Do but consider what an excellent thing sleep is; it is so inestimable a jewel that, if a tyrant would give his crown for an hour's slumber, it cannot be bought; of so beautiful a shape is it, that though a man lie with an Empress, his heart cannot beat quiet till he leaves her embracements to be at rest with the other: yea, so greatly indebted are we to this kinsman of death, that we owe the better tributary, half of our life to him: and there is good cause why we should do so: for sleep is that golden chain that ties health and our bodies together.—Thomas Dekker (The Gull's Hornbook, 1609)
Circadian rhythms are endogenously generated rhythms that approximate the length of a day and a night. They are found in most organisms whose life span is counted in days or greater units, rather than hours.1 They enable the organism to anticipate, rather than passively adjust to, the changes imposed by alternation between day and night. These changes relate to positive functions such as vision and negative effects such as ultraviolet light damage. They also include indirectly generated alternations such as temperature, availability of food and prevalence of predators.
PHYSIOLOGY
In mammals, circadian rhythms are governed by a paired master oscillator in the suprachiasmatic nuclei (SCN), which controls slave oscillators throughout the body.2,3 The SCN determines the individual's free-running circadian period, τ,4 which in man is reported to average 24.2 h in the absence of light cues.5 Our laboratory has observed a range of 23.83-25.00 h in blind individuals.6 Variability within the species, whereby individuals reach peak performance at different times of day, may have conveyed an evolutionary advantage to early human societies. In a normal individual the phase of the SCN oscillator is entrained each day to the external light/dark cycle by information relayed through the retinohypothalamic tract.7,8
In vertebrates, sleep has evolved as a sustained period of physical inactivity during a specific part of the circadian cycle.9 The distribution of rest and activity between the night and the day varies between taxa. Our reptilian ancestors, being exothermic, were active during the day (diurnal), but the endothermic mammals that radiated from these species were nocturnal. From there, various mammals (including most primates) reverted to diurnality through secondary modifications, notably in the visual system. The retina was recolonized by colour-sensitive cones (not required for night vision) and the higher visual centres were rewired.9-13
The parameters within the circadian system that define nocturnality and diurnality are only partly understood.14 As a general rule, nocturnal species have a τ<24 hours, diurnal species have a τ>24 hours.15 In addition, the composition of visual photoreceptors seems important. People who are colour blind through complete lack of cone photoreceptors tend to be photophobic, and on the Micronesian island of Pingelap, where the condition is common, some affected individuals specialize in night fishing.16
GENES AND MOLECULES
Information on how circadian rhythms are generated at molecular level comes mainly from studies in mice. The mechanism depends on tightly controlled concerted coexpression of specific clock genes. Most of these genes are remarkably conserved amongst coelomates17—including insects, molluscs18 and vertebrates—although the precise roles of specific components have drifted during evolution. At the centre of the machinery in mammals are the Period (Per1, Per2 and Per3) and Cryptochrome (Cry1 and Cry2) genes. The protein products of all these oscillate over the 24-hour cycle by inhibiting their own promoters operating in an intricate negative feedback loop.
Permanently attached to these promoters is a heterodimer of two transcriptional activator proteins, CLOCK and BMAL1.19 The undisturbed activation of the Per and Cry promoters leads to the accumulation of Per and Cry mRNA, which in the suprachiasmatic nucleus takes place during the latter part of the day and the early part of the night. Exported to the cytoplasm, these mRNA transcripts are translated into the proteins PER1, PER2 and PER3 and CRY1 and CRY2, the concentrations of which peak during the middle of the night. The production of sufficient quantities of these proteins allows the formation of trimeric complexes between CRY, PER and an additional clock component, casein kinase I (CK I). All combinations of the different PER and CRY types seem to be possible; in addition, the two CK I forms δ and ε are apparently interchangeable. This trimeric complex enters the suprachiasmatic nucleus, where it binds to the CLOCK:BMAL1 complex and prevents its positive effect on the transcription of Per and Cry mRNA.3 Phosphorylation by casein kinase I appears important both in targeting clock gene proteins for transport into the nucleus and in their eventual degradation. This prevents PER and CRY protein from acting on their own promoters, and their temporary absence once again allows the unperturbed action of BMAL1 and CLOCK on the Per and Cry promoters, starting the whole cycle anew. An added twist to this negative feedback loop is a positive one; the PER 2 protein has been shown to be a positive regulator of BMAL1 gene expression, which also cycles whilst the level of its partner CLOCK remains constant.20
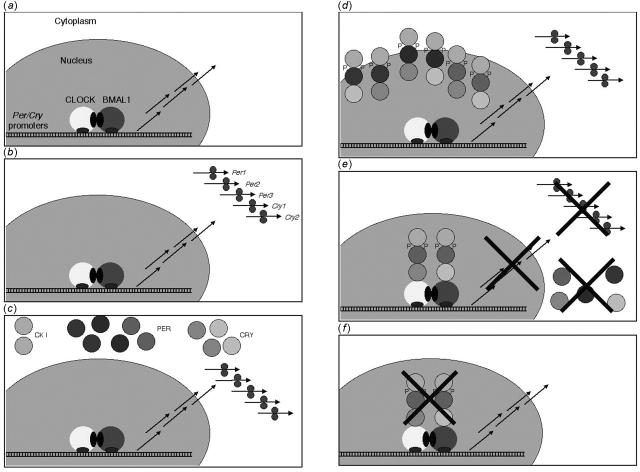
Clearly the mechanisms that regulate each person's τ with such precision are complex (Figure 1), so it is not surprising that mutations in the different clock genes can affect the circadian output. The Clock gene was identified in a mouse strain with a long and unstable circadian period,21 and knockout mice have been produced lacking the different Per and Cry genes and combinations thereof.3 Whereas single knockouts of either one of the Per or Cry genes will alter the circadian period, functional double knockouts of Per1 and Per222 and of Cry1 and Cry223 abolish circadian rhythmicity altogether. The effect of perturbing the expression of Per3 is only to shorten the circadian period.24 A shortened period is also observed in the τ mutant strain of the Syrian golden hamster,25 in which a point mutation in casein kinase ε decreases its ability to phosphorylate both positive and negative regulators of the circadian mechanism.26,27 The net result is likely to be less degradation of the phosphorylated protein.
Figure 1.
The 24-hour cycle of clock gene expression. (a) In the morning, the promoters of the Cry and Per genes are activated by the CLOCK and BMAL1 proteins, producing mRNA transcripts that are exported into the cytoplasm. (b) During the day, ribosomes bind to these mRNA molecules and translate their genetic code into proteins. (c) In the latter part of the day, PER (1, 2 and 3) and CRY (1 and 2) proteins accumulate in the cytoplasm, where CK I (δ and ε) are present constitutively. (d) In the evening, CRY, PER, and CK I molecules bind to each other in different combinations. PER is phosphorylated by CK I, and the complexes enter the nucleus in a coordinated fashion at night. (e) During the rest of the night, the activation of the Cry and Per promoters by CLOCK:BMAL1 is inhibited by the CRY:PER:CK I complex. As a result, production of Cry and Per mRNA is interrupted, which in turn interrupts production of CRY and PER proteins. (f) As both the mRNA and the proteins are short-lived, they will gradually disappear towards the morning. The cycle is complete, and the production of Cry and Per mRNA starts again
SLEEP DISORDERS
Sleep has famously been described as being ‘of the brain, by the brain, and for the brain’.28 Its relation to the circadian clock is less simple to describe. Some disorders of sleep are unrelated to circadian rhythms29; others are undoubtedly related to it, particularly the advanced and delayed sleep phase syndromes.30,31 In these conditions, sleep occurs either abnormally early or abnormally late. This could theoretically be caused either by an abnormal τ or by abnormal timing of the sleep phase within a circadian cycle of normal periodicity.
From what we can tell from their sequence features, their function in in vitro studies, and the rather limited insights we possess from ex vivo material, human clock genes behave in a very similar fashion to murine clock genes. But our secondary reversion to a diurnal lifestyle (which did not happen in the mouse) will have been accompanied by limited but important changes in the clock gene machinery, and these should soon be identifiable through comparative analysis of the respective genomes.
In addition, differences are seen between members of the human race. Polymorphisms in clock genes can be related to circadian parameters, and the most famous finding so far is a large family where advanced sleep phase syndrome seems to be inherited as a single-gene defect.32 The condition manifests itself in this family with advanced melatonin, temperature, and sleep/wake rhythms co-segregating with a missense mutation in the Per2 gene.33 Because of the non-homologous aminoacid substitution, the resulting PER2 protein is phosphorylated less efficiently by casein kinase than the native one—an observation that offers a satisfying mechanistic explanation for the similarity between this phenotype and that of the τ hamster, whose missense mutation in casein kinase I ε results in essentially the same net effect.
MORNING AND EVENING PREFERENCES
It is a common observation that some individuals are most alert in the morning, others in the evening. These preferences can be quantified by the Horne-Östberg (HO) questionnaire, in which a low score indicates evening preference34 and a long τ.35 Katzenberg and co-workers36 studied the relation between HO score and a single-nucleotide polymorphism (SNP) at position 3111 in the Clock gene transcript. Although this locus does not alter the aminoacid sequence of the CLOCK protein, it could nonetheless be important for the stability and translatability of the transcript. Individuals homozygous or heterozygous for the 3111C allele of Clock were reported to have an HO score average of two points higher than 3111T homozygotes. Our laboratory attempted to confirm this finding by a four-pronged approach.6 First, we sought a correlation between the Clock3111 polymorphism and extreme HO score in a population of 484 individuals. Secondly, we sought a correlation with τ as determined in blind subjects with free-running circadian rhythm. Thirdly, we compared the prevalence of this polymorphism in patients with delayed sleep phase syndrome and normal controls. Fourthly, we investigated the expression levels of a reporter gene construct carrying the two different forms of the relevant section of the 3′-UTR of Clock. In none of these studies did we observe any significant effect of this polymorphism.
The gene associated with evening preference that has produced the most interesting results to date in humans is Per3. In a Japanese population, Ebisawa and colleagues37 reported that a rare single-nucleotide polymorphism causing an aminoacid substitution correlated with delayed sleep phase syndrome. Our laboratory has studied a more dramatic genetic polymorphism, initially described but not characterized in Ebisawa's paper, encoding an 18-aminoacid tandem repeat sequence, of which humans have either four or five successive copies in each of their Per3 alleles.38 By comparing HO-characterized subjects whose scores were around the mean for their gender and age group with those with extreme evening and morning preference, we were able to distinguish an excess prevalence of the shorter repeat allele in subjects with extreme evening preference. Extending the study to a cohort of patients with delayed sleep phase syndrome, we showed that the association with the shorter allele was even stronger in this population. Thus, although no physiological studies have formally linked the extremes of evening preference and/or long τ with delayed sleep phase syndrome, it would appear from the convergence of their Per3 genotype that such a study is not only worthwhile but long overdue.
CONCLUSION
In mice, single null mutations in clock genes seem to correlate neatly with circadian phenotype in a monogenic mendelian inheritance pattern. In man, the known human clock gene differences appear merely to predict a greater or lesser propensity. One reason for this difference is that most of the mouse models studied so far have been engineered to abolish the function of a specific gene, rather than carrying a more or less altered form of it. Another is that laboratory rodent strains are highly inbred and thus much more homogeneous with respect to all other clock genes. The human circadian genotype, being a polygenic trait, is more akin to a hand of cards. Most of us will have a hand containing average cards or a balanced mixture of high and low ones. Only the hands that contain predominantly low or high cards will stand out. The great majority of us function normally with our allotted circadian phenotype and its interaction with our environment, much as we are able to deal with other aspects of our genetic inheritance. But the minority who have been dealt a dud hand of the clock genes card game deserve more sympathy and clinical help than they are often accorded.
Our culture trends to associate early waking with virtue and industry, and late sleeping with vice and lassitude. Lack of conformity with this norm is not always a matter of choice: clearly, some people are genetically programmed for an extreme diurnal preference.
References
- 1.Kippert F, Saunders DS, Blaxter ML. Caenorhabditis elegans has a circadian clock. Curr Biol 2002;12: R47-9 [DOI] [PubMed] [Google Scholar]
- 2.Yamazaki S, Numano R, Abe M, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science 2000;288: 682-5 [DOI] [PubMed] [Google Scholar]
- 3.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature 2002;418: 935-41 [DOI] [PubMed] [Google Scholar]
- 4.Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science 1990;247: 975-8 [DOI] [PubMed] [Google Scholar]
- 5.Czeisler CA, Duffy JF, Shanahan TL, Brown EN, Mitchell JF, Rimmer DW, et al. Stability, precision, and near-24-hour period of the human circadian pacemaker. Science 1999;284: 2177-81 [DOI] [PubMed] [Google Scholar]
- 6.Robilliard D, Archer SN, Arendt J, et al. The 3111Clock gene polymorphism is not associated with sleep and circadian rhythmicity in phenotypically characterized human subjects. J Sleep Res 2002;11: 305-12 [DOI] [PubMed] [Google Scholar]
- 7.Panda S, Sato TK, Castrucci AM, et al. Melanopsin (Opn4) requirement for normal light-induced circadian phase shifting. Science 2002;298: 2213-16 [DOI] [PubMed] [Google Scholar]
- 8.Ruby NF, Brennan TJ, Xie X, et al. Role of melanopsin in circadian responses to light. Science 2002;298: 2211-13 [DOI] [PubMed] [Google Scholar]
- 9.Kavanau JL. Origin and evolution of sleep: roles of vision and endothermy. Brain Res Bull 1997;42: 245-64 [DOI] [PubMed] [Google Scholar]
- 10.Menaker M, Tosini G. The evolution of vertebrate circadian system. In: Honma K, Honma S, eds. Circadian Organization and Oscillatory Coupling. Sapporo: Hokaido University Press, 1996
- 11.von Schantz M. The relationship between the retina and the circadian clock in mammals. Med Sci Monit 1998;4(suppl. 1): 35-45 [Google Scholar]
- 12.von Schantz M, Szél Á, van Veen T, Farber DB. Expression of phototransduction genes in the ground squirrel retina. Invest Ophthalmol Vis Sci 1994;35: 2558-66 [PubMed] [Google Scholar]
- 13.Szél Á, Lukats A, Fekete T, Szepessy Z, Röhlich P. Photoreceptor distribution in the retinas of subprimate mammals. J Opt Soc Am A Opt Image Sci Vis 2000;17: 568-79 [DOI] [PubMed] [Google Scholar]
- 14.Jiao YY, Lee TM, Rusak B. Photic responses of suprachiasmatic area neurons in diurnal degus (Octodon degus) and nocturnal rats (Rattus norvegicus). Brain Res 1999;817: 93-103 [DOI] [PubMed] [Google Scholar]
- 15.Aschoff J. Circadian rhythms: influences of internal and external factors on the period measured in constant conditions. Z. Tierpsychol 1979;49: 255-49 [DOI] [PubMed] [Google Scholar]
- 16.Sacks O. The Island of the Colorblind. New York: Alfred A. Knopf, 1996
- 17.Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet 2001;2: 702-15 [DOI] [PubMed] [Google Scholar]
- 18.Constance CM, Green CB, Tei H, Block GD. Bulla gouldiana period exhibits unique regulation at the mRNA and protein levels. J Biol Rhythms 2002;17: 413-27 [DOI] [PubMed] [Google Scholar]
- 19.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell 2001;107: 855-67 [DOI] [PubMed] [Google Scholar]
- 20.Shearman LP, Sriram S, Weaver DR, et al. Interacting molecular loops in the mammalian circadian clock. Science 2000;288: 1013-19 [DOI] [PubMed] [Google Scholar]
- 21.Vitaterna MH, King DP, Chang A-M, et al. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 1994;264: 719-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng BH, Albrecht U, Kassik K, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 2001;105: 683-94 [DOI] [PubMed] [Google Scholar]
- 23.van der Horst GY, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 1999;398: 627-30 [DOI] [PubMed] [Google Scholar]
- 24.Shearman LP, Jin X, Lee C, Reppert SM, Weaver DR. Targeted disruption of the mPer3 gene: subtle effects on circadian clock function. Mol Cell Biol 2000;20: 6269-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ralph MR, Menaker M. A mutation of the circadian system in golden hamsters. Science 1988;241: 1225-7 [DOI] [PubMed] [Google Scholar]
- 26.Lowrey PL, Shimomura K, Antoch MP, et al. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 2000;288: 483-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eide EJ, Vielhaber EL, Hinz WA, Virshup DM. The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase I epsilon. J Biol Chem 2002;277: 17248-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hobson JA. Sleep. New York: Scientific American Library, 1989
- 29.Mignot E. Genetics of narcolepsy and other sleep disorders. Am J Hum Genet 1997;60: 1289-302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dijk DJ, Lockley SW. Integration of human sleep-wake regulation and circadian rhythmicity. J Appl Physiol 2002;92: 852-62 [DOI] [PubMed] [Google Scholar]
- 31.Wijnen H, Boothroyd C, Young MW, Claridge-Chang A. Molecular genetics of timing in intrinsic circadian rhythm sleep disorders. Ann Med 2002;34: 386-93 [DOI] [PubMed] [Google Scholar]
- 32.Jones CR, Campbell SS, Zone SE, et al. Familial advanced sleep-phase syndrome: a short-period circadian rhythm variant in humans. Nat Med 1999;5: 1062-5 [DOI] [PubMed] [Google Scholar]
- 33.Toh KL, Jones CR, He Y, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 2001;291: 1040-3 [DOI] [PubMed] [Google Scholar]
- 34.Horne JA, Östberg O. A self-assessment questionnaire to determine morningness-eveningness in human circadian rhythms. Int J Chronobiol 1976;4: 97-110 [PubMed] [Google Scholar]
- 35.Duffy JF, Rimmer DW, Czeisler CA. Association of intrinsic circadian period with morningness-eveningness, usual wake time, and circadian phase. Behav Neurosci 2001;115: 895-9 [DOI] [PubMed] [Google Scholar]
- 36.Katzenberg D, Young T, Finn L, et al. A CLOCK polymorphism associated with human diurnal preference. Sleep 1998;21: 569-76 [DOI] [PubMed] [Google Scholar]
- 37.Ebisawa T, Uchiyama M, Kajimura N, et al. Association of structural polymorphisms in the human period3 gene with delayed sleep phase syndrome. EMBO Rep 2001;2: 342-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Archer SN, Robilliard D, Skene DJ, et al. A length polymorphism in the circadian clock gene Per3 is linked to delayed sleep phase syndrome and extreme diurnal preference. Sleep 2003;26: 413-15 [DOI] [PubMed] [Google Scholar]