SUMMARY
The recent discovery of the positive-sense ssRNA Orsay virus (OV) as a natural pathogen of the nematode, Caenorhabditis elegans, has stimulated interest in exploring virus-nematode interactions. However, OV infection is restricted to a small number of intestinal cells, even in nematodes defective in their antiviral RNA interference (RNAi) response, and is neither lethal nor vertically transmitted. Using a fluorescent reporter strain of the negative-sense ssRNA vesicular stomatitis virus (VSV), we show that microinjection of VSV particles leads to a dose-dependent, muscle tissue-tropic, lethal infection in C. elegans. Furthermore, we find nematodes deficient for components of the antiviral RNAi pathway, such as Dicer-related helicase 1 (DRH-1), to display hypersusceptibility to VSV infection as evidenced by elevated infection rates, virus replication in multiple tissue types, and earlier mortality. Strikingly, infection of oocytes and embryos could also be observed in drh-1 mutants. Our results suggest that the antiviral RNAi response not only inhibits vertical VSV transmission but also promotes transgenerational inheritance of antiviral immunity. Our study introduces a new, in vivo virus-host model system for exploring arbovirus pathogenesis and provides the first evidence for vertical pathogen transmission in C. elegans.
Keywords: Caenorhabditis elegans, vesicular stomatitis virus, RNA interference, small RNAs, virus-host interactions, vertical transmission, transgenerational inheritance, antiviral immunity
INTRODUCTION
The genetic tractability, ease of culture, and susceptibility to a variety of bacterial and fungal pathogens [1, 2], has made C. elegans attractive for exploring microbe-host interactions. However, due to a lack of convenient experimental systems, relatively few studies have explored virus-C. elegans interactions. Initial studies investigating virus-C. elegans interactions used primary cell cultures and defined an antiviral role for the nematode RNA interference (RNAi) response [3, 4].
RNAi is a highly conserved mechanism of gene silencing that contributes to antiviral defense in insects [5], plants [6], and mammals [7, 8]. In C. elegans, the antiviral RNAi response is initiated after recognition and cleavage of viral double-stranded RNAs by a complex consisting of Dicer-related helicase 1 (DRH-1), DCR-1, and RDE-4, into 23 nucleotide (nt)-long primary small interfering RNAs (siRNAs) [9]. These duplex siRNAs are then loaded into the primary Argonaute protein RDE-1 and one strand of the duplex is lost. These RDE-1-primary siRNA complexes then recruit RNA-dependent RNA polymerase complexes to viral RNA targets where they generate secondary siRNAs termed “22Gs”, which are typically 22 nts long and contain 5′ guanines [10, 11]. These 22Gs then complex with, and guide, secondary Argonautes to complementary viral single-stranded (ss) RNA targets (e.g. mRNAs or genomes) resulting in Argonaute-mediated target cleavage and inhibition of virus replication [9].
Despite initial insights into virus-C. elegans interactions provided by primary cell culture studies, these cultures have limited utility because they are technically-challenging to generate and may not be representative of infection in animals. To investigate virus-C. elegans interactions in vivo, a nematode strain encoding a Flock House virus replicon was created by Lu et al [12]. Although this replicon system has identified host factors, such as DRH-1 [13], that restrict virus replication, it cannot identify nematode factors influencing aspects of the viral life cycle that would only be afforded with a bona fide viral pathogen (e.g. transmission, entry, exit, etc.) [14].
More recently, the positive-sense ssRNA Orsay virus (OV) was described as a natural pathogen of C. elegans [14]. The discovery of OV represents an important step in defining virus-nematode interactions, however, this model also possesses limitations. First, OV infection is limited to 1–6 intestinal cells, even during infection of RNAi-deficient animals [15]. Therefore, identifying antiviral factors specific to non-intestinal tissues with the OV model may be difficult. Second, because OV infection is not lethal [14], scoring the minor pathological features of infection can be challenging. Third, recombinant OV strains expressing fluorescent or quantifiable reporter genes are unavailable, making analysis of OV replication limited to PCR or immunofluorescence-based methods. Finally, because OV is not vertically transmitted [14], this model may be unsuitable for identifying immunity mechanisms guarding against vertical transmission.
Given the shortcomings of current virus-C. elegans model systems, we asked whether an alternative system could be established using the negative-sense ssRNA vesicular stomatitis virus (VSV). VSV is a member of the arboviruses, a group of emerging viral pathogens that are transmitted by arthropods to vertebrate hosts. The wide availability of reverse genetic and immunological tools for VSV [16, 17] makes it convenient for studying arboviral disease mechanisms relevant to human and animal health [18, 19]. Furthermore, because VSV can replicate in C. elegans primary cells [3, 4], we thought it possible that VSV could infect C. elegans animals.
Here we show that microinjection of VSV particles produces a lethal infection in C. elegans. We further show that the susceptibilities of animals to infection, the tissues infected, and animal survival, are dependent upon virus dose, culturing conditions, and host genetic background. We also establish a role for the nematode antiviral RNAi pathway in restricting arbovirus replication and pathogenesis. Finally, we use this model to provide the first evidence for vertical transmission in C. elegans and implicate the antiviral RNAi response in both inhibiting vertical transmission and promoting transgenerational inheritance of antiviral immunity.
RESULTS
Microinjection of VSV into C. elegans results in an infection primarily restricted to muscle tissue
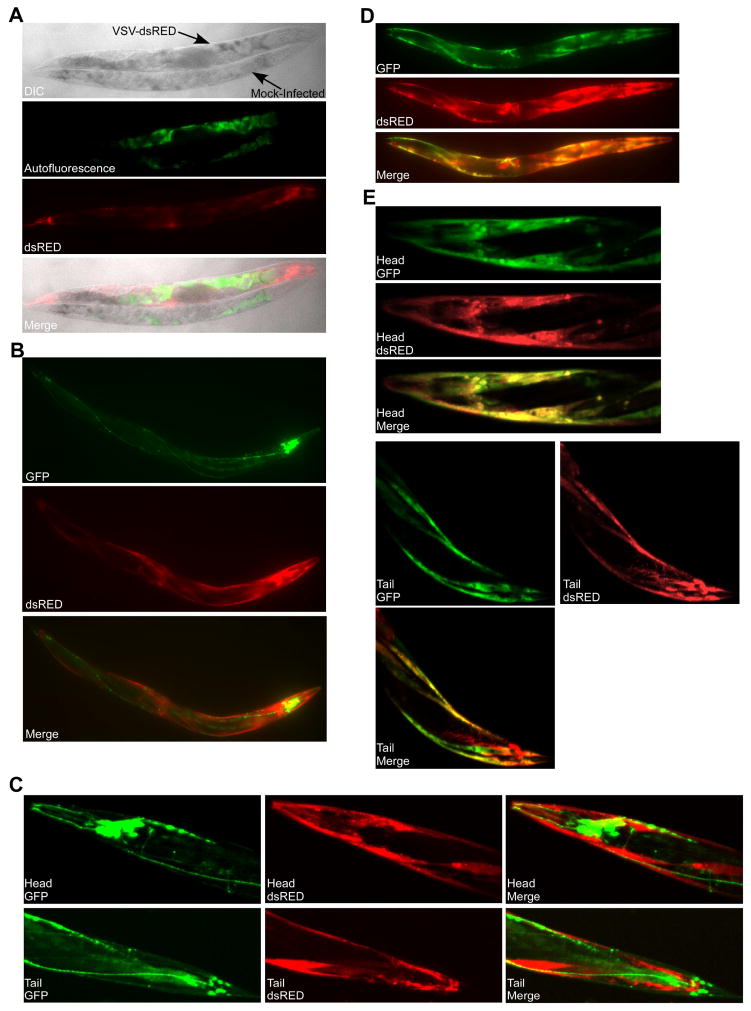
To determine if VSV could infect C. elegans, we microinjected wild-type (N2 strain) adults with a recombinant VSV strain encoding the fluorescent reporter dsRED (VSV-dsRED) [20]. We targeted the body cavity and intestinal tissue just posterior to the terminal bulb of the pharynx for injections. After microinjection, we observed a prominent dsRED signal in animals that was both above background signals in mock-infected animals and significantly different from autofluorescence observed in intestinal tissues (Figure 1A). Microinjection of VSV-dsRED into transgenic animals expressing GFP under a neuronal promoter [21] revealed only a minor overlap of GFP and dsRED signals in head neurons (Figure 1B–C). However, significant overlap in dsRED and GFP signals was observed throughout infected transgenic animals expressing GFP under a muscle-specific promoter ([21]; Figure 1D), with clear infection of body wall muscle in the head and tail (Figure 1E). We could also establish VSV-dsRED infections in the Hawaiian isolate (CB4856) of C. elegans (Figure S1A) and in Caenorhabditis briggsae (Figure S1B). Interestingly, whereas both the N2 and Hawaiian animals displayed infection rates of 85–90%, only ~20% of C. briggsae animals displayed dsRED signal by 72 hours post-infection (hpi), suggesting that C. briggsae may be more resistant to infection (Figure S1C).
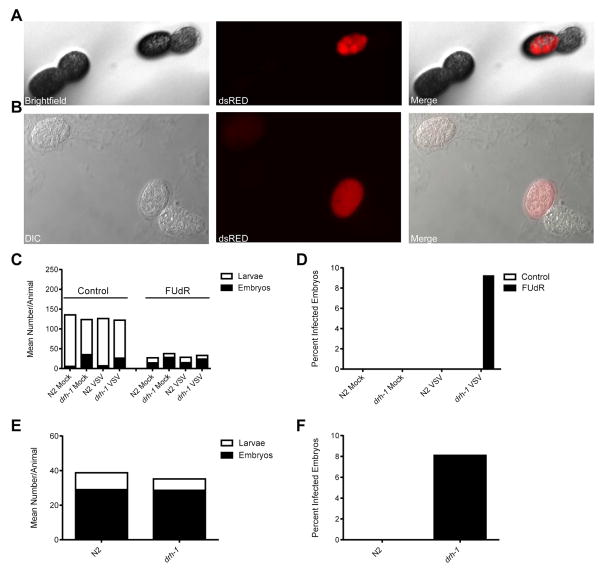
Figure 1. Microinjection of adult C. elegans animals with recombinant vesicular stomatitis virus (VSV) expressing dsRED (VSV-dsRED) results in an infection primarily restricted to muscle tissue.
(A) Differential interference contrast (DIC) and fluorescence micrographs (10X magnification) of N2 adults either mock- or VSV-dsRED-infected. Images were taken 72 hours post-infection (hpi). Green fluorescence indicates autofluorescence signal in the intestine. (B) Fluorescence micrographs (10X magnification) of psng-1::LUC-GFP adult infected with VSV-dsRED 72 hpi. (C) Confocal microscopy fluorescence micrographs (20X magnification) of head and tail regions of a psng-1::GFP-LUC adult infected with VSV-dsRED 72 hpi. (D) Fluorescence micrographs (10X magnification) of pmyo-3::LUC-GFP adult infected with VSV-dsRED 72 hpi. (E) Confocal microscopy fluorescence micrographs (20X magnification) of head and tail regions of a pmyo-3::GFP-LUC adult infected with VSV-dsRED 72 hpi. All infections were carried out using 104 plaque forming units (PFU) of VSV-dsRED per injection. See also Figure S1.
Loss of DRH-1 function results in hypersusceptibility to lethal VSV infection
Previous Flock house virus replicon [13] and OV [9] studies have implicated DRH-1 in the restriction of positive-sense ssRNA virus replication in C. elegans. DRH-1 functions in a similar manner as homologous mammalian RIG-I-like helicases (RLHs) which sense cytosolic viral RNA signatures and subsequently trigger antiviral response programs [9, 22]. While mammalian RLHs and DRH-1 both trigger an antiviral response upon viral RNA recognition, mammalian RLHs trigger the interferon response [23], whereas DRH-1 promotes the initiation of the C. elegans antiviral RNAi response [9, 13].
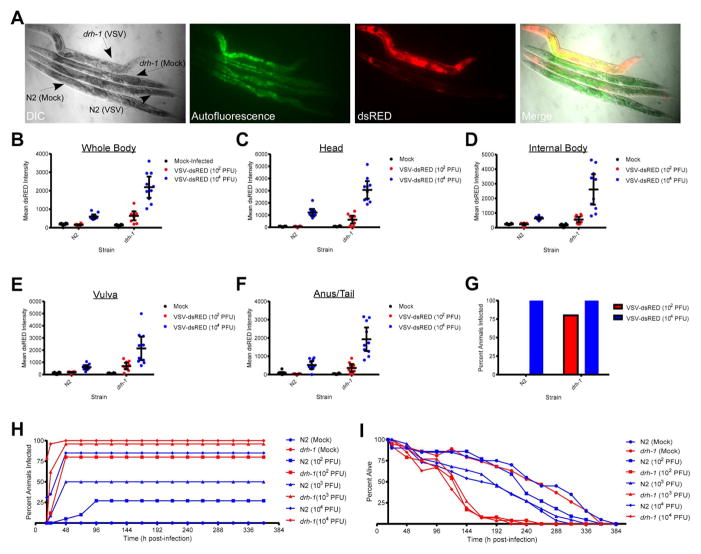
To determine if DRH-1 is involved in sensing negative-sense ssRNA virus infection, we challenged animals carrying a loss-of-function mutation in drh-1 with VSV-dsRED and compared the dsRED pattern to N2 infections. Interestingly, drh-1 worms displayed infection in multiple tissues not involved in N2 infections such as intestinal tissue (Figure 2A). To further compare dsRED signals in N2 and drh-1 strains, we microinjected either a low [102 plaque-forming units (PFU)] or high (104 PFU) dose of VSV-dsRED into either strain and then measured signals in either whole animals (Figure 2B) or specific tissues (Figure 2C–F) 48 hpi. Whole body dsRED measurements indicated that drh-1 animals displayed ~5- and 18-fold higher signals than mock-infected animals at the 102 and 104 PFU doses, respectively. In contrast, dsRED signals in N2 animals were either essentially identical to or ~three-fold higher than mock-infected animals at these respective doses (Figure 2B). Measurement of dsRED signals in individual tissues revealed a similar pattern with drh-1 animals presenting with ~4–10-fold higher signals at the 102 PFU dose and ~17–51-fold higher signals at the 104 PFU dose compared to mock-infected animals. Significant N2 dsRED signals were typically only observed with 104 PFU treatments and were ~3–7-fold higher than mock-infected animals. Animals were scored as infected if their whole body dsRED signals were at least two-fold higher than mock-infected animals by 48 hpi. Using this cutoff, we found that at a dose of 104 PFU, 100% of both N2 and drh-1 animals scored as infected. In contrast, no N2 and 80% of drh-1, animals scored as infected in 102 PFU treatments (Figure 2G). However, when only vulval tissue dsRED signals were used to score infection in 102 PFU treatments, ~30% of N2 animals scored as infected (Figure S2), suggesting that this tissue may be a more reliable indicator of low-level infections. These results suggest that higher viral doses allow for viral replication to reach a threshold at which dsRED signals become detectable by 48 hpi in both N2 and drh-1 strains. However, at lower viral doses, N2 animals may be more capable of suppressing VSV-dsRED replication and hence are not scored as infected by 48 hpi. Importantly, we confirmed that the elevated dsRED signal observed in drh-1 animals reflected bona fide VSV transcription using RT-PCR (Figure S3; [24]).
Figure 2. Loss of Dicer-related helicase 1 (DRH-1) function results in enhanced susceptibility to VSV-dsRED infection.
(A) Differential interference contrast (DIC) and fluorescence micrographs (10X magnification) of N2 or drh-1 adults mock-infected or infected with 104 PFU of VSV-dsRED. Images were taken 72 hpi. Green fluorescence indicates autofluorescence obtained in the intestine. (B–F) Mean dsRED intensity measurements for N2 or drh-1 adults (n = 10/treatment) 48 hpi using the indicated VSV-dsRED dose. Measurements were taken of either the whole body (B) or of the indicated tissues (C–F). Mean dsRED measurements for the entire group (horizontal bar) and 95% confidence intervals (error bars) are shown. (G) Percentage of animals displaying a whole body dsRED signal at least two-fold above mock-infected animals by 48 hpi. (H) Percentage of animals (n = 20–30/treatment) displaying dsRED signal at the indicated times post-infection. (I) Percentage of animals from (H) alive at the indicated times post-infection. See also Figures S2–3.
To examine viral susceptibilities of N2 and drh-1 strains further, we tracked animals that had been microinjected with 102, 103, or 104 PFU of VSV-dsRED for infection rate (Figure 2H) and survival (Figure 2I). The maximal number of infected animals in each injected group was reached by 48 hpi with the exception of N2 animals injected with 102 PFU, which took until 96 hpi (Figure 2H). The 102, 103, and 104 PFU doses resulted in infection of 25%, 50%, and 85% of N2 animals, respectively. In contrast, these doses resulted in 80%, 95%, and 100% infection rates in drh-1 animals (Figure 2H). Given that infection rates in several treatments never achieved 100%, it is likely that horizontal transmission of VSV between adults is inefficient.
We used lifespan assays (Figure 2I) to estimate the time at which 50% of animals in each treatment had died [expressed as lethal time 50 (LT50) in hpi]. We also calculated 95% confidence intervals (CIs) for each LT50 and in cases where 95% CIs of two different LT50 values did not overlap, these two values were deemed statistically different (P<0.05). The LT50 (95% CI) for mock-infected N2 animals [250 hpi (236-265 hpi)] was not significantly different from mock-infected drh-1 mutants [240 hpi (228-252 hpi)]. However, both N2 and drh-1 animals had significantly lower LT50 values when infected with VSV-dsRED at any of the three doses used, suggesting that VSV infection is ultimately lethal. However, N2 animals survived for significantly longer periods of time than drh-1 animals injected with the same viral dose. For example, at a dose of 102 PFU, the LT50 (95% CI) for N2 animals was 220 hpi (210-228 hpi) versus 112 hpi (104-119 hpi) for drh-1 animals. This trend was also observed at the 103 PFU dose, with LT50 (95% CI) for N2 animals being 176 hpi (166-186 hpi) versus 121 hpi (116-127 hpi) for drh-1 animals and at the 104 PFU dose [N2 = 168 hpi (162-174 hpi) versus drh-1 = 106 hpi (99-112 hpi)]. These results suggest that drh-1 animals suffer higher infection rates and reduced survival times compared to N2 animals.
A small RNA response is generated upon VSV infection
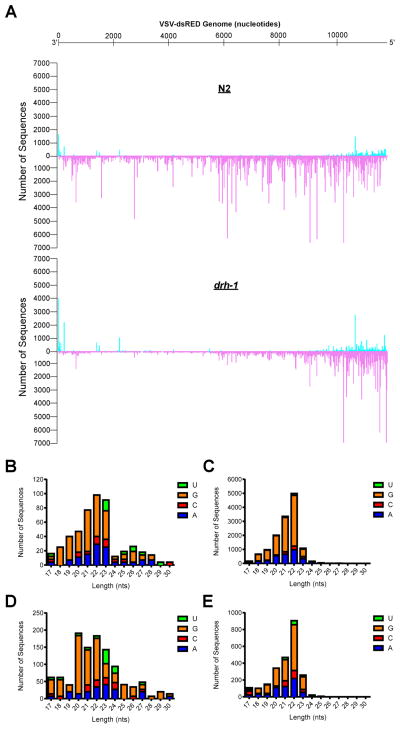
We next used deep-sequencing to determine if small RNAs (17–30 nts in length) were generated in response to VSV-dsRED infection. Small RNAs that mapped to the VSV-dsRED genome are shown in Figure 3A. These RNAs were virtually absent from uninfected animals (data not shown). In N2 libraries, small RNAs corresponding to the antisense (or genomic) strand of VSV-dsRED were ~29-fold more abundant than those mapping to the sense (antigenomic) strand (Figure 3B–C). Both antisense and sense RNAs were characterized by a peak length of 22 nts and a strong preference for G at their 5′ ends, and thus likely represent 22Gs. Consistent with a defect in the initiation of an antiviral RNAi response, ~four-fold fewer viral small RNAs were detected in drh-1 animals than in N2 infections. Furthermore, while both antisense and sense small RNAs from drh-1 animals also displayed a bias for 5′G residues, a clear 22 nt peak length was only observed among antisense small RNAs (Figure 3D–E). In addition, drh-1 antisense small RNAs were only two-fold more abundant than sense RNAs. These results suggest that both N2 and drh-1 animals raise a small RNA response to VSV-dsRED infection but that drh-1 animals may be impaired in generating antisense small RNAs.
Figure 3. Small RNAs produced upon VSV-dsRED Infection of N2 and drh-1 animals.
(A) The number of unique sequences obtained by Illumina high-throughput small RNA sequencing that match a given position in the VSV-dsRED genome in infected N2 or drh-1 animals is shown. The number of unique sequences in sense and antisense orientation are shown on the positive (cyan) and negative (purple) y-axis, respectively. The negative-sense ssRNA VSV-dsRED genome (in 3′-to-5′ orientation) is depicted above. (B–C) Features of sense (B) and antisense (C) small RNAs (length and identity of first nucleotide) cloned from VSV-dsRED-infected N2 animals. (D–E) Features of sense (D) and antisense (E) small RNAs cloned from VSV-dsRED-infected drh-1 animals.
Development of a luciferase assay for VSV replication in C. elegans
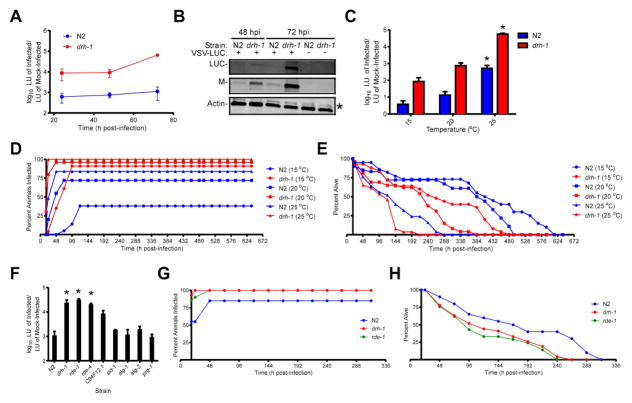
To further characterize VSV infection dynamics, we created a simple, quantitative assay to assess VSV replication. To do this, we employed a recombinant VSV strain encoding a firefly luciferase (LUC) gene under a viral promoter (VSV-LUC; [25]) along with chemiluminescent LUC assays to measure VSV gene expression. LUC activity detected from VSV-LUC infections closely mirrors virion production and serves as a convenient and sensitive assay for virus production [24]. To test the utility of these assays, we microinjected groups of N2 or drh-1 animals with VSV-LUC and collected equal numbers of animals at various times post-infection. Although light unit (LU) signals from infected N2 animals were ~600-fold higher than from mock-infected animals by 24 hpi, these signals only increased by ~two-fold by 72 hpi (Figure 4A). In contrast, LU signals from VSV-LUC-infected drh-1 animals were ~8800-fold higher than from mock-infected animals by 24 hpi and these signals increased by ~7-fold by 72 hpi. Trends observed in LUC assays were further confirmed by immunoblotting (Figure 4B). While we were unable to detect LUC protein from infected N2 lysates, we did detect a small amount of VSV Matrix structural protein (Figure 4B). In contrast, abundant LUC and Matrix proteins were detected in VSV-LUC-infected drh-1 lysates (Figure 4B). Collectively, these data show that LUC assays provide a sensitive and convenient method to assess VSV replication in C. elegans.
Figure 4. Characterization of the effect(s) of nematode culturing temperature and host genetic background on VSV replication and host survival.
(A) Luciferase (LUC) assay [in arbitrary light units (LU)] of adult animals (n =15/treatment/experiment) infected with 104 PFU of VSV-LUC. LU detected from infected animals were divided by LU obtained from mock-infected animals of the same strain. Data represent means (± SEM). (B) Immunoblot of lysates from (A) for LUC, VSV Matrix (M), and cellular actin proteins. The asterisk indicates a non-specific band in the actin immunoblot. (C) LUC assay of lysates from adult animals (n = 10/treatment/experiment) infected with 104 PFU of VSV-LUC and cultured at the indicated temperatures until 72 hpi. Data represent means (+ SEM) and asterisks indicate treatments that are significantly different (P<0.05) from 15°C treatments within strains. (D) Percentage of animals (n = 20–30/treatment) displaying dsRED signal at the indicated times post-infection with 104 PFU VSV-dsRED. (E) Percentage of animals from (D) alive at the indicated times post-infection. (F) LUC assay of lysates from adult animals (n =10/treatment/experiment) infected with 104 PFU of VSV-LUC until 72 hpi. LU detected from infected animals were divided by LU obtained from mock-infected N2 animals. Data represent means (+ SEM) and asterisks indicate treatments that are significantly different (P<0.05) than N2 treatments. (G) Percentage of animals (n = 20–30/treatment) displaying dsRED signal at the indicated times post-infection with 104 PFU VSV-dsRED. (H) Percentage of animals from (G) alive at the indicated times post-infection.
VSV replication and infection and mortality rates are temperature-dependent
To determine if temperature influences VSV replication, we measured LU signals from VSV-LUC-infected animals cultured at 15, 20, or 25°C. LU signals were significantly higher when either N2 or drh-1 animals were cultured at 25°C as compared to 15°C (P<0.05). Although not statistically different, LU signals from 20°C incubations trended towards being higher than signals from 15°C incubations (Figure 4C).
To investigate if temperature affected infection or mortality rates, we challenged animals with VSV-dsRED and then monitored animals for infection (Figure 4D) and survival (Figure 4E). Interestingly, incubation temperatures appeared to affect both the maximum percentage of animals infected and the timing of when these values were reached. For example, use of 15, 20, and 25°C incubation temperatures resulted in maximal infection rates of 38, 72, and 84% of N2 animals by 120, 72, and 48 hpi, respectively (Figure 4D). A similar trend was observed for drh-1 animals (albeit with higher infection rates) such that 91, 95, and 100% of animals incubated at 15, 20, and 25°C scored as infected by 96, 48, and 24 hpi, respectively (Figure 4D).
Examination of survival rates suggested that LT50 values decrease with increasing temperature (Figure 4E). For example, LT50 (95% CI) values for infected N2 animals cultured at 15°C [408 hpi (403-413 hpi)], 20°C [384 hpi (380-388 hpi)], and 25°C [124 hpi (116-131 hpi)] were all significantly different from one another. The LT50 (95% CI) values for infected drh-1 animals cultured at 15°C [257 hpi (239-274 hpi)], 20°C [191 hpi (177-205 hpi)], and 25°C [94 hpi (86-101 hpi)] also decreased with increasing temperature. Collectively, these results suggest that VSV replication and infection rates increase with increasing temperature while animal survival rates decrease.
Downstream components of the antiviral RNAi pathway are also required to restrict VSV infection
We next wanted to examine if strains with loss of function mutations in other RNAi pathway components display VSV hypersusceptibility. Using our LUC-based assays for viral replication, we found that, in addition to drh-1 animals, the RNAi-defective strains, rde-1 [26] and rde-4 [26], also displayed significantly higher (P<0.05) VSV-LUC replication than N2 animals (Figure 4F), suggesting that these downstream components are also required to restrict viral replication. Although LU signals in RNAi-defective C04F12.1 mutants [27] were ~8-fold higher than N2 animals, these values did not reach statistical significance (P=0.1). Furthermore, strains deficient for SID-1 (required for systemic spreading of exogenous RNAi signals [28]), ALG-1 and ALG-2 (Argonautes involved in the microRNA pathway [29]), and PRG-1 (an Argonaute required for the Piwi-interacting RNA pathway [30]) did not display altered VSV-LUC susceptibilities (P>0.05).
We next asked if the enhanced viral replication observed in rde-1 animals correlated with increased infection rates and reduced survival rates. While only 85% of N2 animals scored positive for infection, all drh-1 and rde-1 animals displayed dsRED signal by 48 hpi (Figure 4G). Furthermore, the LT50 (95% CI) values for both drh-1 [119 hpi (105-133 hpi)] and rde-1 [105 hpi (90-121 hpi)] animals were significantly lower than that of N2 animals [168 hpi (164-172 hpi)] but did not significantly differ from one another (Figure 4H). These data suggest that RNAi pathway components downstream of DRH-1 are also required for combating VSV infection.
DRH-1 is required for full immunity to vertical virus transmission
During our lifespan assays, we noticed that when we used NGM plates containing the DNA synthesis inhibitor, Fluorodeoxyuridine (FUdR), dsRED-positive embryos could often be observed with a fluorescence stereo zoom microscope on plates containing VSV-dsRED-infected drh-1 animals (Figure 5A). FUdR has been shown to induce sterility and prevent egg hatching [31] and thus is useful when tracking adult nematodes over extended incubation periods. Importantly, FUdR does not impede VSV replication [32] and thus is not expected to directly affect viral replication. We further confirmed dsRED signals in embryos using differential interference contrast and fluorescence microscopy (Figure 5B). Furthermore, dsRED signals could be observed in oocytes (Figure S4A) and embryos (Figure S4B) within infected drh-1 animals, suggesting that VSV-dsRED was entering and infecting germline tissues. We further confirmed VSV transcription in dsRED-positive embryos using RT-PCR (Figure S5).
Figure 5. DRH-1 is required for full immunity to vertical virus transmission.
(A) Brightfield and fluorescence micrographs (112X magnification) taken with a fluorescence stereo zoom microscope of a dsRED-positive embryo laid by an adult drh-1 animal infected with VSV-dsRED. (B) Differential interference contrast and fluorescence micrographs (40X magnification) of dsRED-positive embryos laid by an adult drh-1 animal infected with VSV-dsRED. (C) Mean number (+SEM) of larvae or dead embryos laid by mock- or VSV-dsRED-infected animals (n = 10 animals/treatment) 48 hpi when cultured on normal nematode growth media (control) or media containing Fluorodeoxyuridine (FUdR). (D) Percentage of total embryos from (C) displaying dsRED signal. (E) Mean number of larvae or dead embryos laid by VSV-dsRED-infected animals (n = 20 animals/treatment) 48 hpi when cultured on plates containing FUdR. (F) Percentage of total embryos from (E) displaying dsRED signal. See also Figures S4–6.
Given our inability to detect dsRED-positive embryos in our initial experiments using normal NGM plates, we asked whether FUdR treatment might influence vertical transmission of VSV-dsRED. When cultured on control plates, both mock- and VSV-dsRED-infected N2 and drh-1 animals produced a similar number of total progeny, suggesting that there were no virus- or strain-dependent differences. Similar results were obtained when injected animals were cultured on FUdR plates, albeit the total progeny sizes were reduced compared to control plates and a greater proportion of progeny were embryos (Figure 5C). Despite these reduced brood sizes, dsRED-positive embryos were only detected on FUdR plates containing drh-1 animals (Figure 5D). Because only ~9% of embryos laid by infected drh-1 animals displayed dsRED signal, we wanted to confirm that this was not simply a “jackpot” event that only occurred on drh-1 plates by chance. Therefore, we repeated these experiments using larger numbers of N2 and drh-1 adults and plated all VSV-dsRED-infected animals onto FUdR plates. Despite similar brood sizes between N2 and drh-1 strains (Figure 5E), dsRED-positive embryos were again only detected on drh-1 plates (Figure 5F).
We next asked if direct germline injection of VSV-dsRED might alter either overall or germline tissue-specific infection rates in N2 or drh-1 animals after culturing animals on FUdR. Interestingly, both strains displayed similar overall infection rates when either challenged by somatic or germline VSV-dsRED injections. In contrast, higher germline infection rates were observed when VSV-dsRED was directly injected into the germline (as oppose to the soma) for both N2 (0 versus 6%) and drh-1 (24 versus 74%) animals (Figure S6A). These data suggest that direct injection of VSV-dsRED particles into the germline may overwhelm germline antiviral defenses that might otherwise be protective when VSV-dsRED must first spread from somatic tissues.
We next asked whether FUdR was still required to observe germline VSV-dsRED infection after direct challenge of the germline of drh-1 animals. Strikingly, we observed similar germline infection rates in animals cultured on either control or FUdR-containing medium (Figure S6B). These data suggest that FUdR does not influence germline infection rates when VSV-dsRED is directly injected into the germline.
Collectively, these results show that VSV can be vertically transmitted to offspring, and that germline immunity to VSV infection can be influenced by the site of injection, the presence of FUdR in culturing medium, and DRH-1 function.
Inheritance of antiviral immunity after VSV infection
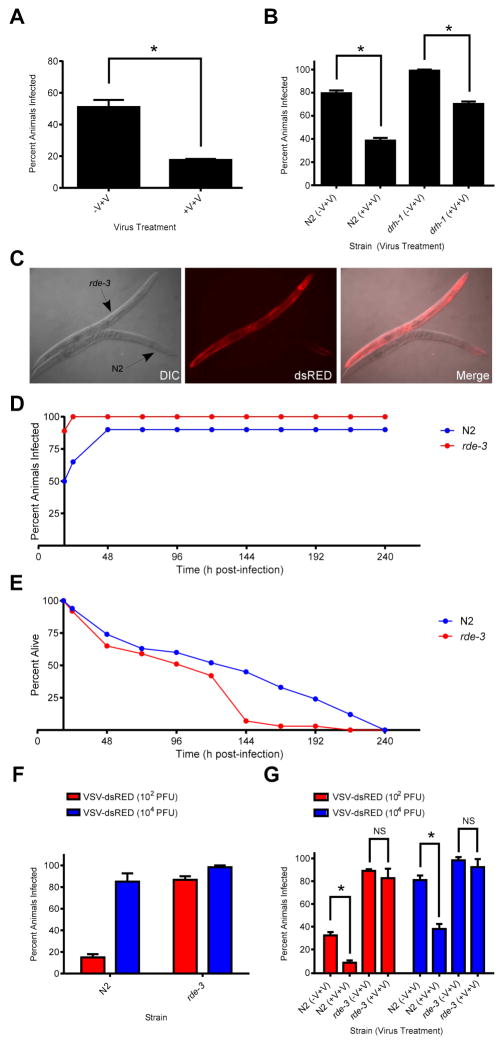
Although there is clear evidence for the transgenerational inheritance of RNAi responses generated by expression of foreign transgenes or the Flock House virus replicon in C. elegans [33, 34], inheritance of antiviral RNAi responses to OV remain controversial. Therefore, we were interested to determine if transgenerational immunity could be observed with our VSV model. To address this, we collected embryos from N2 animals that had either been mock- or VSV-dsRED-infected, allowed the resultant progeny to develop to adulthood, and then challenged these progeny with VSV-dsRED. As shown in Figure 6A, animals whose mother was previously exposed to VSV-dsRED exhibited a significantly lower infection rate than animals whose mother had been mock-infected (P<0.01). We then asked whether this inherited protection against infection could also be observed in drh-1 animals. Interestingly, we found that both N2 and drh-1 progeny animals displayed significantly reduced infection rates when they derived from infected mothers as opposed to mock-infected mothers (P<0.05) (Figure 6B).
Figure 6. Prior exposure of parental animals to virus infection confers protective immunity to their progeny that is dependent on RDE-3.
(A) Percentage of progeny animals collected from mock- (−V+V) or VSV-dsRED-infected (+V+V) mothers displaying dsRED signal 96 hpi with 103 PFU of VSV-dsRED. (B) Percentage of N2 or drh-1 progeny animals collected from mock- (−V+V) or VSV-dsRED-infected (+V+V) mothers displaying dsRED signal 72 hpi with 104 PFU of VSV-dsRED. (C) Differential interference contrast (DIC) and fluorescence micrographs (10X magnification) of N2 or rde-3 adults infected with 104 PFU of VSV-dsRED. Images were taken 72 hpi. (D) Percentage of animals displaying dsRED signal at the indicated times post-infection with 104 PFU VSV-dsRED. (E) Percentage of animals from (D) alive at the indicated times post-infection. (F). Percentage of N2 or rde-3 animals displaying dsRED signal at 72 hpi after challenge with the indicated doses of VSV-dsRED. (G) Percentage of N2 or rde-3 progeny animals collected from mock- (−V+V) or VSV-dsRED-infected (+V+V) mothers displaying dsRED signal 72 hpi after challenge with either 102 or 104 PFU of VSV-dsRED. All quantitative experiments used 20–30 animals/treatment and data in bar graphs represent means (+ SEM). Where statistical analyses were performed, asterisks indicate treatments that are significantly different (P<0.05) from control (−V+V) treatments.
In C. elegans, heritable antiviral immunity is thought to be mediated by small RNAs [34, 35], and our deep-sequencing results revealed a reduced, but not absent, small RNA response against VSV-dsRED in drh-1 animals. Therefore, we thought it possible that the residual small RNA response (consisting primarily of 22Gs) in drh-1 animals might be still capable of mediating a protective effect in resulting progeny. To address this, we made use of animals with an inactivating mutation in the rde-3 gene, which encodes a putative nucleotidyl transferase required for 22G biogenesis [36, 37]. When challenged with 104 PFU of VSV-dsRED, rde-3 animals displayed hypersusceptibility to infection as evidenced by infection of multiple tissues (Figure 6C), elevated infection rates (Figure 6D), and significantly reduced LT50 (91 versus 123 hpi) values compared to N2 animals (Figure 6E). Higher infection rates of rde-3 animals were not only observed with higher doses (104 PFU) but also at lower (102 PFU) doses, further confirming the hypersusceptibility of rde-3 animals to infection (Figure 6F). We then performed transgenerational immunity assays with N2 and rde-3 animals in which progeny from mock- or VSV-dsRED-infected mothers were challenged with either 102 or 104 PFU of VSV-dsRED and then scored for infection 72 h later. While N2 progeny derived from infected mothers displayed significantly reduced infection rates compared to those from mock-infected mothers when challenged with either low or high viral doses (P<0.05), this protective effect was lost in rde-3 animals (P>0.05) (Figure 6G). Collectively, our results suggest that VSV infection of adult animals can lead to a heritable antiviral response that is dependent upon RDE-3 function.
DISCUSSION
An advantage of using VSV-dsRED microinjection for viral studies in C. elegans is that it allows the direct delivery of a known quantity of virus into an animal and the ability to score for infection and tissue involvement in real-time. The infection of RNAi-deficient mutants has revealed the ability of VSV to not only replicate in muscle tissue (as in N2 infections) but also in various tissues throughout the animal. The muscle-tropic nature of N2 infections was surprising given that in mouse models, VSV is typically neurotropic, although other tissues, including muscle, can become involved [19]. VSV neurotropism in mice might reflect tissue-specific differences in antiviral responses [19], and so it is possible that in C. elegans, muscle tissue antiviral responses may be less robust than in other tissues.
While previous studies have reported enhanced nodavirus replication in animals defective for RNAi pathway components such as DRH-1 [9, 13], RDE-1 [14], RDE-4 [14], it was unclear whether these factors contributed to the restriction of negative-sense ssRNA virus infection in vivo. A mammalian homolog of DRH-1, RIG-I, is required to initiate an interferon response to VSV infection in vertebrates [38]. The enhanced susceptibility of drh-1 animals to VSV suggests that DRH-1 may act in an analogous manner, but instead initiate an antiviral response through the RNAi pathway. Our examination of small RNA responses in VSV-infected nematodes revealed a clear reduction in antisense 22Gs targeting VSV in drh-1 mutants compared to N2 animals. A reduction in antisense 22Gs has also been described during OV infection of drh-1 animals, suggesting that small RNAs targeting viral transcripts contribute significantly to the restriction of virus replication [9]. Interestingly, in both N2 and drh-1 animals there were clear “hotspots” of small RNAs mapping to the 5′ end of the VSV genome. This has also been observed during small RNA responses against VSV in Drosophila, and has been attributed to the presence of self-complementary snapback defective interfering particles derived from 5′ end of the genome [39]. Curiously, although Drosophila Dicer proteins cleave these VSV snapback particles, the resulting siRNAs are, for unknown reasons, not loaded into Argonautes [39]. It has been postulated that these interfering particles may serve as a RNA decoy, allowing VSV evasion of RNAi machinery [39, 40]. It will be interesting to determine if VSV produces defective interfering particles in C. elegans and whether small RNAs targeting these particles are loaded into RDE-1.
Although SID-1 is required for the systemic spread of exogenous RNAi signals [28], sid-1 mutants were as susceptible to VSV as N2 animals. This is consistent with previous virus studies in C. elegans [3, 33]. Whether another, unidentified, RNA transporter is required for mounting systemic RNAi responses to infection is unknown.
Interestingly, we found that VSV could infect the germline of drh-1 animals and be vertically transmitted when somatically-injected animals were placed on FUdR-containing plates. How this DNA synthesis inhibitor might compromise germline immunity to infection is unknown. However, FUdR treatment can modulate resistance to thermal, hypertonic, and proteotoxic stresses, likely through alteration of gene expression [40]. Therefore, FUdR-induced inhibition of germline gene expression programs and/or stress responses that normally serve antiviral roles may promote germline infection and vertical transmission. However, because VSV could efficiently establish germline infection after direct injection into the germline in the absence of FUdR, these drug-induced effects may only be required to sensitize the germline to infection as the virus spreads from the soma. Direct injection of VSV into the germline may simply overwhelm germline antiviral responses, even without modulation by FUdR treatment. Importantly, no evidence was found for vertical transmission of other intracellular C. elegans pathogens such as microsporida [41] and OV [14]. Therefore, our model provides new opportunities to study vertical transmission in C. elegans.
Previous reports have demonstrated that administration of a mild stress (e.g. temporary starvation) to C. elegans can lead to enhanced protection of offspring from these same stressors, a phenomenon termed “transgenerational hormesis” [42, 43]. In some cases, this phenomenon can be mediated by small RNAs [43]. A previous report found transgenerational silencing of a Flock House virus transgene [34] and Sterken et al. [35] reported that OV-infected N2, but not RNAi-deficient, animals could transmit a heritable, protective antiviral response to their progeny. In contrast, Ashe et al. [33] did not find evidence for transgenerational immunity after OV infection. We found that prior exposure of N2 or drh-1 animals to VSV led to significant reductions in the infection rates of their progeny when challenged with VSV. We were initially surprised by the inheritance of an antiviral response in drh-1 animals, which is in contradiction with Sterken et al. [35]. However, because antiviral small RNA production in drh-1 animals was reduced, but not eliminated, it is possible that these residual small RNAs may still confer protection to offspring. We attempted to detect small RNAs targeting VSV in the progeny from naïve or VSV-exposed mothers but were unable to observe significant quantities of viral small RNAs in these deep-sequencing experiments. It is unclear if viral small RNAs exist in progeny animals at too low a level for detection by our methods or whether the heritable antiviral effect is not mediated by small RNAs. We favor the former scenario as rde-3 animals are completely defective in 22G biogenesis [37], and we found no evidence for transgenerational antiviral immunity in rde-3 animals. The ability of C. elegans to use a heritable antiviral response to protect offspring from infection represents a fascinating adaptation that warrants further investigation.
The susceptibility of C. elegans to VSV provides a genetically-tractable in vivo model system to explore negative-sense ssRNA virus biology and antiviral immunity. Our system will nicely complement in vivo models that have been established for VSV in Drosophila and mice [19], giving investigators the rare opportunity to study a single virus in disparate hosts.
EXPERIMENTAL PROCEDURES
Cell, virus, and worm culture
BHK and BSC-40 cells (from American Type Culture Collection) were cultured as described [24] in Dulbecco’s Modified Eagles Medium (DMEM; Invitrogen). VSV stocks were prepared as described [24] and resuspended in DMEM. Viral and nematode strains used in the study are described in Supplemental Experimental Procedures. Unless otherwise indicated, NGM plates were incubated at 25°C for the duration of experiments.
Microinjections
Microinjections used young adults and pulled capillary needles secured onto a Nikon TE-200 microscope equipped with a micromanipulator and regulated pressure source. Needles containing either DMEM (mock-infections) or virus resuspended in DMEM were used for microinjections. Doses of VSV (in PFU) represent estimates based on VSV titration on BSC-40 cells and a dispensed volume of 10 nL during microinjections. Injected worms were immediately placed on NGM plates seeded with OP50 Escherischia coli. Where indicated, NGM also contained 50 μg/mL FUdR (Sigma).
RNA isolation and RT-PCR
Total RNA was purified from adult worms or embryos at the indicated times as described [44]. VSV (+)-sense transcription and C. elegans actin gene transcription were analyzed by RT-PCR as described [24].
Immunoblotting
Antibodies used for immunoblotting included: mouse anti-LUC (Invitrogen), rabbit anti-actin (Abcam), and mouse anti-VSV Matrix (Dr. Douglas Lyles (Wake Forest School of Medicine, Winston–Salem, NC). Immunoblots were performed as described [24].
Lifespan, LUC, and Transgenerational Immunity Assays, Microscopy, and Small RNA Cloning and Analysis
Supplementary Material
Acknowledgments
We thank Dr. Rita Sharma and Ms. Tauny Tambolleo for excellent technical assistance. D.B.G. was supported by fellowships from the Natural Sciences and Engineering Research Council of Canada and Alberta Innovates Health Solutions. C.C.M. was funded by the National Institutes of Health (Grant 5R37GM058800) and is a Howard Hughes Medical Institute Investigator. N.S. was funded by the National Institutes of Health (Grant AI060025). The authors declare that no competing interests exist.
Footnotes
Supplemental Information includes six figures (S1–S6) and Supplemental Experimental Procedures.
AUTHOR CONTRIBUTIONS
Conception and design: DBG, TI, WG, NS, CCM. Acquisition of data: DBG, TI, LL. Analysis and interpretation of data, DBG, TI, LL, WG, NS, CCM. Drafting or revising the article: DBG, TI, LL, WG, NS, CCM.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhang R, Hou A. Host-Microbe Interactions in Caenorhabditis elegans. ISRN Microbiol. 2013;2013:356451. doi: 10.1155/2013/356451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen LB, Troemel ER. Microbial pathogenesis and host defense in the nematode C. elegans. Curr Opin Microbiol. 2015;23:94–101. doi: 10.1016/j.mib.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schott DH, Cureton DK, Whelan SP, Hunter CP. An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2005;102:18420–18424. doi: 10.1073/pnas.0507123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilkins C, Dishongh R, Moore SC, Whitt MA, Chow M, Machaca K. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature. 2005;436:1044–1047. doi: 10.1038/nature03957. [DOI] [PubMed] [Google Scholar]
- 5.Gammon DB, Mello CC. RNA interference-mediated antiviral defense in insects. Curr Opin Insect Sci. 2015;8:111–120. doi: 10.1016/j.cois.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szittya G, Burgyan J. RNA interference-mediated intrinsic antiviral immunity in plants. Curr Top Microbiol Immunol. 2013;371:153–181. doi: 10.1007/978-3-642-37765-5_6. [DOI] [PubMed] [Google Scholar]
- 7.Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, Voinnet O. Antiviral RNA interference in mammalian cells. Science. 2013;342:235–238. doi: 10.1126/science.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Lu J, Han Y, Fan X, Ding SW. RNA interference functions as an antiviral immunity mechanism in mammals. Science. 2013;342:231–234. doi: 10.1126/science.1241911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashe A, Belicard T, Le Pen J, Sarkies P, Frezal L, Lehrbach NJ, Felix MA, Miska EA. A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity. Elife. 2013;2:e00994. doi: 10.7554/eLife.00994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 11.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 12.Lu R, Maduro M, Li F, Li HW, Broitman-Maduro G, Li WX, Ding SW. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005;436:1040–1043. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu R, Yigit E, Li WX, Ding SW. An RIG-I-Like RNA helicase mediates antiviral RNAi downstream of viral siRNA biogenesis in Caenorhabditis elegans. PLoS Pathog. 2009;5:e1000286. doi: 10.1371/journal.ppat.1000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felix MA, Ashe A, Piffaretti J, Wu G, Nuez I, Belicard T, Jiang Y, Zhao G, Franz CJ, Goldstein LD, et al. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011;9:e1000586. doi: 10.1371/journal.pbio.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franz CJ, Renshaw H, Frezal L, Jiang Y, Felix MA, Wang D. Orsay, Santeuil and Le Blanc viruses primarily infect intestinal cells in Caenorhabditis nematodes. Virology. 2014;448:255–264. doi: 10.1016/j.virol.2013.09.024. [DOI] [PubMed] [Google Scholar]
- 16.Whelan SP, Ball LA, Barr JN, Wertz GT. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci U S A. 1995;92:8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawson ND, Stillman EA, Whitt MA, Rose JK. Recombinant vesicular stomatitis viruses from DNA. Proc Natl Acad Sci U S A. 1995;92:4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang G, Gao X, Gould EA. Factors responsible for the emergence of arboviruses; strategies, challenges and limitations for their control. Emerg Microbes Infect. 2015;4:e18. doi: 10.1038/emi.2015.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hastie E, Cataldi M, Marriott I, Grdzelishvili VZ. Understanding and altering cell tropism of vesicular stomatitis virus. Virus Res. 2013;176:16–32. doi: 10.1016/j.virusres.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duntsch CD, Zhou Q, Jayakar HR, Weimar JD, Robertson JH, Pfeffer LM, Wang L, Xiang Z, Whitt MA. Recombinant vesicular stomatitis virus vectors as oncolytic agents in the treatment of high-grade gliomas in an organotypic brain tissue slice-glioma coculture model. J Neurosurg. 2004;100:1049–1059. doi: 10.3171/jns.2004.100.6.1049. [DOI] [PubMed] [Google Scholar]
- 21.Kern A, Ackermann B, Clement AM, Duerk H, Behl C. HSF1-controlled and age-associated chaperone capacity in neurons and muscle cells of C. elegans. PLoS One. 2010;5:e8568. doi: 10.1371/journal.pone.0008568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo X, Zhang R, Wang J, Ding SW, Lu R. Homologous RIG-I-like helicase proteins direct RNAi-mediated antiviral immunity in C. elegans by distinct mechanisms. Proc Natl Acad Sci U S A. 2013;110:16085–16090. doi: 10.1073/pnas.1307453110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roers A, Hiller B, Hornung V. Recognition of Endogenous Nucleic Acids by the Innate Immune System. Immunity. 2016;44:739–754. doi: 10.1016/j.immuni.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Gammon DB, Duraffour S, Rozelle DK, Hehnly H, Sharma R, Sparks ME, West CC, Chen Y, Moresco JJ, Andrei G, et al. A single vertebrate DNA virus protein disarms invertebrate immunity to RNA virus infection. Elife. 2014;3 doi: 10.7554/eLife.02910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cureton DK, Massol RH, Saffarian S, Kirchhausen TL, Whelan SP. Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog. 2009;5:e1000394. doi: 10.1371/journal.ppat.1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 27.Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, Rual JF, Kennedy S, Dybbs M, Bertin N, Kaplan JM, et al. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308:1164–1167. doi: 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- 28.Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–2459. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- 29.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 30.Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitchell DH, Stiles JW, Santelli J, Sanadi DR. Synchronous growth and aging of Caenorhabditis elegans in the presence of fluorodeoxyuridine. J Gerontol. 1979;34:28–36. doi: 10.1093/geronj/34.1.28. [DOI] [PubMed] [Google Scholar]
- 32.Levine S, Olson W. Nucleic acids of measles and vesicular stomatitis viruses. Proc Soc Exp Biol Med. 1963;113:630–631. doi: 10.3181/00379727-113-28446. [DOI] [PubMed] [Google Scholar]
- 33.Ashe A, Sarkies P, Le Pen J, Tanguy M, Miska EA. Antiviral RNA Interference against Orsay Virus Is neither Systemic nor Transgenerational in Caenorhabditis elegans. J Virol. 2015;89:12035–12046. doi: 10.1128/JVI.03664-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rechavi O, Minevich G, Hobert O. Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell. 2011;147:1248–1256. doi: 10.1016/j.cell.2011.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sterken MG, Snoek LB, Bosman KJ, Daamen J, Riksen JA, Bakker J, Pijlman GP, Kammenga JE. A heritable antiviral RNAi response limits Orsay virus infection in Caenorhabditis elegans N2. PLoS One. 2014;9:e89760. doi: 10.1371/journal.pone.0089760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen CC, Simard MJ, Tabara H, Brownell DR, McCollough JA, Mello CC. A member of the polymerase beta nucleotidyltransferase superfamily is required for RNA interference in C. elegans. Curr Biol. 2005;15:378–383. doi: 10.1016/j.cub.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 37.Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D, Jr, Mello CC. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 39.Sabin LR, Zheng Q, Thekkat P, Yang J, Hannon GJ, Gregory BD, Tudor M, Cherry S. Dicer-2 processes diverse viral RNA species. PLoS One. 2013;8:e55458. doi: 10.1371/journal.pone.0055458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson EN, Corkins ME, Li JC, Singh K, Parsons S, Tucey TM, Sorkac A, Huang H, Dimitriadi M, Sinclair DA, et al. C. elegans lifespan extension by osmotic stress requires FUdR, base excision repair, FOXO, and sirtuins. Mech Ageing Dev. 2016;154:30–42. doi: 10.1016/j.mad.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Troemel ER, Felix MA, Whiteman NK, Barriere A, Ausubel FM. Microsporidia are natural intracellular parasites of the nematode Caenorhabditis elegans. PLoS Biol. 2008;6:2736–2752. doi: 10.1371/journal.pbio.0060309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jobson MA, Jordan JM, Sandrof MA, Hibshman JD, Lennox AL, Baugh LR. Transgenerational Effects of Early Life Starvation on Growth, Reproduction, and Stress Resistance in Caenorhabditis elegans. Genetics. 2015;201:201–212. doi: 10.1534/genetics.115.178699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rechavi O, Houri-Ze’evi L, Anava S, Goh WS, Kerk SY, Hannon GJ, Hobert O. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell. 2014;158:277–287. doi: 10.1016/j.cell.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu W, Shirayama M, Conte D, Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.