Summary
Nager acrofacial dysostosis is a rare craniofacial syndrome characterized by facial anomalies (cleft palate, external ear abnormalities, and micrognathia) and limb defects due to haploinsufficiency of SF3B4 (or SAP49), a protein involved in pre-mRNA splicing. We describe a 31-year-old female patient with Nager syndrome who presented to our institution with synchronous, bilateral breast cancer. She was treated with neoadjuvant hormonal therapy, bilateral mastectomies, and adjuvant chemotherapy with dose-dense doxorubicin, cyclophosphamide, and paclitaxel (ddAC-T). No other germline mutations were identified in a 29-gene panel of known breast cancer-associated genes. To our knowledge, this is the first report of cancer in a patient with Nager syndrome. Somatic inactivating mutations of SF3B4 occur occasionally in breast cancer, and these findings support the idea that SF3B4 is a tumor suppressor. This study illustrates the importance of evaluating the risk of chronic disease in individuals with rare inherited disorders.
Keywords: Nager syndrome, breast cancer, cancer, SF3B4, SAP49, spliceosome, splicing
Nager syndrome, also known as Nager acrofacial dysostosis, is a rare disorder with an incidence of ~3 per 1,000,000 characterized by pre-axial upper limb anomalies and mandibulofacial dysostosis [1]. Fewer than 100 cases of Nager syndrome have been reported [2]. Infants with Nager syndrome frequently have respiratory and feeding problems that may require gavage feeding or gastrostomy tube placement. The high perinatal mortality rate (approximately 11 percent) is related to respiratory compromise [3]. However, advances in care now allow many individuals to survive childhood. It is unknown whether these adults have a greater risk of diseases in adulthood.
Most cases of Nager syndrome are sporadic, although both autosomal dominant and recessive inheritance are reported [4 5]. Mutations in SF3B4 (splicing factor 3B, subunit 4) leading to haploinsufficiency are identified in about 60 percent of affected individuals [6–8]; this is consistent with an older report describing a child diagnosed with Nager syndrome with a deletion of chromosomal region 1q12-q21, which contains the SF3B4 gene [9]. SF3B4 encodes spliceosome-associated protein 49 kD (SAP49), a component of the pre-mRNA spliceosome complex. Xenopus and zebrafish models of SF3B4 mutation demonstrates impaired neural crest differentiation [10 11], which appears to be due to increased apoptosis of neural crest rather than a proliferative defect [10]. In mice embryos, SF3B4 is expressed at a high level in fore limbs, hind limbs, and somites during early stages, consistent with its role in skeletal development [12]. Mutations in the splicing machinery, including the closely related splicing factor SF3B1, are a major cause of human myelodysplasia [13 14]. However, the risk of malignancy in patients with Nager syndrome is unknown.
A 31-year-old female with sporadic Nager syndrome presented with a non-tender left breast lump and nipple inversion. Consistent with this diagnosis, she had micrognathia, midface retrusion, downslanting palpebral fissures, and abnormal ears. She had a prior history of multiple craniofacial surgeries and hearing loss, and a mutation in SF3B4: c.1060dupC (Figure 1). This mutation has previously been reported to be associated with Nager syndrome [6], and results in a frameshift mutation that replaces an arginine with a proline and extends translation by 132 amino acids (p.Arg354Profs*132). There was a family history of postmenopausal breast cancer in two maternal great aunts but no other family history of breast or ovarian cancer. Diagnostic mammogram with left ultrasound-guided biopsy revealed invasive ductal carcinoma. Left axillary lymph node biopsy demonstrated tumor cells with the same receptor statuses as the primary tumor. The tumor was grade 2, estrogen receptor positive, progesterone receptor positive, and negative for human epidermal growth factor receptor 2 (HER2). A staging CT of chest, abdomen, and pelvis additionally demonstrated enlarged lymph nodes in the right axilla without any other signs of systemic disease. Biopsy of the right axillary nodes revealed invasive ductal carcinoma, also estrogen receptor positive, progesterone receptor positive, and HER2 negative. Diagnostic imaging of the right breast was unremarkable. Genetic testing with a panel of 29 known cancer risk genes did not reveal any other deleterious germline mutations, including in BRCA1, BRCA2, PALB2, TP53, PTEN (Comprehensive Cancer Panel, GeneDx). The patient was diagnosed with bilateral breast cancer, clinical stage T3 N1 M0 left, and T0 N1 M0 right. Treatment with curative intent was recommended.
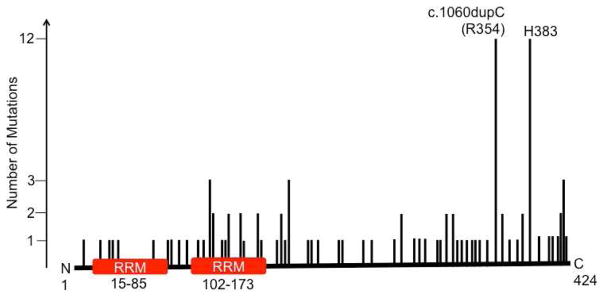
Figure 1. Map of SF3B4 mutations in cancer.
This schematic summarizes the locations of missense mutations in SF3B4. This figure is modified from data in the Cosmic database. Our patient has a c.1060dupC mutation, which results in an elongated protein. RRM = RNA recognition motif.
Due to proximity of the left axillary lymph nodes with the axillary vein, initial treatment was neoadjuvant endocrine therapy. She received anastrozole and goserelin for four 28-day cycles followed by bilateral modified radical mastectomies, revealing final pathologic stages of ypT2 N3 M0 (left; 10 of 14 lymph nodes involved) and ypT0 N0 M0 (right; 0/13 lymph nodes). Subsequently, she received adjuvant chemotherapy with doxorubicin, cyclophosphamide, and paclitaxel, interrupted briefly by Mediport infection. Following chemotherapy, she was treated with comprehensive chest wall radiation. She is now receiving hormonal therapy with anastrozole and goserelin and is participating in a trial investigating a novel agent. She recovered well from initial therapy and is currently doing well.
To the best of our knowledge, this is the first report of cancer in a patient with Nager syndrome. The patient’s youth, lack of identifiable risk factors or family history, and the presence of bilateral cancer suggests that Nager syndrome confers an elevated risk. This risk has not been recognized, possibly because there are fewer than 100 cases per year of Nager syndrome, and historically many individuals died in childhood. Analysis of cBioPortal [15] and Cosmic (cancer.sanger.ac.uk) [16] databases revealed that SF3B4 mutations have been discovered cancers of the bladder, breast, colorectal, and liver. However, this has not been identified as a driver and is infrequent. Interestingly, TCGA reports 10 cancers with the same c.1060dupC mutation in our patient as well as 2 other mutations at this site (Figure 1); 9 of these are breast cancers, 2 colorectal, and 1 ovarian. The c.1060dupC mutation causes a frameshift that inactivates SF3B4. There are also 12 cases of frameshift mutations at c.1147 (H383). These observations support the ideas that SF3B4 is a tumor suppressor whose haploinsufficiency can be tumorigenic, and that SF3B4 mutations are rare drivers for cancer growth.
The removal of introns from pre-mRNA by the spliceosome is essential for translation into functional proteins. SF3B4 is a subunit of the splicing factor SF3b complex, which is a component of both the U2 snRNP (contributes to the major spliceosome) and the U11/U12 di-snRNP (contributes to the minor spliceosome) [17]. SF3B4 directly binds both the pre-mRNA and the U2 snRNP, suggesting that it plays a role in helping the U2 snRNP bind to the branchpoint sequence of the pre-mRNA [18 19]. Inactivation of SF3B4 likely disrupts U2 binding to the branchpoint, which prevents efficient splicing. Splicing abnormalities are common in cancer and can occur either by mutation of splicing factors or splice sites [20 21]. Mutations are reported in 5′ and 3′ splice sites and branch sites, as well as exonic and intronic splicing enhancers and silencers. For example, in breast cancer, a mutation in the BRCA1 exonic splicing enhancer results in skipping of exon 18 and production of non-functional BRCA1 [22]. An example oncogenic splicing factors is the SR family of RNA-binding proteins that are essential for splicing and are commonly mutated in cancer [21]. When splicing is impaired, it is possible that certain un-spliced mRNAs encode proteins that exert dominant negative effects on signaling pathways and promote carcinogenesis. Another explanation for the possible association between Nager syndrome and cancer is that spliceosome mutations prevent cell differentiation. Cancer cells are often thought of as being de-differentiated, and replicative immortality is a hallmark of cancer, providing a possible mechanism of susceptibility to cancer. Given the known role of improper splicing on cancer risk and the unusual presentation of synchronous, bilateral breast cancer in a pre-menopausal woman, this patient’s cancer is most likely due to germline mutation in SF3B4.
Looking forward, this case leads to important questions to be addressed. First, how does mutated SF3B4 affect splicing and does it preferentially disrupt expression of tumor suppressors such as p53 or BRCA1? This could be addressed by employing RNA sequencing in SF3B4-null cells. Second, does Nager syndrome confer an increased risk of chronic diseases such as cancer? Now that care has improved and these individuals commonly survive to adulthood, cohort studies are needed to address this question. Furthermore, animal model experiments could be used to establish causation. For example, the aforementioned mouse model of SF3B4 mutation could be crossed with a mouse breast cancer model to assess whether mutation of SF3B4 confers a greater risk of developing breast cancer.
In conclusion, this study reports a case of Nager syndrome in a young woman with synchronous, bilateral breast cancer with no other cause. These data strongly support the idea that SF3B4 mutations increase risk of breast cancer. Moreover, it illustrates the importance of evaluating the risk of chronic disease in individuals with rare inherited disorders.
Acknowledgments
This work was supported in part by P30 CA014520. RAD is in the University of Wisconsin Medical Scientist Training Program (T32GM008692) and an ICTR TL1 trainee (supported under NIH awards UL1TR000427 and TL1TR000429). The content is the responsibility of the authors and does not necessarily represent the views of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lansinger Y, Rayan G. Nager syndrome. J Hand Surg Am. 2015;40(4):851–4. doi: 10.1016/j.jhsa.2014.10.064. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 2.Schlieve T, Almusa M, Miloro M, et al. Temporomandibular joint replacement for ankylosis correction in Nager syndrome: case report and review of the literature. J Oral Maxillofac Surg. 2012;70(3):616–25. doi: 10.1016/j.joms.2011.02.053. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 3.Ho AS, Aleshi P, Cohen SE, et al. Airway management in Nager Syndrome. Int J Pediatr Otorhinolaryngol. 2008;72(12):1885–8. doi: 10.1016/j.ijporl.2008.09.007. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 4.Lowry RB. The Nagar syndrome (acrofacial dysostosis): evidence for autosomal dominant inheritance. Birth Defects Orig Artic Ser. 1977;13(3C):195–202. [PubMed] [Google Scholar]
- 5.Hall BD. Nager acrofacial dysostosis: autosomal dominant inheritance in mild to moderately affected mother and lethally affected phocomelic son. Am J Med Genet. 1989;33(3):394–7. doi: 10.1002/ajmg.1320330321. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 6.Bernier FP, Caluseriu O, Ng S, et al. Haploinsufficiency of SF3B4, a component of the pre-mRNA spliceosomal complex, causes Nager syndrome. Am J Hum Genet. 2012;90(5):925–33. doi: 10.1016/j.ajhg.2012.04.004. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czeschik JC, Voigt C, Alanay Y, et al. Clinical and mutation data in 12 patients with the clinical diagnosis of Nager syndrome. Hum Genet. 2013;132(8):885–98. doi: 10.1007/s00439-013-1295-2. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 8.Petit F, Escande F, Jourdain AS, et al. Nager syndrome: confirmation of SF3B4 haploinsufficiency as the major cause. Clin Genet. 2014;86(3):246–51. doi: 10.1111/cge.12259. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 9.Waggoner DJ, Ciske DJ, Dowton SB, et al. Deletion of 1q in a patient with acrofacial dysostosis. Am J Med Genet. 1999;82(4):301–4. doi: 10.1002/(sici)1096-8628(19990212)82:4<301::aid-ajmg5>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 10.Devotta A, Juraver-Geslin H, Gonzalez JA, et al. Sf3b4-depleted Xenopus embryos: A model to study the pathogenesis of craniofacial defects in Nager syndrome. Dev Biol. 2016 doi: 10.1016/j.ydbio.2016.02.010. published Online First: Epub Date. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.An M, Henion PD. The zebrafish sf3b1b460 mutant reveals differential requirements for the sf3b1 pre-mRNA processing gene during neural crest development. Int J Dev Biol. 2012;56(4):223–37. doi: 10.1387/ijdb.113383ma. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruiz-Lozano P, Doevendans P, Brown A, et al. Developmental expression of the murine spliceosome-associated protein mSAP49. Dev Dyn. 1997;208(4):482–90. doi: 10.1002/(SICI)1097-0177(199704)208:4<482::AID-AJA4>3.0.CO;2-E. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida K, Sanada M, Shiraishi Y, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64–9. doi: 10.1038/nature10496. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 14.Makishima H, Visconte V, Sakaguchi H, et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood. 2012;119(14):3203–10. doi: 10.1182/blood-2011-12-399774. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forbes SA, Beare D, Gunasekaran P, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue):D805–11. doi: 10.1093/nar/gku1075. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarn WY, Steitz JA. Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge. Trends Biochem Sci. 1997;22(4):132–7. doi: 10.1016/s0968-0004(97)01018-9. [DOI] [PubMed] [Google Scholar]
- 18.Champion-Arnaud P, Reed R. The prespliceosome components SAP 49 and SAP 145 interact in a complex implicated in tethering U2 snRNP to the branch site. Genes Dev. 1994;8(16):1974–83. doi: 10.1101/gad.8.16.1974. [DOI] [PubMed] [Google Scholar]
- 19.Golas MM, Sander B, Will CL, et al. Molecular architecture of the multiprotein splicing factor SF3b. Science. 2003;300(5621):980–4. doi: 10.1126/science.1084155. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 20.Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J Cell Sci. 2006;119(Pt 13):2635–41. doi: 10.1242/jcs.03053. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 21.Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64(21):7647–54. doi: 10.1158/0008-5472.CAN-04-1910. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 22.Mazoyer S, Puget N, Perrin-Vidoz L, et al. A BRCA1 nonsense mutation causes exon skipping. Am J Hum Genet. 1998;62(3):713–5. doi: 10.1086/301768. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]