Abstract
Post-translational modifications (PTMs) are one of the main contributors to the diversity of proteoforms in the proteomic landscape. In particular, protein phosphorylation represents an essential regulatory mechanism that plays a role in many biological processes. Protein kinases, the enzymes catalyzing this reaction, are key participants in metabolic and signaling pathways. Their activation or inactivation dictate downstream events: what substrates are modified and their subsequent impact (e.g., activation state, localization, protein-protein interactions (PPIs)). The biomedical literature continues to be the main source of evidence for experimental information about protein phosphorylation. Automatic methods to bring together phosphorylation events and phosphorylation-dependent PPIs can help to summarize the current knowledge and to expose hidden connections. In this chapter, we demonstrate two text mining tools, RLIMS-P and eFIP, for the retrieval and extraction of kinase-substrate-site data and phosphorylation-dependent PPIs from the literature. These tools offer several advantages over a literature search in PubMed as their results are specific for phosphorylation. RLIMS-P and eFIP results can be sorted, organized, and viewed in multiple ways to answer relevant biological questions, and the protein mentions are linked to UniProt identifiers.
Keywords: Bioinformatics, Phosphorylation, Post-translational Modification, Protein-protein Interaction, Text Mining
1. Introduction
Post-translational modifications (PTMs) are an important contributor to protein diversity. PTMs play a pivotal role in protein function, regulating activity, localization, and protein-protein interactions (PPIs), and therefore disruptions in PTMs can lead to disease [1]. In particular, protein phosphorylation is an essential regulatory mechanism in many biological processes. Proteins can be phosphorylated at different and/or multiple positions, most commonly on serine, threonine, and tyrosine residues. Protein kinases, the enzymes catalyzing the phosphorylation reaction, play a key role in regulating these events and have become therapeutic targets for drug design in multiple diseases [2–4]. However, few drugs targeting kinases have been completely successful in the clinic mainly due to the conserved nature of kinases. Consequently, many of the available inhibitors lack sufficient selectivity for effective clinical application. The identification and characterization of kinase-substrate interactions are keys to improve the approaches to targeted drug development [5].
The scientific literature contains a wealth of protein phosphorylation data derived both from traditional experiments that focus on a small number of proteins and from high-throughput experiments that attempt to assess the phosphorylation state of the whole proteome [6]. Researchers frequently query PubMed or specialized databases to gain access to this information. Similarly, database biocurators collect literature, and read and extract the most salient information relevant to their domain. Given the continuing increase of the size of the PubMed database, finding or collecting information that is spread across this vast knowledge pool remains challenging. Automatic methods to bring this data together can help to summarize the current knowledge and to expose hidden connections. For example, one article might describe that phosphorylation of a protein at a given site is implicated in a particular disease, and another article might describe a kinase that phosphorylates the site, leading to the connection of the kinase to the disease, which could be investigated further. Text mining tools have evolved considerably in number and quality and are being used to address a variety of research questions in the biomedical domain; for recent reviews see [7–9].
iProLINK (integrated Protein Literature, INformation and Knowledge) [10] offers a portfolio of text mining tools and annotated corpora developed by our group. Some of these are intended for developers to serve as modules in specific steps of their text mining pipelines (e.g., iXtractR [11] for relation extraction, and iSimp for sentence simplification [12]). Others are applications for biomedical researchers and biocurators to facilitate the exploration of the literature about proteins (pGenN [13], eFIP [14,15], eGIFT [16], and RLIMS-P [17,18]) and microRNAs (miRTex [19]) (Table 1).
Table 1.
Text mining tools available in iProLINK.
| Tool | Description | Bioentities/Relations | Standard Used |
|---|---|---|---|
| pGenN | Identifies plant gene name mentions in Medline abstracts |
|
|
| eGIFT | Identifies informative terms (iTerms) and documents relevant to a gene/protein (abstract level) |
|
|
| miRTex | Identifies miRNA-target relations as well as miRNA-gene and gene-miRNA regulation relations in Medline abstracts |
|
|
| RLIMS-P | Identifies information relevant to protein phosphorylation: kinase, substrate and sites. (abstract and full-length PMC open access articles) |
|
|
| eFIP | Identifies phosphorylation-dependent protein-protein interactions (abstract and full-length PMC open access articles) |
|
|
Among these applications, RLIMS-P and eFIP facilitate the extraction of phosphorylation information from the literature and therefore are the focus of this book chapter.
RLIMS-P is a rule-based information extraction system that identifies kinase, substrate, and site relations in the scientific literature (including PubMed abstracts and PMC open access (OA) full-length articles). For example, the tuple <Akt, CHK1, Ser280> is extracted by RLIMS-P from the following sentence:
“CHK1 is directly phosphorylated by Akt at Ser280, a modification that results in cytoplasmic sequestration” [20].
Since these three entities (kinase, substrate and site) are rarely co-mentioned in the same sentence, RLIMS-P employs techniques that combine information found in different sentences. The kinase or substrate names detected could correspond to individual proteins (e.g., Crm1), protein complexes (e.g., CDK1-cyclin-B), or a group of related proteins (e.g., Src kinases), whereas a site could be a residue type (e.g., serine, threonine, and tyrosine), a specific residue (e.g., Ser-391), or a protein region or domain (e.g., C-terminal domain) [18]. RLIMS-P has been benchmarked with multiple corpora [17]. The F-scores (harmonic mean between precision and recall), based on a collection of sections derived from 100 full-text articles, have previously been reported to be 0.88, 0.91, and 0.92 for kinases, substrates, and sites, respectively [17]. In addition, RLIMS-P integrates GNormPlus [21] to link the detected kinase and substrate names to UniProt identifiers whenever possible.
eFIP builds on RLIMS-P by first detecting mentions of protein phosphorylation (kinase, substrate, and site), but adds detection of protein-protein interactions (PPIs) involving the phosphorylated protein. The types of PPIs captured include interactions between two proteins, or interactions between a protein and a protein complex, protein region, or protein class. Once the phosphorylation and PPI mentions are detected, the second step is to identify a possible relation between the two events. The evaluation of eFIP on full-length articles achieved an F-measure of 0.84 on 100 article sections [14]. Selected data from RLIMS-P and eFIP has been integrated in iPTMnet (http://proteininformationresource.org/iPTMnet/) and is actively used in the curation of proteoforms in the Protein Ontology [22].
This chapter demonstrates how to use RLIMS-P and eFIP to uncover information about protein phosphorylation and phosphorylation-dependent PPIs from the literature.
2. Materials
2.1. Web Sites
iProLINK: http://proteininformationresource.org/iprolink
2.2. General aspects of the RLIMS-P and eFIP interfaces
Input
Both the RLIMS-P and eFIP web sites allow the input of keywords or phrases that can be combined with Boolean operators (AND, OR, NOT) in the same way as building a PubMed query. Similarly, MeSH terms (controlled vocabulary used to index Medline abstracts) can be included in the search (e.g., “Alzheimer Disease” [Mesh]). The input is sent to the PubMed web site and relevant PMIDs are retrieved. The PMIDs are then used to query a backend database that hosts pre-processed results for PubMed abstracts and full-length PMC OA documents by RLIMS-P or eFIP. In both systems, you have the option to restrict the search to a particular organism of interest (Figure 1A 3, Figure 2A). You can also select to exclude review articles if you are only interested in research articles, and/or query only abstracts (Figure 1A 4). eFIP also supports searches based on protein roles (kinases, substrates, interacting partners) for protein names. Alternatively, a list of PMIDs or PMCIDs, delimited by comma, space or listed in new lines, can be entered (Figure 1A 5, Figure 2A).
Fig. 1.
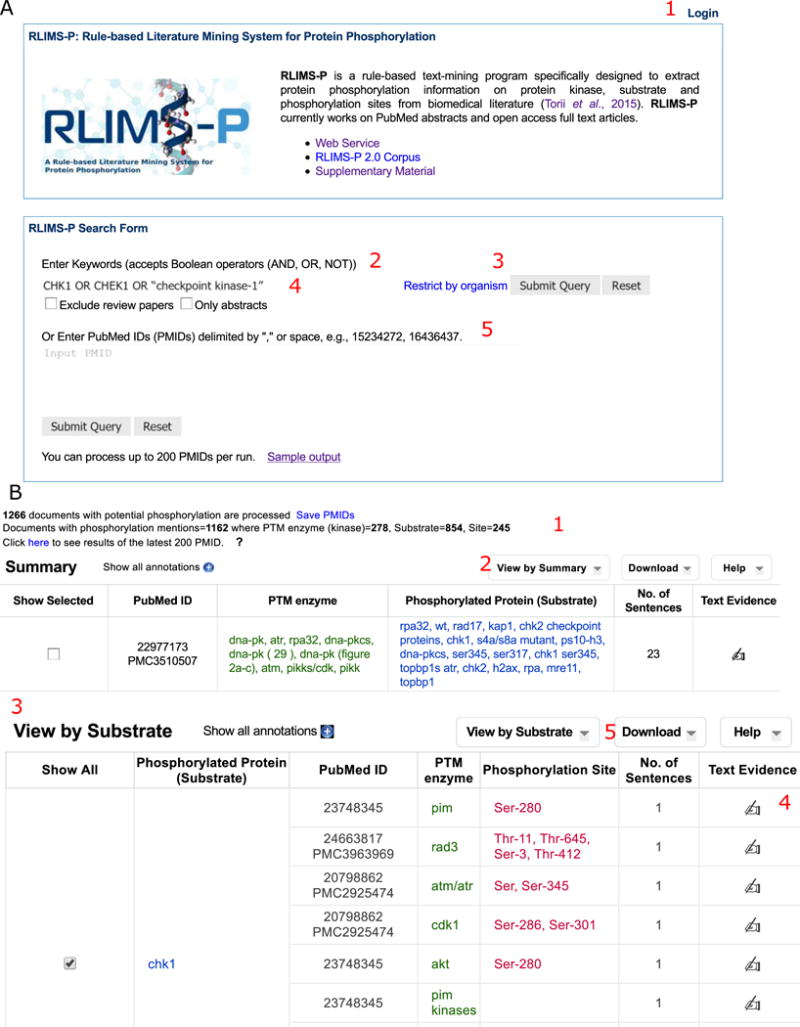
RLIMS-P for extraction of kinase-substrate-site information about CHK1. A. RLIMS-P homepage, showing the different functionalities: login capability (1), input query options such as keywords (2) or PubMed IDs (5) and search options (3, organism restriction or 4, exclusion of review articles or only abstracts). B. Partial display of RLIMS-P results for CHK1 search with summary statistics (1), and tables with “View by Summary” (2), and “View by Substrate” views (3). The “Text Evidence” (4) column provides links to the text evidence page. Text mining results can be downloaded in CSV format (5)
Fig. 2.
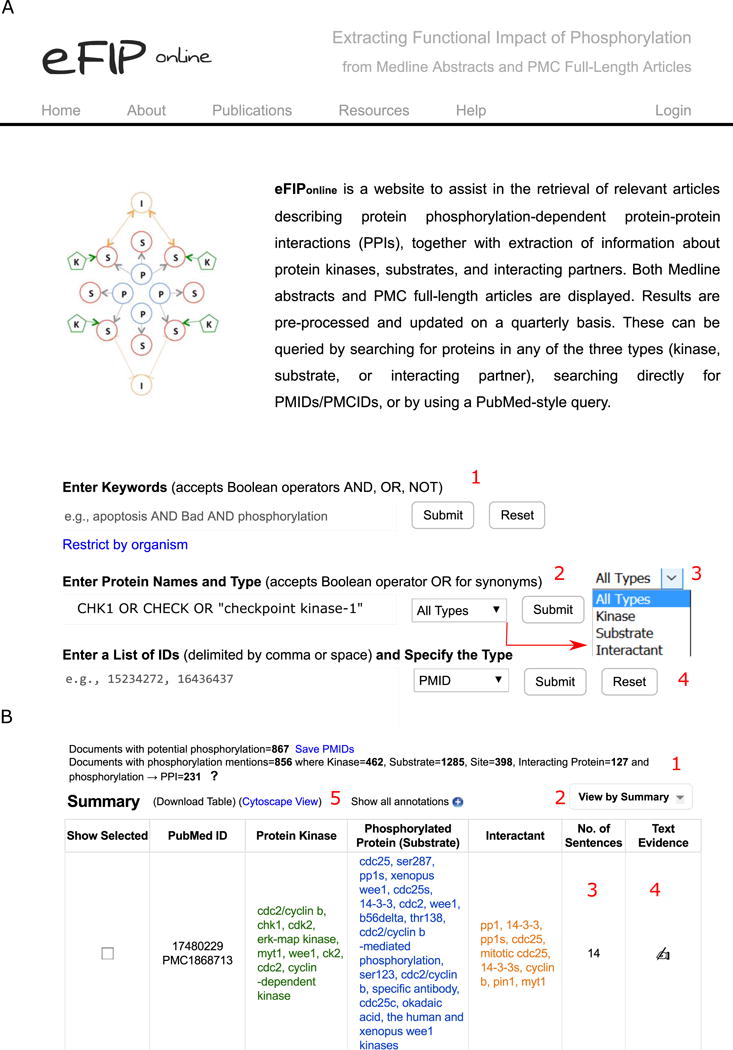
eFIP for extraction of phosphorylation-dependent PPI information about CHK1. A. eFIP homepage, showing the different functionalities: input query options such as keywords (1), protein names (2), or PubMed/PMC IDs (4) and search options for organism restriction and protein type including substrate, kinase or interactant (3). B. Partial display of eFIP results for CHK1 search with summary statistics (1), and table with “View by Summary” (2). The columns “No. of Sentences” (3) and “Text Evidence” (4) provide links to the text evidence pages. Text mining results can be downloaded in CSV format and be viewed in Cytoscape (5)
Results
The RLIMS-P result page presents summary statistics of the retrieved results (Figure 1B 1), listing separately the number of documents with potential phosphorylation information (i.e., those with the word “phosphorylation” or similar ones) and those with phosphorylation information according to RLIMS-P (i.e., there is at least one substrate identified). In addition, eFIP shows the summary statistics for interactants detected (Figure 2B 1).
Editing capabilities
To unlock editing capabilities, user registration and login are required (Figure 1A 1, Figure 2A, see Note 1). Edited results can be downloaded.
Cytoscape
eFIP offers a graphical view of the text mining results, displaying the protein entities as nodes and their relations as edges. The node names correspond to the protein entities in the result table, with some of the longer names abbreviated. The graph can be saved in PNG and XGMML-beta (Cytoscape compatible) format (Figure 6 2). Substrates, kinases, and interactants are represented as nodes with red circles, green pentagons, and orange circles, respectively. Interactions that are enabled or enhanced by phosphorylation are depicted as edges using solid orange lines with pointed arrowheads, whereas those that are decreased or inhibited are depicted by dashed orange lines with T-type arrowheads (Figure 6).
Fig. 6.
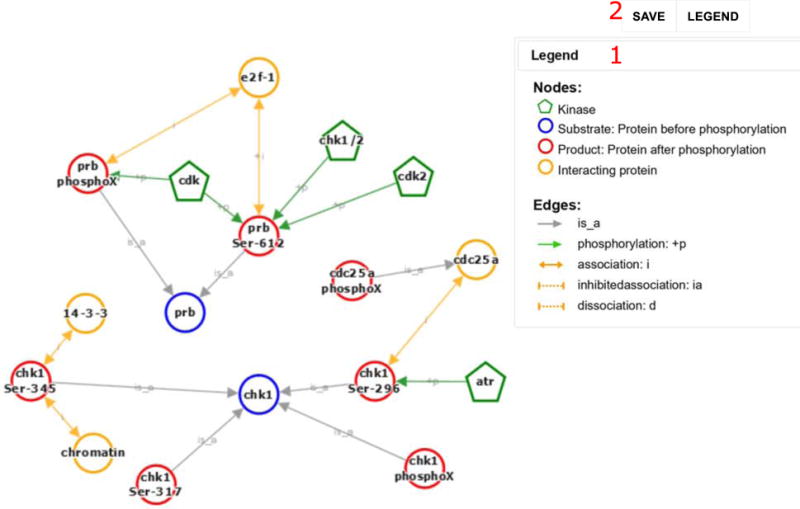
The Cytoscape representation for the phosphorylation and PPI events extracted from articles with PMID 12676962, 17380128, and 20639859. Events extracted are illustrated with nodes and edges; see legend for details (1). The graph can be exported (2)
3. Methods
For illustration purposes, we will showcase RLIMS-P and eFIP tool usage with examples from the Checkpoint kinase-1 protein, commonly referred to as CHK1 or CHEK1. This protein is a serine/threonine-specific protein kinase. It coordinates the DNA damage response (DDR) and cell cycle checkpoint response [23]. Activation of CHK1 results in the initiation of cell cycle checkpoints, cell cycle arrest, DNA repair and cell death to prevent damaged cells from progressing through the cell cycle [24]. A recent review article by Goto et al. [25] describes the regulation of CHK1 via phosphorylation, its substrates and the functional impact. To validate the approach, we compare the output of our text mining tools with the knowledge in the review article when applicable. We illustrate in the following text a variety of examples of RLIMS-P and eFIP usage via specific biological questions.
3.1 How to find kinases acting on a given substrate. What sites are phosphorylated?
Is CHK1 phosphorylated? If so, which sites? By what kinases? To answer these questions, we will use the RLIMS-P website (http://proteininformationresource.org/rlimsp, Figure 1A). The goal in this case is to find the articles mentioning CHK1 as a substrate, as we are interested in its phosphorylation sites. To achieve the most comprehensive result, it is recommended to include the different names by which CHK1 is known (e.g., CHEK1, Checkpoint kinase-1). If you are not familiar with the variety of names that are used for your protein of interest, you can check in a reference curated source, such as UniProt [26] or Entrez [27]. For this case, we will use the query (Figure 1A 2):
CHK1 OR CHEK1 OR “checkpoint kinase-1”
Go to RLIMS-P website and enter this query in the box and submit. Results are returned as shown in Figure 1B. Information on the top of the page summarizes the general statistics for the search results (Figure 1B 1), including the number of articles with potential protein phosphorylation mentions and the number of kinase, substrate, and site mentions (see Note 2).
Display results by “Substrate.” The results from the search in RLIMS-P include articles where the keywords are mentioned and which are about protein phosphorylation. The default table view is a summary listing the kinase and substrate mentions for each PMID. To obtain the subset where CHK1 is the phosphorylated protein, choose the option “View by Substrate” from the pull-down menu (Figure 1B 2) (see Note 3).
Find CHK1 as substrate. The table in Figure 1B 3 is now substrate centric. Next, we have to find CHK1 in the substrate column. As shown in this table, there are many articles describing phosphorylation of CHK1 (where CHK1 acts as a substrate). In addition, the kinases that phosphorylate CHK1 and the phosphorylation sites can now be easily identified in the columns “PTM enzyme” and “Phosphorylation Site,” respectively.
Validate and summarize the information. When the results are viewed by substrate (as shown in Figure 1B 3), all the phosphorylation sites on a substrate are shown. Now continue with our example by looking for CHK1 as substrate. The “No. of Sentences” column provides quick access to evidence sentences with color-coded highlighting of kinase (green), substrate (blue), and site (red) mentions (see Figure 4 bottom panel). This page is almost the same as the page linked out through icons in the “Text Evidence” column (Figure 1B 4), except that it restricts its sentence display to those where the information tuples are directly derived. To validate the information, the evidence can also be viewed by clicking on the icon in the “Text Evidence” column (Figure 1B 4), which will take you to the evidence page (Figure 3A). The evidence page presents a table summarizing the data extracted from the article with links to the source sentences (Figure 3A 2), a block showing the relevant sentences from the text (abstract or full text) with color-coding highlighting (Figure 3A 3), and the normalization table, which suggests UniProt identifiers for the kinases and substrates detected (Figure 3A 3–4). Results can be filtered by specific sections of the article (e.g., figure legends, result section, abstract, etc., see Figure 3A 1). If a user is logged in, he or she can validate individual information tuples by clicking on the check or “X” next to the annotation to agree or disagree, respectively (Figure 3B 1). The example shown in Figure 3B demonstrates the agreement on data extracted for phosphorylation of Ser-280 on Chk1 by PIM kinases. User can add additional information in the comment box, in this case, the more specific kinase PIM1 (Figure 3B). In addition, the “Add Annotation” (Figure 3B 2) allows addition of manually curated information tuples. Furthermore, the normalization table becomes editable after user logs in (Figure 3B 3–4).
Fig. 4.
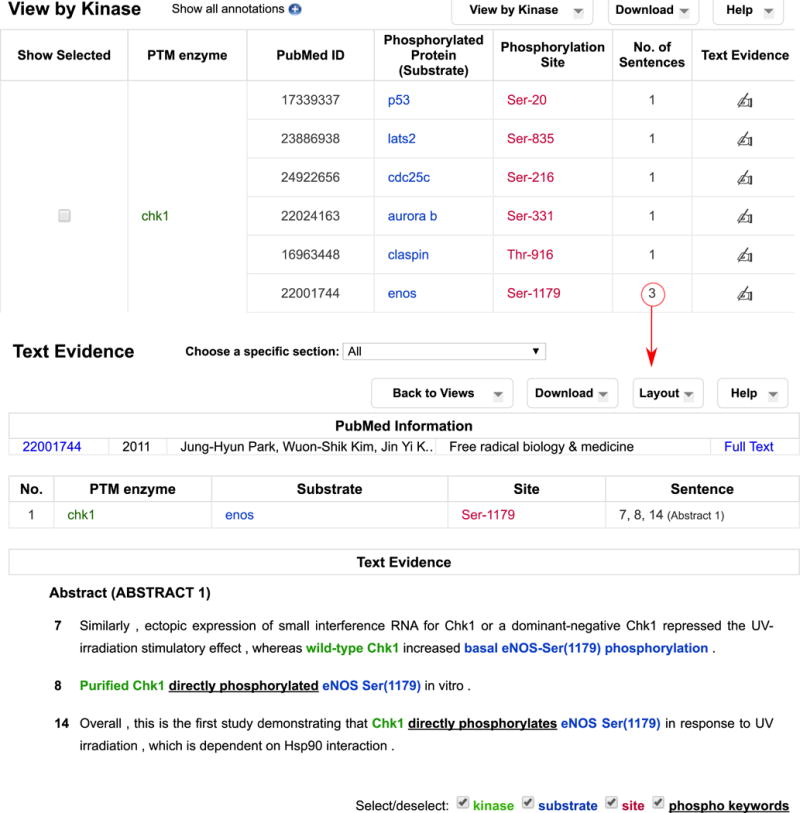
RLIMS-P “Kinase view” partial results for CHK1 search. The “No. of sentences” column provides a quick link to evidence sentences for the specific annotation
Fig. 3.

Analysis of CHK1 phosphorylation text evidence for PMID:23748345. A. RLIMS-P text evidence view. The information can be filtered by the different sections of the article when applicable (1). The table shows kinase-substrate-site data, with each row displaying a unique information tuple with the sentence number and section source (2). The text panel on the right (3) contains the evidence text with the sentence numbers. The kinase, substrate, and site are color coded. The gene normalization table (4) shows possible UniProt identifiers for the kinases and substrates mentioned in the table. B. RLIMS-P table view when editing capability is unlocked. New columns appear: “Comment” for adding notes, and “Validation” for accepting/rejecting the annotation (1). Missing annotations can be added (2). Normalization data can be validated as well (3). If needed an auto-filled UniProt search can be triggered by clicking on the search icon with “UniProt” link in the “Name” column (this icon appears after hovering over anywhere in a given row) (4). Clicking on the link leads to the corresponding UniProt entries [26]. C. Partial view of the downloaded CSV format file. The file includes PMID, substrate, kinase, site, and evidence sentence
Another way to review the RLIMS-P results is to download them in CSV format, which could be done on a single article or on the selected collective result by clicking the Download button in the right corner of the Results page (Figure 1B 5). The file can be opened in Excel (Figure 3C) where you can filter or sort the information as needed. For example, you can download all results and then filter to i) show those where CHK1 is the substrate and ii) hide rows with “Blank” information in the “PTM enzyme” and “Site” columns (Figure 3C). The file contains the evidence sentences to assist you in validating the results (see Note 3).
The results can be summarized as in Table 2. RLIMS-P found all the sites and kinases cited in the review by Goto et al. [25], and in addition, RLIMS-P found an article describing a kinase not listed in that review, namely PIM1 (bold in Table 2).
Table 2.
CHK1 phosphorylation sites with kinases validated from RLIMS-P results (species non-specific)
| Site | Kinase | PMIDs |
|---|---|---|
| Ser-280 | P90 RSK AKT PIM1 |
19406993, 15710331, 22481935, 15107605, 12062056, 22357623, 23748345 |
| Ser-286 | CDK1 CDK2 |
20798862, 19837665, 22686412, 18983824, 16629900 |
| Ser-296 | CHK1 | 20639859, 22357623, 22686412, 23068608, 20053762 |
| Ser-301 | CDK1 CDK2 |
20798862, 19837665, 22686412, 18983824, 16629900 |
| Ser-317 | ATR ATM |
21730979, 19625493, 20798862, 18723495, 20062519, 16629900, 16547171 |
| Ser-345 | ATR ATM |
21289283, 20798862, 20976184, 15107605, 15159397, 22357623, 16629900, 22357623, 23383325, 16547171, 23422000, 11687578, 20053762, 17210576, 20609246 |
3.2 How to find all substrates for a given kinase
Because CHK1 is itself a kinase, we can easily identify all substrates of CHK1 by choosing the “View by Kinase” option (Figure 4). A variety of substrates are identified here under the column “Phosphorylated Protein (Substrate).” This column can be sorted using the arrow next to the title “Phosphorylation Protein (Substrate)” so that the information regarding the same substrate is brought together. Table 3 shows the summary of substrates of CHK1 and detected phosphorylation sites. Based on the number of articles linked to the substrates, CDC25 proteins seem to be the most widely studied CHK1 substrates.
Table 3.
Substrates of CHK1 and phosphorylation sites. These are manually validated results from RLIMS-P output. n/a indicates that the phosphorylated site is not described in the RLIMS-P output
| CHK1 Substrates | Site | PMIDs |
|---|---|---|
| AURKB | Ser-331 | 22024163, 23321637 |
|
| ||
| BLM | Ser-646 | 20719863 |
|
| ||
| BRCA2 | Thr-3387 | 24627786, 18317453 |
|
| ||
| CDC10 | n/a | 24006488 |
|
| ||
| CDC2 | Tyr-15 | 24996846, 11479224 |
|
| ||
| CDC25 | Ser-287 | 9744884 |
| Ser-99 | 10198041 | |
| n/a | 9923681, 11133168, 15272308, 9774107, 10469601, 17912454 | |
|
| ||
| CDC25A | Ser-123 | 12399544, 12759351 |
| Ser-178,Thr-507 | 14559997 | |
| Ser-73 | 12110582 | |
| Ser-75 | 12759351 | |
| Ser-76 | 14681206, 20348946, 18480045, 21252624, 20798862 | |
| Thr-504 | 15272308 | |
| n/a | 12110582, 12399544, 20609246, 18414041, 24022480, 19244340, 21851590, 15272308, 19638579, 23272087, 21347609, 9278511, 18480045 | |
|
| ||
| CDC25B | Ser-230,Ser-563 | 17003105 |
| n/a | 9278511, 10713667, 20798862 | |
|
| ||
| CDC25C | Ser-216 | 14681223, 9278511, 10676638, 11027648, 24922656, 15282313, 10557092, 22623962, 23874958, 20700484 |
| n/a | 18272544, 10090724, 9278511, 11479224, 11925443, 15220526, 10681541, 22941630, 20798862, 21347609, 11278490, 10068474, 24038466 | |
|
| ||
| CDK1 | n/a | 20798862 |
|
| ||
| CDKN1A | n/a | 21791608 |
|
| ||
| CDKN1C | n/a | 21791608 |
|
| ||
| Ser-296 | 23068608, 20053762, 21289283, 24996846 | |
| CHK1 | Ser-317,Ser-345 | 21851590 |
| n/a | 14681223, 15371427, 23548269, 23593009, 19421147 | |
|
| ||
| CK1D | n/a | 23861943 |
|
| ||
| CK2 | n/a | 15225637 |
|
| ||
| CLP1 | n/a | 22918952 |
|
| ||
| CLSPN | Thr-916 | 16963448 |
|
| ||
| Ser-80 | 22792081 | |
| CRB2 | Thr-73 | 22792081 |
|
| ||
| CSNK1D | Ser-328, Ser-331,Thr-397 | 23861943 |
| Ser-328,Ser-331,Ser-370,Thr-397 | 23861943 | |
| Ser-328,Thr-329,Ser-331,Ser-361,Ser-382 | 23861943 | |
|
| ||
| E2F6 | n/a | 23954429 |
|
| ||
| ENOS3 | Ser-1179 | 22001744 |
|
| ||
| ERRFI1 | Ser-251 | 22505024 |
|
| ||
| FANCD2 | n/a | 21926477 |
|
| ||
| FANCE | Thr-346,Ser-374 | 17296736 |
|
| ||
| H2AFX | Ser-345 | 24913641 |
|
| ||
| H2AX | Thr-16 | 20639511 |
|
| ||
| KAP1 | Ser-473 | 21851590 |
|
| ||
| LATS2 | Ser-408 | 21118956 |
| Ser-835 | 23886938 | |
|
| ||
| MAD2 | n/a | 23454898 |
|
| ||
| MDMX | Ser-367 | 16511572 |
|
| ||
| p33 (ING1b) | Ser-126 | 17585055 |
|
| ||
| p50 | n/a | 22152481 |
|
| ||
| PDS1 | n/a | 11390356, 17671432 |
|
| ||
| RAD51 | n/a | 18317453 |
|
| ||
| RAD9 | n/a | 24376897 |
|
| ||
| RASSF1 | Ser-184 | 24197116 |
|
| ||
| RB1 | n/a | 17380128 |
|
| ||
| RELA | Ser-612 | 15970704 |
| Thr-505 | 17962807 | |
|
| ||
| RPA1 | n/a | 16412704 |
|
| ||
| SETMAR | n/a | 25024738 |
| Ser-495 | 22231448 | |
|
| ||
| SYK | Ser-295 | 22585575 |
|
| ||
| TAU | n/a | 23550703 |
|
| ||
| TLK1 | n/a | 12660173 |
| Ser-695 | 24376897, 12955071 | |
|
| ||
| TP53 | Ser-20 | 15467443, 17339337 |
| Ser-23 | 23152407 | |
| n/a | 15659650, 11599922, 23272087 | |
|
| ||
| TP73 | Ser-47 | 14585975 |
|
| ||
| WEE1 | Ser-549 | 11251070 |
3.3 How to find the interacting partners of phosphorylated proteins
In our examples in the following text, we address i) how phosphorylation on CHK1 affects its interaction with other proteins, ii) how PPIs are affected by proteins phosphorylated by CHK1, and iii) how phosphorylation of other proteins affect their interaction with CHK1. eFIP is capable of identifying the impact of phosphorylation, e.g., whether the phosphorylation enables the binding to a partner or inhibits the binding.
Go to the eFIP homepage (http://proteininformationresource.org/efip)
Enter the following protein names in the “Enter Protein Names and Type” query box: CHK1 OR CHEK1 OR “checkpoint kinase-1” and click Submit (Figure 2A 2). Note that the search can be restricted to retrieve results with CHK1 as a substrate, a kinase or an interactant (Figure 2A 3).
Select “Substrate View”. After submission, the result page (Figure 2B) displays the data in a summary view (as a list of entities detected that are grouped by PMID). Similar to RLIMS-P, by selecting “Substrate view” the information can be grouped by phosphorylated substrate, so that we can check the PPIs for phosphorylated CHK1.
-
Select “Kinase view” to investigate phosphorylation-dependent PPIs for CHK1 substrates. Review results for CHK1 as kinase, and check the information for interactant with its associated text evidence. Figure 5A depicts the text evidence for PMID: 14559997. In this particular case, the phosphorylation of CDC25A on Ser-178 and Thr-507 by CHK1 promotes the binding to 14-3-3 proteins. In addition to highlighting kinases, substrates, and sites using the same color scheme as RLIMS-P, interactants are highlighted in orange.
You can also check for information about CHK1 as interactant, using the “Interactant view.”
Download the result table. Similar to RLIMS-P, eFIP results can be downloaded in CSV format by using the “Download Table” link in the upper left corner of the results table (Figure 2B). Table 4 provides a summary of results where CHK1 participated in a phosphorylation-dependent PPI either as the phosphorylated substrate or as the interactant. Table 5 provides a summary of phosphorylation-dependent PPIs where CHK1 acts as the kinase.
Fig. 5.
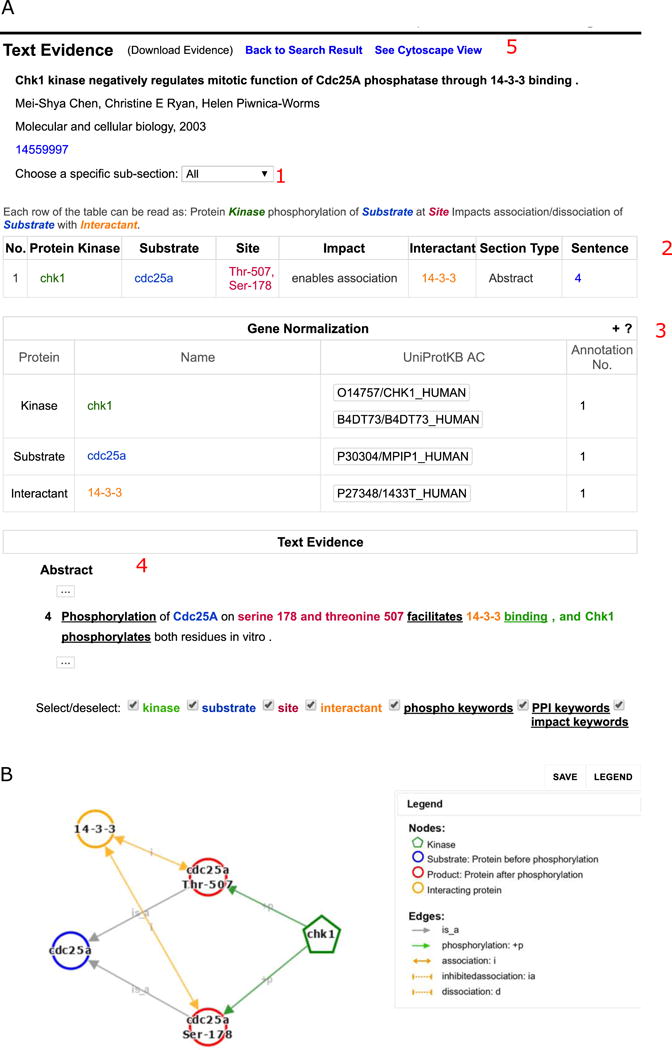
Analysis of CHK1 eFIP text evidence for PMID:14559997. A. The results can be filtered by section of the article (1). The table with extracted results shows the substrate, kinase (if known), phosphorylation site, the impact (inhibit, enhance, unknown association/dissociation), the interactant, the section of the article where the information comes from and the sentence number in that section (2). The gene normalization table (3) shows possible UniProt identifiers for the kinase, substrate, and interactant mentioned in the table. The text evidence panel (4) contains the evidence sentences with section title and sentence numbers. B. The Cytoscape representation of the relations extracted in the article
Table 4.
Summary of eFIP results for phosphorylation-dependent PPI involving CHK1
| Substrate | PMID | Kinase | Site | Impact | PPI | Interactant |
|---|---|---|---|---|---|---|
| 12676962 | Ser-345 | enables | association | 14-3-3 | ||
|
| ||||||
| 12415000 | Ser-345 | enables | association | RAD24 (14-3-3 homolog) | ||
|
| ||||||
| 15585577 | enables | association | RAD25 (14-3-3 homolog) | |||
| CHK1 | 20639859 | CHK1 | Ser-296 | enables | association | CDC25A |
| 12676962 | Ser-345 | enables | association | chromatin | ||
|
| ||||||
| 16360315 | enables | dissociation | chromatin | |||
|
| ||||||
| 23593009 | Thr-125 | inhibits | association | RAD9 | ||
|
| ||||||
| 23593009 | Thr-143 | enables | association | RAD9 | ||
|
| ||||||
| CRB2 | 22792081 | increases | association | CHK1 | ||
|
| ||||||
| 15707391 | Thr-916, Ser-945 | enables | association | CHK1 | ||
| CLSPN | 22792081 | increases | association | CHK1 | ||
| 12766152 | unknown | association | CHK1 | |||
| 12545175 | unknown | association | CHK1 | |||
Table 5.
Summary of eFIP results for CHK1 substrates with phosphorylation-dependent PPIs
| Substrate | PMID | Kinase | Site | Impact | PPI | Interactant |
|---|---|---|---|---|---|---|
| BRCA2 | 18317453 | CHK1 | enables | association | RAD51 | |
|
| ||||||
| CDC25 | 23166842 | CHK1 | increases | association | SCFBETATr CP |
|
| 22806395 | enables | association | RAD24 (14-3-3 homolog) | |||
|
| ||||||
| CDC25A | 14559997 | CHK1 | Thr-507,Ser-178 | enables | association | 14-3-3 |
| 15272308 | Thr-504 | enables | association | 14-3-3 | ||
| 15272308 | Thr-504 | inhibits | association | Cdk1-cyclin A | ||
| Cdk1-cyclin B | ||||||
| Cdk2-cyclin E | ||||||
|
| ||||||
| CDC25B | 20798862 | CHK1 | Ser-309, Ser-323 | enables | association | 14-3-3 |
|
| ||||||
| CDC25C | 23874958 | CHK1 | Ser-216 | increases | association | 14-3-3beta |
| 20798862 | Ser-216 | enables | association | 14-3-3 | ||
|
| ||||||
| RB1 | 17380128 | CHK1 | Ser-612 | enables | association | E2F1 |
|
| ||||||
| RAD51 | 18317453 | CHK1 | enables | association | BRCA2 | |
3.4 Visualization of phosphorylation and interaction events in Cytoscape
eFIP also supports visual exploration of phosphorylation interaction networks using Cytoscape [28], which depicts in one graph a network of kinase-substrate relations, as well as PPI relations, including both the enhancement and inhibition of an interaction. Therefore, the phosphorylation-dependent interactions described in Subheading 3.3 can be displayed in Cytoscape.
Cytoscape view from text mining PMID evidence page
-
1
Go to the eFIP homepage.
-
2
Search for PMID:14559997. Enter the PMID in the search box (Figure 2A 4) and submit.
-
3
Open the “Text Evidence” page. Click on the “hand” icon in the last column (Figure 2B 4) to see the text evidence (Figure 5A).
-
4
Click on “See Cytoscape View”. The link to the Cytoscape view is at the top right of the evidence table (Figure 5A 5). The Cytoscape view for this example is shown in Figure 5B. CHK1 phosphorylates CDC25A at two residues. The phosphorylated residues enable interaction with 14-3-3.
Cytoscape view for multiple articles (see Note 4)
-
5
Go to the eFIP homepage.
-
6
Conduct query. For this case, enter the PMIDs 12676962, 17380128, and 20639859 separated by commas in the search box (Figure 2A 4) and then submit.
-
7
Open “Cytoscape View.” The link to Cytoscape is on the top left of the result table (Figure 2B 5). The Cytoscape view for this example is shown in Figure 6.
Acknowledgments
This work was supported by grants from the National Institutes of Health: R01GM080646 and U01HG008390.
Footnotes
The “Login” link is located in the upper-right corner of the webpage. When you click it, it will ask you to either enter your credentials or sign up. Select sign up and complete the information needed. After the registration, an automatic email will be sent out to explain details on how to log into RLIMS-P or eFIP.
For our CHK1 query, we obtained 1266 articles with potential protein phosphorylation mentions, with 278 kinase mentions, 854 substrate mentions, and 245 site mentions as of 05/27/2016. Note that if you query PubMed instead of RLIMS-P with the same query, it retrieves many more articles, 2781 as of 05/27/2016, many of which are not relevant to CHK1 phosphorylation at all. In PubMed,finding the subset where CHK1 is phosphorylated could then only be achieved by manual inspection, whereas in RLIMS-P, selecting the appropriate view will enable quick access to the most relevant set.
The text mining results should not be assumed to be completely correct. There is a possibility of encountering false positive results or of missing relevant data. The different tools provide their own metrics of performance, and it is important to be aware of them when using the tools. In addition, one should consider reviewing the substrate names thoroughly, as textual variants of CHK1 presently appear as separate substrates. We are currently working on improving the consistency and grouping of the substrate and kinase names.
The Cytoscape view provides an overview of the text mining results in graphical format. However, if the output includes multiple articles the number of nodes and edges may become overwhelming. The current version of Cytoscape used in eFIP does not allow hiding or displaying selected nodes. To see only a selected subset, one could either retrieve data for selected PMIDs, or alternatively, download a desktop version of Cytoscape to read the saved XGMML-beta (Cytoscape compatible) file (Figure 6 2).
References
- 1.Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–520. doi: 10.1093/nar/gku1267. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steelman LS, Martelli AM, Cocco L, Libra M, Nicoletti F, Abrams SL, McCubrey JA. The therapeutic potential of mTOR inhibitors in breast cancer. Br J Clin Pharmacol. 2016 doi: 10.1111/bcp.12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamaoka K. Janus kinase inhibitors for rheumatoid arthritis. Curr Opin Chem Biol. 2016;32:29–33. doi: 10.1016/j.cbpa.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Ma H. Protein kinase profiling assays: a technology review. Drug Discov Today Technol. 2015;18:1–8. doi: 10.1016/j.ddtec.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 5.de Oliveira PS, Ferraz FA, Pena DA, Pramio DT, Morais FA, Schechtman D. Revisiting protein kinase-substrate interactions: toward therapeutic development. Sci Signal. 2016;9(420):re3. doi: 10.1126/scisignal.aad4016. [DOI] [PubMed] [Google Scholar]
- 6.Ross KE, Arighi CN, Ren J, Huang H, Wu CH. Construction of protein phosphorylation networks by data mining, text mining and ontology integration: analysis of the spindle checkpoint. Database (Oxford) 2013;2013:bat038. doi: 10.1093/database/bat038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleuren WW, Alkema W. Application of text mining in the biomedical domain. Methods. 2015;74:97–106. doi: 10.1016/j.ymeth.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 8.Zhu F, Patumcharoenpol P, Zhang C, Yang Y, Chan J, Meechai A, Vongsangnak W, Shen B. Biomedical text mining and its applications in cancer research. J Biomed Inform. 2013;46(2):200–211. doi: 10.1016/j.jbi.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Huang CC, Lu Z. Community challenges in biomedical text mining over 10 years: success, failure and the future. Brief Bioinform. 2016;17(1):132–144. doi: 10.1093/bib/bbv024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Z-Z, Mani I, Hermoso V, Liu H, Wu CH. iProLINK: an integrated protein resource for literature mining. Comput Biol and Chem. 2004;28(5–6):409–416. doi: 10.1016/j.compbiolchem.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Peng Y, Torii M, Wu CH, Vijay-Shanker K. A generalizable NLP framework for fast development of pattern-based biomedical relation extraction systems. BMC Bioinf. 2014;15:285. doi: 10.1186/1471-2105-15-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng Y, Tudor C, Torii M, Wu C, Vijay-Shanker K. iSimp: A Sentence Simplification System for Biomedical Text. International Conference on Bioinformatics and Biomedicine (BIBM2012) 2012:211–216. [Google Scholar]
- 13.Ding R, Arighi CN, Lee JY, Wu CH, Vijay-Shanker K. pGenN, a gene normalization tool for plant genes and proteins in scientific literature. PLoS One. 2015;10(8):e0135305. doi: 10.1371/journal.pone.0135305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tudor CO, Ross KE, Li G, Vijay-Shanker K, Wu CH, Arighi CN. Construction of phosphorylation interaction networks by text mining of full-length articles using the eFIP system. Database. 2015;2015:bav020. doi: 10.1093/database/bav020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tudor CO, Arighi CN, Wang Q, Wu CH, Vijay-Shanker K. The eFIP system for text mining of protein interaction networks of phosphorylated proteins. Database. 2012;2012:bas044. doi: 10.1093/database/bas044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tudor CO, Schmidt CJ, Vijay-Shanker K. eGIFT: mining gene information from the literature. BMC Bioinf. 2010;11:418. doi: 10.1186/1471-2105-11-418. doi:1471-2105-11-418 [pii]10.1186/1471-2105-11-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torii M, Arighi CN, Li G, Wang Q, Wu CH, Vijay-Shanker K. RLIMS-P 2.0: A Generalizable Rule-Based Information Extraction System for Literature Mining of Protein Phosphorylation Information. IEEE/ACM Trans Comput Biol Bioinform. 2015;12(1):17–29. doi: 10.1109/TCBB.2014.2372765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torii M, Li G, Li Z, Oughtred R, Diella F, Celen I, Arighi CN, Huang H, Vijay-Shanker K, Wu CH. RLIMS-P: an online text-mining tool for literature-based extraction of protein phosphorylation information. Database. 2014;2014:bau081. doi: 10.1093/database/bau081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li G, Ross KE, Arighi CN, Peng Y, Wu CH, Vijay-Shanker K. miRTex: A text mining system for miRNA-Gene relation extraction. PLoS Comput Biol. 2015;11(9) doi: 10.1371/journal.pcbi.1004391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu N, Lao Y, Zhang Y, Gillespie DA. Akt: a double-edged sword in cell proliferation and genome stability. J Oncol. 2012;2012:951724. doi: 10.1155/2012/951724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei CH, Kao HY, Lu Z. GNormPlus: An Integrative Approach for Tagging Genes, Gene Families, and Protein Domains. Biomed Res Int. 2015;918710(10):25. doi: 10.1155/2015/918710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Natale DA, Arighi CN, Blake JA, Bult CJ, Christie KR, Cowart J, D’Eustachio P, Diehl AD, Drabkin HJ, Helfer O, Huang H, Masci AM, Ren J, Roberts NV, Ross K, Ruttenberg A, Shamovsky V, Smith B, Yerramalla MS, Zhang J, AlJanahi A, Celen I, Gan C, Lv M, Schuster-Lezell E, Wu CH. Protein Ontology: a controlled structured network of protein entities. Nucleic Acids Res. 2014;42:21. doi: 10.1093/nar/gkt1173. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, Elledge SJ. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science (New York, NY) 1997;277(5331):1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 24.McNeely S, Beckmann R, Bence Lin AK. CHEK again: revisiting the development of CHK1 inhibitors for cancer therapy. Pharmacol Ther. 2014;142(1):1–10. doi: 10.1016/j.pharmthera.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 25.Goto H, Kasahara K, Inagaki M. Novel insights into Chk1 regulation by phosphorylation. Cell Struct Funct. 2015;40(1):43–50. doi: 10.1247/csf.14017. [DOI] [PubMed] [Google Scholar]
- 26.UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–212. doi: 10.1093/nar/gku989. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coordinators NR. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016;44(D1):D7–D19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N. Integrated models of biomolecular interaction networks. Genome Res. 13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]


