ABSTRACT
Peroxisome biogenesis disorders (PBDs) are metabolic disorders caused by the loss of peroxisomes. The majority of PBDs result from mutation in one of 3 genes that encode for the peroxisomal AAA ATPase complex (AAA-complex) required for cycling PEX5 for peroxisomal matrix protein import. Mutations in these genes are thought to result in a defect in peroxisome assembly by preventing the import of matrix proteins. However, we show here that loss of the AAA-complex does not prevent matrix protein import, but instead causes an upregulation of peroxisome degradation by macroautophagy, or pexophagy. The loss of AAA-complex function in cells results in the accumulation of ubiquitinated PEX5 on the peroxisomal membrane that signals pexophagy. Inhibiting autophagy by genetic or pharmacological approaches rescues peroxisome number, protein import and function. Our findings suggest that the peroxisomal AAA-complex is required for peroxisome quality control, whereas its absence results in the selective degradation of the peroxisome. Thus the loss of peroxisomes in PBD patients with mutations in their peroxisomal AAA-complex is a result of increased pexophagy. Our study also provides a framework for the development of novel therapeutic treatments for PBDs.
KEYWORDS: AAA ATPase complex, autophagy, peroxisomes, peroxisome biogenesis disorder, PEX1, PEX5, PEX26, pexophagy, selective autophagy, Zellweger spectrum disorder
Introduction
In mammalian cells, damaged and superfluous organelles are turned over by the degradation process called selective macroautophagy (herein referred to as selective autophagy). This process involves the sequestration of the substrate by a double-membrane sequestering organelle, the phagophore, which matures into an autophagosome; the latter fuses with lysosomes to degrade its contents.1 One mechanism by which cytoplasmic substrates are selectively targeted to phagophores for degradation is through the ubiquitination of proteins on the target organelle (for a review see refs. 2, 3). In this mechanism, the cytosol-facing proteins of damaged or superfluous organelles are ubiquitinated, followed by the recruitment of a group of cytosolic proteins called autophagy receptors that bind to the ubiquitinated organelle. These autophagy receptors then bridge the substrates to phagophores by binding to the phagophore membrane factor MAP1LC3/LC3.4
What is unclear, however, is the mechanism by which damaged organelles are selectively ubiquitinated, while healthy organelles are protected from degradation. A potential mechanism for monitoring organelle health in some organelles may involve their protein import machinery. Here, damaged organelles that can no longer import their proteins are ubiquitinated and degraded, whereas healthy ones are able to prevent ubiquitination, or have the ability to deubiquitinate their proteins. This import-competence hypothesis is best exemplified by mitochondria. Damaged mitochondria may be degraded by the PINK1-PARK2-mediated mitophagy pathway, leading to the autophagic degradation of mitochondria (for a review see ref. 5). In normal or healthy mitochondria, PINK1 is imported into the mitochondrial inner membrane where it is rapidly degraded by a member of the rhomboid family of intramembrane serine proteases, PARL.6 However, damaged mitochondria that no longer can import proteins into the inner membrane accumulate PINK1 on the outer membrane. PINK1 then phosphorylates various proteins and recruits the E3 ubiquitin ligase PARK2,7-9 which ubiquitinates mitochondrial outer membrane proteins to signal for mitophagy.
Peroxisomes are another organelle degraded by selective autophagy, through a process referred to as pexophagy, whereby ubiquitinated peroxisomes are recognized by autophagy receptors SQSTM1/p62 and NBR1, which target them to phagophores for degradation.10,11 These single membrane-bound organelles that are involved in both catabolic and anabolic reactions of lipids and reactive oxygen species are unique from other single membrane-bound organelles in that they can import all of their matrix (lumen) proteins directly from the cytosol.12 Most peroxisomal matrix proteins contain a peroxisomal targeting sequence-1 (PTS1) that is recognized by the shuttling receptor PEX5.13 PEX5 binds and escorts PTS1-containing proteins to peroxisomes by docking and inserting into the membrane through its interaction with its membrane receptors, PEX14 and PEX13. After the matrix protein is translocated into the peroxisomal lumen, PEX5 is ubiquitinated by the peroxisomal E3 ubiquitin ligase complex consisting of PEX2-PEX10-PEX12. Ubiquitinated PEX5 is then subsequently removed from the membrane by the peroxisomal AAA-complex made up of PEX26, PEX1, and PEX6, by a similar mechanism as that of the AAA ATPase VCP/p97.14-16
Loss of peroxisomal AAA-complex function is the most common mutation for an autosomal recessive group of disorders called peroxisome biogenesis disorders (PBDs).17 Mutation in the AAA-complex results in a decrease in peroxisome number and function (for a review see refs. 18, 19). However, since the AAA-complex is required to remove the matrix protein receptor PEX5 following its ubiquitination, we hypothesize that the loss of the AAA-complex can result in an increase in ubiquitinated PEX5 on peroxisome membranes that can then signal pexophagy. In other words, the peroxisomal AAA-complex promotes the stability of import-competent peroxisomes by limiting the accumulation of ubiquitinated PEX5 on the peroxisome surface. If this is the case, it suggests that most PBDs are not due to the inability to form peroxisomes, but rather due to the loss of the ability to prevent their degradation. Here, we present evidence to support this hypothesis, and provide a framework for a possible therapeutic treatment of patients with PBDs resulting from a defect in the peroxisomal AAA-complex.
Results
Depleting AAA-complex function results in the loss of peroxisomes
Because the peroxisomal AAA-complex is required to remove the ubiquitinated form of the shuttling peroxisomal matrix protein receptor, PEX5, from the peroxisomal membrane, we hypothesized that the AAA-complex may also be involved in peroxisome quality control. To test this hypothesis, we first established whether an acute loss of the peroxisomal AAA-complex leads to a change in peroxisome number. Our strategy was to use RNAi to deplete cells of PEX1 or PEX26, 2 of the 3 proteins that make up the peroxisomal AAA-complex, and examine the fate of peroxisomes. To identify peroxisomes, cells were fixed and immunostained for the endogenous peroxisomal membrane protein ABCD3/PMP70.
When compared with nontargeting siRNA (siCTRL), the knockdown of PEX1 (siPEX1) or PEX26 (siPEX26) resulted in a visible decrease in total ABCD3 fluorescent signal (Fig. 1A). To quantify the change in peroxisomes, we quantified the number of peroxisomes within the entire cell and normalized it to the volume of the cell, which we refer to as “ABCD3 density” (see methods). Quantification of the density of peroxisomes (number of ABCD3 puncta/μm3) (Fig. 1B) indicated that cells depleted of PEX1 or PEX26 had significantly lower peroxisome numbers in comparison to siCTRL cells. This decrease in peroxisome numbers is unlikely due to a defect in peroxisome biogenesis as the depletion of 2 peroxisome biogenesis factors, PEX14 (Fig. 1 siPEX14) or PEX16 (Fig. S1), resulted only in a slight but not significant decrease in peroxisome numbers compared with siCTRL, 3 d after siRNA treatment. This observation is consistent with a previous report, which showed that the depletion of the peroxisome biogenesis factor PEX19 also did not cause a noticeable change in peroxisome abundance within the initial 3 d of depletion by siRNA. 20
Figure 1.

Depletion of AAA-complex components result in the loss of peroxisomes. (A) Representative fluorescence images of HeLa cells transfected with nontargeting siRNA (siCTRL), and siRNA against PEX14 (siPEX14), PEX1 (siPEX1), and PEX26 (siPEX26) as indicated. Cells were fixed and immunostained for ABCD3 48 h post-transfection. Scale bars: 50 μm. (B) Graph of the ABCD3 density (number of ABCD3 puncta per volume of each cell [number/μm3]), of at least 30 cells per trial (n = 3) ± standard deviation in (A). Asterisks represent p-values compared with siCTRL: *p < 0.05. (C) Total cell lysates were prepared from siRNA-treated HeLa cells (as in A) and immunoblotted (IB) with the indicated antibodies.
To further validate the loss of peroxisomes upon the depletion of the peroxisomal AAA-complex components, PEX1 or PEX26, the endogenous levels of ABCD3 and PEX14 were examined by immunoblot analysis. In support of the immunofluorescence microscopy data (Fig. 1A), the immunoblot analysis showed a drastic decrease in ABCD3 and PEX14 in cells depleted of PEX1 and PEX26 (Fig. 1C). Together, these results suggest that the loss of AAA-complex function results in the loss of peroxisomes, which is not due to a decrease in peroxisome biogenesis.
Autophagy mediates the loss of peroxisomes in AAA-complex depleted cells
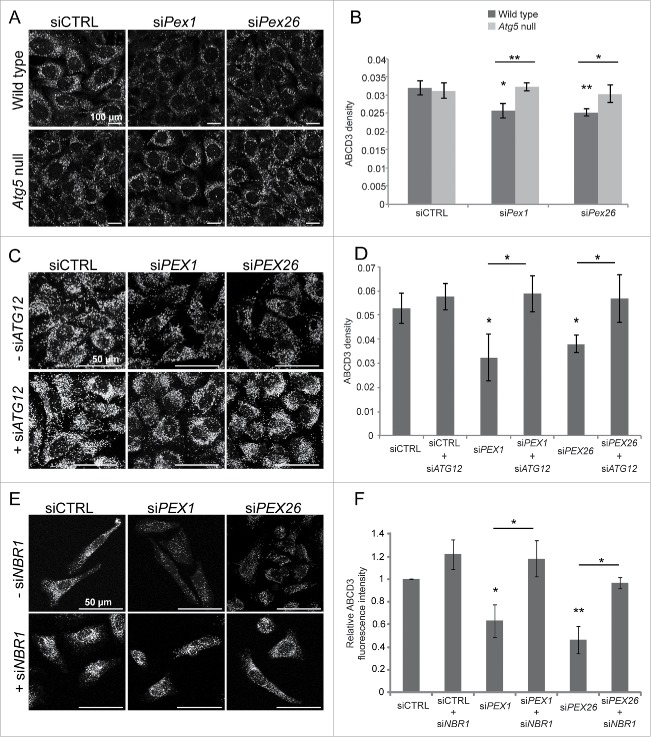
We next examined whether the loss of peroxisomes in cells depleted of AAA-complex components was due to pexophagy. To test this, we examined whether depleting PEX1 or PEX26 expression in the autophagy-deficient atg5−/− mouse embryonic fibroblast (MEF) cell line results in a change in peroxisome number. In the wild-type MEFs, a decrease in ABCD3 density was observed when PEX1 or PEX26 expression was depleted compared with control siRNA-treated cells (Fig. 2A, B). However, this decrease was abrogated in the Atg5 null MEFs, suggesting that the loss of peroxisomes observed in the wild-type cells upon PEX1 or PEX26 depletion requires autophagy.
Figure 2.
Pexophagy mediates the loss of peroxisomes in AAA-complex-depleted cells. (A) Representative fluorescence images of wild-type and Atg5 null MEFs transfected with either nontargeting siRNA (siCTRL), siRNA against Pex1 (siPex1) or Pex26 (siPex26) as indicated, and immunostained for endogenous ABCD3. Scale bars: 100 μm. (B) Graph of the average ABCD3 density per cell (number/μm3) of at least 30 cells per trial (n = 3) ± standard deviation in (A). Asterisks represent p-values compared with siCTRL unless otherwise indicated: *p < 0.05, **p < 0.01. (C) Representative fluorescence images of HeLa cells transfected with nontargeting siRNA (siCTRL), siRNA against PEX1 (siPEX1), and PEX26 (siPEX26), with or without siRNA against ATG12 (siATG12) as indicated and immunostained for ABCD3. Scale bars: 50 μm. (D) Graph of the average ABCD3 density per cell of at least 30 cells per trial (n = 3) ± standard deviation in (C). Asterisks represent p-values compared with siCTRL unless otherwise indicated: *p < 0.05. (E) Same as in (C) but cells cotransfected with or without siRNA against NBR1 (siNBR1) and immunostained for ABCD3. Scale bars: 50 μm. (F) Graph of the average ABCD3 fluorescence intensity within the entire cell normalized to siCTRL of 30 cells per trial (n = 3) ± standard deviation in (E). Asterisks represent p-values of statistics relative to siCTRL unless otherwise indicated: *p < 0.05, **p < 0.01.
To test whether autophagy was also required for the loss of peroxisomes in HeLa cells deficient in the AAA-complex (Fig. 1), we co-transfected HeLa cells with siRNA against the autophagy factor ATG12 (Fig. 2C). Immunoblot analysis confirmed that our siRNA against ATG12 resulted in about 90% knockdown of ATG12 expression (Fig. S2A). Quantification of the ABCD3 density showed that the knockdown of the autophagy factor ATG12 prevented the loss of peroxisomes in cells also depleted of PEX1 or PEX26, as their ABCD3 densities were similar to the control siRNA-treated cells (Fig. 2D). Together, these results suggest that the loss of peroxisomes in cells depleted of the peroxisomal AAA-complex is mediated by autophagy.
Peroxisomes in AAA-complex depleted cells are degraded by selective autophagy
Peroxisomes can be degraded either by bulk degradation of the cytoplasm during general autophagy, or by the selective targeting of peroxisomes to autophagosomes by a process called pexophagy (for a review see ref. 1). Recently, we showed that NBR1 is a key autophagy receptor in pexophagy that is recruited to ubiquitinated peroxisomes.10 If peroxisomes in AAA-complex-deficient cells are being degraded by pexophagy, we reasoned that depleting NBR1 expression in PEX1- or PEX26-deficient cells should prevent the loss of peroxisome numbers. Thus, to determine whether the loss of the AAA-complex leads to pexophagy, we depleted HeLa cells of both NBR1 and a component of the AAA-complex and analyzed the cells for a change in the number of peroxisomes. Immunoblotting confirmed that our siRNA against NBR1 efficiently depleted NBR1 expression in HeLa cells (Fig. S2B). We have previously found that depleting NBR1 results in an increase in peroxisomes that are clustered.10 For this reason, we quantified the total ABCD3 fluorescence intensity from the entire cell to compare the amount of peroxisomes within these cells. When NBR1 was co-depleted with the AAA-complex, peroxisomes were no longer diminished compared with cells depleted of PEX1 or PEX26 alone (Fig. 2E, F). Collectively, these results demonstrate that the loss of peroxisome numbers in cells depleted of a component of the peroxisomal AAA-complex is due to pexophagy, and not due to nonselective degradation of the cytoplasm by autophagy.
Ubiquitinated PEX5 accumulates on the membrane of AAA-complex depleted peroxisomes
Since pexophagy induced by AAA-complex depletion required the ubiquitin-binding autophagy receptor NBR1 (Fig. 2E, F), it suggests that ubiquitination is a signal for AAA-complex-deficient pexophagy. In mammalian cells, the accumulation of ubiquitinated PEX5 has recently been reported to signal pexophagy.21,22 To determine whether the loss of AAA-complex function leads to an accumulation of ubiquitinated proteins on peroxisomes, we looked for any change in ubiquitinated PEX5 in PEX1- and PEX26-depleted cells. Here, HeLa cells depleted of PEX14, PEX1, or PEX26 were transfected with RFP-SKL and GFP-Ubiquitin (GFP-Ub) plasmids (Fig. 3A). RFP-SKL is a red fluorescent protein tagged to the PTS1 consisting of the tripeptide Ser-Lys-Leu that is recognized by PEX5 to target it to peroxisomes. RFP-SKL was expressed in these cells to increase the targeting of endogenous PEX5 to peroxisomes by increasing the matrix import load on these cells. Furthermore, the lysosomotropic agent, chloroquine, was used to prevent peroxisome loss and thus increase the accumulation of ubiquitinated proteins. Total cell lysates from these cells were immunoblotted with PEX5 antibodies (Fig. 3A). In cells treated with siPEX1 or siPEX26, an increase in higher molecular weight bands corresponding to modification by a single GFP-Ubiquitin and possibly multiple were observed compared with siCTRL- and siPEX14-treated cells (Figs. 3A, B). Nonspecific bands (highlighted by asterisks in Fig. S3A) were confirmed by overexpressing a PEX5-MYC construct in HeLa cells and probing for PEX5. To determine whether there was an increase in the ubiquitination of other PMPs in AAA-complex depleted cells, we also examined ABCD3, the most abundant membrane protein on peroxisomes.23 We found that there was no detectable increase in ubiquitinated ABCD3 in cells depleted of AAA-complex function compared with siCTRL- or siPEX14-treated cells (Fig. 3C).
Figure 3.
PEX5 accumulates on the peroxisomal membrane in a ubiquitinated form and signals for pexophagy in cells depleted of peroxisomal AAA-complex components. (A) Immunoblots of the total cell lysates for PEX5 from HeLa cells transfected with plasmids encoding GFP-ubiquitin (GFP-Ub) and RFP-SKL after 2 d of treatment with siRNA as indicated. Cells were also treated with 10 μm chloroquine for 24 h to prevent peroxisome loss. The bracket represents modified PEX5; the arrow indicates PEX5, whereas the asterisks (*, **) indicate nonspecific bands. (B) Graph of the relative densitometry reading of GFP-Ubx-PEX5:PEX5 ratio; where GFP-Ubx-PEX5 represent the higher molecular weight bands indicated in (A). The average (n = 3) ± standard deviation for each condition is shown. Asterisks represent p-values of statistics relative to siCTRL: *p < 0.05. A.U., arbitrary units. (C) Same as (A) but immunoblot for ABCD3. The top dashed arrow represents modified ABCD3; the bottom solid arrow indicates ABCD3. (D) Representative fluorescence images of HeLa cells transfected with siRNA as indicated over 2 consecutive d and immunostained for ABCD3 and PEX5. Scale bars: 20 μm. (E) Graph of the average density of PEX5 punctate structures within a cell (the number of PEX5 puncta per volume of cell [μm3]) of 30 cells per trial (n = 3) ± standard deviation in (D). Asterisks represent p values compared with siCTRL: *p < 0.05. (F) Subcellular fractionation of HeLa cells transfected with GFP-ubiquitin (GFP-Ub) and RFP-SKL plasmids after 2 d of treatment with siRNA and chloroquine as in (A). The post nuclear homozygous lysates (WCL) were fractionated into cytosol and membrane fractions, and immunoblotted with the indicated antibodies. The top dashed arrow represents modified PEX5; the bottom solid arrow indicates PEX5, whereas the asterisk (*) indicates nonspecific bands. (G) Representative fluorescence images of HeLa cells transfected with nontargeting siRNA (siCTRL), siRNA against PEX1 (siPEX1), and PEX26 (siPEX26), with or without siRNA against PEX5 (siPEX5) as indicated and immunostained for ABCD3. Scale bars: 50 μm. (H) Graph of the average ABCD3 density per cell of at least 30 cells per trial (n = 3) ± standard deviation in (G). Asterisks represent p-values: *p < 0.05, **p < 0.01.
Next, we asked whether ubiquitinated PEX5 resided on peroxisomes. First, using immunofluorescence imaging, we found that in siCTRL- or siPEX14-treated cells, PEX5 appeared mainly cytosolic. However, in siPEX1- or siPEX26-treated cells, PEX5 appeared in punctate structures that colocalized with the peroxisome membrane marker ABCD3 (Fig. 3D, E). To further validate the observation of PEX5 accumulation on peroxisomes in AAA-complex-deficient cells, we performed subcellular fractionation on patient-derived wild-type and PEX1-G843D fibroblasts (Fig. S3B). The PEX1G843D mutation is the most common PBD mutation caused by a missense allele resulting in a glycine to aspartic acid replacement in residue 843, and is usually presented as a mild form of PBD.24 In the wild-type cells, the majority of PEX5 was found localized in the cytosol, whereas in the PEX1-G843D fibroblast cells PEX5 was found in the membrane fraction (Fig. S3B). Interestingly, no PEX5 expression was observed in patient-derived PEX1 null cells (Fig. S3C).
To determine whether the PEX5 accumulating on peroxisome membranes under AAA-complex-depletion conditions was ubiquitinated, we performed subcellular fractionation on HeLa cells depleted of PEX14, PEX1, or PEX26 and transfected with RFP-SKL and GFP-Ub plasmids and treated with chloroquine (Fig. 3F). Here, we found that in the membrane fractions of the siPEX1 and siPEX26 cells, when probing for PEX5, there was an increase in higher molecular weight bands corresponding to modification by a single GFP-Ub compared with siCTRL- and siPEX14-treated cells. Finally, to determine whether ubiquitinated PEX5 is required for pexophagy in AAA-complex-deficient cells, HeLa cells were co-depleted of PEX5 along with PEX1 or PEX26. We argue that if PEX5 ubiquitination signals pexophagy, then depleting the cells of PEX5 should prevent pexophagy. Indeed, in cells lacking PEX5 expression, the absence of AAA-complex function did not cause a loss of peroxisomes (Fig. 3G, H). Immunoblotting confirmed that our siRNA against PEX5 efficiently depleted PEX5 expression in HeLa cells (Fig. S2C). Together, these observations suggest that under conditions of AAA-complex defects, ubiquitinated PEX5 accumulates on the peroxisomal membrane to signal for pexophagy.
Peroxisomal AAA-complex depletion induces autophagosome formation
To understand the mechanism by which a loss of the peroxisomal AAA-complex function induces the autophagic machinery, we examined whether the depletion of PEX1 or PEX26 expression can induce autophagosome formation. We hypothesized that autophagic machinery may be upregulated to account for the rapid turnover of peroxisomes in the peroxisomal AAA-complex-deficient cells. To determine whether autophagy is upregulated, we looked for a change in the number of MAP1LC3B-positive punctate structures within the cells depleted of peroxisomal AAA-complex components. MAP1LC3B/LC3B is a ubiquitin-like cytosolic protein that is covalently attached to phosphatidylethanolamine on nascent phagophores; thus, an increase in MAP1LC3B punctate structures is an indication of the induction of autophagosome formation.25 To monitor changes in autophagosome formation, we quantified the density of GFP-MAP1LC3B-positive structures within cells transiently expressing GFP-MAP1LC3B. Imaging of GFP-MAP1LC3B-expressing HeLa cells (Fig. 4A) treated with siRNA against PEX14 did not cause a change in the number of MAP1LC3B-positive structures compared with the control nontargeting siRNA (Fig. 4B). However, cells depleted of the AAA-complex components showed a significant increase in MAP1LC3B-positive structures compared with control siRNA-treated cells. This was further confirmed by probing for endogenous MAP1LC3B in immunofluorescence images (Fig. 4C, D), and also by immunoblot, where there was an increase in the MAP1LC3B-II:MAP1LC3B-I ratio in siPEX1 or siPEX26 cells (Fig. 4E, F). To further demonstrate that the autophagic machinery was upregulated, we also examined levels of NBR1, SQSTM1, ABCD3, and the mitochondrial outer membrane marker TOMM20. In PEX1 and PEX26 knockdown cells, the expression levels of NBR1, SQSTM1 and ABCD3 decreased. However, TOMM20, a mitochondrial protein, remained unchanged, suggesting an increase in pexophagy, but not other forms of selective autophagy (e.g., mitophagy) (Fig. 4E).
Figure 4.

Peroxisomal AAA-complex deficiency induces autophagosome formation. (A) Representative fluorescent images of HeLa cells cotransfected with a GFP-MAP1LC3-encoding plasmid and siRNA as indicated. Cells were fixed and the GFP fluorescence was visualized by confocal microscopy. Scale bars: 10 μm. (B) Graph of the relative GFP-MAP1LC3B density (number of GFP-MAP1LC3B puncta per volume of each cell (μm3) normalized to siCTRL) of at least 30 cells per trial (n = 3) ± standard deviation in (A). Asterisks represent p values compared with siCTRL: *p < 0.05, **p < 0.01. (C) Representative fluorescence images of HeLa cells transfected with siRNA as indicated, fixed and immunostained for endogenous MAP1LC3B. Individual cells are outlined. Scale bars: 10 μm. (D) Graph of the relative MAP1LC3B density normalized to control siRNA of at least 30 cells per trial (n = 3) ± standard deviation in (C). Asterisks represent p values compared with siCTRL: **p < 0.01. (E) Immunoblots of total cell lysates as in (C) probed with antibodies as indicated. Arrowhead in NBR1 panel indicates the band of interest. (F) Graph of the ratio of MAP1LC3B-II:MAP1LC3B-I compared with siCTRL from (E: MAP1LC3B) as determined by densitometry for each siRNA treatment. The average (n = 3) ± standard deviation for each condition is shown. Asterisks represent p-values of statistics relative to siCTRL: *p < 0.05.
Autophagy inhibitors rescue peroxisome number in PEX1-mutated PBD fibroblasts
Mutations in the peroxisomal AAA-complex make up approximately 65% to 85% of all diagnosed cases of PBDs.17,26,27 Because cells depleted of the peroxisomal AAA-complex lose peroxisomes by pexophagy, we postulated that chemically inhibiting autophagy should increase peroxisome numbers in cells deficient in peroxisomal AAA-complex function. To test this hypothesis, we treated 2 PEX1 mutant PBD patient-derived fibroblast cell lines: a PEX1 null genotype, and a PEX1-G843D genotype, with various autophagy inhibitors and examined the cells for changes in peroxisome numbers. Now, all commercially available small molecule autophagy inhibitors indirectly or directly have multiple targets besides a component of autophagy.25 Therefore, we used 3 different small molecules that have different autophagy inhibition properties. The 3 autophagy inhibitors used were: LY294002, a phosphoinositide 3-kinase and phosphatidylinositol 3-kinase (PtdIns3K) inhibitor that targets autophagosome formation; bafilomycin A1, a vacuolar-type H+-ATPase inhibitor that targets autolysosome formation; and chloroquine, a lysosomal lumen alkalinizer that targets the end of the pathway (for a review see ref. 28). To assess whether inhibiting autophagy can recover peroxisome numbers in the PEX1 mutant cell lines, we quantified the peroxisome density from the immunofluorescence images of cells immunostained for ABCD3, and also by immunoblot analysis (Fig. 5).
Figure 5.
Autophagy inhibitors improve peroxisome number and matrix protein import in PEX1-mutated PBD fibroblasts. (A) Representative fluorescence images of ABCD3 in wild-type, PEX1-G843D, and PEX1 null fibroblasts cells that were mock treated, or treated with bafilomycin A1 (2 nM final), chloroquine (20 μM final), or LY294002 (5 mM final) for 24 hours. Scale bars: 100 μm. (B) Graph of the peroxisome density, or ABCD3 puncta per cell, normalized to the mock-treated wild-type fibroblasts of at least 30 cells per trial in (A). The average (n = 3) ± standard deviation for each condition is shown. * = p-values of statistics relative to mock-treated wild-type fibroblasts: *p < 0.05, **p < 0.01. † = p-values of statistics relative to mock-treated PEX1-G843D fibroblasts: †p < 0.05. ‡= p-values of statistics relative to mock-treated PEX1 null fibroblasts: ‡p < 0.05, ‡‡p < 0.01. ns, nonsignificant. (C) Immunoblotted total cell lysates were prepared from fibroblasts treated as in (A) (as indicated) for 24 h. Blots were immunostained with PEX14 and GAPDH antibodies. Relative intensity of the PEX14 bands relative to GAPDH are shown below each band. (D) Representative GFP fluorescence images of PEX1-G843D homozygous fibroblasts stably expressing GFP-PTS1 (PEX1-G843D-PTS1) that were treated with inhibitors as in (A). Scale bars: 50 μm. (E) Graph of the density of GFP-PTS1 puncta per cell normalized to mock-treated cells as in (D). The average (n = 3) ± standard deviation for each condition is shown. Asterisks represent p-values relative to nontreated cells: **p < 0.01.
For ease of comparison between the different cells and treatments, we present the quantification of peroxisome numbers from Fig. 5A as a relative density of ABCD3 structures with respect to the untreated wild-type cells. These results showed a significant increase in peroxisome numbers in both the PEX1-G843D and PEX1 null fibroblast cells when treated with all 3 autophagy inhibitors separately for 24 h (Fig. 5A, B). The increase in peroxisome numbers upon inhibiting autophagy was verified by immunoblot analysis for PEX14, where an increase in PEX14 was seen following 24 h treatments with autophagy inhibitors (Fig. 5C). Of note, the untreated PEX1-G843D cell line containing about 60% of peroxisomes compared with wild type showed a significant increase in peroxisomes to levels similar to the wild type when treated with any of the autophagy inhibitors (Fig 5B: PEX1-G843D). These results suggest that peroxisome numbers in mutant PEX1 cell lines can be increased by chemically inhibiting autophagy.
Autophagy inhibitors increase matrix protein import in PBD fibroblasts
One hallmark of peroxisomal mutant AAA-complex cells is the presence of ghost peroxisomes, which are peroxisomal membrane structures that lack the enzymatic peroxisomal matrix proteins.29 For this reason, the peroxisomal AAA-complex is thought to be required for the import of peroxisomal matrix proteins.30-32 However, it also has been shown that PEX5 can target to peroxisomes independently of the peroxisomal AAA-complex,33 suggesting that AAA-complex-deficient peroxisomes may still be able to import matrix proteins.
Based on our findings that the depletion of AAA-complex components resulted in an increase in ubiquitinated PEX5 (Fig. 3) and pexophagy (Fig. 2 and Fig. 4), we reasoned that peroxisomes in AAA-complex-deficient cells are rapidly degraded. Therefore, the ghost peroxisomes that are typically observed in AAA-complex-deficient cells are likely newly formed peroxisomes that have not yet imported matrix proteins. In such a case, inhibiting autophagy should increase the half-life of peroxisomes, allowing them time to import matrix proteins into peroxisomes and, thus, improve cellular peroxisomal function, because peroxisomal matrix proteins carry out the majority of the peroxisomal enzymatic functions.18,19 To test this hypothesis, we used a PEX1-G843D-PTS1 human fibroblast cell line, which is a PEX1-G843D cell line that stably expresses the peroxisomal matrix marker GFP-PTS1.34 In these cells, GFP-PTS1 was primarily cytosolic, suggesting disrupted matrix protein import (Fig. 5D: mock). When these PEX1-G843D-PTS1 fibroblasts were treated with the autophagy inhibitors for 24 h, we observed a significant redistribution of GFP-PTS1 from the cytosol to punctate structures (Fig. 5D). This was further validated by the quantification of the punctate structures (Fig. 5E).
Low doses of chloroquine improve peroxisomal function without compromising cellular viability
Next we examined whether autophagy inhibitors can improve not only peroxisome numbers, but also their function in PBD patient cell lines. However, since autophagy is an essential cellular process required for various cellular functions including development and cell metabolism, completely inhibiting autophagy will be detrimental for cell viability. Hence, drug concentrations that completely abolish autophagy will not be a viable therapeutic treatment for PBDs. Yet, we reasoned that downregulating autophagy, without abolishing it, may be sufficient to decrease the rate of peroxisome loss without compromising cell viability. Therefore, we next examined whether modulating autophagy activity using the thoroughly characterized and FDA-approved autophagy inhibitor chloroquine can improve peroxisome number and function without compromising cellular viability.
To examine cell viability, we performed a MTT (3–3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide) assay (for a review see ref. 35), which is based on the conversion of the formazan salt into formazan crystals, and reflects mitochondrial activity. Since mitochondrial activity is constant for most viable cells,35 this assay has the ability to measure cytotoxic effects of drugs on cells. Using the MTT assay, we determined 5 μM of chloroquine to be the optimal concentration to treat cells up to 96 h without significantly affecting the viability of wild-type human fibroblasts (Fig. 6A). This concentration also did not cause cell death in 2 different PEX1-G843D mutant cell lines, but instead it appeared to have improved the viability of the homozygous mutant cells after 6 d of chloroquine treatment compared with mock-treated cells (Fig. S4A). To determine whether this improvement in cell viability correlated with an increase in peroxisome numbers, we treated the PEX1-G843D cell line stably expressing GFP-SKL (PEX1-G843D-PTS1)36 with 5 μM chloroquine for 48 and 72 h. We found that this low chloroquine concentration increased peroxisome numbers as indicated by the increase in ABCD3-positive structures (Fig. 6B, C). This improvement in peroxisome numbers was further confirmed by immunoblot analysis of whole cell lysates from cells treated with 5 μM and 10 μM chloroquine for 72 h, which showed that under both concentrations of chloroquine, an increase in various peroxisomal markers, including ABCD3, PEX14, and CAT (catalase), was observed (Fig. 6D). Yet, there was less SQSTM1 accumulation at 5 μM chloroquine compared with 10 μM, suggesting that autophagy was not completely abolished at 5 μM, because SQSTM1 is degraded by autophagy.25 Interestingly, increases in peroxisomal markers were also observed for the PEX1 null cell line (Fig. 6D).
Figure 6.
Chloroquine improves peroxisome number and protein import without compromising cellular viability. (A) Wild-type fibroblasts were treated with various concentrations of chloroquine as indicated for 24 to 96 h. Viability was determined by MTT assay35 at 540 nm, and absorbance values for each respective concentration at a particular time point were normalized to mock-treated cells at that same time point to obtain the viability (% control, mock). (B) Representative fluorescence images of PEX1-G843D-PTS1 fibroblasts mock treated or treated with 5 μM chloroquine for 48 or 72 h as indicated, then fixed and stained for ABCD3. Scale bars: 100 μm. (C) Graph of the average ABCD3 density normalized to mock-treated cells per trial (n = 3) ± standard deviation in (B). Asterisks represent p-values of statistics relative to mock-treated fibroblasts: *p < 0.05. (D) Total cell lysates were prepared from wild-type, PEX1 null, and PEX1-G843D fibroblasts mock treated or treated with 5 or 10 μM chloroquine as indicated for 72 h, and immunoblotted with the indicated antibodies. Relative intensity of the PEX14 bands relative to GAPDH are shown below each band. (E) Representative GFP fluorescence images of PEX1-G843D-PTS1 cells mock treated or treated with 5 μM chloroquine for 0, 48 or 72 h. Scale bars: 100 μm. (F) Graph of the average GFP-PTS1 density normalized to the mock-treated fibroblasts of 30 cells per trial (n = 3) ± standard deviation in (E). **p < 0.01 relative to nontreated fibroblasts.
We also determined whether this low chloroquine concentration could improve matrix protein import by examining the ability of GFP-PTS1 to be imported into peroxisomes. In the PEX1-G843D-PTS1 cells, an increase in GFP-PTS1 punctate structures was observed when cells were treated either with 5 μM or 10 μM chloroquine at 24, 48, and 72 h compared with untreated cells (Fig. 6E, F). This increase was observed even after 15 d (Fig. S4B,C). However, no increase in GFP-PTS1 structures was observed in cells treated with 1 μM chloroquine. Similarly, no increase in either peroxisome numbers or GFP-PTS1 import was observed in untreated cells (Fig. S4D,E). This confirms that the improvement in ABCD3 punctate structures and GFP-PTS1 import in the PEX1-G843D-PTS1 cells was due to chloroquine treatment.
Next we examined whether this partial inhibition of autophagy could improve peroxisomal biochemical function in the PEX1 mutants including PEX1 null and PEX1-G843D patient cell lines. Very long chain fatty acids (VLCFA) are exclusively metabolized in peroxisomes, and are elevated in AAA-complex deficient patients.18 To investigate whether autophagy inhibitors can improve (decrease) VLCFA levels, we quantified their levels in several PBD cell lines using liquid chromatography-tandem mass spectrometry (LC-MS/MS). All PBD mutant primary fibroblasts, including the PEX1 mutant cell lines, showed elevated C26:0 lysophosphatidylcholine (lysoPC) levels (Fig. 7A) as expected. However, when several different PEX1-G843D mutant human fibroblast cells were treated with 5 μM chloroquine for 6 d, a significant reduction in C26:0 lysoPC levels was observed (Fig. 7A). This decrease in C26:0 lysoPC upon chloroquine treatment of PEX1-G843D mutant fibroblast cells could be the result of either a decrease in VLCFA synthesis, or improvement in peroxisomal β-oxidation activity. To distinguish between the 2 possibilities, we also treated a wild-type and a PEX16 null PBD cell line. This PEX16 null line has previously been shown to lack any detectable peroxisomal membrane structures.37 We argue that if chloroquine is inhibiting VLCFA synthesis, then a similar reduction in VLCFA should be observed in all cells, including the peroxisome-deficient PEX16 null cell line. However, as seen in Fig. 7A, the VLCFA (C26:0 lysoPC) was not significantly lower in either the PEX16 null or the wild-type cells following chloroquine treatment. In fact, a small increase was observed in the wild-type cells. Furthermore, we also observed that the levels of C16:0 through C22:0 (Fig. 7B and Table S1) did not decrease in chloroquine-treated cells, suggesting that the decrease in the VLCFA levels is unlikely due to a decrease in its synthesis (i.e., elongation from C16:0 and/or C20). Instead, these results suggest that chloroquine not only increases peroxisome numbers and peroxisomal matrix protein content, but also decreases VLCFA accumulation.
Figure 7.

Chloroquine decreases VLCFA accumulation in PEX1-mutated fibroblasts. (A) Graph of the average VLCFA (C26:0 lysophosphatidylcholine, lysoPC) levels (pmol) in wild-type, PEX16 null, 2 different PEX1 null (PEX1 null-A, PEX1 null-B), PEX1-G843D hemizygous and PEX1-G843D homozygous primary fibroblast cell lines grown with a vehicle mock control (water) or with 5 μM chloroquine for 6 d. Asterisks represent p-values relative to mock-treated fibroblasts: **p < 0.01; ns, nonsignificant. (B) Graph of the average LCFA (C18:2 lysophosphatidylcholine, lysoPC) levels (pmol) in the same conditions as in (A) in chloroquine-treated cells compared with the mock-treated cells. Asterisks represent p-values relative to mock-treated fibroblasts: **p < 0.01; ns, nonsignificant.
Discussion
Now, there is no cure for PBDs, and only symptomatic treatments are available. Mutations in the peroxisomal AAA-complex make up most of all PBD cases. 17 We find here that upon the acute depletion of the peroxisomal AAA-complex, peroxisomes are rapidly lost when compared with cells deficient in other peroxisomal proteins, such as PEX14 or PEX16 (Fig. 1, S1). This loss of peroxisomes is mediated by autophagy, which is upregulated in cells deficient in peroxisomal AAA-complex components (Fig. 2). This finding suggests that depressing autophagy activity may prevent the loss of peroxisomes in patient cells without a functional AAA-complex. Indeed, we find that inhibiting autophagy with small chemical molecules such as chloroquine in AAA-complex-deficient PEX1 mutant PBD patient cell lines not only restored peroxisome numbers, but also improved peroxisomal function (Figs. 6, 7).
Interestingly, bafilomycin A1 and chloroquine, both of which neutralize the pH of lysosomes, were more effective than the PtdIns3K inhibitor LY294002 in improving PTS1 import. One possible explanation is that LY294002 is not highly specific for autophagy, as it can inhibit other lipid kinases that are involved in other functions such as cell survival, proliferation and metabolism.38 Alternatively, it is also possible that bafilomycin A1 and chloroquine may have other indirect effects on peroxisomes other than autophagy by perturbing the acidification of lysosomes.25 Furthermore, if bafilomycin A1 and chloroquine simply prevent lysosomes acidification (for a review see ref. 28), then it can be postulated that treating cells with these agents will result in sequestration of peroxisomes within autophagosomes and autolysosomes, and thus these peroxisomes will not be accessible by the cytosol for peroxisomal function. However, our observations show that most of the peroxisomes are cytosolic and not sequestered, as they are able to import matrix proteins. This observation suggests that these lysosomotropic drugs do not prevent the increase in free peroxisomes. One possible explanation for this observation is that the formation of autophagosomes may be saturated, thus allowing the newly formed peroxisomes to remain free in the cytosol.
Our finding that the peroxisomal AAA-complex is a regulator of peroxisome quality control is not exclusive to the mammalian cell. During the preparation of this manuscript, similar results were elegantly reported for S. cerevisiae.39 Similar to what we observed in the mammalian system, the loss of peroxisomal AAA-complex components results in the enhancement of pexophagy, and an increase in ubiquitinated Pex5. However, it appears that ubiquitination of peroxisomal protein(s) is not necessary for pexophagy in S. cerevisiae. This mechanistic difference between the 2 systems likely results from a dissimilarity by which the 2 systems selectively target cytosolic substrates to phagophores. Peroxisomes in S. cerevisiae are targeted to nascent phagophores by the autophagy receptor ATG36, which does not bind ubiquitin; whereas in the mammalian cells, pexophagy requires the ubiquitin-binding autophagy receptors NBR1 and SQSTM1.11,40-42
Although the 2 systems may differ in the targeting of peroxisomes to phagophores, the role of the peroxisomal AAA-complex in pexophagy is likely similar, in that they both act as a peroxisome quality control system by removing a pexophagy signal. Based on our findings, we propose that in mammalian cells, the AAA-complex prevents the degradation of peroxisomes by removing ubiquitinated peroxisomal proteins, which are the pexophagy signal in mammalian cells (Fig. 8). Recent studies examining early peroxisome biogenesis genes suggest that immature peroxisomal structures are degraded by pexophagy.43,44 However, our work suggests that the degradation of AAA-complex-deficient peroxisomes is not simply due to its inability to form mature peroxisomes, but the inability of the peroxisome to recycle its matrix protein import complex, namely PEX5. We find that inhibiting AAA-complex function results in the accumulation of ubiquitinated PEX5 on peroxisomes (Fig. 3), and the induction of autophagy (Fig. 4). However, inhibiting autophagy resulted in an increase in peroxisome numbers and matrix protein import (Figs. 5, 6). Because the peroxisomal AAA-complex is required to complete the recycling of PEX5 in a ubiquitin-dependent fashion,32 and ubiquitination of peroxisomes leads to pexophagy,11 we propose that the peroxisomal AAA-complex is a peroxisomal quality control factor that prevents pexophagy by removing ubiquitinated proteins from peroxisomes. Thus, the loss of its function will result in the accumulation of ubiquitinated PEX5 (and perhaps other peroxisomal membrane proteins) on the peroxisomal membrane, which then promotes the recruitment of autophagy receptors such as NBR1 and SQSTM1 to signal its degradation (Fig. 8B). Therefore, inhibiting autophagy with chemical inhibitors, such as chloroquine, will result in the accumulation of these AAA-complex-deficient peroxisomes, which are able to import matrix proteins required for peroxisomal function (Fig. 8C).
Figure 8.

A model for AAA-dependent pexophagy. The peroxisomal AAA-complex prevents ubiquitin-dependent pexophagy and enables peroxisomal matrix protein import. (A) Cells under basal conditions possess their peroxisomal AAA-complex, consisting of PEX1, PEX6 and PEX26, allowing for the efficient removal of ubiquitinated PEX5. Import of matrix proteins is represented by -PTS1. (B) Defect in any of the AAA-complex genes results in the loss of AAA-complex function and the accumulation of ubiquitinated PEX5. This results in the recruitment of autophagy receptors, NBR1 and SQSTM1, which then results in targeting of peroxisomes to phagophores for degradation via pexophagy. (C) Inhibiting pexophagy with chloroquine (CQ) prevents the degradation of peroxisomes, increasing their half-life, and allowing for the import of matrix proteins. This increase in peroxisome numbers and their subsequent increase in import of peroxisomal matrix enzymes results in improved peroxisomal function.
A corollary of this postulation is that matrix protein import signals the degradation of the peroxisomes by accumulating ubiquitinated PEX5 on the membrane. However, peroxisomes that express and recruit the peroxisomal AAA-complex to peroxisomes can overcome this degradation signal by removing the ubiquitinated proteins. This implies that the ability of peroxisomes to recruit the AAA ATPases (PEX1, PEX6) is critical for their survival. In fact, recent cryo-electron microscopy structure studies show that the AAA ATPases toggle on and off peroxisomes to remove ubiquitinated proteins.14-16 This suggests that peroxisomes must constantly recruit the AAA ATPases to maintain longevity. Therefore, it is possible that one of the signals for pexophagy in conditions such as hypoxia or amino acid starvation22,45,46 may be either the removal of, or prevention of localization of, AAA ATPases to peroxisomes to initiate the degradation of an individual peroxisome or the bulk degradation of peroxisomes.
Finally, as mutation in the peroxisomal AAA-complex makes up the majority of PBD cases, our study suggests that use of autophagy inhibitors for some PBDs may be a viable treatment that can be used with other treatments to lessen the progressive nature of the moderate and mild forms of PBDs. We have previously shown that peroxisomal function in mutant PEX1-G843D cells can be restored by chemical chaperones that might improve the folding of PEX1G843D.34 However, autophagy inhibitors may be used with other PEX1 mutations where the mutation does not lead to misfolding of the protein. Our finding that the peroxisomal AAA-complex is required for peroxisomal quality control, whereby its dysfunction leads to increased degradation of peroxisomes by autophagy, suggests that small chemical molecules with autophagy inhibition properties may be a therapeutic strategy for treating PBDs caused by AAA-complex dysfunction. As both chemical chaperones and autophagy inhibitors work on different aspects of the PEX1G843D mutation, we foresee the possibility of a combined therapy for treating mild forms of AAA-complex-deficient PBDs. Future experiments will involve determining the effectiveness of these inhibitors in a long-term period in vivo using a PEX1-G844D mouse model, which has been reported to recapitulate many features of PBD patients.47
Materials and methods
Plasmids and siRNA
GFP-MAP1LC3 was a gift from Dr. Yoshinori Ohsumi (Tokyo Institute of Technology, Yokohama, Japan). GFP-UbG76V (GFP-Ub) was purchased from Addgene (11928; deposited by Nico Dantuma). PEX5-MYC was a gift from Dr. Suresh Subramani (University of California, San Diego). siRNA directed against PEX26 (CAAGACCCAGCCAAUCAAA), ATG12 (GUGGGCAGUAGAGCGAACA), PEX1 (CCAAGCAACUUCAGUCAAA), PEX14 (GAACUCAAGUCCGAAAUU), NBR1 (GGAGUGGAUUUACCAGUUAUU), PEX16 (UGACGGGAUCCUACGGAAG), and PEX5 (GGCAGAGAAUGAACAAGAACUAUUA) were custom synthesized by Sigma. The control non-targeting siRNA was purchased (Sigma, SIC001).
Antibodies
Primary antibodies used in this study include: rabbit polyclonal anti-ATG12 (1:500; Cell Signaling Technology, 2010); rabbit polyclonal anti-CAT (1:5000; Calbiochem, 219010); mouse monoclonal anti-GAPDH-HRP (1:10,000; Novus Biologicals, NB300–328H); rabbit polyclonal anti-MAP1LC3B (1:1000 for immunoblotting; Novus Biologicals, NB100–2220, and 1:500 for immunofluorescence; Cell Signaling, 2775); mouse monoclonal anti-MYC (1:1000; BD Biosciences, 3800–1); mouse monoclonal anti-NBR1 (1:1000 for immunoblotting; Abnova, H00004077-M01); mouse monoclonal anti-SQSTM1 (1:10,000; BD Biosciences, 610832); mouse monoclonal anti-PEX1 (1:500; BD Transduction Laboratories, 611719); rabbit polyclonal anti-PEX14 (1:3000; Cedarlane, 10594–1-AP); mouse polyclonal anti-PEX26 (1:250; Novus Biologicals, NBP1–32743); rabbit monoclonal anti-ABCD3 (1:5000 for immunoblotting or 1:2000 for immunofluorescence; Abcam, ab3421); and rabbit polyclonal anti-TOMM20 (FL-145, 1:10,000; Santa Cruz Biotechnology, sc-11415). The anti-PEX5 antibody used for immunofluorescence was a gift from Dr. Gabrielle Dodt (University of Tübingen, Tübingen, Germany) and was used at 1:500. Secondary antibodies used for western blotting include goat anti-rabbit IgG-HRP (Santa Cruz Biotechnology, sc-2030) and goat anti-mouse IgG-HRP (Cedarlane, CLCC30007) and both were used at 1:10,000. Secondary antibodies used for immunofluorescence include goat anti-rabbit IgG, DyLight™ 594 (Thermo Scientific, PI35561) and goat anti-mouse IgG, DyLight™ 488 (Thermo Scientific, PI35553) and both were used at 1:1000.
The rabbit polyclonal anti-PEX5 antibody used for immunoblotting (1:1000) was generated by immunizing the New Zealand white rabbit with full-length PEX5–6xHIS (accession #NM_001131023) by standard protocol (Cedarlane, Burlington, Ontario). In brief, the plasmid pGEX-4T-GST-PEX5–6xHIS was constructed and transformed into Escherichia coli BL21 competent cells for prokaryotic expression. The recombinant GST-PEX5–6xHIS tagged PEX5 was expressed and purified first using a glutathione sepharose 4b column (GE Healthcare, 17075605). PEX5–6xHIS was eluted from the column by treating the cells with thrombin (GE Healthcare, 27–0846–01) to cleave it from the GST tag using an internal thrombin cleavage site. PEX5–6xHis was further purified using a Ni-NTA column (Qiagen, 30210).
Reagents
Hydroxychloroquine sulfate (chloroquine) was purchased from Sigma (H0915). Bafilomycin A1 was purchased from Santa Cruz Biotechnology (sc-201550A). LY294002 was purchased from Cedarlane (BML-ST420–0005). Tris hydrochloride (Tris-HCl) was purchased from FisherBrand (BP1521). 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) was purchased from BioShop (HEP002.500). Sodium dodecyl sulfate (SDS) was purchased from Bio-Rad (161–0416). Sucrose was purchased from Bioshop (SUC507.1). Potassium chloride (KCl) and ethylene glycol tetraacetic acid (EGTA) were purchased from Sigma (94505 and E3889, respectively). Magnesium chloride (MgCl2) was purchased from BioShop (MAG516.100) Ethylenediaminetetraacetic acid (EDTA) was purchased from Fisher Scientific (SS412–1). Authentic standards of lyso-PCs (20-lyso-PC, 855777; 22:0-lyso-PC, 855779; 24:0-lyso-PC, 855800; 26:0-lyso-PC 855810 and 16:0-Lyso PAF, 878119) and the tetradeuterated internal standard (26:0 D4 lyso PC, 860389) were purchased from Avanti Polar Lipids. The tetradeuterated internal standard (16:0 D4 lyso PAF, 860389) was purchased from Cayman Chemical Co. HPLC grade solvents (methanol, A956–4, acetonitrile A955–4) were purchased from Fisher Scientific. Chloroform (650498) and formic acid (94318) were purchased from Sigma-Aldrich.
Cell culture
HeLa cells were purchased from American Type Culture Collection (CCL-2). Wild-type and Atg5 null MEFs were a gift from Dr. Noboru Mizushima (Tokyo Medical and Dental University). The patient-derived wild-type and all patient-derived PEX1 PBD patient cell lines (PEX1 null, PEX1-G843D homozygous and hemizygous, PEX1-G843D-PTS1) were a gift from Dr. Nancy Braverman (McGill University, Montreal, Quebec, Canada). The PEX16 null line (GM6231) was obtained from Coriell Biorepository (Camden, NJ, USA). All cells were grown in Dulbecco's modified Eagle's medium (Fisher, 53695) supplemented with 10% fetal bovine serum (Wisent, 08–150) and 0.1% 200 mM L-glutamine (Fisher, SH3003401) and were cultured at 5% CO2 in a 37°C incubator. Cells were routinely tested for mycoplasma (FroggaBio, 25235).
For immunofluorescence, cells were grown on #1 glass coverslips (VWR, CA-89015725). Plasmids and siRNA were transfected using Optimem (Life Technologies, 31985070) and Lipofectamine-2000 (Life Technologies, 11668019) according to the manufacturer's instructions. For simultaneous siRNA knockdown and plasmid overexpression, siRNA alone was transfected on the first day, and siRNA and plasmids (0.5 μg DNA per 500 μL) were cotransfected on the second day. Cells were imaged 48 h after plasmid/siRNA transfection. Coverslips were mounted onto microscope slides (Fisher, 22037247) using Dako Fluorescent Mounting Medium (Dako Canada, 34538) and dried overnight at room temperature.
Microscope image processing and acquisition
For immunofluorescence, cells were fixed using 3.7% paraformaldehyde (Cedarlane, 15710) in phosphate-buffered saline (PBS; Bio Basic Inc., PD8117) for 15 min and washed twice with warm PBS. Cells were permeabilized using 0.1% Triton X-100 (BioShop, TRX506.500) in PBS for 20 min. Cells were then incubated in blocking buffer (10% fetal bovine serum in 1X PBS) for 1 h and incubated with the appropriate primary and secondary antibodies in blocking buffer, each for 1-h incubations at room temperature.
Laser-scanning confocal microscopy was performed on a Zeiss LSM710 (NA: 0.55, WD: 26 mm) with a 63x/1.4 Plan-Apochromat oil immersion objective at 37°C (Hospital for Sick Children, Toronto, Ontario, Canada). The microscope has 6 laser lines (405, 458, 488, 514, 561, 633 nm) and 3 channels. For the quantification of ABCD3 structures, images were acquired in Z-sections of 0.5 to 1.0 μm thickness using ZEN 2009 (Zeiss Enhanced Navigation) acquisition software. For quantification of the total fluorescence intensity, an open pinhole was used. GFP and Dylight 488 signals were acquired using a 488-nm argon laser with a 493- to 565-nm bandpass filter. Dylight 594 signal was acquired using a 594-nm diode laser with a 600- to 700-nm bandpass.
For each experiment set, all images were taken with the same acquisition setting and on the same day. This method allows for direct comparison between the different conditions. All images were acquired with a setting for optimal signal-to-noise ratio and minimal saturated pixel. Brightness/contrast of images were adjusted merely for presentation purposes, and all images of the same data set were adjusted to the same degree. Images on the figures are representative images that best demonstrate the quantified results.
Quantification
Images were analyzed using Volocity software (Perkin Elmer). At least 30 cells were quantified for each trial (n = 3) and averaged, and were normalized against the control when necessary. For quantification of peroxisome density, first, the number of peroxisomal structures (ABCD3) and the volume of the cell were quantified using the ‘find object’ algorithm in Volocity in cells of interest from a Z-stack image. The density of peroxisomes was calculated by dividing the number of peroxisomal structures by the volume of each cell (μm3). The same method was done for quantifying GFP-PTS1 density. The total fluorescence intensity of ABCD3 was measured from an image acquired with an open pinhole, corrected for background, and divided by the area of the cell.
Statistical analysis
Statistics were performed on data that had been generated in triplicate using an unpaired Student t Test. For all experiments, a p-value ≥ 0.05 was considered not significant (ns); 0.01–0.05 was considered significant (*), and < 0.01 was considered very significant (**). All statistics and error bars (representing standard deviation, SD) shown are representative of independent experiments. All statistical analyses were performed on all available data for each experiment.
Western blots
Cells were washed twice with PBS and lysed with ice cold lysis buffer (100 mM Tris, pH 9, buffer containing 1% SDS [Bio-Rad, 161–0416]) supplemented with 1X protease inhibitor cocktail (BioShop, PIC002.1), and the lysate was heated at 95°C with vortexing for 30 min, then centrifuged at 13,000 x g for 10 min. The protein concentration in the supernatant was determined by BCA assay (VWR, CA82601–004), equivalent sample amounts (10–20 μg) were subjected to 10–15% SDS-PAGE, and protein was transferred to 0.45-μm PVDF membrane (BioRad, 1620177). Membranes were blocked in 2.5% skim milk (BioShop, SK1400.1) in PBST (1x PBS, 0.05% Tween-20 [BioShop, 1M23298]). Membranes were probed with the appropriate primary and HRP-conjugated secondary antibodies in 2.5% skim milk. Proteins were visualized using either Amersham ECL Prime Western Blot Detection Reagent (VWR, CA89168–782) or EZ-ECL (FroggaBio, EZ-ECL-500). Protein density was measured using ImageJ, an imaging processing and analysis software, and was normalized to the density of the GAPDH loading control.
MTT assay
For a review on the MTT assay see ref. 35 Cells were treated with the tetrazolium dye MTT 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Sigma-Aldrich, M2128) at a final concentration of 0.5 mg/mL, and incubated for 4 h at 5% CO2 in a 37°C incubator. The formazan reaction product was dissolved in dimethyl sulfoxide (Fisher Scientific, BP231–100). The quantity of formazan was measured by recording the absorbance values at 540 nm using a plate reading spectrophotometer (Molecular Devices VersaMax 190) and analyzed using SoftMax Pro (Hospital for Sick Children, Toronto, Ontario).
LC-MS/MS analysis
Cells were washed with 1X DPBS (Thermo Fisher Scientific, 14190–144), trypsinized, and centrifuged at 1000 x g for 5 min and cell concentration was determined using a haemocytometer. Cells were homogenized in 1X DPBS. An extraction solution of methanol (Fisher Scientific, A456–4) containing 10 ng each of the internal standards, 2H4–16:0-lyso-PAF (Cayman Chemical, 360906) and 2H4–26:0-lyso-PC (Avanti Polar Lipids, Inc., 860389) was added to 50 µg protein cell extract in a glass tube. The samples were incubated on a shaker at room temperature for 1 h. The samples were transferred to Corning Costar spin-X centrifuge tube filters (VWR, 29442–760) and centrifuged for 5 min. The filtrates were then transferred to autosampler Verex vials (Phenomenex, AR0–9992–13) for analysis by LC-MS/MS (Research Institute of the MUHC and McGill University, Montreal, Quebec).
A Waters TQD interfaced with an Acquity UPLC system was used for positive ion electrospray (ESI)-MS/MS ionization. Elution of lyso-PCs was detected by monitoring multiple reaction monitoring (MRM) transitions representing fragmentation of [M+H]+ to m/z 104. Chromatographic resolution was achieved via the use of a 2.1 × 50 mm, 1.7 µm Waters Acquity UPLC BEH column. The solvent systems used were: mobile phase A = 54.5% water (Fisher Scientific, W6–4):45% acetonitrile (Fisher Scientific, A955–4):0.5% formic acid (Sigma, 94318–250ML-F) and mobile phase B = 99.5% acetonitrile:0.5% formic acid with both solutions containing 2 mM ammonium acetate (Fisher Scientific, A639–500). Injections of extracts dissolved in methanol were made with initial solvent conditions of 85% mobile phase A:15% mobile phase B. The gradient used was from 15% to 100% mobile phase B over a period of 2.5 min, held at 100% mobile phase B for 1 min before reconditioning the column back to 85% mobile phase A:15% mobile phase B at a solvent rate of 0.7 ml/min. Quantification of analytes was determined by generating standard curves with authentic standards.
Membrane fractionation
Cells were washed with cold 1X PBS and lysed using subcellular fractionation buffer (250 mM sucrose, 20 mM HEPES, pH 7.4, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA) with added 1 mM dithiothreitol (Fisher Scientific, R0861) and 1X protease inhibitor cocktail before use. Lysates were passed through a 26.5-G needle (VWR, CABD305111) 10 times, left on ice for 20 min, and spun at 845 x g for 5 min at 4°C. A fraction was taken for the whole cell lysate, and the remaining supernatant was then spun using an ultracentrifuge at 105,000 x g for 30 min at 4°C. The pellet (membrane fraction) was resuspended in lysis buffer (100 mM Tris, pH 9, buffer containing 1% SDS) and the supernatant (cytosol fraction) was transferred to a fresh microcentrifuge tube. All samples were frozen at -80°C and later analyzed via immunoblot.
Supplementary Material
Abbreviations
- AAA-complex
AAA ATPase complex
- ABCD3
ATP binding cassette subfamily D member 3
- ATG5
autophagy-related 5
- ATG12
autophagy-related 12
- CAT
catalase
- ECL
enhanced chemiluminescence
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GFP
green fluorescent protein
- GFP-MAP1LC3
green fluorescent protein tagged to MAP1LC3 (see MAP1LC3)
- GFP-PTS1
green fluorescent protein tagged to PTS1 (see PTS1)
- GFP-Ub
green fluorescent protein tagged to ubiquitin
- HRP
horseradish peroxidase
- IgG
immunoglobulin
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- LysoPC
lysophosphatidylcholine
- MAP1LC3B
microtubule-associated protein 1 light chain 3 β
- MEF
mouse embryonic fibroblast
- MTT
3–3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide
- MYC
v-myc avian myelocytomatosis viral oncogene homolog
- NBR1, NBR1
autophagy cargo receptor
- PBDs
peroxisome biogenesis disorders
- PBS
phosphate-buffered saline
- PEX1
peroxisomal biogenesis factor 1
- PEX5
peroxisomal biogenesis factor 5
- PEX6
peroxisomal biogenesis factor 6
- PEX14
peroxisomal biogenesis factor 14
- PEX16
peroxisomal biogenesis factor 16
- PEX26
peroxisomal biogenesis factor 26
- PINK1
PTEN induced putative kinase 1
- PtsIns3K
phosphatidylinositol 3-kinase
- PTS1
peroxisome targeting sequence-1
- RFP
red fluorescent protein
- RFP-SKL
red fluorescent protein tagged to serine-lysine-leucine
- SDS
sodium dodecyl sulfate
- SKL
serine-lysine-leucine
- SQSTM1
sequestosome 1
- TOMM20
translocase of outer mitochondrial membrane 20
- Ub
ubiquitin
- VLCFA
very long chain fatty acids
Acknowledgements
We would like to thank Yuqing Wang, Rong Hua and Nicholas Demers for critical commentary on the manuscript and Catherine Argyriou for graciously maintaining and sending all of the wild type and PBD patient-derived cell lines. J.H.B. holds the Pitblado Chair in Cell Biology at the Hospital for Sick Children. K.B.L. holds the Restracomp Award (Hospital for Sick Children), the Ontario Graduate Scholarship (University of Toronto), the OSOTF Hilda and William Courtney Clayton Pediatric Research Fund Award (University of Toronto), the OSOTF Hayden Hantho Award (University of Toronto), the OSOTF Dr. Joe A. Connolly Memorial Award (University of Toronto), and the Dorothy Sterling Dow Walsh Award (University of Toronto). P.K.K. is a recipient of the Ontario Early Research Award.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
The work presented in this manuscript is funded by a research grant from the Canada Institute of Health Research to P.K.K. This paper is dedicated to the memory of Dr. Eliana Chan, a bright young scientist who pursued her love for science to the very end.
References
- [1].Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ [Internet] 2005; 12 Suppl 2:1542-52. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16247502; http://dx.doi.org/ 10.1038/sj.cdd.4401765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Okamoto K. Organellophagy: eliminating cellular building blocks via selective autophagy. J Cell Biol [Internet] 2014; 205:435-45. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24862571; PMID: 24862571; http://dx.doi.org/ 10.1083/jcb.201402054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol [Internet] 2014; 16:495-501. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24875736; PMID: 24875736; http://dx.doi.org/ 10.1038/ncb2979 [DOI] [PubMed] [Google Scholar]
- [4].Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy [Internet] 2011; 7:279-96. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21189453; PMID: 21189453; http://dx.doi.org/ 10.4161/auto.7.3.14487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron [Internet] 2015; 85:257-73. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25611507; http://dx.doi.org/ 10.1016/j.neuron.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol [Internet] 2010; 191:933-42. Available from: http://jcb.rupress.org/content/191/5/933 PMID: 21115803; http://dx.doi.org/ 10.1083/jcb.201008084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, Youle RJ. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol [Internet] 2014; 205:143-53. Available from: http://jcb.rupress.org/content/205/2/143; PMID: 24751536; http://dx.doi.org/ 10.1083/jcb.201402104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, Hofmann K, Alessi DR, Knebel A, Trost M, Muqit MMK. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J [Internet] 2014; 460:127-41. Available from: http://www.biochemj.org/content/460/1/127; PMID: 24660806; http://dx.doi.org/ 10.1042/BJ20140334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, et al.. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature [Internet] 2014; 510:162-6. Available from: http://www.nature.com/nature/journal/vnfv/ncurrent/full/nature13392.html [DOI] [PubMed] [Google Scholar]
- [10].Deosaran E, Larsen KB, Hua R, Sargent G, Wang Y, Kim S, Lamark T, Jauregui M, Law K, Lippincott-Schwartz J, et al.. NBR1 acts as an autophagy receptor for peroxisomes. J Cell Sci [Internet] 2013; 126:939-52. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23239026; PMID: 23239026 http://dx.doi.org/ 10.1242/jcs.114819 [DOI] [PubMed] [Google Scholar]
- [11].Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A [Internet] 2008; 105:20567-74. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19074260; http://dx.doi.org/ 10.1073/pnas.0810611105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hettema EH, Distel B, Tabak HF. Import of proteins into peroxisomes. Biochim Biophys Acta - Mol Cell Res [Internet] 1999; 1451:17-34. Available from: http://www.sciencedirect.com/science/article/pii/S0167488999000877; http://dx.doi.org/ 10.1016/S0167-4889(99)00087-7 [DOI] [PubMed] [Google Scholar]
- [13].Platta HW, Hagen S, Reidick C, Erdmann R. The peroxisomal receptor dislocation pathway: to the exportomer and beyond. Biochimie [Internet] 2014; 98:16-28. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24345375; PMID: 24345375; http://dx.doi.org/ 10.1016/j.biochi.2013.12.009 [DOI] [PubMed] [Google Scholar]
- [14].Blok NB, Tan D, Wang RY-R, Penczek PA, Baker D, DiMaio F, Rapoport TA, Walz T. Unique double-ring structure of the peroxisomal Pex1/Pex6 ATPase complex revealed by cryo-electron microscopy. Proc Natl Acad Sci U S A [Internet] 2015; 112:E4017-25. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26170309; PMID: 26170309; http://dx.doi.org/ 10.1073/pnas.1500257112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ciniawsky S, Grimm I, Saffian D, Girzalsky W, Erdmann R, Wendler P. Molecular snapshots of the Pex1/6 AAA+ complex in action. Nat Commun [Internet] 2015; 6:7331. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26066397; PMID: 26066397; http://dx.doi.org/ 10.1038/ncomms8331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gardner BM, Chowdhury S, Lander GC, Martin A. The Pex1/Pex6 complex is a heterohexameric AAA+ motor with alternating and highly coordinated subunits. J Mol Biol [Internet] 2015; 427:1375-88. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25659908; PMID: 25659908; http://dx.doi.org/ 10.1016/j.jmb.2015.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Steinberg SJ, Dodt G, Raymond G V, Braverman NE, Moser AB, Moser HW. Peroxisome biogenesis disorders. Biochim Biophys Acta [Internet] 2006; 1763:1733-48. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17055079; http://dx.doi.org/ 10.1016/j.bbamcr.2006.09.010 [DOI] [PubMed] [Google Scholar]
- [18].Braverman NE, Raymond G V, Rizzo WB, Moser AB, Wilkinson ME, Stone EM, Steinberg SJ, Wangler MF, Rush ET, Hacia JG, et al.. Peroxisome biogenesis disorders in the Zellweger spectrum: An overview of current diagnosis, clinical manifestations, and treatment guidelines. Mol Genet Metab [Internet] 2015; 117:313-21. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26750748; PMID: 26750748; http://dx.doi.org/ 10.1016/j.ymgme.2015.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Waterham HR, Ferdinandusse S, Wanders RJA. Human disorders of peroxisome metabolism and biogenesis. Biochim Biophys Acta [Internet] 2015; 1863:922-33. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26611709; http://dx.doi.org/ 10.1016/j.bbamcr.2015.11.015 [DOI] [PubMed] [Google Scholar]
- [20].Jones JM, Morrell JC, Gould SJ. PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins. J Cell Biol [Internet] 2004; 164:57-67. Available from: http://www.ncbi.nlm.nih.gov/pubmed/14709540; PMID: 14709540; http://dx.doi.org/ 10.1083/jcb.200304111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nordgren M, Francisco T, Lismont C, Hennebel L, Brees C, Wang B, Van Veldhoven PP, Azevedo JE, Fransen M. Export-deficient monoubiquitinated PEX5 triggers peroxisome removal in SV40 large T antigen-transformed mouse embryonic fibroblasts. Autophagy [Internet] 2015; 11:1326-40. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26086376; PMID: 26086376; http://dx.doi.org/ 10.1080/15548627.2015.1061846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhang J, Tripathi DN, Jing J, Alexander A, Kim J, Powell RT, Dere R, Tait-Mulder J, Lee J-H, Paull TT, et al.. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat Cell Biol [Internet] 2015; 17:1259-69. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26344566; PMID: 26344566; http://dx.doi.org/ 10.1038/ncb3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kamijo K, Taketani S, Yokota S, Osumi T, Hashimoto T. The 70-kDa peroxisomal membrane protein is a member of the Mdr (P-glycoprotein)-related ATP-binding protein superfamily. J Biol Chem [Internet] 1990; 265:4534-40. Available from: http://www.ncbi.nlm.nih.gov/pubmed/1968461; PMID: 1968461 [PubMed] [Google Scholar]
- [24].Steinberg SJ, Raymond G V, Braverman NE, Moser AB. Peroxisome Biogenesis Disorders, Zellweger Syndrome Spectrum [Internet] In: Pagon RA, Adam MP, Ardinger HH, Bird TD, Dolan CR, Fong C-T, Smith RJH, Stephens K, editors GeneReviews(®). Seattle, WA: University of Washington, Seattle; 1993. Available from: http://www.ncbi.nlm.nih.gov/books/NBK1448/ [Google Scholar]
- [25].Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al.. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy [Internet] 2012; 8:445-544. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22966490; http://dx.doi.org/ 10.4161/auto.19496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Waterham HR, Ebberink MS. Genetics and molecular basis of human peroxisome biogenesis disorders. Biochim Biophys Acta [Internet] 2012. [cited 2016 Dec 21]; 1822:1430-41. Available from: http://linkinghub.elsevier.com/retrieve/pii/S0925443912000932; PMID: 22871920; http://dx.doi.org/ 10.1016/j.bbadis.2012.04.006 [DOI] [PubMed] [Google Scholar]
- [27].Braverman NE, Raymond G V., Rizzo WB, Moser AB, Wilkinson ME, Stone EM, Steinberg SJ, Wangler MF, Rush ET, Hacia JG, et al.. Peroxisome biogenesis disorders in the Zellweger spectrum: An overview of current diagnosis, clinical manifestations, and treatment guidelines. Mol Genet Metab 2016; 117:313-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yang Y, Hu L, Zheng H, Mao C, Hu W, Xiong K, Wang F, Liu C. Application and interpretation of current autophagy inhibitors and activators. Acta Pharmacol Sin [Internet] 2013; 34:625-35. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23524572; PMID: 23524572; http://dx.doi.org/ 10.1038/aps.2013.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Fujiki Y, Nashiro C, Miyata N, Tamura S, Okumoto K. New insights into dynamic and functional assembly of the AAA peroxins, Pex1p and Pex6p, and their membrane receptor Pex26p in shuttling of PTS1-receptor Pex5p during peroxisome biogenesis. Biochim Biophys Acta [Internet] 2012; 1823:145-9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22079764; PMID: 22079764; http://dx.doi.org/ 10.1016/j.bbamcr.2011.10.012 [DOI] [PubMed] [Google Scholar]
- [30].Kiel JAKW, Emmrich K, Meyer HE, Kunau W-H. Ubiquitination of the peroxisomal targeting signal type 1 receptor, Pex5p, suggests the presence of a quality control mechanism during peroxisomal matrix protein import. J Biol Chem [Internet] 2005; 280:1921-30. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15536088; PMID: 15536088; http://dx.doi.org/ 10.1074/jbc.M403632200 [DOI] [PubMed] [Google Scholar]
- [31].Okumoto K, Misono S, Miyata N, Matsumoto Y, Mukai S, Fujiki Y. Cysteine ubiquitination of PTS1 receptor Pex5p regulates Pex5p recycling. Traffic [Internet] 2011; 12:1067-83. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21554508 ; http://dx.doi.org/ 10.1111/j.1600-0854.2011.01217.x [DOI] [PubMed] [Google Scholar]
- [32].Platta HW, El Magraoui F, Schlee D, Grunau S, Girzalsky W, Erdmann R. Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J Cell Biol [Internet] 2007; 177:197-204. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17452527; PMID: 17452527; http://dx.doi.org/ 10.1083/jcb.200611012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Miyata N, Fujiki Y. Shuttling mechanism of peroxisome targeting signal type 1 receptor Pex5: ATP-independent import and ATP-dependent export. Mol Cell Biol [Internet] 2005; 25:10822-32. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16314507; http://dx.doi.org/ 10.1128/MCB.25.24.10822-10832.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhang R, Chen L, Jiralerspong S, Snowden A, Steinberg S, Braverman N. Recovery of PEX1-Gly843Asp peroxisome dysfunction by small-molecule compounds. Proc Natl Acad Sci U S A [Internet] 2010. [cited 2014 Mar 28]; 107:5569-74. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid equals;2851769&tool=pmcentrez&rendertype=abstract; PMID: 20212125; http://dx.doi.org/ 10.1073/pnas.0914960107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sumantran VN. Cellular chemosensitivity assays: an overview. Methods Mol Biol [Internet] 2011. [cited 2016 Oct 3]; 731:219-36. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21516411; PMID: 21516411 [DOI] [PubMed] [Google Scholar]
- [36].Zhang R, Chen L, Jiralerspong S, Snowden A, Steinberg S, Braverman N. Recovery of PEX1-Gly843Asp peroxisome dysfunction by small-molecule compounds. Proc Natl Acad Sci U S A [Internet] 2010; 107:5569-74. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20212125; PMID: 20212125; http://dx.doi.org/ 10.1073/pnas.0914960107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Brocard CB, Boucher KK, Jedeszko C, Kim PK, Walton PA. Requirement for microtubules and dynein motors in the earliest stages of peroxisome biogenesis. Traffic [Internet] 2005; 6:386-95. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15813749 ; http://dx.doi.org/ 10.1111/j.1600-0854.2005.00283.x [DOI] [PubMed] [Google Scholar]
- [38].Gharbi SI, Zvelebil MJ, Shuttleworth SJ, Hancox T, Saghir N, Timms JF, Waterfield MD. Exploring the specificity of the PI3K family inhibitor LY294002. Biochem J [Internet] 2007; 404:15-21. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17302559; PMID: 17302559; http://dx.doi.org/ 10.1042/BJ20061489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Nuttall JM, Motley AM, Hettema EH. Deficiency of the exportomer components Pex1, Pex6, and Pex15 causes enhanced pexophagy in Saccharomyces cerevisiae. Autophagy [Internet] 2014; 10:835-45. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24657987; PMID: 24657987; http://dx.doi.org/ 10.4161/auto.28259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Motley AM, Nuttall JM, Hettema EH. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J [Internet] 2012; 31:2852-68. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22643220; PMID: 22643220; http://dx.doi.org/ 10.1038/emboj.2012.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yamashita S, Abe K, Tatemichi Y, Fujiki Y. The membrane peroxin PEX3 induces peroxisome-ubiquitination-linked pexophagy. Autophagy [Internet] 2014; 10:1549-64. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25007327; PMID: 25007327; http://dx.doi.org/ 10.4161/auto.29329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell [Internet] 2005; 16:1593-605. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15659643; PMID: 15659643; http://dx.doi.org/ 10.1091/mbc.E04-11-1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Knoops K, Manivannan S, Cepinska MN, Krikken AM, Kram AM, Veenhuis M, van der Klei IJ. Preperoxisomal vesicles can form in the absence of Pex3. J Cell Biol [Internet] 2014; 204:659-68. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24590171; PMID: 24590171;http://dx.doi.org/ 10.1083/jcb.201310148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].van der Zand A, Gent J, Braakman I, Tabak HF. Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell [Internet] 2012; 149:397-409. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22500805; PMID: 22500805; http://dx.doi.org/ 10.1016/j.cell.2012.01.054 [DOI] [PubMed] [Google Scholar]
- [45].Walter KM, Schönenberger MJ, Trötzmüller M, Horn M, Elsässer H-P, Moser AB, Lucas MS, Schwarz T, Gerber PA, Faust PL, et al.. Hif-2α promotes degradation of mammalian peroxisomes by selective autophagy. Cell Metab [Internet] 2014; 20:882-97. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25440060; PMID: 25440060; http://dx.doi.org/ 10.1016/j.cmet.2014.09.017 [DOI] [PubMed] [Google Scholar]
- [46].van Zutphen T, Ciapaite J, Bloks VW, Ackereley C, Gerding A, Jurdzinski A, Allgayer de Moraes R, Zhang L, Wolters JC, Bischoff R, et al.. Malnutrition-associated liver steatosis and ATP depletion is caused by peroxisomal and mitochondrial dysfunction. J Hepatol [Internet] 2016; 65(6):1198-1208. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27312946; PMID: 27312946 [DOI] [PubMed] [Google Scholar]
- [47].Hiebler S, Masuda T, Hacia JG, Moser AB, Faust PL, Liu A, Chowdhury N, Huang N, Lauer A, Bennett J, et al.. The Pex1-G844D mouse: A model for mild human Zellweger spectrum disorder. Mol Genet Metab [Internet] 2014; 111:522-32. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24503136; PMID: 24503136; http://dx.doi.org/ 10.1016/j.ymgme.2014.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.