Abstract
The cytokine IL-6 acts via a specific receptor complex that consists of the membrane-bound IL-6 receptor (mIL-6R) or the soluble IL-6 receptor (sIL-6R) and glycoprotein 130 (gp130). In this study, we investigated the role of IL-6R components in asthma. We observed increased levels of sIL-6R in the airways of patients with allergic asthma as compared to those in controls. In addition, local blockade of the sIL-6R in a murine model of late-phase asthma after OVA sensitization by gp130–fraction constant led to suppression of Th2 cells in the lung. By contrast, blockade of mIL-6R induced local expansion of Foxp3-positive CD4+CD25+ Tregs with increased immunosuppressive capacities. CD4+CD25+ but not CD4+CD25– lung T cells selectively expressed the IL-6R α chain and showed IL-6–dependent STAT-3 phosphorylation. Finally, in an in vivo transfer model of asthma in immunodeficient Rag1 mice, CD4+CD25+ T cells isolated from anti–IL-6R antibody–treated mice exhibited marked immunosuppressive and antiinflammatory functions. IL-6 signaling therefore controls the balance between effector cells and Tregs in the lung by means of different receptor components. Furthermore, inhibition of IL-6 signaling emerges as a novel molecular approach for the treatment of allergic asthma.
Introduction
Allergic asthma is a chronic inflammatory disease characterized by airway inflammation and airway hyperresponsiveness (AHR) that affects about 10% of the population in the US (1). The development of allergic immune responses in asthma is mediated by CD4+ effector T cells producing Th2 cytokines (2–4). These Th2 cells and their signature cytokines, IL-4, IL-5, IL-9, and IL-13, are thought to play a key pathogenic role in asthma (5–8).
Although allergic asthma is associated with the presence of mucosal Th2 cells, it is not entirely clear which factors are responsible for priming of T cells to differentiate into Th2 effector cells in this disease. However, it has been recently shown in experimental asthma that IL-4 produced by invariant natural killer T cells might be responsible for this priming (9). Another possibility is that, upon antigen exposure, the activation of APCs results in the release of factors which are responsible for the activation and differentiation of T cells. In this respect, the cytokine IL-6 might be of interest, since it has been demonstrated that IL-6 derived from APCs is able to induce initial IL-4 production in naive CD4+ spleen T cells via activation of the transcription factor NFATc2, thereby polarizing these cells into Th2 cells (10–12).
IL-6 is a proinflammatory cytokine that plays a central role in host defense against infections and tissue injury (13–18). IL-6 elicits cellular actions by binding to the membrane-bound IL-6 receptor (mIL-6R), then recruiting 2 membrane-spanning glycoprotein 130 (gp130) molecules into tetra- or hexametric structures, thereby forming the active IL-6R complex. However, neither IL-6 nor IL-6R alone can bind or activate gp130. Homodimerization of the 2 gp130 molecules leads to intracellular activation of the nonreceptor protein tyrosine kinases Src and JAK as well as members of the STAT family of transcription factors (19–23). While gp130 is expressed ubiquitously, the expression of mIL-6R is restricted to the membrane of hepatocytes and hematopoietic cells. However, cells lacking mIL-6R can also respond to IL-6 via the soluble IL-6 receptor (sIL-6R), since the IL-6/sIL-6R complex can activate target cells expressing gp130 in a process termed IL-6 trans-signaling (24–31). Interestingly, recent studies in peritonitis models suggest that such IL-6 trans-signaling is of crucial importance in the transition from acute to chronic phases of inflammatory processes (25).
In this study we delineate different roles for sIL-6R and mIL-6R in asthma. In fact, we found increased levels of sIL-6R in the airways of patients with allergic asthma as compared to those of controls. In addition, local blockade of sIL-6R in a murine model of late-phase asthma after OVA sensitization by gp130–fraction constant (gp130-Fc) led to suppression of Th2 cells in the lung. By contrast, local treatment in the lung with anti–IL-6R antibody induced Th1-CD4+ cells as well as Foxp3-positive, IL-6R–positive CD4+CD25+ Tregs with immunosuppressive functions in the lung in vivo. Our results therefore demonstrate different roles for sIL-6R and mIL-6R in experimental asthma; sIL-6R regulates Th2 cell functions in CD4+CD25– T effector cells lacking the mIL-6R chain. By contrast, mIL-6R controls the cell fate at the beginning of T cell differentiation by directing CD4+ naive cells toward Th2 pathways and inhibiting Treg differentiation in the lung. Blockade of IL-6 signaling thus emerges as a promising approach for the treatment of Th2-dependent inflammatory processes such as allergic asthma in humans.
Results
Increased levels of sIL-6R in the bronchoalveolar lavage fluid of patients with allergic asthma.
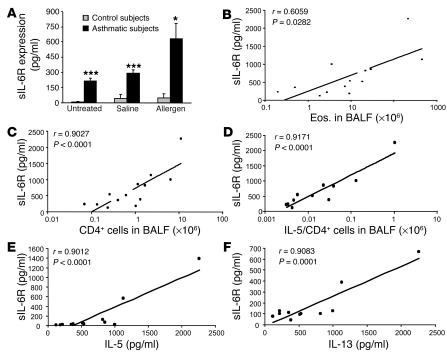
Whereas IL-6 is known to be increased in bronchoalveolar lavage fluid (BALF) obtained from patients with allergic asthma after allergen challenge (32), diagnostic levels of sIL-6R in BALF have not been determined in this disease. To analyze levels of sIL-6R in asthma, we determined sIL-6R levels in BALF of patients with allergic asthma before and after allergen challenge. As shown in Figure 1A, baseline concentrations of sIL-6R in BALF of asthmatic patients before allergen challenge were significantly increased compared to those of control subjects. Furthermore, 24 hours after antigen challenge, sIL-6R concentrations in BALF of asthmatics were increased as compared to baseline levels and significantly higher as compared to those in BALF of control patients. A positive correlation (r = 0.90; P < 0.0001) was found between the number of CD4+ cells in BALF of asthmatic subjects after allergen challenge and BALF concentration of sIL-6R (Figure 1C). Specifically, sIL-6R levels positively correlated with the number of IL-5–producing CD4+ T cells (r = 0.91; P < 0.0001) (Figure 1D), the levels of IL-5 and IL-13 proteins (r = 0.90; P < 0.0001) (Figure 1, E and F, respectively), and, to a lesser extent, with the number of eosinophils (Figure 1B) (r = 0.60; P = 0.028) in BALF after allergen challenge. This suggests a potential role for sIL-6R in controlling Th2 function in asthmatic subjects after allergen challenge.
Figure 1.
sIL-6R is increased in BALF of asthmatic patients as compared to control subjects, and its levels correlate with the number of IL-5–producing CD4+ T cells in BALF after allergen challenge. (A) sIL-6R was measured before (untreated) and 24 hours after allergen or saline challenge in control subjects (gray bars) or in subjects with asthma (black bars). In the asthmatic patients, sIL-6R levels were increased at baseline and were further increased after allergen challenge. *P < 0.05; ***P < 0.001. (B–D) In these patients, a positive correlation was found between the levels of sIL-6R and the number of CD4+ T cells (r = 0.9027; P < 0.0001) after allergen challenge (C). Furthermore, the value of sIL-6R positively correlated with the number of CD4+ T cells producing IL-5 in BALF (r = 0.9171; P = 0.0001) (D), while a lower correlation was found between sIL-6R and the number of eosinophils (Eos.) in BALF (r = 0.6059; P = 0.0282) (B). Furthermore, the value of sIL-6R in the airways of asthmatic subjects after allergen challenge correlates positively with the respective sIL-6R of IL-5 (E) and IL-13 (F) in BALF.
Blockade of sIL-6R by use of gp130-Fc inhibits Th2 cytokine production in experimental asthma by downregulating GATA-3 levels.
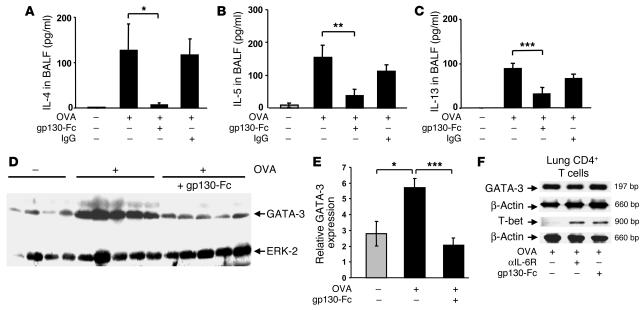
Asthma is characterized by increased infiltration of the airways with CD4+ Th2 cells, which produce signature cytokines such as IL-4, IL-5, and IL-13. Since the above data in humans suggests a potential correlation between sIL-6R levels in BALF and Th2 cytokine production in asthma, we next analyzed Th2 cytokine levels in a model of experimental asthma upon blockade of sIL-6R function in vivo. Accordingly, we used a murine model of late-phase asthma induced by OVA sensitization and subsequent intranasal (i.n.) OVA challenge and determined Th2 cytokine production in the lung upon specific blockade of sIL-6R using the chimeric protein gp130-Fc: a treatment that does not affect signaling via mIL-6R (24–32). Selective blockade of sIL-6R in the lung led to a significant downregulation of IL-4, IL-5, and IL-13 levels in BALF of OVA-sensitized mice as compared to that of OVA-sensitized and -challenged, untreated or IgG-treated control mice (Figure 2A, P = 0.03; Figure 2B, P = 0.006; Figure 2C, P = 0.00067). Since GATA-3 is the master transcription factor regulating expression of the Th2 cytokines IL-4, IL-5, and IL-13 in asthma (33, 34), we next analyzed GATA-3 expression by Western blot analysis and found that gp130-Fc treatment downregulated GATA-3 in the lungs of treated mice (Figure 2, D and E; P = 0.016 for OVA-treated versus saline-treated mice; P = 0.00056 for OVA/gp130-Fc–treated versus OVA-sensitized and -challenged, untreated mice). This was not accompanied by decreased GATA-3 expression per CD4+ lung T cell, as shown by RT-PCR of 105 CD4+ lung T cells (Figure 2F). We therefore conclude that the decreased GATA-3 in the lung must be due to a decrease of total CD4+ T cells. Taken together, these data indicate that sIL-6R signaling controls the function of Th2 cells in experimental asthma. Moreover, treatment with anti–IL-6R antibody as well as gp130-Fc led to an upregulation of T-bet mRNA levels in the CD4+ lung cells of OVA-sensitized mice, indicating an upregulation of the Th1 phenotype in the lungs of treated mice (Figure 2F). The consequence of these immunological changes was a downregulation of the total number of eosinophils in BALF of treated mice (Figure 3C; P = 0.0002 for anti–IL-6R antibody–treated mice and P = 0.0001 for gp130-Fc–treated mice).
Figure 2.
Local blockade of sIL-6R by gp130-Fc downregulates IL-4, IL-5, and IL-13 levels and reduces GATA-3 expression in experimental asthma. BALB/c mice were sensitized and challenged with OVA whereas control mice were given saline. Some OVA-sensitized mice received additional treatment with gp130-Fc to block sIL-6R function in vivo, as indicated. gp130-Fc treatment was associated with a significant decrease in IL-4 (A), IL-5 (B), and IL-13 (C) levels in BALF of OVA-sensitized mice. *P < 0.05; **P < 0.01; ***P < 0.001. Data represent mean values ± SEM from 5 mice per group. (D) Total lung proteins were isolated from gp130-Fc–treated mice and untreated control mice at day 28 and analyzed by Western blot analysis after immunoblotting with a monoclonal antibody directed against GATA-3. Furthermore, ERK2 expression was determined on the same blot after membrane stripping and incubation with an anti–ERK-2 antibody. Each lane in the Western blot was loaded with 50 μg proteins isolated from different mice (saline, n = 4; OVA, n = 5; OVA + gp130-Fc, n = 5). Quantification of Western blots by densitometry is reported in E and shows decreased GATA-3 expression after i.n. delivery of gp130-Fc in OVA-sensitized and -challenged mice (*P < 0.05; ***P = 0.00056). (F) RT-PCR for GATA-3 and T-bet in 105 lung CD4+ T cells per group after total RNA extraction. Both anti–IL-6R antibody and gp130-Fc treatment led to upregulation of T-bet, while GATA-3 remained unchanged in lung OVA-specific CD4+ T cells.
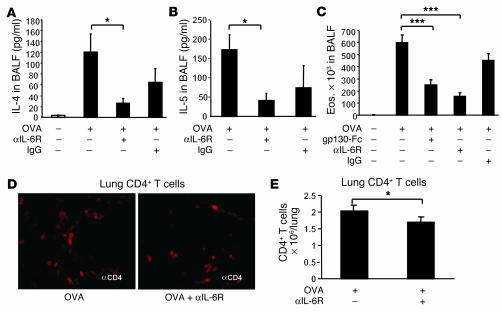
Figure 3.
Antiinflammatory mechanism of local treatment with anti–IL-6R antibodies. Blockade of IL-6R led to a downregulation of IL-4 (P = 0.05) (A), and, at higher doses (100 μg/day), of IL-5 (P = 0.029) (B) in the lungs of treated mice. These findings were accompanied by downregulation of the total number of eosinophils (P = 0.0002) in BALF (C) and CD4+ cells (P = 0.05) in the airways of treated mice (D and E). (D) CD4+ lung cells were stained by using monoclonal anti-CD4 antibodies (BD), and immunohistochemistry was performed as previously described (34). Pictures were taken with an Olympus inverted microscope connected to a digital camera. Original magnification, ×400. (E) The number of CD4+ lung cells obtained from 1 lung after CD4 isolation is reported for different groups. *P < 0.05; ***P < 0.001.
Antiinflammatory mechanism of local treatment with anti–IL-6R antibodies.
It has been previously reported that pulmonary DCs secrete higher amounts of IL-6 as compared to spleen DCs. In addition, pulmonary DCs induce naive T cells to secrete IL-4, which induces a strong Th2 polarization in the lung (12). We then asked whether blockade of mIL-6R would decrease IL-4 and the other Th2 cytokines. As shown in Figure 3A, blockade of IL-6R led to a specific downregulation of IL-4 (P = 0.05) and, at higher doses, IL-5 (Figure 3B; P = 0.029) in the lungs of treated mice. This was accompanied by downregulation of the total number of eosinophils in BALF (Figure 3C; P = 0.0002 for OVA-sensitized and -challenged, untreated mice versus OVA plus anti–IL-6R–treated mice) and total CD4+ lung cells (Figure 3, D and E; P = 0.05) in the airways of treated mice, indicating that blockade of mIL-6R interferes with early events during Th2 differentiation rather than after Th2 cells have developed.
Local treatment with anti–IL-6R antibodies ameliorates AHR in OVA-sensitized mice.
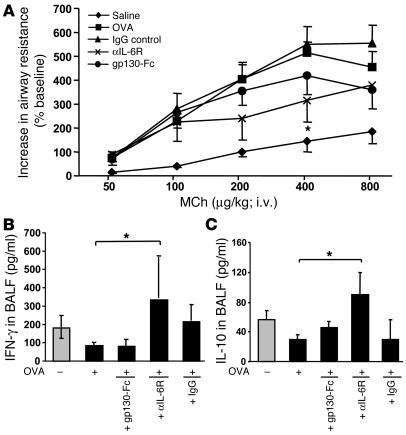
Next we determined whether blockade of sIL-6R with gp130-Fc would affect allergic AHR and compared such effects with those observed following blockade of both mIL-6R and sIL-6R with anti–IL-6R antibodies. Accordingly, we subjected 6 to 8 OVA-sensitized mice per group to i.n. treatment with the fusion protein gp130, anti–IL-6R antibodies, IgG control antibodies, or saline and assessed AHR by invasive plethysmography after intravenous methacholine (MCh) challenge at day 28. As shown in Figure 4A, OVA-sensitized mice showed an increase in AHR compared with saline-treated control mice after challenge. Furthermore, it was found that anti–IL-6R treatment (P = 0.049, as compared to IgG treatment; Figure 4A) reduced AHR more than gp130-Fc–treatment in OVA-sensitized mice, which indicates that anti–IL-6R antibody treatment has a beneficial effect on AHR.
Figure 4.
Blockade of mIL-6R through i.n. application of anti–IL-6R antibodies ameliorates AHR and induces IFN-γ and IL-10 levels in BALF in a mouse model of asthma after OVA sensitization. (A) Eight to 10 BALB/c mice per group were sensitized to OVA-alum (OVA-sensitized mice) followed by local treatment with OVA alone or treatment with OVA plus either gp130-Fc, IgG, or anti–IL-6R antibody. Control mice were sensitized with saline-alum and exposed to saline aerosol (saline). Transpulmonary resistance was performed 24 hours after the last local treatment in all mice, as specified in Methods. Dose-response curves to MCh were obtained after administering indicated doses of intravenous MCh. OVA-sensitized mice reacted with an increase of airway resistance at low doses of MCh as compared to that of mice given saline. Anti–IL-6R–treated, OVA-sensitized mice were more protected from the development of AHR compared to untreated (P = 0.049) or IgG-treated, OVA-sensitized mice. Moreover, blockade of sIL-6R by gp130-Fc was less effective compared to anti–IL-6R antibody treatment. (B and C) Local anti–IL-6R antibody treatment induced significant release of IFN-γ (P = 0.048) (B) and IL-10 (P = 0.020) (C) in BALF of OVA-senstitized mice as compared to untreated, OVA-sensitized mice (5 < n < 15). Mean values ± SEM are shown. *P < 0.05.
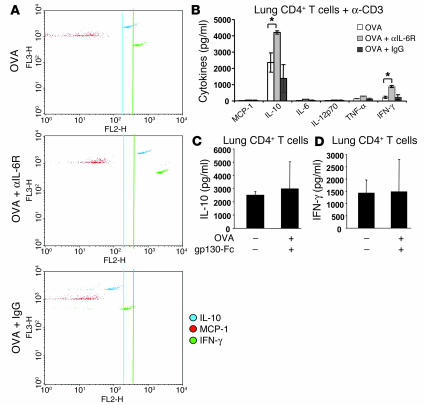
Anti–IL-6R antibody treatment induces IL-10 and IFN-γ production by lung CD4+ T lymphocytes.
We next analyzed cytokines in BALF and found that treatment with anti–IL-6R antibodies but not gp130-Fc was associated with significant induction of IFN-γ (P = 0.048) and IL-10 levels (P = 0.020) in BALF of OVA-sensitized mice (Figure 4, B and C, respectively). These results are consistent with previous reports showing enhanced ability of pulmonary DCs isolated from IL-6–/– mice to induce a Th1 response (12). In fact, we found that a blockade of IL-6R led to an upregulation of T-bet, the Th1 signature transcription factor in lung CD4+ cells (Figure 2F).
Further, in isolated CD4+ T cells from the lungs of treated or untreated, OVA-sensitized mice, after overnight cell culture, Cytometric Bead Array (CBA; BD Biosciences — Clontech) analysis was performed, using CD4+ T cell supernatants. It was found that CD4+ T cells isolated from the lungs of anti–IL-6R, antibody–treated (Figure 5, A and B) but not gp130-Fc–treated mice (Figure 5, C and D; as assessed by ELISA) secreted significantly increased amounts of IL-10 (Figure 5, A and B, for anti–IL-6R antibodies, P = 0.023; Figure 4C for gp130-Fc–treated mice) and IFN-γ (Figure 5, A and B, for anti–IL-6R antibodies, P = 0.013; Figure 5D for gp130-Fc–treated mice) after T cell receptor–dependent stimulation, as compared to IgG-treated or OVA-sensitized and -challenged, untreated mice. In contrast, levels of the Th2 type chemokine monocyte chemoattractant protein-1 (MCP-1) were not upregulated upon anti–IL-6R antibody treatment (Figure 5A). Taken together, these data suggest that blockade of mIL-6R causes induction of IL-10 and IFN-γ production by lung CD4+ T lymphocytes.
Figure 5.
IL-10–producing CD4+ T cells in the lungs of anti–IL-6R antibody–treated mice. (A and B) CD4+ T cells were isolated from the lung of treated or untreated mice and cultured overnight in the presence of anti-CD3 antibodies. CBA was performed on the CD4+ T cell supernatants. CD4+ T cells isolated from the lung of anti–IL-6R antibody–treated mice secreted increased amounts of IL-10 and IFN-γ as compared to those of OVA-sensitized and -challenged, untreated or IgG-treated mice (P = 0.023 and P = 0.013 for IL-10 and IFN-γ, respectively). Levels of the Th2-type chemokine MCP-1 were not upregulated upon anti–IL-6R antibody treatment, however. (C and D) By contrast, lung CD4+ cells isolated from mice treated i.n. with gp130-Fc did not show changes either in IL-10 (C) or IFN-γ production (D).
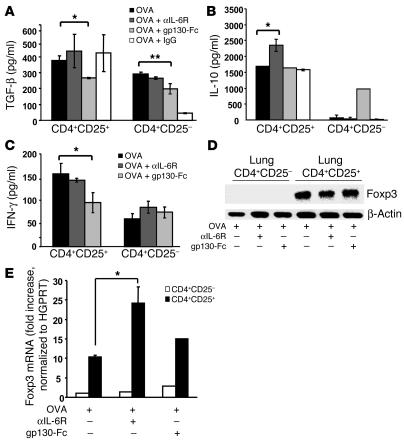
Increased release of IL-10 in Foxp3+ CD4+CD25+ Tregs isolated from the lungs of anti–IL-6R–treated mice.
Since IL-10 is a cytokine with potent antiinflammatory activities that is produced by subsets of Tregs (35–37), we next determined whether CD4+CD25+ T cells from the lung produce IL-10 upon anti–IL-6R antibody treatment. Accordingly, we separated CD4+CD25+ lung T cells from CD4+CD25– lung T cells and analyzed cytokine release. As shown in Figure 6B, CD4+CD25+ T cells isolated from anti–IL-6R antibody–treated, OVA-sensitized mice released significantly increased amounts of IL-10 per cell as compared to CD4+CD25+ T cells isolated from OVA-sensitized and -challenged, untreated (P = 0.0034) or IgG-treated mice. In addition to IL-10, CD4+CD25+ lung cells also produced TGF-β, suggesting their potential regulatory activity (Figure 6A; TGF-β released from CD4+CD25– versus CD4+CD25+ lung cells; P = 0.019 for anti–IL-6R antibody–treated mice; P = 0.004 for OVA-sensitized and -challenged, untreated mice). In contrast, CD4+CD25– lung T cells produced little or no IL-10 (Figure 6B), although they did produce TGF-β (Figure 6A). Moreover, CD4+CD25+ as well as CD4+CD25– cells isolated from the lungs of gp130-Fc–treated mice released significantly less TGF-β as compared to those isolated from the lungs of untreated mice (P = 0.05 for CD4+CD25+ and P = 0.0035 for CD4+CD25–), indicating a lower T regulatory inductive capability of the lung effector T cells isolated from gp130-Fc–treated mice (38). Furthermore, we analyzed IFN-γ production in CD4+CD25+ and CD4+CD25– cells and found that both cell populations released IFN-γ. However, CD4+CD25+ lung cells isolated from gp130-Fc–treated mice released less IFN-γ as compared to CD4+CD25+ cells isolated from OVA-sensitized and -challenged, untreated mice (Figure 6C).
Figure 6.
Increased release of IL-10 from Foxp3+ CD4+CD25+ Tregs isolated from the lungs of anti–IL-6R–treated mice. CD4+CD25+ T cells and CD4+CD25– T cells were isolated from lung cells in the different experimental groups, after which cytokine production was analyzed. The purity of the CD4+CD25+ cell population was 95–98% as determined by FACS analysis during cell sorting. (A–C) Lung CD4+CD25+ T cells isolated from OVA-sensitized, anti–IL-6R antibody–treated mice released increased amounts of IL-10 (B) per cell as compared to CD4+CD25+ T cells from OVA-sensitized and -challenged, untreated or OVA-sensitized, IgG-treated mice. In contrast, lung CD4+CD25– T cells produced little IL-10 (B) but more TGF-β (A) and some IFN-γ (C). n = 6. By contrast, CD4+CD25+ isolated from gp130-Fc–treated mice released less IL-10 (B), IFN-γ (C), and TGF-β (A), while the CD4+CD25– isolated from the same mice released the same amount of IL-10 (B) and less TGF-β (A). (D) Expression of Foxp3 on CD4+CD25+ lung T cells upon anti–IL-6R antibody treatment. CD4+CD25+ and CD4+CD25– lung T cells from untreated or anti–IL-6R antibody–treated, OVA-sensitized mice were separated as described above. This was followed by RNA extraction and analysis of Foxp3 or β-actin expression by RT-PCR. (E) Real-time PCR for Foxp3 in CD4+CD25+ and CD4+CD25– cells is reported as the ratio of Foxp3 to HGPRT. Anti–IL-6R antibody treatment led to a significant upregulation of Foxp3 as compared to OVA treatment. This experiment was performed 3 times in duplicate. *P < 0.05; **P < 0.01.
Moreover, as CD4+CD25+ Tregs in the lung are known to specifically express the forkhead family member transcription factor Foxp3, we finally determined Foxp3 expression in CD4+CD25+ lung T cells (39–43). It was found that CD4+CD25+ lung T cells but not CD4+CD25– lung T cells express Foxp3 (Figure 6D). Furthermore, anti–IL-6R antibody treatment but not gp130-Fc treatment led to an upregulation of Foxp3 expression in CD4+CD25+ T cells, as assessed by real-time PCR (Figure 6E; P = 0.04 for CD4+CD25+ cells isolated from the lungs of OVA-sensitized and -challenged, untreated versus anti–IL-6R antibody–treated mice). These results indicated specific activation of Foxp3-expressing CD4+CD25+ lung Tregs releasing IL-10 and TGF-β, especially after local treatment with anti–IL-6R antibodies.
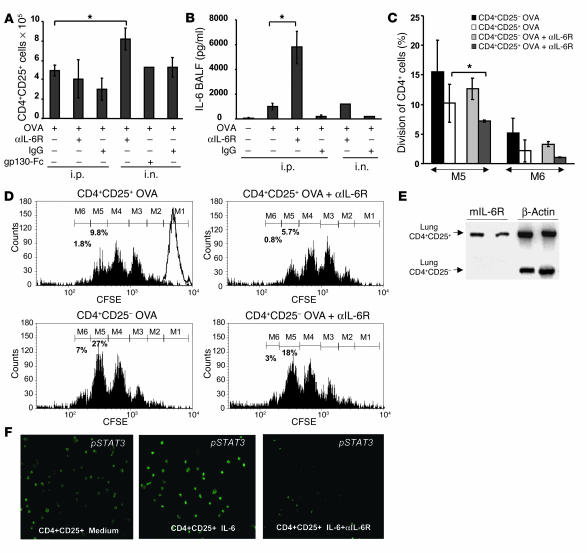
Expansion and augmented regulatory function of CD4+CD25+ lung Tregs upon local anti–IL-6R antibody treatment.
To determine whether anti–IL-6R antibody treatment modulates the expansion and immunosuppressive function of CD4+CD25+ lung Tregs, we next analyzed the number of CD4+CD25+ cells in the lungs of untreated and treated OVA-sensitized mice by cell sorting. As shown in Figure 7A, the number of CD4+CD25+ Tregs was increased significantly in the lungs upon local anti–IL-6R antibody treatment but not upon gp130-Fc treatment in OVA-sensitized mice (P = 0.034). In contrast, systemic blockade of IL-6R function before sensitization with OVA at day 0 by i.p. administration of anti–IL-6R antibody treatment did not increase the number of CD4+CD25+ Tregs in the lungs of OVA-sensitized mice. Furthermore, IL-6R blockade was accompanied by a significant increase in IL-6 levels in BALF of treated mice (Figure 7B; P = 0.02 OVA-treated mice versus i.p. anti–IL-6R antibody–treated mice), which suggests that i.p. application of anti–IL-6R antibodies leads to a negative feedback mechanism that increases IL-6 levels.
Figure 7.
Increased number and augmented immunosuppressive function of CD4+CD25+ T cells in the lungs of anti–IL-6R–treated, OVA-sensitized mice. (A) i.n. but not i.p. anti–IL-6R antibody treatment after OVA sensitization and challenge led to an induction of CD4+CD25+ T cell number in the lung (*P = 0.057). (B) IL-6 levels were increased in BALF of OVA-sensitized mice as compared to those of saline-treated mice. i.p. but not i.n. injection of anti–IL-6R antibodies led to a further increase of IL-6 in the airways. (C) CD4+CD25+ T cells isolated from the lungs of anti–IL-6R antibody–treated (i.n.) mice inhibited proliferation of CFSE-labeled target CD4+ spleen T cells more efficiently compared to CD4+CD25+ T cells isolated from the lungs of OVA-sensitized and -challenged, untreated mice. Mean values ± SEM; n = 5 mice per group; *P < 0.05. (D) Histograms of a representative cell population of CD4+ spleen cells labeled with CFSE and coincubated for 4 days with either CD4+CD25+ or CD4+CD25– cells isolated from the lungs of different groups. Percentages indicate the number of spleen CD4+/CFSE-labeled cells at day 4 (M1, 20 hours). (E) RT-PCR for the IL-6R α chain shows selective expression on Foxp3+ CD4+CD25+ lung T cells but not CD4+CD25– lung T cells. One representative experiment out of 3 is shown. (F) phospho–STAT-3 (pSTAT3) immunostaining in CD4+CD25+ cells. Spleen CD4+CD25+ T cells were incubated either with medium alone (left panel), with 20 ng/ml of IL-6 (middle panel), or with IL-6 (20 ng/ml) and anti–IL-6R antibodies (10 μg/ml) (right panel). Magnification, ×200.
The above data demonstrate that local rather than systemic blockade of IL-6 signal transduction leads to an expansion of CD4+CD25+ Tregs in the lung. We next analyzed the regulatory potential of these cells in coculture experiments with spleen target cells. In these studies, CD4+CD25+ lung T cells were incubated with CFSE-labeled, primary CD4+ spleen T cells in the presence of antigen-presenting cells and anti-CD3 antibodies. As shown in Figure 7, C and D, CD4+CD25+ lung T cells isolated from locally anti–IL-6R–treated, OVA-sensitized mice exhibited an increased suppressive capacity against spleen target cells as compared to CD4+CD25+ Tregs derived from untreated, OVA-sensitized mice (P = 0.033), as shown by the decreased number of CFSE+ cells that could be recovered at day 4 either at the fifth mitosis (M5) or at M6. Taken together, these data suggest that local blockade of IL-6 signal transduction in the lung results in both expansion and augmentation of immunosuppressive capacities of CD4+CD25+ Tregs.
Selective expression of the IL-6Rα chain in lung CD4+CD25+ T lymphocytes.
Since the above data suggested that IL-6 signal transduction in CD4+CD25+ lung Tregs involves signaling via mIL-6R rather than sIL-6R, we determined in subsequent studies the expression of mIL-6R mRNA in such cells. This approach was important in understanding whether IL-6 would act directly on CD4+CD25+ Tregs via mIL-6R. Interestingly, we found that the IL-6R α chain is selectively expressed on CD4+CD25+ lung T cells but not on CD4+CD25– lung T cells, which suggests a direct effect of IL-6 via mIL-6R on CD4+CD25+ Tregs (Figure 7E).
Anti–IL-6R antibodies antagonize IL-6–induced STAT-3 phosphorylation in CD4+CD25+ Tregs.
To start to investigate the mechanism of action of IL-6 on CD4+CD25+ Treg function, we incubated CD4+CD25+ Tregs with IL-6 alone or with anti–IL-6R antibodies for 36 minutes. IL-6 induced phosphorylation and nuclear translocation of STAT-3 (44) in CD4+CD25+ cells, which could be reversed by coincubation with anti–IL-6R antibodies (Figure 7F), demonstrating functional activation of IL-6R in CD4+CD25+ cells.
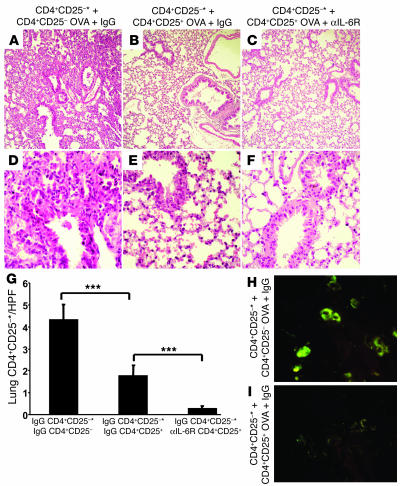
CD4+CD25+ T cells from anti–IL-6R–treated mice inhibit CD4+CD25– T cell–induced inflammation in an adoptive transfer model in Rag1 knockout mice.
In a final series of experiments, we aimed at characterizing the immunosuppressive properties of CD4+CD25+ T lymphocytes upon anti–IL-6R antibody treatment in vivo. To demonstrate the functional importance of CD4+CD25+ Tregs in asthma, we first isolated CD4+CD25+ T cells and CD4+CD25– T cells from the spleens of OVA-sensitized mice treated i.n. during the challenge phase with either anti–IL-6R or IgG control antibodies. We then cotransferred i.p. CD4+CD25+ T cells from either IgG or anti–IL-6R antibody–treated mice or CD4+CD25– T cells from IgG-treated mice together with equal numbers of CFSE-labeled CD4+CD25– T cells from OVA-sensitized mice into Rag1 knockout mice. As shown in Figure 8H, Rag1–/– mice receiving CD4+CD25– T cells showed CFSE positive cells in the lung, indicating homing of the labeled T cells. Moreover, mice cotransferred with CD4+CD25+ T cells from IgG-treated mice showed a marked decrease in the number of CFSE-positive cells (Figure 8, G–I) in the lung, suggesting inhibition of effector T cell proliferation by CD4+CD25+ T cells (Figure 8G). Moreover, CD4+CD25+ cells isolated from anti–IL-6R antibody–treated mice showed increased immunosuppressive capability as compared to those isolated from IgG-treated mice (P = 0.00040). Such inhibition was accompanied by reduced levels of IL-4 in BALF of reconstituted Rag1 knockout mice (data not shown). Histologically, Rag1–/– mice receiving CD4+CD25+ T cells were protected from the development of airway inflammation (Figure 8, A–F; 891 ± 1070 or 305 cells/mm2 for CD4+CD25– reconstituted mice; –231 ± 682 or 179 cells/mm2 for CD4+CD25+ IgG-treated and CD4+CD25– IgG-treated mice; 88 ± 407 or 162 cells/mm2 for CD4+CD25– and anti–IL-6R antibody–treated mice). These data indicate an antiinflammatory effect similar to that of CD4+CD25+ cells derived from either IgG-treated or anti–IL-6R antibody–treated mice. Taken together, these results indicate that the number of CD4+CD25+ T cells is important in inducing immunosuppressive functions on allergic airway inflammation under in vivo conditions.
Figure 8.
CD4+CD25+ T cells from OVA-sensitized mice can inhibit CD4+CD25– T cell–induced experimental asthma in Rag1–/– mice. (A–F) Cotransfer of CD4+CD25+ and CD4+CD25– spleen T cells into Rag1–/– mice. CD4+CD25+ and CD4+CD25– T cells were isolated from the spleens of OVA-sensitized and -challenged mice given IgG-control antibodies or anti–IL-6R antibodies. CFSE-labeled CD4+CD25– spleen T cells from OVA-sensitized mice (5 × 105; CSFE-labeled indicated with asterisks) were cotransferred i.p. with either 5 × 105 unlabeled CD4+CD25– T cells (A and D) or CD4+CD25+ T cells (B, C, E, and F) into immunocompromised Rag1 knockout mice. Cotransfer of CD4+CD25+ T cells from anti–IL-6R–treated (C and F) or IgG-treated (B and E) mice suppressed allergic airway inflammation induced by transfer of CD4+CD25– T cells from OVA-sensitized mice. Magnification, ×100 (A–C), ×400 (D–F). Mice receiving only CD4+CD25– T cells showed CFSE-positive cells in the lung (H), whereas mice receiving CD4+CD25+ T cells as well showed a marked decrease in the number of CFSE-positive cells (G and I), suggesting a CD4+CD25+ T cell–mediated suppression of effector CD4+CD25– T cell proliferation. Results in G were obtained by calculating the average number of CFSE-positive cells per high power field (HPF) (n = 30). ***P = 0.001.
Discussion
Although previous studies demonstrated increased production of IL-6 in patients with allergic asthma as compared to that in controls (32), it remained unclear whether IL-6 signaling plays a role in the effector phase of allergic airway inflammation and whether antagonizing the function of IL-6 can be used for the therapy of allergic airway inflammation and AHR. Furthermore, the potential mechanisms of action of such therapy remained to be elucidated. Here we demonstrate that IL-6 signaling controls the balance between effector T cells and Tregs in experimental asthma by means of different receptor components. Specifically, our data suggest that IL-6 trans-signaling via sIL-6R supports Th2 lung T cell cytokine production, whereas IL-6 signaling via mIL-6R suppresses the development and functional activity of Foxp3+ CD4+CD25+ Tregs in the lung and induces the early development of lung CD4+ naive T cells via IL-4. Consistent with this concept, blockade of mIL-6R signaling induced expansion and immunosuppressive capacities of CD4+CD25+ lung Tregs in vivo and induced CD4+ IFN-γ–producing cells in the lung, ameliorating AHR in experimental asthma. These Tregs are similar to those previously described in the spleen (45, 46). Although, in our study, Tregs isolated from the lung, in contrast to those isolated from the spleen, produced IFN-γ in addition to TGF-β and IL-10, we think that additional experiments are required to define whether 2 different populations of Tregs are present in the CD4+CD25+ lung Treg population used in this study.
IL-6 elicits cellular actions by binding to the membrane-bound α chain of its receptor (mIL-6R α chain). This event is followed by recruitment of 2 membrane-spanning gp130 molecules into tetra- or hexametric structures, thereby forming the active IL-6R complex. In addition, a soluble form of the mIL-6R α chain can mediate signal transduction in cells lacking mIL-6R by a mechanism called trans-signaling. In both cases, binding of IL-6 to the α chain leads to homodimerization of the 2 gp130 molecules and intracellular activation of Src and JAK, which in turn phosphorylate STAT3 (19–23) and activate IL-6 target genes. The functional importance of IL-6 signaling in T lymphocytes has been underlined by the finding that such signaling augments T cell activation and T helper 2 cell polarization in vitro and in vivo.
In initial studies on IL-6 signaling in asthma, we observed an increased production of sIL-6R in BALF of patients with allergic asthma at baseline as compared to that of control subjects and a further increase after allergen challenge. Interestingly, sIL-6R levels strongly correlated with the number of CD4+/IL-5–producing cells along with IL-5 and IL-13 levels in BALF of asthmatics after allergen challenge, suggesting that sIL-6R levels contribute to Th2 cell development in asthma. To further extend the above findings, we took advantage of a model of asthma in mice associated with Th2-mediated airway inflammation and AHR (28). To understand the immunopathological role of IL-6 in this model, during the challenge phase, we selectively blocked either sIL-6R via gp130-Fc or sIL-6R plus mIL-6R via anti–IL-6R antibodies. In the first therapeutic approach, we used a chimeric gp130-Fc fusion protein that competes with gp130 and thereby specifically blocks IL-6 trans-signaling. Consistent with the above data in humans, it was found that specific suppression of sIL-6R signaling via administration of gp130-Fc suppresses Th2 T cell development and subsequent production of effector cytokines in experimental asthma in vivo. Specifically, blockade of sIL-6R signaling in experimental asthma led to downregulation of the master transcription factor of Th2 cells, GATA-3, and suppression of IL-4, IL-5, and IL-13 production. Collectively, these data indicate an important role for IL-6 trans-signaling via sIL-6R in controlling Th2 T cell function in asthma.
In contrast to gp130-Fc treatment, local administration of anti–IL-6R antibodies led to downregulation of IL-4 and induction of IFN-γ production by CD4+ lung T cells as well as amelioration of AHR, suggesting the existence of important IL-6–dependent but sIL-6R–independent signaling events at the beginning of Th2 differentiation in the lung in experimental asthma. These results are consistent with a Th2-promoting potential of pulmonary DCs due to the inhibition of Th1 differentiation caused by IL-6 production (12). Moreover, we identified mIL-6R as being responsible for the Th1 inhibition, since blockade of the IL-6R with antibodies significantly induced IFN-γ in the airways. By contrast, sIL-6R might control the number of Th2 cells, since IFN-γ was not induced by this treatment (Figure 5C). We further demonstrated that anti–IL-6R antibody treatment led to the simultaneous increase of CD4+CD25+ lung T cells releasing IL-10. Such cells were increased in number in the airways of mice given anti–IL-6R antibodies locally, suggesting that mIL-6R signaling controls expansion of CD4+CD25+ lung Tregs. This concept was underlined by the finding that CD4+CD25+ but not CD4+CD25– lung T cells expressed mRNA for mIL-6R. Thus, whereas CD4+CD25+ lung T cells are susceptible to signaling via mIL-6R, CD4+CD25– effector T cells in the lung may respond to sIL-6R signaling only.
CD4+CD25+ Tregs are known to regulate innate and adaptive immune responses and to possess potent antiinflammatory capacity in autoimmune and chronic inflammatory diseases such as experimental autoimmune encephalitis, diabetes, and allergy in vivo (42–44). Although such Tregs are known to be produced in the thymus after birth, recent evidence suggests that they can also develop in the lung (45). However, the signaling pathways that control Treg development in the lung are still poorly understood. Our data provide evidence for a key role of IL-6 signaling via mIL-6R in preventing Treg responses in experimental asthma. Specifically, we found that local application of anti–IL-6R antibodies leads to expansion of Foxp3+ CD4+CD25+ lung T cells (39–46), producing IL-10. Thus, signaling via mIL-6R present on T cells apparently suppresses the function of CD4+CD25+ Tregs in the lung. In addition, we demonstrate that IL-6R is functionally active in CD4+CD25+ T cells, since we have shown phosphorylation and activation of STAT-3 upon culture of these cells with IL-6, which is inhibited by anti–IL-6R antibody coincubation. These data extend other studies showing that IL-6 production by spleen DCs enhances effector T cell responses by overcoming CD4+CD25+ Treg suppression (37). However, our data suggest that this process occurs in the peripheral immune system in the lung in vivo and involves signaling via mIL-6R. The functional relevance of this concept for asthma was underlined by adoptive transfer studies in which CD4+CD25+ spleen T cells were able to suppress CD4+CD25– T cell–induced allergic airway inflammation in Rag1 knockout mice.
In summary, IL-6 signaling in the lung tightly controls the critical balance between effector and Treg function in the lung via differential signaling events involving sIL-6R and mIL-6R, respectively. Inhibition of IL-6 signaling results in beneficial effects in experimental asthma by simultaneously suppressing Th2 cell development via sIL-6R signaling and skewing naive CD4+ cells to a Th1 pathway and/or expanding Tregs via mIL-6R signaling. Although these data are obtained from an experimental model of asthma, they suggest a potential therapeutic utility of antibodies against the IL-6R as a novel molecular approach for the induction of local T regulatory responses in patients with allergic asthma.
Methods
Patients with allergic asthma.
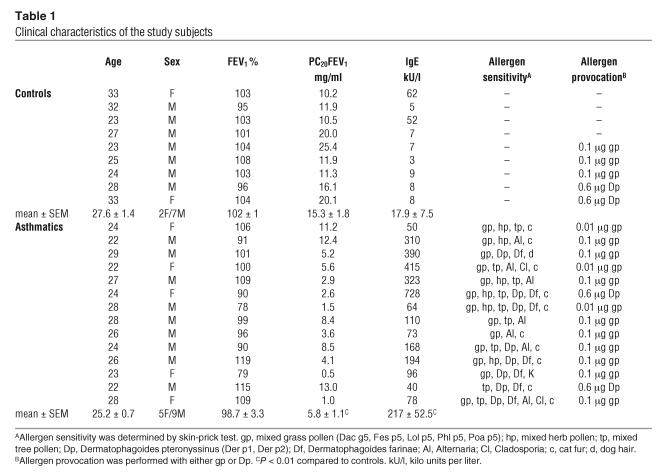
Nine healthy volunteers and 14 patients with mild asthma participated in the study. The subject characteristics are described in Table 1. All patients had mild allergic asthma as defined in the International Consensus Report on Diagnosis and Treatment of Asthma (47). Subjects with asthma met the American Thoracic Society criteria for the diagnosis of asthma. They had histories of intermittent wheezing with reversible airflow obstruction, and asthma had previously been diagnosed by independent physicians. Each patient had a positive skin-prick test, defined as a greater than 4 mm diameter skin wheal response to 1 or more of 8 common allergens (Dermatophagoides pteronyssinus, Dermatophagoides farinae, mixed grass pollen, mixed tree pollen, dog hair, feather, cat fur, Alternaria; allergen extractions from ALK-Abelló). The allergen extract used for segmental allergen challenge was the one which produced the largest wheal response on skin-prick testing, and the chosen concentration was one-tenth the dilution in saline that elicited a 3 mm diameter skin wheal response. Bronchial hyperresponsiveness and concentration of histamine producing a 20% fall in forced expiratory volume in 1 second (PC20FEV1) were determined as described (48). Patients were using only salbutamol when required for relief of symptoms. None was treated with corticosteroids, sodium cromoglycate, theophylline, or antileukotriens. The controls had no history of allergic or other diseases and had negative skin-prick tests, normal IgE (≤ 100 IU/ml), normal lung function tests, and no bronchial hyperresponsiveness (PC20FEV1 > 8 mg/ml). All study subjects were nonsmokers, and no subject had experienced acute bronchitis within 4 weeks prior to the investigations. All patients gave their written consent after being fully informed about the purpose and nature of the studies, which were approved by the Ethical Committee of Hannover Medical School.
Table 1.
Clinical characteristics of the study subjects
Segmental allergen challenge.
Segmental allergen challenge was performed as previously described (48). Briefly, all subjects (14 patients with mild asthma and 9 controls) received nebulized salbutamol (1.25 mg), atropine (0.5 mg s.c.), and midazolam (2–8 mg i.v.) prior to bronchoscopy. The bronchoscope (P30; Olympus) was wedged into the inferior lingular bronchus, and a bronchoalveolar lavage (BAL) was performed with 5 × 20 ml sterile saline. The instrument was passed into the superior lingular bronchus, and 10 ml saline solution was instilled as a control challenge. Finally, the bronchoscope was passed to the medial segment of the middle lobe, and 10 ml allergen solution was instilled. After 24 hours, subjects (all asthmatics and 5 controls) were bronchoscoped again with the same premedication, and the superior lingular bronchus and the medial middle bronchus were lavaged with 100 ml saline. BALF samples were filtered through a 100 μm filter and centrifuged, and the supernatant was stored at –80°C. Differential cell counts were performed from cytospin slides with 300 cells per slide being counted after staining with May-Grünwald-Giemsa stain.
Flow cytometric detection of intracellular cytokines and ELISA in BALF T cells.
Intracellular cytokine detection of BALF-derived T cells was performed as previously described (49). Briefly, BALF cells isolated from 11 patients were resuspended in RPMI 1640 supplemented with 10% fetal calf serum (Biochrom AG) (1 × 106 cells/ml). Cells were cultured in 24-well flat-bottom plates (Nunc) and stimulated with phorbol 12-myristate 13-acetate (PMA) (10 ng/ml) and ionomycin (1 μM) in the presence of monensin (2.5 μM). After incubation for 4 hours at 37°C in a humidified atmosphere (5% CO2), cells were washed in PBS, and half of the cells were fixed in 1 ml 4% ice-cold paraformaldehyde (Riedel-de Haën) for 10 minutes. After a further wash in PBS, these cells were resuspended in 100 μl portions with saponin buffer and 0.01 M N-2-hydroxyethylpiperazine-N′-ethane sulfonic acid buffer (SERVA Electrophoresis). Anti-CD4+ FITC (1 mg/ml; Medac International) and PE-labeled antibodies against IFN-γ or IL-5 (10 ml, dilution 1:30; BD Biosciences — Pharmingen) were added to the cell suspension and incubated for 30 minutes in the dark. Nonspecific FITC-PE–labeled antibodies (BD; Medac International) were used as controls. Flow cytometric analysis was performed with scatter gates on lymphocyte fractions, using a flow cytometer (FACSCalibur; BD). The human ELISA-kits (Duo-sets) for IL-5 and IL-13 were purchased from R&D Systems.
Allergen sensitization and challenge and anti–IL-6R antibody treatment.
Female BALB/c mice (6–8 weeks of age) were maintained under specific pathogen-free conditions and received i.p. injections of 100 μg OVA (Calbiochem) complexed with alum (Sigma-Aldrich) on days 0 and 14, as previously described (24, 50). On days 25, 26, and 27, mice underwent anesthesia with avertine (1 mg tribromethanol/ml t-amylalcohol; 2.5% in PBS) before receiving 50 μg OVA in saline. Treated mice received 50 μg of anti–IL-6R antibodies. Control animals received i.p. saline complexed with alum on days 0 and 14 and an i.n. dose of 50 μl saline on days 25, 26, and 27 (24, 50).
To assess the role of IL-6 on airway inflammation and AHR, blocking antibodies and control antibodies were given by i.n. administration. Mice received 50 μg of anti–IL-6R antibodies (rat anti-mouse; Chugai Pharmaceutical Inc.); control IgG (rat anti-mouse, Sigma-Aldrich; rat IgG1 isotype control, R&D Systems) or gp130-Fc (obtained from K.J. Kallen, University of Kiel, or R&D Systems) by i.n. administration on days 25–27. The antibody was given 30 minutes before the i.n. OVA challenge. On day 28, airway plethysmography was performed followed by BAL, as described below.
Additional experiments were performed to assess the effect of anti–IL-6R antibody treatment i.p. before the first i.p. sensitization with OVA. In this set of experiments, mice were treated with an additional dose of anti–IL-6R antibody i.p. on day 0 (1 mg of anti–IL-6R antibodies) or IgG as control antibodies. All mice were housed in ventilated cages equipped with microisolator lids. All experiments were undertaken with approved license (number 177-07-991-32) from the Ethical Committee of the Region Rheinland-Pfalz (Mainz, Germany).
Assessment of airway reactivity by invasive body plethysmography.
AHR in mice was evaluated by using an invasive, whole-body plethysmograph, as previously described (51–53). Lung resistance (RL) was measured in untreated, OVA-sensitized mice and control mice as well as in IgG-, anti–IL-6R antibody–, or fusion protein gp130-Fc–treated OVA-sensitized mice, as described previously (29). In brief, dose response curves to MCh were obtained in anesthetized mice after administering increasing doses of intravenous MCh (33–1,000 μg/kg). Data were expressed as mean values of RL ± SEM.
Collection and analysis of BAL.
Twenty-four hours after the last i.n. challenge with either OVA or saline, BAL of the right lung was performed with 0.75 ml saline 4 times. Total BALF was collected and cells were stained with trypan blue and counted using a 50 μl aliquot. Samples were centrifuged at 680 g for 5 minutes, and cell pellets were resuspended in 1 ml PBS. Cytospins were made by centrifugation at 22.86 g for 5 minutes. Eosinophils were detected by staining according to May-Grünwald-Giemsa. The cytospins were analyzed with an Olympus microscope, using a ×40 objective. The supernatants were frozen and subsequently analyzed by ELISA.
Isolation and analysis of lung CD4+ T cells.
Lungs were removed from mice, transported in RPMI 1640 medium (with NaHCO3, glutamine tested for endotoxin; Biochrom AG), and cut into small pieces (1–2 mm2) with a forceps. Tissue pieces were suspended in Dulbecco’s PBS (5 ml per lung) containing 300 U/ml of collagenase type II (Worthington Biochemical Corp.) and 0.001% DNase (Roche Diagnostics). The suspension was incubated at 37°C for 1 hour with constant shaking. The digest was filtered through a 70-μm nucleopore filter to remove tissue fragments, centrifuged (300 g, 10 minutes), and the cell pellet washed once with RPMI 1640, 100 units/ml penicillin, and 100 μg/ml streptomycin. Erythrocytes were removed from lung cell suspension by hypotonic lysis in ammonium chloride and potassium chloride (ACK) buffer. After 2 washes in PBS, the cell pellet was resuspended in MACS buffer (PBS/EDTA with 0.5% BSA; Sigma-Aldrich). The CD4+ lung T cells were directly purified from isolated lung cell suspension by incubation at 4°C for 15 minutes with 10 μl of anti-mouse CD4 L3T4 microbeads per 107 total lung cells and positively sorted in a multiparameter magnetic cell sorter system (MACS; Miltenyi Biotech). After 3 washes in complete medium, the CD4-positive cells were cultured with plate-bound or soluble (as indicated) anti-CD3 antibodies (CD3 clone 145-2C11, 5 μg/ml; BD Biosciences — Pharmingen) for 24 hours at a density of 106 cells/ml. The supernatant was analyzed for cytokine production by the BD Cytometric Bead Array System (BD Biosciences — Clontech) or ELISA.
Isolation of CD4+CD25+ cells from the lung.
In additional experiments, the resulting CD4+ lung cells (purity 97%) were further separated by cell sorting into CD4+CD25+ and CD4+CD25– T cells. Briefly, isolated lung CD4+ T cells from BALB/c OVA-sensitized and -challenged (untreated) in vivo, anti–IL-6R antibody–treated, and IgG-treated mice were stained with 1 μg CD25-PE/1 × 106 (Clone PC61; BD Biosciences) cells at 4°C for 30 minutes in the dark. Cells were then washed once in PBS and kept on ice. CD4+CD25+ cells were sorted on a BD FACSVantage SE System, and analysis was done with Cell Quest Pro software (BD Biosciences). Dead cells and debris were excluded from data analysis through appropriate forward scatter channel and SSC gating. For each sample, 20,000 events were collected. Markers and regions were set according to control staining. In 3 experiments performed either in duplicated or triplicated conditions, sorted CD4+CD25+ lung cells (105 cells/well) or CD4+CD25– lung cells (105 cells/well) were cultured overnight with 2 μg/ml of soluble anti-CD3 antibodies (BD Biosciences — Pharmingen). Supernatants were then collected from cell suspensions, and IL-10 and TGF-β were measured by ELISA. During sorting we also measured the number of CD4+CD25+ cells isolated per group. In additional experiments, we isolated CD4+CD25+ cells from the lung as described below for the spleen.
Functional studies of CD4+CD25+ lung cells.
In 3 independent experiments, CD4+CD25+ lung cells were purified after staining isolated CD4+ lung T cells with CD25-PE antibody. This was followed by incubation with anti-PE beads and immunomagnetic separation. CD4+CD25+ lung cells (105 cells/well) or CD4+CD25– lung cells (105 cells/well) were then cocultured with CFSE-labeled (Molecular Probes Inc.) target primary CD4+ spleen cells (105 cells/well) isolated from untreated mice and antigen-presenting cells (A20 a murine B lymphoma cell line; 104 cells/well) in the presence of soluble anti-CD3 antibodies (2.5 μg/ml) overnight. After 20 hours, the starting levels of incorporated fluorescence was recorded (M1). The proliferation of the target spleen CD4+(CFSE+) cells was determined as the percentage of daughter cells that would reach generation M5 or M6. Two to 3 pools with 2 lungs per pool were analyzed for each experimental group.
Functional studies on spleen CD4+CD25+ cells after IL-6 or anti–IL-6R antibody treatment.
To perform functional studies on CD4+CD25+ cells, we isolated CD4+CD25+ spleen cells, as described below, from 7-week-old BALB/c mice. In each experiment, we isolated CD4+CD25+ cells from 12 mice and performed functional studies on CD4+CD25+ cells incubated overnight with IL-6 (20 ng/ml) either alone, with 10 μg/ml of anti–IL-6R antibodies, or with 10 μg/ml of IgG normal mouse control antibodies.
Then the question arose as to whether anti–IL-6R antibody treatment would influence cytokine release induced by IL-6 in CD4+CD25+ and CD4+CD25– cells. To study this possibility, we incubated either CD4+CD25+ or CD4+CD25– cells overnight with medium alone or with medium and IL-6 (20 ng/ml) alone or with IL-6 and 10 μg/ml of anti–IL-6R antibodies in the presence of soluble anti-CD3 antibodies (2.5 μg/μl). On the following day, we collected supernatants and measured IL-10 and TGF-β by ELISA.
pSTAT-3 immunostaining of CD4+CD25+ spleen cells.
Fifty-thousand CD4+CD25+ or CD4+CD25– spleen cells were incubated for 30 minutes with IL-6 (20 ng/ml) alone or with 10 μg/ml of anti–IL-6R antibodies in the presence of soluble anti-CD3 antibodies (2.5 μg/μl) in duplicate. After that, cells were frozen for detection of pSTAT-3 activation. For immunodection of pSTAT-3, cells were fixed in 2% PFA in PBS for 10 minutes and washed twice in PBS. Cells were then permeabilized for 4 minutes in permeabilization buffer (0.2% Triton X-100 in PBS) and washed twice in TBS buffer (0.05 M Tris hydrochloride). After 40 minutes incubation in blocking buffer (3% BSA, 0.05% Tween 20 in TBS), the rabbit polyclonal anti–pSTAT-3 (Tyr705) antibody (Cell Signaling Technology) was applied (1:50) in blocking buffer and incubated overnight at 4°C. Detection of positive cells was performed after cells were incubated with a biotinylated goat anti-rabbit antibody (Vector Laboratories Inc.; 1:200 in blocking buffer) for 30 minutes. Finally the Cy2-streptavidine (1:500 in PBS) was added, followed by incubation for 60 minutes, then washing in PBS. Cells were analyzed with an inverted fluorescence microscope (Olympus) at both ×200 and ×400 magnifications.
Reconstitution of Rag1–/– mice with CD4+CD25+ and CD4+CD25– cells.
To demonstrate that isolated CD4+CD25+ cells would be able to inhibit the development of the asthmatic reaction, CD4+CD25+ T cells were isolated from the spleen of OVA-senstitized, IgG- or anti–IL-6R antibody–treated mice as described below. Briefly, CD4+ spleen cells were isolated by positive magnetic selection according to the manufacturer’s instructions (Dynal Biotech; 107 Dynabeads CD4-L3T4/ml; 25 μl beads for 2.5 × 106 cells) for 20 minutes at 4°C under shaking conditions. Afterwards, the CD4 beads were detached by using mouse CD4 DETACHaBEAD (Dynal Biotech). Briefly, 10 μl of DETACHaBEAD were incubated with 106 CD4+ T cells under rotating conditions for 45 minutes at room temperature. The resulting CD4+ lung T cells were incubated with 10 μl (1 μg) of anti–CD25-PE/1 × 106 (Clone 7D4; BD Biosciences) cells at 4°C for 20 minutes in the dark. Cells were then washed once in PBS/EDTA/0.5% BSA and kept on ice. CD4+CD25+ cells were isolated with a kit (CD25 Microbead Kit, mouse; Miltenyi Biotec) by immunomagnetic isolation after incubation with anti-PE magnetic beads. Sorted CD4+CD25+spleen cells (5 × 105 cells/mouse) or CD4+CD25– cells (5 × 105 cells/mouse) were separately injected i.p. in Rag1–/– mice previously aerosolized once with 1% OVA aerosol for 30 minutes. Specifically, mice received i.p. either unlabeled CD4+CD25– cells (5 × 105 cells/mouse) from OVA-sensitized and -challenged mice treated i.n. with IgG control antibodies or CD4+CD25+ cells (5 × 105 cells/mouse) from OVA-sensitized and -challenged and IgG- or anti–IL-6R antibody–treated mice. Next all mice were injected i.p. with 5 × 105 of CFSE-labeled CD4+CD25– derived from OVA-sensitized and -challenged mice treated i.n. with a control antibody IgG. The next 3 days after reconstitution, mice were aerosolized with 1% OVA, and 24 hours later BALF was performed and lungs stored at –80°C for analysis of CFSE labeling. For CFSE analysis, paraffin slides were fixed in 2% PFA in PBS for 10 minutes and washed twice in PBS, mounted in an aqueous mounting medium (Vector Laboratories Inc.), and analyzed at ×400 with an Olympus IX70 microscope connected to a digital camera (Sony Power HAD, 3CCD color video camera DXC-950P; Sony Corp.). Images were then transferred to a computer and analyzed through a soft imaging system analysis program.
Histological assessment of airway inflammation.
Images were imported (×20; image width: 640 μm each) into Photoshop (version 7.0; Adobe Systems Inc.). The hematoxylin-stained darker nuclei around the bronchi were counted around a mask layered over the image. A 100-μm margin was then placed around the bronchus and the mean gray level (corresponding to the number of cells, as determined using a calibration curve) determined using the histogram command in Photoshop. Then the rest of the image was quantified in the same way, with the exclusion of large empty areas (corresponding to vessels and/or other neighboring bronchi). This background gray level corresponded to the cellular density of the lung parenchyma and was then subtracted from the peribronchial cell density in each image. This was performed for every image (5 < n > 12 per group). The results are reported as the subtracted peribronchiolar area (corresponding to the inflammatory cell infiltrate around the bronchi) and are given as mean ± SD and SEM.
Isolation of RNA from CD4+ T cells.
Sorted and cultured CD4+CD25+ and CD4+CD25– lung cells were immediately frozen after culture until RNA was isolated following the manufacturer’s instructions (RNeasy Micro Kit; QIAGEN). Briefly, cells were disrupted directly in the plastic wells by adding 350 μl of lysis buffer containing β-mercaptoethanol. Cells were then homogenized with a rotor for 30 seconds and mixed with an equal volume (350 μl) of 70% ethanol. The samples were then applied to an RNeasy MinElute Spin Column (QIAGEN) in a 2 ml collection tube. Closed tubes were then centrifuged for 15 seconds at 8,000 g. The column was washed several times, and the eluate was collected by applying 14 μl of RNAse-free water to the column. RNA concentration was determined using a densitometer.
RT-PCR from CD4+ lung cells.
Six μl of total RNA from either CD4+CD25+ or CD4+CD25– cells was then reverse transcribed by use of the RevertAid 1st Strand cDNA Synthesis Kit for RT-PCR (M-MuLV RT; MBI Fermentas GmbH). The resulting cDNA was then used as a template for the PCR. For Foxp3 analysis we used the following primers: 5′-GGCGAAAGTGGCAGAGAGGTAT-3′ and 5′-AAGACCCCAGTGGCAGCAGAA-3′; the RNA for the IL-6R α chain was amplified by using 5′-ACACACTGGTTCTGAGGGAC-3′ sense and 5′-TACCACAAGGTTGGCAGGTG-3′ antisense primers; T-bet antisense 5′-TGCCCCGCTTCCTCTCCAACCAA-3′, sense 5′-TGCCTGCAGTGCTTCTAACA-3′; GATA-3 antisense CTGGAGGAGGAACGCTAATG-3′, sense GGTTGAAGGAGCTGCTCTTG-3′. The primers for actin were: 5′-TGACGGGGTCACCCACACTGTGCCCATCTA-3′ and 5′-CTAGAAGCATTTGCGGTGGACGATGGAGGG-3′. The PCR program was as follows: 93°C for 3 minutes, 32 cycles of 93°C for 30 seconds, 60°C (56°C for GATA-3) for 30 seconds (1 minute for T-bet), and 72°C for 1 minute followed by a final extension period of 10 minutes at 72°C. PCR products were analyzed on 1.5% agarose gels.
Real-time analysis of Foxp3 mRNA.
To eliminate amplifications from contaminating genomic DNA, the following primers were designed to span an intron/exon boundary and thus to anneal specifically to cDNA: Foxp3 forward 5′-CTTATCCGATGGGCCATCCTGGAAG-3′, reverse 5′-TTCCAGGTGGCGGGGTGGTTTCTG-3′; HGPRT forward 5′-GTTGGATACAGGCCAGACTTTGTTG-3′, reverse 5′-GAGGGTAGGCTGGCCTATAGGCT-3′.
Real-time analysis of Foxp3 mRNA was performed on an iCycler (Bio-Rad Laboratories) using the IQ SYBR Green Supermix (Bio-Rad Laboratories). After normalization of the data according to the expression of HGPRT mRNA, relative expression levels of Foxp3 mRNA were calculated.
CBA.
CD4+ T cells isolated from the lungs of untreated or anti–IL-6R antibody– or IgG-treated, OVA-sensitized mice were incubated overnight in the presence of anti-CD3 antibodies. The supernatants were then analyzed by FACS using CBA (Mouse Inflammation Kit; BD Biosciences — Pharmingen) in accordance with the manufacturer’s instructions. Briefly, 6 bead populations with distinct fluorescence intensities were coated with capture antibodies specific for IL-6, IL-10, MCP-1, IFN-γ, TNF-α, and IL-12p70 proteins. The 6 bead populations were mixed together to form the CBA, which was resolved in the FL3 channel of a flow cytometer. The capture beads, PE-conjugated detection antibodies, and recombinant standards or test samples were incubated together to form sandwich complexes. Following acquisition of samples data using a flow cytometer, the sample results were generated in graphical and tabular format using CBA Analysis Software (BD).
ELISA.
Mouse IL-4, IL-5, IL-6, IFN-γ, and IL-10 were detected using a specific sandwich ELISA (OptEIA; BD Biosciences — Pharmingen). Murine IL-13 was detected using an ELISA kit (R&D Systems). TGF-β analysis was performed by using purified rat anti-mouse, anti-human, and anti-pig TGF-β1 as capture antibody (BD Biosciences — Pharmingen) and biotinylated rat anti-mouse, anti-human, and anti-pig TGF-β1 (BD Biosciences — Pharmingen) polyclonal antibody. Recombinant TGF-β was purchased from R&D. To activate TGF-β, 200-μl samples were pretreated with 10 μl of 1N HCl for 30 minutes at 37°C. Samples were then neutralized by adding 10 μl of 1 N NaOH.
Protein extraction and Western blot analysis.
Tissue proteins were extracted as previously described (39). Briefly, tissue was homogenized in PBS and protein extracted in the presence of protease inhibitors (6.75% aprotinin, 312 μg/ml trypsin inhibitor) and detergent (Nonidet). Protein concentrations were determined by spectrophotometry with the Bio-Rad protein assay (Bio-Rad Laboratories). Equal amounts of protein extract were added to the electrophoresis sample buffer. After being boiled, proteins were separated by 10% or 15% SDS-PAGE, transferred to nitrocellulose membranes, and incubated in blocking solution (5% milk in PBS/0.05% Tween 20) for 1 hour at room temperature; they were subsequently exposed to 1 μg/ml of antibodies against GATA-3 and ERK-2 (Santa Cruz Biotechnology Inc.) in blocking solution. Specific binding was visualized with the ECL Western Blotting Detection System (Amersham Biosciences) according to the manufacturer’s instructions after 1-hour incubation with the corresponding secondary HPR-conjugated antibody (1:2000 in blocking solution; Amersham Biosciences).
Statistical analysis.
Differences were evaluated for significance (P < 0.05) by the Student’s 2-tailed t test for independent events (Microsoft Excel, version 2000; Microsoft Corp.). The coefficient of correlations was calculated by statistical analysis using Microsoft Excel. Data are given as mean values ± SEM. For the analysis of RL data, the multivariate analysis MANOVA (Wilks’ Lambda) was used.
Acknowledgments
The authors thank J.M. Drazen for critical reading of the manuscript and M. Schipp, C. Lux, and P. Scholtes for their technical help. This work is supported by a Deutsche Forschung Gemeinschaft (DFG) grant.
Footnotes
Nonstandard abbreviations used: AHR, airway hyperresponsiveness; BAL, broncho-alveolar lavage; BALF, bronchoalveolar lavage fluid; CBA, Cytometric Bead Array; Fc, fraction constant; gp130, glycoprotein 130; i.n., intranasal(ly); M5, fifth mitosis (M6); MCh, methacholine; MCP-1, monocyte chemoattractant protein-1; mIL-6R, membrane-bound IL-6 receptor; PC20FEV1, concentration of histamine producing a 20% fall in forced expiratory volume in 1 second; sIL-6R, soluble IL-6 receptor.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Holgate ST. The epidemic of allergy and asthma. Nature. 1999;402:B2–B4. doi: 10.1038/35037000. [DOI] [PubMed] [Google Scholar]
- 2.Robinson DS, et al. Activation of CD4+ T cells, increased Th2-type mRNA expression, and eosinophil recruitment in bronchoalveolar lavage after allergen inhalation challenge in patients with atopic asthma. J. Allergy Clin. Immunol. 1993;92:313–324. doi: 10.1016/0091-6749(93)90175-f. [DOI] [PubMed] [Google Scholar]
- 3.Robinson DS, et al. Predominant Th2-like bronchoalveolar T-lymphocyte population in atopic asthma. N. Engl. J. Med. 1992;326:298–304. doi: 10.1056/NEJM199201303260504. [DOI] [PubMed] [Google Scholar]
- 4.Tang C, Rolland JM, Ward C, Quan B, Waters EH. IL-5 production by broncho-alveolar lavage and peripheral blood mononuclear cells in asthma and atopy. Eur. Respir. J. 1997;10:624–632. [PubMed] [Google Scholar]
- 5.Ying S, et al. T cells are the principal source of interleukin-5 mRNA in allergen-induced rhinitis. Am. J. Respir. Cell Mol. Biol. 1993;4:356–360. doi: 10.1165/ajrcmb/9.4.356. [DOI] [PubMed] [Google Scholar]
- 6.Neurath MF, Finotto S, Glimcher LH. The role of Th1/Th2 polarization in mucosal immunity. Nat. Med. 2002;8:567–573. doi: 10.1038/nm0602-567. [DOI] [PubMed] [Google Scholar]
- 7.Zhu Z, et al. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J. Clin. Invest. 1999;103:779–788. doi: 10.1172/JCI5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Temann UA, Ray P, Flavell RA. Pulmonary overexpression of IL-9 induces Th2 cytokine expression, leading to immune pathology. J. Clin. Invest. 2002;109:29–39. doi:10.1172/JCI200213696. doi: 10.1172/JCI13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akbari O, et al. Essential role of NKT cells producing IL-4 and IL-13 in the development of allergen-induced airway hyperreactivity. Nat. Med. 2003;9:582–588. doi: 10.1038/nm851. [DOI] [PubMed] [Google Scholar]
- 10.Rincon M, Anguita J, Nakamura T, Fikrig E, Flavell RA. Interleukin (IL)-6 directs the differentiation of IL-4-producing CD4+ T cells. J. Exp. Med. 1997;185:461–469. doi: 10.1084/jem.185.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diehl S, et al. Induction of NFATc2 expression by interleukin 6 promotes T helper type 2 differentiation. J. Exp. Med. 2002;196:39–49. doi: 10.1084/jem.20020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dodge IL, Carr MW, Cernadas M, Brenner MB. IL-6 production by pulmonary dendritic cells impedes Th1 immune responses. J. Immunol. 2003;170:4457–4464. doi: 10.4049/jimmunol.170.9.4457. [DOI] [PubMed] [Google Scholar]
- 13.Peters M, Müller A, Rose-John S. Interleukin-6 and soluble interleukin-6 receptor: direct stimulation of gp130 and hematopoiesis. Blood. 1998;92:3495–3504. [PubMed] [Google Scholar]
- 14.Romano M, et al. Role of IL-6 and its soluble receptor in induction of chemokines and leukocyte recruitment. Immunity. 1997;5:315–325. doi: 10.1016/s1074-7613(00)80334-9. [DOI] [PubMed] [Google Scholar]
- 15.Rose-John S, Heinrich PC. Soluble receptors for cytokines and growth factors: generation and biological function. Biochem. J. 1994;300:281–290. doi: 10.1042/bj3000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teague TK, Marrack P, Kappler JW, Vella AT. IL-6 rescues resting mouse T cells from apoptosis. J. Immunol. 1997;158:5791–5796. [PubMed] [Google Scholar]
- 17.Naka T, Nishimoto N, Kishimoto T. The paradigm of IL-6: from basic science to medicine. Arthritis. Res. 2002;4:S233–S242. doi: 10.1186/ar565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choy EH, et al. Therapeutic benefit of blocking interleukin-6 activity with an anti-interleukin-6 receptor monoclonal antibody in rheumatoid arthritis. Arthritis Rheum. 2002;46:3143–3150. doi: 10.1002/art.10623. [DOI] [PubMed] [Google Scholar]
- 19.Schuringa JJ, Dekker LV, Vellenga E, Kruijer W. Sequential activation of Rac-1, SEK-1/MKK-4 and PKCdelta is required for interleukin-6 induced STAT3 Ser-727 phosphorylation and transactivation. J. Biol. Chem. 2001;29:27709–27715. doi: 10.1074/jbc.M009821200. [DOI] [PubMed] [Google Scholar]
- 20.Minami M, et al. STAT3 activation is a critical step in gp130-mediated terminal differentiation and growth arrest of a myeloid cell line. Proc. Natl. Acad. Sci. U. S. A. 1996;93:3963–3966. doi: 10.1073/pnas.93.9.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novotny-Diermayr V, Zhang T, Gu L, Cao X. Protein kinase C delta associates with the interleukin-6 receptor subunit glycoprotein (gp) 130 via Stat3 and enhances Stat3-gp130 interaction. J. Biol. Chem. 2002;277:49134–49142. doi: 10.1074/jbc.M206727200. [DOI] [PubMed] [Google Scholar]
- 22.Fukada T, et al. Two signals are necessary for cell proliferation induced by a cytokine receptor gp130: involvement of STAT3 in anti-apoptosis. Immunity. 1996;5:449–460. doi: 10.1016/s1074-7613(00)80501-4. [DOI] [PubMed] [Google Scholar]
- 23.Akira S. IL-6-regulated transcription factors. Int. J. Biochem. Cell Biol. 1996;29:1401–1418. doi: 10.1016/s1357-2725(97)00063-0. [DOI] [PubMed] [Google Scholar]
- 24.Chow D, He X, Snow AL, Rose-John S, Garcia KC. Structure of an extracellular gp130 cytokine receptor signaling complex. Science. 2001;291:2150–2155. doi: 10.1126/science.1058308. [DOI] [PubMed] [Google Scholar]
- 25.Hurst SM, et al. IL-6 and its soluble receptor orchestrate a temporal switch in the pattern of leukocyte recruitment seen during acute inflammation. Immunity. 2001;14:705–714. doi: 10.1016/s1074-7613(01)00151-0. [DOI] [PubMed] [Google Scholar]
- 26.Broide DH, et al. Cytokines in symptomatic asthma airways. J. Allergy Clin. Immunol. 1992;89:958–967. doi: 10.1016/0091-6749(92)90218-q. [DOI] [PubMed] [Google Scholar]
- 27.Jones SA, Horiuchi S, Topley N, Yamamoto N, Fuller GM. The soluble interleukin 6 receptor: mechanisms of production and implications in disease. FASEB J. 2001;15:43–58. doi: 10.1096/fj.99-1003rev. [DOI] [PubMed] [Google Scholar]
- 28.Tamura T, et al. SIL-6R triggers osteoclast formation by IL-6. Proc. Natl. Acad. Sci. U. S. A. 1993;90:11924–11928. doi: 10.1073/pnas.90.24.11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kallen KJ. The role of transsignaling via the agonistic sIL-6R in human diseases. . Biochim. Biophys. Acta. 2002; 1592:323–343. doi: 10.1016/s0167-4889(02)00325-7. [DOI] [PubMed] [Google Scholar]
- 30.Rose-John S, Neurath MF. IL-6 trans-signaling: the heat is on. Immunity. 2004;20:2–4. doi: 10.1016/s1074-7613(04)00003-2. [DOI] [PubMed] [Google Scholar]
- 31.Atreya R, et al. Blockade of IL-6 trans-signaling suppresses T cell resistance against apoptosis in chronic intestinal inflammation: Evidence in Crohn’s disease and experimental colitis in vivo. Nat. Med. 2000;6:583–588. doi: 10.1038/75068. [DOI] [PubMed] [Google Scholar]
- 32.Yokoyama A, et al. Circulating levels of soluble interleukin-6 receptor in patients with bronchial asthma. Am. J. Respir. Crit. Care. Med. 1997;156:1688–1691. doi: 10.1164/ajrccm.156.5.9610070. [DOI] [PubMed] [Google Scholar]
- 33.Zhang DH, et al. Inhibition of allergic inflammation in a murine model of asthma by expression of a dominant-negative mutant of GATA-3. Immunity. 1999;11:473–482. doi: 10.1016/s1074-7613(00)80122-3. [DOI] [PubMed] [Google Scholar]
- 34.Finotto S, et al. Treatment of allergic airway inflammation and hyperresponsiveness by local antisense-induced blockade of GATA-3 expression. J. Exp. Med. 2001;193:1247–1260. doi: 10.1084/jem.193.11.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maloy KJ, Powrie P. Tregs in the control of immune pathology. Nat. Immunol. 2001;2:816–822. doi: 10.1038/ni0901-816. [DOI] [PubMed] [Google Scholar]
- 36.Belkaid Y, Piccirillo CA, Mendez S, Shevach EM, Sacks DL. CD4+CD25+Tregs control Leishmania major persistence and immunity. Nature. 2002;420:502–507. doi: 10.1038/nature01152. [DOI] [PubMed] [Google Scholar]
- 37.Pasare C, Medzhitov R. Toll pathway-dependent blockade of CD4+CD25+ T cell-mediated suppression by dendritic cells. Science. 2003;299:1033–1036. doi: 10.1126/science.1078231. [DOI] [PubMed] [Google Scholar]
- 38.Fantini MC, et al. TGF-beta induces a regulatory phenotype in CD4+CD25– T cells through Foxp3 induction and down-regulation of Smad7. J. Immunol. 2004;172:5149–5153. doi: 10.4049/jimmunol.172.9.5149. [DOI] [PubMed] [Google Scholar]
- 39.Powrie F, Maloy KJ. Regulating the regulators. Science. 2003;299:1030–1031. doi: 10.1126/science.1082031. [DOI] [PubMed] [Google Scholar]
- 40.Hori S, Nomura T, Sakaguchi S. Control of Treg development by the transcription factor Foxp3. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- 41.Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for Scarfin in CD4+CD25+ T regulatory cells. Nat. Immunol. 2003;4:337–342. [PubMed] [Google Scholar]
- 42.Fontenot JD, Gavin MA, Rudensky YA. Foxp3 programs the development and function of CD4+CD25+ regulatory cells. Nat. Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 43.O’Garra A, Vieira P. Twenty-first century Foxp3. Nat. Immunol. 2003;4:304–306. doi: 10.1038/ni0403-304. [DOI] [PubMed] [Google Scholar]
- 44.Croker BA, et al. SOCS3 negatively regulates IL-6 signaling in vivo. Nat. Immunol. 2003;4:507–509. doi: 10.1038/ni931. [DOI] [PubMed] [Google Scholar]
- 45.Akbari O, et al. Antigen-specific Tregs develop via the ICOS-ICOS-ligand pathway and inhibit allergen-induced airway hyperreactivity. Nat. Med. 2002;8:1024–1032. doi: 10.1038/nm745. [DOI] [PubMed] [Google Scholar]
- 46.Stock P, et al. Induction of T helper type 1-like regulatory cells that express Foxp3 and protect against airway hyper-reactivity. Nat. Immunol. 2004;5:1149–1156. doi: 10.1038/ni1122. [DOI] [PubMed] [Google Scholar]
- 47.National Heart, Lung, Blood Institute. International consensus report on diagnosis and treatment of asthma. Eur. Respir. J. 1992;5:601–641. [PubMed] [Google Scholar]
- 48.Erpenbeck VJ, et al. Local release of eosinophil peroxidase following segmental allergen provocation in asthma. Clin. Exp. Allergy. 2003;33:331–336. doi: 10.1046/j.1365-2222.2003.01580.x. [DOI] [PubMed] [Google Scholar]
- 49.Krug N, et al. Cytokine profile of broncho-alveolar lavage derived CD4+, CD8+ and gd T cells in people with asthma after segmental allergen challenge. Am. J. Respir. Cell Mol. Biol. 2001;25:125–131. doi: 10.1165/ajrcmb.25.1.4194. [DOI] [PubMed] [Google Scholar]
- 50.Henderson WR, Jr, et al. Blockade of CD49d (α4 integrin) on intrapulmonary but not circulating leukocytes inhibits airway inflammation and hyperresponsiveness in a mouse model of asthma. J. Clin. Invest. 1997;100:3083–3092. doi: 10.1172/JCI119863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Sanctis GT, et al. T-lymphocytes regulate genetically determined airway hyperresponsiveness in mice. Nat. Med. 1997;3:460–462. doi: 10.1038/nm0497-460. [DOI] [PubMed] [Google Scholar]
- 52.De Sanctis GT, et al. Quantitative locus analysis of airway hyperresponsiveness in A/J and C57BL/6J mice. Nat. Genet. 1995;11:150–154. doi: 10.1038/ng1095-150. [DOI] [PubMed] [Google Scholar]
- 53.Finotto S, et al. Development of spontaneous airway changes consistent with human asthma in mice lacking T-bet. Science. 2002;295:336–338. doi: 10.1126/science.1065544. [DOI] [PubMed] [Google Scholar]