Abstract
Chronic hepadnavirus infections cause liver damage with ongoing death and regeneration of hepatocytes. In the present study we set out to quantify the extent of liver turnover by measuring the clonal proliferation of hepatocytes by using integrated viral DNA as a genetic marker for individual hepatocyte lineages. Liver tissue from woodchucks with chronic woodchuck hepatitis virus (WHV) infection was assayed for randomly integrated viral DNA by using inverse PCR. Serial endpoint dilution of viral–cell junction fragments into 96-well plates, followed by nested PCR and DNA sequencing, was used to determine the copy number of specific viral cell junctions as a measure of the clonal distribution of infected cell subpopulations. The results indicated that the livers contained a minimum of 100,000 clones of >1,000 cells containing integrated DNA, representing at least 0.2% of the hepatocyte population of the liver. Because cells with integrated WHV DNA comprised only 1–2% of total liver cells, it is likely that the total number of clones far exceeds this estimate, with as much as one-half of the liver derived from high copy clones of >1,000 cells. It may be inferred that these clones have a strong selective growth or survival advantage. The results provide evidence for a large amount of hepatocyte proliferation and selection having occurred during the period of chronic WHV infection (≈1.5 years) in these animals.
Keywords: endpoint dilution, inverse PCR, hepatocyte clones
Chronic hepadnavirus infection is associated with chronic inflammatory disease of the liver that may progress to cirrhosis and hepatocellular carcinoma (HCC) (1). Hepatocytes, the target of infection, are generally long lived with a lifetime thought to exceed 6 months and a liver turnover rate possibly in excess of a year (2–6). Histologic evidence reveals an increase in hepatocyte destruction during chronic infections (7) with a shortening of hepatocyte lifetime. For example, in woodchucks, chronic woodchuck hepatitis virus (WHV) infection is associated with, on average, an ≈10-fold increase in the proliferating cell nuclear antigen staining index of hepatocytes, involving ≈0.5–1% of the liver cell population (8). We estimate that this level of turnover in chronically infected woodchucks might represent 6–12 turnovers of the liver cell population per year. Because surviving hepatocytes can divide to replace dying cells, an elevated rate of random cell death caused by the antiviral immune response should lead to changes in the hepatocyte population, with some hepatocyte lineages dying out and others expanding on a random basis (9). In addition, such an environment provides the opportunity for the expansion of hepatocyte variants with a selective advantage either in growth or survival.
During the course of an infection, hepadnavirus DNA is integrated into host DNA [e.g., during repair of double-stranded breaks by nonhomologous end joining (10)]. In chronically WHV-infected woodchucks 2–3 years of age, integrations are stable and present in at least 1–5% of the hepatocyte population (11). Moreover, in a study of recovery from transient WHV infections, we showed that the unique viral–cell junctions of integrated DNA could be used to track clonal proliferation of hepatocytes (9).
In an early study of clonal proliferation, performed by using Southern blot hybridization, cirrhotic nodules in the liver of hepatitis B virus carriers contained clones of at least 100,000 cells (12, 13). Large dysplastic foci have also been reported to be clonal (14). What is less clear is the extent of clonal proliferation in the livers of WHV and hepatitis B virus carriers with liver disease without macronodular cirrhosis. We therefore used inverse, endpoint dilution PCR to detect and quantify clonal proliferation of cells with integrated WHV DNA in woodchucks in which viral replication, and presumably new integrations, had been suppressed for up to a year by antiviral therapy. Populations of clonally expanded cells were detected ranging up to 26,000 cells in size. The entire liver was estimated to contain a minimum of 100,000 clones of >1,000 cells containing integrated DNA. We estimate that 1–2% of liver cells contain integrated viral DNA and almost half of these cells were present as high copy cell clones. If the population of cells with integrated DNA reflects the characteristics of hepatocytes in general, then chronic hepatitis produces a remarkable degree of cell proliferation and clonal selection in the infected liver.
Materials and Methods
Tissue Specimens. The collection of liver biopsies and autopsy tissue from woodchucks infected with WHV at 3 days of age was described in table 2 of ref. 15. Liver biopsies were initially collected at 14–16 months of age. These animals were then subjected to antiviral therapy with clevudine starting 3 weeks later and, in some cases, gene therapy with a single dose of adenovirus vectors expressing β-gal, or a combination of TNF-α and IFN-γ, administered 5 weeks after the initiation of clevudine therapy. Clevudine was administered for a total of 38 weeks, and, during that time, levels of WHV covalently closed circular DNA (cccDNA) dropped ≈100-fold, facilitating the detection of integrated DNA. Clevudine has been shown to be nontoxic in animals and nonmutagenic in standard assays (Bukwang Pharm, Seoul). In addition, clevudine is not incorporated into cellular DNA (16), and its triphosphate is not a substrate for human DNA polymerases α, β, γ, or δ (17). Therefore, clevudine is not expected to influence cell proliferation in the liver. Woodchucks receiving clevudine alone included wc370 and wc372, whereas wc359 and wc371 received the β-gal expression vector, and wc366 received a combination of TNF-α and IFN-γ expression vectors. HCC was detected in all five woodchucks at autopsy. Autopsies were performed 48, 52, 33, 34, or 52 weeks after initiation of clevudine therapy for woodchucks wc359, wc366, wc370, wc371, and wc372, respectively. At the time of the autopsies, the woodchucks ranged in age from 22 to 28 months. Samples of liver and HCC collected at the autopsy were stored frozen or processed for immunohistochemistry and in situ hybridization as described in ref. 15.
DNA Extraction. DNA was initially extracted from 10- to 150-mg wet weight samples of liver and HCC collected at autopsy. The samples were disrupted in 2.5 ml of 10 mM Tris·HCl, pH 7.6/1 mM EDTA/0.1% Triton X-100 by using a Dounce homogenizer, and nuclei were collected by centrifugation for 1 min at 11,000 × g in a microcentrifuge. The pellet was suspended in 0.5 ml of 50 mM NaCl/25 mM Tris·HCl, pH 7.6/5 mM EDTA containing 0.1% SDS and 1 mg of proteinase K per ml, incubated for 2 h at 55°C, and then extracted with a 1:1 mixture of phenol and chloroform. Total nucleic acids were collected by ethanol precipitation. Analyses of the liver samples were summarized in Table 1.
Table 1. Repeated virus-cell junctions in chronically WHV-infected woodchuck livers.
| Fraction of total DNA analyzed
|
Total viral—cell junctions
|
No. of distinct viral—cell junctions (predicted clone size, no. of cells)*
|
Percent as multicopy clones
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Woodchuck | 1 | 2 | 3 | 4 | 5 | 6 | 8 | 9 | |||
| 359 | 0.051 | 58 | 46 | 4 (39) | 1 (78) | 21 | |||||
| 366 | 0.003 | 89 | 44 | 9 (663) | 2 (1,658) | 1 (2,653) | 1 (2,985) | 51 | |||
| 370 | 0.008 | 33 | 28 | 1 (242) | 1 (364) | 15 | |||||
| 371 | 0.049 | 75 | 45 | 10 (41) | 1 (82) | 1 (84) | 40 | ||||
| 372 | 0.009 | 54 | 37 | 2 (213) | 1 (532) | 1 (851) | 20 | ||||
The numbers below the line indicate the number of times a particular integration site was detected. For instance, for woodchuck 366, there were nine sites that were detected twice and were predicted to have a clone size of 663 cells.
For the remaining studies, DNA was isolated from 0.5- to 3-mg wet weight samples of liver. The entire sample was suspended in proteinase K/SDS digestion buffer, and the nuclear pelleting step was eliminated. Subsequent steps were essentially as described above. Alternatively, some samples of liver were extracted by using the Promega Wizard Genomic DNA Purification Kit according to the manufacturer's instructions.
Cellular DNA concentrations were determined by using realtime PCR to quantify the single-copy woodchuck hcr gene (18). Cellular DNAs extracted from 103 to 105 cells were assayed in 20-μl reactions of SYBR Green Supermix (Bio-Rad) with primers extending from nucleotides 3820–3844 (5′-CTC TGG TCA AAT GTT AAC CAC TCA G-3′) and nucleotides 4034–4012 (5′-CTC TGT GAA TAA ACC CTT CTG GA-3′) of the hcr locus (GenBank entry X13234). The reactions were subjected to thermocycling in a Bio-Rad iCycler equipped with an optical detector by using the conditions 95°C for 3 min, 40 cycles of 95°C for 10 sec, and 60°C for 30 sec. Known amounts of a purified PCR product of the hcr gene were used to generate a standard curve.
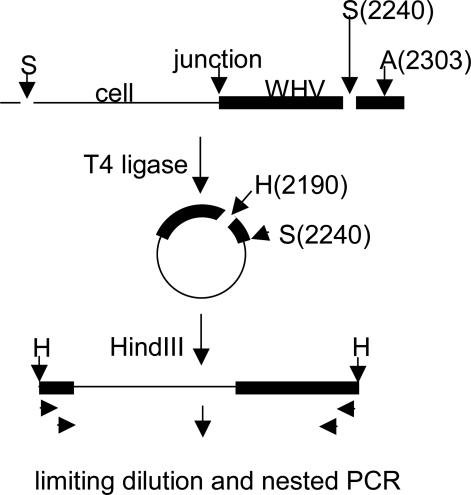
Inverse PCR. Linear viral DNA molecules created by in situ priming (19) appear to preferentially integrate as compared with relaxed circular viral DNA (20, 21). In the plus-strand orientation, the left end of the terminally redundant linear DNA extends to the 5′ end of the pregenomic RNA [about nucleotide 1935, numbering according to Galibert et al. (22)] and the right side to the 5′ end of the minus strand DNA, which maps ≈9 nucleotides downstream. In this study, inverse PCR (Fig. 1) was carried out for detection of viral–cell junctions at or near the left end of in situ primed DNA as described in ref. 9. Briefly, liver DNA was digested with SacI, which cleaves in the core gene at nucleotide 2240 in the viral genome and at upstream sites in cell DNA, and SacI was then inactivated by heating at 80°C for 15 min. The DNA was then incubated with T4 DNA ligase (400 units/ml) for 2 h at room temperature. The ligase was inactivated for 15 min at 80°C, and the circularized DNA was digested with HindIII, which cleaves at nucleotide 2190. The efficiency of the inversion reaction was estimated to be as high as 80% when tested by inversion of a portion of the hcr gene bounded by SacI sites 892 nucleotides apart. Digestion with AflII, which cleaves at nucleotide 2303, was carried out by inclusion of AflII (100 units/ml) in the first PCR mix and an initial incubation for 30 min at 37°C. Although full-length WHV cccDNA is too large to amplify with our reaction conditions, AflII cleavage prevents PCR amplification of defective cccDNA molecules with large deletions, except those lacking the AflII site, which accounted for ≈15% of the total products of inverse PCR.
Fig. 1.
Inverse PCR assay for detection of viral–cell junctions. DNA fragments containing viral–cell junctions were released by SacI digestion (S), ligated to produce circular DNAs, and converted to linear DNAs by digestion with HindIII (H). These fragments were diluted to microtiter plates, digested with AflII (A), and amplified by nested PCR by using primers specific to viral sequences at the left and right ends (arrows).
Unless noted, DNA samples were assayed in 96-well microtiter plates at a dilution at which each PCR would produce on average one or fewer amplicons. Aliquots of the larger DNA samples extracted from the 10- to 150-mg samples of liver and HCC were distributed in appropriate amounts, as determined by an initial titration, to each well of a 96-well plate. One-quarter to 1/14th of the DNA samples extracted from the 0.5- to 3-mg liver samples were assayed in 3-fold serial dilutions, 12 wells per dilution. Nested PCR was carried out by using primer sets S1 (primers WHV2185–2167 and WHV2195–2216) and S2 (primers WHV2165–2145 and WHV2217–2245), as described in ref. 9. PCR products were subjected to electrophoresis in 1.3% agarose gels. Bands were cut from the gels, recovered by using the QIAEX II (Qiagen, Valencia, CA) agarose extraction kit, and sequenced for detection of the viral–cell junction by using WHV2165–2145 as the primer (9). Viral–cell junctions were identified by using the wisconsin package 10.2 (Genetics Computer Group, Madison, WI) and cellular sequences present at each junction were aligned by using sequencher (Gene Code, Ann Arbor, MI). The size of each clone defined by a repeated viral cell junction was taken to be the number of those viral cell junctions at the highest dilution times the dilution factor.
Immunohistochemistry and in Situ Hybridization. Sections of ethanol/acetic acid fixed liver tissue were subjected to immunohistochemistry for detection of WHV core antigen and in situ hybridization for detection of WHV nucleic acids as described in refs. 9, 15, and 23.
Results
To assay for clonally expanded cells with integrated WHV DNA, we first examined DNA extracted from pieces of liver containing >1.5 × 106 cells (10–150 mg wet weight). After inversion (Fig. 1), aliquots were diluted and distributed for nested PCR to 96-well microtiter plates so that on average each well produced ≈1 or fewer amplicons. Sequencing of a subset of the reaction products revealed that the majority contained viral–cell junctions, with the remainder apparently arising from defective cccDNA molecules lacking the AflII site at nucleotide 2303. Despite the fact that <10% of each sample was analyzed, and in some cases <1%, every sample contained several viral–cell junctions that were represented more than once (Table 1), indicating clonal proliferation of cells containing these integration sites. After sequencing, blast searches were done to determine whether expansion was associated with integration near a host oncogene.
Flanking DNA in one of the cell clones in wc366 [predicted clone size of 1,658 cells (Table 1)] was homologous to nucleotides 421 to 244 in the win locus (GenBank entry AY173019). WHV DNA integration in the win locus is associated with transcriptional activation of the distal Nmyc2 gene (24), which is not normally expressed in the liver. Studies from Buendia and colleagues (refs. 24 and 25; reviewed in ref. 26) have revealed transcriptional activation of Nmyc2 in ≈80% of HCCs arising in woodchucks chronically infected with WHV, raising the possibility that this clone contained transformed cells. No other viral–cell junctions were mapped to win. Among 309 sequences from the experiment in Table 1, only 17 other distinct integrations occurred in regions with homology to known genes, including one in a cell clone with a size of 2,985 cells with integration at a site homologous to the gene for α-2-macroglobulin (wc366), one in a clone with a size of 41 cells with integration at a site homologous to TRPV3, a member of the vanilloid receptor family (wc371), two in clones with a size of 663 and 41 cells, respectively, with integrations at sites homologous to apolipoprotein B (wc366 and wc371, respectively), and one in a clone with a size of 41 cells with integration at a site homologous to ribosomal DNA (wc371). Integration into ribosomal DNA was also found with wc359, wc366, and wc372. In subsequent experiments, integration in this region was also detected in wc370. Four independent integrations in ribosomal DNA were downstream of the SacI site at nucleotide 10603 and five were upstream of the SacI site at nucleotide 12016 (GenBank entry BK000954) of this repeated gene [≈200 copies in the mouse (27)]. No viral–cell junction fragments were found in the HCCs in these five woodchucks by using the same inverse PCRs (data not shown), indicating that clonally proliferating cells were not simply metastases of existing tumors. Thus, with one exception, none of the integration sites were found to be near known oncogenes. Because a SacI site is not nearby, our assay would not have detected integration in the 3′ UTR of Nmyc-2, which with win is the most common site of integration in woodchuck HCC (reviewed in ref. 26).
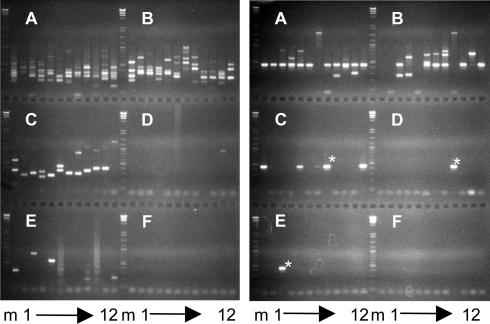
Experiments were next carried out to differentially detect large cell clones in 35 liver fragments (≈0.5–3 mg wet weight) from wc370, wc371, and wc372, containing from 6 × 104 to 1.4 × 106 cells (see Tables 3–5, which are published as supporting information on the PNAS web site). Samples were extracted, and each was assayed by serial dilution inverse PCR. Repeated or possibly repeated bands were selected from each for sequencing. A simple pattern of PCR products with a clearly repeating fragment representing a clone of ≈4,000 cells is shown in Fig. 2 Right (wc371.1a). A more complex pattern, with no highly repeated bands, is shown in Fig. 1 Right (wc371.5a). Sequencing of bands that appeared to be repeated revealed four clones of ≈120 cells.
Fig. 2.
Analysis of liver fragments for clonal expansion of cells by serial dilution. DNA was extracted from pieces of liver containing 8.7 × 104 cells (wc371 liver fragment 1a, Right) and 1.1 × 105 cells (wc371 liver fragment 5a, Left) based on DNA recovery and assuming a woodchuck genome size of 2.5 pg. Following SacI cleavage, ligation, and HindIII digestion, aliquots were distributed to microtiter trays for nested PCR, followed by gel electrophoresis. Lanes A1 to A12 were from reactions containing in combination 2.5% (Right) or 22% (Left) of the total DNA, and subsequent groups of 12 contained serial 3-fold dilutions of the DNA. The marker (m) at the left and middle of each panel is HindIII cut bacteriophage λ-DNA mixed with a 1-kbp DNA ladder. Three occurrences of common band in the wc371 liver fragment 1a are marked with asterisks.
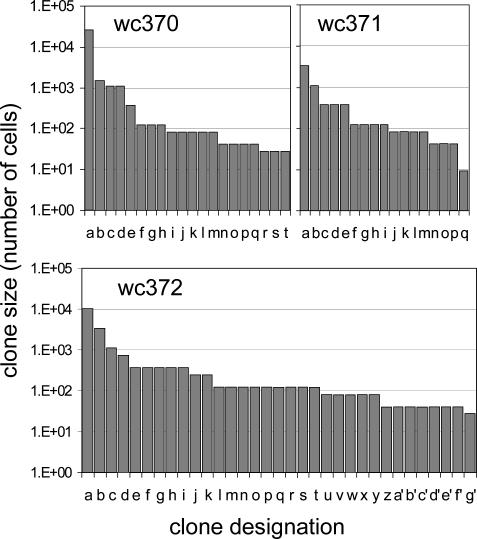
A summary of the analyses of the liver fragments is presented in Fig. 3 and Table 2. Fragments from wc370 contained four clones of ≥1,000 cells in a sample of 2.5 × 106 cells, and these clones in combination represented 1.2% of the liver cell population analyzed. wc371 had two clones of >1,000 cells in a sample of 5.0 × 105 cells that represented 0.9% of the population, and wc372 had three clones of 1,000 cells or greater in a sample of 3.0 × 106 cells that represented 0.5% of the liver cell population. Thus, high-copy cell clones that could be detected by using integrated WHV DNA represented a significant population of the liver. blast searches did not reveal integration into known oncogenes for any of the multicopy clones in Fig. 3. This finding was also true for a total of 555 sequenced viral–cell junctions detected only once in this experiment and in that described in Table 1.
Fig. 3.
Size distribution of cell clones detected by end point dilution inverse PCR. Clones exceeding 27 cells in size were detected by end point dilution inverse PCR, as illustrated in Fig. 2. The results obtained with 15 liver fragments from wc370, 5 from wc371, and 15 from wc372, containing in total 2.5 × 106, 5.0 × 105, and 3.0 × 106 cells, respectively, are summarized in the three panels.
Table 2. Summary of analyses of small liver fragments.
| No. of viral-cell junctions (cell clones) with size, cells
|
Viral-cell junctions per cell
|
||||
|---|---|---|---|---|---|
| Woodchuck | <27* | 27-1,000 | >1,000 | Cells analyzed | |
| 370 | 22,284 | 1,381 (16) | 29,892 (4) | 2.5E + 06 | 2.2E - 02 |
| 371 | 2,121 | 2,039 (15) | 4,375 (2) | 5.0E + 05 | 1.7E - 02 |
| 372 | 13,455 | 4,857 (30) | 14,217 (3) | 3.0E + 06 | 1.1E - 02 |
Summarized data are from Tables 3-5.
PCR products detected only once. Data are corrected for frequency of products from cccDNA.
Previous studies have reported the presence of foci of altered hepatocytes (28–31), from which HCC is thought to emerge in WHV-infected woodchucks and hepatitis B virus-infected humans. These foci are distinguished from normal hepatocytes by distinct or subtle morphological changes in hematoxylin/eosin staining or in the periodic acid/Schiff reagent reaction (29, 30). In addition, variations in viral core antigen staining have been reported in chronically WHV-infected woodchuck (8, 32) and hepatitis B virus-infected human livers (33–43), and some but not all clusters of cells with reduced levels of antigen staining have an obviously altered appearance. Whether such foci of variation in antigen staining or appearance represent clonal outgrowths of hepatocyte variants or local effects is not known. To determine whether the woodchuck livers that we analyzed showed these previously reported focal heterogeneities, we performed analysis of liver tissue by WHV core antigen staining and detection of WHV nucleic acids by in situ hybridization.
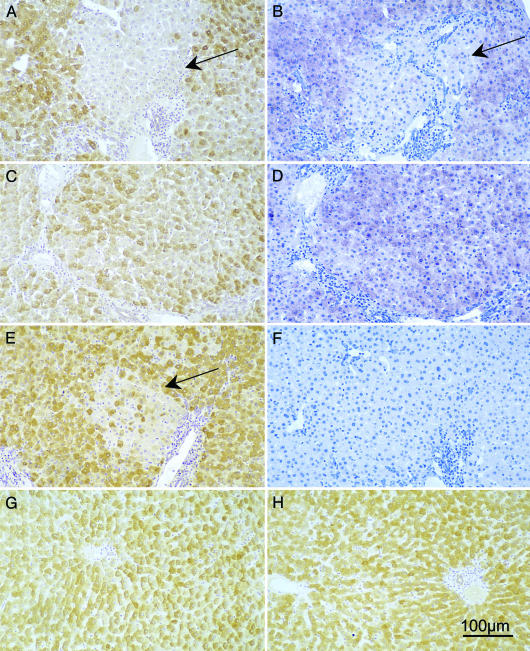
Liver biopsies taken before clevudine therapy, at ≈15 months of age, were used for this analysis. As illustrated in Fig. 4, distinct foci, small clusters, and scattered hepatocytes with low intensity or undetectable WHV core antigen staining and nucleic acid signals could be seen to account for as many as one-half of the hepatocytes in wc359 and wc370 (Fig. 4 A–E), infected at 3 days of age, as compared with the more uniform viral signals at the peak of a transient infection of a woodchuck that was ≈18 months old at the time of infection (Fig. 4 G and H). A similar pattern was seen in the biopsies from wc366, wc371, and wc372. This heterogeneity is consistent with the outgrowth of phenotypically distinct cell clones already one year before the time at which samples were taken for analysis of integrated DNA.
Fig. 4.
Analysis of liver tissue for variability in levels of WHV infection. Consecutive sections of liver biopsies collected from wc359 (A, B, and F) and wc370 (C–E) at ≈15 months of age were analyzed for the distribution of hepatocytes expressing WHV core antigen (A, C, and E) and nucleic acids (B and D). F shows a plasmid (negative) control for hybridization specificity. A and B show consecutive liver sections from wc359 containing a patch of cells with low or undetectable WHV core antigen and nucleic acids (arrows). C and D similarly show consecutive sections of liver wc370 with variability in levels of WHV core antigen and nucleic acids across the lobule. E shows an area of wc370 with low levels of WHV core antigen. For comparison, G and H show WHV core antigen detection in liver tissue collected 7 and 11 weeks after infection during a transient infection (wc38; ref. 23) that was fully resolved by 21 weeks after infection. Magnification: ×120. (Scale bar, 100 μm.)
Discussion
In this study we attempted to obtain information about cumulative liver cell growth and turnover during chronic hepatitis in WHV-infected woodchucks. Our strategy was to examine the amount of hepatocyte proliferation as reflected by the clonal expansion of cells containing integrated viral DNA, assuming that each viral–cell junction corresponded to an individual cell. This strategy was limited by the fact that our assay detected integrated DNA in only 1–2% of liver cells (Table 2, column 6), whereas cells with no integrated DNA or other integrated DNAs would not be detected, depending on specific features of the integration site. By enumerating high copy viral–cell junctions, we observed a broad distribution of hepatocyte clone sizes between 27 and 26,000 cells, indicating a high amount of proliferation of some hepatocytes. High copy clones (>1,000 cells) accounted for almost one-half of the cells containing integrated viral DNA (Table 2, columns 2–4). Moreover, if the presence of integrated DNA is simply a genetic marker of infected hepatocyte lineages rather than a cause of clonal expansion, then almost half of the entire liver might consist of high-copy cell clones. Some interpretations of the clonal distribution we observed are considered below.
Clonal Expansion by Random Killing of Hepatocytes Accompanied by Random Division of Other Hepatocytes. Some clonal expansion of hepatocytes during liver turnover is to be expected, even if those cell clones that expand are entirely normal. During hepatocyte killing that accompanies chronic hepatitis, some hepatocyte lineages would be eliminated and replaced by expansion of other lineages. Because expanding cell clones are also subject to killing, expansion would be slow and the population of any one liver cell clone would undergo random fluctuations in size. We have calculated that for clones of 1,000 cells or more to be produced by this process, ≈124 complete random turnovers of the liver would be required (Supporting Text, which is published as supporting information on the PNAS web site). Proliferating cell nuclear antigen staining indices for hepatocytes in chronically WHV-infected livers are on average ≈0.5–1% (8), and if that corresponds to a rate of hepatocyte killing of 1.5–3% per day (44) sustained for an entire year, it would account for only 6–12 turnovers of the liver cell population. Thus, although smaller cell clones could be the result of random turnover, the presence of high-copy clones requires alternative explanations.
Clonal Expansion of Transformed Hepatocytes. Hepatocyte clones might be produced from variant hepatocytes that do not require a regenerative stimulus for cell division. The growth of such clones would therefore not depend on liver cell turnover. High-copy clones with detectable integrated DNA were observed at a frequency of 1 per ≈500,000 cells (Table 2, columns 3 and 4) or ≈100,000 such clones per liver. Typically, woodchucks with chronic WHV infection develop one or a only a few independent HCC nodules, which contain integrated DNA (32), so we infer that the vast majority of the high-copy cell clones we observed were not fully malignant but could represent cells that had acquired premalignant changes. For one clone, WHV DNA was integrated into the win locus, a common integration site in WHV, and it is possible that other high-copy clones expanded because of undetected integrations in Nmyc-2 or win (24–26). The idea that expansion of high-copy clones was frequently due to insertional mutagenesis by the integrations that we detected, however, is at odds with the fact that common regions for integration in cellular DNA were detected for only 2 of the 106 clones of 27 or more cells. In addition, with the exception of the single integration at win, no sites of integration related to known tumor suppressors or oncogenes were identified.
Clonal Expansion Due To Resistance to Killing and/or Selective Regeneration. High-copy cell clones might also be produced during liver turnover from hepatocytes with a selective advantage for surviving the cytotoxic immune response or for responding to regenerative signals. The growth of such cell clones would depend on hepatocyte turnover, and such clones would increase in size much more rapidly than clones with no selective advantage. For example, a hepatocyte that was resistant to immune killing, or one that regenerated with twice the probability of that of a normal liver cell, would require only 9.2 liver turnovers to drive its expansion to produce 10,000 cells (Supporting Text and Fig. 5, which is published as supporting information on the PNAS web site). A stronger advantage would require even less liver turnover. Thus, a selective survival or regeneration advantage during liver turnover seems adequate to explain the existence of even the highest copy cell clones we observed. A selective advantage during turnover might be due to, for example, an inability to express viral antigens or display them on MHC molecules, a premalignant change conferring hyperresponsiveness to regenerative signals, or a natural propensity for some rare infected cells to act as stem cells.
Finally, selective clonal expansion does not seem to be a normal component of postnatal growth of the liver (45). Nonetheless, we would like to point out that because we are unable to perform our analyses on an uninfected woodchuck, our results do not prove that the high degree of hepatocyte clonality we observed was actually a consequence of chronic hepatitis. However, with this caveat, our results imply a highly dynamic process of hepatocyte proliferation during chronic infection, with strong selective pressures remodeling the liver by providing opportunities for the emergence of hepatocytes with a growth or survival advantage. This process may be a major factor in the pathogenesis of chronic hepadnavirus infections to primary hepatocellular carcinoma.
Supplementary Material
Acknowledgments
We thank C. Aldrich (Fox Chase Cancer Center), B. H. Zhang (University of New Mexico), and C. A. Scougall (University of Adelaide and Institute of Medical and Veterinary Science) for excellent technical assistance; John Taylor and Christoph Seeger (Fox Chase Cancer Center), Frank Chisari, Luca Guidotti, and Stefan Wieland (The Scripps Research Institute, La Jolla, CA), Raymond Schinazi (Emory University, Atlanta), and Joe Grisham (University of North Carolina, Chapel Hill) for a critical reading of the manuscript; and Drs. Raymond Schinazi and Yung-Chi Cheng (Yale University, New Haven, CT) for helpful discussions of antiviral agents. Parts of this work were carried out by W.S.M. while on sabbatical in the laboratory of J.S. in the University of New Mexico School of Medicine. This work was supported by grants from the National Institutes of Health (J.S. and W.S.M.) of the U.S. Public Health Service, by a project grant from the National Health and Medical Research Council of Australia (A.R.J.), and by an appropriation from the Commonwealth of Pennsylvania.
Author contributions: W.S.M., A.R.J., and J.S. designed research; W.S.M., A.R.J., and J.S. performed research; W.S.M., A.R.J., and J.S. analyzed data; and W.S.M., A.R.J., and J.S. wrote the paper.
Abbreviations: cccDNA, covalently closed circular DNA; HCC, hepatocellular carcinoma; WHV, woodchuck hepatitis virus.
References
- 1.Beasley, R. P. (1982) Hepatology 2, 21S–26S. [Google Scholar]
- 2.Vemura, R. P., Aragona, E. & Gupta, S. (1992) Hepatology 16, 968–973. [DOI] [PubMed] [Google Scholar]
- 3.Seki, S., Sakaguchi, H., Kawakita, N., Yanai, A., Kuroki, T., Mizoguchi, Y., Kobayashi, K. & Monna, T. (1991) Hepatology 14, 781–788. [DOI] [PubMed] [Google Scholar]
- 4.MacDonald, R. A. (1960) Arch. Int. Med. 107, 335–343. [DOI] [PubMed] [Google Scholar]
- 5.Grisham, J. W. (1962) Cancer Res. 22, 842–849. [PubMed] [Google Scholar]
- 6.Koukoulis, G., Rayner, A., Tan, K.-H., Williams, R. & Portmann, B. (1992) J. Pathol. 166, 359–368. [DOI] [PubMed] [Google Scholar]
- 7.Chisari, F. V. & Ferrari, C. (1995) Ann. Rev. Immunol. 13, 29–60. [DOI] [PubMed] [Google Scholar]
- 8.Mason, W. S., Cullen, J., Moraleda, G., Saputelli, J., Aldrich, C. E., Miller, D. S., Tennant, B., Frick, L., Averett, D., Condreay, L. D. & Jilbert, A. R. (1998) Virology 245, 18–32. [DOI] [PubMed] [Google Scholar]
- 9.Summers, J., Jilbert, A. R., Yang, W., Aldrich, C. E., Saputelli, J., Litwin, S., Toll, E. & Mason, W. S. (2003) Proc. Natl. Acad. Sci. USA 100, 11652–11659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bill, C. & Summers, J. (2004) Proc. Natl. Acad. Sci. USA 101, 11135–11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Summers, J. & Mason, W. S. (2004) Proc. Natl. Acad. Sci. USA 101, 638–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yasui, H., Hino, O., Ohtake, K., Machinami, R. & Kitagawa, T. (1992) Cancer Res. 52, 6810–6814. [PubMed] [Google Scholar]
- 13.Aoki, N. & Robinson, W. S. (1989) Mol. Biol. Med. 6, 395–408. [PubMed] [Google Scholar]
- 14.Ochiai, T., Urata, Y., Yamano, T., Yamagishi, H. & Ashihara, T. (2000) Hepatology 31, 615–621. [DOI] [PubMed] [Google Scholar]
- 15.Zhu, Y., Cullen, J. M., Aldrich, C. E., Saputelli, J., Miller, D., Seeger, C., Mason, W. S. & Jilbert, A. R. (2004) Virology 327, 26–40. [DOI] [PubMed] [Google Scholar]
- 16.Balakrishna Pai, S., Liu, S. H., Zhu, Y. L., Chu, C. K. & Cheng, Y. C. (1996) Antimicrob. Agents Chemother. 40, 380–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao, G. Q., Liu, S. H., Chou, E., Kukhanova, M., Chu, C. K. & Cheng, Y. C. (1996) Biochem. Pharmacol. 51, 941–947. [DOI] [PubMed] [Google Scholar]
- 18.Moroy, T., Etiemble, J., Bougueleret, L., Hadchouel, M., Tiollais, P. & Buendia, M. A. (1989) Oncogene 4, 59–65. [PubMed] [Google Scholar]
- 19.Strapans, S., Loeb, D. D. & Ganem, D. (1991) J. Virol. 65, 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong, S. S., Jensen, A. D., Chang, C. J. & Rogler, C. E. (1999) J. Virol. 73, 1492–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang, W. & Summers, J. (1999) J. Virol. 73, 9710–9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galibert, F., Chen, T. N. & Mandart, E. (1982) J. Virol. 41, 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kajino, K., Jilbert, A. R., Saputelli, J., Aldrich, C. E., Cullen, J. & Mason, W. S. (1994) J. Virol. 68, 5792–5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fourel, G., Couturier, J., Wei, Y., Apiou, F., Tiollais, P. & Buendia, M. A. (1994) EMBO J. 13, 2526–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei, Y., Fourel, G., Ponzetto, A., Silvestro, M., Tiollais, P. & Buendia, M. A. (1992) J. Virol. 66, 5265–5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seeger, C. & Mason, W. S. (1999) in Persistent Viral Infections, eds. Ahmed, R. and Chen, I. (Wiley, New York) pp. 607–621.
- 27.Grozdanov, P., Georgiev, O. & Karagyozov, L. (2003) Genomics 82, 637–643. [DOI] [PubMed] [Google Scholar]
- 28.Govindarajan, S., Conrad, A., Lim, B., Valinluck, B., Kim, A. M. & Schmid, P. (1990) Arch. Pathol. Lab. Med. 114, 1042–1045. [PubMed] [Google Scholar]
- 29.Radaeva, S., Li, Y., Hacker, H. J., Burger, V., Kopp-Schneider, A. & Bannasch, P. (2000) J. Hepatol. 33, 580–600. [DOI] [PubMed] [Google Scholar]
- 30.Li, Y., Hacker, H., Kopp-Schneider, A., Protzer, U. & Bannasch, P. (2002) J. Hepatol. 37, 478–485. [DOI] [PubMed] [Google Scholar]
- 31.Yang, D., Alt, E. & Rogler, C. E. (1993) Cancer Res. 53, 2020–2027. [PubMed] [Google Scholar]
- 32.Jacob, J. R., Sterczer, A., Toshkov, I. A., Yeager, A. E., Korba, B. E., Cote, P. J., Buendia, M. A., Gerin, J. L. & Tennant, B. C. (2004) Hepatology 39, 1008–1016. [DOI] [PubMed] [Google Scholar]
- 33.Hirohashi, S., Shimosato, Y., Ino, Y. & Kishi, K. (1982) J. Natl. Cancer Inst. 69, 565–568. [PubMed] [Google Scholar]
- 34.Nayak, N. C., Dhar, A., Sachdeva, R., Mittal, A., Seth, H. N., Sudarsanam, D., Reddy, B., Wagholikar, U. L. & Reddy, C. R. (1977) Int. J. Cancer 20, 643–654. [DOI] [PubMed] [Google Scholar]
- 35.Chu, C. M., Yeh, C. T., Chien, R. N., Sheen, I. S. & Liaw, Y. F. (1997) J. Clin. Microbiol. 35, 102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamothe, F., Laurencin-Piche, J., Cote, J., Guevin, R., Viallet, A. & Richer, G. (1976) Gastroenterology 71, 102–108. [PubMed] [Google Scholar]
- 37.Ray, M. B., Desmet, V. J., Bradburne, A. F., Desmyter, J., Fevery, J. & De Groote, J. (1976) Gastroenterology 71, 462–467. [PubMed] [Google Scholar]
- 38.Kojima, M., Udo, K., Takahashi, Y., Yoshizawa, H., Tsuda, F., Itoh, Y., Miyakawa, Y. & Mayumi, M. (1977) Gastroenterology 73, 664–667. [PubMed] [Google Scholar]
- 39.Hsu, H.-Y., Lin, Y.-H., Chang, M.-H., Su, I.-J. & Chen, D.-S. (1988) Hepatology 8, 378–382. [DOI] [PubMed] [Google Scholar]
- 40.Omata, M., Afroudakis, A., Liew, C.-T., Ashcavai, M. & Peters, R. L. (1978) Gastroenterology 75, 1003–1009. [PubMed] [Google Scholar]
- 41.Suzuki, K., Uchida, T., Horiuchi, R. & Shikata, T. (1985) Cancer 56, 321–327. [DOI] [PubMed] [Google Scholar]
- 42.Gowans, E. J. & Burrell, C. J. (1985) J. Clin. Pathol. 38, 393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gowans, E. J., Burrell, C. J., Jilbert, A. R. & Marmion, B. P. (1985) Intervirology 24, 220–225. [DOI] [PubMed] [Google Scholar]
- 44.Zhu, Y., Yamamoto, T., Cullen, J., Saputelli, J., Aldrich, C. E., Miller, D. S., Litwin, S., Furman, P. A., Jilbert, A. R. & Mason, W. S. (2001) J. Virol. 75, 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kennedy, S., Rettinger, S., Flye, M. W. & Ponder, K. P. (1995) Hepatology 22, 160–168. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.