Abstract
We have developed a new ambient-ionization mass spectrometric technique named laser desorption/ionization droplet delivery mass spectrometry (LDIDD-MS). LDIDD-MS permits high-resolution, high-sensitivity imaging of tissue samples as well as measurements of both single-cell apoptosis and live-cell exocytosis. A pulsed (15 Hz) UV laser beam (266 nm) is focused on a surface covered with target analytes to trigger their desorption and ionization. A spray of liquid droplets is simultaneously directed onto the laser-focused surface region to capture the ionized analytes and deliver them to a mass spectrometer. The approach of rapid and effective capturing of molecules after laser desorption/ionization allows the limit of detection for the amino acid lysine to be as low as 2 amol under ambient ionization conditions. Two-dimensional maps of the desorbed/ionized species are recorded by moving the sample on an XY translational stage. The spatial resolution for imaging with LDIDD-MS was determined to be 2.4 μm for an ink-printed pattern and 3 μm for mouse brain tissue. We applied LDIDD-MS to single-cell analysis of apoptotic HEK cells. Differences were observed in the profiles of fatty acids and lipids between healthy HEK cells and those undergoing apoptosis. We observed upregulation of phosphatidylcholine (PC) with a relatively shorter carbon chain length and downregulation of PC with a relatively longer carbon chain length. We also applied LDIDD-MS for a real-time direct measurements of live-cell exocytosis. The catecholamine dopamine and trace amines (phenethylamine and tyramine) were detected from live PC12 cells without damaging them.
Graphical abstract

Mass spectrometric imaging has provided the opportunity to identify the distribution and localization of molecules in cells and tissue samples for metabolomic and proteomic analyses.1–4 Various ionization and desorption methods have been used,5 including laser-based approaches, such as matrix-assisted laser desorption ionization (MALDI)1 and laser desorption ionization (LDI);6 ion beam-based ionization, such as secondary ion mass spectrometry (SIMS);7 plasma-based ionization, such as direct analysis in real time (DART);8 as well as spray-based approaches, such as desorption electrospray ionization (DESI).9 Recently, new desorption/ionization methods combining the laser- and spray-based approaches have been introduced, including electrospray-assisted laser desorption ionization (ELDI),10 laser ablation electrospray ionization (LAESI),11 infrared laser-assisted desorption electrospray ionization (IR LADESI),12 laser electrospray mass spectrometry (LEMS),13 infrared matrix-assisted laser desorption electrospray ionization (IR-MALDE-SI),14,15 and laser-induced acoustic desorption/electrospray ionization (LIAD/ESI).16 However, the implementation of single-cell analysis with these approaches remains challenging because of limited sensitivity and spatial resolution. Here, we report a new ambient-ionization mass spectrometric technique, which we call laser desorption/ionization droplet delivery mass spectrometry (LDIDD-MS). The technique allows the imaging of tissue samples with high resolution and sensitivity as well as the measurement of both single-cell apoptosis and live-cell exocytosis.
For LDIDD-MS, we utilized a focused, pulsed UV laser for desorption and ionization of target molecules deposited on a surface. We achieved a spatial resolution of a few micrometers by focusing the output of the UV laser, which was constrained by the far-field difffraction limit. Under ambient conditions, effective delivery of the desorbed molecular ions was critical for high sensitivity. To minimize the loss of the desorbed molecular ions, we sprayed liquid droplets directly onto the laser-irradiated region to deliver the molecular ions to a mass spectrometer inlet. As the droplets were so close to the laser desorption/ionization region, we also achieved high sensitivity in capturing and delivering ions. The combination of UV photoionization and electrospray ionization increased the signal. To demonstrate the imaging power of LDIDD-MS, we conducted high-resolution imaging of animal tissue and single-cell analysis.
Application of mass spectrometry for live-cell analysis is limited by its destructive nature and the difficulty in collecting and introducing biological molecules from live cells to a mass spectrometer. Masujima and co-workers17,18 used mass spectrometry to analyze secreted molecules from a live single cell by introducing a nanospray tip near the surface of a cell and bringing the tip to a mass spectrometer inlet for electrospray of the contents aspirated in the nanospray tip. However, this approach allows only low-throughput and single time-point measurements because the measurement is performed through two separate steps of sampling and spraying of analytes. The application of this method for live-cell analysis is especially limited for molecules that are rapidly degraded once secreted from cells or for continuous or kinetic analysis of cell secretion.
We overcame this obstacle with LDIDD-MS by directly sampling the cell culture solution’s surface that contained the secreted molecules from cells. We illustrate the advantages of the direct liquid-phase sampling capability of LDIDD-MS by the real-time measurement of live-cell secretion from PC12 cells induced to carry out exocytosis.
EXPERIMENTAL SECTION
Chemicals and Sample Preparation
Caffeine, lysine, phenanthrene, phenylalanine, bradykinin, cytochrome c, poly-D-lysine, phenethylamine, and tyramine were purchased from Sigma-Aldrich (St. Louis, MO). HPLC-grade methanol and water were purchased from Fisher Scientific (Nepean, ON, Canada). Mouse brain tissue (C57BL/6J females at 9 weeks) was purchased from The Jackson Laboratory (Bar Harbor, ME) and cryosectioned at a 25-μm thickness with a 550 M Cryostat microtome. The sliced tissues were individually mounted on glass plates and dried for approximately 10 min in a vacuum desiccator prior to imaging. Cell culture medium and reagents including Dulbecco’s Modified Eagle Medium (DMEM), live-cell imaging solution, heat-inactivated fetal bovine serum, horse serum, calf serum, penicillin, streptomycin, and L-glutamine were purchased from Life Technology (Rockville, MD).
LDIDD-MS
The LDIDD-MS system was built by combining a pulsed laser and an apparatus for generating droplets. A Thermo Scientific LTQ Orbitrap XL Hybrid Ion Trap-Orbitrap was the chosen mass spectrometer. The cone and capillary voltages were set to 44 and 60 V, respectively. The capillary temperature was set to 275 °C. A voltage of 5 kV was applied to the metal tips of a syringe infused with solvent for droplet delivery. A DCR-11 pulsed Nd:YAG laser (Spectra Physics, CA) with a fundamental wavelength of 1064 nm was used to generate UV pulses. An HG-2 harmonic generator and PHS-1 harmonic separator were used to generate and separate the output of the UV laser at 266 nm. The UV laser beam was fired at 15 Hz and was aligned and focused using a 4 cm focal length lens on the sample surface, typically a glass slide with a target sample of interest. Dried N2 gas was supplied at 80 psi to promote nebulization and generation of aerosol droplets and propulsion to sample surface as well as delivery of analytes to the mass spectrometry inlet. A stream of liquid droplets generated by nebulizing N2 gas and electrospray was tilted at 50° to 55° downward and directed onto the laser-irradiated region to capture desorbed molecules and molecular ions from the substrate and deliver them to the inlet (extended capillary at a length of 9 cm) of the mass spectrometer. The electrospray source was placed at a distance of 3–5 mm from the focused laser spot. The mixture of methanol and water (1:1, v/v) was used as a solvent for droplets except for the live-cell analysis, where pure water was used. The solvent flow rate and pressure of N2 gas supply were adjusted such that desorption solely by the liquid droplets was minimized. The optimal condition for LDIDD-MS imaging was a flow rate of 0.5 μL/min and a N2 pressure of 80 psi. The diameter of the electrospray on the sample surface was around 200 μm. The diameter of the focused laser beam at 266 nm was ideally 1.4 μm and practically 2–3 μm. The distance between the mass spectrometer inlet and the substrate was kept at 1 mm. Test samples of caffeine or rhodamine B were used to ensure desorption of target molecules. The lateral distance between the mass spectrometer inlet and focused laser spot was adjusted to maximize collected ion intensity ranging between 5 and 8 mm. A custom-built X-Y motorized translational mechanical stage was used for constructing two-dimensional mass spectrometric images. The stage was operated in stop-and-go mode for sweeping the laser beam across the target sample at step sizes of 100 and 5 μm. The dwell time per pixel was typically 2 s and was increased up to 3 s for detecting low abundant molecules. The collected mass spectrometry data were imported to Biomap imaging software (Novartis Institutes for BioMedical Research, Basel, Switzerland) to construct images.
LDIDD-MS Single- and Live-Cell Analyses
All cells were purchased from American Type Culture Collection (Manassas, VA). The HEK 293T cell line was cultured in high-glucose DMEM supplemented with 10% heat-inactivated fetal bovine serum as well as 1% penicillin and streptomycin. The cells were seeded onto an 8 mm round coverslip for single-cell analysis. Prior to mass spectrometric analysis, the cells were rinsed two times with 100 mM ammonium acetate solution. Line scanning on isolated HEK 293T cells ensured the acquisition of mass spectra from a single cell. PC12 cells were used for analyses of live-cell secretions. PC12 cells were cultured in DMEM (11965) supplemented with 4.5 g/L of glucose, 2 mM L-glutamine, 5% horse serum, and 5% fetal bovine serum. Immediately prior to live-cell analysis, the PC12 cells were washed twice with phosphate-buffered saline and transferred into live-cell imaging medium (Life Technologies, Rockville, MD), which contained 20 mM HEPES buffer to maintain physiological pH under ambient conditions. The cells were mounted on the mass spectrometry imaging stage equipped with a temperature controller maintained at 37 °C. Secretion from the live PC12 cells was induced by treatment with 50 mM KCl. Incubating the cells in propidium iodide solution, and confirming the lack of stained cells, assessed the viability and damage of the cells. The mass spectra were acquired from the average of ~180 laser shots per cell for the single cell experiment and ~30 shots for the live-cell experiment. The error bars represent the standard deviation from triple measurements unless otherwise indicated.
Targeted Analysis of Secreted Molecules from PC12 Cells
PC12 cells grown in a T75 cell-culture flask were induced for neurotransmitter release by immersion in 10 mL of HBSS buffer containing 50 mM KCl. The buffer was centrifuged at 2000g for 5 min to remove any debris. Sample cleanup was performed with Strata-X-CW 33 μm weak cation exchange 1 mL columns with 1 mg of sorbent (Phenomenex, Torrance, CA) by passing dropwise in the following sequence through the column: 1 mL of methanol, 1 mL of H2O, 10 mL of sample, 1 mL of H2O, and 1 mL of methanol. The column was dried under vacuum for 5 min before the final sample was eluted with 300 μL of methanol containing 5% formic acid. An analytical standard sample was prepared containing 0.2 μM phenethylamine and 0.2 μM tyramine in 2 mM ammonium bicarbonate buffer. The LC-MS analyses are described in detail in the Supporting Information.
RESULTS AND DISCUSSION
Characterization of LDIDD
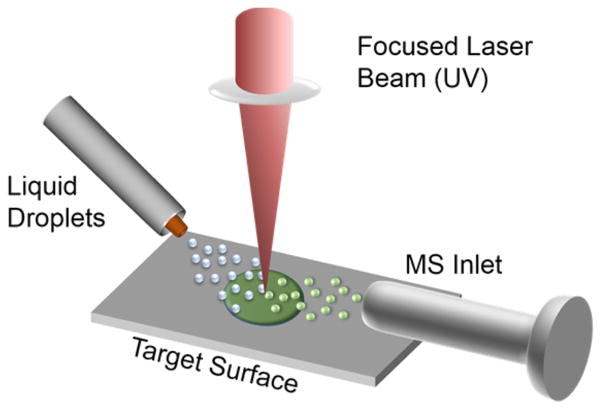
Figure 1 shows a schematic of the LDIDD-MS system. The LDIDD-MS approach relied on a pulsed (~8 ns) 266 nm laser set to a 15 Hz repetition rate for desorption/ionization of target molecules on a surface. Liquid droplets formed by a nebulizing N2 gas flow were directly sprayed onto the laser-beam spot on the surface. Molecules that were desorbed, and in some cases ionized by the laser pulse, were captured in the liquid droplets and carried to the mass spectrometer.
Figure 1.
Schematic of experimental setup for desorption/ionization droplet delivery mass spectrometry (LDIDD-MS). Focused and pulsed UV laser-irradiated target molecules on a surface for desorption and ionization. The desorbed ions are captured in liquid droplets that were directed onto the laser-irradiated region. The resulting splash of smaller droplets was delivered to a mass spectrometer.
The UV laser is known to induce fragmentation of the molecules.19 We expected a fraction of the analytes to be fragmented. However, we found little or no fragmentation of molecules with LDIDD-MS. We suggest that this lack of fragmentation can be attributed to the cage effect20 of the liquid droplet. The solvent cluster surrounding solute molecules works as a cage for trapping the fragments of solute molecules and cools them, leading to the recombination of the fragments by modifying the distribution of the excessive energy during photofragmentation.21
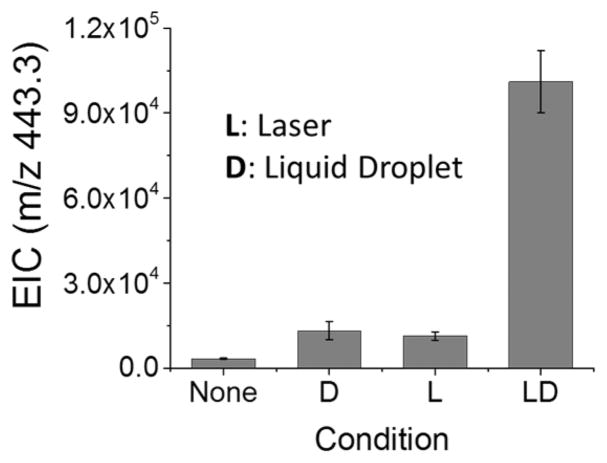
To explore the mechanism of desorption and delivery of the desorbed ions to the mass spectrometer inlet, we operated LDIDD-MS under different settings by turning on and off the laser and liquid droplets. The analyte we selected was rhodamine B, which possesses a net charge, so the different settings of the laser and the droplet influenced only desorption and delivery. We adjusted a set of parameters for the droplets that included solvent flow rate (0.3 μL/min), droplet incident angle (55°), and nebulizing gas pressure (80 psi) to minimize desorption by the droplets.22 When there was only a stream of droplets or the focused and pulsed laser, denoted as D and L, the ion signal intensity of rhodamine B remained at 1.3 × 104 and 1.1 × 104 (au), respectively. When both laser and stream of droplets were turned on, denoted as LD, the ion intensity increased to 1.0 × 105 (au) (Figure 2). This result indicates that the droplets on their own did not effectively desorb ions for mass spectrometry. Although the laser was operated at a sufficiently high power of 50 μJ per pulse at 15 Hz, the intensity of the rhodamine B ions was much less compared to that when both the laser and droplet stream were on. This showed that the desorbed ions were not effectively delivered to a mass spectrometry inlet. However, when both the laser and stream of droplets were on, the ion intensity was much improved. These studies demonstrate that liquid droplets sprayed directly on the laser desorption region and splashed from the surface are a very effective method for capturing and delivering analytes desorbed by the UV laser.
Figure 2.
Desorption and delivery mechanism by LDIDD-MS applied to rhodamine B showing the extracted ion current (EIC) under different conditions. D: Only droplets of (1:1) methanol–water at a flow rate of 0.3 μL/min with nebulization gas pressure (N2, 80 psi); L: only laser (266 nm, 50 μJ/pulse, 15 Hz repetition rate); and LD: both laser and liquid droplets.
There are two possible ways of ionizing analytes with LDIDD: (1) photoionization caused by the UV laser at 266 nm and (2) electrospray ionization from charged droplets generated by applying 5 kV to the syringe that is infused with a 1:1 (v/v) mixture of methanol and water. We explored whether mechanism 1 or 2 dominated the ionization of different molecules. The signal intensities of the selected molecules of bradykinin, caffeine, and phenanthrene were measured under four different conditions, as shown in Figure 3. “None” indicates both laser and voltage for electrospray were off; “L” denotes laser on and voltage off; “V” denotes laser off and voltage on; and “LV” denotes both laser and voltage on. Figure 3A shows normalized signal intensities of the bradykinin +2 charge state at m/z 530 under these four conditions. The signal intensity was largest for the case of LV. For caffeine (Figure 3B), the signal intensity of LV was still dominant. L provided a reasonable signal intensity of approximately 20%, suggesting that photoionization was an effective method for ionization, whereas the combined ionization by both laser and electrospray was five times more effective than the laser alone.
Figure 3.
Ionization mechanism of LDIDD-MS. Molecules were analyzed under four different conditions to assess the effect of photoionization by UV laser and electrospray ionization by voltage when applied to the droplets. The letters on the x-axis refer to the four different conditions: None (laser and voltage off), L (laser on and voltage off), V (laser off and voltage on), and LV (laser and voltage on). Normalized ion intensities of (A) + 2 charge state of bradykinin, (B) caff eine, and (C) phenanthrene (m/z 178.08 for M+ and m/z 179.8 for [M + H]+). The power of the laser was 50 μJ/pulse with a 15 Hz firing rate. A 5 kV voltage was applied for electrospray ionization.
For phenanthrene (Figure 3C), which is known to be difficult to ionize by electrospray ionization,23 only the L condition showed a higher signal compared to the LV condition, indicating that photoionization plays a dominant role for the ionization of polycyclic aromatic hydrocarbons. In cases where the laser was turned off, we cannot exclude that the low signal intensity might be caused by the lack of desorption and not just a lack of photoionization.
The combined effect of photoionization by the UV laser and electrospray ionization boosted the ionization of analytes in an analyte-dependent manner. Photoionization without electro-spray ionization was still effective for the molecules hardly ionizable with a soft ionization technique, such as phenanthrene. This synergistic effect can be partly attributed to the additional supply of protons from the photodissociation of water by the UV laser in liquid droplets.
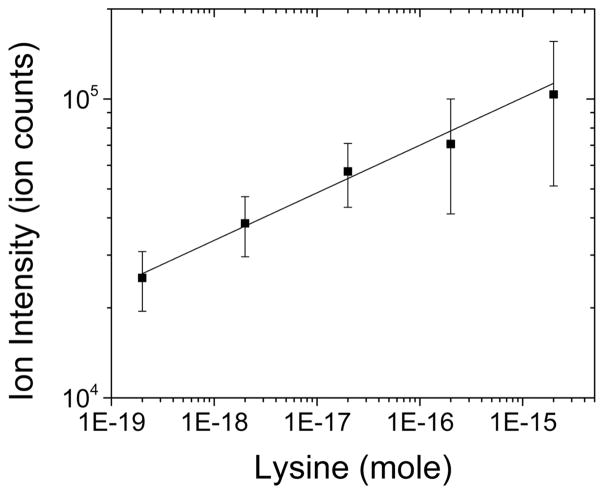
Figure 4 shows the sensitivity and linearity of the LDIDD-MS ion signal for lysine. The signal intensity was linearly proportional to the amount of lysine over 5 orders of magnitude, illustrating that LDIDD-MS is suitable for quantitative analysis. The detection limit for lysine was 2 amol using a 3 dB criterion. This low limit of detection results from an effective ionization and delivery of molecules by LDIDD-MS.
Figure 4.
Plot of the ion count versus amount of lysine. The ion intensity was found to be proportional to the amount of lysine over a concentration range of 5 orders of magnitude. The minimum detection limit of LDIDD-MS for lysine was 2 amol. The solid line is a linear fit to the data points (R2 = 0.98).
Spatial Resolution of LDIDD
Figure 5 presents images acquired by LDIDD-MS. Figure 5A shows the scanned MS signal from a microcontact-printed rhodamine B ink pattern, demonstrating the spatial resolution of this technique. The spatial resolution, defined as the distance required for the signal intensity to rise from 20 to 80% of the maximum intensity,24,25 was calculated to be 2.4 μm from Figure 5B. Figure 5C is an optical microscopy image of a sliced mouse brain. Images of sliced mouse brain tissue were taken by LDIDD-MS at 200 μm pixel size (Figure 5D–H) and 5 μm pixel size (Figure 5K and L). Panels D–H in Figure 5 show mass spectrometry images for sulfatide C24:1, phosphoserine (36:1), phosphatidylinositol (38:4), phosphoserine (34:1), and sulfitides C24:1-OH, respectively. The detected molecular species exhibited a heterogeneous distribution across different regions in a coronal section of a mouse brain. Sulfatide C24:1 at m/z 888.6 was detected with high abundance in the midbrain and hippocampus region and low abundance in the corpus callosum and superior colliculus. Phosphoserine (36:1) at m/z 788.5 was homogeneously distributed in the entire brain except for the corpus callosum. Phosphatidylinositol (38:4) at m/z 885.6 was found to be highly abundant in the hippocampus, entorhinal cortex, and midbrain regions. Phosphoserine (34:1) at m/z 760.6 was detected uniformly in the entire brain section. Sulfitides C24:1-OH at m/z 904.6 was detected with high abundance in the superior colliculus and midbrain region and low in corpus callosum.
Figure 5.
Imaging by LDIDD-MS. (A) A rhodamine B ink pattern made with microcontact printing as imaged by LDIDD-MS. (B) The spatial resolution defined by the distance required for the intensity to rise from the base level of 20% to 80% of the maximum was measured to be 2.4 μm. (C) H&E stained image of a mouse brain tissue slice. Mass spectrometric images acquired at negative mode for (D) m/z 888.6 identified as sulfatide C24:1, (E) m/z 788.5 identified as phosphoserine (36:1), (F) m/z 885.6 identified as phosphatidylinositol (38:4), (G) m/z 760.6 identified as phosphoserine (34:1), and (H) m/z 904.6 identified as sulfitides C24:1-OH. (I) Optical image of sliced mouse brain tissue and (J) H&E stained image of the red-square region in (I). (K) m/z 888.6 identified as sulfatide C24:1 and (L) m/z 906.4 identified as sulfitides C24:1-OH. The minimum distinguishable distance was around 7 μm, as shown in the inset. (M) Ion intensity profile for m/z 906.4 corresponding to the red line in (L) inlet. The spatial resolution defined by the distance required for the intensity to rise from the base level of 20% to 80% of the maximum was measured to be 3 μm. The scale bar for (C–H) is 1 mm and (J–L) is 50 μm.
Panels K and L in Figure 5 show the distribution of sulfatide C24:1 for m/z 888.6 and sulfitides C24:1-OH for m/z 906.4 at 5 μm spatial resolution. Phosphatidylinositol (38:4) at m/z 888.6 exhibited a high intensity in the midbrain region but was low in the parieto-temporal lobe. Sulfitides C24:1-OH at m/z 906.4 was detected with a higher intensity than that of phosphatidylinositol (38:4). A few speckles of m/z 906.4 were observed, indicating the existence of a single or group of cells exhibiting high abundance of the phosphatidylinositol (38:4). The minimum distinguishable distance in the tissue imaging was around 7 μm. The spatial resolution for the tissue imaging based on the 20–80% rule was measured to be 3 μm (Figure 5M). The identification of the mass peaks using a tandem MS analysis are provided in Figures S1–S5.
Single-Cell LDIDD-MS
The high spatial resolution and sensitivity of LDIDD-MS allow for the analysis of single cells, which we demonstrated by examining the apoptotic process of HEK 293T cells, a model system for studying apoptosis.26–28 We induced apoptosis in HEK 293T cells by applying 35 mM ethanol for 20 min to the cell culture. Strong differences were apparent in both bright-field images and in the changes in the types and intensities of the peaks in the mass spectra (Figure 6). The cell shrinkage in Figure 6B was a typical morphological change during apoptosis.29 Panels E and F in Figure 6 illustrate large differences in the regulation of lipids during apoptosis.
Figure 6.
MS analysis of single HEK 293T cells that have undergone apoptosis after being treated with 35 mM ethanol for 20 min. Representative phase-contrast images of (A) an untreated cell and (B) an apoptosis-induced cell with a yellow line to denote where the scanning was performed; (C, D) Relative amounts of total ion current acquired from a LDIDD-MS scan of the untreated and apoptosis-induced cells, respectively; (E, F) Mass spectra of the untreated and apoptosis-induced cells, respectively. The scale bar shown in (A–D) is 10 μm.
Figure 7 shows the intensities of PC with different carbon chain lengths as a function of the progression of apoptosis. The identification of these PCs with a tandem MS analysis are provided in Figures S6–S8. The intensities of PC (33:1) at m/z 768.55 and PC (34:1) at m/z 782.57 with a shorter carbon chain length increased whereas PC (36:1) at m/z 810.60 with a longer carbon chain length decreased. Table 1 summarizes the detected molecules and their regulation in 293T cell apoptosis. The tandem MS analysis for the molecules are provided in Figures S5–S10. To the best of our knowledge, the upregulation of PCs with a shorter length carbon chain and downregulation of PC with a longer length carbon chain upon apoptosis has not previously been reported. This shortening of the PC carbon chain may lead to an increase of membrane curvature,30 which may provide a clue for the mechanism of cell body shrinkage and cell membrane blebbing observed during apoptosis. Further studies are needed to address the mechanism of the carbon chain length change during apoptosis. This result shows that this technique is readily applicable for single-cell analysis under ambient conditions.
Figure 7.

Normalized intensities of PC with different carbon chain lengths in HEK 293T cells as a function of time after treatment with 35 mM ethanol. Note that the intensities of relatively shorter carbon chain PCs gradually increased whereas the intensities of longer carbon chain PCs gradually decreased. The error bars represent the standard deviation of 28 cells.
Table 1.
Summary of the Detected Molecules and Their Regulation in 293T Cell Apoptosis
| compound | ion | observed m/z | theoretical m/z | mass error (ppm) | fold changea |
|---|---|---|---|---|---|
| PC(32:0) | [M + K]+ | 772.52415 | 772.5253 | 1.5 | 2.54 |
| PC (33:1) | [M + Na]+ | 768.55008 | 768.5514 | 1.7 | 40.5 |
| PC (34:1) | [M + Na]+ | 782.56593 | 782.5670 | 1.4 | 51.8 |
| PC (36:1) | [M + Na]+ | 810.59706 | 810.5983 | 1.5 | 0.053 |
| PC (36:1) | [M + K]+ | 826.57076 | 826.5723 | 1.8 | 0.15 |
The fold change was defined by the ratio of the EIC of each compound after apoptosis over the EIC before apoptosis. The EIC was normalized to the total ion current of each spectrum.
SIMS has been employed for the analysis of cell membrane lipid and metabolites in the cell membrane.31–33 Ewing and coworkers have reported the difference in the composition of phosphatidylcholine lipid at the junction between two cells.32 The localization of vitamin E in the neuronal cell membrane at the subcellular level was reported by Sweedler and coworkers.33 Although SIMS has demonstrated its usefulness for lipid analysis at high resolution, SIMS has limitations including operation under high vacuum and detection of only low mass species. In contrast, our work has proved the capability of analyzing cell lipids at the single-cell level with LDIDD-MS under ambient conditions.
Real-Time Measurement of Samples in Liquid Phase and Detection of Live-Cell Secretions
Mass spectrometry is destructive by nature so it can only be applied to nonviable dead cells. The mass spectrometric analysis of aqueous samples using femtosecond laser vaporization-based mass spectrometry was previously reported by Levis and co-workers,34 demonstrating that the solution-phase conformation of proteins was preserved during laser vaporization. We explored the application of LDIDD-MS for mass spectrometric analysis for detecting secreted molecules from live cells.
To first test that it was possible to detect molecules in aqueous solutions by LDIDD-MS, we focused the fundamental wavelength of the Nd:YAG laser at 1064 nm on the surface of a liquid sample. The laser ablated a small amount of liquid that contained dissolved analytes. The ablated plume of liquid was captured in the droplet stream and delivered to the inlet of a mass spectrometer. Figure S11 presents representative mass spectra taken directly from phenylalanine, bradykinin, and cytochrome c dissolved in water. The +7 to +10 charge states of cytochrome c indicate that the LDIDD mass spectrum obtained from the liquid sample detected the naturally folded structure of this protein.35
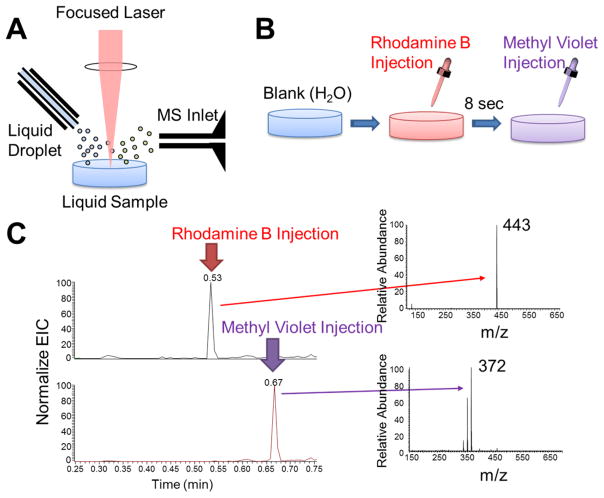
Figure 8 demonstrates a real-time measurement of liquid-phase samples by applying LDIDD-MS to aqueous solutions containing drops of different dyes added one after the other. The setup is illustrated in Figure 8A. Rhodamine B at a concentration of 10 μg/mL was injected into the water container with 3 mL volume followed by an injection of methyl violet at the same concentration with a time delay of 8 s (Figure 8B). At each injection, the corresponding mass peak of the dye was detected (Figure 8C). After detection, the intensity of the mass signal decreased because the dyes diffused throughout the water container. The temporal resolution of the real-time analysis of the liquid sample in Figure 8 was 60 ms. The temporal resolution can be pushed further to the limit of the scanning rate of a mass spectrometer if the sample concentration is high enough to be detectable by LDIDD-MS. With the particular mass spectrometer used for the present studies, the achievable maximum temporal resolution was 12 ms. This test demonstrates the capability of real-time measurement of samples in liquid phase.
Figure 8.
(A) Experimental setup for real-time liquid-phase mass spectrometric analyses by LDIDD-MS. (B) Rhodamine B and methyl violet dye solutions were sequentially injected in water with a delay of 8 s. (C) The corresponding ion intensity of rhodamine B (m/z 443) and methyl violet (m/z 372) as a function of time.
Next, LDIDD-MS was applied to track real-time secretion from live cells. We used a neuronal cell line, PC12, because the secretion of different molecules from this cell line, including neurotransmitters, neuropeptides, and proteins, upon depolarization is well-characterized.36–38 PC12 cells, grown in cell-culture medium, were washed with PBS and transferred into a salt solution containing 20 mM HEPES buffer that maintained the pH at 7.4. The PC12 cells were placed onto a temperature-controlled stage maintained at 37 °C. A 50 mM KCl solution induced depolarization of these neuronal cells.39 Figure 9A presents the setup for the real-time measurement of cell secretions. Panels B and C in Figure 9 are the LDIDD mass spectra obtained before and after depolarization, respectively. Because the PC12 cells were washed with PBS prior to treatment, the mass spectrum before KCl treatment shows only background peaks (Figure 9B).
Figure 9.
LDIDD-MS of secreted molecules from live PC12 cells with (A) the experimental setup and (B, C) mass spectra before and after (120 s) treatment with 50 mM KCl solution to cause depolarization-induced exocytosis, respectively. Several secreted molecules were detected from the PC12 cells, including phenethylamine (m/z 122.09676), tyramine (m/z 138.09177), and dopamine (m/z 154.08685). Note that the y-axis scale for (B) is ten times smaller than for (C).
After the depolarization of PC12 cells by treating with KCl solution, several mass peaks were detected (Figure 9C), for example, at m/z 122, 134, and 154. We hypothesized the two analytes at m/z 122 and 134 to be molecules involved in trace amine metabolism,40 closely related to synthesis pathways of canonical PC12 catecholamines, and the m/z 154 peak to be dopamine. By analyzing a larger sample of secreted molecules obtained from a bulk culture of PC12 cells with multiple reaction monitoring (MRM) liquid chromatography–mass spectrometry (LC–MS), we identified the two peaks of m/z 122 and 134 to be phenethylamine and tyramine (Figures S12 and S13). These molecules have previously been shown to coexist in dopaminergic neurons.40 The exocytosis measurement in the present work was carried out by collecting exocytosis events averaged over approximately two seconds from a group of cells. A summary of the detected secreted molecules is provided in Table 2. Although the identity of dopamine (m/z 154) was not confirmed by the MRM LC-MS assay because of its low signal intensity, we presumed the m/z 154 peak to be dopamine because the molecule is known to be released from PC12 cells upon depolarization,37,41,42 and the observed mass neatly matches with the theoretical mass of dopamine with a small mass error at 0.3 ppm (Table 2).
Table 2.
List of Catecholamines Detected from the Measurement of the Secreted Molecules from PC12 Cells Using LDIDD-MS
| compound | theoretical m/z ([M + H]+) | observed m/z ([M + H]+) | mass error (ppm) | confirmation of identity |
|---|---|---|---|---|
| phenethylamine | 122.09697 | 122.09676 | 1.7 | MRM LC-MSb |
| tyramine | 138.09188 | 138.09177 | 0.8 | MRM LC-MSb |
| dopamine | 154.08680 | 154.08685 | 0.3 | no |
The identity of the detected compound was confirmed with multiple reaction monitoring (MRM) LC–MS (see the Supporting Information for further detail and MRM LC-MS data).
After the LDIDD-MS analysis, the cells were incubated in propidium iodide solution, which is widely used for the assessment of membrane damage and cell death.43,44 We did not observe any indications of cell death or damage (data not shown). These results demonstrate that LDIDD-MS can measure live-cell secretion profiles in real time. A distinct difference of LDIDD-MS compared to previous approaches of mass spectrometric analysis of live-cell secretion, including that by Masujima and co-workers,17,18 is that the collection and delivery of analytes in liquid phase was performed in one step. This makes LDIDD-MS suitable for real-time kinetic analysis of live-cell secretion or evolution of chemical reactions in liquid phase.
CONCLUSIONS
We have developed a new form of ambient ionization mass spectrometry using a UV laser for desorption and ionization of analytes and electrospray liquid droplets to deliver the ionized analytes to a mass spectrometer. This combination exhibits a synergistic effect of increased ionization efficiency caused by photoionization from the UV laser pulse and electrospray ionization. High spatial resolution of imaging was achieved at around 2.4 μm for an ink-printed pattern and 3 μm for mouse brain tissue. Effectively capturing and delivering analytes under ambient conditions achieved a detection limit of 2 amol of lysine. The mass signal was found to be linear with concentration over 5 orders of magnitude. This high sensitivity and spatial resolution of LDIDD made it possible to acquire mass spectra from a single cell, as we did in analyzing apoptosis in single HEK 293T cells. We were able to observe at the single-cell level the up- and downregulation of different PC lipids with various carbon chain lengths as apoptosis progressed. We also demonstrated the capability of LDIDD-MS to measure analytes in a solution in real time. Live-cell secretions, including neurotransmitters from PC12 cells, were detected with LDIDD-MS when PC12 cells were depolarized with a concentrated KCl solution.
Thus, LDIDD-MS proves to be a new ambient ionization method for high-resolution mass spectrometry imaging with high sensitivity that requires minimal sample preparation and bypasses the need for a matrix for ionization. Furthermore, the capability of analyzing live-cell secretions in real time allows for the collection of temporal information on a cell’s secreted molecules.
Supplementary Material
Acknowledgments
We thank Karolina Krasińska from the Vincent Coates Foundation Mass Spectrometry Laboratory, Stanford University Mass Spectrometry (http://mass-spec.stanford.edu) for MRM LC-MS and help with analysis. This work was supported by National Institutes of Health (NIH 1R21DA039578-01) and the Institute for Basic Science (IBS-R013-D1). E.T.J. was supported by the Swedish Research Council through grant 2015-00406. This work was also supported in part by NIH P30 CA124435 utilizing the Stanford Cancer Institute Proteomics Shared Resource.
Footnotes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.anal-chem.6b00881.
Protocols for LC–MS analyses with MRM, tandem MS analysis of lipid, mass spectra of the liquid-phase samples analyzed by LDIDD-MS, and MRM LC–MS analysis results of neurotransmitters from PC12 cells (PDF)
References
- 1.Walch A, Rauser S, Deininger SO, Höfler H. Histochem Cell Biol. 2008;130:421–434. doi: 10.1007/s00418-008-0469-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esquenazi E, Yang YL, Watrous J, Gerwick WH, Dorrestein PC. Nat Prod Rep. 2009;26:1521–1534. doi: 10.1039/b915674g. [DOI] [PubMed] [Google Scholar]
- 3.McDonnell LA, Heeren RMA. Mass Spectrom Rev. 2007;26:606–643. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- 4.Nimesh S, Mohottalage S, Vincent R, Kumarathasan P. Int J Mol Sci. 2013;14:11277–11301. doi: 10.3390/ijms140611277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monge ME, Harris GA, Dwivedi P, Fernàndez FM. Chem Rev. 2013;113:2269–2308. doi: 10.1021/cr300309q. [DOI] [PubMed] [Google Scholar]
- 6.Vidova V, Novák P, Strohalm M, Pòl J, Havlìček Vr, Volný M. Anal Chem. 2010;82:4994–4997. doi: 10.1021/ac100661h. [DOI] [PubMed] [Google Scholar]
- 7.Williams P. Annu Rev Mater Sci. 1985;15:517–548. [Google Scholar]
- 8.Cody RB, Laramee JA, Durst HD. Anal Chem. 2005;77:2297–2302. doi: 10.1021/ac050162j. [DOI] [PubMed] [Google Scholar]
- 9.Wiseman JM, Ifa DR, Zhu YX, Kissinger CB, Manicke NE, Kissinger PT, Cooks RG. Proc Natl Acad Sci U S A. 2008;105:18120–18125. doi: 10.1073/pnas.0801066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang MZ, Hsu HJ, Lee LY, Jeng JY, Shiea JT. J Proteome Res. 2006;5:1107–1116. doi: 10.1021/pr050442f. [DOI] [PubMed] [Google Scholar]
- 11.Nemes P, Vertes A. Anal Chem. 2007;79:8098–8106. doi: 10.1021/ac071181r. [DOI] [PubMed] [Google Scholar]
- 12.Rezenom YH, Dong J, Murray KK. Analyst. 2008;133:226–232. doi: 10.1039/b715146b. [DOI] [PubMed] [Google Scholar]
- 13.Brady JJ, Judge EJ, Simon K, Levis RJ. Proc SPIE. 2010;7568:7568R. [Google Scholar]
- 14.Cochran KH, Barry JA, Muddiman DC, Hinks D. Anal Chem. 2013;85:831–836. doi: 10.1021/ac302519n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sampson JS, Murray KK, Muddiman DC. J Am Soc Mass Spectrom. 2009;20:667–673. doi: 10.1016/j.jasms.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng SC, Cheng TL, Chang HC, Shiea J. Anal Chem. 2009;81:868–874. doi: 10.1021/ac800896y. [DOI] [PubMed] [Google Scholar]
- 17.Fujii T, Matsuda S, Tejedor ML, Esaki T, Sakane I, Mizuno H, Tsuyama N, Masujima T. Nat Protoc. 2015;10:1445–1456. doi: 10.1038/nprot.2015.084. [DOI] [PubMed] [Google Scholar]
- 18.Masujima T. Anal Sci. 2009;25:953–960. doi: 10.2116/analsci.25.953. [DOI] [PubMed] [Google Scholar]
- 19.Rockwood S, Reilly JP, Hohla K, Kompa KL. Opt Commun. 1979;28:175–178. [Google Scholar]
- 20.Edwards JO. Progress in Inorganic Chemistry, Inorganic Reaction Mechanisms. Vol. 13 John Wiley & Sons; 2009. [Google Scholar]
- 21.Bačić Z. In: Theory of Atomic and Molecular Clusters: With a Glimpse at Experiments. Jellinek J, editor. Springer; Berlin, Heidelberg: 1999. pp. 54–85. [Google Scholar]
- 22.Venter A, Sojka PE, Cooks RG. Anal Chem. 2006;78:8549–8555. doi: 10.1021/ac0615807. [DOI] [PubMed] [Google Scholar]
- 23.Moriwaki H, Imaeda A, Arakawa R. Anal Commun. 1999;36:53–56. [Google Scholar]
- 24.Jungmann JH, MacAleese L, Buijs R, Giskes F, de Snaijer A, Visser J, Visschers J, Vrakking MJJ, Heeren RMA. J Am Soc Mass Spectrom. 2010;21:2023–2030. doi: 10.1016/j.jasms.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 25.Whitby JA, Ostlund F, Horvath P, Gabureac M, Riesterer JL, Utke I, Hohl M, Sedlacek L, Jiruse J, Friedli V, Bechelany M, Michler J. Adv Mater Sci Eng. 2012;2012:1. [Google Scholar]
- 26.Chang H, Ma YG, Wang YY, Song Z, Li Q, Yang N, Zhao HZ, Feng HZ, Chang YM, Ma J, Yu ZB, Xie MJ. J Cell Physiol. 2011;226:1660–1675. doi: 10.1002/jcp.22497. [DOI] [PubMed] [Google Scholar]
- 27.Mao WP, Ye JL, Guan ZB, Zhao JM, Zhang C, Zhang NN, Jiang P, Tian T. Toxicol In Vitro. 2007;21:343–354. doi: 10.1016/j.tiv.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 28.Verbsky J, Majerus PW. J Biol Chem. 2005;280:29263–29268. doi: 10.1074/jbc.M503366200. [DOI] [PubMed] [Google Scholar]
- 29.Elmore S. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szule JA, Fuller NL, Rand RP. Biophys J. 2002;83:977–984. doi: 10.1016/s0006-3495(02)75223-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorey DR, Morrison GH, Chandra S. Anal Chem. 2001;73:3947–3953. doi: 10.1021/ac0103266. [DOI] [PubMed] [Google Scholar]
- 32.Ostrowski SG, Van Bell CT, Winograd N, Ewing AG. Science. 2004;305:71–73. doi: 10.1126/science.1099791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monroe EB, Jurchen JC, Lee J, Rubakhin SS, Sweedler JV. J Am Chem Soc. 2005;127:12152–12153. doi: 10.1021/ja051223y. [DOI] [PubMed] [Google Scholar]
- 34.Brady JJ, Judge EJ, Levis RJ. Proc Natl Acad Sci U S A. 2011;108:12217–12222. doi: 10.1073/pnas.1105673108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Going CC, Xia Z, Williams ER. Analyst. 2015;140:7184–7194. doi: 10.1039/c5an01710f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gualandris A, Jones TE, Strickland S, Tsirka SE. J Neurosci. 1996;16:2220–2225. doi: 10.1523/JNEUROSCI.16-07-02220.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Westerink R, Ewing AG. Acta Physiol. 2008;192:273–285. doi: 10.1111/j.1748-1716.2007.01805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Margioris AN, Markogiannakis E, Makrigiannakis A, Gravanis A. Endocrinology. 1992;131:703–709. doi: 10.1210/endo.131.2.1353443. [DOI] [PubMed] [Google Scholar]
- 39.Kilbourne EJ, McMahon A, Sabban EL. J Neurosci Methods. 1991;40:193–202. doi: 10.1016/0165-0270(91)90068-b. [DOI] [PubMed] [Google Scholar]
- 40.Burchett SA, Hicks TP. Prog Neurobiol. 2006;79:223–246. doi: 10.1016/j.pneurobio.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 41.Zhu WH, Conforti L, Millhorn DE. Am J Physiol. 1997;273:C1143–1150. doi: 10.1152/ajpcell.1997.273.4.C1143. [DOI] [PubMed] [Google Scholar]
- 42.Koshimura K, Tanaka J, Murakami Y, Kato Y. Metab, Clin Exp. 2003;52:922–926. doi: 10.1016/s0026-0495(03)00059-3. [DOI] [PubMed] [Google Scholar]
- 43.Steinkamp JA, Lehnert BE, Lehnert NM. J Immunol Methods. 1999;226:59–70. doi: 10.1016/s0022-1759(99)00053-8. [DOI] [PubMed] [Google Scholar]
- 44.Liao TT, Jia RW, Shi YL, Jia JW, Wang L, Chua H. J Environ Sci Health, Part A: Toxic/Hazard Subst Environ Eng. 2011;46:1769–1775. doi: 10.1080/10934529.2011.624016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.