Abstract
Drosophila melanogaster is a well-established model organism to understand biological processes and study human diseases at the molecular-genetic level. The central nervous system (CNS) of Drosophila larvae is widely used as a model to study neuron development and network formation. This has been achieved by using various genetic manipulation tools such as microinjection to knock down certain genes or over-express proteins for visualizing the cellular activities. However, visualization of an intact-live neuronal response in larva's Central Nervous System (CNS) is challenging due to robust digging/burrowing behaviour that impedes neuroimaging. To address this problem, dissection is used to isolate and immobilize the CNS from the rest of the body. In order to obtain a true physiological response from the Drosophila CNS, it is important to avoid dissection, while the larva should be kept immobilized. In this paper, a series of microfluidic clamps were investigated for intact immobilization of the larva. As a result, an optimized structure for rapid mechanical immobilization of Drosophila larvae for CNS imaging was determined. The clamping and immobilization processes were characterized by imaging and movement measurement of the CNS through the expression of genetically encoded Calcium sensor GCaMP5 in all sensory and cholinergic interneurons. The optimal structure that included two 3D constrictions inside a narrowed channel considerably reduced the internal CNS capsule movements. It restricts the CNS movement to 10% of the motion from a glued larva and allows motion of only 10 ± 30 μm over 350 s immobilization which was sufficient for CNS imaging. These larva-on-a-chip platforms can be useful for studying CNS responses to sensory cues such as sound, light, chemosensory, tactile, and electric/magnetic fields.
I. INTRODUCTION
Drosophila melanogaster (normally known as fruit fly or vinegar fly) is a model organism with a simple cellular and neuronal system and short developmental period. Its transparent body in the embryonic and larval stages makes it an ideal tool for genetic studies using advanced live imaging methods. Therefore, D. melanogaster is a great model organism for studying human biology and diseases at the molecular-genetic level. It is used to gain understanding of neurobiological development and physiological, behavioral, biochemical, or genetic processes at the molecular level that can then be co-related with disease mechanisms in mammals or humans.1–3
Similar to other insects, the Drosophila life cycle passes through embryonic, larval, and pupal stages prior to becoming an adult fly. As a model organism, each developmental stage of the Drosophila has specific applications. Drosophila has been a powerful tool for geneticists and developmental biologists due to its short generation time, small size, and significant similarity to human genome. At the larval stage, Drosophila contains different types of sensory neurons, which are formed in a sectional arrangement (Fig. 1). They allow the larvae to sense various environmental cues (e.g., mechanical, visual, and chemical) and transmit signals to the central nervous system (CNS) to generate response signals for stereotypic motor behaviors. This simple neural circuit is used for studying numerous developmental-genetic and neurobiological problems primarily through deploying surgical, histological, transgenic, and behavioral methods.4,5
FIG. 1.
Schematic of a 3rd instar larva expressing green fluorescent protein (GFP) in all cholinergic neurons as driven by Cha-Gal4, UAS-GFP transgenes.
The larva is a mobile organism and is constantly moving and seeking food. However, in a variety of live imaging protocols, the larva should be completely immobilized. Additionally, the digging and burrowing behaviour exhibited by the larva even when it is physically held stationary makes the high-resolution neuroimaging study of the larva's CNS very challenging. This challenge is associated with the anterior structure of Drosophila that includes the cephalo-pharyngeal skeleton (CPS) and sets of skeletal muscles, which are segmentally connected to the body wall. The CPS is actuated by specialized muscles to cause digging into food substrates.6 In order to perform in vivo imaging in Drosophila, the digging and burrowing movement has to be eliminated and the whole-larva body should be made completely stationary during the imaging.
Although conventional immobilization protocols, which use dissection or anaesthetic drugs7 (chloroform and isofluorane), efficiently stop the CPS motion, they also attenuate neural activities and affect animals' neurophysiological status. The larvae can also be attached to a substrate using an adhesive or glue in order to immobilize without anaesthetics. However, adhesive attachment is an irreversible method, which does not allow one to perform further behavioural or developmental studies on the mobile larva. Ideally, immobilization has to be performed in a simple and reversible manner while still allowing sensory stimulus to affect the larva. Recently, miniaturized microfluidic devices have been developed for immobilization of microorganisms, and these devices could be used for this purpose.
Microfluidic lab-on-chip devices are increasingly being used for studying model organisms such as Caenorhabditis elegans8–10 and Drosophila.11–14 These devices have the ability to automate immobilization of these small organisms and can be applied for in vivo visualization and tracking of cellular and physiological responses. Microfluidic-based immobilization techniques for C. elegans have been well developed using chemical (CO2) or mechanical [tapering microchannels or encapsulation using deflectable polydimethylsiloxane (PDMS) membranes] approaches.15–19
Furthermore, various microfluidic devices have also been developed for improving the throughput of the assays performed on embryos20–27 such as microinjection,20–24 self-assembly of eggs and morphogenesis,25,26 and developmental studies.27 However, very few devices have been developed for on-chip larval studies. Immobilization of Drosophila larvae is more complex than other small model organisms such as C. elegans due to the stronger forces that the larva is capable of generating. The strong forces are a result of the size and the number of muscles in the larva compared to C. elegans. In addition, the internal organs of the larva can loosely move inside its hemolymph-filled body cavity even if the outer body is completely immobilized by encapsulation. This makes the visualization of organs such as the CNS, brain, and gut even more difficult. Moreover, the Drosophila larvae use a peristaltic type of motion as compared to other micro model organisms such C. elegans, which also makes its immobilization more complex.
Recently, mechanical encapsulation,11 CO2 anaesthetic exposure,12 and mechanical clamp13 have been used to immobilize the body wall of the Drosophila larva in order to visualize the internal organ of interest such as CNS inside the body. They allow whole-larva body compression inside the chip so that neuronal transport processes,11 sensory neuron regeneration upon injury,12 and cardiac activities can be visualized. Mechanical encapsulation can efficiently reduce the larva's motion for a short period. However, it cannot allow one to capture high-resolution images of the internal organs such as CNS over long period. The CO2 exposure combined with mechanical capsulation can increase the duration of the immobilization. However, the use of anaesthetic leads to spurious neurobehavioral responses and the use of encapsulation prevents the exposure of the larva to external sensory stimulus.
To address the abovementioned challenges associated with microfluidic-based Drosophila larva immobilization techniques, we recently developed a 3D mechanical clamp to immobilize the larva for imaging the neurological response of D. melanogaster to the auditory stimulus.13 However, a systematic study on immobilization of the Drosophila larva has not been performed. Here, a systematic analysis of various mechanical constrictions incorporated into microfluidic channels was conducted in order to optimize the process of rapid loading and mechanical immobilization of Drosophila larvae with proper orientation for CNS imaging. The immobilization designs were studied by quantitative imaging and movement assessment of the CNS through expression of genetically encoded Calcium sensor GCaMP5.28 Our devices are engineered to first stop the larva's whole body locomotion and then immobilize the internal organs of interest such as CNS that may move due to ongoing motor movements and the resulting internal hemolymph displacements. Our larva-lab-on-a-chip platform will also be useful for studying CNS responses to sensory cues including sound, chemosensory, light, and electric/magnetic fields.
II. MATERIALS AND METHODS
A. Animal preparation
Larvae of the genotype w, Cha-Gal4/CyO; UAS-GCaMP5/TM3, Sb were used for CNS activity imaging in response to external stimulations. In this protocol, hetereozygotes and homozygotes were not separated before testing. Expression of the GCaMP5 GECI was conducted using the Gal4/UAS system.29 Through standard fly crosses, a stable fly stock was created containing two transgenes, namely: (1) Cha-Gal4—A promoter sequence of CholineAcetyltransferase (Cha) driving the expression of the Gal4 transcription factor30 and (2) UAS-GCaMP5—a transgene contains the binding sites for the Gal4 transcription factor.28 Thus, in the Cha-Gal4/CyO; UAS-GCaMP5/TM3 strain, all sensory and central neurons that express the CholineAcetyltransferase gene express the GCaMP5 calcium sensor. The GCaMP calcium sensor is circularly permuted protein containing the Green Fluorescent Protein (GFP), Calcium binding protein called Calmodulin, and the M13 (Calmodulin binding) peptide.31 Influx of Ca2+ during neuronal activity triggers a conformational change of GCaMP so that solvent access to the chromophore is prevented, thus resulting in a higher level of fluorescence.32 GCaMP5 is a recently developed high signal-to-noise ratio calcium sensor.28 This genotype was generated through a standard genetic crossing scheme. Larvae in the 3rd instar stage were isolated using a fine brush and washed with distilled water and dried on a tissue paper before loading into the chips.
B. Experimental setup
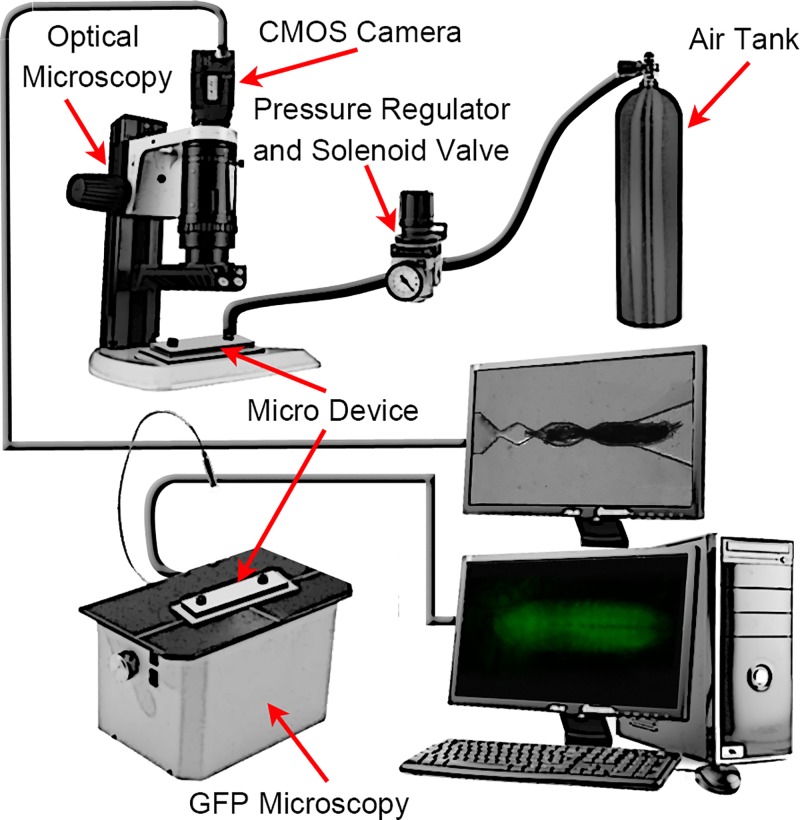
The experimental setup (Fig. 2) consisted of four modules, i.e., pneumatic system, optical system, the microfluidic devices, and image acquisition and processing software. The fluidic system, which was used to introduce the larva into the device and pressurize it during imaging, consisted of a pressurized air tank regulated at 400 kPa pressure with a regulator (2000 series regulator, ARO, Ingersoll Rand, USA) and a solenoid valve (S10 MM-30-12-3, 3-way normally closed, Pneumadyne, Inc., USA). The optical system and digital camera (Flea3 FLs-U3-32S2C, Point Grey Research, Inc., Canada) were used to image and record the immobilization process. The dissecting microscope (S8 APO, Leica, Canada) with low magnification was used to image and record the loading of the larvae and the fluorescent microscope (Model 500 LumaScope, Etaluma, Inc., CA) with higher magnification (40× objective) was used to record the CNS motions in GFP mode. The image acquisition software (flyCapture2©, Etaluma© and Labview©) was used for image recording and data analysis. The microfluidic device was mounted on the dissection microscope to observe the animal loading and record the whole-larva body movement. A fluorescent microscope (Excitation 457–493 nm; Emission 508–552 nm) was used to record CNS movements.
FIG. 2.
The experimental setup used for larva loading, sequential image recording, and processing. The experimental setup consisted of four modules, i.e., fluidic unit (air tank, pressure regulator and solenoid valve), optical system (optical microscope, CMOS camera, and GFP microscope), microfluidic devices (Micro device), and image acquisition software (ImageJ).
C. Device fabrication
The devices for immobilization of Drosophila larvae were fabricated by casting Polydimethylsiloxane (PDMS, Sylgard SYLGARD® 184, Dow Corning, USA) on 3D printed master mold. First, the 3D mold was designed in Solidworks software and converted into a STL format. A 3D printer (Projet HD 3000 from 3D Systems, Material: VisiJet® EX200 Plastic) was used to print the design from the STL file in hard plastic part. Next, 1 cm silicone tubes (3/16″ID × 5/16″OD, Cole-Parmer Canada Inc.) were placed on the reservoirs as the inlet/outlet. Subsequently, the uncured PDMS (10:1 ratio base:agent) was poured on the mold and then cured at room temperature for 24 h. Finally, the cured PDMS was peeled off and bonded to the glass slides (80 s, 50 W, plasma oxygen) to form a complete device.
D. Data acquisition and processing
Image sequences captured for each larva were quantified by Labview© software to measure the movement of the whole body and CNS movement during the time that larva tried to release itself from the trap. The RGB image sequences (2 f/s) for each video were converted to 8-bit grey scale images. Then, the pixel value of 255 was assigned for the larva's body or its CNS, while the background was adjusted to the pixel value of 0. Subsequently, the pixel centre of mass of both larva's body and its CNS was measured in the entire image stack and recorded.
III. RESULTS AND DISCUSSION
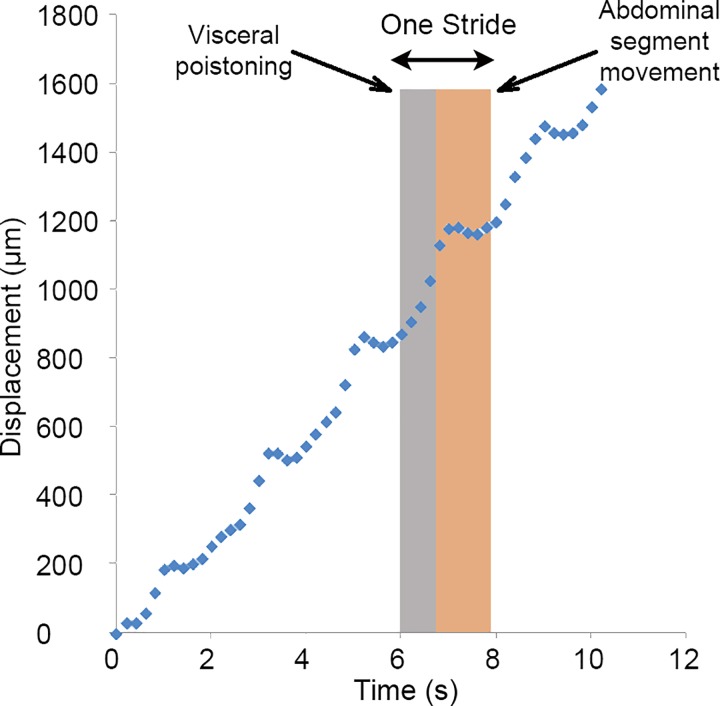
The Drosophila larva is highly mobile and its internal organs cannot be sharply imaged without complete immobilization. The locomotion of the larva follows a characteristic pattern which can be observed by tracking the larva's centre of the mass (CM) over time as shown in Fig. 3. As described in Sec. II D, the CM of the larvae was determined by recording an image sequence (0.2 s intervals) of the larvae's locomotion inside a 2 mm × 2 mm first. Next, the images were converted to binary images with the larvae as white (255) and the background was black (0). Then, the CM of larva was calculated by finding the centre of the white region using ImageJ. The larva crawls using a repetitive cycle of motion called strides, where each stride is composed of two phases.2 In the first phase called visceral pistoning movement, the head, tail, and gut move slowly forward due to the internal muscle contractions in the head and tail, while most of the abdominal segments remain firmly attached to the substrate. This results in the forward motion of the centre of mass of the larva as shown in Fig. 3. In the second phase, the tail and head are attached to the substrate and the peristaltic motion of the body wall moves the abdominal segments in the direction of the crawling. The CM of the larva remains nearly stationary in this phase as shown in Fig. 3. A suitable device that will immobilize the larva for imaging has to stop or interrupt this pistoning motion of the larva. Furthermore, the internal organs of interest such as the brain or the CNS capsule that need to be imaged are loosely attached to the outer body wall and are free to move inside the hemolymph-filled body cavity. Therefore, even if the outer body is completely immobilized by encapsulation, the contraction of internal muscles due to the digging and burrowing behaviour can induce hemolymph motion resulting in movement of the loosely held organs. Consequently, a desired immobilization system should be able to not only immobilize whole-larva body movements but also inhibit the motion of internal organs such as CNS. In the rest of the paper, a set of microfluidic clamps for immobilizing and in-vivo imaging of Drosophila larva's internal organs such as CNS have been examined and characterized.
FIG. 3.
Linear periodic crawling of the 3rd instar D. melanogaster larva inside a channel. Crawling is composed of repetitive cycles of motion called strides. The strides can be divided into two phases: (i) movement of the center of mass and (ii) abdominal segment movement.
A. Narrowed channel immobilization
Imaging Drosophila larva is challenging due to the loose movement of the internal organs inside the hemolymph-filled body cavity even if the outer body is completely immobilized. Any immobilization mechanism should first be capable of inhibiting whole-larval body motion. The first design considered was a tapered (guide) channel with a constriction (narrow channel) at its end, which is similar to that used to immobilize C. elegans.16 The design of the chip consisted of an inlet port for animal loading into the device [“inlet” in Fig. 4(a)], a channel for transporting the animal towards the narrowed channel that was designed to immobilize the larvae for imaging and an outlet for ejecting the tested animal upon completion of each experiment. The inlet and narrowed channels were connected via a guide channel. The gate at the end of the narrowed channel was designed such that it allows only the head of the larva to pass through the narrowed channel. The cross section of the tapered channel was 600 × 500 μm2 (width × depth) at its wide end and reduced to 200 × 500 μm2 at its narrow end. A further section of the channel with a uniform cross section of 200 × 500 μm2 and a length of 500 μm forms the constriction gate [see the side view in Fig. 4(a)]. This dimension of the channel enables us to compress and flatten a 700 μm × 3 mm (diameter × length) larva into the constriction gate for body immobilization and CNS imaging.
FIG. 4.
(a) Schematic design of the narrowed channel chip (not to scale)—top view (top image) and side view (bottom). (b) Steps to load a 3rd instar Drosophila larva using the narrowed channel chip. (1)–(4) Larva swam freely into the trap and (5) and (6) larva was pneumatically moved into the trap and immobilized. (c) The movement of the center of mass (CM) for ten different larvae over time, inside the narrowed channel. (d) The release time of ten different 3rd instar larvae from the narrowed channel trap. By defining a threshold of 500 μm on the larva's CM displacement, the releasing time of each larva was calculated. In average, the narrowed channel was not able to immobilize the larvae more than ∼20 s. Scale bar = 400 μm for all figures.
In order to test the design, a 3D printed master mold was fabricated and PDMS was cast and cured on top of it to form the channel structure as discussed in Sec. II. Then, this structure was bonded with a glass slide (75 mm × 25 mm) to form the complete device. Drosophila larvae (3rd instar) were washed from the food medium onto a Petri dish with DI water. Next, by using a soft brush, a larva was picked from the dish and loaded into the chip at the inlet. The larva was pneumatically (0.2 bar pressure pulse) moved to the guide channel near the entrance region of the narrow channel as shown in Fig. 4(b-1). The pressure was removed to allow the larva orient and crawl voluntarily into the trap as shown in the sequence images in Figs. 4(b-2)–4(b-4). The voluntary movement allowed the head of the larva to orient forward and also ensured proper axial orientation for suitable imaging of the CNS. The self-orientation process usually took up to 30 s but robustly created the desired orientations after immobilization. In rare cases, it was observed that the larvae attempted to exit from the inlet channel. After achieving the proper orientation, the animal was then pneumatically pushed further inside the trap using a 0.8 bar continuous pressure which was stopped when the head of the larvae reached the end of the trap [Figs. 4(b-5) and 4(b-6)] which completely immobilized the outer body of the animal.
After loading and immobilizing the 3rd instar larvae (n = 10) inside the narrowed channel, image sequences (2 f/s) of the whole-larva body movements were recorded until they could release themselves from the trap. Then, the movement of the CM of the larvae (representing their locations) was calculated by using a custom-made image processing code in the LabVIEW© software. Using these data, the horizontal movement of the CM vs. time was plotted for all 10 larvae as shown in Fig. 4(c). The data reveal that the displacement of the CM from the original position increased over time for all larvae, which indicated that the larvae actively attempted to release themselves from the trap. This behaviour caused erratic back and forth motions of the CM of the larvae which would induce movement of the internal organs such as the CNS, making it difficult to obtain good images of this organ with the current design. The back and forth motion is similar to the digging and burrowing behaviour of the larva.
In order to quantify the ability of the immobilization mechanism to hold the larva in place for imaging purposes, the time that the larva was present in the field of view of a CNS imaging setup (FOV of ∼1000 × 800 μm2 in 40× objective lens) was calculated. Such quantification is useful in comparison of the various immobilization schemes to identify the best performing ones. In a typical imaging setup for whole-CNS imaging, the proper field of view is ∼1000 × 800 μm2 as the CNS is approximately 600 μm long and 200 μm wide. Therefore, a CM movement of more than 500 μm in either direction was considered out-of-range and the time that this movement happened was designated as the release time. A total of ten 3rd instar larvae were analysed and their release times are plotted in Fig. 4(d).
The results showed that the narrow channel was capable of immobilizing the body of the whole-larva for only 20 s on an average. However, the required time for high resolution CNS imaging when the larva is exposed to external stimulus is usually more than 1 min.13 Therefore, the narrow channel design was found to be not suitable for whole-larva body immobilization for imaging purposes. An alternate design which could improve the whole body immobilization was required. Also, the longitudinal contraction of the abdominal muscle segments, which was a critical mechanism in larva locomotion and digging behaviour, was found to be the crucial factor that facilitated the release of the larva from the trap. A modified configuration which could impede the segmental contraction, in addition to compressing its body, might be more suitable for immobilization.
B. 2D segmental pinning
Unlike C .elegans, which has rounded cylindrical body shape, Drosophila larva consists of a mouth hook, two thoracic segments (T2 and T3), seven abdominal segments (A1–7), and a tail on its body. Muscle contraction patterns in abdominal segments cause a pistoning action that results in the Drosophila larva's forward and reverse crawling. The narrow channel immobilization approach demonstrated that disruption of this muscle contraction is critical if the larval body is to be immobilized. Therefore, the second device was designed in order to introduce segmental constrictions along the body designed to disrupt transmission of abdominal constriction along the body of the larva. Similar to the first chip, the second design consisted of an inlet port, inlet channel, a channel with 2D segmental pinning for immobilization, a gate to only allow the head to pass through it, and an outlet for ejecting the tested animal [schematically shown in Fig. 5(a)]. The key design modification here was addition of three equispaced 2D protrusions to the narrowed channel that served as pinning points for the larva. The spacing between the protrusions was closely matched with abdominal segment lengths on the larva's body [indicated in Fig. 5(a)]. Therefore, when the larva was loaded into the trap, the segmental protrusions would latch into the larva's abdominal segments and potentially restrict larva's locomotion more efficiently.
FIG. 5.
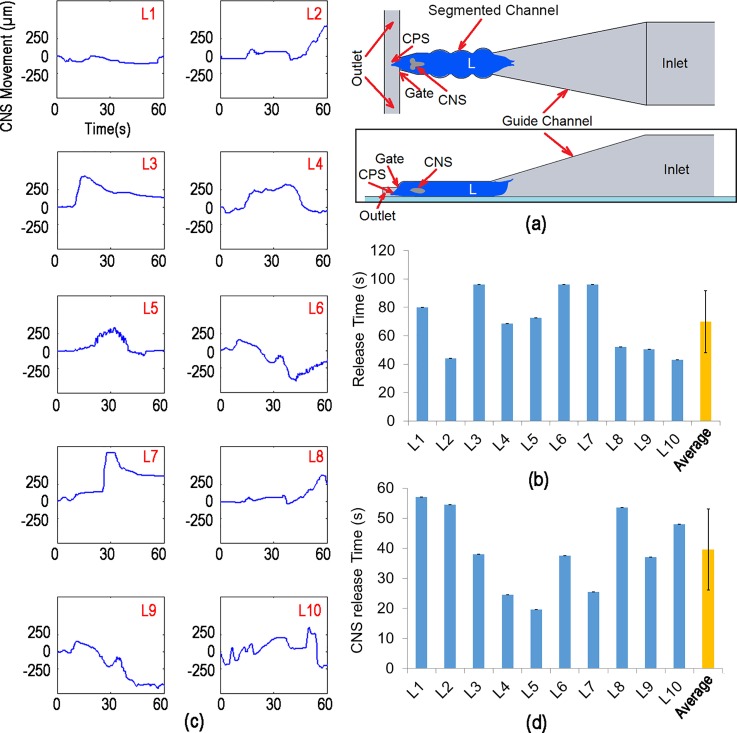
(a) Schematic design of the immobilization channel with 2D segmental pinning structure (not to scale)—top view (top image) and side view (bottom image). (b) The releasing time of ten different 3rd instar larvae from the 2D segmented channel. The segmented channel could improve the immobilization time from the average of ∼20 s (in narrowed channel) to ∼70 s. (c) The CM movement of the larva's CNS for 10 different larvae over time, inside the 2D segmented channel. (d) The CNS resealing time of ten different 3rd instar larvae from the narrowed channel with the 2D segmental pinning structure. By defining a threshold of 200 μm on the larva's CNS displacement, the CNS releasing time of each larva was calculated. In average, the 2D segmented channel was not able to immobilize the larvae's CNS more than ∼40 s; while the whole-larva body was immobilized.
Similar to the previous characterization on the narrowed channel (Sec. II A), ten 3rd instar Drosophila larvae were loaded and immobilized into the segmented channel one at the time in order to characterize the device. Next, the image sequence (2 f/s) of whole-larva body was recorded until larva could release itself from the trap. Subsequently, the displacement of the larva's CM along the x-axis vs. time was calculated. Finally, the release time of each larva was obtained as shown in Fig. 5(b).
The results indicate that the average immobilization time for the segmented channel was ∼70 s, which is a significant improvement over the narrow channel design. This time is generally sufficient for typical stimuli responsive assays where the larva is exposed to a stimulus and the response of the neural system is recorded. However, it was also found that this immobilization design does not fully immobilize the CNS of the larva even though the outer body shell was immobilized. In order to characterize the movement of the CNS, ten 3rd instar larvae of the genotype ChaT-Gal4/CyO; UAS-GCaMP5G/TM3 (Bloomington 56500) that have genetically engineered calcium indicators of neural activity were immobilized in this chip and imaged under a fluorescent microscope. The image sequence (2 f/s) of CNS was recorded until larva left the field of the view (700 μm length and 500 μm width). Next, the CNS movements of the larvae were calculated by using the custom-made image processing code on LabVIEW© software which is shown in Fig. 5(c).
The results show that even though the outer bodies of the larvae were immobilized, the CNS was capable of moving by ∼250 μm or more in some cases, which may be due to local muscle contractions. Although the CNS movement was dampened by the segmental protrusions in comparison to the narrowed channel, its movement is still significant to affect imaging of internal organs. In order to quantify the immobilization for CNS imaging, the release time was obtained when the displacement of CNS was greater than 200 μm from its original location. The results plotted in Fig. 5(d) showed that the narrowed channel with 2D segmental pinning was able to immobilize the CNS for only ∼40 s. Therefore, compared to the narrow channel, the 2D segmented channel allowed us to immobilize whole-larva body more effectively by using the indented segments in the trap. Those protrusions could latch the muscle abdominal segments and consequently improve the larva immobilization. However, this design was still not able to completely immobilize the CNS for more than 40 s.
C. 3D segmental pinning
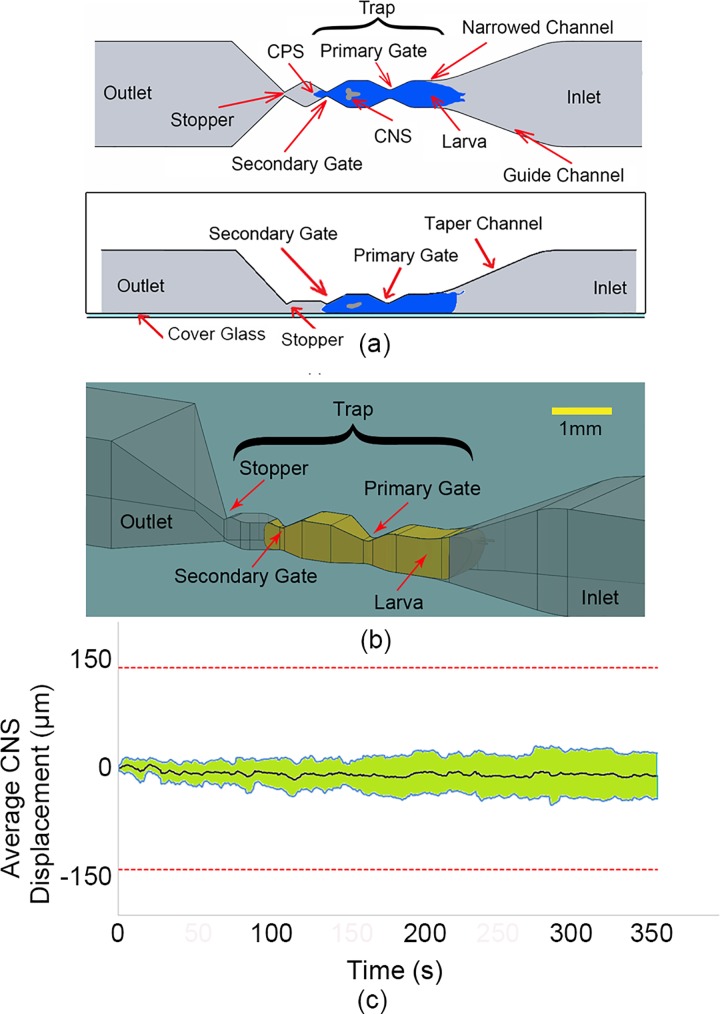
The previous design using 2D segmental protrusions in the narrow channels provided an effective way to immobilize the body of the larva sufficiently for imaging. However, due to the digging and burrowing behaviour exhibited by the larva, the internal organs such as the CNS capsule, heart, and gut could freely move inside the hemolymph-filled body cavity, regardless of the outer body immobilization. Therefore, a new design that incorporates 3D segmental protrusions was developed to further curtail the borrowing or digging induced motion, thereby preventing the movements of internal organs inside the larva for longer period, which was the ultimate goal of the immobilization device. The 3D segmental pinning design [schematically shown in Figs. 6(a) and 6(b)] consisted of an inlet port for animal loading into the device, a channel for transporting the animal towards the narrowed trap that was designed to immobilize the larvae for imaging and an outlet for ejecting the tested animal upon completion of each experiment. The trap consisted of a narrowed channel (770 × 700 μm2 cross-section with 500 μm length), primary (200 μm width and 450 μm depth) and secondary gates (100 μm width and 425 μm depth), and a stopper (100 μm width and 100 μm depth). The size of a 3rd instar larva is approximately 1 mm × 3.5 mm (diameter × length), which indicated that larva's body was compressed from ∼1 mm diameter to ∼0.7 mm (30%) by loading into this trap. The primary and secondary gates were designed to pin the 3rd instar larvae at two locations on its body while the rest of it was encapsulated in the narrowed channel. The dimension of the secondary gate was designed such that only the nose region of the immobilized 3rd instar larva could protrude through the gate. This stopper was used to prevent the larvae from escaping the trap when a small pressure (0.3 bar) was sustained on the posterior side for complete immobilization.
FIG. 6.
(a) and (b) Schematic design of the 3D segmental pinning chip—top view (top image) and side view (bottom). (c) The CM movement of the larva's CNS for 10 different larvae over time, inside the 3D segmental pinning chip. The black line shows the average CNS movement of 10 larvae, while the two blue lines indicate the standard deviation for each average data point.
Similar to the previous characterization on the narrowed channel and 2D segmental pinning (Secs. II A and II B), ten 3rd instar Drosophila larvae were loaded into the 3D segmental pinning chip and immobilized one at the time in order to characterize the device. Next, the image sequence (2 f/s) of the whole-larva body was recorded for 350 s. The image processing showed that the whole body motion of larvae was almost non-existent which indicated that the chip was able to completely immobilize the larvae during the imaging process.
In order to study the movement of the CNS, when the larva is immobilized inside the 3D segmental pinning chip, ten 3rd instar larvae were immobilized in this chip and imaged under a fluorescent microscope. Next, the image sequence (2 f/s) of the CNS was recorded for 350 s and the CNS movements were calculated by using the custom-made image processing code on LabVIEW© software. The longitudinal motions of the CNS are plotted in Fig. 6(c). The results indicated that the device could keep the CNS completely inside the field of view (700 × 500 μm2). Nevertheless, the CNS had minor forward or backward movements inside the field of view. The upper and lower lines (blue) in the graph represent the standard deviation of the data points obtained from the 10 larvae tested. The black line shows the average CNS motion for the larvae, the fluctuation of which is under 10 μm over 350 s. The larvae were unloaded for further analysis such as viability test and behavioural studies. To unload the larva, a Tygon tube was connected from the inlet to a Petri dish. Then, a 2 bar pressure pulse with a duration of 1 s was applied from the outlet to the inlet. This process was suitable to release the larvae from the trap and wash them out from the device to a Petri dish.
In comparison to the narrowed channel and 2D segmental pinning design, the 3D segmental pinning design significantly improved the whole-larva body immobilization. In addition, the 3D pinning design was successfully able to restrict the motion of the CNS to 10 ± 30 μm over 350 s which is a first for live un-anaesthetised imaging of these animals. It is important to note that this movement is ∼10% of the CNS movement when the larva is glued onto a substrate—a method that is commonly used. It also uses only mechanical ad completely reversible means for immobilization. A possible explanation for such robust immobilization could be that the design of the primary and secondary gates efficiently prevented the wave of muscle contraction from building up which prevented any motion of the internal organs. As a result, the 3D segmental pinning design allowed us to successfully immobilize Drosophila larva with minimal internal CNS movements for its subsequent live neuronal imaging under various external stimulations.
Although the various immobilization designs described in this paper could either partially or completely immobilize both internal and external movement of the larvae, such a confinement may affect their viability due to the high mechanical stress on the body.3 In order to study the impact of mechanical stress caused by immobilization mechanisms on the viability of the larvae, a set of experiments was performed. A total of forty five 3rd instar larvae were picked from their food vial, washed with DI water one at a time, and prepared as described in Sec. II A. Next, the larvae were loaded into the microfluidic devices (15 larvae for each design) using the loading method described in Sec. III A. The larvae were kept in the channel until they could release themselves from the immobilization channel. The total time that the larvae were immobilized and were kept inside the loading channel was 350 s. In the case of larvae loaded into the 3D segmental pinning chip, they were taken off from trap after 350 s, by applying a 2 bar pressure pulse with a duration of 1 s from the outlet to the inlet. Subsequently, the unloaded larvae were transferred into the food media (one food well per chip) to study the viability of the larvae. In order to compare the result with a control sample, 15 fresh larvae were placed into the DI water bath for 350 s and then transferred into the food media. The viability test was repeated three times, and the results are shown in Fig. 7. The viability study of the larvae after 7 days indicated that the narrow channel and 2D segmental pinning chip did not have a significant effect on the viability of the larvae in comparison with the control. However, the viability of the larvae loaded into the 3D segmental pinning chip was ∼40% lower and statistically significant (p < 0.005) from the control group as shown in Fig. 7. This reduction in viability might be due to two potential reasons. One reason is that the 3D segmental pinning design induced higher mechanical stress on the larvae due to the 3D mechanical pins. The other reason could be because of the unloading mechanism. The larvae could release themselves from the traps inside the narrow channel and 2D segmental pinning design. However, pressure of 2 bars was required to unload the larvae from the trap in the 3D segmental pinning design, which caused higher mechanical stress on the larvae.
FIG. 7.
The viability test of the larvae loaded into the microfluidics devices. The impact of the 3D segmental pinning chip on the viability if the larvae are more significant compared to other narrowed channel and segmented channel design.
D. Comparison with other immobilization methods
Currently, dissection, anesthetization, and gluing are three common methods for the immobilization of the Drosophila larvae. The first two methods could significantly affect the neuronal response of the larvae. The dissection of the larvae could allow one to firmly immobilize the larva and remove undesired organs from the larva's body for capturing a clear image form the organ of interest. However, it is associated with a large amount of damage to the tissues, axons, and neurons. Therefore, it could not be used for live imaging and neuronal studies of the larva, while the larva is exposed to external stimulation. Furthermore, once dissected there is a limited time window for an assay before the responses degrade. Alternatively, anaesthesia such as ether and chloroform can be used to immobilize the larva. Although this technique is effective and keeps the larva intact, the use of anaesthesia will significantly affect and could alter the functioning of the neuronal response.
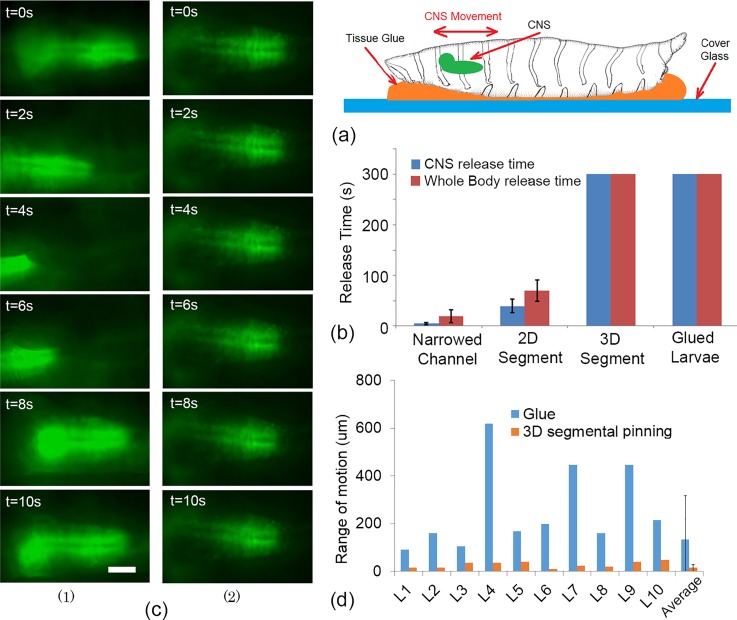
Gluing of the larva on to a substrate is another widely used method that could be used for imaging the neuronal response of the larva. Tissue glue and double side tape are two common adhesives used. In order to compare the performance of the immobilization designs described in this paper with the gluing technique, a number of larvae (n = 10) were glued to glass substrates using tissue glue (Nexcare™ Skin Crack Care, Catalog No. 112, UPC 00051131861015) as shown in Fig. 8(a). Then, displacement of larvae's whole body and the CNS has been recorded and was compared [see Fig. 8(b)] to the three microfluidic immobilization designs that were investigated in this paper. The results showed that the whole body of the larvae could be immobilized completely with almost non-existent net displacement by gluing. Additionally, the CNS of the larvae glued to the substrate stayed within the field of the view; however, the CNS had significant motion with an average range of approximately 300 μm. Although the 2D segmental pinning design could more effectively immobilize the CNS in comparison to the narrowed channel, the duration of the immobilization (∼40 s) would not be enough for many in vivo assays.13 In contrast, 3D segmental pinning design could strongly immobilize the whole body movement and keep the CNS inside the field of view with an average range of motion of only 40 μm.
FIG. 8.
(a) Schematic of the larva immobilization using tissue glue. A droplet of tissue glue has been placed on a cover glass and the larva was placed on the droplet. After a few minutes, the larva will be glued to the substrate. (b) A comparison of the whole body releasing time and CNS releasing time for three microfluidics chips and the gluing technique. The results were compared over 300 s of larvae (n = 10) immobilization. (c) The time sequences images (10 s) of the larvae's CNS, when they were immobilized by using (1) gluing technique and (2) 3D segmental pinning chip. The CNS experienced a large number of the random forward/backward motions, which reduced the resolution of the images in some farms significantly. The scale bar is 100 μm. (d) The domain of the pistonic CNS's motion for 10 larvae that were immobilized by tissue glue and the 3D segmental pinning chip. The image sequence of the CNSs has been captured in an 1100 μm × 700 μm field of view. In average, the domain of the CNS motion, when the larvae were immobilized by gluing technique, was significantly large. This could lead to different challenges for catering high-resolution time-lapse images of the CNS.
In order to achieve high quality images of the internal organs over long periods of time, their complete immobilization is essential. Although the gluing method could immobilize the body wall and keep the desired organ in the defined field of view, this might not be sufficient for capturing high resolution images of the internal organs such as CNS. The image sequences of the larvae's CNS, when they were glued to the substrate, showed a large number of random pistonic motion. These motions could significantly reduce the resolution of the images. The time sequence images of the CNS for the gluing technique and 3D segmental pinning chip are shown in Fig. 8(c). To compare the performance of the 3D segmental pinning chip and gluing technique on the range of the larvae's CNS motion, the location of the CNS (its centre of mass) has been recorded in the 1100 μm × 700 μm field of the view, while they were immobilized in the 3D segmental pinning chip or glued to the substrate. Next, the average and the standard deviation of the CNS location in the x-axis have been calculated. Since the larvae could not release themselves from neither 3D segmental pinning chip or glue, their CNS experience only a random pistonic motion in the field of view, which resulted in almost non-existant motion in both methods. However, the range of larvae's CNS motion in the 3D segmental pinning chip was significantly smaller than the gluing technique. This was determined by comparing the standard deviation of the CNS motion along the x-axis as shown in Fig. 8(d) for 10 larvae. Consequently, only the 3D segmental pinning chip was effective and completely immobilized the CNS of the Drosophila larvae for high resolution imaging, with minimum tissue damage (compared to dissection techniques and gluing if they want to be used for further study and viability assays) and neuronal effects (compared to both dissection and anesthetize techniques) on the larvae. Additionally, the microfluidic based design could potentially allow one to apply this technique for various high throughput live imaging protocols, while the larvae could be exposed to various external stimulates such as sound, chemical, light, and electric fields.
IV. CONCLUSION
The ability to immobilize internal organs such CNS of the Drosophila larva without dissection can potentially allow imaging of the organs at a high resolution, without losing the true physiological responses. This could be valuable in the study of biological processes and disease mechanisms and in drug discovery. However, the robust digging/burrowing motion in larva's behaviour makes the intact-live study of neuronal response challenging. In this paper, a series of microfluidic clamps for intact immobilization of the larva were investigated. An optimized structure for rapid mechanical immobilization of Drosophila larvae for CNS imaging was developed. The optimal design had 3D segmental pinning and was able to restrict CNS movement to the maximum of 10 ± 30 μm. The stability of the CNS inside 3D segmental pinning chip allowed one to visualize neuronal activities using a genetically encoded calcium indicator probe (GCaMP5G) without the use of any anaesthetic drugs that could affect animals' neurophysiological status. However, the viability of the larvae load into the 3D segmental pinning chip was reduced in comparison to the control. Nevertheless, we anticipate that our intact larva-on-a-chip will also be useful for a variety of assays that involve Calcium imaging, optogenetic, and electrophysiological, auditory response approaches.
ACKNOWLEDGMENTS
The authors gratefully acknowledge financial support from the Natural Science and Engineering Research Council of Canada (NSERC) through their Discovery Program, Discovery Accelerator Supplement Award, The Canada Research Chairs Program, and the Ontario Ministry of Research and Innovation through their Early Researchers Award.
References
- 1. Tickoo S. and Russell S., Curr. Opin. Pharmacol. 2, 555–560 (2002). 10.1016/S1471-4892(02)00206-0 [DOI] [PubMed] [Google Scholar]
- 2. Heckscher E. S., Lockery S. R., and Doe1 C. Q., J. Neurosci. 32(36), 12460–12471 (2012). 10.1523/JNEUROSCI.0222-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bier E., Nat. Rev. Genet. 6, 9–23 (2005). 10.1038/nrg1503 [DOI] [PubMed] [Google Scholar]
- 4. Duffy J. B., Genesis 34, 1–15 (2002). 10.1002/gene.10150 [DOI] [PubMed] [Google Scholar]
- 5. Kohsaka H., Okusawa S., Itakura Y., Fushiki A., and Nose A., Dev. Growth Differ. 54, 408–419 (2012). 10.1111/j.1440-169X.2012.01347.x [DOI] [PubMed] [Google Scholar]
- 6. Schoofs A., Niederegger S., van Ooyen A., Heinzel H., and Spiess R., J. Insect Physiol. 56, 695–705 (2010). 10.1016/j.jinsphys.2009.12.008 [DOI] [PubMed] [Google Scholar]
- 7. Mondal S., Ahlawat S., and Koushika S. P., J. Visualized Exp. 67, 3780 (2012) 10.3791/3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rezai P., Salam S., Selvaganapathy P., and Gupta B. P., in Integrated Microsystems, edited by Iniewski K. ( CRC Press, 2011), pp. 581–608. [Google Scholar]
- 9. Hulme S. E. and Whitesides G. M., Angew. Chem. Int. Ed. Engl. 50, 4774–4807 (2011). 10.1002/anie.201005461 [DOI] [PubMed] [Google Scholar]
- 10. Rezai P., Siddiqui A., Selvaganapathy P. R., and Gupta B. P., Lab Chip 10(2), 220–226 (2010). 10.1039/B917486A [DOI] [PubMed] [Google Scholar]
- 11. Mondal S., Ahlawat S., Rau K., Venkataraman V., and Koushika S. P., Traffic 12, 372–385 (2011). 10.1111/j.1600-0854.2010.01157.x [DOI] [PubMed] [Google Scholar]
- 12. Ghannad-Rezaie M., Wang X., Mishra B., Collins C., and Chronis N., PLoS One 7, e29869 (2012). 10.1371/journal.pone.0029869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ghaemi R., Rezai P., Iyengar B. G., and Selvaganapathy P. R., Lab Chip 15(4), 1116–1122 (2015). 10.1039/C4LC01245C [DOI] [PubMed] [Google Scholar]
- 14. Ardeshiri R. et al. , “ Cardiac screening of intact Drosophila melanogaster larvae under exposure to aqueous and gaseous toxins in a microfluidic device,” RSC Adv. 6(70), 65714–65724 (2016). 10.1039/C6RA14159E [DOI] [Google Scholar]
- 15. Chokshi T. V., Ben-Yakar A., and Chronis N., Lab Chip 9, 151–157 (2009). 10.1039/B807345G [DOI] [PubMed] [Google Scholar]
- 16. Hulme S. E., Shevkoplyas S. S., Apfeld J., Fontana W., and Whitesides G. M., Lab Chip 7, 1515–1523 (2007). 10.1039/b707861g [DOI] [PubMed] [Google Scholar]
- 17. Chung K., Crane M. M., and Lu H., Nat. Methods 5, 637–643 (2008). 10.1038/nmeth.1227 [DOI] [PubMed] [Google Scholar]
- 18. Gilleland C. L., Rohde C. B., Zeng F., and Yanik M. F., Nat. Protoc. 5, 1888–1902 (2010). 10.1038/nprot.2010.143 [DOI] [PubMed] [Google Scholar]
- 19. Zeng F., Rohde C. B., and Yanik M. F., Lab Chip 8, 653–656 (2008). 10.1039/b804808h [DOI] [PubMed] [Google Scholar]
- 20. Fakhoury J. R., Sisson J. C., and Zhang X. J., Microfluid. Nanofluid. 6, 299–313 (2009). 10.1007/s10404-009-0405-x [DOI] [Google Scholar]
- 21. Zappe S., Fish M., Scott M. P., and Solgaard O., Lab Chip 6, 1012–1019 (2006). 10.1039/b600238b [DOI] [PubMed] [Google Scholar]
- 22. Zhang X., Scott M. P., Quate C. F., and Solgaard O., “ Microoptical characterization of piezoelectric vibratory microinjections in Drosophila embryos for genome-wide RNAi screen,” Microelectromech. Syst., J. 15(2), 277–286 (2006). 10.1109/JMEMS.2006.872242 [DOI] [Google Scholar]
- 23. Delubac D., Highley C. B., Witzberger-Krajcovic M., Ayoob J. C., Furbee E. C., Minden J. S., and Zappe S., “ Microfluidic system with integrated microinjector for automated Drosophila embryo injection,” Lab Chip 12(22), 4911–4919 (2012). 10.1039/c2lc40104e [DOI] [PubMed] [Google Scholar]
- 24. Ghaemi R. and Selvaganapathy P. R., “ A microfluidic microinjection of drosophila embryo in a format using compliant mechanism and electrokinetic dosage control,” in The 19th International Conference on Miniaturized Systems for Chemistry and Life Sciences, Gyeongju, Korea, 25–29 October 2015. [Google Scholar]
- 25. Dagani G. T., Monzo K., Fakhoury J. R., Chen C. C., Sisson J. C., and Zhang X., Biomed. Microdevices 9, 681–694 (2007). 10.1007/s10544-007-9077-z [DOI] [PubMed] [Google Scholar]
- 26. Levario T. J., Zhan M., Lim B., Shvartsman S. Y., and Lu H., Nat. Protoc. 8, 721–736 (2013). 10.1038/nprot.2013.034 [DOI] [PubMed] [Google Scholar]
- 27. Lucchetta E. M., Munson M. S., and Ismagilov R. F., Lab Chip 6, 185–190 (2006). 10.1039/b516119c [DOI] [PubMed] [Google Scholar]
- 28. Akerboom J., Chen T. W., Wardill T. J., Tian L., Marvin J. S., Mutlu S., Calderon N. C., Esposti F., Borghuis B. G., Sun X. R., Gordus A., Orger M. B., Portugues R., Engert F., Macklin J. J., Filosa A., Aggarwal A., Kerr R. A., Takagi R., Kracun S., Shigetomi E., Khakh B. S., Baier H., Lagnado L., Wang S. S., Bargmann C. I., Kimmel B. E., Jayaraman V., Svoboda K., Kim D. S., Schreiter E. R., and Looger L. L., J. Neurosci. 32, 13819–13840 (2012). 10.1523/JNEUROSCI.2601-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brand A. H. and Perrimon N., Development 118(2), 401–415 (1993). [DOI] [PubMed] [Google Scholar]
- 30. Salvaterra P. M. and Kitamoto T., Gene Expression Patterns 1(1), 73–82 (2001). 10.1016/S1567-133X(01)00011-4 [DOI] [PubMed] [Google Scholar]
- 31. Nakai J., Ohkura M., and Imoto K., Nat. Biotechnol. 19(2), 137–141 (2001). 10.1038/84397 [DOI] [PubMed] [Google Scholar]
- 32. Akerboom J., Rivera J. D., Guilbe M. M., Malavé E. C., Hernandez H. H., Tian L., Hires S., Marvin J. S., Looger L. L., and Schreite E. R., J. Biol. Chem. 284(10), 6455–6464 (2009). 10.1074/jbc.M807657200 [DOI] [PMC free article] [PubMed] [Google Scholar]