ABSTRACT
Peroxisome biogenesis disorders (PBDs) is a group of diseases caused by mutations in one of the peroxins, proteins responsible for biogenesis of the peroxisomes. In recent years, it became clear that many peroxins (e.g., PEX3 and PEX14) play additional roles in peroxisome homeostasis (such as promoting autophagic degradation of peroxisomes or pexophagy), which are often opposite to their originally established functions in peroxisome formation and maintenance. Even more interesting, the peroxins that make up the peroxisomal AAA ATPase complex (AAA-complex) in yeast (Pex1, Pex6 and Pex15) or mammals (PEX1, PEX6, PEX26) are responsible for the downregulation of pexophagy. Moreover, this might be even their primary role in human: to prevent pexophagy by removing from the peroxisomal membrane the ubiquitinated peroxisomal matrix protein import receptor, Ub-PEX5, which is also a signal for the Ub-binding pexophagy receptor, NBR1. Remarkably, the peroxisomes rescued from pexophagy by autophagic inhibitors in PEX1G843D (the most common PBD mutation) cells are able to import matrix proteins and improve their biochemical function suggesting that the AAA-complex per se is not essential for the protein import function in human. This paradigm-shifting discovery published in the current issue of Autophagy has raised hope for up to 65% of all PBD patients with various deficiencies in the AAA-complex. Recognizing PEX1, PEX6 and PEX26 as pexophagy suppressors will allow treating these patients with a new range of tools designed to target mammalian pexophagy.
KEYWORDS: ATPase, peroxin, peroxisome, peroxisome biogenesis disorder, PEX1, PEX5, PEX6, PEX15, PEX26, pexophagy
Peroxisomes are the organelles that carry out many important functions in human metabolism. As a consequence, inherited mutations in many of the peroxisomal genes can lead to peroxisomal disorders. Most of the peroxisomal disorders can be grouped into either single peroxisomal enzyme deficiencies (PEDs) or peroxisome biogenesis disorders (PBDs) characterized by multiple abnormalities due to a defect in the formation or maintenance of peroxisomes. The latter 2 functions are supported by the peroxisome biogenesis factors or peroxins encoded by the PEX genes. The bi-allelic recessive mutations in 14 PEX genes, including PEX1, PEX6 and PEX26 that encode the subunits of the peroxisomal AAA ATPase complex (AAA-complex), have been reported to cause most of the PBDs in human (for a review, see ref. 1). Interestingly, mutations in the AAA-complex genes, PEX1 (48.5%), PEX6 (13.1%) and PEX26 (3.4%), are the most common among PBD patients and account for 65% of all the PBD cases.2
Until recently, it was widely accepted that the AAA-complex participates mainly in the import of the peroxisomal matrix proteins, because the cells from corresponding PBD patients have membrane remnants of the peroxisomes with properly localized peroxisomal membrane proteins. Precisely, the heteromeric complex of PEX1 and PEX6 ATPases recruited to the peroxisomal membrane by PEX26 (Pex15 in yeast or APEM9 in plants) drives the ATP- and ubiquitin-dependent release of the ubiquitinated peroxisomal matrix protein import receptor, Ub-PEX5, from the peroxisomal membrane to the cytosol. Such dislocation of Ub-PEX5 from peroxisomes and its deubiquitination (mediated by Pex6-bound Ubp15 in yeast) are required for receptor recycling and repeated rounds of import of the peroxisomal matrix proteins that contain the peroxisomal targeting signal 1 (PTS1). It was also proposed that the ATPase-mediated removal of PEX5 from the peroxisomes might serve as a pulling force for PTS1-cargo translocation across the peroxisomal membrane (i.e., export driven import). For reviews on these topics, see refs. 3, 4
Before the peroxisomal matrix protein import function of the AAA-complex dominated the peroxisome field, a beautiful set of studies in yeast suggested that Pex1, Pex6 and ATP hydrolysis were required for the heterotypic fusion (at priming and docking steps) of small peroxisomal vesicles, P1 and P2, which are the earliest precursors of mature peroxisomes.5,6 Such a fusion was later proposed to be required for the assembly of a complete peroxisomal translocon, which imports matrix proteins, since each of the 2 peroxisomal vesicles carried only half of the peroxins involved in peroxisomal matrix protein import.7 However, subsequent studies did not confirm the existence of half-translocons in the peroxisomal membrane remnants of pex1 and pex6 cells. Instead, these remnants contained a complete translocon but no matrix proteins supporting an essential role of Pex1 and Pex6 in peroxisomal matrix protein import.8,9 The 2 established functions of Pex1 and Pex6 in (1) fusion of the pre-peroxisomal vesicles and (2) export of the ubiquitinated Pex5 from the peroxisomal membrane (for peroxisomal matrix protein import) are analogous to the roles of 2 other AAA ATPases, NSF (Sec18 in yeast)—in membrane fusion, and VCP/p97 (Cdc48 in yeast)—in export of the ubiquitinated proteins from the ER membrane. The homohexameric complexes of NSF and VCP have distinct structural features that might help to model the molecular function(s) of the heterohexameric Pex1-Pex6 complex. However, despite the fact that the first structures of the Pex1-Pex6 complex were solved recently, an accurate prediction of its molecular role(s) in the cell must await further studies (for a review, see ref. 10).
Recently, the yeast AAA-complex was implicated in the 3rd function: prevention of pexophagy.11 Pexophagy is the selective autophagic degradation of peroxisomes. This process is necessary for removal (from the cytosol) and recycling (in the lysosomes or vacuoles) of superfluous and/or damaged peroxisomes (for a review, see ref. 12). Interestingly, lack of Pex1, Pex6 or Pex15 triggers degradation of the peroxisomal membrane remnants that depends on the yeast pexophagy receptor, Atg36. It is remarkable that depletion of Pex1 in yeast first causes the peroxisomal matrix protein import defect before it results in pexophagy. However, the buildup of Ub-Pex5 in the peroxisomal membrane is not a trigger for pexophagy in yeast.11 In this issue of Autophagy, Peter Kim and colleagues report that in human cells the AAA-complex also prevents pexophagy mediated (in this case) by the Ub-binding pexophagy receptor, NBR1.13 In contrast to yeast pexophagy, this process in the AAA-complex-depleted human cells depends on the accumulation of Ub-PEX5 in the peroxisomal membrane. In this regard, the role of this protein is consistent with 2 other recent reports that identified mammalian Ub-PEX5 as an important pexophagic signal.14,15
Strikingly, the loss of a functional AAA-complex in human cells does not per se abrogate peroxisome formation or peroxisomal matrix protein import—rather, the enhanced pexophagy does. Law et al. have elegantly shown that if you titrate the excessive pexophagy with autophagic inhibitors, such as chloroquine, the fibroblasts of a patient with the most common PBD mutation, PEX1G843D (that causes a mild form of PBD), will not only restore their peroxisome number but also PTS1-protein import and peroxisomal β-oxidation of very long-chain fatty acids.13 Apparently, the recycling of Ub-PEX5 is not essential for peroxisomal matrix protein import in human. The export of Ub-PEX5 from the peroxisomal membrane via the AAA-complex is more important for the prevention of pexophagy than the recycling of PEX5 and PTS1-protein import (Fig. 1). Indeed, in contrast to yeast, the mammalian cells use Ub as a degradation signal for many selective autophagy pathways, including pexophagy.16 Therefore, a continuous removal of the ubiquitin and/or ubiquitinated proteins from the intracellular organelles, such as peroxisomes, must be an essential housekeeping mechanism. In addition, if human (but not yeast) cells have an excess of cytosolic PEX5, it could explain why the PEX5 recycling function of the AAA-complex is secondary in human.
Figure 1.
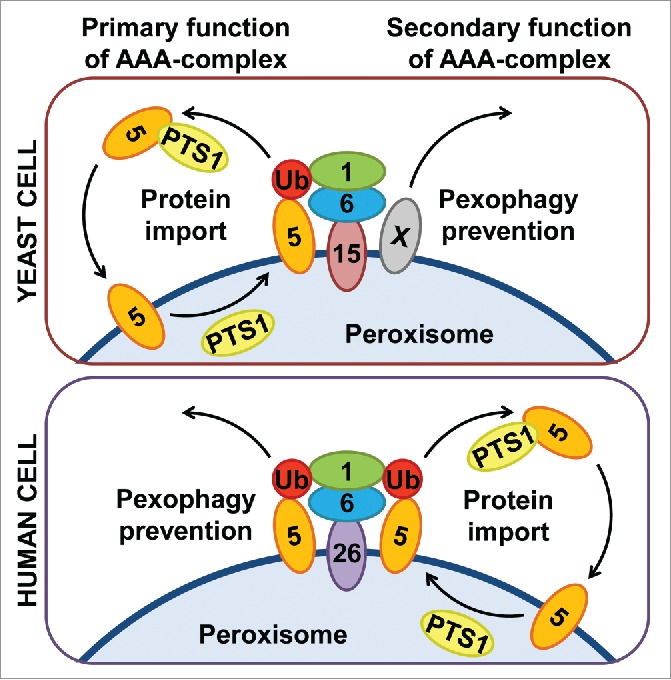
The primary role of the human peroxisomal AAA-complex is to prevent pexophagy. Both yeast (Pex1, Pex6 and Pex15) and human (PEX1, PEX6 and PEX26) AAA-complexes extract Ub-PEX5 from the peroxisomal membrane. However, while the main purpose of such extraction in yeast cells is Pex5 recycling for repeated rounds of PTS1-protein import, the extraction of Ub-PEX5 in human cells is essential to prevent pexophagy. Since the yeast AAA-complex also prevents pexophagy, but independent of Pex5, there must be another pexophagic signal (protein X) removed from the peroxisomal membrane by the AAA-complex in yeast.
Next, it will be interesting to see how this pexophagy prevention function of the human AAA-complex is terminated under pexophagy-inducing conditions. A possibility exists that the AAA-complex or its single component is degraded ahead of the peroxisomes when pexophagy is induced. Such degradative inactivation of the AAA-complex would allow Ub-PEX5 to recruit NBR1 to the peroxisomal membrane and trigger pexophagy. Recently, it was demonstrated that both PEX1 and PEX6 contain evolutionarily-conserved Atg8-interacting motifs.17 Moreover, for plant PEX6, the interaction with Atg8 was also shown experimentally, suggesting that at least PEX6 (if not both peroxisomal ATPases) might indeed be a substrate of the selective autophagic degradation that contributes to the induction of pexophagy from yeast to human.
Currently, there is no curative therapy for the PBDs (for reviews of symptomatic therapy, see refs. 18, 19). Recognition of PEX1, PEX6 and PEX26 as pexophagy suppressors opens up an exciting opportunity to cure the majority of PBD patients. The Kim group has already reported that daily addition of a small, nontoxic amount of chloroquine (5 μM) in the growth medium to suppress autophagic pathways supported PTS1-protein import in PEX1G843D fibroblasts for up to 15 d in cell culture.13 This result suggests that targeting of the human pexophagy-specific proteins could potentially provide even better therapeutic outcomes (stronger inhibition of pexophagy leading to even better peroxisomal matrix protein import, combined with intactness of other autophagic pathways enabling even greater cell survival). However, such an ideal target would also have to be dispensable for peroxisome biogenesis and metabolism. Unfortunately, a protein satisfying all of these criteria is unknown yet.
Interestingly, the effects described by Law et al. for chloroquine (increased peroxisome number, matrix protein import and biochemical function) in PEX1G843D cells were previously reported for: (1) nonspecific chemical chaperones, such as 4-phenylbutyrate (4-PBA), trimethylamine N-oxide (TMAO), glycerol and arginine,20-22 and (2) PEX1G843D-specific chaperones, such as acacetin diacetate (AD), aminoalkyl bisindolylmaleimide (GF109203x) and structurally related Ro 31–8220.21 Despite the fact that other mechanisms were proposed to explain the pharmacological effects of these molecules (e.g., increased transcription of ABCD2/ALDR and PEX11A/PEX11α genes in response to 4-PBA20 or competitive and reversible binding of AD, GF109203x and Ro 31–8220 to the ATP binding sites21 on PEX1G843D), many of them also modulate autophagy. For example, 4-PBA inhibits ER stress-induced autophagy,23 arginine attenuates IFNG (interferon gamma)-induced autophagy,24 and AD, GF109203x and Ro 31–8220 might all inhibit protein kinase C21 required for pexophagy signaling in yeast25 and, most probably, in mammalian cells (at least under hypoxic conditions).26,27 Therefore, it is possible that previous successes in the pharmacological induction of peroxisomes in patient fibroblasts with mild AAA-complex deficiencies were also due to attenuation of pexophagy.
In the future, it will be important to examine the effectiveness of chloroquine treatment in restoring the AAA-complex defects in vivo. Luckily, the murine model of human PEX1G843D deficiency, the PEX1G844D mouse, has been developed recently.28 This model recapitulates many features of the human disorder, including the responsiveness of murine PEX1G844D cells to chemical chaperones. Therefore, Kim and colleagues are poised to test if their paradigm-shifting discovery (that a primary role of the human AAA-complex is to prevent pexophagy) can change the way we treat up to 65% of PBD patients who display excessive pexophagy.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by NIH grants DK106344 and GM119571.
References
- [1].Waterham HR, Ferdinandusse S, Wanders RJ. Human disorders of peroxisome metabolism and biogenesis. Biochim Biophys Acta 2016; 1863(5):922-33; PMID:26611709; http://dx.doi.org/ 10.1016/j.bbamcr.2015.11.015 [DOI] [PubMed] [Google Scholar]
- [2].Waterham HR, Ebberink MS. Genetics and molecular basis of human peroxisome biogenesis disorders. Biochim Biophys Acta 2012; 1822(9):1430-41; PMID:22871920; http://dx.doi.org/ 10.1016/j.bbadis.2012.04.006 [DOI] [PubMed] [Google Scholar]
- [3].Fujiki Y, Nashiro C, Miyata N, Tamura S, Okumoto K. New insights into dynamic and functional assembly of the AAA peroxins, Pex1p and Pex6p, and their membrane receptor Pex26p in shuttling of PTS1-receptor Pex5p during peroxisome biogenesis. Biochim Biophys Acta 2012; 1823(1):145-9; PMID:22079764; http://dx.doi.org/ 10.1016/j.bbamcr.2011.10.012 [DOI] [PubMed] [Google Scholar]
- [4].Grimm I, Saffian D, Platta HW, Erdmann R. The AAA-type ATPases Pex1p and Pex6p and their role in peroxisomal matrix protein import in Saccharomyces cerevisiae. Biochim Biophys Acta 2012; 1823(1):150-8; PMID:21963882; http://dx.doi.org/ 10.1016/j.bbamcr.2011.09.005 [DOI] [PubMed] [Google Scholar]
- [5].Titorenko VI, Chan H, Rachubinski RA. Fusion of small peroxisomal vesicles in vitro reconstructs an early step in the in vivo multistep peroxisome assembly pathway of Yarrowia lipolytica. J Cell Biol 2000; 148(1):29-44; PMID:10629216; http://dx.doi.org/ 10.1083/jcb.148.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Titorenko VI, Rachubinski RA. Peroxisomal membrane fusion requires two AAA family ATPases, Pex1p and Pex6p. J Cell Biol 2000; 150(4):881-6; PMID:10953011; http://dx.doi.org/ 10.1083/jcb.150.4.881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].van der Zand A, Gent J, Braakman I, Tabak HF. Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell 2012; 149(2):397-409; PMID:22500805; http://dx.doi.org/ 10.1016/j.cell.2012.01.054 [DOI] [PubMed] [Google Scholar]
- [8].Knoops K, de Boer R, Kram A, van der Klei IJ. Yeast pex1 cells contain peroxisomal ghosts that import matrix proteins upon reintroduction of Pex1. J Cell Biol 2015; 211(5):955-62; PMID:26644511; http://dx.doi.org/ 10.1083/jcb.201506059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Motley AM, Galvin PC, Ekal L, Nuttall JM, Hettema EH. Reevaluation of the role of Pex1 and dynamin-related proteins in peroxisome membrane biogenesis. J Cell Biol 2015; 211(5):1041-56; PMID:26644516; http://dx.doi.org/ 10.1083/jcb.201412066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tan D, Blok NB, Rapoport TA, Walz T. Structures of the double-ring AAA ATPase Pex1-Pex6 involved in peroxisome biogenesis. FEBS J 2016; 283(6):986-92; PMID:26476099; http://dx.doi.org/ 10.1111/febs.13569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nuttall JM, Motley AM, Hettema EH. Deficiency of the exportomer components Pex1, Pex6, and Pex15 causes enhanced pexophagy in Saccharomyces cerevisiae. Autophagy 2014; 10(5):835-45; PMID:24657987; http://dx.doi.org/ 10.4161/auto.28259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nazarko TY, Farré JC. Molecular machines involved in pexophagy In: Brocard C, Hartig A, editors. Molecular machines involved in peroxisome biogenesis and maintenance. Springer-Verlag Wien; 2014. p. 481-506. [Google Scholar]
- [13].Law KB, Bronte-Tinkew D, Di Pietro E, Snowden A, Jones RO, Moser A, Brumell JH, Braverman N, Kim PK. The peroxisomal AAA ATPase complex prevents pexophagy and development of peroxisome biogenesis disorders. Autophagy 2017; 13(5); http://dx.doi.org/ 10.1080/15548627.2017.1291470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nordgren M, Francisco T, Lismont C, Hennebel L, Brees C, Wang B, Van Veldhoven PP, Azevedo JE, Fransen M. Export-deficient monoubiquitinated PEX5 triggers peroxisome removal in SV40 large T antigen-transformed mouse embryonic fibroblasts. Autophagy 2015; 11(8):1326-40; PMID:26086376; http://dx.doi.org/ 10.1080/15548627.2015.1061846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhang J, Tripathi DN, Jing J, Alexander A, Kim J, Powell RT, Dere R, Tait-Mulder J, Lee JH, Paull TT, et al.. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat Cell Biol 2015; 17(10):1259-69; PMID:26344566; http://dx.doi.org/ 10.1038/ncb3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Khaminets A, Behl C, Dikic I. Ubiquitin-Dependent And Independent Signals In Selective Autophagy. Trends Cell Biol 2016; 26(1):6-16; PMID:26437584; http://dx.doi.org/ 10.1016/j.tcb.2015.08.010 [DOI] [PubMed] [Google Scholar]
- [17].Xie Q, Tzfadia O, Levy M, Weithorn E, Peled-Zehavi H, Van Parys T, Van de Peer Y, Galili G. hfAIM: A reliable bioinformatics approach for in silico genome-wide identification of autophagy-associated Atg8-interacting motifs in various organisms. Autophagy 2016; 12(5):876-87; PMID:27071037; http://dx.doi.org/ 10.1080/15548627.2016.1147668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Klouwer FC, Berendse K, Ferdinandusse S, Wanders RJ, Engelen M, Poll-The BT. Zellweger spectrum disorders: clinical overview and management approach. Orphanet J Rare Dis 2015; 10:151; PMID:26627182; http://dx.doi.org/ 10.1186/s13023-015-0368-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Braverman NE, Raymond GV, Rizzo WB, Moser AB, Wilkinson ME, Stone EM, Steinberg SJ, Wangler MF, Rush ET, Hacia JG, et al.. Peroxisome biogenesis disorders in the Zellweger spectrum: An overview of current diagnosis, clinical manifestations, and treatment guidelines. Mol Genet Metab 2016; 117(3):313-21; PMID:26750748; http://dx.doi.org/ 10.1016/j.ymgme.2015.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wei H, Kemp S, McGuinness MC, Moser AB, Smith KD. Pharmacological induction of peroxisomes in peroxisome biogenesis disorders. Ann Neurol 2000; 47(3):286-96; PMID:10716247; http://dx.doi.org/ 10.1002/1531-8249(200003)47:3%3c286::AID-ANA3%3e3.0.CO;2-B [DOI] [PubMed] [Google Scholar]
- [21].Zhang R, Chen L, Jiralerspong S, Snowden A, Steinberg S, Braverman N. Recovery of PEX1-Gly843Asp peroxisome dysfunction by small-molecule compounds. Proc Natl Acad Sci U S A 2010; 107(12):5569-74; PMID:20212125; http://dx.doi.org/ 10.1073/pnas.0914960107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Berendse K, Ebberink MS, Ijlst L, Poll-The BT, Wanders RJ, Waterham HR. Arginine improves peroxisome functioning in cells from patients with a mild peroxisome biogenesis disorder. Orphanet J Rare Dis 2013; 8:138; PMID:24016303; http://dx.doi.org/ 10.1186/1750-1172-8-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kim DS, Li B, Rhew KY, Oh HW, Lim HD, Lee W, Chae HJ, Kim HR. The regulatory mechanism of 4-phenylbutyric acid against ER stress-induced autophagy in human gingival fibroblasts. Arch Pharm Res 2012; 35(7):1269-78; PMID:22864750; http://dx.doi.org/ 10.1007/s12272-012-0718-2 [DOI] [PubMed] [Google Scholar]
- [24].Xia X, Che Y, Gao Y, Zhao S, Ao C, Yang H, Liu J, Liu G, Han W, Wang Y, et al.. Arginine Supplementation Recovered the IFN-γ-Mediated Decrease in Milk Protein and Fat Synthesis by Inhibiting the GCN2/eIF2α Pathway, Which Induces Autophagy in Primary Bovine Mammary Epithelial Cells. Mol Cells 2016; 39(5):410-7; PMID:27025389; http://dx.doi.org/ 10.14348/molcells.2016.2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Manjithaya R, Jain S, Farré JC, Subramani S. A yeast MAPK cascade regulates pexophagy but not other autophagy pathways. J Cell Biol 2010; 189(2):303-10; PMID:20385774; http://dx.doi.org/ 10.1083/jcb.200909154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chen JL, Lin HH, Kim KJ, Lin A, Forman HJ, Ann DK. Novel roles for protein kinase Cdelta-dependent signaling pathways in acute hypoxic stress-induced autophagy. J Biol Chem 2008; 283(49):34432-44; PMID:18836180; http://dx.doi.org/ 10.1074/jbc.M804239200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Walter KM, Schönenberger MJ, Trötzmüller M, Horn M, Elsässer HP, Moser AB, Lucas MS, Schwarz T, Gerber PA, Faust PL, et al.. Hif-2α promotes degradation of mammalian peroxisomes by selective autophagy. Cell Metab 2014; 20(5):882-97; PMID:25440060; http://dx.doi.org/ 10.1016/j.cmet.2014.09.017 [DOI] [PubMed] [Google Scholar]
- [28].Hiebler S, Masuda T, Hacia JG, Moser AB, Faust PL, Liu A, Chowdhury N, Huang N, Lauer A, Bennett J, et al.. The Pex1-G844D mouse: a model for mild human Zellweger spectrum disorder. Mol Genet Metab 2014; 111(4):522-32; PMID:24503136; http://dx.doi.org/ 10.1016/j.ymgme.2014.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]


