Abstract
Molecular markers are needed for enhancing the development of elite sweetpotato (Ipomoea batatas (L.) Lam) cultivars with a wide range of commercially important traits in sub-Saharan Africa. This study was conducted to estimate the heritability and determine trait correlations of storage root yield, dry matter, starch and β-carotene content in a cross between ‘New Kawogo’ × ‘Beauregard’. The study was also conducted to identify simple sequence repeat (SSR) markers associated with these traits. A total of 287 progeny and the parents were evaluated for two seasons at three sites in Uganda and genotyped with 250 SSR markers. Broad sense heritability (H2) for storage root yield, dry matter, starch and β-carotene content were 0.24, 0.68, 0.70 and 0.90, respectively. Storage root β-carotene content was negatively correlated with dry matter (r = −0.59, P < 0.001) and starch (r = −0.93, P < 0.001) content, while storage root yield was positively correlated with dry matter (r = 0.57, P = 0.029) and starch (r = 0.41, P = 0.008) content. Through logistic regression, a total of 12, 4, 6 and 8 SSR markers were associated with storage root yield, dry matter, starch and β-carotene content, respectively. The SSR markers used in this study may be useful for quantitative trait loci analysis and selection for these traits in future.
Keywords: sweetpotato, SSR marker, heritability, yield
Introduction
Sweetpotato (Ipomoea batatas (L.) Lam) is a key food, feed, nutrition and income security crop in the developing world (FAOSTAT 2012). Sweetpotato can be produced under marginal conditions with minimal input requirements (Karyeija et al. 1998). As a result, it is considered one of the most affordable sources of carbohydrates, vitamin A, vitamin C, fiber, and minerals particularly in the sub-Saharan Africa (SSA) region (Hotz et al. 2012, Low et al. 2007, Woolfe 1992). Vitamin A deficiency (VAD) and related diseases account for 1.8% of deaths globally that occur mainly in the developing world (Black 2003).
Unfortunately, farmers in the SSA region predominantly grow low yielding, white- or cream-fleshed landrace varieties of sweetpotato that are typically less nutritious (Low et al. 2007, Woolfe 1992, Yada et al. 2010a). Average storage root yield in this region is less than 5.0 metric tons/ha, far below the global average root yield of 13.3 metric tons/ha (FAOSTAT 2012). The landraces and released orange-fleshed sweetpotato (OFSP) varieties have lower β-carotene content than those varieties being produced in the developed world (Mwanga et al. 2009, Yencho et al. 2008).
Genetic improvement of sweetpotato for storage root yield, dry matter, starch and β-carotene content has been difficult. This is partly attributed to the complex genome of sweetpotato, which makes breeding very difficult (Chang et al. 2009). Sweetpotato is a highly heterozygous autohexaploid (2n = 6× = 90) crop (Cervantes-Flores et al. 2008a, Nishiyama et al. 1975) with high levels of sporophytic self-incompatibility (Gurmu et al. 2013, Martin 1965). High levels of cross-incompatibility also occur in this clonally propagated crop (Gurmu et al. 2013, Martin 1970), and this limits the levels of hybridization that can be exploited for crop improvement.
Another limitation to attaining genetic gains in sweetpotato is that yield, dry matter, starch and β-carotene content and other important agronomic traits are quantitatively inherited (Cervantes-Flores et al. 2011, Chang et al. 2009). These traits are controlled by multiple genes with high levels of genotype × environment interactions that further complicate variety development (Adebola et al. 2013, Grüneberg et al. 2005). Also, storage root dry matter and β-carotene content are negatively correlated, making it difficult to select for high levels of both traits in the same background (Grüneberg et al. 2005).
Sweetpotato improvement has generally been done by mass selection (Jones et al. 1987). This method, though simple to implement, has not been effective for the improvement of complex sweetpotato traits. The development of new genomics tools to facilitate sweetpotato improvement would enable breeders to overcome some of the inherent genetic barriers to sweetpotato breeding. Though sweetpotato genomics research has lagged behind compared to other crops (Cervantes-Flores et al. 2008a), modern breeding methods will be critical to unlock its full potential.
Most molecular genetic studies in sweetpotato to date have used amplified fragment length polymorphism (AFLP) markers (Vos et al. 1995) and random amplified polymorphic DNA (RAPD) markers. These dominant markers (i.e. they only amplify the dominant allele and cannot differentiate between dominant homozygous and heterozygous individuals) have been used in sweetpotato genetic linkage analysis (Cervantes-Flores et al. 2008a, Kriegner et al. 2003, Mwanga et al. 2002, Zhao et al. 2013). These markers have also been used in quantitative trait loci (QTL) analysis of root-knot nematode resistance, and storage root dry matter, starch and β-carotene content (Cervantes-Flores et al. 2008b, 2011, Chang et al. 2009, Zhao et al. 2013). However, none of these QTLs have been used for sweetpotato improvement by marker assisted selection (MAS) due to the inherent limitations of the marker technology.
In the absence of high-density genetic linkage maps for sweetpotato, discriminant analysis and logistic regression have been employed to identify molecular markers linked to key traits. AFLP markers linked to root-knot nematode resistance (Mcharo et al. 2005b), sweetpotato virus disease (SPVD) resistance (Mcharo et al. 2005a, Miano et al. 2008) and storage root β-carotene content (Mcharo and LaBonte 2010) were identified by breeders. However, to the best of our knowledge, none of these markers are being used in a MAS breeding program.
Presently, there is no reference genome sequence for cultivated hexaploid sweetpotato. However, through de novo whole-genome sequencing, the genome sequence of the diploid I. trifida, the close ancestor of cultivated was recently published (Hirakawa et al. 2015). This genome sequence for I. trifida, in addition to other genomic resources, will be important for developing the more versatile but currently limited co-dominant SSR and single nucleotide polymorphism (SNP) markers for sweetpotato improvement. While some SSR markers have been used in sweetpotato diversity analysis (Koussao et al. 2014, Veasey et al. 2008, Yada et al. 2010b, 2015), no sole SSR-based linkage maps and QTL of sweetpotato traits have been reported.
Earlier research reported genetic linkage maps and QTL for sweetpotato storage root dry matter content (Zhao et al. 2013) and starch content (Yu et al. 2014) based on a combination of AFLP and SSR markers. Only three QTL for dry matter were reported to be co-localized with SSR markers in the study. Two QTL for starch content were co-localized with SSR markers in the study (Yu et al. 2014). Recent studies associated a total of 32, 16, and 17 EST and mRNA based SSR markers with starch, β-carotene content and starch composition in a natural sweetpotato population (Zhang et al. 2016). To date, no sweetpotato SNP discovery or mapping studies have been reported.
This study was conducted to understand the genetic basis of storage root yield, dry matter, and starch and β-carotene production in sweetpotato and to identify SSR markers linked to these traits for future use in sweetpotato genetic improvement.
Materials and Methods
Plant material
The population used in this study consisted of 287 F1 progeny from a bi-parental cross between ‘New Kawogo’ (NK) and ‘Beauregard’ (B) developed at NaCRRI (0°32′N, 32°35′E, 1,150 m.a.s.l) in Uganda (Yada et al. 2015). ‘New Kawogo’ (female) is a weevil resistant, SPVD resistant, high dry matter content and white-fleshed released Ugandan landrace (Mwanga et al. 2001, Stevenson et al. 2009). ‘Beauregard’ (male) is a weevil and SPVD susceptible, low dry matter content and orange-fleshed (high β-carotene content) popular US cultivar (Rolston et al. 1987). This mapping population and the parents have been maintained in a screen house at NaCRRI.
Genomic DNA extraction and SSR genotyping of mapping population
Genomic DNA was extracted from the young leaves (ca. 100 g) of each progeny at the NaCRRI Biosciences laboratory using a modified C-TAB method (Doyle and Doyle 1990) as described in detail by Yada et al. (2015).
Genomic DNA samples were shipped from NaCRRI to North Carolina State University for genotyping through the DHL Express Courier Service. DNA concentrations were measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, Delaware). A total of 405 published expressed sequence tag (EST) SSR primers (Buteler et al. 1999, Hu et al. 2004, Schafleitner et al. 2010, Wang et al. 2011) were designed and screened for polymorphism among the parents and ten randomly selected progeny. The primers were redesigned by addition of an M13 tail universal primer sequence (TGTAAAACGACGGCCAGT) to the 5′ end of the forward primer sequence and then synthesized by Eurofins Genomics (Huntsville, Alabama). The complementary M13 sequences were fluorescently labeled with VIC (green), 6FAM (blue), NED (yellow) and PET (red) tags from Applied Biosystems (Foster City, California) for automated detection of polymerase chain reaction (PCR) amplification products. Out of the 405 primers, 250 were polymorphic among the parents and selected progeny and subsequently used for genotyping the whole population.
The PCR reaction volume was 10 μl. The reaction consisted of 3.0 μl (20–40 ng/μl) DNA template, 1.0 μl 10X PCR buffer, 1.0 μl MgCl2 (15 mM), 0.8 μl DNTPs (10 mM) mix, 0.2 μl forward primer (1.0 μM), 1.0 μl reverse primer (1.0 μM), 0.5 μl M13 primer (1.0 μM), 0.1 μl Taq polymerase (50 U/μl) and 2.4 μl PCR water. The PCR conditions were as follows; one hold at 94.0°C for 4 min, followed by first 15 cycles of 94.0°C denaturation for 30 sec, 55.0°C annealing for 30 sec, and 72.0°C extension for 1 min, then followed by 25 cycles of 94.0°C for 30 sec, 50.0°C annealing for 30 sec, and 72.0°C extension for 1 min, followed by two holds at 72.0°C for 7 min, and at 4.0°C for infinite time.
The PCR amplifications were performed using an Eppendorf Mastercycler (Eppendorf AG, Hamburg, Germany). The PCR products were sized by capillary electrophoresis using an ABI3730xl Genetic Analyzer (Applied Biosystems, Foster City, California) and data were analyzed using GeneMarker 2.2.0 (SoftGenetics, State College, Pennsylvania). The allele scores were converted to binary data, that is 1 (allele present) and 0 (allele absent).
Field trials and storage root yield determination
The phenotyping trials were conducted for two seasons at three sites during 2012. In Uganda, there are typically two rainy seasons in a year. For both seasons, the 287 NKB progeny and parents were planted in a randomized complete block design with 3 replications. The clones were planted on 1.5 meter long ridged rows (plots) on 1.0 m centers. Each plot contained five plants spaced 30 cm apart within the row. The trials were conducted under rain fed conditions and weeded three times.
The three trial sites in Uganda were located at the National Semi-Arid Resources Research Institute (NaSARRI) (1°32′N, 33°27′E), the Ngetta Zonal Agricultural Research and Development Institute (NgeZARDI) (2°202′N, 33°62′E), and the National Crops Resources Research Institute (NaCRRI) (0°32′N, 32°35′E). NaSARRI is located in eastern Uganda, 1,140 meters above sea level (m.a.s.l.), with annual mean temperature of 26.0°C, mean annual bi-modal rainfall of between 800 mm and 1,150 mm, and has sandy loam soils, and pH 5.2–6.0. NgeZARDI is located in northeastern Uganda, has an altitude of 1,180 m.a.s.l., annual mean temperature of 30.4°C, and mean annual rainfall of 1,319 mm, and has sandy loam soils, pH 5.5. NaCRRI is located in central Uganda and has an altitude of 1,150 meters above sea level, annual mean temperature of 22.2°C, and mean annual bimodal rainfall of 1,270 mm and has sandy clay loam soils, pH 4.9–5.0.
The first (2012A) trials were planted in June, 2012 and harvested in November, 2012, while the second (2012B) trials were planted in November, 2012 and harvested in May, 2013. The trials were harvested at 5–6 months after planting to ensure ample time for pest and disease infestation.
At harvest, data were collected on vine weight, marketable storage root weight (MRW) and non-marketable storage root weight (NMRW). Total storage root yield in tons per hectare was computed as: RYTha = (MRW + NMRW)/Net plot area in m2) × 10.
Dry matter, starch and β-carotene content analyses
Storage root samples were brought to NaCRRI and a sub-sample from each plot was washed and processed. For the oven method of storage root dry matter determination, approximately 100 g of fresh storage root samples of each clone were sliced and weighed (DMF) and then dried in paper bags in an oven at 65°C for 72 hrs to a constant weight. The samples were immediately weighed after drying (DMD). Storage root dry matter content was calculated as DM = (DMD/DMF) × 100.
Another sub-sample (100 g) of the fresh storage root was cut transversely into 2–3 discs, weighed and freeze-dried using a vacuum freeze dryer (True-Ten Industrial Co., Taichung City, Taiwan). The freeze-dried samples were weighed and then milled (mesh size 0.425 mm) using a Thomas Wiley Mini-Mill (Thomas Scientific, Swedesboro, New Jersey). The milled samples were sent to the International Potato Center (CIP), Ghana, for storage root chemistry profiling using near-infrared reflectance spectroscopy (NIRS).
The NIRS calibration equations for β-carotene, dry matter, starch, iron, zinc, protein, and glucose and fructose content were developed at CIP, Lima, Peru (Zum Felde et al. 2009). For the scanning, approximately 2.0 g of each milled sweetpotato sample was transferred into a small quartz window-clad cylindrical cell (internal diameter; 35 mm, and depth 9 mm). The samples were scanned in the Rapid Content Analyzer (RCA) of the NIRS instrument (FOSS NIRSystems, Höganäs, Sweden). Spectral data were collected by measurement of diffuse reflectance from the samples in the near-infrared region of the electromagnetic spectrum using a monochromator (XDS 1000, Höganäs, Sweden).
Samples were scanned and the results recorded as log (1/R) at 2 nm increments and averaged. The results were processed using ISscans, version 4.2.0 (FOSS NIRSystems, Höganäs, Sweden), with the average spectrum of each sample used for analysis. The spectral data were then analyzed with Win ISI II software, version 1.04 (FOSS NIRSystems, Höganäs, Sweden) using calibration equations developed by Zum Felde et al. (2009). The calibration equation and cross validation employed had strong positive correlations with the reference data of 216 to 422 sweetpotato samples. The calibration coefficients of determination (R2c) values were 0.98 and 0.97 for β-carotene and starch, respectively. The verification process also yielded high coefficients of determination (R2cv) of 0.8 to 0.96 for the traits.
Data analysis
The trait means and ranges were obtained through PROC MEANS and analysis of variance (ANOVA) for the traits was conducted by the generalized linear mixed model procedure, PROC GLIMMIX (SAS 9.4, SAS Institute, Cary, North Carolina). The analyses were done using 284 progeny and the 2 parents as some progeny were lost in the field. The parents and progeny were treated as fixed effects while block, site and season were random effects. For each trait, the overall least significant (LS mean) of parents was compared with that of progeny (Gtype) across sites and seasons. The trait LS mean of individual progeny and individual parents (Gen(Gtype)) across sites and seasons were also compared. All the LS means were compared using the Turkey-Kramer grouping (alpha = 0.05). The genotype LS means were plotted to assess the level of transgressive segregation for each trait in the population. Trait LS means were later ranked to select the best performing progeny that have cultivar release potential.
The trait phenotypic and genotypic variances were analyzed by PROC MIXED and PROC IML and used to compute broad sense heritability estimates of storage root yield, dry matter, and starch and β-carotene content. For estimating the genotypic trait correlations among storage root yield, dry matter, starch and β-carotene content, we first computed restricted maximum likelihood (REML) based covariance estimates between traits by PROC MIXED, followed by calculation of the genotypic covariance (CovG) and variance components (VG1 and VG2) for pairs of traits using PROC IML (Holland 2006). Then pairwise trait genetic correlation (RG) was calculated as:
Marker trait association was done using regression analysis of best linear unbiased predictions (BLUPs) values of storage root yield, dry matter, starch and β-carotene content against SSR allele profiles for the genotypes. Trait BLUPs were predicted using PROC MIXED. Then, marker loci and trait BLUP regression analysis was implemented by PROC GLIMMIX with genotypes as fixed effects. Marker selection was based on a probability level (alpha = 0.05). A marker locus was considered to have significant association with a trait if p < 0.05 for the regression analysis.
Results
ANOVA of storage root yield, dry matter, starch and β-carotene content
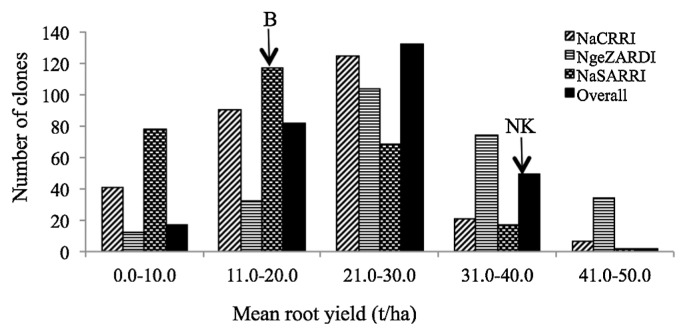
Individual progeny and parents (Gen(Gtype) differed significantly in their mean storage root yields (Table 1). Mean storage root yield ranged from 2.0–48.4 t/ha (Fig. 1) in progeny NKB285 and NKB216, respectively. The overall mean storage root yields of ‘New Kawogo’ and ‘Beauregard’ were 30.2 t/ha and 18.6 t/ha, respectively. Site storage root yield means were also significantly different. The mean storage root yields per site were 22.1 t/ha, 29.2 t/ha and 19.4 t/ha for NaCRRI, NgeZARDI and NaSARRI, respectively. However, the overall mean storage root yield of progeny was not significantly different from the overall mean storage root yield of the parents (Gtype) (P = 0.4139). The overall mean storage root yield of the parents was 24.4 t/ha, while the overall mean storage root yield of the progeny was 22.7 t/ha. For storage root yield, there was a significant genotype by environment interaction (P < 0.0001).
Table 1.
ANOVA of storage root yield and dry matter content of the progeny and parents of the ‘New Kawogo’ × ‘Beauregard’ mapping population
| Source | Yield | Dry matter content | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| DF | SS | MS | F Value | Pr > F | DF | SS | MS | F Value | Pr > F | |
| Sitea | 2 | 4713.4 | 2356.7 | 7.4 | 0.0016 | 2 | 459.6 | 229.8 | 1.04 | 0.3735 |
| Gtypeb | 1 | 100.4 | 100.4 | 0.7 | 0.4139 | 1 | 204.0 | 204.0 | 8.78 | 0.0031 |
| Gen (Gtype)c | 284 | 299719.0 | 1055.4 | 7.0 | <.0001 | 284 | 14034.0 | 49.4 | 2.13 | <.0001 |
| Site*Gtype d | 2 | 1254.7 | 627.4 | 4.2 | 0.0155 | 2 | 5.9 | 3.0 | 0.13 | 0.8807 |
| Site*Gen (Gtype)e | 567 | 180847.0 | 319.0 | 2.1 | <.0001 | 565 | 21328.0 | 37.7 | 1.63 | <.0001 |
| Block (Site*Season) | 15 | 59178.0 | 3945.2 | 26.2 | <.0001 | 15 | 56347.0 | 3756.5 | 161.71 | <.0001 |
| Residual | 3965 | 596194.0 | 150.4 | 3328 | 77306.0 | 23.2 | ||||
DF = degrees of freedom, SS = sum of squares, MS = mean sum of squares (SS/DF).
Mean squares tests the significant effect of locations.
Tests the significant effect of overall least significant mean of the parents vs that of the progeny across sites and seasons.
Tests the significant effect of least significant means of individual genotypes (parents and progeny) across sites and seasons.
Tests the significant effect of site on overall parent and progeny least significant means.
Test the significant effect of genotype × environment interaction.
Fig. 1.
Distribution of mean storage root yield in the progeny and parents of the ‘New Kawogo’ (NK) × ‘Beauregard’ (B) mapping population, bars represent the number of clones in each class of mean storage root yield scores averaged over seasons for sites (NaCRRI, NgeZARDI and NaSARRI), and averaged over seasons and sites for the overall mean.
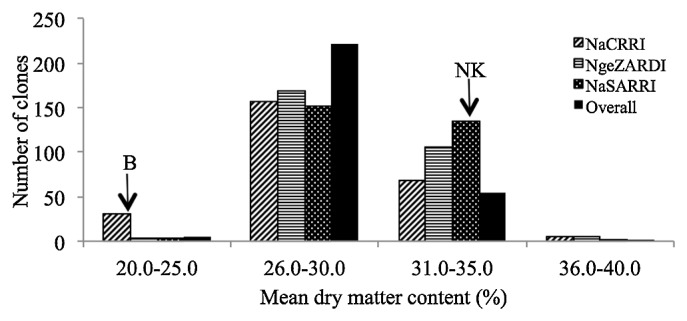
The mean storage root dry matter content of individual progeny and parents differed significantly (Table 1) and ranged from 24.0–35.7% (Fig. 2) in NKB254 and NKB275, respectively. The mean storage root dry matter content of ‘New Kawogo’ was 31.6% whereas that of ‘Beauregard’ was 21.4%. The overall parental mean storage root dry matter content and that of progeny (Gtype) were significantly different and were 26.5% and 29.1%, respectively. Site main effects were not significant for dry matter content.
Fig. 2.
Distribution of mean storage root dry matter content in the progeny and parents of the ‘New Kawogo’ (NK) × ‘Beauregard’ (B) mapping population, bars represent the number of clones in each class of mean storage root dry matter content scores averaged over seasons for sites (NaCRRI, NgeZARDI and NaSARRI), and averaged over seasons and sites for the overall mean.
The individual progeny and parents had significantly different mean storage root starch content (Table 2). The mean starch content (%) of ‘New Kawogo’ and ‘Beauregard’ was 73.8 and 53.2, respectively. The mean starch content of parents was not significantly different from that of the progeny. Just like for dry matter, the mean storage root starch content was not different at the sites as mean starch content (%) of 65.1, 66.1 and 64.1 for NaCRRI, NgeZARDI and NaSARRI, respectively were observed. There was no genotype by environment interaction for starch content in this population (P = 0.0814).
Table 2.
ANOVA of storage root β-carotene and starch content of the progeny and parents of the ‘New Kawogo’ × ‘Beauregard’ mapping population
| Source | β-carotene content | Starch content | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| DF | SS | MS | F Value | Pr > F | DF | SS | MS | F Value | Pr > F | |
| Sitea | 2 | 326.6 | 163.3 | 2.7 | 0.0770 | 2 | 198.6 | 99.3 | 0.4 | 0.7100 |
| Gtypeb | 1 | 978.0 | 978.0 | 31.3 | <.0001 | 1 | 317.0 | 317.0 | 3.3 | 0.0700 |
| Gen (Gtype)c | 284 | 117739.0 | 414.6 | 13.3 | <.0001 | 284 | 98799.0 | 347.9 | 3.6 | <.0001 |
| Site*Gtyped | 2 | 4.6 | 2.3 | 0.1 | 0.9285 | 2 | 145.4 | 72.7 | 0.8 | 0.4708 |
| Site*Gen (Gtype)e | 566 | 26301.0 | 46.5 | 1.5 | <.0001 | 567 | 59709.0 | 105.3 | 1.1 | 0.0814 |
| Block (Site*Season) | 15 | 9086.5 | 605.8 | 19.4 | <.0001 | 15 | 60377.0 | 4025.1 | 41.7 | <.0001 |
| Residual | 3420 | 106708.0 | 31.2 | 3650 | 352270.0 | 96.5 | ||||
DF = degrees of freedom, SS = sum of squares, MS = mean sum of squares (SS/DF).
Mean squares tests the significant effect of locations.
Tests the significant effect of overall least significant mean of the parents vs that of the progeny across sites and seasons.
Tests the significant effect of least significant means of individual genotypes (parents and progeny) across sites and seasons.
Tests the significant effect of site on overall parent and progeny least significant means.
Test the significant effect of genotype × environment interaction.
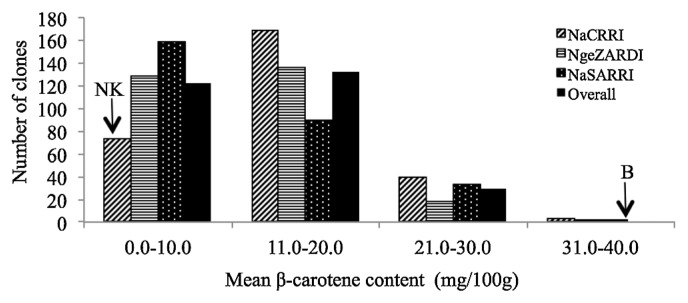
The mean storage root β-carotene content of individual progeny and parents were significantly different (Table 2). The mean β-carotene contents of the progeny ranged from 4.2–30.5 mg/100 g (Fig. 3) in NKB221 and NKB92, respectively. The mean storage root β-carotene content of ‘New Kawogo’ was 4.3 mg/100 g while that of ‘Beauregard’ was 31.7 mg/100 g on a fresh weight basis. Mean storage root β-carotene content at sites was not significantly different.
Fig. 3.
Distribution of mean storage root β-carotene content in the progeny and parents of the ‘New Kawogo’ (NK) × ‘Beauregard’ (B) mapping population, bars represent the number of clones in each class of mean storage root β-carotene content scores averaged over seasons for sites (NaCRRI, NgeZARDI and NaSARRI), and averaged over seasons and sites for the overall mean.
Heritability and trait correlation analysis
Trait heritability estimates are shown in Table 3. High broad sense heritability (H2 = 0.90) was observed for β-carotene content. The broad sense heritability estimate of starch content was 0.70 and that of dry matter content was 0.68. As is expected, the broad sense heritability of yield was low (H2 = 0.24). From our genotypic trait correlations of storage root yield, dry matter, starch and β-carotene content (Table 4), we observed a high positive genetic correlation between storage root starch and dry matter content (r = 0.77, P < 0.001). Storage root yield was positively correlated with dry matter (r = 0.57, P = 0.029) and starch (r = 0.41, P = 0.008) content. On the other hand, storage root β-carotene content was negatively correlated with dry matter (r = −0.59, P < 0.001) and starch content (r = −0.93, P < 0.001).
Table 3.
Broad sense heritability estimates for storage root yield, dry matter, starch and β-carotene content in the progeny of the ‘New Kawogo’ × ‘Beauregard’ mapping population
| Trait | H2 | SEa |
|---|---|---|
| Root yield | 0.24 | 0.02 |
| Dry matter content | 0.68 | 0.08 |
| Starch content | 0.70 | 0.03 |
| β-carotene content | 0.90 | 0.02 |
Standard error.
H2 = Broad sense heritability estimate.
Table 4.
Genetic correlation analysis of storage root yield, dry matter, starch and β-carotene content in the progeny of the ‘New Kawogo’ × ‘Beauregard’ mapping population
| Yield | Dry matter | Starch | β-carotene | |
|---|---|---|---|---|
| Yield | 1.00 | 0.57* | 0.41* | −0.06ns |
| Dry matter | 1.00 | 0.77* | −0.59* | |
| Starch | 1.00 | −0.93* | ||
| β-carotene | 1.00 |
Significant at p < 0.05.
Not significant at p < 0.05.
Transgressive segregation analysis
A number of progeny exhibited transgressive segregation (i.e. progeny that fall beyond parental phenotypes for a trait) for storage root yield, dry matter, and starch content. Many progeny performed better than the higher parent for these traits. Mean genotype storage yields for the three sites were normally distributed. Similarly, the overall mean genotype storage root yields across sites and seasons were also normally distributed. A total of 51 genotypes showed transgressive segregation for root yield (Fig. 1).
The overall mean root dry matter content of genotypes was also normally distributed (Fig. 2). The mean dry matter content for most of the genotypes was in the range of 21– 30%. A total of 21 genotypes had higher mean dry matter contents than ‘New Kawogo’ (31.6%), the higher parent. A similar distribution was observed for starch content and a total of 14 genotypes showed transgressive segregation for starch content (data not shown). The mean genotype β-carotene content distribution was skewed to the left for sites and the overall mean (Fig. 3). The mean β-carotene content of most genotypes was below 20.0 mg/100 g. No genotype had a higher overall mean β-carotene content than ‘Beauregard’ (31.7 mg/100 g), the higher parent.
SSR marker trait association
A total of 405 published expressed sequence tag (EST) SSR primers were designed for use in this study. Of these markers, 250 were polymorphic among the parents and selected progeny and used to genotype the population. Detailed description of the markers and polymorphic information was previous reported (Yada et al. 2015).
The marker trait regression analysis associated several marker loci with storage root yield, dry matter, and starch and β-carotene content (Table 5). A total of 12 SSR markers were highly associated with variation in storage root yield and explained 69.6% of the storage root yield variance in this mapping population. Whereas, four SSR markers were significantly associated with storage root dry matter content and explained 15.8% of total dry matter content variance in the population. The level of marker trait association was slightly lower for the dry matter content markers.
Table 5.
Association of SSR markers with the best linear unbiased predictions (BLUPS) of storage root yield (YLD), dry matter (DM), starch (STCH) and β-carotene (BC) of genotypes, highlighted markers are associated with multiple traits
| Trait | Marker | Number of alleles | R2 | Total variance (%) | BLUP_ Trait Mean | F Value | ProbF |
|---|---|---|---|---|---|---|---|
| YLD | IBS62 | 5 | 0.091 | 69.6 | 0.138 | 5.47 | <.0001 |
| IbJ175 | 5 | 0.073 | 0.132 | 4.27 | 0.0009 | ||
| IBS72 | 4 | 0.069 | 0.218 | 4.98 | 0.0007 | ||
| IbJ263 | 4 | 0.064 | 0.101 | 4.42 | 0.0018 | ||
| IBSSR01 | 3 | 0.060 | 0.138 | 5.85 | 0.0007 | ||
| IBS103 | 5 | 0.058 | 0.141 | 3.35 | 0.0060 | ||
| IbE4 | 4 | 0.052 | 0.114 | 3.75 | 0.0055 | ||
| IbJ10a | 4 | 0.052 | 0.126 | 3.55 | 0.0076 | ||
| IbU4 | 4 | 0.051 | 0.180 | 3.59 | 0.0071 | ||
| IbU10 | 3 | 0.042 | 0.138 | 4.08 | 0.0074 | ||
| IBR19 | 3 | 0.042 | 0.165 | 4.06 | 0.0076 | ||
| IBCIP2 | 4 | 0.042 | 0.138 | 3.04 | 0.0179 | ||
|
| |||||||
| DM | IBS56 | 3 | 0.044 | 15.8 | 0.006 | 4.17 | 0.0066 |
| IbL16 | 5 | 0.042 | 0.002 | 2.42 | 0.0364 | ||
| IBS126 | 4 | 0.041 | 0.005 | 2.83 | 0.0251 | ||
| IBSSR01 | 3 | 0.031 | 0.002 | 2.98 | 0.0319 | ||
|
| |||||||
| STCH | IBCIP2 | 4 | 0.069 | 32.3 | 0.011 | 5.06 | 0.0006 |
| IBS56 | 3 | 0.060 | 0.051 | 5.8 | 0.0007 | ||
| IBS62 | 5 | 0.056 | 0.011 | 3.25 | 0.0072 | ||
| IBS48 | 3 | 0.050 | 0.033 | 4.76 | 0.0030 | ||
| IBS109 | 2 | 0.045 | 0.015 | 4.21 | 0.0062 | ||
| IbJ175 | 5 | 0.043 | 0.011 | 2.41 | 0.0369 | ||
|
| |||||||
| BC | IBS62 | 5 | 0.074 | 37.8 | 0.006 | 4.39 | 0.0007 |
| IBS56 | 3 | 0.058 | 0.048 | 5.54 | 0.0011 | ||
| IBS85 | 4 | 0.050 | 0.108 | 3.48 | 0.0087 | ||
| Ib242 | 4 | 0.050 | 0.006 | 3.64 | 0.0066 | ||
| IBS48 | 4 | 0.046 | 0.050 | 4.34 | 0.0052 | ||
| IBSSR02 | 3 | 0.044 | 0.006 | 4.25 | 0.0059 | ||
| IbY58 | 2 | 0.036 | 0.006 | 5.12 | 0.0065 | ||
| IBCIP2 | 4 | 0.020 | 0.009 | 3.48 | 0.0085 | ||
A total of six (explaining 32.3% variance) and eight (explaining 37.8% variance) markers were significantly associated with variation in starch and β-carotene content, respectively. Some SSR markers were associated with multiple traits and could be useful for future selection of multiples traits in a genotype when fine mapped. For example, markers IBCIP2 and IBS62 were associated with variation in storage root yield, starch and β-carotene content. Similarly, marker IBS56 was associated with storage root dry matter, starch and β-carotene content. Then, marker IBSSR01 was associated with storage root yield and dry matter content. Marker IbJ175 was associated with storage root yield and starch content, and IBS48 was associated with variation in starch and β-carotene content.
From the ranking of the trait LS means of progeny, we identified a total of 16 progeny that had high performance for these traits and have been advanced for variety trials (Table 6). Progeny NKB3 and NKB15 were of special interest for their high storage root yield, dry matter, and starch and β-carotene content. Though the dry matter content for these orange-fleshed genotypes were 28.0% and 28.1%, respectively, which is slightly lower than the preferred 30% and above in SSA, their yields were impressive. NKB135 and NKB254 had high storage root yield and β-carotene content but had relatively low dry matter content.
Table 6.
Performance of the top 16 progeny with cultivar release potential across sites and seasons ranked using mean storage root yield (YLD), dry matter (DM), starch (STCH) and β-carotene (BC) content, highlighted clones are candidate clones with preferred storage root yield, dry matter and β-carotene content in sub-Saharan Africa
| Genotype | Mean | Ranke | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| YLDa | DMb | STCHc | BCd | YLD | DM | STCH | BC | |
| NKB216 | 48.4 | 29.0 | 71.9 | 9.7 | 1 | 138 | 30 | 187 |
| NKB9 | 41.0 | 30.4 | 70.2 | 7.4 | 2 | 63 | 68 | 250 |
| NKB254 | 40.4 | 24.3 | 52.4 | 29.1 | 3 | 285 | 285 | 3 |
| NKB177 | 40.1 | 30.6 | 72.3 | 9.3 | 4 | 55 | 23 | 200 |
| NKB168 | 38.8 | 29.1 | 70.8 | 8.7 | 5 | 134 | 50 | 218 |
| NKB93 | 38.5 | 30.9 | 71.5 | 6.2 | 6 | 44 | 37 | 272 |
| NKB21 | 38.3 | 29.4 | 72.6 | 9.7 | 7 | 112 | 22 | 188 |
| NKB153 | 38.1 | 29.5 | 71.2 | 10.0 | 8 | 105 | 45 | 175 |
| NKB105 | 37.9 | 28.5 | 68.6 | 14.0 | 9 | 178 | 113 | 94 |
| NKB114 | 36.8 | 25.2 | 68.3 | 6.8 | 10 | 282 | 120 | 263 |
| NKB15 | 36.5 | 28.1 | 62.2 | 20.7 | 11 | 208 | 235 | 31 |
| NKB193 | 36.4 | 30.3 | 70.9 | 11.5 | 12 | 69 | 46 | 131 |
| NKB3 | 36.1 | 28.0 | 63.5 | 17.8 | 13 | 213 | 219 | 56 |
| NKB201 | 35.9 | 30.4 | 68.2 | 12.0 | 14 | 60 | 121 | 122 |
| NKB5 | 35.8 | 29.7 | 73.1 | 5.5 | 15 | 100 | 19 | 280 |
| NKB135 | 35.7 | 25.6 | 61.6 | 20.9 | 16 | 277 | 239 | 29 |
YLD = Genotype least significant mean storage root yield (t/ha) averaged over three sites and two seasons.
DM = Genotype least significant mean storage root dry matter content (%) averaged over three sites and two seasons.
STCH = Genotype least significant mean storage root starch content (%) averaged over three sites and two seasons.
BC = Genotype least significant mean storage root β-carotene content (mg/100 g) averaged over three sites and two seasons.
Ranking of 286 genotypes (284 progeny and 2 parents) in descending order for storage root yield, dry matter, β-carotene and starch content.
Discussion
The improvement of storage root yield, dry matter, β-carotene content, and pest and disease resistance are the key priorities for sweetpotato research in SSA (Fuglie 2007). This research addresses four of these key traits for enhancing food security in the region. The yield of the progeny is typical of a segregating population of a diverse bi-parental cross. ‘New Kawogo’ is a white fleshed, high yielding, high dry matter and starch content released landrace in Uganda (Mwanga et al. 2001). In contrast, ‘Beauregard’ is a popular low dry matter and starch content, orange-fleshed US cultivar (Rolston et al. 1987). The low yield of ‘Beauregard’ under Ugandan conditions could be attributed to the different climatic conditions and a number of biotic stresses such as SPVD and sweetpotato weevils in Uganda relative to the US where the cultivar was developed.
The range in progeny performance in storage root yield, dry matter, starch and β-carotene content is typical of a segregating population of a wide cross. The F1 progeny in sweetpotato generated from controlled and polycross nurseries segregate in a similar pattern due to the heterogeneous nature of sweetpotato genotypes (Grüneberg et al. 2009). Similar trait ranges in progeny performance have been observed from breeding trials in Uganda (Mwanga et al. 2009). Also, Tumwegamire et al. (2011) observed genotype starch content range of 30.0–73.3% and dry matter content range of 5.5–36.7% in the east African sweetpotato germplasm evaluated in Uganda.
Sweetpotato is highly sensitive to variations in environmental conditions (Ngeve 1993). Our study showed a significant (P < 0.0001) genotype × environment interaction for storage root yield. Earlier studies showed considerable genotype × environment interaction for storage root yield of nine clones of diverse origin and a local check evaluated in Peru (Grüneberg et al. 2005). Similarly, significant genotype × environment interactions were reported for sweetpotato storage root yields of selected landrace varieties in Uganda (Osiru et al. 2009) and in 28 breeding lines in South Africa (Adebola et al. 2013). Just like in other sites, significant genotype × environment interaction was observed for storage root yield performance of nine introductions and two local genotypes in Turkey (Caliskan et al. 2007).
This means that, selection for storage root yield should be done in multiple sites and in high and low yielding environments for the benefit of SSA sweetpotato farmers who grow their crop with minimal inputs. Storage root yield in these environments could be influenced by soil type, nutrient levels, prevailing pests and diseases and genotype photo-sensitivity. The advantage of selecting for yield in both low and high yielding environments was demonstrated in wheat selection (Ud-Din et al. 1992), where they found a low genetic correlation coefficient (r = 0.20 ± 0.16) for yields between irrigated and non-irrigated wheat fields and recommended testing in both environments.
In contrast to storage root yield, like in our study, a relatively small amount of genotype × environment interactions were observed when dry matter, starch and β-carotene content for nine clones of diverse origins and a local check were evaluated at multiple sites in Peru (Grüneberg et al. 2005). No genotype × environment interaction was observed for β-carotene content as well for 42 orange-fleshed hybrids and a local check evaluated in advanced yield trials at multiple sites in India (Vimala et al. 2011b). Therefore, selection for dry matter and β-carotene content can be done in one or two locations in the early stages of breeding to save breeding resources for screening other environmentally sensitive traits in multiple locations.
Broad sense heritability estimates in this study were high for storage root β-carotene content, moderate for starch and dry matter content and low for root yield like in other studies. Heritability estimates for storage root yield, dry matter content, starch and β-carotene content in the east African sweetpotato germplasm were 0.5, 0.64, 0.44 and 0.94, respectively (Tumwegamire et al. 2011). A lower heritability estimate of 0.25 was reported for sweetpotato storage root yield (Jones 1986). Afuape et al. (2014) reported the heritability of β-carotene content to be 0.98 close to what has been found in our research. The high heritability estimate is in agreement with inheritance of β-carotene that was reported to be controlled by about six genes acting in an additive mode (Hernandez et al. 1965). Therefore, it is possible to breed for high β-carotene content cultivars since β-carotene is a highly heritable trait in sweetpotato.
Moderately high heritability estimates for starch and sugar content of greater than 0.33 and 0.77 on general combining ability and specific combining ability basis, respectively were reported in a North Carolina II (NCII) design population (Todd 2013). The broad sense heritability for storage root dry matter content from full-sib families evaluated in Louisiana, USA was 0.9 (Courtney et al. 2008) and the broad sense heritability for dry matter content from a diallel cross population in Uganda was 0.7 (Shumbusha et al. 2014). Jones (1986) reported the broad sense heritability of dry matter content in sweetpotato of 0.65, close to what we observed in this research. Thus, significant genetic gains can be made in root dry matter and starch content through breeding.
Negative correlations observed between β-carotene and dry matter and starch content in our study are consistent with other findings. Storage root dry matter and starch content were reported to be negatively correlated with β-carotene content in sweetpotato in earlier studies (Cervantes-Flores et al. 2011, Grüneberg et al. 2005, Vimala et al. 2011a). This negative correlation has been a major challenge in the development of desired orange-fleshed cultivars in SSA. Sweetpotato farmers and consumers in SSA mostly prefer high dry matter cultivars (ca. >30%), yet it has been difficult to breed deep orange fleshed cultivars with high dry matter content. The recently released orange-fleshed cultivars in Uganda, NASPOT 9 O (DM = 30.1% DM) and NASPOT 10 O (DM = 30.5%) have lower β-carotene contents of 314.5 μg/g and 246.1 μg/g, respectively compared to those released in the US (Mwanga et al. 2009). The exact biochemical mechanism for the negative correlation between starch and β-carotene biosynthesis in sweetpotato needs to be investigated. Despite this negative correlation, Cervantes-Flores et al. (2011) identified AFLP-based QTL (e46m6015, e32m3724 and e42m3812) linked to storage root dry matter, starch and β-carotene content in ‘Beauregard’. This could offer an opportunity for improving both traits through MAS.
However, just like in our study, strong positive correlations were found between sweetpotato storage root starch and dry matter content (Cervantes-Flores et al. 2011, Hall and Smittle 1983, Ma et al. 2009). This would be expected since 60% of root dry matter in sweetpotato consists of starch (Woolfe 1992). While it is possible to select clones for high yield, dry matter and starch content at the same time, the negative correlation of these traits with β-carotene content makes it difficult to develop high dry matter and high yielding orange-fleshed cultivars, the preferred types in SSA. The positive correlation of storage root yield with starch and dry matter content can easily be exploited to develop high starch content and high yielding industrial cultivars as the utilization of sweetpotato is diversified in SSA with time.
We observed significant transgressive segregation for storage root yield, dry matter content and starch content. The distribution of progeny means demonstrated the quantitative nature of the inheritance of these traits. No single genotype exhibited transgressive segregation for all the four traits in this study. This means that most of these transgressives will be used as parents for specific traits for population improvement. Transgressive segregation was reported for storage root dry matter, starch and β-carotene content in the ‘Tanzania’ × ‘Beauregard’ population (Cervantes-Flores et al. 2011). Transgressive segregation has been attributed to the accumulation in an offspring of complementary alleles at multiple loci inherited from parents (Rick and Smith 1953, Tanksley 1993). The transgressive segregation observed in our study could therefore be due to the accumulation of favorable alleles in the progeny from this diverse bi-parental cross.
Since index selection is superior to selection in successive stages by tandem and independent culling (Wricke and Weber 1986), the rank based selection of the top performing genotypes in our study is useful. The top 16 genotypes, with medium to high storage root yield, dry matter, starch and β-carotene content will be advanced for possible variety release and also for use as parental genotypes for population improvement. From our study, progeny NKB3 and NKB15 had high storage root yield, dry matter, and starch and β-carotene content (Table 6). These orange-fleshed genotypes NKB3 (DM = 28.0%, BC = 17.8 mg/100 g) and NKB15 (DM = 28.1%, BC = 20.7 mg/100 g) had slightly similar trait attributes to those preferred by farmers in SSA. Progeny NKB135 and NKB254 had high storage root yield and β-carotene content, but low dry matter and starch content and could be potential parental genotypes for future population improvement of storage root yield and β-carotene content.
The development of diverse novel products from sweetpotato will need cultivars that combine multiple traits in one background. A good example is the high dry matter and starch content purple-fleshed Japanese sweetpotato cultivars for production of alcoholic beverage ‘Pa-Puru’ (Saigusa and Ohba 2006). The development of such multiple trait cultivars will require the application of genomics tools, as some of the traits are complex and negatively correlated.
Our study has showed that it is possible to combine high storage root yield, dry matter content and β-carotene content into one background by using diverse parents. It has always been difficult to develop high dry matter content orange-fleshed cultivars because of the negative correlation of these traits. To succeed, breeding programs could combine many (20–50) diverse parental genotypes varying in β-carotene content, dry matter and starch content and pest and disease resistance in polycross nurseries to generate populations for selection based on combining ability.
The SSR markers are still pivotal in the genomic improvement of sweetpotato. The marker trait association in our study identified 30 SSR markers significantly associated with storage root yield, dry matter, starch and β-carotene content. One major limitation of SSR markers in this study was the amplification of one locus at a time resulting in fewer marker allele points compared to AFLP markers that normally analyze large numbers of polymorphic loci simultaneously and generate many markers for analysis. Secondly, the level of marker trait association was low for most of the markers that showed significant association (P = 0.05).
We also observed that, the SSR markers accounted for 69.6% of the total variance of storage root yield in this population though the heritability of storage root yield was low (H2 = 0.24). On the contrary, the SSR markers accounted for 15.8% of the total variance in the highly heritable storage root dry matter content (H2 = 0.68) in this population. This disparity in marker trait association in this study can be attributed to the insufficient number of SSR markers used in this study. A recent study designed 1476 EST and mRNA SSR markers and mapped 214 of these markers in a natural sweetpotato population identified 63 SSR associated with storage root dry matter and starch content (Zhang et al. 2016). In this study, the amount of dry matter content variance explained per marker ranged from 0.01592 in marker SIP285 to 0.2805 in marker SIP029. This confirmed the observation that the small number of markers caused disparity in marker trait association for storage root yield and dry matter in our study.
Nonetheless, we have demonstrated that, SSR markers are associated with variation in these key sweetpotato traits. These SSR markers may not be used for selection of progeny for these traits at this point as more research is needed to map the QTL of these traits. However, these 30 SSR markers could be used by breeders to genotype their sweetpotato germplasm for selecting diverse parents for improvement of storage root yield, dry matter, starch, and β-carotene content.
Earlier marker trait associations of dry matter, starch and β-carotene content were done through QTL and discriminant analysis with dominant markers (Cervantes-Flores et al. 2011, Mcharo and LaBonte 2010, Zhao et al. 2013). Cervantes-Flores et al. (2011) mapped 12 QTL of storage root starch content, 13 QTL of dry matter content and 8 QTL of β-carotene content using AFLP markers. Meanwhile, 27 QTL of storage root dry matter content were also mapped using a combination of AFLP markers and SSR markers (Zhao et al. 2013). These AFLP markers were highly significantly associated with the above traits due to the high density of AFLP markers on the linkage maps. However, a major limitation of the AFLP markers is their dominant inheritance in genomes, making it impossible to differentiate between hetero zygotes and homozygotes, thus limiting the utility of these QTL in the improvement of autohexaploid sweetpotato.
Finally, our study demonstrated that genetic gains in sweetpotato improvement for storage root yield, dry matter, starch and β-carotene content could be made within a diverse cross. SSR markers could be accurately used to tag traits in sweetpotato. However, we recommend that, more sweetpotato SSR markers be developed and associated with storage root yield, dry matter, starch and β-carotene content in this ‘New Kawogo’ × ‘Beauregard’ population using QTL analysis. We also recommend that, through genotyping by sequencing (GBS) of the ‘New Kawogo’ × ‘Beauregard’ population, single nucleotide polymorphism (SNP) marker should be mined and used for developing high density SNP and SSR based linkage maps for fine mapping of QTL of storage root yield traits. This will pave the way for marker assisted breeding (MAB) in sweetpotato for developing high storage root yielding cultivars with high dry matter, starch and β-carotene content for improved nutrition and incomes in SSA.
Acknowledgement
This research was funded by the McKnight Foundation Collaborative Crop Research Program (CCRP) and the Norman E. Borlaug Leadership Enhancement in Agriculture Program (Borlaug LEAP). Dr. Consuelo Arellano of the Department of Statistics, NCSU is appreciated for providing assistance in statistical data analysis.
Literature Cited
- Adebola, P.O., Shegro, A., Laurie, S.M., Zulu, L.N. and Pillay, M. (2013) Genotype × environment interaction and yield stability estimate of some sweetpotato [Ipomoea batatas (L.) Lam] breeding lines in South Africa. J. Plant Breed. Crop Sci. 5: 182–186. [Google Scholar]
- Afuape, S.O., Nwankwo, I.I.M., Omodamiro, R.M., Echendu, T.N.C. and Toure, A. (2014) Studies on some important consumer and processing traits for breeding sweetpotato for varied end-uses. Am. J. Exp. Agric. 4: 114–124. [Google Scholar]
- Black, R. (2003) Micronutrient deficiency: An underlying cause of morbidity and mortality. Bull. World Health Organ. 81: 79. [PMC free article] [PubMed] [Google Scholar]
- Buteler, M.I., Jarret, R.L. and LaBonte, D.R. (1999) Sequence characterization of microsatellites in diploid and polyploid Ipomoea. Theor. Appl. Genet. 99: 123–132. [Google Scholar]
- Caliskan, M.E., Erturk, E., Sogut, T., Boydak, E. and Arioglu, H. (2007) Genotype × environment interaction and stability analysis of sweetpotato (Ipomoea batatas) genotypes. N. Z. J. Crop Hortic. Sci. 35: 87–99. [Google Scholar]
- Cervantes-Flores, J.C., Yencho, G.C., Kriegner, A., Pecota, K.V., Faulk, M.A., Mwanga, R.O.M. and Sosinski, B. (2008a) Development of a genetic linkage map and identification of homologous linkage groups in sweetpotato using multiple-dose AFLP markers. Mol. Breed. 21: 511–532. [Google Scholar]
- Cervantes-Flores, J.C., Yencho, G.C., Pecota, K.V., Sosinski, B. and Mwanga, R.O.M. (2008b) Detection of quantitative trait loci and inheritance of root-knot nematode resistance in sweetpotato. J. Am. Soc. Hortic. Sci. 133: 844–851. [Google Scholar]
- Cervantes-Flores, J.C., Sosinski, B., Pecota, K.V., Mwanga, R.O.M., Catignani, G.L., Truong, V.D., Watkins, R.H., Ulmer, M.R. and Yencho, G.C. (2011) Identification of quantitative trait loci for dry-matter, starch, and β-carotene content in sweetpotato. Mol. Breed. 28: 201–216. [Google Scholar]
- Chang, K., Lo, H., Lai, Y., Yao, P., Lin, K. and Hwang, S. (2009) Identification of quantitative trait loci associated with yield-related traits in sweet potato (Ipomoea batatas). Bot. Stud. 50: 43–55. [Google Scholar]
- Courtney, M., Mcharo, M., LaBonte, D.R. and Gruneberg, W. (2008) Heritability estimates for micronutrient composition of sweetpotato storage roots. HortScience 43: 1382–1384. [Google Scholar]
- Doyle, J.J. and Doyle, J.L. (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13–15. [Google Scholar]
- FAOSTAT (2012) Food and Agricultural Organization of the United Nations, Production statistics (http://faostat.fao.org/) Accessed August 2013.
- Fuglie, K.O. (2007) Priorities for sweetpotato research in developing countries: Results of a survey. HortScience 42: 1200–1206. [Google Scholar]
- Grüneberg, W.J., Manrique, K., Zhang, D. and Hermann, M. (2005) Genotype × environment interactions for a diverse set of sweetpotato clones evaluated across varying ecogeographic conditions in Peru. Crop Sci. 45: 2160–2171. [Google Scholar]
- Grüneberg, W.J., Mwanga, R., Andrade, M., Dapaah, H. (2009) Sweetpotato breeding. In: Andrade, M., Barker, I., Cole, D., Dapaah, H., Elliott, H., Fuentes, S., Grüneberg, W.J., Kapinga, R., Kroschel, J., Labarta, R.et al. (eds.) Thiele G Unleashing the potential of sweetpotato in sub-Saharan Africa: current challenges and way forward Nairobi, CIP-SSA. [Google Scholar]
- Gurmu, F., Hussein, S. and Laing, M. (2013) Self- and cross-incompatibilities in sweetpotato and their implications on breeding. Aust. J. Crop Sci. 7: 2074–2078. [Google Scholar]
- Hall, M. and Smittle, D. (1983) Industrial-type sweetpotatoes: A renewable energy source for Georgia. University of Georgia College of Agriculture Experiment Stations; Research Report 429, June 1983. [Google Scholar]
- Hernandez, T.P., Hernandez, T., Constanin, R.J. and Miller, J.C. (1965) Inheritance of methods of rating flesh color in Ipomoea batatas. Proc. Am. Soc. Hort. Sci. 87: 387–390. [Google Scholar]
- Hirakawa, H., Okada, Y., Tabuchi, H., Shirasawa, K., Watanabe, A., Tsuruoka, H., Minami, C., Nakayama, S., Sasamoto, S., Kohara, M.et al. (2015) Survey of genome sequences in a wild sweet potato, Ipomoea trifida (H.B.K.) G. Don. DNA Res. 22: 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland, J.B. (2006) Estimating genotypic correlations and their standard errors using multivariate restricted maximum likelihood estimation with SAS Proc MIXED. Crop Sci. 46: 642–654. [Google Scholar]
- Hotz, C., Loechl, C., Lubowa, A., Tumwine, J.K., Ndeezi, G., Masawi, A.N., Baingana, R., Carriquiry, A., De Brauw, A., Meenakshi, J.V.et al. (2012) Introduction of β-carotene-rich orange sweetpotato in rural Uganda resulted in increased Vitamin A intakes among children and women and improved Vitamin A status among children. J. Nutr. 142: 1871–1880. [DOI] [PubMed] [Google Scholar]
- Hu, J., Nakatani, M., Mizuno, K. and Fujimura, T. (2004) Development and characterization of microsatellite markers in sweetpotato. Breed. Sci. 58: 177–188. [Google Scholar]
- Jones, A. (1986) Sweetpotato heritability estimates and their use in breeding. HortScience 21: 14–17. [Google Scholar]
- Jones, A., Dukes, P.D. and Schalk, J.M. (1987) Sweetpotato breeding. In: Breeding vegetable Crops. AVI Pub Co. [Google Scholar]
- Karyeija, R.F., Gibson, R.W. and Valkonen, J.P.T. (1998) The significance of sweet potato feathery mottle virus in subsistence sweetpotato production in Africa. Plant Dis. 82: 4–15. [DOI] [PubMed] [Google Scholar]
- Koussao, S., Gracen, V., Asante, I., Danquah, E.Y., Ouedraogo, J.T., Baptiste, T.J., Jerome, B. and Vianney, T.M. (2014) Diversity analysis of sweet potato (Ipomoea batatas [L.] Lam) germplasm from Burkina Faso using morphological and simple sequence repeats markers. Afr. J. Biotechnol. 13: 729–742. [Google Scholar]
- Kriegner, A., Cervantes-Flores, J.C., Burg, K., Mwanga, R.O.M. and Zhang, D. (2003) A genetic linkage map of sweetpotato (Ipomoea batatas (L.) Lam.) based on AFLP markers. Mol. Breed. 11: 169–185. [Google Scholar]
- Low, J.W., Arimond, M., Osman, N., Cunguara, B., Zano, F. and Tschirley, D. (2007) A food-based approach introducing orange-fleshed sweetpotatoes increased vitamin A intake and serum retinol concentrations in young children in rural Mozambique. J. Nutr. 137: 1320–1327. [DOI] [PubMed] [Google Scholar]
- Ma, D., Li, Q., Li, X., Li, H., Tang, Z., Hu, L., Cao, Q., Xie, Y. and Wang, X. (2009) Selection of parents for breeding edible varieties of sweetpotato with high carotene content. Agric. Sci. China 8: 1166–1173. [Google Scholar]
- Martin, F.W. (1965) Incompatibility in sweetpotato: a review. Econ. Bot. 19: 406–415. [Google Scholar]
- Martin, F.W. (1970) Self- and interspecific incompatibility in the Convolvulaceae. Bot. Gaz. 131: 139–144. [Google Scholar]
- Mcharo, M., LaBonte, D.R., Mwanga, R.O.M. and Kriegner, A. (2005a) Associating molecular markers with virus resistance to classify sweetpotato genotypes. J. Am. Soc. Hortic. Sci. 130: 355–359. [Google Scholar]
- Mcharo, M., LaBonte, D.R., Clark, C., Hoy, M. and Oard, J.H. (2005b) Molecular marker variability for southern root-knot nematode resistance in sweetpotato. Euphytica 144: 125–132. [Google Scholar]
- Mcharo, M. and LaBonte, D.R. (2010) Multivariate selection of AFLP markers associated with β-carotene in sweetpotatoes. Euphytica 175: 123–132. [Google Scholar]
- Miano, D.W., LaBonte, D.R. and Clark, C.A. (2008) Identification of molecular markers associated with sweetpotato resistance to sweetpotato virus disease in Kenya. Euphytica 160: 15–24. [Google Scholar]
- Mwanga, R.O.M., Odongo, B., Ocitti p’Obwoya, C., Gibson, R.W., Smit, N.E.J.M. and Carey, E.E. (2001) Release of five sweetpotato cultivars in Uganda. HortScience 36: 385–386. [Google Scholar]
- Mwanga, R.O.M., Kriegner, A., Cervantes-Flores, J.C., Zhang, D.P., Moyer, J.W. and Yencho, G.C. (2002) Resistance to sweetpotato chlorotic stunt virus and sweetpotato feathery mottle virus is mediated by two separate recessive genes in sweetpotato. J. Am. Soc. Hortic. Sci. 127: 798–806. [Google Scholar]
- Mwanga, R.O.M., Odongo, B., Niringiye, C., Alajo, A., Kigozi, B., Makumbi, R., Lugwana, E., Namakula, J., Mpembe, I., Kapinga, R.et al. (2009) ‘NASPOT 7’, ‘NASPOT 8’, ‘NASPOT 9 O’, ‘NASPOT 10 O’, and ‘Dimbuka-Bukulula’ sweetpotato. HortScience 44: 828–832. [Google Scholar]
- Ngeve, J.M. (1993) Regression analysis of genotype × environment interaction in sweetpotato. Euphytica 71: 231–238. [Google Scholar]
- Nishiyama, I., Miyazaki, T. and Sakamoto, S. (1975) Evolutionary autoploidy in the sweet potato (Ipomoea batatas (L.) Lam.) and its progenitors. Euphytica 24: 197–208. [Google Scholar]
- Osiru, M.O., Olanya, O.M., Adipala, E., Kapinga, R. and Lemaga, B. (2009) Yield stability analysis of Ipomoea batatas L. cultivars in diverse environments. Aust. J. Crop Sci. 3: 213–220. [Google Scholar]
- Rick, C.M. and Smith, P.G. (1953) Novel variation in tomato species hybrids. Am. Nat. 87: 359–373. [Google Scholar]
- Rolston, L.H., Clarke, C.A., Cannon, J.M., Randle, W.M., Riley, E.G., Wilson, P.W. and Robbins, M.L. (1987) “Beauregard” sweetpotato”. HortScience 22: 1338–1339. [Google Scholar]
- Saigusa, N. and Ohba, R. (2006) Healthy alcoholic beverage “Pa-Puru” from purple-fleshed sweetpotato. Sweetpotato Research Front Newsletter (SPORF). National Agricultural Research Center for Kyushu Okinawa Region (KONARC), Japan. [Google Scholar]
- SAS Institute Inc. (2013) Step-by step programming with base SAS 9.4 Cary, NC: SAS Inc. [Google Scholar]
- Schafleitner, R., Tincopa, L.R., Palomino, O., Rossel, G., Robles, R.F., Alagon, R., Rivera, C., Quispe, C., Rojas, L., Pacheco, J.A.et al. (2010) A sweetpotato gene index established by de novo assembly of pyrosequencing and Sanger sequences and mining for gene-based microsatellite markers. BMC Genomics 11: 604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumbusha, D., Tusiime, G., Edema, R., Gibson, P., Adipala, E. and Mwanga, R.O.M. (2014) Inheritance of root dry matter content in sweetpotato. Afr. Crop Sci. J. 22: 69–78. [Google Scholar]
- Stevenson, P.C., Muyinza, H., Hall, D.R., Porter, E.A., Farman, D.I., Talwana, H. and Mwanga, R.O.M. (2009) Chemical basis for resistance in sweetpotato Ipomoea batatas to the sweetpotato weevil Cylas puncticollis. Pure Appl. Chem. 81: 141–151. [Google Scholar]
- Tanksley, S.D. (1993) Mapping polygenes. Annu. Rev. Genet. 27: 205–233. [DOI] [PubMed] [Google Scholar]
- Todd, S.M. (2013) Application of near infrared spectroscopy to study inheritance of sweetpotato composition traits. Ph.D. Dissertation, Department of Horticultural Science, North Carolina State University, Raleigh, NC. [Google Scholar]
- Tumwegamire, S., Kapinga, R., Rubaihayo, P.R., LaBonte, D.R., Grüneberg, W.J., Burgos, G., zum Felde, T., Carpio, R., Pawelzik, E. and Mwanga, R.O.M. (2011) Evaluation of dry matter, protein, starch, sucrose, β-carotene, iron, zinc, calcium, and magnesium in East African sweetpotato [Ipomoea batatas (L.) Lam] germplasm. HortScience 46: 348–357. [Google Scholar]
- Ud-Din, N., Carver, B.F. and Clutter, A.C. (1992) Genetic analysis and selection for wheat yield in drought-stressed and irrigated environments. Euphytica 62: 89–96. [Google Scholar]
- Veasey, E.A., Borges, A., Rosa, M.S., Queiroz-Silva, J.R., Bressan, E.A. and Peroni, N. (2008) Genetic diversity in Brazilian sweetpotato (Ipomoea batatas (L.) Lam, Solanales, Convolvulaceae) landraces assessed with microsatellite markers. Genet. Mol. Biol. 31: 725–733. [Google Scholar]
- Vimala, B., Nambisan, B. and Hariprakash, B. (2011a) Retention of carotenoids in orange-fleshed sweetpotato during processing. J. Food Sci. Technol. 48: 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimala, B., Sreekanth, A., Binu, H. and Grüneberg, W. (2011b) Variability in 42 orange-fleshed sweet potato hybrids for tuber yield and carotene & dry matter content. Gene Conserve 40: 190–200. [Google Scholar]
- Vos, P., Hogers, R., Bleeker, M., Reijans, M., van de Lee, T., Hornes, M., Frijters, A., Pot, J., Peleman, J., Kuiper, M.et al. (1995) AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Li, J., Luo, Z., Huang, L., Chen, X., Fang, B., Li, Y., Chen, J. and Zhang, X. (2011) Characterization and development of EST-derived SSR markers in cultivated sweetpotato (Ipomoea batatas). BMC Plant Biol. 11: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wricke, G. and Weber, W.E. (1986) Quantitative genetics and selection. Walter de Gruyter, Berlin. [Google Scholar]
- Woolfe, J.A. (1992) Sweetpotato—an untapped food resource. Cambridge University Press, New York. [Google Scholar]
- Yada, B., Tukamuhabwa, P., Villordon, A., Alajo, A. and Mwanga, R.O.M. (2010a) An on-line database of sweetpotato germplasm collection in Uganda. HortScience 45: 153. [Google Scholar]
- Yada, B., Tukamuhabwa, P., Wanjala, B., Kim, D., Skilton, R.A., Alajo, A. and Mwanga, R.O.M. (2010b) Characterization of Ugandan sweetpotato germplasm using fluorescent labeled simple sequence repeat markers. HortScience 45: 225–230. [Google Scholar]
- Yada, B., Guedira, G.B., Alajo, A., Ssemakula, G.N., Mwanga, R.O.M. and Yencho, G.C. (2015) Simple sequence repeat marker analysis of genetic diversity among progeny of a biparental mapping population of sweetpotato. HortScience 50: 1143–1147. [Google Scholar]
- Yencho, G.C., Pecota, K.V., Schultheis, J.R., VanEsbroeck, Z-P., Holmes, G.J., Little, B.E., Thornton, A.C. and Truong, V.D. (2008) ‘Covington’ sweetpotato. HortScience 43: 1911–1914. [Google Scholar]
- Yu, X., Zhao, N., Li, H., Jie, Q., Zhai, H., He, S., Li, Q. and Liu, Q. (2014) Identification of QTLs for starch content in sweetpotato (Ipomoea batatas (L.) Lam.). J. Integr. Agric. 13: 310–315. [Google Scholar]
- Zhang, K., Wu, Z., Tang, D., Lv, C., Luo, K., Zhao, Y., Liu, X., Huang, Y. and Wang, J. (2016) Development and identification of SSR markers associated with starch properties and β-carotene content in the storage root of sweet potato (Ipomoea batatas L.) Front. Plant Sci. 7: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, N., Yu, X.X., Jie, Q., Li, H., Li, H., Hu, J., Zhai, H., He, S.Z. and Liu, Q.C. (2013) A genetic linkage map based on AFLP and SSR markers and mapping of QTL for dry-matter content in sweetpotato. Mol. Breed. 32: 807–820. [Google Scholar]
- Zum Felde, T., Burgos, G., Espinoza, J., Eyzaguirre, R., Porras, E. and Grüneberg, W. (2009) Screening for β-carotene, iron, zinc, starch, individual sugars and protein in sweetpotato germplasm by near-infrared reflectance spectroscopy (NIRS). 15th Triennial Symposium of the International Society for Tropical Root Crops, Lima, Peru, November 2–6, 2009. [Google Scholar]