdown of these enzymes inhibits insulin release in INS-1 832/13 cells and in human pancreatic islets [14].
The role of long chain acyl CoA synthetases (ACSLs) in insulin secretion has not been extensively studied. ACSLs convert fatty acids into acyl-CoAs that are incorporated into triglyceride, phospholipids, and cholesterols [15, 16] and also undergo β-oxidation to produce energy [17, 18]. The active molecule available for β-oxidation is not fatty acid itself but fatty acyl-CoA ester catalyzed by ACSLs. Fatty acyl-CoA esters are involved in various cellular functions including protein transport, enzyme activation, protein acylation and cell signaling [19–27] including in vesicle fusion and membrane interaction [20, 24]. Five isozymes of ACSL (ACSL1, ACSL3, ACLS4, ACSL5, and ACSL6) are known in mammalian cells [28, 29]. The five ACSLs in mammalian cells are divided into two sub-families depending on amino acid sequence similarity and substrate specificity. ACSL1, ACSL5 and ACSL6 are in a sub-family preferring saturated fatty acids with broad chain length as substrates [15, 28, 29]. ACSL3 and ACSL4 are in the other sub-family preferring highly unsaturated fatty acids as substrates [30–32]. ACSL3 accepts a wider range of unsaturated fatty acids than ACSL4, which highly prefers arachidonate and eicosapentaenoate. Excluding the pancreatic islet, which has not been extensively studied, the tissue distribution of the various ACSL isozymes is as follows: ACSL1 with broad substrate specificity is expressed abundantly in the liver, heart, and adipose tissue in which various fatty acids are used for energy production and storage [33]. ACSL3 and ACSL6 are expressed highly in brain, and the amounts of mRNA that encode these two ACSLs change in the course of the development of rat brain [30, 34]. ACSL4 is expressed abundantly in tissues responsible for steroid synthesis, especially adrenal gland and ovary [31]. ACSL5 utilizes a wide range of fatty acids and exists most abundantly in the small intestine to conjugate CoA to dietary fatty acids to form triglycerides that are absorbed into the circulation [19, 30].
Except for one study of the rat beta cell line INS-1 832/13 [35], the ACSLs in pancreatic insulin cells have not been studied. Because little is known about ACSLs in the pancreatic insulin-producing cell, we studied the expression of all five known mammalian ACSLs in human pancreatic islets and in the INS-1 832/13 cell line. We found that ACSL3 and ACSL4 were the only ACSLs present in both human pancreatic islets and INS-1 832/13 cells. Each of these ACSLs was concentrated in insulin secretory granules and mitochondria of INS-1 832/13 cells, but more concentrated in the insulin secretory granules than in the mitochondria and not present or present at very low concentrations in other intracellular organelles. Gene silencing of ACSL3 and ACSL4 in INS-1 832/13 cells and in human pancreatic islets inhibited glucose-stimulated insulin release implicating them in insulin secretion. We also studied the effect of gene silencing of ACSLs on the fatty acid composition of phospholipids in the INS-1 832/13 cell line.
MATERIALS AND METHODS
Materials
The antibody against ACSL1 (catalog no. 4047S) was from Cell Signaling (Danvers, MA USA). Antibodies against ACSL3 (catalog no. ab51959), ACSL4 (catalog no. ab137525), ACSL5 (catalog no. ab104892), ACSL6 catalog no. ab154094) and catalase (catalog no. ab1877) were from Abcam (Cambridge, MA USA). The antibody against VAMP2 (catalog no. V1389) was from Sigma-Aldrich (St. Louis, MO USA). The antibody against TGN38 (catalog no. NBP1-03495) was from Novus Biologicals (Littleton, CO USA). All of the primary antibodies were raised in rabbits. The secondary antibody Goat anti-Rabbit IgG (catalog no. 31460) was from Thermo-Fisher (Rockford, IL USA). INS-1 832/13 cells were from Chris Newgard (Duke University). PIM(S) Prodo Islet Media (Cat # S001GMP) was from Prodo Laboratories (Aliso Viejo, CA). Cells and media. The INS-1 832/13 cells were grown and maintained in RPMI 1640 cell culture medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin and 100 μg/ml streptomycin and 1 mM pyruvate and 50 μM beta-mercaptoethanol (INS-1 medium) as described previously [13, 14]. The 293-T cells were purchased from Clontech, USA (cat no. 632180) and maintained in DMEM containing 10% fetal bovine serum, 100 units/ml penicillin and 100 μg/ml streptomycin.
shRNA Vector construction and generation of constitutive cells lines
The shRNA expression vector used in this study is a modified version of pSilencer 2.1- U6 hygromycin vector (Ambion, Life Technologies) that was previously described [36]. The shRNA target sequences for INS-1 832/13 cells were identified by using the computer program siRNA Wizard v 3.1 (Invivogen, USA) using rat mRNA sequences of ACSL1 (NM_012820), ACSL3 (NM_057107.1), ACSL4 (NM_053623), ACSL5 (NM_053607) and ACSL6 (NM_130739.1) and human mRNA sequences for ACSL4 (NM022977.2) ACSL3 (NM004457.3). The oligonucleotide sequences used to target different ACSLs genes are listed in Supplemental Table 1. The DNA of the plasmids containing various ACSL shRNAs was sequenced to confirm the intended target locations.
For generation of constitutively shRNA-expressing rat insulinoma cell lines, the INS-1 832/13 cells were plated in 6-well plates. Twenty-four hours later the cells were transfected with shRNA expression vectors along with pCMV-Tol2 vector using Lipofectamine2000 (Life Technologies, USA) [14, 36]. On the next day the complete growth media was replaced with media containing 150 μg/ml of Hygromycin B (Invivogen, USA) and further incubated for several days with the selection media changed every 4 days. After 15 days colonies appeared that were trypsinized and further plated in 10-cm cell culture dishes. The selected cell populations were maintained in selective media at all times and processed for total RNA isolation, western blot or enzyme assays. In a similar way a control cell line was also generated in which the scrambled shRNA sequences was cloned into the above vector. The scrambled shRNA sequence (ACTACCGTTGTTATAGGTG) was from Ambion, Life Technologies, and the cell line was named pHyg-C.
RNA isolation and Real Time RT-PCR
Approximately 30 mg of liver sample was sliced into small pieces and homogenized in 1.5 ml Eppendorf tube with the Pellet Pestle Cordless Motor in a 1.5 ml microfuge test tube in 600 μl of RLT buffer provided with the Qiagen kit. The total RNA was isolated using RNeasy mini kit (Qiagen, USA) following manufacturer’s protocol. For isolation of total RNA from human islets, the human islets were received from the Integrated Islet Distribution Program sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases (City of Hope, CA, USA) and maintained for 2–3 hours in PIM(S) islet medium or longer when the islets were targeted with virus containing shRNAs. The islet medium was removed and the pellet of human islets was lysed with 300 μl of RLT buffer and total RNA was isolated as described above. Pancreatic islets from eight rats were isolated as described previously [7, 37–39] and lysed in 300 μl of RLT buffer and processed for total RNA isolation as described above. For isolating RNA from shRNA-mediated knockdown INS-1 832/13 cells, the cells were grown in 10 cm cell culture dishes in the presence of Hygromycin. At about 80% cell confluency the monolayer was washed two times with PBS and lysed with 600 μl of RLT buffer and total RNA was isolated as above.
The RNA samples were quantified and 2 μg of total RNA was reverse transcribed using the RETRO-script kit (Ambion, Life Technologies) in the presence of oligo(dT) primers. Real time quantitative PCR was performed using the BioRad MyIQ instrument using SYBR Premix ExTaq (Takara Bio, USA), gene specific forward and reverse primers and Fluorescein reagent (Cat. 170–8780, BioRad). The standard curve was calculated using cDNA from the pHygC cell line, human liver, human islets, rat liver or rat islets. The values were normalized using the expression of either the rat glutamate dehydrogenase 1 (Glut1) gene or the human glutamate dehydrogenase 1 (GLUD1) gene as a reference gene as previously described [14, 37]. The qRT-PCR primers used to detect human and rat ACSLs are listed in Supplemental Table 2.
Generation of Lentiviruses with different shRNAs
Before generating Lentiviruses containing shRNAS, we identified the most potent shRNA targets in 293T cells that displayed maximum lowering of mRNAs and their cognate proteins. Several shRNA targets were cloned into the pSilencer 2.1 U6 Puro (Life technologies, USA) vector as described above and puromycin-selected 293T cells were tested for mRNA levels and respective protein expressions. The most potent shRNA sequences were again synthesized with BamHI and EcoRI restriction enzyme ends and cloned directionally into the pLVX1-shRNA1 vector (Clontech, USA). The pseudopackaged Lentiviruses were recovered using the Lenti-X shRNA Expression System (Cat # 632177, Clontech, USA) following the manufacturer’s protocol. In brief, the pLVX1-shRNA vector DNA along with packaging mix was transfected into 293T cells and incubated at 37°C. After 48 h the supernatant was collected, clarified of cell debris and the viral titer was measured. To measure the virus titer the supernatant was serially diluted and overlaid onto INS-1 832/13 cells plated in 6-well plates the day before. After incubating the plates at 37°C for an hour, the media was aspirated and replaced with complete growth medium containing 500 ng/ml Puromycin. The plates were incubated for several days with medium changes. After 10 days the medium was aspirated and 1.0 ml of staining solution (0.01% crystal violet, 19% methanol) was added to each well. After a 1 h incubation at room temperature the staining solution was discarded and plaques were gently washed with water to remove excess dye. Plaques were air dried and counted to assign viral titers. Solutions with high viral titers were stored at −80°C in small aliquots for future use.
In order to generate a constitutive human cell line expressing shRNA directed against human ACSL4 (NM_022977.2) and ACSL3 (NM_04457.3), the identified shRNA sequences were cloned in the pSilencer 2.1-U6 Puromycin vector (Life Technologies, USA). The shRNA expressing vector along with pCMV-Tol2 plasmid DNA were transfected into 293T cells as described above. After 24 h the media was replaced with complete media containing 1 μg/ml of Puromycin (Invivogen, USA) and further incubated for several days with selective media changed every 4 days. After 15 days the selected cells were further grown in 10-cm cell culture dishes and processed for total RNA isolation as well as immunoblot analysis. Similarly a control cell line was generated using the above scrambled shRNA sequences and named as pPuro-C. The shRNAs showing the strongest suppression of ACSL3 and ACSL4 levels were used in experiments of insulin release in human pancreatic islets as described above.
Homogenates of cells and protein concentration
For dilution to the optimal concentrations of cell protein to use for immunoblot analysis, it was practical to homogenize INS-1 832/13 cells (or human islets at a concentration of ~ 7,000 islets/ml) in KMSH solution (5 mM potassium Hepes buffer, pH 7.5, 220 mM mannitol and 70 mM sucrose) with or without 1 mM dithiothreitol [37, 40] to produce a homogenate with about 2 mg whole-cell protein/ml that was diluted as needed at the time of use. Protease inhibitor cocktail (ThermoScientific, Catalog no. 78415) was included in the homogenization solution. Rat pancreatic islets were isolated as previously described [8, 38, 41] and homogenized in the KMSH solution described above, except that the concentration of rat pancreatic islets was about ~ 3,600 islets/ml of homogenizing solution. The protein concentrations of whole-cell homogenates and supernatant fractions used for measurements of enzyme activity and immunoblot analysis were measured by the Bradford method.
Subcellular fractionation of INS-1 832/13 cells
Plates (150 mm) of monolayers of INS-1 832/13 cells were placed on ice and quickly washed twice with cold phosphate buffered saline. Cells were scraped from the plates into a 1.5 ml microfuge test tube and centrifuged at 600 × g for 20 s. All subsequent steps were at 4°. To obtain insulin secretory granules subcellular fractions were prepared with slight modifications of our [38, 41–44] and others’ [45–48] methods. The cell pellet was suspended in 1 ml of KMSH containing 0.5 mM EGTA, 0.5 mM EDTA and protease inhibitor (per one or two 150 mm plates of cells) and homogenized with 40 strokes up and down in a Potter Elvehjem homogenizer. The method was designed to obtain pure insulin secretory granules free from mitochondria and resulted in slight contamination of mitochondria with insulin secretory granules and no contamination of insulin secretory granules with mitochondria [13, 14]. The homogenate was centrifuged at 600 × g for 10 min and the resulting supernatant fraction was centrifuged at 12,000 × g for 10 min to generate a mitochondrial pellet. The supernatant fraction from this centrifugation was centrifuged at 20,800 × g for 20 min to generate the insulin secretory granule pellet (Exps. in Figure 6). The pellet was washed up to three times with the KMSH homogenization solution by centrifugation at 20,800 × g for 20 min. The resulting insulin secretory granules were judged to be >90% pure [13, 14].
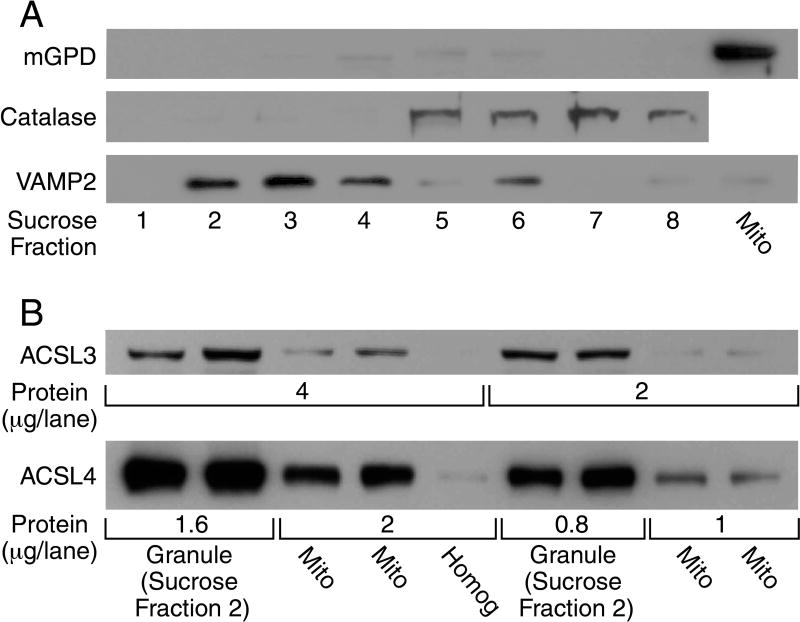
Figure 6. ACSL3 and ACSL4 are concentrated in insulin secretory granules and less so in mitochondria and not present in other subcellular compartments of beta cells.
Subcellular fractions of INS-1 832/13 cells were obtained as described in the Material and Methods section to further purify the sample. Both ASCL3 and ACSL4 were found to be absent in a 21-K-130K subcellular fraction containing trans-Golgi network, microsomes, endoplasmic reticulum and plasma membrane. A. Sucrose gradient centrifugation of insulin secretory granules (ISG) obtained by differential centrifugation further separates the insulin secretory granule marker VAMP2 from the peroxisomal marker catalase and the mitochondrial marker mGPD. Insulin secretory granules obtained by differential centrifugation (a 600 × g × 10 min post-nuclei/cell debris fraction were centrifuged at 15,000 × g for 10 min to remove mitochondria. The resulting supernatant fraction was centrifuged at 35,000 × g for 20 min to remove trans-Golgi network, endoplasmic reticulum, microsomes and plasma membranes which gives a pellet containing 90% pure insulin secretory granules. This pellet was resuspended and applied to a sucrose gradient as described under Experimental Procedures. Equal volumes of the sucrose gradient fractions were used for immunoblot analysis and show complete separation of the insulin secretory granule marker VAMP2 from the peroxisomal marker catalase and the mitochondrial marker mGPD (Panel A, Lanes 1–8). Mitochondria (Mito) prepared by a low speed centrifugation (5,000 × g for 10 min and washed three times) to minimize contamination with insulin granules show a large presence of mGPD and almost no VAMP2 (lane 9). B. ACSL3 and ACSL4 are concentrated in insulin secretory granules and are present at a lower concentration in mitochondria. Immunoblots for ACSL3 and ACSL4 of two protein amounts/lane using the insulin secretory granule sucrose gradient fraction 2 shown in Lane 2 of Panel A and the very pure mitochondria shown in Lane 9 of Panel A. Using the same lane configuration as in panel B an immunoblot probed with antibody to TGN38, a trans-Golgi marker, showed none of the marker whatsoever confirming that the ISG sucrose fraction 2 and the very pure mitochondrial fraction contain no trans-Golgi or endoplasmic reticulum. (Immunoblot not shown as it was blank.)
Insulin secretory granules used in some experiments were further purified on a sucrose gradient by suspending the insulin secretory granule pellet obtained by differential centrifugation in 200 μl homogenization solution and overlaying it on top of a discontinuous gradient prepared by layering 300 μl volumes of 60%, 50%, 45%, 40%, 30% and 20% (w/v) sucrose in a Beckman centrifuge test tube (11 mm × 34 mm). The test tube was centrifuged at 75,000 × g in a TLS-55 swinging bucket rotor in a Beckman TL-100 centrifuge at 4° for 3 h. Following this cen trifugation, 250 μl fractions were collected from the top of the gradient down and placed in separate test tubes. Each of the fractions was diluted with 1.75 ml of KMSH solution and centrifuged again at 100,000 × g for 30 min in a TLA-100.2 fixed angle rotor. The resultant pellets containing the various organelles were resuspended in 30 or 50 μl KMSH solution and used for further analyses.
To further purify a low density membrane fraction containing plasma membrane, endoplasmic reticulum and trans-Golgi network the post-insulin secretory granules (20,800 × g × 20 min) supernatant fraction described above was centrifuged at 35,000 × g for 10 min. The resulting supernatant fraction was centrifuged at 130,000 × g for 1 h. The pellet from this centrifugation was used for immunoblot analysis.
To obtain very pure mitochondria and insulin granules separated from one another, yield was sacrificed by obtaining a mitochondrial pellet centrifuged at a low force and an insulin granule pellet centrifuged at higher than usual forces after the mitochondrial pellet was removed. The whole cell homogenate post 600 × g for 10 min post-nuclei/cell debris supernatant fraction was centrifuged at 5,000 × g for 10 min to give a mitochondrial pellet that was washed three times before use. This pellet contained a high concentration of the mitochondrial marker mitochondrial glycerol phosphate dehydrogenase (mGPD) (Figure 6 lane 9) and virtually none of the insulin granule marker VAMP2 (Figure 6 lane 9). To obtain a very pure insulin granule fraction the homogenate 600 × g × 10 min post-nuclei/cell debris supernatant fraction was centrifuged at 15,000 × g for 10 min. The resulting supernatant fraction was centrifuged at 35,000 × g for 20 min and the resulting pellet was resuspended in 200 μl of KMSH solution and further purified by sucrose gradient centrifugation as described above. The sucrose fraction 2 (second from the top of the gradient) contained a high concentration of VAMP2 and no mGPD and no peroxisomal marker catalase (Figure 6) and was used as a very pure insulin granule fraction in immunoblot analysis in Figure 6.
Immunoblot analysis
All immunoblots were performed with total cell lysates homogenized in KMSH solution. Unless described otherwise approximately 10 μg of protein was usually loaded per lane and resolved with SDS-10% PAGE. Blots were blocked with 5% powdered milk in Tris-buffered saline with Tween (TBST) 0.05% and incubated with dilutions of anti-ACSL antibodies recommended by the manufacturer (usually 1:1,000–1:2,000) in 5% blocking buffer overnight at 4°C with shaking. The next day the membrane was washed with TBS-tween 0.05% and incubated with peroxidase-conjugated goat anti-rabbit secondary antibody at a dilution of 1:18000 (Cat no. 31460, ThermoScientific) and incubated at room temp for 3 h. The membrane was washed with TBS-tween 0.05% and signal was developed by using Chemiluminescent HRP substrate (Catalog no. WBKLS0100, Millipore). To perform immunoblots for actin, the membrane was treated with restore western blot stripping buffer (Fisher Scientific) and again blotted with actin antibody to show protein load as previously described [8].
Acyl-CoA synthase enzyme activity
Total Acyl CoA activity was monitored in ACLS3 and ACSL4 knockdown cells following a previously described protocol [49]. In brief, the confluent monolayer was harvested and washed three times with cold PBS. The cell pellet was homogenized in KMSH containing protease inhibitor cocktail (Roche, USA) and the protein concentration was determined with a BCA assay. Approximately 20 μg of protein was incubated in a final volume of 100 μl of a mixture of 8 mM MgCl2, 150 mM KCl, 5 mM DTT, 0.1% Triton-X-100, 0.5 mM CoA, 0.05 mM [5,6,8,9,11,12,14,15-3H(N)]arachidonate (specific radioactivity 0.1mCi/mmol) or [9,10-3H]palmitate (specific radioactivity 0.2 mCi/mmol), 10 mM ATP and 100 mM Tris-HCl buffer pH 7.8. The reaction mixture was incubated at 37°C for 20 min. The reaction was stopped by adding 500 μl of isopropyl alcohol:hexane:1 M sulfuric acid (40:10:1 ratio) and the reaction mixture was transferred to a 12 × 75 mm glass test tube. After adding 250 μl H2O and 1.0 ml heptane the test tubes were vortexed and kept at room temp to separate the layers. After 5 min the top layer was aspirated and discarded and 1.0 ml heptane was added again to the bottom layer, mixed and allowed to set for a few minutes to separate the layers. The top layer was aspirated and discarded again and approximately 200 μl from the bottom layer was transferred into a scintillation vial to measure tritium incorporation. The reaction was performed in triplicate and the average of three reactions is presented.
Insulin release from INS-1 832/13 cells
The INS-1 832/13 cells containing either control shRNA or target shRNA were tested for insulin release as previously described [14, 37]. In brief, approximately 0.5 × 106 INS-1 832/13 cells were plated in INS-1 medium in 24 well plates in quadruplicate. The next day the medium was replaced with the same medium containing 5 mM glucose and the cells were maintained at 37°C. After 24 h the media was replaced with Krebs-Ringer bicarbonate solution containing 3 mM glucose and 0.5% BSA and incubated for 2 h. The cells were washed once with Krebs-Ringer HEPES buffer BSA solution and further incubated with 1 ml of the same buffer for 1 h at 37°C in the presence or absence of 11.1 mM glucose. Samples of the incubation solution (0.5 ml) were collected from each well and briefly centrifuged to get rid of any possible floating cells. An aliquot of the supernatant fraction was saved for measurement of insulin by RIA using rat insulin as a standard. The monolayer of cells in each well was processed for protein concentration as previously described [14, 40].
Insulin release from human pancreatic islets
After human islets were received, the islets were incubated in PIM(S) supplemented with 100 units/ml penicillin and 100 μg/ml streptomycin in a CO2 incubator. After 2–3 h of incubation the islets were divided into four parts. Two parts were infected with lentiviruses at a multiplicity of infection of 3.0 in the presence of 4 μg/ml polybrene containing lentivirus carrying shRNA directed against human ACLS3 or ACSL4. A third part was incubated with lentivirus containing a scrambled shRNA and a fourth part was incubated with no virus. The islets were incubated at 37°C with occasional gentle shaking. After 2–3 h the islets were transferred to 40 mm petri dishes and supplemented with 5 ml of islet culture medium and incubated at 37°C for 3 days in a CO2 incubator. Islets were then used for the insulin release assay and for isolation of total RNA to quantify ACLS3 or ACSL4 mRNA levels by qRT-PCR. In brief, for insulin release studies, seven human islets of equal size were handpicked and placed in each replicate glass vial containing basal buffer (0 mM glucose in Krebs Ringer bicarbonate buffer, pH 7.3, and 0.5% bovine serum albumin) and stimulation buffer (16.7 mM glucose in Krebs Ringer bicarbonate buffer bovine serum albumin). The vials were incubated at 37°C and after 1 h the medium (0.5 ml) was collected from each vial and centrifuged to remove possible floating cells as described above. Insulin in the medium was quantified by RIA using human insulin as a standard.
Lipid content analysis
The ACSL4 shRNA 2201 carrying INS-1 cell line was maintained in selective media and grown in 150 mm dish. The cells were washed with PBS and processed for lipid analysis as previously described [1, 12, 13].
RESULTS
Immunoblot analyses of ACSL isoforms in pancreatic beta cells and human and rat pancreatic islets
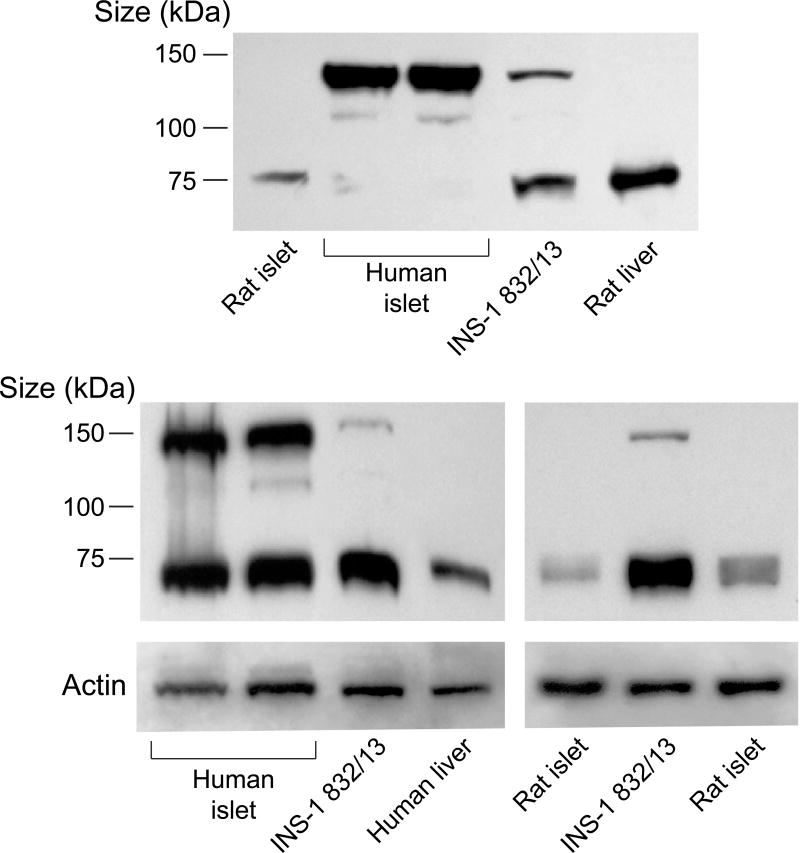
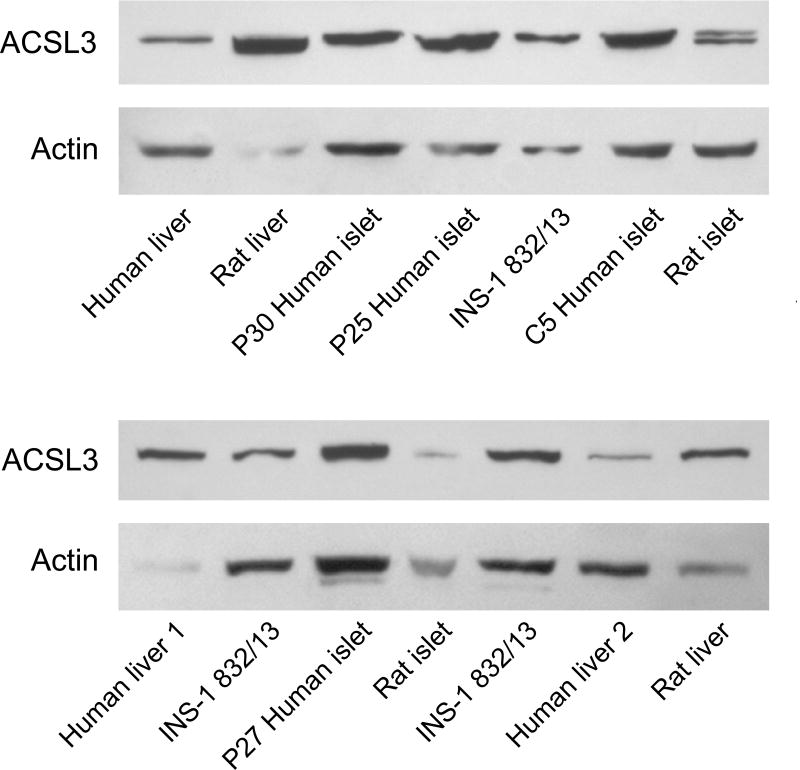
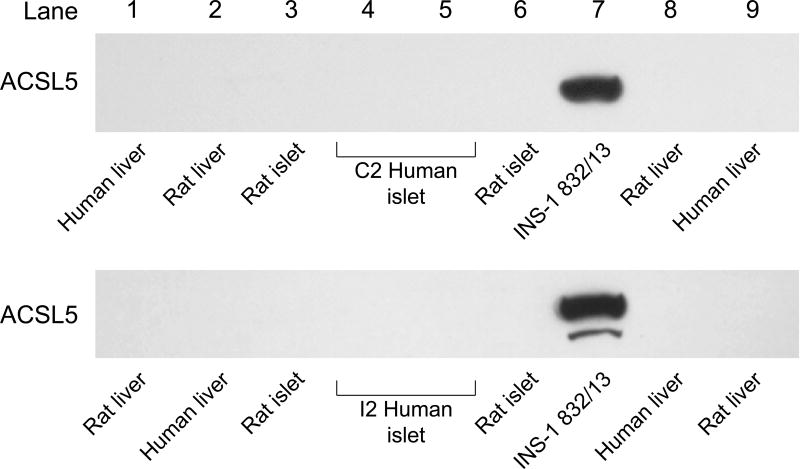
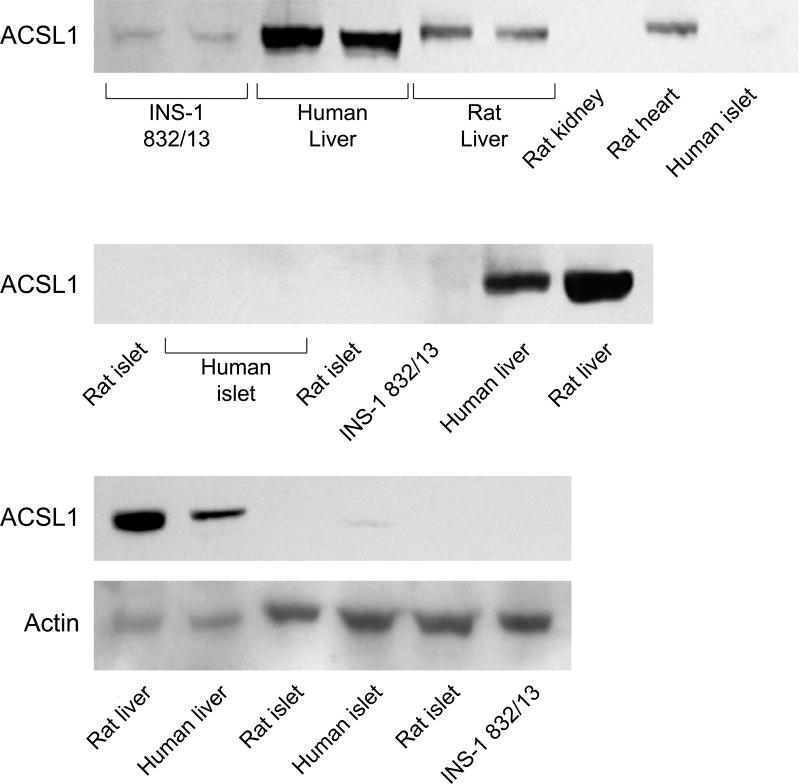
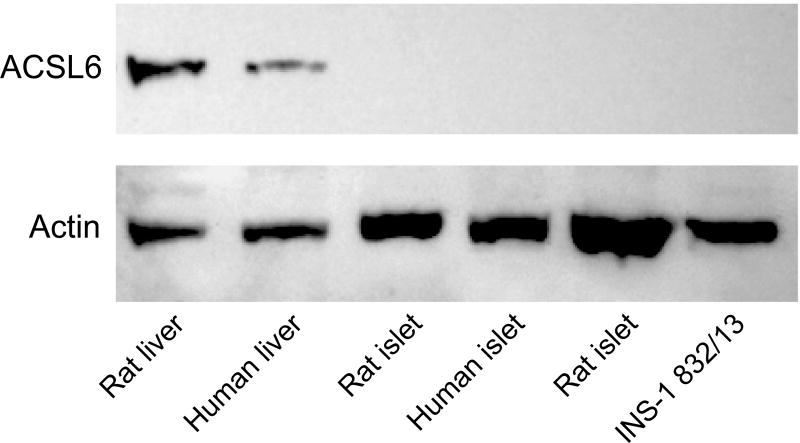
ACSL4 (~74 kDa) protein was detected in human pancreatic islets, rat pancreatic islets, INS-1 832/13 cells and human liver and rat liver (Figure 1). It is reported that ACSL4 can often form a tight homodimer which migrates as a 140 kDa protein in polyacrylamide gel electrophoresis of adrenal cell proteins [50, 51]. We observed both a 140 kDa dimer and a 75 kDa monomer of ACSL4 in human pancreatic islets and less prominently in INS-1 832/13 cells (Figure 1). The ACSL3 protein was detected in human pancreatic islets, rat pancreatic islets, INS-1 832/13 cells and in human and rat liver (Figure 2). The ACSL3 protein was also seen in three additional human pancreatic islet samples (Results with these additional samples are not shown.). The ACSL5 protein was detected only in INS-1 832/13 cells when examined by immunoblot analysis (Figure 3). It was not detected in human liver, human or rat pancreatic islets or rat liver (Figure 3). The ACSL1 protein was detected in human liver, rat liver and rat heart and barely detectable in one sample of INS-1 832/13 cells (Figure 4, top panel). Except for a very faint signal in a single human islet sample (Figure 4, lowest panel), ACSL1 protein was not seen in the other human pancreatic islets or rat pancreatic islets that we analyzed (Figure 4, middle and lowest panels). Four additional samples of human pancreatic islets did not show ACSL1. Additional immunoblots showed no ACSL1 protein in subcellular fractions of insulin secretory granules, mitochondria (12,000 × g pellet) and peroxisomes (20,000 × g pellet) derived from INS-1 832/13 cells (Data not shown.). The ACLS6 protein was detected only in human liver and rat liver but not in human or rat pancreatic islets or INS-1 832/13 cells (Figure 5).
Figure 1. ACSL4 is present in human and rat pancreatic islets, INS-1 832/13 cells and human and rat liver.
Two immunoblots with 15 μg of whole cell protein/lane with anti-ACSL4 antibody and anti-β-actin to show equal loading across lanes from INS-1 832/13 (INS-1) cells and human and rat tissues. Human and rat islet samples are from two different individuals. ACSL4 shows up as a dimer as has been previously reported in other cell types [50, 51] (≈ 140 kDa) as well as a dimer and a monomer (≈ 75 kDa) in human islets (to various degrees probably dependent on solubilization of samples) and less prominently in INS-1 832/13 cells.
Figure 2. ACSL3 is present in human and rat pancreatic islets, INS-1 832/13 cells and human and rat liver.
Two immunoblots with 20 μg of whole cell protein/lane and anti-ACSL3 antibody and anti-β-actin antibody to show relative loading of protein/lane are shown. In several non-beta cell samples, such as liver, used as a positive control, where the actin band is faint (due to the probable poor condition of the original samples beyond our control), the ACSL3 band is relatively more dense indicating the greater abundance of ACSL3 in the sample.
Figure 3. ACSL5 protein is detectable in INS-1 832/13 cells, but not in liver or pancreatic islets of humans or rats.
Immunoblot with 15 μg of whole cell protein/lane.
Figure 4. ACSL1 protein is not present in human and rat pancreatic islets and INS-1 832/13 cells and is present in human liver and rat liver.
Immunoblots with 15 μg whole cell protein/lane. Liver was used as a positive control. In the lowest panel the actin band is faint in rat liver and human liver and probably due to the poor condition of the samples beyond our control, but the ACSL1 band is dense indicating a high concentration of ACSL1 in liver as is known.
Figure 5. ACSL6 protein is not present in human or rat pancreatic islets or INS-1 832/13 cells, but is present in human liver and rat liver.
Immunoblot with 15 μg whole cell protein/lane. Actin bands show equal loading of protein across lanes.
ACSL3 and ACSL4 are concentrated in insulin secretory granules and mitochondria
Further analyses were carried out to discern the intracellular locations of the two predominant ACSL isoforms, ASCL3 and ACSL4, found in INS-1 832/13 cells and human and rat pancreatic islets. Immunoblot analyses of subcellular fractions of INS-1 832/13 cells indicated that both of these ACSL isoforms were concentrated in insulin secretory granules and mitochondria (Figure 6) and were not detectable in lower density subcellular fractions, such as endoplasmic reticulum, microsomes, trans-Golgi network and plasma membrane. That is ACSL3 and ACSL4 were not present in a 20 × g − 130,000 × g subcellular fraction that we previously showed contained these lower density organelles [14], but they were present in higher density fractions, such as insulin secretory granules and mitochondria that can be found in pellets centrifuged at low forces [14]. To discern the relative amounts of these ACSL isoforms in insulin secretory granules and mitochondria, we prepared very pure insulin secretory granules by further purification with sucrose gradient centrifugation of an insulin secretory granule fraction obtained by differential centrifugation that was already 90% pure insulin secretory granules. Fractions 2 and 3 of the sucrose gradient contained the insulin secretory granule marker VAMP2 and were free of the mitochondrial marker mitochondrial glycerol phosphate dehydrogenase (mGPD) and the peroxisome marker catalase (Figure 6A). Mitochondria that were free of insulin granules were prepared by centrifuging a post-nuclei/cell debris supernatant fraction at a low force that precipitated a mitochondrial pellet (as described under Materials and Methods) that contained a high concentration of the mitochondrial marker mGPD and was free of the insulin granule marker VAMP2 as shown in lane 9 of Figure 6A. Figure 6B shows that both ACSL3 and ACSL4 were present in insulin secretory granules and mitochondria of the cells, but each of these isoforms was more concentrated in insulin secretory granules than in mitochondria. An immunoblot using the same lane configuration as in Figure 6B and an antibody to TGN38, a marker for the trans-Golgi network and endoplasmic reticulum, was completely blank indicating the lower weight subcellular fractions were not contaminating the insulin granules and mitochondrial fractions and confirming the presence of the ACSl3 and ACSL4 isoforms in insulin granules and mitochondria (This immunoblot that showed a negative result is not shown.).
ACSL enzyme activity in INS-1 832/13 cells and human pancreatic islets
ACSL enzyme activity in whole cell homogenates of INS-1 832/13 cells and human pancreatic islets was measured with palmitate and arachidonate as substrates. In INS-1 832/13 cells the enzyme activity with arachidonate as a substrate was 1.8 ± 0.04 (mean ± SE) fold the enzyme activity with palmitate as a substrate (Table 1). This is consistent with the observation that ACSL3 and ACSL4 are the predominate ACSLs in INS-1 832/13 cells. Both of these enzymes prefer the unsaturated fatty acid arachidonate as a substrate over the saturated fatty acid palmitate as a substrate. However, ACSL3 is reported to accept a wider range of unsaturated fatty acids than ACSL4. ACSL4 is reported to highly prefer arachidonate as a substrate [15, 31]. When arachidonate and palmitate were used as substrates, the ACSL enzyme activities in INS-1 832/13 cells were 2.4 and 1.1 fold, respectively, the activities in human pancreatic islets. ACSLs in human pancreatic islets preferred palmitate over arachidonate as a substrate with an arachidonate/palmitate enzyme activity ratio of 0.84 ± 0.06 (mean ± SE) (Table 1). The differences between human pancreatic islets and INS-1 832/13 cells might be explained by the fact that human islets contain only 60–80% beta cells.
Table 1.
ACSL enzymes in INS-1 832/13 pancreatic beta cells prefer the unsaturated fatty acid arachidonate over the saturated fatty acid palmitate as a substrate. In human pancreatic islets that are composed of beta and non-beta cells, the fatty acid preferences are approximately equal.
| ACSL Enzyme Activity (nmol product formed/min/mg protein) Enzyme Substrate | Activity Ratio Arachidonate/Palmitate (mean ± SE) | ||
|---|---|---|---|
| Arachidonate | Palmitate | ||
| INS-1 832/13 Cells | 5.05 ± 0.32 (12) | 2.76 ± 0.26 (12) | 1.8 ± 0.04 (12) |
| Human Islets | 2.08 ± 0.29 (3) | 2.46 ± 0.52 (3) | 0.84 ± 0.06 (3) |
ACSL enzyme activity in INS-1 832/13 cells is concentrated in insulin secretory granules and in mitochondria
Insulin secretory granules and mitochondria were purified by differential centrifugation as described in the Materials and Methods section and in the legend to Figure 6. ACSL enzyme activity was four-fold more concentrated in insulin secretory granules and two-fold more concentrated in mitochondria compared to the homogenate of whole cells (Table 2).
Table 2. ACSL enzyme activity in pure beta cells is concentrated in insulin secretory granules and mitochondria.
ACSL enzyme activity was measured with palmitate as a substrate as described under Material and Methods. The specific enzyme activity of the homogenate of whole cells is defined as 1.
| Cellular Fraction | Relative Specific ACSL Activity/mg Protein |
|---|---|
| Homogenate of whole cells | 1 |
| Insulin Secretory Granules | 4.1 |
| Mitochondria | 1.9 |
shRNA-mediated gene silencing of various ACSLs and insulin release
In most of the experiments two or more target shRNAs for the same gene were selected to generate stable cell lines from INS-1 832/13 cells often with two cell lines selected for a single target. As we have observed previously with stable shRNA-mediated knockdown cell lines when glucose-stimulated insulin release was inhibited, the order of knockdown was generally mRNA > protein > stimulated insulin release [7]. Virus-delivered shRNAs were used to knock down ACSL3 and ACSL4 in order to also study the effects of their knockdown on glucose-stimulated insulin release in human pancreatic islets.
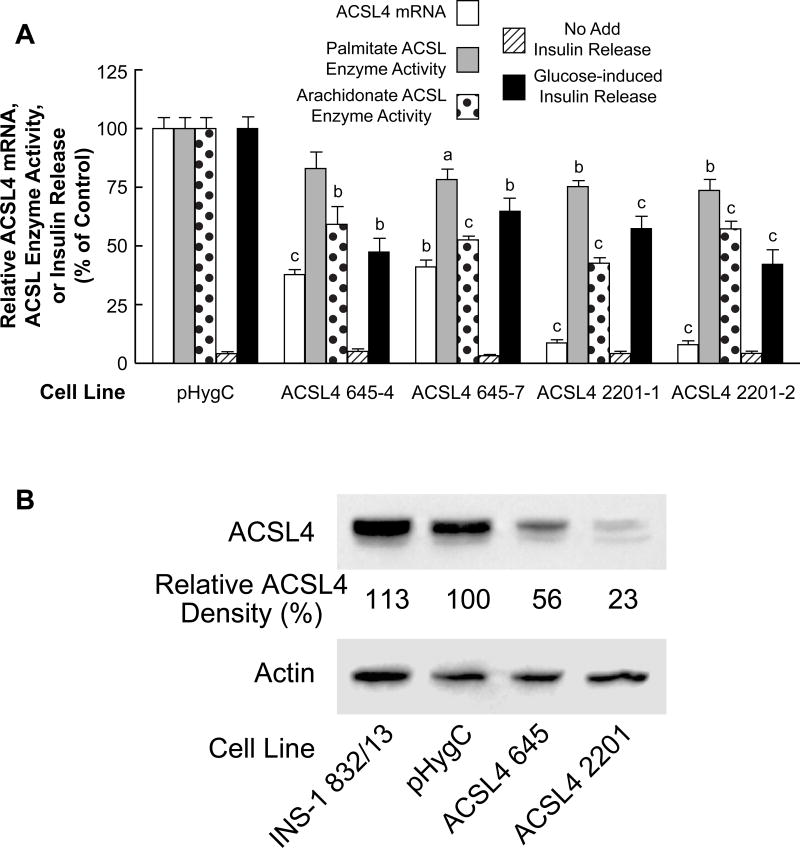
ACSL4 knockdown
ACSL4 shRNAs 645 and 2201 significantly knocked down ACLS4 mRNA (Figure 7A) and ACLS4 protein as judged from immunoblot analysis (Figure 7B) in INS-1 832/13-derived cell lines. The ACSL4 protein was decreased more in the ACSL4 2201 cell lines than in the ACSL4 645 cell lines. When tested for inhibition of glucose-stimulated insulin release, both cell lines showed ~ 50% reduction in insulin release (Figure 7A). Consistent with the known property of ACSL4 preferring the unsaturated fatty acid arachidonate over the unsaturated palmitate as a substrate, the knockdown of ACSL enzyme activity measured in the presence of arachidonate exceeded the knockdown of ACSL enzyme activity with palmitate as a substrate (Figure 7A).
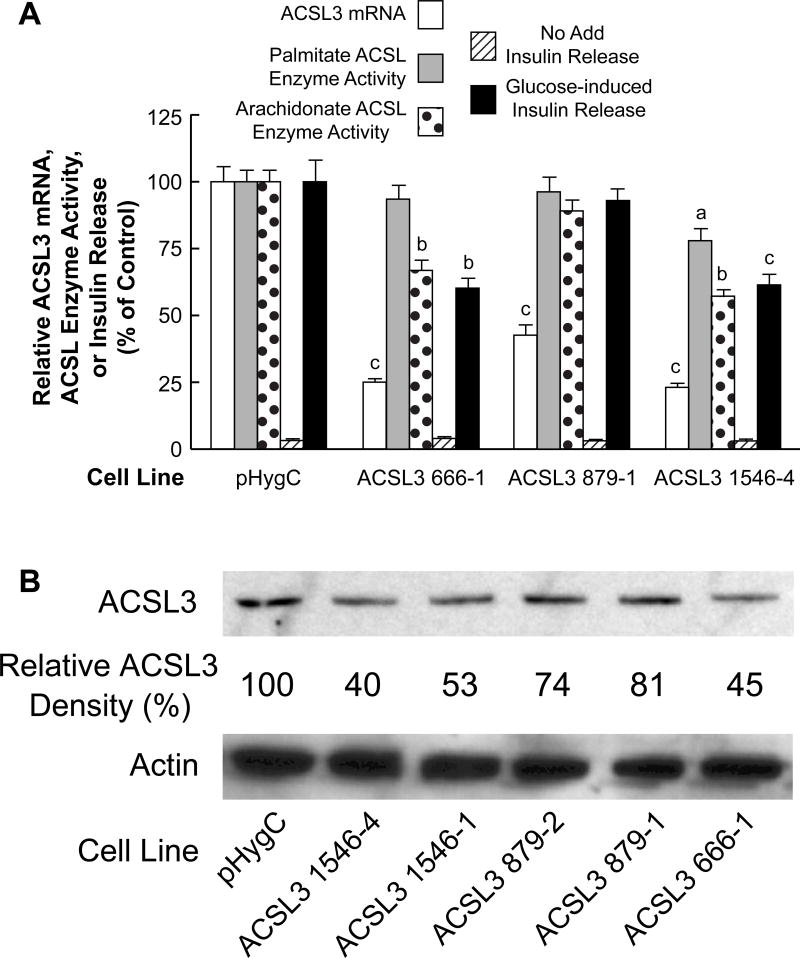
Figure 7. Stable knockdown of ACSL4 mRNA and ACSL4 protein in the INS-1 832/13 cell line inhibits arachidonate ACSL enzyme activity more than palmitate ACSL enzyme activity and inhibits glucose-stimulated insulin release.
A. mRNA values are the mean ± SE of three cell preparations. Insulin release values are the mean ± SE of 12–29 replicate incubations of cells incubated in the presence of 11.1 mM glucose for 1 h. Glucose-stimulated insulin release of the HygC control cell line that contains a non-targeting shRNA sequence was 20.4 mU +/− 1.4 (mean +/− SE insulin/mg cell protein/h. ACSL enzyme activity was measured with arachidonate or palmitate as a substrate. ACSL enzyme activities of the HygC control were similar to the values of the parent INS-1 832/13 cell line shown in Table 1 with the activity with arachidonate equal to 1.8 fold that with palmitate as a substrate (N = ≥ 6 for each condition). All values are expressed relative to the HygC control values equal to 100%. ap < 0.05, bp < 0.01 and cp < 0.001 vs control. B. Immunoblot with 10 μg protein/lane. The density of each band is shown in percent relative to the pHygC control cell line. Actin bands show relatively equal loading of protein across lanes.
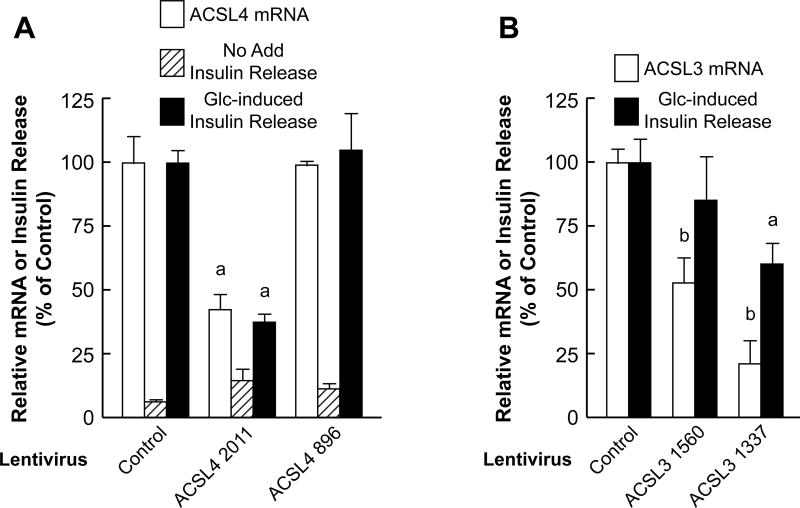
To examine the role of ACLS4 in human pancreatic islets we delivered shRNA using Lentivirus. To this end, we first examined several shRNA targets in 293-T cells after cloning into the pSilencer 2.1 U6 Puro vector and drug selection. We identified two shRNAs; ACSL4 896 and ACSL4 2011 which exhibited the most potent mRNA inhibition (~85%) when tested by qRT-PCR (data not shown). These 293-T cells were also confirmed to contain significantly less ACSL4 protein when examined by immunoblot assay (data not shown). These two shRNA sequences were cloned into the pLVX1-shRNA1 vector (Clontech, USA) and pseudotyped viruses were recovered. The ACSL4 2011 shRNA displayed ~55% ACSL4 mRNA knockdown whereas ACSL4 896 shRNA did not lower ACSL4 mRNA and thus served as an additional control. Inhibition of insulin release in human pancreatic islets was observed only with the ACSL4 2011 shRNA (60%) and this correlated with the ACSL4 mRNA knockdown (Figure 8A).
Figure 8. Knockdown of ACSL4 mRNA or ACSL3 mRNA with virus-delivered shRNA inhibits glucose-stimulated insulin release in human pancreatic islets.
A. Islets were infected with Lentivirus containing shRNAs that targeted ACSL4 mRNA and three days later ACSL4 mRNA and insulin release in the absence or presence of 16.7 mM glucose were measured. Glucose-induced insulin release in the presence of the Hyg nontargeting vector control was 263 ± 18 μ Units insulin/7 islets/1 h (mean ± SE, n = 6 replicates per condition) with experiments with islets from three donors and is expressed as 100%. ap < 0.01 vs control. B. Islets were infected with Lentivirus that targets ACSL3 mRNA and three days later ACSL3 mRNA and glucose-stimulated insulin release were measured. Relative mRNA values are the mean ± SE from four donors. Glucose-stimulated insulin release was calculated by subtracting insulin values in the presence of no glucose from values in the presence of 16.7 mM glucose. Net glucose-stimulated insulin release in the presence of the HygC nontargeting vector was 758 +/− 68 μU insulin/7 islets/1 h (means ± SE, N = 6-10) with batches of islets from four separate donors and is expressed as 100%. ap < 0.05 and bp < 0.01 vs control.
ACSL3 knockdown
Beta cell lines ACSL3 661 and ACSL3 1546 showed the largest knockdown of ACSL3 mRNA and ACSL3 protein and the largest knockdown of glucose-stimulated insulin release (Figures 9A and 9B). Cell line ACSL3 879 showed only about 50% knockdown of ACSL3 mRNA and 20% knockdown of ACSL3 protein and no knockdown of ACSL enzyme activity and glucose-stimulated insulin release (Figures 9A and 9B). ACSL3 in liver and other body tissues is believed to prefer a wider range of unsaturated fatty acid substrates than ACSL4 that prefers the unsaturated fatty acid arachidonate [15, 32]. In agreement with this idea, the ACSL3 knockdown cell lines showed slightly more inhibition of ACSL enzyme activity with arachidonate as a substrate than with the saturated fatty acid palmitate as a substrate in INS-1 832/13 cells (Figure 9A) and proportionately not as large inhibition of ACSL enzyme activity with arachidonate as a substrate compared to the ACSL4 knockdown cell lines (Figure 7A).
Figure 9. Stable knockdown of ACSL3 mRNA and ACSL protein in the INS-1 832/13 cell line inhibits arachidonate ACSL enzyme activity more than palmitate ACSL enzyme activity and inhibits glucose-stimulated insulin release.
A. Conditions are the same as in Figure 7A. Glucose-stimulated insulin release of the control was 19.0 mU +/− 1.3 (mean +/− SE) insulin/mg cell protein/h. B. Immunoblot showing knockdown of ACSL3 protein and with actin to show relative loading of cell protein across lanes. Relative densities of ACSL3 protein bands are shown. Conditions are the same as in Figure 7B. ap < 0.05, bp < 0.01 and cp < 0.001 vs control.
Lentiviruses containing shRNA targeting ACSL3 mRNA were generated similarly to those targeting ACSL4 in order to assess their effects on glucose-stimulated insulin release in human pancreatic islets. ACSL3 1337 shRNA knocked down ACSL3 mRNA 79% and inhibited glucose-stimulated insulin release by 40%. ACSL3 shRNA 1560 decreased ACSL3 mRNA by only 48% and, similarly to our previous studies when an mRNA is decreased only 50% or less [7], this was insufficient to significantly inhibit glucose-stimulated insulin release (Figure 8B).
No inhibition of insulin release with knockdown of the expression of Acsl5, Acsl1 and Acsl6 genes
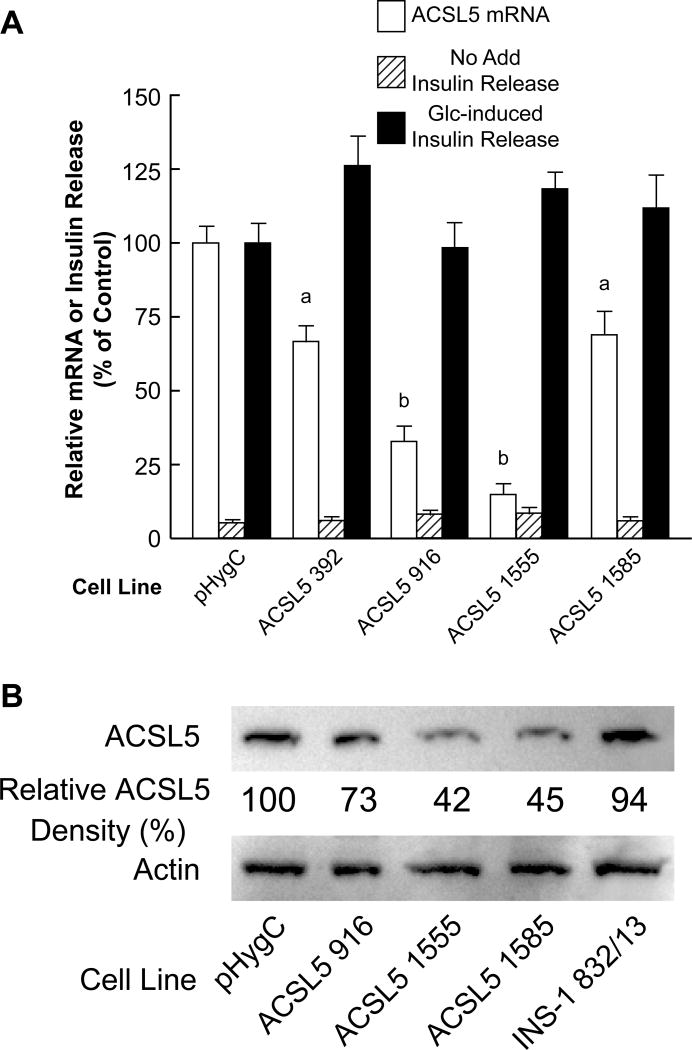
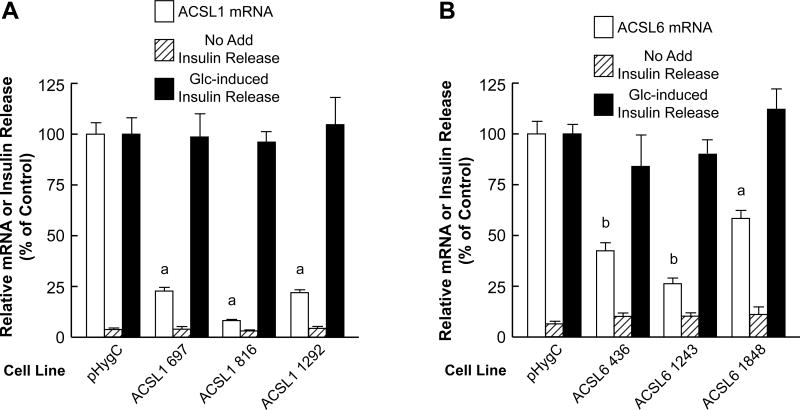
Acsl5 mRNA and ACSL5 protein (Figure 10), Acsl1 mRNA (Figure 11A), and Acsl6 mRNA (Figure 11B) were knocked down in INS-1 832/13-derived cells. However, severe knockdown of these three mRNAs did not significantly affect glucose-stimulated insulin release (Figures 10 and 11).
Figure 10. Severe stable knockdown of ACSL5 mRNA and ACSL5 protein in the INS-1 832/13 cell line does not inhibit glucose-stimulated insulin release.
A. Conditions are the same as in Figure 7A. B. Immunoblot showing knockdown of ACSL5 protein and with actin to show loading of cell protein across lanes. Relative densities of protein bands are shown. Conditions are the same as in Figure 7B. ap < 0.05 and bp < 0.001 vs control.
Figure 11. Severe stable knockdown of ACSL1 mRNA or ACSL6 mRNA in the INS-1 832/13 cell line does not inhibit glucose-stimulated insulin release.
Conditions are the same as in Figure 7. A. ap < 0.001 vs control. B. ap < 0.01 and bp < 0.001 vs control.
Knockdown of ACSL4 changes the pattern of certain fatty acids in phospholipids in INS-1 832/13 cells
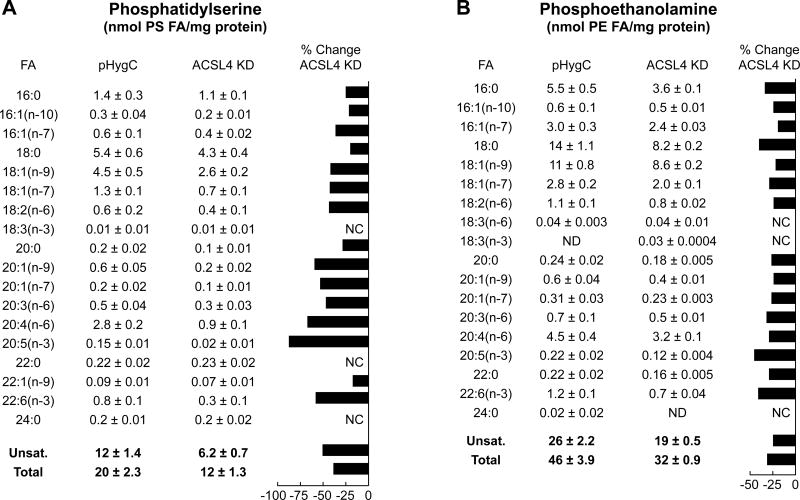
The ACSL4 2201 cell line was chosen to study the effect of ACSL knockdown on the patterns of fatty acids in two phospholipids relevant to insulin secretion. As Figure 7A shows, ACSL4 is capable of activating palmitate, an unsaturated fatty acid, even though it is reported to be more specific for unsaturated fatty acids and highly specific for the unsaturated fatty acid arachidonate. Knockdown of ACSL4 in the INS-1 832/13 cell line lowered the ACSL enzyme activity with palmitate as a substrate, albeit proportionately less than the inhibition of the enzyme activity with arachidonate as a substrate (Figure 7A). Figure 12 shows the effects of knockdown of ACSL4 on fatty acid side chains in phosphatidylserine and phosphatidylethanolamine. Although the data show that knockdown of ACSL4 decreased the contents of saturated and unsaturated 18 carbon fatty acid chains that are potential elongation products of 16 carbon acyl-CoAs, the percentage decreases of elongation of the longer chain 18–22 carbon unsaturated fatty acids was proportionally greater. For example, the level of eicosapentaenoic acid (EPA) (20:5 (n-3)) was decreased 80% in phosphatidylserine and 50% in phosphatidylethanolamine. ACSL4 is reported to favor arachidonate (20:4 (n-6)) and EPA (20:5 (n-3)) as substrates. EPA (20:5 (n-3)) can be metabolized to 22:6 (n-3) (docosahexaenoic acid (DHA)) by two different pathways. One pathway occurs by the elongation of 20:5 (n-3) (EPA) to 22:5 (n-3) followed by desaturation of 22:5 (n-3) to 22:6 (n-3) (DHA). 22:5 (n-3) can also be elongated to 24:5 (n-3) followed by desaturation of 24:5 (n-3) to 24:6 (n-3) followed by oxidation of 24:6 (n-3) to 22:6 (n-3) (DHA). The levels of phosphatidylserine and phosphatidylethanolamine containing DHA were decreased 63% and 42%, respectively, by ACSL4 knockdown (Figures 12A and 12B).
Figure 12. Knockdown of ACSL4 alters patterns of fatty acids in phosphatidylserines and phosphatidylethanolamines in INS-1 832/13 cells.
The ACSL knockdown cell line ACSL4 2201 (ACSL KD) and the control HygC cell line containing a scrambled RNA were stimulated with 11.1 mM glucose for 45 min and harvested for measurements of phosphatidylserine (A) and phosphatidylethanolamine (B). Results are the mean +/− SE of 6 replicate incubations for each condition performed on 2 separate days. The percent change in a phospholipid with ACSL4 or ACSL3 knockdown compared to a very low control fatty acid value or no change was not calculated (NC).
DISCUSSION
ACSLs in INS-1 832/13 beta cells and human and rat pancreatic islets and insulin release
We found that of the five known ACSLs present in mammals, ACSL1, ACSL3, ACSL4, ACSL5 and ACSL6, only ACSL3 and ACSL4 (Figures 1 and 2) were present in rat beta cells (the INS-1 832/13 cell line) and in human and rat pancreatic islets (Figures 1–5). We used shRNA gene silencing knockdown of ACSL3 and ACSL4 mRNA and/or protein and enzyme activity to generate stable cell lines from INS-1 832/13 cells and lentivirus to knock down ACSL3 and ACSL4 in human pancreatic islets. This inhibited glucose-stimulated insulin release by about 50% in INS-1 832/13-derived cell lines and human pancreatic islets (Figures 7–9), suggesting that these two ACSLs play important roles in insulin secretion. Knockdown of ACSL1, ACSL5 and ACSL6 mRNA and/or protein in INS-1 832/13 cells did not inhibit glucose-stimulated insulin release (Figures 10 and 11). Since ACSL5 was detected only in the INS-1 832/13 cell line and its knockdown did not inhibit glucose-stimulated insulin release (Figure 3), and it is not present in normal pancreatic islets, it probably arose in the INS-1 832/13 cell line during the derivation of the cell line. Because the majority of our study was completed before the results of the only other study of ACSLs in INS-1 832/13 cells [35] was published, the results of the two studies independently partially confirm one other. As in the other study, we observed, that although ACSL4 and ACSL5 were each present in INS-1 832/13 cells only gene knockdown of ACSL4 (Figure 7), but not knockdown of ACSL5 mRNA (Figure 10) in INS-1 832/13 cells, inhibited glucose-stimulated insulin secretion. Unlike the other study, we found ACSL3 was highly expressed in normal human pancreatic islets, rat pancreatic islets and INS-1 832/13 cells (Figure 2). We also observed that knockdown of ACSL3 in the INS-1 832/13 cells (Figure 9) and human pancreatic islets (Figure 8) inhibited glucose-stimulated insulin release. The expression of ACSL3 in normal pancreatic islets in addition to in INS-1 832/13 cells and that its knockdown in the INS-1 832/13 cells and human islets inhibited glucose-stimulated insulin release indicates that ACSL3 as well as ACSL4 is important for normal insulin secretion.
ACSL enzyme activity in beta cells and human pancreatic islets
Both ACSL3 and ACSL4 have been reported to prefer unsaturated fatty acids over saturated fatty acids as enzyme substrates, but do accept saturated fatty acids as substrates. ACSL4 has been reported to strongly prefer the unsaturated fatty acid arachidonate as a substrate over other unsaturated fatty acids, but ACSL3 is reported to be less specific in its selectivity and accepts a wide range of unsaturated and saturated fatty acids as substrates [30–32]. When whole cell homogenates are used in assays of ACSL enzyme activity, the assay shows the sum total of the activities of all of the ACSL enzymes in the sample that utilize a particular fatty acid substrate. Consistent with these ideas when Vmax concentrations of the unsaturated fatty acid arachidonate and the saturated fatty acid palmitate were used in assays of enzyme activity using INS-1 832/13 cells, the assayable enzyme activity with arachidonate as a substrate was 1.8 fold higher than with palmitate as a substrate (Table 1). In contrast, ACSLs in human pancreatic islets showed a slight preference for palmitate over arachidonate as a substrate with an arachidonate/palmitate ASCL activity ratio of 0.84 (Table 1). It must be kept in mind that unlike the INS-1 832/13 cell line where all cells are beta cells, human pancreatic islets contain only 60–80% beta cells. Consistent with ACSL4 showing a specificity for arachidonate as an enzyme substrate, in INS-1 832/13 cells knockdown of ACSL4 inhibited ACSL enzyme activity with arachidonate as a substrate more than with palmitate as a substrate (Figure 7A). In the ACSL3 knockdown cells the inhibition of ACSL enzyme activity with arachidonate as the substrate the inhibition was only slightly more than with palmitate as the substrate (Figure 9A). This is consistent with the idea that ACSL3, although preferring unsaturated fatty acids over saturated fatty acids as substrates, has less of a preference for arachidonate than does ACSL4 [30–32].
ACSL3 and ACSL4 are concentrated in insulin secretory granules and mitochondria
When immunoblots were used to analyze the intracellular locations of ACSL3 and ACSL4, these showed that both ACSLs were most concentrated in pure insulin secretory granules purified by differential centrifugation followed by density gradient centrifugation (Figure 6B). Each of these ACSLs was also concentrated in mitochondria, but at a lower concentration than in insulin secretory granules (Figure 6B). Although ACSL4 has been reported to be present in liver peroxisomes, we were unable to detect either ACSL in any other subcellular fraction of INS-1 832/13 cells including peroxisomes (identified by the marker enzyme catalase) (Figure 6A), trans-Golgi network, plasma membrane or cytosol (See legend of Figure 6B). The presence of each ACSL in insulin secretory granules implicates these ACSLs in insulin secretion.
Knockdown of ACSL4 changes phospholipids in beta cells
We also observed that knockdown of ACSL4 in INS-1 832/13 cells changed the composition of fatty acids in phospholipids, particularly fatty acids that are elongation products EPA. These results interconnect with other observations about phospholipids, especially phosphatidylserine [13, 14] in insulin granules. We previously observed that fatty acids in phospholipids, particularly in phosphatidylserine in insulin secretory granules, acutely undergo extensive remodeling [1, 13, 14]. Furthermore, knockdown or inhibition of enzymes, such as pyruvate carboxylase, acetyl-CoA carboxylase and fatty acid synthase involved in supplying precursors for the synthesis of fatty acids in phospholipids in pancreatic beta cells inhibits glucose-stimulated insulin release [1, 7, 12, 18]. Also, knockdown of P4 ATPase (flippase) enzymes involved in translocation of phosphatidylserines and phosphoethanolamines across the lipid bilayer in insulin secretory granules and the plasma membrane in pancreatic beta cells inhibits glucose-stimulated insulin secretion [14].
CONCLUSIONS
Conclusions
The current results support the idea that ACSL4 and ACSL3 participate in insulin secretion by modifying characteristics of fatty acids in insulin granules as well as in mitochondria.
Supplementary Material
Highlights ACSL paper.
Acyl-CoA synthetases were studied in INS-1 832/13 cells and human pancreatic islets
ACSL3 and ACSL4 were present in INS-1 832/13 cells, human and rat pancreatic islets
ACSL3 and ACSL4 were concentrated in insulin secretory granules > mitochondria
Gene silencing of ACSL3 and ACSL4 inhibited glucose-stimulated insulin release
Gene silencing lowered various phosphatidylserines and phosphatidylethanolamines
Acknowledgments
Funding: This work was supported by the National Institutes of Health [grant number DK28348] and the Nowlin Family Trust of the InFaith Community Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: ACC1, acetyl-CoA carboxylase 1; ACC2 acetyl-CoA carboxylase 2; ACSL, acyl-CoA synthetase. EPA, eicosapentaenoic acid
Conflict of interest: The authors declare that they have no conflicts of interest with the contents of this article.
References
- 1.MacDonald MJ, Dobrzyn A, Ntambi J, Stoker SW. The role of rapid lipogenesis in insulin secretion: Insulin secretagogues acutely alter lipid composition of INS-1 832/13 cells. Arch Biochem Biophys. 2008;470:153–162. doi: 10.1016/j.abb.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turk J, Wolf BA, Lefkowith JB, Stump WT, McDaniel ML. Glucose-induced phospholipid hydrolysis in isolated pancreatic islets: quantitative effects on the phospholipid content of arachidonate and other fatty acids. Biochim Biophys Acta. 1986;879:399–409. doi: 10.1016/0005-2760(86)90232-8. [DOI] [PubMed] [Google Scholar]
- 3.Berne C. The metabolism of lipids in mouse pancreatic islets. The biosynthesis of triacylglycerols and phospholipids. Biochem J. 1975;152:667–673. doi: 10.1042/bj1520667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berne C, Andersson A. Lipid metabolism of isolated mouse pancreatic islets maintained in culture at different glucose concentrations. Ups J Med Sci. 1981;86:55–61. doi: 10.3109/03009738109179210. [DOI] [PubMed] [Google Scholar]
- 5.Hallberg A, Andersson A. Effects of starvation on phospholipid metabolism of pancreatic islets. Diabetes Res. 1984;1:105–110. [PubMed] [Google Scholar]
- 6.Vara E, Tamarit-Rodriguez J. Glucose stimulation of insulin secretion in islets of fed and starved rats and its dependence on lipid metabolism. Metabolism. 1986;35:266–271. doi: 10.1016/0026-0495(86)90212-x. [DOI] [PubMed] [Google Scholar]
- 7.Hasan NM, Longacre MJ, Stoker SW, Boonsaen T, Jitrapakdee S, Kendrick MA, Wallace JC, MacDonald MJ. Impaired anaplerosis and insulin secretion in insulinoma cells caused by siRNA-mediated suppression of pyruvate carboxylase. J Biol Chem. 2008;283:28048–28059. doi: 10.1074/jbc.M804170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacDonald MJ, Longacre MJ, Stoker SW, Kendrick MA, Thonpho A, Brown LJ, Hasan NM, Jitrapakdee S, Fukao T, Hanson MS, Fernandez LA, Odorico J. Differences between human and rodent pancreatic islets: low pyruvate carboxylase, ATP citrate lyase and pyruvate carboxylation; high glucose-stimulated acetoacetate in human pancreatic islets. J Biol Chem. 2011;286:18383–18396. doi: 10.1074/jbc.M111.241182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martins EF, Miyasaka CK, Newsholme P, Curi R, Carpinelli AR. Changes of fatty acid composition in incubated rat pancreatic islets. Diabetes Metab. 2004;30:21–27. doi: 10.1016/s1262-3636(07)70085-x. [DOI] [PubMed] [Google Scholar]
- 10.Farfari S, Schulz V, Corkey B, Prentki M. Glucose-regulated anaplerosis and cataplerosis in pancreatic beta-cells: possible implication of a pyruvate/citrate shuttle in insulin secretion. Diabetes. 2000;49:718–726. doi: 10.2337/diabetes.49.5.718. [DOI] [PubMed] [Google Scholar]
- 11.Flamez D, Berger V, Kruhøffer M, Orntoft T, Pipeleers D, Schuit FC. Critical role for cataplerosis via citrate in glucose-regulated insulin release. Diabetes. 2002;51:2018–2024. doi: 10.2337/diabetes.51.7.2018. [DOI] [PubMed] [Google Scholar]
- 12.MacDonald MJ, Hasan NM, Dobrzyn A, Stoker SW, Ntambi JM, Liu X, Sampath H. Knockdown of pyruvate carboxylase or fatty acid synthase lowers numerous lipids and glucose-stimulated insulin release in insulinoma cells. Arch Biochem Biophys. 2013;532:23–31. doi: 10.1016/j.abb.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacDonald MJ, Ade L, Ntambi JM, Ansari IH, Stoker SW. Characterization of phospholipids in insulin secretory granules and mitochondria in pancreatic beta cells and their changes with glucose stimulation. J Biol Chem. 2015;290:11075–11092. doi: 10.1074/jbc.M114.628420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ansari IH, Longacre MJ, Paulusma CC, Stoker SW, Kendrick MA, MacDonald MJ. Characterization of P4 ATPase phospholipid translocases (flippases) in human and rat pancreatic beta cells: Their gene silencing inhibits insulin secretion. J Biol Chem. 2015;290:23110–23123. doi: 10.1074/jbc.M115.655027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ago H, Miyano M. Structure basis of the unidirectional catalysis of long chin fatty acyl-CoA synthetase. – Toward the vectorial movement across the membrane. In: Morikawa K, Tate S, editors. Functional and Structural Biology on the Lipo-network. Transworld Research Network; Kerala, India: 2006. pp. 95–115. [Google Scholar]
- 16.Theisen MJ, Misra I, Saadat D, Campobasso N, Miziorko HM, Harrison DHT. 3-Hydroxy-3-methylglutaryl-CoA synthase intermediate complex observed in “real-time”. Proc Natl Acad Sci USA. 2004;101:16442–16447. doi: 10.1073/pnas.0405809101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Overath P, Pauli G, Schairer HU. Fatty acid degradation in Escherichia coli. An inducible acyl-CoA synthetase, the mapping of old-mutations, and the isolation of regulatory mutants. Eur J Biocem. 1969;7:559–574. [PubMed] [Google Scholar]
- 18.Ishikawa M, Tsuchiya D, Oyama T, Tsunaka Y, Morikawa K. Structural basis for channelling mechanism of a fatty acid beta-oxidation multienzyme complex. EMBO J. 2004;23:2745–2754. doi: 10.1038/sj.emboj.7600298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glick BS, Rothman JE. Possible role for fatty acyl-coenzyme A in intracellular protein transport. Nature. 1987;326:309–312. doi: 10.1038/326309a0. [DOI] [PubMed] [Google Scholar]
- 20.Pfanner N, Glick BS, Arden SR, Rothman JE. Fatty acylation promotes fusion of transport vesicles with Golgi cisternae. J Cell Biol. 1990;110:955–961. doi: 10.1083/jcb.110.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai JC, Liang BB, Jarvi EJ, Cooper AJ, Lu DR. Differential effects of fatty acyl coenzyme A derivatives on citrate synthase and glutamate dehydrogenase. Res Commun Chem Pathol Pharmacol. 1993;82:331–338. [PubMed] [Google Scholar]
- 22.Færgeman NJ, Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signaling. Biochem J. 1997;323:1–12. doi: 10.1042/bj3230001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bronfman M, Orellana A, Morales MN, Bieri F, Waechter F, Stäubli W, Bentley P. Potentiation of diacylglycerol-activated protein kinase C by acyl-coenzyme A thioesters of hypolipidaemic drugs. Biochem Biophys Res Commun. 1989;159:1026–1031. doi: 10.1016/0006-291x(89)92211-0. [DOI] [PubMed] [Google Scholar]
- 24.McLaughlin S, Aderem A. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends Biochem Sci. 1995;20:272–276. doi: 10.1016/s0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- 25.Gordon JI, Duronio RJ, Rudnick DA, Adams SP, Gokel GW. Protein N-myristoylation. J Biol Chem. 1991;266:8647–8650. [PubMed] [Google Scholar]
- 26.Hertz R, Magenheim J, Berman I, Bar-Tana J. Fatty acyl-CoA thioesters are ligands of hepatic nuclear factor-4alpha. Nature. 1998;392:512–516. doi: 10.1038/33185. [DOI] [PubMed] [Google Scholar]
- 27.van Aalten DMF, DiRusso CC, Knudsen J, Wierenga RK. Crystal structure of FadR, a fatty acid-responsive transcription factor with a novel acyl coenzyme A-binding fold. EMBO J. 2000;19:5167–5177. doi: 10.1093/emboj/19.19.5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iijima H, Fujino T, Minekura H, Suzuki H, Kang MJ, Yamamoto T. Biochemical studies of two rat acyl-CoA synthetases, ACS1 and ACS2. Eur J Biochem. 1996;242:186–190. doi: 10.1111/j.1432-1033.1996.0186r.x. [DOI] [PubMed] [Google Scholar]
- 29.Oikawa E, Iijima H, Suzuki T, Sasano H, Sato H, Kamataki A, Nagura H, Kang MJ, Fujino T, Suzuki H, Yamamoto TT. A novel acyl-CoA synthetase, ACS5, expressed in intestinal epithelial cells and proliferating preadipocytes. J Biochem. 1998;124:679–685. doi: 10.1093/oxfordjournals.jbchem.a022165. [DOI] [PubMed] [Google Scholar]
- 30.Fujino T, Kang MJ, Suzuki H, Iijima H, Yamamoto T. Molecular characterization and expression of rat acyl-CoA synthetase 3. J Biol Chem. 1996;271:16748–16742. doi: 10.1074/jbc.271.28.16748. [DOI] [PubMed] [Google Scholar]
- 31.Kang MJ, Fujino T, Sasano H, Minekura H, Yabuki N, Nagura H, Iijima H, Yamamoto TT. A novel arachidonate-preferring acyl-CoA synthetase is present in steroidogenic cells of the rat adrenal, ovary, and testis. Proc Natl Acad USA. 1997;94:2880–2884. doi: 10.1073/pnas.94.7.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soupene E, Kuypers FA. Mammalian long-chain acyl-CoA synthetases. Exp Biol Med (Maywood) 2008;233:507–521. doi: 10.3181/0710-MR-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki H, Kawarabayasi Y, Kondo J, Abe T, Nishikawa K, Kimura S, Hashimoto T, Yamamoto T. Structure and regulation of rat long-chain acyl-CoA synthetase. J Biol Chem. 1990;265:8681–8685. [PubMed] [Google Scholar]
- 34.Fujino T, Yamamoto T. Cloning and functional expression of a novel long-chain acyl-CoA synthetase expressed in brain. J Biochem. 1992;111:197–203. doi: 10.1093/oxfordjournals.jbchem.a123737. [DOI] [PubMed] [Google Scholar]
- 35.Klett EL, Chen S, Edin ML, Li LO, Ilkayeva O, Zeldin DC, Newgard CB, Coleman RA. Diminished acyl-CoA synthetase isoform 4 activity in INS 832/13 cells reduces cellular epoxyeicosatrienoic acid levels and results in impaired glucose-stimulated insulin secretion. J Biol Chem. 2013;288:21618–21629. doi: 10.1074/jbc.M113.481077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balciunas D, Wangensteen KJ, Wilber A, Bell J, Geurts A, Sivasu-bbu S, Wang X, Hackett PB, Largaespada DA, McIvor RS, Ekker SC. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2006;2:e169. doi: 10.1371/journal.pgen.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown LJ, Longacre MJ, Hasan NM, Kendrick MA, Stoker SW, MacDonald MJ. Chronic reduction of the cytosolic or mitochondrial NAD(P)P-malic enzyme does not affect insulin secretion in a rat insulinoma cell line. J Biol Chem. 2009;284:35359–35367. doi: 10.1074/jbc.M109.040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacDonald MJ. Feasibility of a mitochondrial pyruvate malate shuttle in pancreatic islets: Further implication of cytosolic NADPH in insulin secretion. J Biol Chem. 1995;270:20051–20058. [PubMed] [Google Scholar]
- 39.MacDonald MJ. Metabolism of the insulin secretagogue methyl succinate by pancreatic islets. Arch Biochem Biophys. 1993;300:201–205. doi: 10.1006/abbi.1993.1028. [DOI] [PubMed] [Google Scholar]
- 40.MacDonald MJ, Smith AD, III, Hasan NM, Sabat G, Fahein LA. Feasibility of pathways for transfer of acyl groups from mitochondria to the cytosol to form short chain acyl CoAs in the pancreatic beta cell. J Biol Chem. 2007;282:30596–30606. doi: 10.1074/jbc.M702732200. [DOI] [PubMed] [Google Scholar]
- 41.MacDonald MJ, Longacre MJ, Langberg E-C, Tibell A, Kendrick MA, Fukao T, Ostenson C-G. Decreased levels of metabolic enzymes in pancreatic islets of patients with type 2 diabetes. Diabetologia. 2009;52:1087–1091. doi: 10.1007/s00125-009-1319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rana RS, Kowluru A, MacDonald MJ. Secretagogue-responsive and – unresponsive pools of phosphatidylinositol in pancreatic islets. Arch Biochem Biophys. 1986;245:411–416. doi: 10.1016/0003-9861(86)90232-8. [DOI] [PubMed] [Google Scholar]
- 43.Rana RS, Kowluru A, MacDonald MJ. Enzymes of phospholipid metabolism in rat pancreatic islets: subcellular distribution and the effect of glucose and calcium. J Cell Biochem. 1986;32:143–150. doi: 10.1002/jcb.240320206. [DOI] [PubMed] [Google Scholar]
- 44.Kowluru A, Rana RS, MacDonald MJ. Phospholipid methyltransferase activity in pancreatic islets: activation by calcium. Arch Biochem Biophys. 1985;242:72–81. doi: 10.1016/0003-9861(85)90481-3. [DOI] [PubMed] [Google Scholar]
- 45.Kowluru A, Rabaglia ME, Muse KE, Metz SA. Subcellular localization and kinetic characterization of guanine nucleotide binding proteins in normal rat and human pancreatic islets and transformed β cells. Biochim Biophys Acta. 1994;1222:348–359. doi: 10.1016/0167-4889(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 46.Wang Z, Thurmond DC. Differential phosphorylation of RhoGDI mediates the distinct cycling of Cdc42 and Rac1 to regulate second-phase insulin secretion. J Biol Chem. 2010;285:6186–6197. doi: 10.1074/jbc.M109.072421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spurlin BA, Thurmond DC. Syntaxin 4 facilitates biphasic glucose-stimulated insulin secretion from pancreatic beta-cells. Mol Endocrinol. 2006;20:183–193. doi: 10.1210/me.2005-0157. [DOI] [PubMed] [Google Scholar]
- 48.Rhodes CJ, Brennan SO, Hutton JC. Proalbumin to albumin conversion by a proinsulin processing endopeptidase of insulin secretory granules. J Biol Chem. 1989;264:14240–14245. [PubMed] [Google Scholar]
- 49.Askari B, Kanter JE, Sherrid AM, Golej DL, Bender AT, Liu J, Hsueh WA, Beavo JA, Coleman RA, Bornfeldt KE. Rosiglitazone inhibits acyl-CoA synthetase activity and fatty acid partitioning to diacylglycerol and triacylglycerol via a peroxisome proliferator-activated receptor-gamma-independent mechanism in human arterial smooth muscle cells and macrophages. Diabetes. 2007;56:1143–1152. doi: 10.2337/db06-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith ME, Saraceno GE, Capani F, Castilla R. Long-chain acyl-CoA synthetase 4 is regulated by phosphorylation. Biochem Biophys Res Commun. 2013;430:272–277. doi: 10.1016/j.bbrc.2012.10.138. [DOI] [PubMed] [Google Scholar]
- 51.Hisanaga Y, Ago H, Nakagawa N, Hamada K, Ida K, Yamamoto M, Hori T, Arii Y, Sugahara M, Kuramitsu S, Yokoyama S, Miyano M. Structural basis of the substrate-specific two-step catalysis of long chain fatty acyl-CoA synthetase dimer. J Biol Chem. 2004;279:31717–31726. doi: 10.1074/jbc.M400100200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.