Abstract
The creation of physiologically-relevant human cardiac tissue with defined cell structure and function is essential for a wide variety of therapeutic, diagnostic, and drug screening applications. Here we report a new scalable method using Faraday waves to enable rapid aggregation of human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) into predefined 3D constructs. At packing densities that approximate native myocardium (108–109 cells/ml), these hiPSC-CM-derived 3D tissues demonstrate significantly improved cell viability, metabolic activity, and intercellular connection when compared to constructs with random cell distribution. Moreover, the patterned hiPSC-CMs within the constructs exhibit significantly greater levels of contractile stress, beat frequency, and contraction-relaxation rates, suggesting their improved maturation. Our results demonstrate a novel application of Faraday waves to create stem cell-derived 3D cardiac tissue that resembles the cellular architecture of a native heart tissue for diverse basic research and clinical applications.
Keywords: human induced pluripotent stem cells, cardiac regenerative medicine, cardiomyocytes, sound wave cellular patterning
Graphical Abstract
Schematic summary of the bioengineering approach to create 3D cardiac tissue analogues. A: Culture and cardiomyocyte (CM) differentiation of human induced pluripotent stem cells (hiPSCs). B: Faraday wave patterning of hiPSC-CMs in fibrin prepolymer, generating highly-packed 3D cell construct. C: Cell encapsulation in 3D fibrin hydrogel and culture in vitro, leading to the formation of inter-connected cell bands that exhibit physiologically-relevant CM density and contractile function.
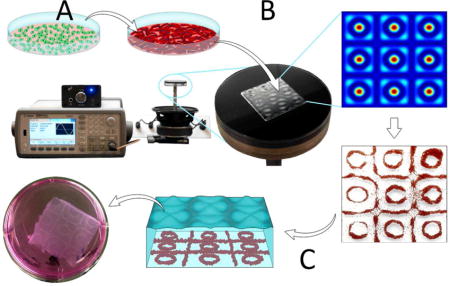
1. Introduction
The generation of three-dimensional (3D) functional tissues that can restore the structure and/or function of damaged myocardium is a central goal in cardiac regenerative medicine [1,2]. In addition, the creation of high-fidelity in vitro tissue models may improve our understanding of various biological processes including heart development, myocardial damage, and disease [2–5]. The creation of 3D tissue constructs that mimic native myocardium requires an appropriate selection of cell source and biomaterial that resembles the native tissue structure and support cell viability, function, electromechanical integration with host tissue, and vascularization [6].
The intricate structure of native myocardium is characterized by closely-packed cardiomyocytes (CMs) (e.g., 108–9 cells/cm3 in adult rat myocardium)[7] that contract synchronously via the orchestrated propagation of an electrical signal generated by specialized pacemaker cells. The challenge in restoring the structure and function of damaged heart tissue is due, at least in part, to the complexity of recreating the natural myocardial architecture. In particular, the ability to create 3D cardiac tissues with high packing density of functional CMs that are organized in a pre-defined pattern is critical to mimic the native tissue in maintaining CM viability, intercellular connections, and contractile function [8–10].
To date, majority of technologies that enable control over spatial cell arrangement in 3D tissue constructs are based on assembly of cell-encapsulating microscale hydrogels [11–14], seeding cells in scaffolds with defined architecture[15–17], and additive manufacturing / 3D bioprinting techniques [15,18,19]. While these techniques have been successfully used in a wide range of applications including bone and skin regeneration [20–22], gene delivery [23,24], and cell differentiation [21,25], they are not yet able to achieve spatially-controlled, physiologically-relevant cell packing densities, comparable to that in the native cardiac tissue [7] for cardiovascular applications.
Using the Faraday wave principle to induce patterning in liquid medium, we recently demonstrated rapid and dynamic aggregation of microscale objects (e.g., cell spheroids) into diverse sets of geometric configurations at the air-liquid interface [26,27]. In this study, by applying Faraday waves to the fibrin prepolymer, human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) were rapidly patterned (< 10 secs) into ordered, closely-packed, symmetric 3D constructs. Within the high cell density regions, hiPSC-CMs demonstrated significantly improved viability, intercellular connections, and contractile function (force and motion), in comparison to constructs with random cell distribution. Our results support the feasibility of creating 3D cardiac tissue constructs that approximate native human tissues in their cell density, structure, and function for diverse basic research and clinical applications.
2. Results
2.1 Faraday Wave Patterning of hiPSC-CMs
Experimental setup designed for this study is shown in Figure 1A and SI Figure 1. Altering hydrodynamic drag force fields tuned by Faraday wave frequency generated a variety of particle aggregation patterns (SI Figure 2). Numerical modeling of hydrodynamic drag exerted on the particles demonstrated that regions with minimum force potential overlapped with the nodal pattern of the Faraday waves (Figure 1B), resulting in an aggregation pattern consisting of circles and squares (top-down view, Figure 1C, SI Movie 1). The force potential field penetrated inside the liquid layer and decayed exponentially (cross-sectional view Figure 1D–E). Particles within the force potential field were driven away from the regions with the higher to lower force potential and closely packed together into a multilayer structure. The maximum force potential was on the surface of substrate at z=0 (Figure 1D). For this reason, we waited for 1 minute for CMs to settle down at the bottom of container, before starting the wave-patterning. Difference in force potential decreased by an increase in the thickness of the liquid medium (Figure 1D–F). Using the Equation (1) in Methods, we predicted the difference in max and min force potential (green curve in Figure 1B) as a function of liquid bath thickness. This modeling showed that for three different Faraday wavelengths (λ = 7, 13.33, and 20 mm), the force potential decreased exponentially as the liquid thickness increased (Figure 1E).
Figure 1.
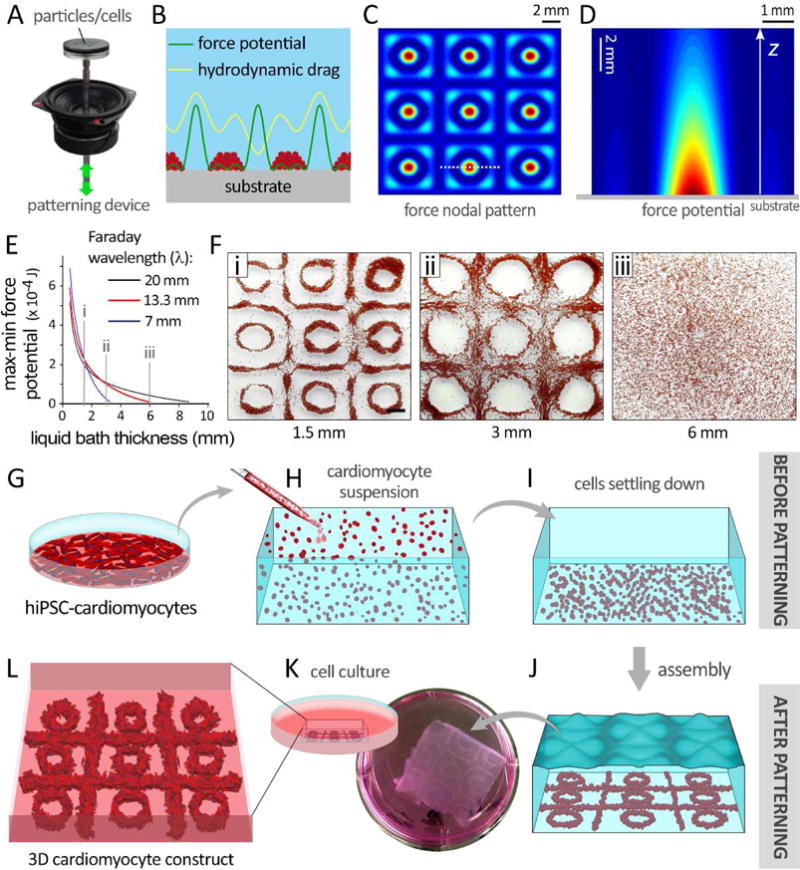
Faraday wave patterning of particles / cardiomyocytes (CMs) resulting in an aggregation pattern consisting of circles and squares. A–E: Numerical simulation of force potential for 20-μm particles (mimicking iPSC-CMs), exposed to the Faraday waves. A: The set up used for patterning the particles/cells. B: Demonstration of hydrodynamic force and corresponding force potential profiles generated by standing waves in a fluid microenvironment. Hydrodynamic drag force packed the cells at the minimum force potential regions. C: Top-down view of the force potential nodal pattern of Faraday waves simulated according to equation (1). D–E: The force potential field inside the liquid bath as a function of liquid thickness and distance from substrate, z (cross-sectional view). F: Evaluating the model by patterning particles at three different liquid thickness values (i–iii, 1.5, 3, and 6 mm). G: hiPSC-CMs preparation, H: suspension of single cells within fibrinogen prepolymer, and I: settlement of CMs at the bottom of the bath. J: CM patterning into specific 3D configurations by applying Faraday waves. K: Schematic (left) and actual (right) demonstration of fibrin gel encapsulating patterned CMs. L: Patterned CMs in fibrin scaffolds were maintained in culture media.
Our modeling was experimentally validated by formation of more loosely packed patterns of plastic beads at deeper liquid baths (1.5 to 6 mm, Figure 1F, i–iii). Based on this data, a fibrin prepolymer solution at the thickness of 1.5 mm was used to yield a compact hiPSC-CM aggregation with minimal cell dispersion (Figure 1G–L). hiPSC-CMs aggregated at the nodal patterns of the Faraday waves, where the lowest force potentials existed. Further in vitro culture of the patterned scaffolds gave rise to formation of self-organized, closely-packed 3D constructs (Figure 1K, L).
As control, random patterning with no waves generated cell clusters that showed spotty distribution within the dish (Figure 2 A–D). Application of Faraday waves (i.e., ordered patterning) resulted in formation of multiple (6–7) stacking layers of closely-packed cells (“band”) (Figure 2 E–G) and a small number of unpatterned cells in the “gap” areas (Figure 2 H). Following 5 days of culture, aggregated cells established an elongated morphology and formed packed, continuous tissue constructs in ordered patterns (Figure 2 I–L). As quantified by optical microscopy at day 0, a significantly greater cell packing density was obtained within the cell bands (8,000 cells/mm2) in comparison to those in random patterns and in the gap areas (n=10, Figure 2 M).
Figure 2.
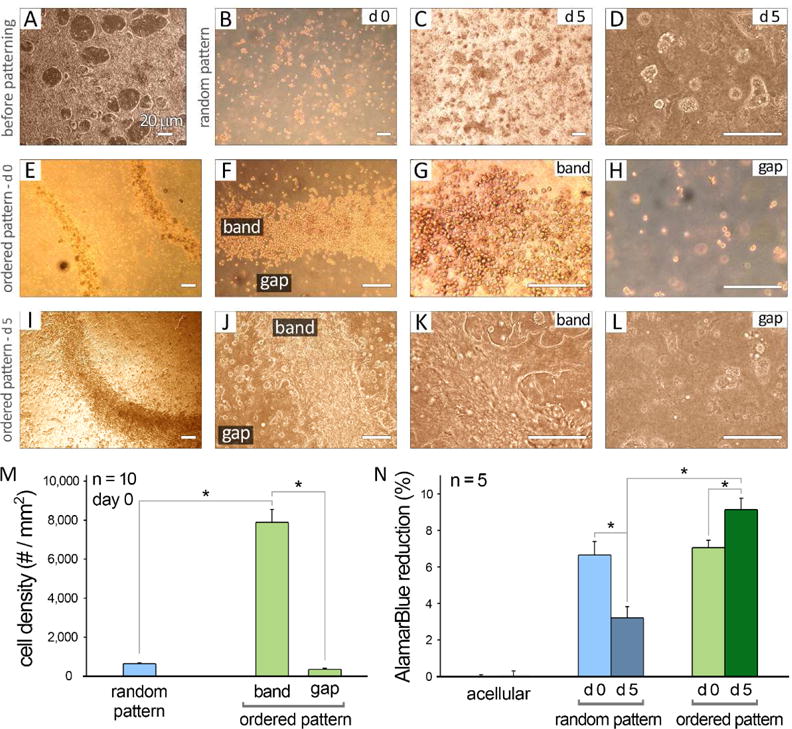
Optical microscopy of human iPSC-CMs in 2D culture prior to patterning (A), and after patterning to either random (B–D) or ordered structures (E–L) at days 0 (d 0) and 5 (d 5) in culture. Scale bars show 20 μm. M: Quantification of cell packing density in various experimental groups at day 0 of culture (n = 10). N: AlamarBlue® reduction percentage was measured for different groups at days 1 and 5 in culture (n = 5). Hydrogels without cells (acellular) were used as control. Error bars are S.E.M. *: P<0.05 was considered statistically significant.
To quantify hiPSC-CM survival after encapsulation in 3D fibrin hydrogels, we performed an Alamar Blue™ reduction assay and showed that highly-packed cells exhibited a significant increase in cell metabolic activity from day 0 (right after patterning) to day 5 of culture (n=5/group/time point, Figure 2 N). This was in contrast to randomly distributed cells in 3D fibrin gels (random pattern) showing significantly decreased cell metabolism.
2.2 Scanning Electron Microscopy of Patterned Constructs
Scanning electron microscopy of hiPSC-CMs in random and ordered pattern groups after 5 days in culture provided a detailed view of cells within the bands versus randomly dispersed cells (SI Figure 3). hiPSC-CMs within the bands were closely-packed while cells in the gap areas were sparse. This is further illustrated by cells at the periphery of the band that were oriented indiscriminately to both perpendicular and parallel directions to the band axis (as indicated by yellow and green arrows in panel E).
2.3 Immunohistochemical Analysis of Patterned Constructs
Cells encapsulated in random, cell band, and gap areas within 3D fibrin gels were immunostained for multiple cardiac markers at day 5 of culture (Table 1, Figure 3). Confocal microscopy and 3D reconstruction demonstrated the continuous presence of interconnected hiPSC-CMs at a vertical height of ~300 μm for the band area within fibrin hydrogel (SI Figure 4 and SI Movie 2–3). While cells in the random patterns exhibited disorganized α-actinin expression and sarcomeric pattern (Figure 3B–C), a majority of the hiPSC-CMs in the bands showed elongated shape, strong α-actinin expression (Figure 3D–F), and organized sarcomeric structure (white arrows, Figure 3F and SI Figure 5). α-actinin+ cells in the gap areas were sparse and disorganized similar to those in the random patterns (Figure 3G–I). This was further quantified for both total cell density and the α-actinin expressing cell density at day 5 in culture (n=10, Figure 3J). Moreover, CMs within the band regions showed significantly higher levels of α-actinin expression compared with the other two groups (n=10, Figure 3K).
Table 1.
List of antibodies used for the immunohistochemical analysis of hiPSC-derived cardiomyocyte and non-cardiomyocyte populations.
| Target | Antibody | Company — Catalog number |
|---|---|---|
| cardiomyocytes | oactinin | Sigma-Aldrich (A7811) |
| intercellular connection | connexin-43 | Abcam (ab63851) |
| nuclei | DAPI | Thermo Fisher (R37606) |
| endothelial cells | CD31 | Abcam (ab28364) |
| mesenchyme/fibroblasts | vimentin | Abcam (Ab92547) |
| smooth muscle cells | smooth muscle myosin heavy chain (SM-MHC) | Abcam (ab53219) |
| cell membrane | wheat germ agglutinin (WGA) | Thermo Fisher (W11261) |
| undifferentiated iPSCs | nanog | Abcam (ab21624) |
| endoderm | alpha fetoprotein (AFP) | Santa Cruz Biotechnology (sc-8108) |
| neurons | MAB377 (NeuN) | Sigma-Aldrich (MAB377) |
Figure 3.
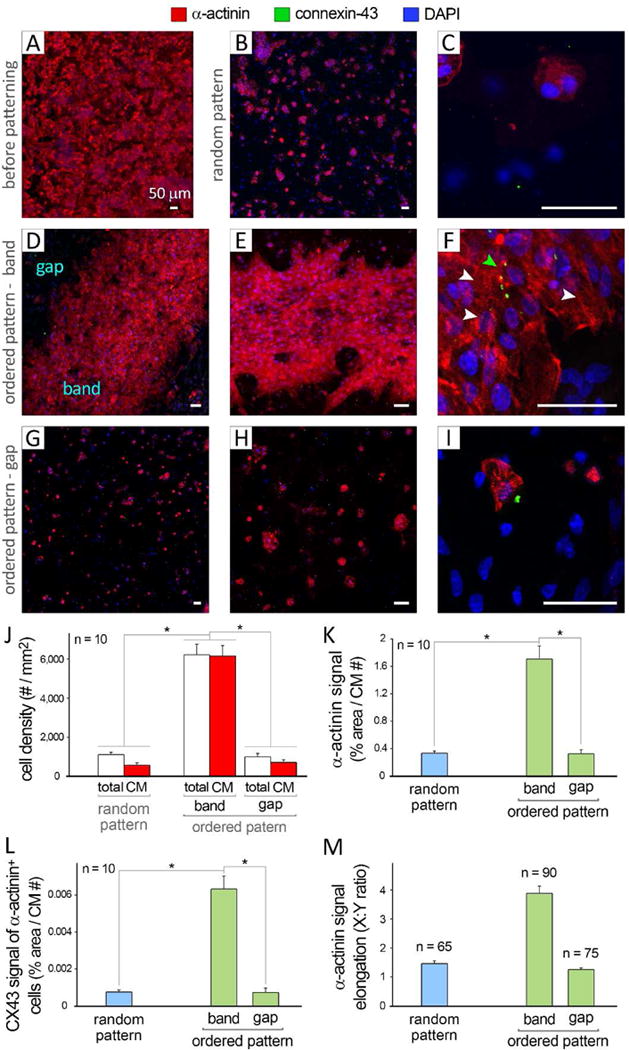
Immunohistochemical analysis of human iPSC-CMs in 2D culture prior to patterning (A) and after patterning to random (B–C) and ordered (D–I) structures at day 5 in culture. Ordered patterns consisted of closely-packed CMs in the “band” regions and randomly distributed CMs in the “gap” areas. Scale bars show 50 μm. Green arrow in panel F highlights CX43+ cells. J: Quantification of packing density of total cells and CMs in various experimental groups at day 5 of culture, based on immunostaining images (n = 10). K: Quantification of α-actinin signal in different groups, normalized by CM number (n = 10). L: Quantification of CX43 signal of CMs, normalized by CM number (n = 10). M: Quantification of α-actinin signal elongation (X:Y ratio) among different groups (n = 65–90). Error bars are SEM. *: P<0.05 was considered statistically significant.
Co-immunostaining for connexin-43 (CX43, green) revealed a significantly greater number of intercellular connections in CMs residing in the cell bands in comparison to the gap and random pattern groups (green arrow in Figure 3F and SI Figure 5, and quantified in Figure 3L) (n=10). Densely packed CMs in the bands demonstrated a more elongated α-actinin expression (X:Y ratio, n=65, 90, and 75 for random, band, and gap samples, respectively) (Figure 3M).
To examine the purity of the hiPSC-CM encapsulated in the band area, we assessed for the presence of non-CMs by immunostaining for mesenchyme/fibroblasts (vimentin, Figure 4A–J), smooth muscle cells (smooth muscle myosin heavy chain, SM-MHC), undifferentiated iPSCs (nanog) (Figure 4K–Q), endothelial cells (CD31), neurons (MAB377, NeuN), and endoderm (alpha fetoprotein, AFP) in the band and gap areas (SI Figure 6). As quantified in Figure 4J,Q (n=10), the random pattern group showed the highest proportion of mesenchymal/fibroblasts (i.e., vimentin+ α-actinin−) and SM-MHCs, while the cell band areas showed the lowest. To account for varying cell density among different groups, signal quantifications were normalized by dividing by the cell number (Figure 4J,Q). Interestingly, no cell positive for either NeuN, nanog, or CD31 was detected among all groups. The only exception was the detection of an extremely small population of AFP-expressing cells in all three conditions (SI Figure 6).
Figure 4.
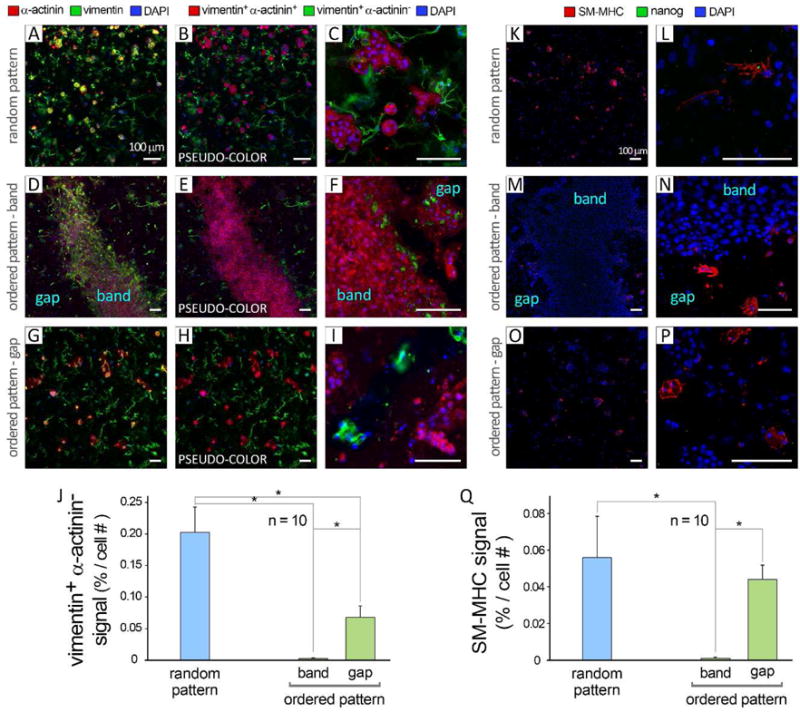
Immunohistochemical analysis of patterned human iPSC-derived cells (non-cardiomyocyte population) at day 5 in culture. A: Randomly patterned cells stained for cardiomyocytes (α-actinin, red), mesenchyme/fibroblasts (vimentin, green), and nuclei (DAPI, blue). Panels B–C show “pseudo colors” for vimentin+ α-actinin+ (red), vimentin+ α-actinin− (green) and DAPI (blue) signals, obtained by digital subtraction of red from green color in (A). D and G: Original confocal images of band and gap regions in ordered pattern group, respectively. E–F and H–I: Corresponding processed images with pseudo colors for band and gap, respectively. Scale bars show 100 μm. J: Quantification of vimentin+ α-actinin− (red) signal in various experimental groups at day 5 of culture, based on immunostaining images (n = 10). K–P: Immunostaining of random (K–L) and ordered (M–P) pattern groups for smooth muscle cell (smooth muscle myosin heavy chain, SM-MHC, red), undifferentiated iPSCs (nanog, green), and nuclei (DAPI blue). Q: Quantification of SM-MHC signal normalized by total cell number (n = 10). Error bars are SEM. *: P<0.05 was considered statistically significant.
2.4 Functional Assessment of iPSC-CMs
To assess the functional properties of iPSC-CMs in 3D fibrin hydrogels, we first measured the compressive modulus of the hydrogels and found that this was roughly 239 ± 43 Pa (see SI). To determine the level of contraction of hiPSC-CMs, we performed both qualitative (Figure 5 A–C, SI Movie 4–12) and quantitative (n=10, Figure 5 D–G) video-assisted motion analysis on beating CMs at day 5 after Bioacoustic patterning. The hiPSC-CMs in the cell bands showed significantly greater levels of tangential stress (an index of contractile force), maximum contraction-relaxation rate (velocity), and beating rate, when compared to the cells in random and gap areas. Moreover, while synchronous sequential contractions were observed for CMs from adjacent areas within the cell bands, random and gap CMs showed nonsynchronous beating (Figure 5 H–K, SI Movie 4–12).
Figure 5.
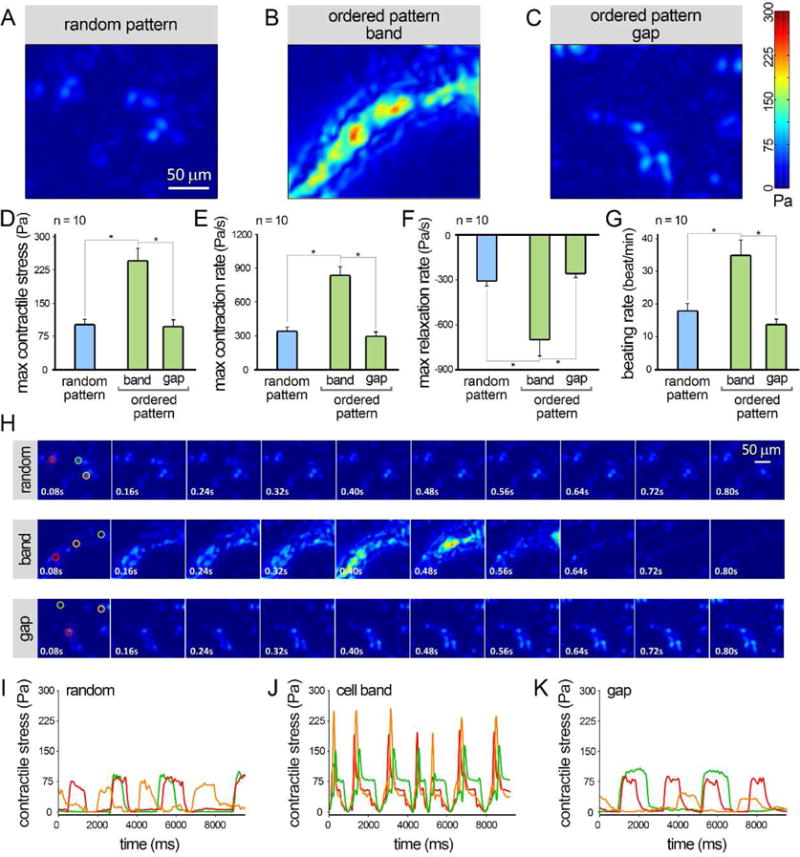
Contractile function analysis of the human iPSC-CMs encapsulated in 3D fibrin hydrogels at day 5 of culture. A–C: Heatmaps of max contractile stress in random pattern (A) and in ordered pattern — band (B) and gap (C) regions, obtained from motion analysis of CMs. D–G: Quantification of max contractile stress (D), max contraction and relaxation rates (E–F), and beat per minute (G) for various groups (n = 10). H: Representative temporal series of contractility distribution profile throughout one contraction cycle of hiPSC-CMs. I–K: Contractile stress signals captured from 3 arbitrary spots from each experimental group, demonstrating synchronous sequential contractions of CMs from adjacent areas within the cell bands, versus nonsynchronous beating behavior of random and gap CMs. Error bars are S.E.M. *: P<0.05 was considered statistically significant.
3. Discussion
A variety of techniques have been employed to create 3D cardiac tissue constructs from pluripotent stem cells. Earlier approaches have seeded high concentrations of stem cell-derived CMs onto the surface of pre-fabricated scaffolds, with the expectation that these CMs would penetrate into the scaffold to create 3D cellular construct. Examples include seeding CMs at densities of 0.03–3 ×108 cells/ml onto alginate scaffolds [28], 4×107 cells/ml onto gelatin mesh [29,30], and 2×106 cells/ml onto gelatin-collagen scaffolds [31]. In more recent years, scaffold-free methods such as spheroids (e.g., hanging drop at 1–6×104 cells/ml) [32–35] or monolayer CM sheets (e.g., 1.2×107 cells/cm2) [36] have also been tried to create CM constructs. However, current technology supports the use of 3D hydrogels to encapsulate CMs prior to polymerization. Examples of this approach include casting fibrin gels containing CMs at 5×106 cells/ml [37] or 3D printing of rat myoblasts encapsulated in decellularized tissue at 1–5×106 cells/ml [18]. While majority of these techniques create cellular scaffolds, the lack of normal contractile function in these engineered tissues is due to the fact that the cell densities of these constructs are significantly lower than that of the native myocardium.
Other approaches have aimed to simultaneously pattern multiple cardiac cells [35,38,39]. Saini et al. used micropatterning technique to create 3D cardiac microtissues by encapsulating CMs and cardiac fibroblasts in gelatin methacrylate (1:1 or 2:1 CM to fibroblast ratio, ~25×106 total cells/ml). By precisely controlling of the geometrical features and aspect ratio of the microtissues, they demonstrated that the co-culture condition improved CM network formation, structure, and contractile function [38,39]. In another study, geometric confinement and condensation of hiPSCs, induced by polyethylene glycol micropatterns, were used to self-organize functional hiPSC-CM microchambers [39]. Remarkably, the biophysical cues provided by different patterns generated spatially distinct cell densities via cell condensation, resulting in CM differentiation of hiPSCs in the center, while cells turned into myofibroblasts on the perimeter [39].
Here, we applied Faraday standing waves with differential force potential fields in a liquid layer to bioengineer highly-packed hiPSC-CMs with defined geometric pattern prior to fibrin hydrogel encapsulation. We previously demonstrated that microscale bio-entities can be aggregated into highly diverse geometric patterns at the air-liquid interface by tuning frequency, amplitude and boundary condition of Faraday waves [27]. However, those studies only demonstrated the generation of 2D monolayer structures. It remained unclear if this approach involving bottom up tissue engineering is capable of generating 3D constructs. Here, we demonstrate the feasibility of utilizing Faraday waves to generate 3D patterns by tuning the hydrodynamic drag force inside the liquid layer (SI Movie 2–3). The regions of minimum force potential generated by Faraday waves acted as traps to aggregate individual hiPSC-CMs into closely-packed 3D constructs (Figure 1).
We also developed a theoretical model (Equation 1 in Methods) to predict the distribution of the force potential under various frequency, amplitude, and cellular physical properties (i.e., cell size and density) and liquid properties (i.e., kinematic viscosity and density) (Figure 1 B–E). Furthermore, we showed that the relative position of particles (e.g., beads or cells) to the bottom of the liquid bath (z, Figure 1D) and the thickness of liquid bath (Figure 1E–F) were the key factors in determining the diffusivity of the constructs. Increase in the liquid layer thickness was associated with a decrease in force potential difference, resulting in formation of more loosely packed constructs. By allowing the iPSC-CMs to re-distribute themselves at the bottom of the fibrinogen prepolymer according to the force potential differences, we created 3D tissue constructs with significantly greater cell packing density.
The geometry of cellular architecture is dependent on the nodal configuration of Faraday waves which can be highly diverse [27]. Faraday waves usually take symmetric and periodic patterns, mainly determined by the wave vibrational frequency and amplitude as well as the boundary geometry of liquid container. Therefore, only symmetric cellular patterns can be generated by this technique. The feature size of the Faraday wave pattern is determined by half wavelength of the Faraday waves and can be tuned by the vibrational frequency according to the dispersion equation of Faraday waves [40]. The feature size of the cellular architecture (i.e., the cell band width) is also determined by both Faraday wavelength and the number of cells in the system. In terms of spatial resolution, the cellular bands can be as narrow as the width of a single line of packed cells (SI Figure 7). The overall size of the patterned construct is usually determined by the size of the liquid chamber (we have successfully patterned in chamber sizes of 10 to 30 mm). The optimal particle size at which maximum aggregation can be achieved, depends on the wave field, the particle density, the fluid thickness, and the fluid properties [41]. Patterning particles/cells at smaller sizes becomes challenging as hydrodynamic drag force (the driving force for patterning) decreases linearly with the radius of particle and hence, can be balanced by the Brownian motion. Another limitation inherent to this technique is the specific range of hydrogel viscosity (< 10 mPa.s) that is required for inducing the cell patterning and subsequent gelation in timely manner. Following the addition of thrombin to the fibrinogen, there is a narrow time window (~4 minutes) to apply the Faraday waves and generate the desired pattern, before the polymer becomes viscous.
While a wide variety of patterns were created by this approach (SI Figure 2), the specific CM aggregation pattern selected in this study consisted of a more complex and diverse combination of forms and geometries (lines-squares and circles) (Figure 1). We plan to study the potential effect of other geometric patterns on hiPSC-CM viability and function in our future work. Moreover, applying mechanical conditioning to promote CM alignment within the highly-packed patterns [42] can advance this technology to engineering biomimetic 3D cardiac scaffolds. This patterning approach can also be applied to other cell types (e.g., primary rat hepatocytes) to generate similar organized structures (SI Figure 8), suggesting the broad applicability of this technique to generate diverse cellular/tissue constructs.
The 3D packing density of hiPSC-CMs quantified immediately after patterning (day 0) was within the range of that of native myocardium (108–109 cells/ml). An in vitro CM density of 8,000 cells/mm2 can be converted to volumetric density of ~0.4 × 109 cells/ml, taking into account thickness of each iPSC-CM layer (~20 μm) (SI Figure 9). Remarkably, this packing density remained high following 5 days of in vitro culture (6,000 cells/mm2 or ~0.3 × 109 cells/ml) (Figure 3J). The slight reduction in cell density over time could be attributed to: 1) expansion of CM size and morphology after plating (Figure 2I–J), 2) migration of a small number of cells from the highly-populated cell bands towards the gap areas, and 3) expansion of a small number of non-CMs in the bands. It is interesting to note that cells found at the periphery of bands were largely non-CMs (e.g., vimentin+ α-actinin− or SM-MHC+ α-actinin− cells) (Figure 4D–F and M,N) suggesting their exclusion from the cell bands which may be explained by the differences in cell size/mass [41,43].
While a number of studies have shown that it is challenging to maintain viable, metabolically active CMs within 3D scaffold systems [2,28,44], we showed, in this study, that ordered patterning significantly improved hiPSC-CMs survival for 5 days in 3D culture (Figure 2N). This was confirmed by immunohistochemical analysis (Figure 3) that demonstrated greater cell density and intercellular gap junctions in the ordered pattern group. Moreover, CMs within the bands exhibited more elongated shape and partial organization of sarcomeric structure as demonstrated by α-actinin staining (white arrows in Figure 3F and SI Figure 5). 3D culture of such thick (>1mm) 3D constructs with high cell packing densities for longer duration may require utilizing a perfusion bioreactor or incorporating porosity/vasculature within the construct in order to alleviate the limited oxygen/nutrient diffusion.
Our finding that α-actinin+ CMs constituted ~98% of cells within the cell band region was highly encouraging given our interest in generating interconnected cardiac tissues. Noticeably greater levels of CX43 expression within the band versus gap or random conditions was expected, considering the significant differences in the CM packing densities. Interestingly, we noted that CM purity was significantly lower in random (46%) and gap (66%) regions. The higher CM enrichment within the cell bands can be attributed to significantly greater cell density which may foster the development of cell-cell connection (i.e., expression of CX43) within the bands (Figure 3L). We believe this is responsible for enhanced CM survival in our study and perhaps in other studies as well [45]. The presence of primarily non-CMs outside the bands may be a result of exclusion by CMs, a hypothesis that we are actively pursuing. This would explain the observed higher CM density in the bands (Figure 2E–H and Figure 4). Possible mechanisms for the exclusion of non-CMs from the band regions include differential cell size/mass during the Faraday wave patterning process and/or the secretion of repelling signals by CMs to prevent incorporation of non-CMs within the cell bands. Characterization of non-CM cell population revealed that hiPSC-derived mesenchymal/fibroblasts and smooth muscle cells constituted the majority of these cells (Figure 4). No evidence of endothelial or neuronal cells were detected among these groups.
CMs residing in the high-density cell bands exhibited significantly greater contractile stress and faster beating (contraction-relaxation velocity and beating rate), in comparison to gap and randomly patterned CMs (Figure 5A–G). The maximum contractile stress exerted by CMs ranged from ~90–95 Pa in the random and gap regions and further increased (~2.5 folds) to 244 ± 31 Pa in the cell bands. These values can be compared with the contractile stress measured for hiPSC-CMs. For instance, we previously found that the maximum contractile stress of human iPSC-CMs at different stages of maturation (days 30–60) ranged from ~80–120 Pa [46]. This is similar to findings by another group who measured the peak twitch stress levels of ~45–60 Pa for single CMs cultured on 2D fibronectin, laminin, and collagen IV substrates [47]. These numbers can be compared with the contractile stress generated in the mouse heart ranging from ~360 Pa in the early embryo (e10.5), to ~3,000 Pa in neonatal heart, and a maximum of ~6,000–8,000 Pa in the adult heart [48,49]. Additionally, within the bands CMs exhibited ~3X higher contraction/relaxation rates and ~2X greater beating rate than those of gap/random cells (Figure 5E–G). Connected dense CMs within the cell bands also demonstrated synchronized conduction of action potential and contractility along the band axis (Figure 5H–K and SI Movie 4–6). Inducing CM alignment, increasing CM purity, and generating more uniform packing densities within the band, could further improve the contractile function of patterned constructs. While the CMs within the bands showed some signs of being more mature (sarcomeric actinin structure, enhanced contractile function, and intercellular connectivity) than unpatterned cells, they did not exhibit the characteristic elongated morphology of mature CMs (Figure 3 and SI Figure 5).
While a number of other bioengineering approaches are actively being utilized to create patterned cellular constructs, the main advantages of this bioacoustic patterning technique include: i) the unique ability to generate physiologically-relevant cell densities (e.g., 108–9 CMs/ml) in 3D macro-scale structures. in macroscopic (centimeter) scales; ii) in contrast to other bioengineering approaches (e.g., 3D bioprinting [50,51] and micro-organoid assembly [52,53]), this technique allows rapid, simple, and in situ patterning of various cells into diverse, highly-packed 3D patterns. Thus, this Faraday-wave patterning technology enables conducting a variety of basic research studies (e.g., disease/drug modeling) in large-scale models in a high-throughput manner. Moreover, highly-packed CM bands can be dissected out of the construct and used as in vitro models or as cardiac patch for in vivo applications. Main 3D design parameters that can be controlled by this approach include: cell packing density, geometric pattern (shape) of the bands, band width and spacing (gap) (SI Figure 2).
4. Conclusion
In summary, the novel procedure described here provides a unique platform for the preparation of stem cell-derived complex 3D cardiac tissue constructs with predefined cellular architecture and organization. These tissue engineered constructs approximate biologically-relevant CM density and functionality and can be of great benefit to studies aimed at understanding human CM behavior in a more physiological context. In the future, they may enable repair and regeneration of damaged hearts by providing a source of cells and a favorable architecture for therapeutic remuscularization.
5. Experimental Section
Preparation, Culture, and Chemically-Defined CM Differentiation of hiPSCs
The protocols used in this study were approved by the Stanford University Institutional Review Board. Reprogramming of peripheral blood mononuclear cells was performed using a Sendai virus vector as previously described [54]. One month post reprogramming, the obtained hiPSC clones were isolated and cultured on 6-well tissue culture plates (Greiner) coated with growth factor-reduced Matrigel (Corning). E8 pluripotent stem cell culture medium (Life Technologies) was used for iPSC culture.
Once a confluency of ~80–90% in the hiPSC culture was achieved, cells were differentiated into human CMs utilizing a chemically-defined cardiomyocyte differentiation protocol [55]. CDM3 culture media (RPMI 1640 supplemented with recombinant human albumin and ascorbic acid) was used [55]. Differentiation started by treating hiPSCs with a small molecule inhibitor of GSK3B signaling, CHIR99021, in CDM3 media for 2 days. The CHIR factor activates the Wnt signaling pathway. Cells were subsequently treated with CDM3 media supplemented with a Wnt signaling inhibitor, Wnt-C59, for another 2 days. At days 4–8 of differentiation, CDM3 media without any factors was used and changed every other day. To improve the CM purity, cells were glucose-starved by culturing in CDM3 media without glucose, supplemented with 5 mM sodium DL-lactate, for 4 days [56]. For plating hiPSC-CMs into 3D hydrogels, monolayers of beating CMs at day 16–18 of differentiation were dissociated into single cells using TrypLE Express (Life Technologies) for 3–5 minutes.
Preparation of 3D Fibrin Hydrogels — Encapsulation of hiPSC-CMs
We utilized the hydrodynamic drag force, generated by Faraday waves at the liquid surface, to pattern cells into predetermined configuration. A square-shaped liquid container (20 mm × 20 mm × 1.5 mm) with brim-full fibrin prepolymer solution was mounted onto a vertical vibration generator (U56001, 3B Scientific) (SI Figure 1). The container was carefully tuned and balanced using a bubble level (Spirit Level, Hoefer) as a reference. The vibration generator was driven by a power amplifier (Lepai LP-2020A+, Parts Express) and controlled by a function generator (33500B, Keysight Technologies) to create vertical vibration on the liquid layer (Figure 1A). When the vertical vibration exceeded the threshold of the hydrodynamic instability, [57] Faraday waves were generated at the liquid-air interface. The pattern of the hydrodynamic force corresponds to the pattern of the Faraday waves and could be flexibly tuned by the vertically vibrational frequency and amplitude via the function generator [41].
Once dissociated from culture plates, hiPSC-CMs were resuspended in fibrin prepolymer solution (550 μL of 5 mg/ml fibrinogen mixed with 50 μL of 2.5 UN/ml thrombin in HBSS with calcium and magnesium) on ice, at a density of 1.7 × 106 cells/ml. The cellular solution was then cast into the liquid container and gravitationally sedimented (1 min) onto the glass coverslip at the bottom of the chamber (Figure 1G–I). By applying the Faraday waves (10 s for 5 times with intervals of 10 s), randomly-distributed cells at the substrate were driven by the hydrodynamic drag field and patterned into a 3D construct (Figure 1J–L). We chose the circle-square hybrid pattern excited at 127 Hz and 110 mV via a waveform generator for the CM patterning.
The force potential, U, experienced by a CM with a radius R at the substrate exposed to a hydrodynamic drag field can be described as follows:
| (1) |
where ρcell is the density of the cell, ρliq is the density of liquid, ν is the kinematic viscosity of the liquid, h is the height of Faraday wave, λ is the Faraday wavelength, ω is the Faraday wave frequency, k is the Faraday wavenumber, H is the thickness of the liquid, δ is the Stokes characteristic length and ζsh is the sub-harmonic component of deformation of liquid surface. ζh and ζsh describe wave functions and are tuned by ω.
Following the patterning, an additional 25 μl of thrombin (2.5 UN/ml) was added to the cast solution to accelerate the crosslinking of fibrinogen. Crosslinking was completed by incubating the patterned constructs in a tissue culture incubator (37°C) for 10 minutes. Cell encapsulating fibrin scaffolds were next detached from the liquid container and transferred to individual wells of 6-well plates. The 3D cellular constructs were cultured in vitro for 5 days. To inhibit cell-induced fibrin degradation, 1 TIU/mL aprotinin (A1153, Sigma-Aldrich) was added to the culture media for the duration of experiments [58].
Encapsulation of Primary Rat Hepatocytes in 3D Patterned Hydrogels
Primary rat hepatocytes were obtained from Dr. Usta’s Lab (Center for Engineering in Medicine at Massachusetts General Hospital). Isolation of primary rat hepatocytes was conducted as previously described [59] and approved by the Subcommittee on Research Animal Care, Committee on Research at the Massachusetts General Hospital. To prepare the construct, 1 M rat hepatocytes were suspended in the mixture solution of 550 μL fibrinogen (10 mg/mL) and 50 μL thrombin (2.5 UN/mL) and loaded into a liquid container. Once cells settled down (1 min) onto the substrate, Faraday waves were applied to the liquid container at 127 Hz and 110 mV (10 s for 5 times with 10-s intervals) to generate cellular construct. After patterning, another 50 μL thrombin (2.5 UN/mL) was added to the liquid container. Gelation was completed by transferring the liquid container to cell incubator (37°C) for 10 mins.
Optical and Immunohistochemical Analysis of Patterned Constructs
Bright field microscopy of 3D constructs was conducted using a Leica Microscope (DFC 500) at 5, 10, and 20= magnifications. For immunofluorescence staining, 3D fibrin scaffolds seeded with hiPSC-CMs were rinsed (3 × 20 mins with DPBS), fixed with 4% paraformaldehyde (Electron Microscopy Services) for 60 mins on rocker at room temperature, rinsed (3 × 20 mins), and permeabilized with 0.1% saponin (Sigma-Aldrich) for 60 mins at room temperature with gentle agitation. CMs in 3D constructs were next blocked in 10% goat serum (Sigma-Aldrich), 0.1% saponin, and 1% BSA solution (blocking solution) for 3 hrs on rocker at room temperature. Samples were then drained and stained with 1:200 dilution of primary antibodies overnight at 4 °C in the blocking solution. Primary antibodies used: mouse anti-sarcomeric α-actinin (A7811, Sigma-Aldrich), mouse anti-α-smooth muscle actin (A2547, Sigma-Aldrich), rabbit anti-vimentin (ab92547, Abcam), rabbit anti-nanog (ab21624, Abcam), rabbit anti-CD31 (ab28364, Abcam), goat anti-AFP (sc-8108, Santa Cruz Biotechnology), and mouse anti-NeuN (MAB377, Sigma-Aldrich). The next day, constructs were washed (4 × 30 mins) with DPBS on the rocker and then incubated for 3 hrs at room temperature in the dark with 1:200 secondary antibodies in the blocking solution. Secondary antibodies included: Alexa Fluor 488 goat anti-rabbit and anti-mouse, Alexa Fluor 568 goat anti-mouse, Alexa Fluor 546 goat anti-mouse, Alexa Fluor 647 goat anti-rabbit, and Alexa Fluor 594 donkey anti-goat (all Life Technologies). Subsequently, constructs were washed (3 × 30 mins with DPBS) and cell nuclei were stained with NucBlue Fixed Cell Stain (Life Technologies) in DPBS. Confocal imaging was performed with an LSM510Meta Confocal Microscope (Zeiss). 3D reconstruction and panoramic videos of the Z-stacks was generated by the confocal microscope software (ZEN 2012, Carl Zeiss Microscopy GMBH, MA) (SI Figure 4 and SI Movie 2–3). Cell density measurements using optical images were conducted by counting the number of cells in each focal plane (in focus). A second cell density quantification was performed by counting (DAPI-stained) cell nuclei for total cell number and counting α-actinin+ DAPI-stained cells for the CM number. A total number of n=10 samples were analyzed for each quantification.
hiPSC-CM Metabolic Activity Assay
AlamarBlue (Invitrogen), a metabolic activity assay, was used to measure the number of hiPSC-CMs in the 3D constructs as an indicator of cell viability and growth. At different time points in culture, the medium (CDM3) was renewed and AlamarBlue reagent was added to each well at 10% of the culture volume. Cellular constructs were then incubated at 37°C for 4 hrs. Acellular fibrin gels were used for background reference subtraction. The absorbance of 100 μL of medium was read at 550 and 600 nm using a microplate reader (Cytation 5 Cell Imaging Multi-Mode Reader, BioTek Instruments) and the percentage of reduced AlamarBlue was calculated according to the manufacturer’s instructions.
Mechanical Testing of Fibrin Hydrogels
Unconfined compression tests were conducted using an Instron 5944 material testing system (Instron) fitted with a 10 N load cell (Interface). The test set-up consisted of custom-made aluminum compression platens lined with PTFE to minimize friction. All tests were conducted in PBS solution at room temperature. Specimen diameter and thickness were measured using digital calipers and the material testing system’s position read-out, respectively. Before each test, a preload of approximately 1 mN was applied. The upper platen was then lowered at a rate of 1% strain/sec to a maximum strain of 40%. Load and displacement data were recorded at 100 Hz. The compressive modulus was determined for strain range of 10%–20% from linear curve fits of the stress vs. strain curve.
Motion Analysis of Beating Cardiomyocytes
The videos of beating CMs encapsulated in 3D fibrin gels were captured at day 5 of culture, using a Leica Microscope (DFC 500) with 10X objective at a speed of 12.5 frame per second. The raw videos were transformed to 8 bit gray scale and then analyzed using an image velocimetry script in MATLAB (MathWorks) as previously described [46]. Contractile stress values were calculated from these video recordings, based on the stiffness and depth of material, and the cell displacement, which is relative to the reference frames at resting states between contractions.
Statistical Analysis
The number of samples (n) used in each experiment is recorded in the text and shown in figures. All data are presented as mean ± SEM. Cell density and signal quantifications were all conducted in a blind manner. All statistical analysis was assessed using a one-way ANOVA with Tukey’s post hoc evaluation. For all comparisons, P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by NIH Pathway to Independence Award 1K99HL127295-01A1 (to V.S.), CIRM RT3-07798 (to J.C.W.), NIH Progenitor Cell Biology U01 HL099776 (to J.C.W. and S.M.W.), NSF CAREER CBET 1461602, NSF 1547791, R15 HL115556, and DoD W81XWH-15-1-0576 (to U.D.), NIH Director’s Pioneer Award DP1 LM012179-01, and the Endowed Faculty Scholar Award of Lucile Packard Foundation for Children and Child Health Research Institute (to S.M.W.). We thank Dr. A. Olson (Director of the Stanford Neuroscience Microscopy Service) for providing consult in immunohistochemical imaging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
None
References
- 1.Curtis MW, Russell B. Cardiac tissue engineering. J Cardiovasc Nurs. 2009;24:87–92. doi: 10.1097/01.JCN.0000343562.06614.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirt MN, Hansen A, Eschenhagen T. Cardiac Tissue Engineering State of the Art. Circ Res. 2014;114:354–367. doi: 10.1161/CIRCRESAHA.114.300522. [DOI] [PubMed] [Google Scholar]
- 3.Ralphe JC, de Lange WJ. 3D engineered cardiac tissue models of human heart disease: learning more from our mice. Trends Cardiovasc Med. 2013;23:27–32. doi: 10.1016/j.tcm.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mazzoleni G, Di Lorenzo D, Steimberg N. Modelling tissues in 3D: the next future of pharmacotoxicology and food research? Genes Nutr. 2009;4:13–22. doi: 10.1007/s12263-008-0107-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kharaziha M, Memic A, Akbari M, Brafman DA, Nikkhah M. Nano-Enabled Approaches for Stem Cell-Based Cardiac Tissue Engineering. Adv Healthc Mater. 2016;5:1533–1553. doi: 10.1002/adhm.201600088. [DOI] [PubMed] [Google Scholar]
- 6.Vunjak-Novakovic G, Tandon N, Godier A, Maidhof R, Marsano A, Martens TP, Radisic M. Challenges in Cardiac Tissue Engineering. Tissue Eng Part B-Rev. 2010;16:169–187. doi: 10.1089/ten.teb.2009.0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandarim-De-Lacerda CA, Meirelles Pereira LM. Numerical density of cardiomyocytes in chronic nitric oxide synthesis inhibition, Pathobiol. J Immunopathol Mol Cell Biol. 2000;68:36–42. doi: 10.1159/000028113. [DOI] [PubMed] [Google Scholar]
- 8.Radisic M, Euloth M, Yang L, Langer R, Freed LE, Vunjak-Novakovic G. High-density seeding of myocyte cells for cardiac tissue engineering. Biotechnol Bioeng. 2003;82:403–414. doi: 10.1002/bit.10594. [DOI] [PubMed] [Google Scholar]
- 9.Carrier RL, Papadaki M, Rupnick M, Schoen FJ, Bursac N, Langer R, Freed LE, Vunjak-Novakovic G. Cardiac tissue engineering: cell seeding, cultivation parameters, and tissue construct characterization. Biotechnol Bioeng. 1999;64:580–589. doi: 10.1002/(sici)1097-0290(19990905)64:5<580::aid-bit8>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 10.Guven S, Chen P, Inci F, Tasoglu S, Erkmen B, Demirci U. Multiscale assembly for tissue engineering and regenerative medicine. Trends Biotechnol. 2015;33:269–279. doi: 10.1016/j.tibtech.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gurkan UA, Fan Y, Xu F, Erkmen B, Urkac ES, Parlakgul G, Bernstein J, Xing W, Boyden ES, Demirci U. Simple Precision Creation of Digitally Specified, Spatially Heterogeneous, Engineered Tissue Architectures, Adv. Mater. Deerfield Beach Fla. 2013;25:1192–1198. doi: 10.1002/adma.201203261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu F, Wu CAM, Rengarajan V, Finley TD, Keles HO, Sung Y, Li B, Gurkan UA, Demirci U. Three-dimensional magnetic assembly of microscale hydrogels. Adv Mater Deerfield Beach Fla. 2011;23:4254–4260. doi: 10.1002/adma.201101962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zamanian B, Masaeli M, Nichol JW, Khabiry M, Hancock MJ, Bae H, Khademhosseini A. Interface Directed Self Assembly of Cell-Laden Microgels. Small Weinh Bergstr Ger. 2010;6:937–944. doi: 10.1002/smll.200902326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tasoglu S, Kavaz D, Gurkan UA, Guven S, Chen P, Zheng R, Demirci U. Paramagnetic Levitational Assembly of Hydrogels. Adv Mater. 2013;25:1137–1143. doi: 10.1002/adma.201200285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeong WY, Chua CK, Leong KF, Chandrasekaran M. Rapid prototyping in tissue engineering: challenges and potential. Trends Biotechnol. 2004;22:643–652. doi: 10.1016/j.tibtech.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Serpooshan V, Muja N, Marelli B, Nazhat SN. Fibroblast contractility and growth in plastic compressed collagen gel scaffolds with microstructures correlated with hydraulic permeability. J Biomed Mater Res A. 2011;96:609–620. doi: 10.1002/jbm.a.33008. [DOI] [PubMed] [Google Scholar]
- 17.Moore MJ, Friedman JA, Lewellyn EB, Mantila SM, Krych AJ, Ameenuddin S, Knight AM, Lu L, Currier BL, Spinner RJ, Marsh RW, Windebank AJ, Yaszemski MJ. Multiple-channel scaffolds to promote spinal cord axon regeneration. Biomaterials. 2006;27:419–429. doi: 10.1016/j.biomaterials.2005.07.045. [DOI] [PubMed] [Google Scholar]
- 18.Pati F, Jang J, Ha D-H, Won Kim S, Rhie J-W, Shim J-H, Kim D-H, Cho D-W. Printing three-dimensional tissue analogues with decellularized extracellular matrix bioink. Nat Commun. 2014;5:3935–3947. doi: 10.1038/ncomms4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tasoglu S, Demirci U. Bioprinting for stem cell research. Trends Biotechnol. 2013;31:10–19. doi: 10.1016/j.tibtech.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahn S, Yoon H, Kim G, Kim Y, Lee S, Chun W. Designed three-dimensional collagen scaffolds for skin tissue regeneration. Tissue Eng Part C Methods. 2009;16:813–820. doi: 10.1089/ten.tec.2009.0511. [DOI] [PubMed] [Google Scholar]
- 21.Serpooshan V, Julien M, Nguyen O, Wang H, Li A, Muja N, Henderson JE, Nazhat SN. Reduced hydraulic permeability of three-dimensional collagen scaffolds attenuates gel contraction and promotes the growth and differentiation of mesenchymal stem cells. Acta Biomater. 2010;6:3978–3987. doi: 10.1016/j.actbio.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 22.Polo-Corrales L, Latorre-Esteves M, Ramirez-Vick JE. Scaffold Design for Bone Regeneration. J Nanosci Nanotechnol. 2014;14:15–56. doi: 10.1166/jnn.2014.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elangovan S, D’Mello SR, Hong L, Ross RD, Allamargot C, Dawson DV, Stanford CM, Johnson GK, Sumner DR, Salem AK. The Enhancement of Bone Regeneration by Gene Activated Matrix Encoding for Platelet Derived Growth Factor. Biomaterials. 2014;35:737–747. doi: 10.1016/j.biomaterials.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K, Kievit FM, Florczyk SJ, Stephen ZR, Zhang M. 3D Porous Chitosan-Alginate Scaffolds as an In Vitro Model for Evaluating Nanoparticle-Mediated Tumor Targeting and Gene Delivery to Prostate Cancer. Biomacromolecules. 2015;16:3362–3372. doi: 10.1021/acs.biomac.5b01032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Tao R, Wu W, Cao H, Xin J, Li J, Guo J, Jiang L, Gao C, Demetriou AA, Farkas DL, Li L. 3D PLGA scaffolds improve differentiation and function of bone marrow mesenchymal stem cell-derived hepatocytes. Stem Cells Dev. 2010;19:1427–1436. doi: 10.1089/scd.2009.0415. [DOI] [PubMed] [Google Scholar]
- 26.Chen P, Güven S, Usta OB, Yarmush ML, Demirci U. Biotunable acoustic node assembly of organoids. Adv Healthc Mater. 2015;4:1937–1943. doi: 10.1002/adhm.201500279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen P, Luo Z, Güven S, Tasoglu S, Ganesan AV, Weng A, Demirci U. Microscale Assembly Directed by Liquid-Based Template. Adv Mater. 2014;26:5936–5941. doi: 10.1002/adma.201402079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dar A, Shachar M, Leor J, Cohen S. Optimization of cardiac cell seeding and distribution in 3D porous alginate scaffolds. Biotechnol Bioeng. 2002;80:305–312. doi: 10.1002/bit.10372. [DOI] [PubMed] [Google Scholar]
- 29.Li RK, Jia ZQ, Weisel RD, Mickle DAG, Choi A, Yau TM. Survival and Function of Bioengineered Cardiac Grafts. Circulation. 1999;100:63–69. doi: 10.1161/01.cir.100.suppl_2.ii-63. [DOI] [PubMed] [Google Scholar]
- 30.Li RK, Yau TM, Weisel RD, Mickle DAG, Sakai T, Choi A, Jia ZQ. Construction of a bioengineered cardiac graft. J Thorac Cardiovasc Surg. 2000;119:368–375. doi: 10.1016/S0022-5223(00)70193-0. [DOI] [PubMed] [Google Scholar]
- 31.Mei JC, Wu AYK, Wu PC, Cheng NC, Tsai WB, Yu J. Three-dimensional extracellular matrix scaffolds by microfluidic fabrication for long-term spontaneously contracted cardiomyocyte culture. Tissue Eng Part A. 2014;20:2931–2941. doi: 10.1089/ten.tea.2013.0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beauchamp P, Moritz W, Kelm JM, Ullrich ND, Agarkova I, Anson BD, Suter TM, Zuppinger C. Development and Characterization of a Scaffold-Free 3D Spheroid Model of Induced Pluripotent Stem Cell-Derived Human Cardiomyocytes. Tissue Eng Part C Methods. 2015;21:852–861. doi: 10.1089/ten.TEC.2014.0376. [DOI] [PubMed] [Google Scholar]
- 33.Fennema E, Rivron N, Rouwkema J, van Blitterswijk C, de Boer J. Spheroid culture as a tool for creating 3D complex tissues. Trends Biotechnol. 2013;31:108–115. doi: 10.1016/j.tibtech.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 34.Kelm JM, Ehler E, Nielsen LK, Schlatter S, Perriard JC, Fussenegger M. Design of artificial myocardial microtissues. Tissue Eng. 2004;10:201–214. doi: 10.1089/107632704322791853. [DOI] [PubMed] [Google Scholar]
- 35.Iyer RK, Chui J, Radisic M. Spatiotemporal tracking of cells in tissue-engineered cardiac organoids. J Tissue Eng Regen Med. 2009;3:196–207. doi: 10.1002/term.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevens KR, Pabon L, Muskheli V, Murry CE. Scaffold-Free Human Cardiac Tissue Patch Created from Embryonic Stem Cells. Tissue Eng Part A. 2009;15:1211–1222. doi: 10.1089/ten.tea.2008.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan Ye K, Sullivan KE, Black LD. Encapsulation of cardiomyocytes in a fibrin hydrogel for cardiac tissue engineering. J Vis Exp JoVE. 2011;55:3251. doi: 10.3791/3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saini H, Navaei A, Van Putten A, Nikkhah M. 3D Cardiac Microtissues Encapsulated with the Co-Culture of Cardiomyocytes and Cardiac Fibroblasts. Adv Healthc Mater. 2015;4:1961–1971. doi: 10.1002/adhm.201500331. [DOI] [PubMed] [Google Scholar]
- 39.Ma Z, Wang J, Loskill P, Huebsch N, Koo S, Svedlund FL, Marks NC, Hua EW, Grigoropoulos CP, Conklin BR, Healy KE. Self-organizing human cardiac microchambers mediated by geometric confinement. Nat Commun. 2015;6:7413–7423. doi: 10.1038/ncomms8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patterning of particulate films using Faraday waves. Rev Sci Instrum. 2003;74:4063–4070. [Google Scholar]
- 41.Tulloch NL, Muskheli V, Razumova MV, Korte FS, Regnier M, Hauch KD, Pabon L, Reinecke H, Murry CE. Growth of Engineered Human Myocardium with Mechanical Loading and Vascular Co-culture. Circ Res. 2011;109:47–59. doi: 10.1161/CIRCRESAHA.110.237206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomsen MB, Calloe K. Human atrial fibroblasts and their contribution to supraventricular arrhythmia. Physiol Rep. 2016p;4:e12711. doi: 10.14814/phy2.12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saylor JR, Kinard AL. Simulation of particle deposition beneath Faraday waves in thin liquid films. Phys Fluids 1994-Present. 2005;17:047106. [Google Scholar]
- 44.Fukuda K, Yuasa S. Cardiac Regeneration using Stem Cells. CRC Press; New York: 2013. [Google Scholar]
- 45.Yasui K, Kada K, Hojo M, Lee JK, Kamiya K, Toyama J, Opthof T, Kodama I. Cell-to-cell interaction prevents cell death in cultured neonatal rat ventricular myocytes. Cardiovasc Res. 2000;48:68–76. doi: 10.1016/s0008-6363(00)00145-0. [DOI] [PubMed] [Google Scholar]
- 46.Wu H, Lee J, Vincent LG, Wang Q, Gu M, Lan F, Churko JM, Sallam KI, Matsa E, Sharma A, Gold JD, Engler AJ, Xiang YK, Bers DM, Wu JC. Epigenetic Regulation of Phosphodiesterases 2A and 3A Underlies Compromised β-Adrenergic Signaling in an iPSC Model of Dilated Cardiomyopathy. Cell Stem Cell. 2015;17:89–100. doi: 10.1016/j.stem.2015.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez ML, Graham BT, Pabon LM, Han SJ, Murry CE, Sniadecki NJ. Measuring the contractile forces of human induced pluripotent stem cell-derived cardiomyocytes with arrays of microposts. J Biomech Eng. 2014;136:051005. doi: 10.1115/1.4027145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liau B, Zhang D, Bursac N. Functional cardiac tissue engineering. Regen Med. 2012;7:187–206. doi: 10.2217/rme.11.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siedner S, Krüger M, Schroeter M, Metzler D, Roell W, Fleischmann BK, Hescheler J, Pfitzer G, Stehle R. Developmental changes in contractility and sarcomeric proteins from the early embryonic to the adult stage in the mouse heart, J. Physiol. 2003;548:493–505. doi: 10.1113/jphysiol.2002.036509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marga F, Jakab K, Khatiwala C, Shepherd B, Dorfman S, Hubbard Bradley, Colbert S, Forgacs G. Toward engineering functional organ modules by additive manufacturing. Biofabrication. 2012;4:022001. doi: 10.1088/1758-5082/4/2/022001. [DOI] [PubMed] [Google Scholar]
- 51.Hribar KC, Meggs K, Liu J, Zhu W, Qu X, Chen S. Three-dimensional direct cell patterning in collagen hydrogels with near-infrared femtosecond laser. Sci Rep. 2015;5:17203. doi: 10.1038/srep17203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cha HD, Hong JM, Kang TY, Jung JW, Ha DH, Cho DW. Effects of micro-patterns in three-dimensional scaffolds for tissue engineering applications. J Micromechanics Microengineering. 2012;22:125002–125011. [Google Scholar]
- 53.Guillotin B, Guillemot F. Cell patterning technologies for organotypic tissue fabrication. Trends Biotechnol. 2011;29:183–190. doi: 10.1016/j.tibtech.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 54.Churko JM, Burridge PW, Wu JC. Generation of human iPSCs from human peripheral blood mononuclear cells using non-integrative Sendai virus in chemically defined conditions. Methods Mol Biol Clifton NJ. 2013;1036:81–88. doi: 10.1007/978-1-62703-511-8_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, Plews JR, Abilez OJ, Cui B, Gold JD, Wu JC. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11:855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, Hashimoto H, Suzuki T, Yamashita H, Satoh Y, Egashira T, Seki T, Muraoka N, Yamakawa H, Ohgino Y, Tanaka T, Yoichi M, Yuasa S, Murata M, Suematsu M, Fukuda K. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 2013;12:127–137. doi: 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 57.Douady S. Experimental study of the Faraday instability. J Fluid Mech. 1990;221:383–409. [Google Scholar]
- 58.Sacchi V, Mittermayr R, Hartinger J, Martino MM, Lorentz KM, Wolbank S, Hofmann A, Largo RA, Marschall JS, Groppa E, Gianni-Barrera R, Ehrbar M, Hubbell JA, Redl H, Banfi A. Long-lasting fibrin matrices ensure stable and functional angiogenesis by highly tunable, sustained delivery of recombinant VEGF164. Proc Natl Acad Sci U S A. 2014;111:6952–6957. doi: 10.1073/pnas.1404605111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dunn JC, Tompkins RG, Yarmush ML. Long-term in vitro function of adult hepatocytes in a collagen sandwich configuration. Biotechnol Prog. 1991;7:237–245. doi: 10.1021/bp00009a007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


