Abstract
Cone snails in the Virgiconus clade prey on marine worms. Here, we identify six related conotoxins in the O1-superfamily from three species in this clade, Conus virgo, Conus terebra and Conus kintoki. One of these peptides, vi6a, was directly purified from the venom of C. virgo by following its activity using calcium imaging of dissociated mouse dorsal root ganglion (DRG) neurons. The purified peptide was biochemically characterized, synthesized and tested for activity in mice. Hyperactivity was observed upon both intraperitoneal and intracranial injection of the peptide. The effect of the synthetic peptide on DRG neurons was identical to that of the native peptide. Using the vi6a sequence, five other homologs were identified. These peptides define a glycine-rich subgroup of the O1-superfamily from the Virgiconus clade, with biological activity in mice.
1. Introduction
Cone snails (family Conidae) are a highly successful group of venomous marine gastropods found in all the tropical oceans. These predatory molluscs can be sorted into three major groups based on their prey preference: fish-hunting (piscivores), mollusc-hunting (molluscivores) and the largest group, worm-hunting (vermivores) (Kohn, 1959; McIntosh and Jones, 2001; 12, 1997; Olivera et al., 1990). There are probably ~850 cone snail species (Favreau and Stocklin, 2009; Olivera, 2002; Röckel et al., 1995) and all of them possess venom, which contains a complex array of small peptidic molecules called conotoxins/conopeptides (Olivera et al., 1985; Olivera et al., 1990; Teichert et al., 2015; Terlau and Olivera, 2004) that the animal uses for diverse biotic interactions, including prey capture.
The conotoxins are gene products that are initially translated as a precursor (Buczek et al., 2005; Olivera, 1997), which can be divided into three regions: the signal sequence at the N-terminus, the pro region and the mature toxin region at the C-terminus (Colledge et al., 1992; Robinson and Norton, 2014). All conotoxins that share a highly conserved signal sequence can be grouped into a conotoxin gene superfamily; members of a gene superfamily also share a distinct pattern of cysteine residues in their amino acid sequence (Terlau and Olivera, 2004; Woodward et al., 1990). Each conotoxin superfamily can be further subdivided into pharmacological families (Terlau and Olivera, 2004). For example, the O1-conotoxin superfamily comprises the ω-conotoxins, which target voltage-gated calcium channels (Hillyard et al., 1992; Olivera et al., 1985; Zhangsun et al., 2006), the κ-conotoxins, which target voltage-gated potassium channels (Shon et al., 1998; Terlau et al., 1996) and the δ-and the µO-conotoxin families, which target voltage-gated sodium channels at different sites (Leipold et al., 2005; McIntosh et al., 1995; Shon et al., 1995; Terlau et al., 1996; Wilson et al., 2015).
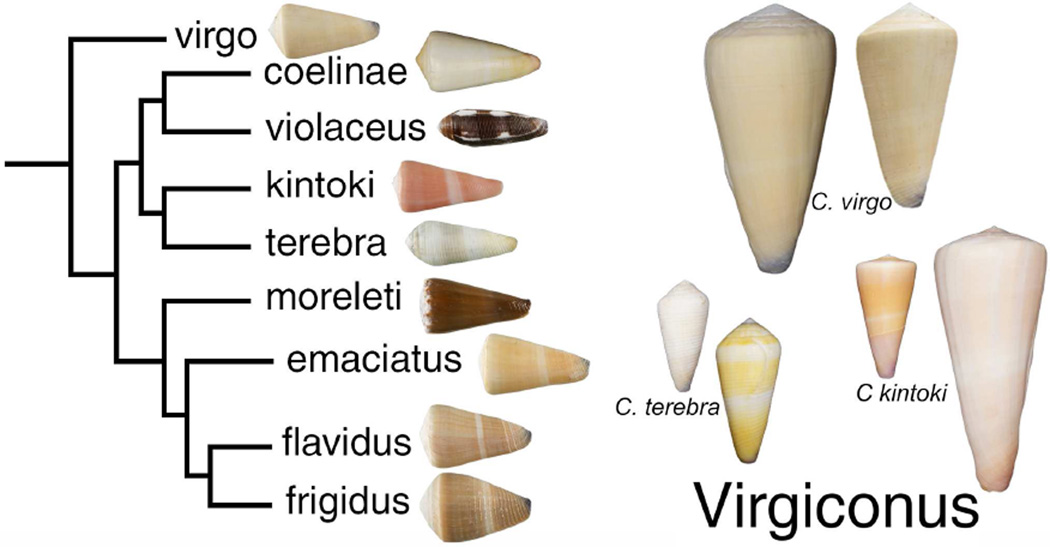
Many members of the O1-conotoxin family that were shown to be biologically active in vertebrates were purified from the venom of fish-hunting cone snails (Terlau and Olivera, 2004). This is not surprising since fish-hunting cone snails evolved their highly expressed conotoxins to potently target their fish prey. However, worm-hunting and mollusc-hunting cone snails do express conotoxins that target specific molecules in vertebrates. We focused this study on cone snails that belong to the vermivorous Virgiconus clade. There are at least nine species, all from the Indo-Pacific region, in this clade (Puillandre et al., 2014). Three species from this clade namely: Conus virgo, Conus terebra and Conus kintoki (shown in Fig. 1) were analyzed; C. virgo and C. terebra are shallow-water species, while C. kintoki is only collected well offshore, at depths between 50–200 meters.
Figure 1. Phylogenetic relationships in the Virgiconus clade.
The snails where the conotoxins reported in this paper were found are shown on the right. The largest specimen in this figure is C. kintoki, which measures 111 mm.
In this paper, six homologous O1-superfamily conotoxins from three cone snail species were identified. One was purified from the venom of C. virgo and we propose to name this conopeptide vi6a. This peptide is active on a subset of mouse dorsal root ganglion (DRG) neurons and elicits a behavioral phenotype in mice upon both intraperitoneal (IP) and intracranial (IC) injection. Homologous peptides were identified from C. terebra and C. kintoki, as well as two additional peptides from C. virgo. These conotoxins are unusually glycine-rich, and define a novel subgroup within the O1-superfamily.
2. Materials and Methods
2.1 Extraction of Crude Venom
Lyophilized C. virgo venom was homogenized in 10 mL of 35% (v/v) CH3CN – 0.1% (v/v) trifluoroacetic acid (TFA) using a glass-PTFE tissue grinder for 5 cycles at 1500 rpm. The homogenate was centrifuged in a Beckmann F0650 rotor at 37,000 × g for 15 min at 5°C and the supernatant containing the crude venom extract was collected.
2.2 Venom fractionation
The crude venom extract was fractionated by Reversed-Phase High Performance Liquid Chromatography (RP-HPLC) using a Vydac C18 preparative column (218TP101522). Elution was done at a flow rate of 7 mL/min and using a two solvent system: Solvent A (0.1% TFA) and Solvent B (90% v/v CH3CN, 0.1% v/v TFA). The elution gradient used was made up of the following steps: 2% – 55% Solvent B for 53 min, 55% – 65% for 5 min and 65% – 100% for 5 min. Solvent B was maintained at 100% for 1 min and then brought down to 10% within 3 min. The subfractionation of the active fraction 15 was done by analytical RP-HPLC using a C18 Vydac monomeric column (238EV54) and using the following gradient: 5% – 10% Solvent B for 5 min, 10% – 25% Solvent B for 45 min. The absorbance was monitored at 220 and 280 nm.
2.3 Reduction and alkylation of the purified peptides
The bioactive peptide from C. virgo was reduced using dithiothreitol (DTT) and was alkylated using 4-vinylpyridine (4-VP). The pH of the solution containing the peptide was buffered to pH 8 using 0.5 M Tris base. The peptide was reduced with 10 mM DTT at 65°C for 30 minutes. A 0.8 µL volume of 4-VP was added to the solution containing the reduced peptide. The alkylation of the reduced peptide was performed in the dark at room temperature for 30 minutes. The reduced and alkylated peptide was purified from the reaction by analytical RP-HPLC using a gradient similar to that previously described for the analytical HPLC purification of fraction 15.
2.4 Peptide Sequencing
The sample was dissolved in 30 µL of solvent B (90% v/v CH3CN, 0.1% v/v TFA). Thirty microliters of the solution were applied to a glass fiber Micro TFA Filter (401111, Applied Biosystems, Foster City, CA) previously treated with 15 µL of Biobrene Plus (400385, Applied Biosystems), dried with Argon, and then sequenced for 34 cycles in a Procise 491 Protein Sequencing System (Applied Biosystems) employing the Pulsed-liquid method.
2.5 Identification and sequencing of O-superfamily conotoxin clones from Conus virgo, Conus terebra and Conus kintoki
RNA from the venom duct dissected from a single specimen each of C. virgo, C. terebra and C. kintoki was isolated using TRIzol® reagent (Invitrogen, Grand Island, NY) following the manufacturer’s recommended protocol. cDNA was synthesized from total RNA using the SMART PCR cDNA synthesis kit (Clontech, Mountain View, CA) according to the manufacturer’s protocols. The first strand cDNA was used as a template for the PCR amplification of O1-superfamily conotoxin genes using the primers Osup+ (5’-ATGAAACTGACGTGYGT-3’) and Osup- (5’-TACGTYCTCTGAATYCACCAGAG-3’). These primers were designed based on the conserved region of the O1-superfamily precursor. The PCR products were gel-purified using PCR product purification kit (Qiagen, Valencia, CA) and ligated into pGEM®-T Easy vector (Promega, Madison, WI) following the manufacturer’s standard protocol. The products from these reactions were transformed into competent E. coli DH10β cells (New England Biolabs, Inc, Ipswich, MA). The colonies were screened by PCR using the Osup+ and the Osup- primers. Positive clones were sequenced using the T7 and SP6 promoter primer sequences (Promega, Madison, WI).
2.6 Solid phase peptide synthesis of vi6a
Linear vi6a was synthesized at a 50-µmol scale using an AAPPTec Apex 396 synthesizer (AAPPTec LLC, Louisville, KY) applying standard solid-phase Fmoc (9-fluorenylmethyloxycarbonyl) protocols. Fmoc-protected amino acids, were purchased from AAPPTec. N-α-Fmoc-O-t-butyl-L-trans-4-hydroxyproline (Hyp) was purchased from EMD Millipore (Darmstadt, Germany). The peptides were assembled on pre-loaded Fmoc-L-Leu-Wang resin (substitution: 0.33 mmol·g−1; Peptides International Inc., Louisville, KY), Fmoc-Gln(Trt)-Wang resin (substitution: 0.57 mmol·g−1, Nova-Biochem, EMD Millipore), Fmoc-Ser(tBu)-Wang resin (substitution: 0.53 mmol·g−1; Peptides International, Louisville, KY). Side-chain protection for each amino acid was: Arg, 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl (Pbf); Asp and Glu, O-tert-butyl (OtBu); Lys, tert-butyloxycarbonyl (Boc) Hyp, Thr and Tyr, tert-butyl (tBu); and Gln and Cys, trytl (Trt). Coupling of each amino acid was achieved using 1 equivalent of 0.4 M benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate (PyBOP) and 2 equivalents of 2 M N,N-diisopropylethyl amine (DIPEA) in N-methyl-2-pyrrolidone (NMP). Standard amino acids were used in ten-fold excess (60-min coupling), with the exception of Arg and Hyp, which were used in five-fold excess (90-min coupling). Fmoc-protecting groups were removed by a 20-min treatment with 20% (v/v) piperidine in dimethylformamide (DMF).
2.7 Oxidative folding of vi6a
The linear vi6a was resuspended in 0.5 mL of 0.01% TFA solution and added to a solution containing: 2.5 mL of 0.2 M Tris·HCl (pH 7.5) + 0.2 mM EDTA, 0.5 mL of 1:1 mixture of 20 mM reduced (GSH) and oxidized (GSSG) glutathione and 1.5 mL of water. The final peptide concentration in the folding mixture was 20 µM. vi6a was folded for 2 hours and the reaction was quenched with 8% (v/v) formic acid. The reaction mixture was separated by RP-HPLC using a Vydac semi-preparative C18 column and employing a linear gradient from 5% to 40% Solvent B for 35 min at a flow rate of 4 mL/min. The eluent was monitored by absorbance at 220nm and at 280 nm. The purity of the folded peptides was assessed by analytical RP-HPLC using the gradient described above, with a flow rate 1 mL/min. The concentration of the folded peptide was determined by UV/Vis spectrophotometry at 280 nm and using an extinction coefficient (ε) of 1420 L mol−1 cm−1. The mass was determined by matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS).
2.8 Co-elution experiment of the native and synthetic vi6a
The co-elution experiments were done using analytical RP-HPLC. One nmol of each of the native and synthetic vi6a was separately injected into the analytical C18 column and eluted using a gradient of 5% – 40% Solvent B for 35 min. A mixture containing 1 nmol of each of the native and synthetic vi6a was also analyzed on the same column using the same gradient.
2.9 Calcium imaging assay
Experimental protocols involving live animals were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Utah. Preparation of the cell cultures from dorsal root ganglia (DRG) and calcium imaging experiments were done as described previously (Smith et al., 2013; Teichert et al., 2012). Briefly, lumbar DRG neurons from wild type C57BL/6 mice were dissociated, pooled and cultured overnight for calcium imaging experiments. Cells were loaded with Fura-2-AM dye one hour before the experiment. During the experiment, the dye inside the cells was excited alternately with 340 nm and 380 nm light. The ratio of the emission at 510 nm from both excitations was measured. The ratio of the fluorescence intensity is a measure of the increase in cytosolic calcium [Ca2+]i resulting from the depolarization caused by the external application of a high concentration of potassium ions, [K+]o. For the purification of the bioactive component from C. virgo venom, a solution containing 20 µM [K+]o and 20 µM veratridine was used to depolarize the DRG neurons. Veratridine prolongs the activation of voltage-gated sodium channels and facilitates the discovery of venom components that may block sodium channels, in addition to those that may act on potassium or calcium channels. This solution was applied every seven minutes. After the third depolarizing pulse, crude C. virgo venom extract or HPLC fractions were incubated with the cells for seven minutes. The fourth depolarizing pulse was applied to identify the effect of the applied venom or fractions on the DRG neurons. Succeeding depolarizing pulses were done to determine the reversibility of the observed effects. A similar protocol was followed for the assessment of the activity of the synthetic vi6a; however, 30 mM [K+]o was used as the depolarizing stimulus and the experiment was performed without veratridine.
2.10 Mouse bioassay
Swiss Webster mice (17 – 19 days; 7 – 10 grams) were injected intracranially (IC) with the peptide in 10 µL normal saline solution (NSS). Mice were observed for aberrant behavior for 3 hours. The systemic effect of vi6a was determined by the intraperitoneal (IP) injection of the peptide into 17 – 19 day old mice. Appropriate concentrations of the peptide were dissolved in 50 µL NSS and injected IP into mice. The mice were then observed for 3 hours. The use of Swiss Webster mice followed protocols that conform to the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the University of Utah Institutional Animal Care and Use Committee.
3. Results
3.1 Purification of a biologically active conotoxin from the venom of Conus virgo
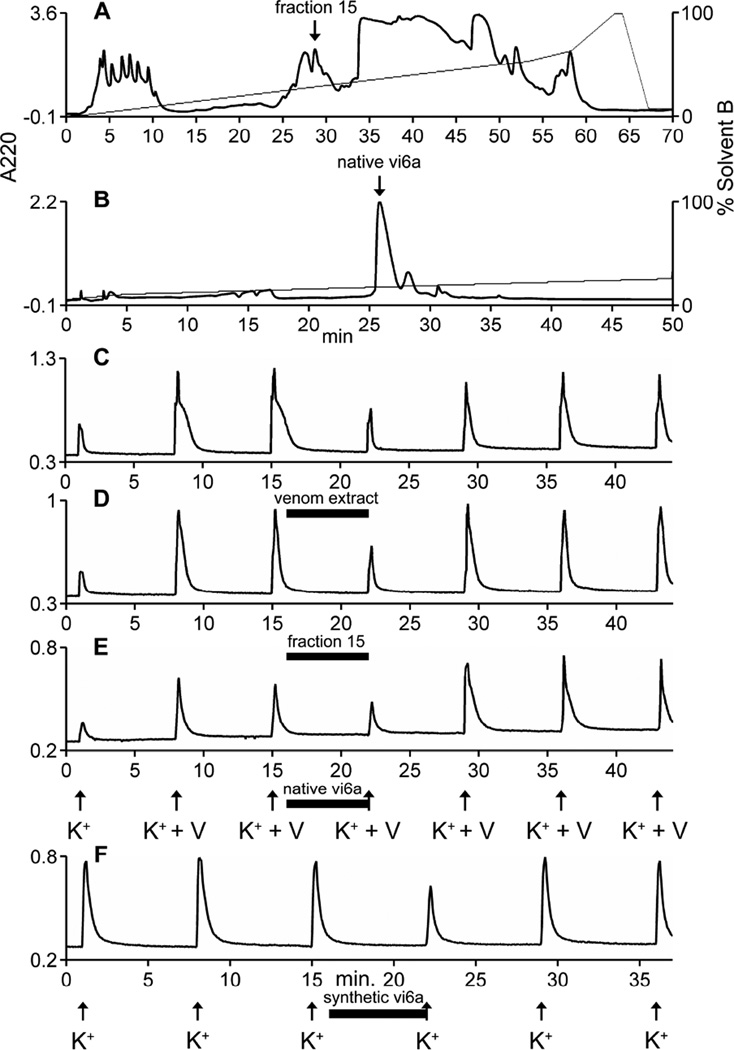
Venom extracted from Conus virgo venom glands was assayed based on its activity in the calcium imaging of dissociated mouse dorsal root ganglion (DRG) neurons. This assay measures the changes in cytosolic calcium [Ca2+]i in response to a stimulus. In this paper, the effect of venom or conotoxins on the changes in [Ca2+] resulting from depolarization by the application of high extracelluar potassium [K+]o is measured. Diverse activities were observed when the venom was subjected to this assay. In some cells, the application of the crude extract of the venom from C. virgo resulted in the inhibition of the increase in [Ca2+]i resulting from a depolarizing pulse of 20 mM [K+]o with 20 µM veratridine. In other cells, the depolarizing pulse following venom application resulted in an increase in [Ca2+]i. Other cells showed a direct response (i.e. a perturbation of the baseline upon venom application). The component responsible for the inhibition of the increase in [Ca2+]i was further purified from the venom using reversed-phase high performance liquid chromatography (RP-HPLC) (Fig 2). Individual HPLC fractions were tested and the inhibitory activity was found in fraction 15. Subfractionation of this fraction resulted in a single major component (Fig. 2B) that retained the inhibitory activity. The bioactive, major fraction in Fig 2B was shown to be homogeneous by linear mode MALDI-MS.
Figure 2. Bioactivity-guided purification of the inhibitory component from C. virgo venom.
HPLC fractions of the C. virgo venom extract was screened for the activity to inhibit the increase in intracellular calcium upon depolarization with 20 mM [K+]o and 20 µM veratridine. Fraction 15, marked by the arrow in A, showed the inhibitory activity. Further subfractionation of the inhibitory fraction yielded a single major active component (native vi6a) indicated by the arrow in B. MALDI-MS in the linear mode of the major peak in B revealed the fraction to contain only vi6a. C – E are the calcium imaging traces of the DRG neurons showing the inhibitory activity. The Y-axis of each graph is the 340/380 nm ratio described in Methods. The arrows represent the 15-second application of 20 mM [K+]o and 20 µM veratridine, and the horizontal bars indicate the time when each sample was present in the bath solution. Panel C shows the inhibitory activity of the crude venom extract; panel D shows the inhibitory activity of fraction 15 and panel E shows the inhibitory activity of the native vi6a. Panel F shows the inhibitory activity of the synthetic peptide. In panel F, the arrows indicate the 15 second application of 30 mM [K+]o. In panels C–F, the horizontal bar indicates the time when each sample was present in the bath solution.
3.2 Biochemical characterization of the inhibitory compound from C. virgo venom
Analysis of the purified fraction (Fig. 2B) by electrospray ionization mass spectrometry (ESI-MS) revealed a homogeneous solution containing a component with a mass of 3114.15 Da. When the component that exhibited the inhibitory activity was reduced and alkylated (see Methods), MALDI-MS in the linear mode yielded a mass of 3754.16 Da. The difference of 640.01 Da between the native compound and the reduced/alkylated compound suggested the presence of 6 cysteine residues that were alkylated by 6 molecules of 4-vinylpyridine (MW 105 Da).
N-terminal sequencing of the active component revealed the peptide sequence to be: DCGGQGEGCYTQOCCOGLRCRGGGTGGGVCQL. This peptide is 32 amino acids long and contains 6 cysteine residues consistent with the number of cysteine residues identified from the comparison of the masses of the native and reduced/alkylated peptides. The arrangement of the cysteine residues of this peptide suggests that it belongs to the O1-conotoxin superfamily. The peptide contains two proline residues that are post translationally modified to hydroxyproline. One striking feature of this peptide sequence is the abundance of glycine residues. Of the 26 non-cysteine residues, 11 are glycine residues.
The peptide sequence assignment was verified by the chemical synthesis of the peptide with the above sequence. Analysis of the folding reaction revealed the formation of a single major component after 2 hours. This major component was purified to >99% purity from the folding reaction using RP-HPLC. MALDI-MS in the reflector mode revealed a mass of 3114.10 Da, which matches the mass of the native peptide previously determined using ESI-MS. Co-elution was carried out to verify whether the synthetic and the native peptides are structurally identical (Fig S1). Separate injections of 1 nmol each of the synthetic and native peptides in an analytical HPLC column at a gradient of 5% – 40% B90 for 35 minutes revealed a similar retention time of ~16 minutes for both peptides. A solution containing 1 nmol each of the native and the synthetic peptides was analyzed using the same HPLC gradient; a single peak eluted at ~16 minutes indicating the co-elution of the native and synthetic peptides (Fig. S1). These experiments validated the sequence assignment and suggest that the folding reaction formed a major component whose disulfide connectivity matches that of the native peptide. We propose to call this peptide vi6a.
3.3 Sequencing of clones from C. virgo, C. terebra, and C. kintoki
Conotoxins are initially translated as larger prepropeptide precursors. The precursor sequence of vi6a was determined using PCR primers designed to hybridize to the conserved region of the O1-superfamily precursor. We also screened cDNA’s extracted from C. terebra, and C. kintoki for the presence of O1-superfamily conotoxins to determine whether these snails express conotoxins with sequences similar to vi6a (these cone snail species, like C. virgo, are members of the Virgiconus clade). The peptides predicted from clones Vi6.1, Vi6.2 and Vi6.4 from C. virgo are shown in Figs. 3 and 4; the predicted toxin from Vi6.1 exactly matches the sequence of vi6a. Thus, the Vi6.1 clone encodes the precursor sequence of vi6a. The screening of the C. terebra cDNA yielded two clones: Tr6.2 and Tr6.3 (Figs. 3 and 4) whose predicted toxins are homologous to vi6a. The clone Kt6.1 (Figs. 3 and 4) from C. kintoki also codes for a homologous peptide. These sequences are compared in Figs.3 and 4. The high degree of similarity in the signal sequences of these precursors indicates that these conotoxins belong to the O1 – superfamily.
Figure 3.
Alignment of conotoxin sequences predicted from clones obtained from C. virgo (Vi), C. terebra (Tr) and C. kintoki (Kt). Vi6.1 is the precursor sequence that codes for vi6a, the peptide purified from the venom of C. virgo. These sequences display a high degree of similarity indicating that they are members of the same gene family. The amino acids that diverge from Vi6.1 are highlighted and conserved amino acids are identified by an asterisk. Vi6.1 is identical to the Conoserver Vi6.1 protein.
Figure 4.
A. Alignment of the precursor sequence of vi6a (Vi6.1) with the precursor sequences of conotoxins identified from C. virgo, C. terebra and C. kintoki. The amino acid sequences of these precursors are highly conserved indicating that these conotoxins belong to the same gene family. The conserved amino acids are marked by an asterisk.
B. Alignment of the precursor sequence of vi6a (Vi6.1) with the precursor sequence of known superfamily conotoxins from the delta conotoxin family (δ-TxVIA) and the omega conotoxin family (ω-GVIA). These precursor sequences have a very high degree of similarity in the signal sequence and a conserved pattern of cysteine residues in the conotoxin region indicating that they belong to the same superfamily. The signal sequence of Vi6.1 is more similar to the signal sequence of ω-GVIA. (ω-GVIA accession number: M84612.1; δ-TxVIA accession number: AF193261.1).
3.4 Biological activity of vi6a
The biological activity of the synthetic vi6a was assessed using the calcium imaging assay. In these experiments, DRG neurons were depolarized by external application of 30 mM [K+]o once every 7 minutes. After the third depolarization, 1 µM vi6a was applied and allowed to incubate with the cells for 6 minutes. Cellular response was assessed upon the fourth application of 30 mM [K+]o immediately following the 1 µM vi6a incubation. Results from 3 experiments using DRG neurons prepared from 3 different mice showed that the synthetic vi6a exhibited the inhibitory activity in 76 ± 10% of the cells. The inhibitory effect of vi6a is observed in small, medium and large diameter DRG neurons. A sample calcium imaging trace showing the inhibitory activity of the synthetic vi6a is shown in Fig 2F.
The peptide, vi6a, was also tested for its in vivo effects upon injection into mice. An intracranial injection (IC) of the peptide dissolved in 10µL normal saline solution (NSS) and injected into 17 – 19 day old mice elicited the behavioral phenotypes shown in Table 1. Mice injected with 0.4 mg/kg vi6a exhibited a hyperactive phenotype manifested by the injected animal (n = 3) continuously running around the cage. Increasing the amount of vi6a to 2.0 mg/kg resulted in the typical hyperactive phenotype; animals would climb the cage, jump and run around.
Table 1.
Biological activity of vi6a injected IC and IP into 17 – 19 day old mice.
| Assay | Dose injected | Effect | Duration |
|---|---|---|---|
| IC injection | 0.4 mg/kg | hyperactivitya | not determined |
| 2.0 mg/kg | hyperactivitya | not determined | |
| IP injection | 0.7 mg/kg | hyperactivitya | 20 – 40 minutes |
| 2.0 mg/kg | hyperactivitya | 40 – 60 minutes | |
| 6.0 mg/kg | hyperactivitya | >2 hours |
These assays used three mice for each dose tested.
Hyperactivity symptoms include continuous running, jumping and climbing in cages.
The systemic effects of vi6a on mice were determined by the intraperitoneal (IP) injection of vi6a (Table 1). Mice between 17 – 19 days old were used for the experiment. Varying amounts of vi6a were dissolved in 50 µL normal saline solution (NSS). IP injection of vi6a (0.7 mg/kg) resulted in the same hyperactive phenotype observed when the peptide was injected IC into mice. Increasing the dose of vi6a to 6.0 mg/kg also elicited the same hyperactive phenotype, which lasted longer than the effects of the lower doses of vi6a.
4. Discussion
In this paper, a subset of conotoxins expressed by cone snails belonging to the worm-hunting Virgiconus clade has been identified. There are at least nine known cone snail species belonging to this clade and these snails are widely distributed across the tropical Indo-Pacific ranging from inter-tidal regions to deep waters. Venoms of species in this clade have not been well characterized. We purified a peptide from C. virgo based on its biological activity in the calcium imaging of dissociated mouse DRG neurons. Using molecular biology techniques, five other related conotoxins from the Virgiconus clade were identified. It seems likely that all species in the Virgiconus clade express related peptides.
We note that Conus virgo has three peptides in this group that have between 8 – 9 amino acid substitutions, and are presumably functionally divergent. However, across species, three sequences (Vi6.1, Tr6.2 and Kt6.1) are much more similar (differing by 2 – 6 amino acids) — these are presumably interspecific variants of peptides with the same molecular target.
The amino acid sequence of vi6a is identical to the amino acid sequence of the predicted toxin contained in precursor virgo 1, previously cloned from the same species by Kauferstein et al. (2005). Since post-translational modifications of amino acids cannot be predicted from the clone, the purification of vi6a from the C. virgo venom and the subsequent biochemical characterization of the peptide (reported in this paper) allowed for the identification of the post translational modification of the prolines at positions 13 and 16 into hydroxyproline. The clone Vi6.1, which we identified from the C. virgo cDNA, is identical to the clone virgo 1.
The pattern of cysteine residues in vi6a matches the cysteine pattern of peptides in the O-conotoxins (i.e. the O1, O2 and O3 superfamilies) (Kaas et al., 2008; Kaas et al., 2012; Puillandre et al., 2012). A comparison of the signal sequences to known O1-superfamily conotoxins such as δ-TxVIA and ω-GVIA (Fig 4B) reveals that there is a high degree of amino acid conservation. An analysis by means of the ConoPrec tool of the ConoServer database (Kaas et al., 2008; Kaas et al., 2012) suggested that all six precursor sequences are most similar (>92% identity) to signal sequences of the O1-superfamily. We conclude that vi6a and other related peptides in the Virgiconus clade are members of the O1-superfamily, and the peptides we have defined in this study comprise a distinctive glycine-rich subgroup within the O1 superfamily.
One of the peptides that we identified, vi6a, has been chemically synthesized and more extensively characterized. As expected from its activity on mouse DRG neurons, vi6a, is active in mammalian systems. The activity of the native peptide observed on dorsal root ganglion cells (see Fig 2), was a decrease in the influx of calcium upon depolarization using high [K+]o, an activity that was also exhibited by the chemically synthesized peptide. Surprisingly however, when the peptide was injected into mice, both IC and IP, it elicited hyperactivity in the injected animals. The juxtaposition of the in vivo excitatory symptomatology and the inhibitory effect on Ca2+ influx in response to high [K+]o in DRG neurons was unexpected and difficult to explain; the excitatory phenotype elicited upon IP injection into mice is particularly noteworthy. Thus, vi6a has a unique profile of biological activity in mice, the molecular basis of which is currently being investigated.
Supplementary Material
Highlights.
A conotoxin, vi6a, was purified from the venom of C. virgo.
vi6a exhibited inhibitory activity in the calcium imaging of dissociated mouse lumbar DRG neurons.
Intracranial and intraperitoneal injection of vi6a resulted in hyperactivity in mice.
Related conotoxins were identified from other members of the Virgiconus clade.
Acknowledgments
This work was supported by a grant from the National Institute of General Medical Science, GM 48677. We thank Joanna Gajewiak for synthesizing the linear vi6a, William Low for MS analyses, Terry Merritt and Roxanne Ghaffarian for helping prepare the manuscript and My Huynh for assembling some of the figures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ETHICAL STATEMENT
The authors of this paper would like to confirm that they have read and accept the Elsevier ethics statement regarding ethical standards in conducting research, writing an article and authorship.
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
References
- 1.Buczek O, Bulaj G, Olivera BM. Conotoxins and the posttranslational modification of secreted gene products. Cell Mol Life Sci. 2005;62:3067–3079. doi: 10.1007/s00018-005-5283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colledge CJ, Hunsperger JP, Imperial JS, Hillyard DR. Precursor structure of ω-conotoxin GVIA determined from a cDNA clone. Toxicon. 1992;30:1111–1116. doi: 10.1016/0041-0101(92)90056-b. [DOI] [PubMed] [Google Scholar]
- 3.Favreau P, Stocklin R. Marine snail venom: use and trends in receptor and channel pharmacology. Curr Opinion in Pharmacology. 2009:594–601. doi: 10.1016/j.coph.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 4.Hillyard DR, Monje VD, Mintz IM, Bean BP, Nadasdi L, Ramachandran J, Miljanich G, Azimi-Zoonooz A, McIntosh JM, Cruz LJ, Imperial JS, Olivera BM. A new. Conus peptide ligand for mammalian presynaptic Ca2+ channels. Neuron. 1992;9:69–77. doi: 10.1016/0896-6273(92)90221-x. [DOI] [PubMed] [Google Scholar]
- 5.Kaas Q, Westermann JC, Halai R, Wang CK, Craik DJ. ConoServer, a database for conopeptide sequences and structures. Bioinformatics. 2008;24:445–446. doi: 10.1093/bioinformatics/btm596. [DOI] [PubMed] [Google Scholar]
- 6.Kaas Q, Yu R, Jin AH, Dutertre S, Craik DJ. ConoServer: updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res. 2012;40:D325–D330. doi: 10.1093/nar/gkr886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kauferstein S, Melaun C, Mebs D. Direct cDNA cloning of novel conopeptide precursors of the O-superfamily. Peptides. 2005;26:261–367. doi: 10.1016/j.peptides.2004.10.027. [DOI] [PubMed] [Google Scholar]
- 8.Kohn AJ. The Ecology of Conus in Hawaii. Ecology Monograph. 1959;29:47–90. [Google Scholar]
- 9.Leipold E, Hansel A, Olivera BM, Terlau H, Heinemann SH. Molecular interaction of delta-conotoxins with voltage-gated sodium channels. FEBS Lett. 2005;579:3881–3884. doi: 10.1016/j.febslet.2005.05.077. [DOI] [PubMed] [Google Scholar]
- 10.McIntosh JM, Hasson A, Spira ME, Gray WR, Li W, Marsh M, Hillyard DR, Olivera BM. A new family of conotoxins that blocks voltage-gated sodium channels. J. Biol. Chem. 1995;270:16796–16802. doi: 10.1074/jbc.270.28.16796. [DOI] [PubMed] [Google Scholar]
- 11.McIntosh JM, Jones RM. Cone venom--from accidental stings to deliberate injection. Toxicon. 2001;39:1447–1451. doi: 10.1016/s0041-0101(01)00145-3. [DOI] [PubMed] [Google Scholar]
- 12.Olivera BM. E.E. Just Lecture, 1996. Conus venom peptides, receptor and ion channel targets, and drug design: 50 million years of neuropharmacology. Molecular biology of the cell. 1997;8:2101–2109. doi: 10.1091/mbc.8.11.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olivera BM. Conus venom peptides: reflections from the biology of clades and species. Annual Review of Ecology, Evolution and Systematics. 2002;33:25–42. [Google Scholar]
- 14.Olivera BM, Gray WR, Zeikus R, McIntosh JM, Varga J, Rivier J, de Santos V, Cruz LJ. Peptide neurotoxins from fish-hunting cone snails. Science. 1985;230:1338–1343. doi: 10.1126/science.4071055. [DOI] [PubMed] [Google Scholar]
- 15.Olivera BM, Rivier J, Clark C, Ramilo CA, Corpuz GP, Abogadie FC, Mena EE, Woodward SR, Hillyard DR, Cruz LJ. Diversity of Conus neuropeptides. Science. 1990;249:257–263. doi: 10.1126/science.2165278. [DOI] [PubMed] [Google Scholar]
- 16.Puillandre N, Bouchet P, Duda TF, Jr, Kauferstein S, Kohn AJ, Olivera BM, Watkins M, Meyer C. Molecular phylogeny and evolution of the cone snails (Gastropoda, Conoidea) Mol Phylogenet Evol. 2014;78:290–303. doi: 10.1016/j.ympev.2014.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puillandre N, Koua D, Favreau P, Olivera BM, Stocklin R. Molecular phylogeny, classification and evolution of conopeptides. J Mol Evol. 2012;74:297–309. doi: 10.1007/s00239-012-9507-2. [DOI] [PubMed] [Google Scholar]
- 18.Robinson SD, Norton RS. Conotoxin Gene Superfamilies. Marin Drugs. 2014:6058–6101. doi: 10.3390/md12126058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Röckel D, Korn W, Kohn AJ. Manual of the Living Conidae. Wiesbaden, Germany: Verlag Christa Hemmen; 1995. [Google Scholar]
- 20.Shon K, Grilley MM, Marsh M, Yoshikami D, Hall AR, Kurz B, Gray WR, Imperial JS, Hillyard DR, Olivera BM. Purification, characterization and cloning of the lockjaw peptide from Conus purpurascens venom. Biochemistry. 1995;34:4913–4918. doi: 10.1021/bi00015a002. [DOI] [PubMed] [Google Scholar]
- 21.Shon K, Stocker M, Terlau H, Stühmer W, Jacobsen R, Walker C, Grilley M, Watkins M, Hillyard DR, Gray WR, Olivera BM. k-Conotoxin PVIIA: a peptide inhibiting the Shaker κ+ channel. J. Biol. Chem. 1998;273:33–38. doi: 10.1074/jbc.273.1.33. [DOI] [PubMed] [Google Scholar]
- 22.Smith NJ, Hone AJ, Memon T, Bossi S, Smith TE, McIntosh JM, Olivera BM, Teichert RW. Comparative functional expression of nAChR subtypes in rodent DRG neurons. Front Cell Neurosci. 2013;225 doi: 10.3389/fncel.2013.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teichert RW, Olivera BM, McIntosh JM, Bulaj G, Horvath MP. The Molecular Diversity of Conoidean Venom Peptides and their Targets: From Basic Research to Therapeutic Applications. In: King GF, editor. Venom to Drugs: Venom as a Source for the Development of Human Therapeutics. London: RSC Publishing; 2015. pp. 163–203. [Google Scholar]
- 24.Teichert RW, Smith NJ, Raghuraman S, Yoshikami D, Light AR, Olivera BM. Functional profiling of neurons through cellular neuropharmacology. Proc Natl Acad Sci U S A. 2012;109:1388–1395. doi: 10.1073/pnas.1118833109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terlau H, Olivera BM. Conus venoms: a rich source of novel ion channel-targeted peptides. Physiological Reviews. 2004;84:41–68. doi: 10.1152/physrev.00020.2003. [DOI] [PubMed] [Google Scholar]
- 26.Terlau H, Shon K, Grilley M, Stocker M, Stühmer W, Olivera BM. Strategy for rapid immobilization of prey by a fish-hunting cone snail. Nature. 1996;381:148–151. doi: 10.1038/381148a0. [DOI] [PubMed] [Google Scholar]
- 27.Wilson MJ, Zhang M-M, Gajewiak J, Azam L, Rivier J, Olivera B, Yoshikami D. Α- and β-subunit composition of voltage-gated sodium channels investigated with μ-conotoxins and the recently discovered μO§-conotoxin GVIIJ. J Neurophysiol. 2015:2289–2301. doi: 10.1152/jn.01004.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woodward SR, Cruz LJ, Olivera BM, Hillyard DR. Constant and Hypervariable Regions in Conotoxin Propeptides. Embo J. 1990;9:1015–1020. doi: 10.1002/j.1460-2075.1990.tb08204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhangsun D, Luo S, Wu Y, Zhu X, Hu Y, Xie L. Novel O-superfamily conotoxins identified by cDNA cloning from three vermivorous Conus species. Chem Biol Drug Des. 2006;68:256–265. doi: 10.1111/j.1747-0285.2006.00443.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.