Abstract
Melanoma is among the most virulent cancers, owing to its propensity to metastasize and its resistance to current therapies. The treatment failure is largely attributed to tumor heterogeneity, particularly subpopulations possessing stem cell-like properties, i.e., melanoma stem-like cells (MSLCs). Evidence indicates that the MSLC phenotype is malleable and may be acquired by non-MSLCs through phenotypic switching upon appropriate stimuli, the so–called “dynamic stemness”. Since the phenotypic characteristics and functional integrity of MSLCs depend on their vascular niche, using a two dimensional (2D) melanoma-endothelium co-culture model, where the MSLC niche is recapitulated in vitro, we identified Notch3 signaling pathway as a micro-environmental cue governing MSLC phenotypic plasticity via pathway-specific gene expression arrays. Accordingly, lentiviral shRNA-mediated Notch3 knockdown (KD) in melanoma cell lines exhibiting high levels of endogenous Notch3 led to retarded/abolished tumorigenicity in vivo through both depleting MSLC fractions, evinced by MSLC marker down-regulation (e.g., CD133 and CD271); and impeding the MSLC niche, corroborated by the attenuated tumor angiogenesis as well as vasculogenic mimicry. In contrast, Notch3 KD affected neither tumor growth nor MSLC subsets in a melanoma cell line with relatively low endogenous Notch3 expression. Thus, Notch3 signaling may facilitate MSLC plasticity and niche morphogenesis in a cell context-dependent fashion. Our findings illustrate Notch3 as a molecular switch driving melanoma heterogeneity, and provide the biological rationale for Notch inhibition as a promising therapeutic option.
Keywords: Notch3, CD133, CD144, CD271, vasculogenic mimicry
Despite improved public education and increased awareness of the danger of sun exposure, the incidence of cutaneous melanoma continues to be on the rise for the past 30 years. Although successful public health campaigns have resulted in early detection and treatment, a significant portion of patients still succumb to metastatic disease, for which no current clinical regimen has proven significant long-term benefits. Melanoma cells are phenotypically and functionally heterogeneous, which is attributed to subpopulations of cells with stem cell-like properties, also known as melanoma stem-like cells (MSLCs) (1). Recent studies have identified and characterized various MSLC subpopulations (including the CD133+, ABCB5+, CD271+, ALDH1A+, CD20+, and JARID1B+ subsets) (2). MSLCs exhibit increased resistance to conventional as well as state-of-the-art pathway-specific targeted therapies (3), and are responsible for clinical relapses, partly owing to their quiescence, upregulated efflux transporters, and heightened ability for DNA damage repair (4). Effective targeting of MSLCs, therefore, represents a crucial step to achieve a cure for melanoma. However, given the diverse MSLC subsets and the notion of a modified “hierarchical”/stochastic model of CSCs, which postulates that several different subpopulations of CSCs co-exist in a given tumor, yet a subset of these may be better suited to survive and proliferate in a given microenvironment, the development of successful all-inclusive targeting strategies may require detailed understanding of the potential lineage and functional relationships among various MSLC fractions. At present, whether the various MSLC markers label the same, similar, or distinct population(s), and whether the various MSLC fractions are hierarchically, spatially, or functionally related remain unclear. Adding to the complexity is the growing awareness of cancer stem-like cell (CSC) plasticity, connoting that progenitor marker-negative cells can evolve into CSCs through proper environment cues or accumulating genetic alterations (5–9). Such “dynamic stemness” is further supported by recent reports where stem-like phenotype can be induced in non-CSCs in response to ectopic expression or up-regulation of stem cell-associated transcription factors (10, 11). Thus, targeting a specific class of cell surface markers may have limited benefit, as the CSCs are heterogeneous and genetically unstable. Ultimately, the success of CSC-targeted therapy may rely on niche-dependent strategies, since the phenotypic characteristics and functional integrity of CSCs depend on their existence within their niche (12–16).
We have recently demonstrated that various MSLCs, i.e., CD133+, CD271+, and ABCB5+ reside in a complex vascular niche, encompassing CD144/VE-cadherin+ melanoma-lined vasculogenic mimicry (VM) channels, mosaic vessels, and authentic endothelial cell (EC)-lined blood vessels (17, 18). VM, originally observed in uveal melanoma (and later in cutaneous melanoma and a myriad of human solid cancers) (19), connotes the periodic acid-Schiff (PAS)-positive, perfusable patterned networks and channels lined by aggressive tumor cells with endothelial phenotype, such as CD144 (VE-cadherin), Tie-1, and Eph2 expression (20–22). Expression gene profiling using microarray analyses reveals that tumor cells capable of VM exhibit a pluripotent, primitive phenotype, suggesting that the VM-engaging tumor cells possess stem cell-like properties with phenotypic plasticity to serve a stromal function (22).
Given the functional co-dependence between the CSCs and their niche, we hypothesize that reciprocal signaling between MSLCs and their vascular niche may govern niche morphogenesis and stem cell homeostasis. However, the underpinning of such a crosstalk remains, by and large, unknown because of the lack of easily manipulable in vitro models that take into account the relevant and pivotal role of the niche environment. To elucidate the critical signaling pathways governing niche micro-environment support of tumor heterogeneity, we developed a simple 2D co-culture system of melanoma cells and ECs that simulates the MSLC niche, where the MSLC phenotypic switch as well as vascular/VM niche morphogenesis are recapitulated in vitro (Fig. 1). Using pathway-specific expression analyses, we identified Notch3 as a candidate that directs “dynamic stemness” and niche morphogenesis. Targeting common niche signals controlling “stemness”, such as Nocth3, represents a novel strategy to eliminate the diverse subsets of pre-existing MSLCs, as well as, the dynamically induced MSLC fractions that may evolve over time. The availability of existing Notch inhibitors currently used for Alzheimer’s disease and many others emerging in the pharmaceutical market makes Notch inhibition a promising, fast-tracked therapeutic option for melanoma.
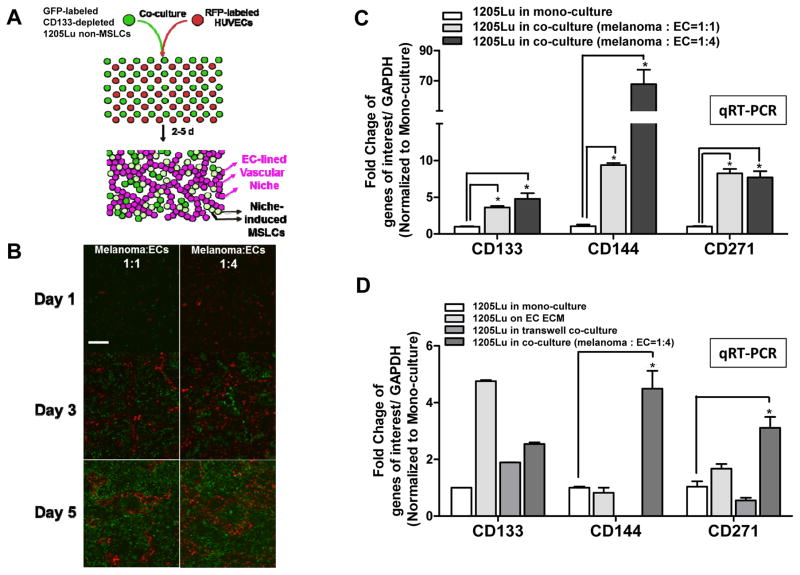
Figure 1.
Two dimensional (2D) melanoma-EC co-culture model recapitulates MSLC niche in vitro. A. Schematic representation of the 2D MSLC niche model in vitro. GFP-labeled CD133− non-MSLCs were co-cultured with RFP-labeled human umbilical vein endothelial cells (HUVECs) at 1:1 or 1:4 ratios for 5 days. B. In this model, ECs aligned to form branching tubular networks, reminiscent of the vascular niche in vivo (Magnification, ×100; scale bar, 200 μm). Co-cultured melanoma cells were then segregated from ECs by flow cytometry. C. MSLC (e.g., CD133 and CD271) and VM (e.g., CD144) markers were up-regulated in co-cultured melanoma cells compared to their mono-culture counter parts using qRT-PCR, simulating “dynamic stemness” and VM morphogenesis in vitro. D. Such a niche-inducing phenomenon was most pronounced when melanoma-EC contact is permitted, while EC extracellular matrix (ECM) or soluble factors in transwell cultures alone exhibited limited/partial effects. *, P < 0.05.
In human, the Notch pathway consists of 4 different transmembrane receptors, Notch1–4, and their membrane-bound ligands, Jagged (Jag1/2) and Delta (Dll1/3/4). Upon ligand binding, sequential proteolytic events, including cleavage by γ-secretase, release the active Notch intracellular domains (NICDs), which then translocate to the nucleus leading to transcriptional activation of the downstream Hes and Hey gene families (23). Overexpression of all 4 Notch receptors during melanoma progression has been reported (23). While the oncogenic functions of Notch1 have been well documented (23), the roles of the other Notch paralogs remain largely unexplored. Only recently Hardy et al. reported that Notch 4 promotes melanoma aggressiveness, including VM and anchorage-independent growth, through Nodal, an embryonic morphogen of the TGF-β superfamily implicated in the maintenance of stem cells (24).
Consistent with this, global γ-secretase inhibitors (GSIs) resulted in melanoma regression through Noxa-mediated apoptosis (25, 26). In another study, Howard et al. identified Notch3 as one of the key mediators of melanoma-EC communication in a co-culture system, whose expression correlates with tumor progression (27). These findings corroborate with our hypothesis that Notch3-mediated melanoma-EC crosstalk regulates MSLC homeostasis and niche morphogenesis. To test our hypothesis, we employed a lentiviral shRNA-mediated loss-of-function approach using 3 independent melanoma cell lines with varying endogenous Notch3 levels in the context of MSLC niche in vitro and in vivo.
Materials and Methods
Cell culture
Isogenic melanoma cell lines derived from different disease stages of tumor progression (obtained from Dr. M. Herlyn at the Wistar Institute, Philadelphia, PA) were cultured as previously described (28). These were comprised of primary vertical growth phase (VGP) melanoma cell lines as well as their metastatic isogenic counterparts, isolated from the same patient as the disease progressed. Red fluorescence protein (RFP)-labeled human umbilical vein endothelial cells (HUVEC) (27) were obtained as a generous gift from Dr. Rhoda Alani (Boston University, Boston, MA) and cultured in EGM-2 (Lonza, Basel, Switzerland).
In vitro 2D melanoma-endothelium co-culture system, recapitulating MSLC niche
Green fluorescence protein (GFP)-labeled 1205Lu melanoma cells (5) were depleted of CD133+ MSLCs using magnetic cell sorting (MACS) technology according to the manufacturer’s protocol (Miltenyi Biotec Inc., Bergisch Gladbach, Germany). CD133− GFP-labeled 1205Lu melanoma cells and RFP-labeled HUVEC cells were plated at ~30% confluence at 1:1 or 1:4 ratios in EGM-2 culture medium. Cells were incubated for five days before segregating into pure populations (GFP vs. RFP), using fluorescence activated cell sorting (FACS). Control mono-cultures were grown under identical conditions. RNA samples were prepared and subjected to the Stem Cell and Notch Signaling PCR Arrays based on the RT2 Profiler PCR Array User Manual (SA Biosciences/Qiagen, Valencia, CA).
Lentiviral constructs and infection
To generate stable Notch3 knockdown (KD) cell lines using lentiviral vector, Notch3 shRNA and control lentiviral particles were generated in HEK293T cells by co-transfecting Notch3 shRNA or scrambled shRNA plasmids (Mission® shRNA, Sigma-Aldrich, St. Louis, MO) and lentiviral packaging mix (Sigma-Aldrich) using Lipofectamine 2000 (Invitrogen, Waltham, MA) according to manufacturer’s instruction. Notch3 stable KD cell lines were achieved by infecting cells with lentiviral particles and followed by selection in puromycin-containing medium (1 μg/ml for 1205Lu; 2 μg/ml for A375 and WM852).
Western blotting
Cells lysates or xenograft tissue homogenates were extracted in RIPA buffer (ThermoFisher Scientific) containing protease inhibitors cocktail (Roche, Basel, Switzerland) and quantified using a BCA protein assay kit (Pierce/ThermoFisher Scientific). Equal amount of proteins were subjected to electrophoresis in 4–12% SDS-PAGE gel (Invitrogen) and transferred onto nitrocellulose membranes. Blots were probed with anti-Notch3 (Cell Signaling Technology, D1118 rabbit mAb, Danvers, MA), anti-CD133 (Miltenyi Biotech Inc., clone W6B3C1), anti-CD271 (Alomone Labs, Jerusalem, Israel), anti-CD144 (Cell Signaling Technology) Abs. Beta-actin or tubulin (Abcam, Cambridge, UK) were used as a loading control. Densitometry analysis was performed using Image J software (http://rsb.info.nih.gov/ij/download.html).
Quantitative real-time polymerase chain reaction (PCR)
RNA from melanoma cells or frozen tumor xenografts was extracted using an RNAeasy kit (Qiagen) and reversely transcribed using the SuperScript III® RT cDNA Synthesis kit (Invitrogen) according to the manufacturer’s protocol. Real-time quantitative PCR (qRT-PCR) was performed subsequently on a StepOnePlus™ Real-Time PCR System (Applied Biosystems/ThermoFisher) using SYBR select master mix (Applied Biosystems/ThermoFisher) and specific primers for CD133, CD144, CD271 and GAPDH. The primer sequences were as follows:
| CD133: | forward 5′-TTCTTGACCGACTGAGAC-3′ reverse 5′-CCAAGCACAGAGGGTCAT-3′ |
| CD144: | forward 5′-ATATGTCAGTGATGACTA-3′ reverse 5′-CTTACCAGGGCGTTCAGG-3′ |
| CD271: | forward 5′-ACTCACTGCACAGACTCT-3′ reverse 5′-GAAGCTTCTCAACGGCTC-3′ |
| GAPDH: | forward 5′-CGACAGTCAGCCGCATCTT-3′ reverse 5′-ACCTTCCCCATGGTGTCTCA-3′ |
All samples were run in triplicate and normalized to the housekeeping gene, GAPDH. Data was analyzed using the 2−ΔΔCt method.
Anchorage-dependent (growth curve) and -independent (soft agar) growth assays
For growth curve assay, cells were plated at 1 × 104/well in 6-well plates and counted after 3, 7 and 10 days using hemocytometer. Soft agar assay was performed as previously described (Hsu et al., 2008). Briefly, cells were suspended in MCDB153/L15 medium (v/v: 4/1) supplemented with 25 μg/ml bovine pituitary extract, 2 ng/ml epidermal growth factor (EGF), 2 μg/ml insulin, 4% fetal bovine serum, and 0.25% agar and plated in triplicate at 1 × 104 cells per well in six-well plates. After 2 weeks, colonies with more than five cells were counted in 20 randomly chosen fields (×100) using an inverted microscope and the percentage of colony formation was calculated. Each experiment was repeated twice with consistency. The represented data was compiled from the independent repeats, and data were analyzed using the Student t test.
In vitro tube forming assay
50 μ Matrigel™ (BD Biosciences, San Jose, CA) was plated in 96-well culture dishes and allowed to polymerize at 37°C for 30 min. 4 × 104 cells per well were then seeded on the Matrigel™ layer and grown for 24 hours. Randomized fields were captured using inverted microscope (Nikon, Tokyo, Japan). For quantitative analysis of tube formation and length, three randomly selected microscopy fields (x 200) were photographed per experimental condition. Tube formation was analyzed by ImageJ software, and data were analyzed using the Student t test.
Immunofluorescence
Frozen melanoma xenograft sections were subjected to double immunofluorescence using standard procedures (29). The primary antibodies used in this study are rabbit anti-hCD144 (human-specific; Cell Signaling Technology), rabbit anti-hCD133 (human-specific; Miltenyi Biotech Inc.), rabbit anti-hCD271 (human-specific; Alomone Labs) and rat anti-mCD31 (mouse-specific; BD Biosciences). The secondary antibodies employed are FITC-conjugated donkey anti-rabbit IgG, AlexaFluor 594-conjugated donkey anti-rat IgG, and AlexaFluor 488-conjugated goat anti-mouse IgG1 (Invitrogen) antibodies. Isotype-matched rabbit, mouse or rat immunoglobulin was used in place of the primary antibody for control. Sections were mounted with VectaShield containing DAPI (Vector Laboratories, Burlingame, CA) and examined under a Nikon Eclipse E400 microscope equipped with FITC and TRITC filters (Nikon) and a Mercury-100W lamp (Chiu Technical Corporation). Host angiogenesis (mCD31+) was quantified by measuring luminal area or number of tubules per field (40 ×) using ImageJ software, and data were analyzed using the Student t test. Quantification of human melanoma–derived VM channels was performed by measuring the hCD144+ area per field (100 ×) using ImageJ software, and data were analyzed using the Student t test.
In vivo tumorigenicity
Mice are maintained under pathogen-free conditions in an American Association for Accreditation of Laboratory Animal Care-accredited facility at the Boston University Medical Center, under the supervision of the Laboratory Animal Science Center and its staff of veterinarians and support personnel. For the effect of Notch3 silencing on tumorigenicity, 2 ×106 1205Lu, A375, or 4 ×106 WM852 cells infected by control and Notch3 shRNA lentiviral constructs were injected subcutaneously in the dorsal skin of each severe combined immune deficient (SCID) mouse (CB17; Taconic Laboratory; 4–5 mice per condition). Tumor growth was monitored twice per week after implantation. Tumor dimensions were measured using calipers and tumor volume was calculated as previously described (29). Melanoma xenografts were harvested when the tumors reached approximately 1 cm3, and subjected to further analyses, including immunofluorescence, qRT-PCR and Western blotting.
Statistical analysis
Differences between two groups were analyzed using the Student t test. Differences between more than two groups were analyzed by one-way ANOVA. Two-sided P values of less than 0.05 were considered significant.
Results
Up-regulation of Notch3 expression in the 2D MSLC niche model in vitro
To study MSLC biology in a tissue context, we developed a simple 2D co-culture system, stimulating the MSLC niche. In this model, GFP-labeled, CD133-depleted, non-MSLC 1205 Lu melanoma cells and RFP-labeled human umbilical vein endothelial cells (HUVECs) were mixed at two different ratios (1:1 vs. 1:4), and co-cultured for 5 days (Fig. 1A). Over time, the endothelial cells aligned themselves to form interconnecting channels/networks around melanoma cells, mimicking the MSLC vascular niche (Fig. 1B). As a consequence, the expression of stem-like cell markers, CD133 and CD271, as well as VM maker, CD144, increased dramatically in sorted melanoma cells from the 2D co-culture system compared to their mono-cultured counterparts (Fig. 1C). These findings demonstrate that the 2D co-culture system recapitulates MSLC niche morphogenesis in vitro. To determine whether MSLC niche induction in vitro is mediated by direct cell-cell contact, extracellular matrix (ECM) or soluble factors secreted by HUVECs in our 2D MSLC model, we examined stem-like cell (i.e., CD133 and CD271) and VM (i.e., CD144) marker expression by qRT-PCR additionally in melanoma cells conditioned by HUVEC ECM and melanoma cells harvested from transwell co-culture with HUVECs. We found that the expression of CD133, CD271 and CD144 was most consistently induced when melanoma-HUVEC contact was permitted (Fig. 1D).
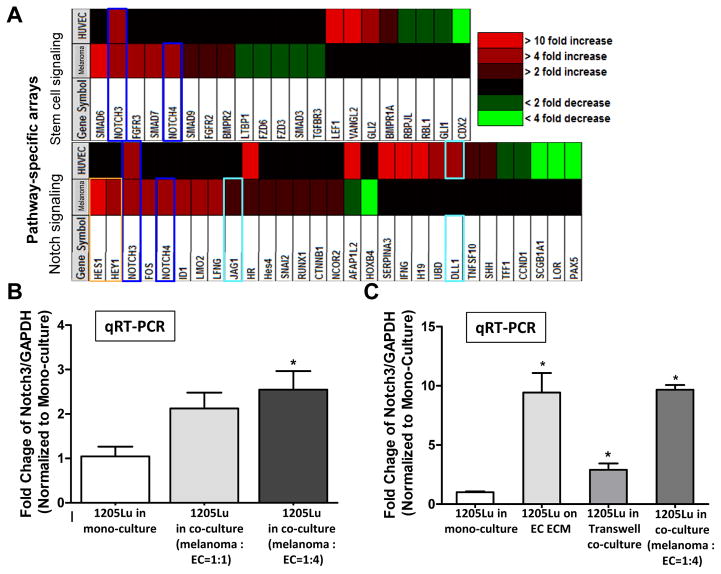
To identify molecular signals driving niche morphogenesis and MSLC induction, flow cytometry-segregated 1205Lu melanoma cells and HUVECs from the 2D MSLC model (mono-cultured counterparts as control; Fig. 2A) were subjected to gene expression profiling using Stem Cell and Notch Signaling Pathway-Specific PCR arrays (SA Biosciences/Qiagen). We found that Notch3 is concurrently up-regulated in both co-cultured melanoma cells and HUVECs from the 2D MSLC model (Fig. 2A). Interestingly, Notch downstream signaling mediators, Hes1 and Hey1, were concomitantly elevated in co-cultured melanoma cells, suggesting the activation of the Notch pathway (Fig. 2A). These results were verified using qRT-PCR (Fig. 2B and data not shown). Similar to MSLC and VM markers induction, Notch3 expression was highly increased when melanoma-HUVEC contact was permitted, while HUVEC ECM and secretory soluble factors conveyed partial effects (Fig. 2C). In addition, Notch4 and Jag1, and Dll1 were also increased in co-cultured melanoma cells, and ECs, respectively. Taking together, our findings corroborate with the hypothesis that the Notch (particularly Notch3)-mediated cell-cell contact between melanoma cells and endothelial cells may contribute to the induction of MSLC phenotype and niche morphogenesis.
Figure 2.
Notch3 up-regulation in co-cultured melanoma cells and ECs in the 2D niche model. Flow cytometry segregated melanoma cells and ECs from the 2D niche model were subjected to Stem cell and Notch signaling pathway-specific PCR arrays (SA Biosciences) using their mono-culture counterparts as control. Notch3 was consistently up-regulated both in co-cultured melanoma cells and ECs. The concomitant up-regulation of Notch downstream effectors, Hes1 and Hey1, further confirmed activated Notch signaling in co-cultured melanoma cells. B. qRT-PCR validated Notch3 induction in co-cultured melanoma cells following sorting by flow cytometry. Data was expressed as fold change of the ratio between Notch3 and GAPDH mRNA in co-cultured melanoma cells, normalized to that of their mono-culture counterpart. C. The magnitude of Notch3 induction in co-cultured melanoma cells was most evident when direct cell-cell contact is allowed, while EC ECM or soluble factors in transwell co-cultures produced only limited/partial result. *, P < 0.05.
Specific and functional Notch3 silencing in melanoma using the lentiviral shRNA approach
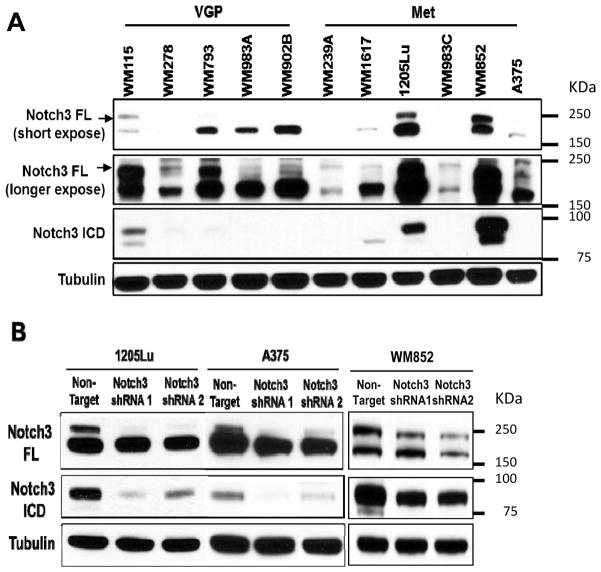
To examine the role of Notch3 in MSLC niche biology, we screened endogenous Notch3 expression in a panel of 11 melanoma cell lines from different stages of tumor progression using Western blotting (Fig. 3A). Based on this screening, most cell lines exhibited low endogenous level of Notch3. Two metastatic melanoma cell lines 1205Lu and WM852 show high endogenous expression of both full-length Notch3 and the active intracellular domain forms (Fig. 3A). Among these cell lines, we chose high Notch3-expressing cell lines, 1205Lu and WM852, as well as one relatively low Notch3-expressing cell line, A375, to explore the functional role of Notch3 with regard to tumor growth, and niche morphogenesis/homeostasis. We generated stable Notch3 KD cells using lentivirus-based shRNAs (Mission® shRNA, Sigma-Aldrich). Western blot analyses confirmed Notch3 KD at the protein level in vitro compared to control cells expressing non-target shRNA (Fig. 3B). Quantitative RT-PCR verified down-regulation of Notch3 message levels in vitro (Fig. S1A). Since members of the Notch gene family are highly homologous, to determine whether Notch3 silencing is specific and functional, we examined the expression of other Notch genes and well-known Notch3 targets, Hes1 and Hes2, using qRT-PCR. Despite slight, statistically non-significant reduction of Notch1 and 4 in WM852 Notch3 shRNA2 cells, no significant non-specific change in other Notch paralogs was noted in all three Notch3 KD cells (Fig. S1B). Notch3 targets, Hes1 and Hes2, were both down-regulated in 1205Lu and WM852 Notch3 KD cells, but in A375 Notch3 KD cells, only Hes2, but not Hes1, was down-regulated (Fig. S1C). These data demonstrated the achievement of specific and functional silencing of Notch3 in melanoma cells.
Figure 3.
Notch3 silencing in melanoma cells using lentiviral shRNA. A. Multiple melanoma cell lines of different stages of tumor progression (i.e., vertical growth phase (VGP) and metastasis (Met) expressed endogenous Notch3 by Western blotting. Tubulin was used as a loading control. B. Using lentiviral shRNA (non-target shRNA as control), significant KD efficiency was achieved in 1205Lu, A375 and WM852 melanoma cell lines as shown by Western blotting.
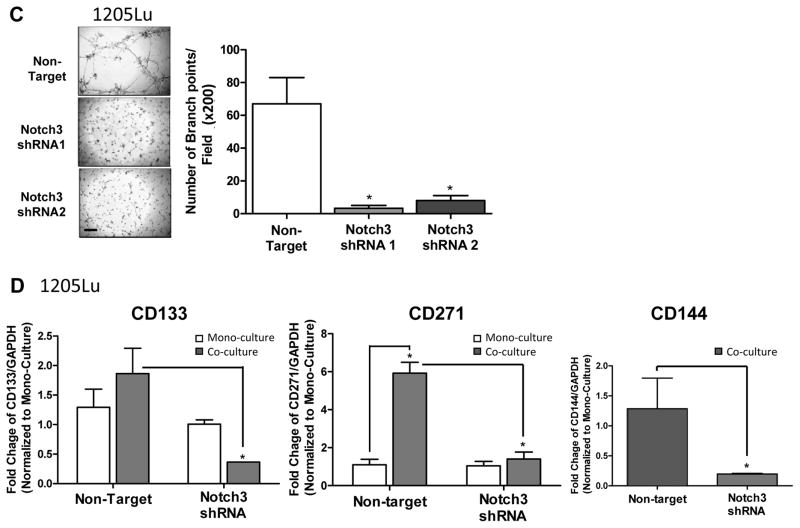
Biological consequences of Notch3 KD in vitro, including cell growth, colony-forming ability, VM/tube formation on Matrigel™, and MSLC niche induction in 2D niche model
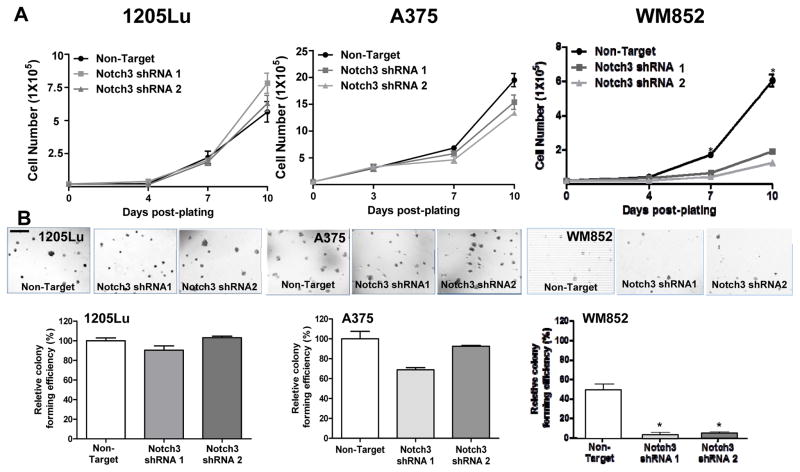
To explore the effect of Notch3 KD on cell proliferation in vitro, we performed conventional as well as anchorage-independent growth assays, using non-target shRNA transduced cells as controls. Notch3 KD in 1205Lu and A375 cells did not modulate cell growth or colony-forming ability (Fig. 4A and B, left and middle panels). In contrast, Notch3 KD led to a significant inhibition in WM852 cell growth as well as colony-forming ability (Fig. 4A and B, right panel). In addition, Notch3 KD in 1205Lu cells resulted in a significant attenuation in VM tubule formation on Matrigel™, as measured by the number of branch point/field (200 ×; Fig. 4C). Both non-target control and Notch3 KD A375 or WM852 cells were unable to form VM tubules in the Matrigel™ tube formation assays (data not shown). These results demonstrated that Notch3 KD exhibits variable cell line-specific inhibitory effects on growth, colony formation, and VM/tubule formation in vitro.
Figure 4.
In vitro consequences of Notch3 KD in melanoma. In 1205Lu melanoma cells, exhibiting high endogenous Notch3 (Fig. 3A), Notch3 KD resulted in no effects on conventional 2D growth (A, left panel) or colony-forming ability in soft agar (B, left panels; Magnification, ×100; scale bar, 500 μm), but significantly inhibited VM in Matrigel™ tube formation assays (C; 200 ×, bar: 100 μm). When incorporated into the 2D niche co-culture model, Notch3 KD hindered EC-mediated MSLC induction and VM phenotypic switch in 1205Lu cells (D). Similarly, Notch3 KD in A375 cells, which demonstrated relatively low endogenous Notch3 expression (Fig. 3A), showed no effect on conventional 2D growth (A, middle panel) or colony-forming efficiency (B, middle panels). In contrast, in another cell lines displaying high baseline Notch3 expression, WM852, Notch3 KD led to significant growth inhibition under conventional culture condition (A, right panel), as well as 3D anchorage-independent growth in soft agar assays (B, right panels). The effect of Notch3 KD in A375 and WM852 cells on VM in in Matrigel™ tube formation assays was assessed; however, both non-target control and Notch3 KD counterparts failed to undergo VM (data not shown). *, P < 0.05.
To determine whether Notch3 KD affects niche morphogenesis in vitro, we incorporated GFP-labeled 1205Lu Notch3 KD cells and RFP-labeled HUVEC cells in our 2D co-culture niche model, and then measured CD133, CD271 and CD144 message levels using qRT-PCR following flow cytometry-sorting (Fig. 4D). Consistent with our hypothesis that Notch3 mediates stem cell phenotype and vascular niche formation, co-culture with HUVECs failed to induce stem cell (e.g., CD133 and CD271), as well as VM (e.g., CD144) marker expression in Notch3 KD 1205Lu cells, while significant up-regulation of these markers was observed in co-cultured non-target control cells. These results highlight a pivotal role of Notch3 in MSLC niche induction and homeostasis.
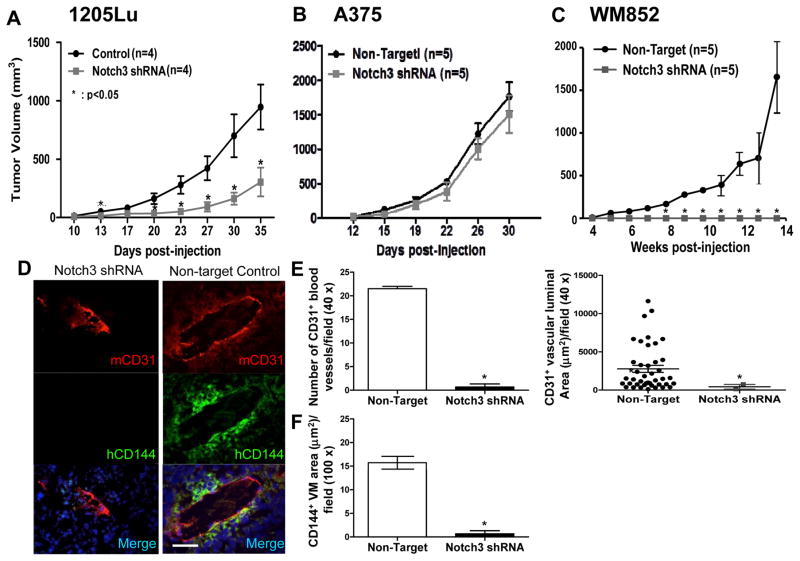
Notch3 KD retards tumor growth by depleting MSLCs and compromising vascular niche in vivo
Given that 1205Lu Notch3 KD shRNA #2, A375 Notch3 KD shRNA #1, and WM852 Notch3 KD shRNA#2 cells exhibited greater Notch3 KD efficiencies (Fig. 3B), we chose to use these cells for tumorigenicity studies. In 1205Lu and WM852 cells, which have high endogenous Notch3 expression, Notch3 KD significantly retarded xenograft growth compared to the non-target control (Fig. 5A and C). Although growth was delayed, 1205Lu Notch3 KD xenografts did eventually develop tumors. On the other hand, Notch3 KD completely abolished tumor growth in WM852 cells as no tumor was observed 14 weeks post inoculation despite high numbers of cells (4 × 106 cells) injected. In contrast, tumor growth of A375, a cell line that exhibits relatively low endogenous Notch3 expression, was not significantly affected by Notch3 KD (Fig. 5B). These findings suggest that Notch3 KD attenuates tumor growth in vivo and the growth regulatory effect of Notch3 depends on the cellular context relevant to its level of endogenous expression. Effective Notch3 KD in 1205 Lu and A375 melanoma xenografts was confirmed both at the mRNA (data not shown) and protein (Fig. 6A and C) levels. Because no tumor xenografts developed from Notch3 KD WM852 cells following 14 weeks of extended observation despite injection of high numbers of cells (4 × 106 cells/mouse), at which time the control tumors grew larger than 1 cm3 and the experiment had to be terminated, further analyses of WM852 xenografts could not be performed.
Figure 5.
In vivo consequences of Notch3 KD in a melanoma xenograft model. In 1205Lu and WM852 melanoma cells, displaying high levels of endogenous Notch3, Notch3 KD retarded/abolished tumorigenicity (A and C). In contrast, in A375 cells, harboring relatively low endogenous Notch3 expression, Notch3 KD failed to affect tumor growth (B). Analyses of the tumor samples using double immunofluorescence coupled with quantification using ImageJ software revealed that Notch3 KD inhibited tumor angiogenesis as well as melanoma-derived VM. Immunofluorescence analysis of 1205Lu xenograft sections double labeled for mCD31+ host blood vessels (red) and hCD144+ VM (green) showed that Notch3 KD xenografts displayed smaller and much less prominent mCD31+ host vessels and abolished VM (D, left panels) compared with non-target control xenografts (D, right panels); magnification, ×400; scale bar, 50 μm. Human melanoma–derived VM vessels (hCD144+) and host angiogenesis (mCD31+) were quantified as measured by the average luminal area or number of tubules per 40 × or 100 × field and revealed a significant reduction in the size and number of mCD31+ host blood vessels (E), as well as hCD144+ VM area (F) in 1205Lu Notch3 KD xenografts compared with controls. Analyses of A375 xenografts failed to show significant difference in tumor angiogenesis and VM between Notch3 KD and control (data not shown). As tumor growth in WM852 cells was completely abolished, no KD xenografts were available for further analyses. *, P < 0.05.
Figure 6.
Effective and functional Notch3 KD depletes MSLCs and VM-engaging melanoma cells in 1205 Lu xenografts. A. Western blot analyses demonstrated that stem cell markers (e.g., CD133 and CD271) as well as VM maker, CD144, were down-regulated in Notch3 KD 1205 Lu tumors, compared to non-target control (A). Consistent with the observed VM inhibition by Notch3 KD above, the expression of Tie-1, a VM-associated gene, was also down-regulated in Nocth3 KD 1205Lu xenografts by qRT-PCR (B). Of note, the efficacy of Notch3 KD was maintained in the tumor xenografts as shown by Western blotting (A). On the other hand, in A375 melanoma cells where no change in tumorigenicity was appreciated following Notch3 KD, the expression of stem-like cell marker, CD271, was not affected (C). Attempts to further validate CD133 and CD144 in Notch3 KD A375 xenografts using Western blot analysis (C8161 lysate included as a positive control) were unsuccessful due to sensitivity issue (Fig. 6C; note that CD133 and CD144 signals were undetectable both in the control and KD xenografts despite maximal loading and prolonged overnight exposure). *, P < 0.05.
To explore the biological significance of Notch3 on niche morphogenesis in vivo, we analyzed tumor vasculatures in human melanoma xenografts using multi-label immunofluorescence (IF; Fig. 5D). Parallel to the data obtained from in vitro tubule/VM formation analyses, CD144+ VM-like melanoma channel formation was significantly hindered in Notch3 KD 1205Lu xenografts compared to their control counterparts (Fig. 5D), quantified by the area of CD144+ channels (Fig. 5F). Consistent with these, CD144 and Tie-1 (a VM-associated gene) expression was significantly down-regulated in Notch3 KD 1205Lu xenografts compared to the control tumors (Fig. 6A and B). Since VM formation in the xenografts is intimately associated with authentic host vessels in keeping with perivascular niche (5, 30), we examined the impact of Notch3 KD on in-grown mouse vessels within the melanoma xenografts. Accordingly, we found that host angiogenesis, characterized by mCD31+ blood vessels, was significantly reduced in Notch3 KD 1205Lu xenografts as measured by the mCD31+ luminal area and number of mCD31+ tubules (Fig. 5E). These results demonstrate that Notch3 KD inhibits tumor angiogenesis as well as VM formation in vivo, both of which are integral components of the MSLC niche. To further analyze Notch3 KD on MSLC homeostasis, we examined expression of stem cell markers, CD133 and CD271, in xenografts by Western blot analysis. We found that protein expression of CD133 and CD271 was reduced in Notch3 KD 1205Lu xenografts compared to their non-target control counterparts (Fig. 6A), suggesting that Notch3 KD in 1205Lu melanoma cells leads to loss of stem-like cell phenotype in vivo. Consistent with the observed differential effect of Notch3 KD on tumorigenicity in A375 melanoma cells, the expression of stem-like cell marker, CD271, was not affected in Notch3 KD A375 xenografts (Fig. 6C). Attempts to further validate CD133 and CD144 in Notch3 KD A375 xenografts using Western blot analysis (C8161 lysate included as a positive control) were unsuccessful due to sensitivity issue (Fig. 6C; note that CD133 and CD144 signals were undetectable both in the control and KD xenografts despite maximal loading and prolonged overnight exposure). Collectively, these data suggest that Notch3 KD may inhibit perivascular niche morphogenesis and MSLC homeostasis through impeding tumor angiogenesis and VM. However, such effects are cell context-dependent.
Discussion
A major gap in MSLC research (or CSC research in general) is the lack of simple and easily reproducible in vitro models that takes into account the relevant and pivotal role of the niche microenvironment. Since the existence in a supportive “niche” (5, 30) is integral to the functional integrity of CSCs, studies on isolated CSCs without the “niche” context are inherently suboptimal. Despite significant advances in mouse xenograft models that provide accessible platforms to study MSLC biology in the mouse host “niche” environment, time and financial constraints as well as the individual host variability negatively impact on their routine use. To facilitate dissecting the niche signals responsible for MSLC “stemness” as a gateway to develop all-encompassing targeting strategies for MSLCs, whether pre-existing or dynamically induced, we developed a robust and easily reproducible 2D in vitro co-culture model system that is simple enough to be manipulated yet sophisticated enough to reflect the complexities inherent to the niche environment. This melanoma-endothelium co-culture niche model simulates MSLC niche, where the vascular/VM niche and “dynamic stemness” are recapitulated. In this model, stem cell (CD133 and CD271) and VM (CD144/VE-cadherin) markers are induced in non-MSLCs upon co-culture with ECs (Fig. 1). Using flow cytometry, various MSLC subsets can be isolated, permitting subsequent expression profiling to identify differentially expressed genes comparing to the non-MSLCs. In addition, this model allows manipulation of single or multiple variables sequentially or simultaneously (with the use of conventional and inducible vectors) in one or both cellular constituents, maximizing the ability to elucidate gene interactions in a defined system that preserves tissue context. More importantly, such a model can be easily adapted for other solid cancers, providing a robust and well-defined experimental system to study CSC-stromal interplay.
Given that Notch signaling mediated cell-cell communication is known to play major roles in cell fate decisions, lineage specification, as well as stem cell homeostasis in multiple organ systems not only during embryonic development but also in adult life (31), it is not surprising that we identified Notch3 as one of the key differentially expressed genes by the dynamically induced CD133+ MSLCs in the 2D niche co-culture model using pathway-specific PCR arrays following segregation by flow cytometry. Accordingly, Notch3 KD in 1205 Lu melanoma cell lines expressing high levels of endogenous Notch3 resulted in retarded/abolished tumorigenicity (Fig. 5A), as well as, attenuated MSLC subsets both in the 2D niche co-culture system in vitro (Fig. 4D) and in the xenograft model in vivo (Fig. 6A). These data support a pivotal role of Notch3 in MSLC induction, maintenance and homeostasis at least in a subset of melanomas. Parallel to our findings, a novel γ-secretase inhibitor (GSI), RO4929097, was shown to inhibit melanoma cell proliferation, colony-forming ability, and 3D spheroid growth in vitro and to suppress tumor-initiating potential in a serial xenotransplantation model (32). In addition, Notch3-signaling enriches CSCs of the lung (33), colorectal (34), hepatocellular (35), and breast (36) cancers, and Notch2/Notch3 antagonists/dominant-negative receptors inhibit tumor growth and decreases tumor-initiating cell frequency in a broad spectrum of epithelial tumors, including breast, lung, ovarian, and pancreatic cancer (37).
Derangements of Notch pathways are commonly observed in human cancers, including melanoma, ovarian, endothelial, cervical, lung, breast, renal, head and neck, prostate, mesothelioma, and various hematological malignancies (38). While the oncogenic functions of Notch1 in melanoma have been well established (39, 40), the roles of the other Notch receptors in melanoma remain substantially uncharted. Only recently, Hardy et al. (24), reported that Notch4 promotes melanoma aggressiveness, including VM and anchorage-independent growth through Nodal, an embryonic morphogen of the TGF-β superfamily implicated in the maintenance of embryonic stem cells. Consistent with this, global Notch inhibition by γ-secretase inhibitors (GSIs) results in reduced tumor initiating potential of melanoma in vitro in soft agar and spheroid formation assays, as well as in vivo in a serial xenotransplantation model (32). In another study, GSI treatment leads to melanoma regression through Noxa-mediated apoptosis in mice (25). Recently, Howard et al.(27, 41), in a co-culture system, identified Notch3 as one of the key mediators of melanoma-EC communication, whose expression correlates with tumor progression, and activated Notch3 signaling results in accelerated melanoma cell migration without affecting tumor cell growth. These findings are in line with our observations that Notch3 KD in melanoma cell lines expressing high endogenous levels of Notch3 abolished VM both in vitro in Matrigel™ tube formation assays (Fig. 4C) and our 2D co-culture niche model (Fig. 4D), and in vivo in tumorigenicity xenograft model (Fig. 5D, 5F, and 6A). Contrarily, Virtanen et al, 2013 reported that blockage of Notch signaling with GSIs, DAPT and dibenzazeoine, or Jag1 neutralizing Ab stabilizes VM in Mel Cher, Mel Kor, and Mel P melanoma cells in mono-culture and in mouse xenografts (42), indicating that Notch signaling attenuates VM. Moreover, Cui et al. (43) showed that Notch3 is down-regulated during melanoma progression and ectopic expression of NICD3 in A375P melanoma cell line leads to growth inhibition through senescence in conventional 2D culture. Correspondingly, our data also demonstrated that Notch3 KD in A375 melanoma cells, exhibiting relatively low endogenous Notch3 expression, failed to affect tumor growth (Fig. 5A), MSLC frequencies (Fig. 6C), and vascular/VM niche in vivo (data not shown). These discrepancies highlight the complexity and the importance of biological/cellular context of Notch signaling in melanoma. Indeed, a plethora of studies have concluded that the biological responses to Notch not only are cell type-, disease stage-, and context-specific, but also depend on the different receptor/ligand pairing. Divergent activities, both oncogenic and tumor suppressor function, have been described (44–46). Aside from the contextual diversity of Notch signaling, other possible explanations for the lack of response to Notch3 KD in A375 melanoma cells are the potential functional redundancy of different Notch receptors, and the possible involvement of signaling pathways other than Notch. In particular, a potential role for Notch4 warrants further investigation as concurrent Notch4 upregulation was observed in co-cultured melanoma cells in our in vitro niche model (Fig. 2A) and Notch4 has been shown previously to control melanoma VM (24).
Notch3 KD abolishes the MSLC niche morphogenesis not only by blocking VM phenotypic switch in 1205Lu melanoma cells, but also by inhibiting tumor angiogenesis as measured by the number and size of CD31+ murine vascular networks (Fig. 5D and E), indicating Notch juxtacrine signaling between melanoma cells and stromal endothelium may mediate tumor angiogenesis. In concordance, Zeng et al. (47) showed that elevated Jag1 in head and neck squamous cell carcinoma triggers Notch activation in neighboring ECs and thereby promotes tumor angiogenesis. Of particular interest, similar Notch3-mediated tumor-stromal crosstalk is recently illuminated in the regulation of tumor dormancy and bone metastasis. Specifically, EC Dll4, induced by angiogenic factors in the tumor mircoenvironment, triggers Notch3 activation in neighboring tumor cells, which then promotes a tumorigenic phenotype permitting escape from dormancy (48, 49). In human breast cancer, interaction with osteoblasts in the bone marrow environment enhances Notch3 expression in the tumor cells, which accelerates bone metastasis (50). In accordance with these, upregulation of Dll1 in ECs in our in vitro niche model (Fig. 2A) suggests similar reciprocal melanoma-EC signaling may be in play.
In summary, we have identified Notch3 as a molecular switch controlling MSLC homeostasis and niche morphogenesis at least in a subset of melanomas. This is especially critical as effective targeting of MSLCs is compounded by their heterogeneous nature and genetic instability. Since Notch3 signaling promotes vascular niche, which in turn drives “dynamic stemness”, Notch3-targeted approaches not only address the diverse pre-existing MSLC subsets by abolishing their vascular niche, but also prevent induction of new MSLCs by blocking “dynamic stemness”. With the availability of existing Notch signaling inhibitors currently available in the clinic for Alzheimer’s disease and many others under development in the pharmaceutical industry, Notch signaling represents a novel and compelling target in melanoma. Of note, a phase 2 clinical trial using a GSI, RO4929097, showed minimal effects against metastatic melanoma likely due to inadequate drug levels since no significant inhibition of interleukin-2 (IL-2), a surrogate pharmacodynamic marker of Notch inhibition, was achieved in the peripheral blood T-cell assays (51). Nevertheless, considering the heterogeneity of melanoma both at clinico-pathologic and molecular/genetic levels, the expectation that a single agent will deliver significant treatment outcome in patients across the board is unrealistic. Nair et al. (52) recently showed that melanoma sensitivity to GSIs is dependent on functional Phosphatase and Tension Homolog (PTEN). Furthermore, it is now appreciated that clinical response to GSIs also relies on endogenous Notch activities of the tumors (53, 54). These studies, like ours, underline the contextual complexity of Notch signaling and may inform prospective genotyping or screening criteria for future patient selection. Combinatorial use of Notch-targeted strategies with cytotoxic agents has shown encouraging clinical efficacies in ovarian cancers (54–57) and nasopharyngeal carcinoma (58). Hence, Notch-targeted approaches in combination with conventional or current state-of-the-art pathway-specific targeting strategies poise tremendous potentials as new therapeutic regimens in treating melanoma.
Supplementary Material
Acknowledgments
Grant Support: This work was supported by the National Institutes of Health, National Cancer Institute grant no. R01CA138649, and a Research Scholars Award from Department of Dermatology, Boston University School of Medicine, awarded to M-Y. Hsu.
Dr. Meenhard Herlyn (Wistar Institute, Philadelphia, PA) is acknowledged for generously providing melanoma cell lines and reagents.
Footnotes
Disclosure/Duality of Interest: No potential conflicts of interest were disclosed.
Supplementary information is available at Laboratory Investigation’s website.
References
- 1.Abel EV, Aplin AE. Finding the root of the problem: the quest to identify melanoma stem cells. Front Biosci (Schol Ed) 2011;3:937–945. doi: 10.2741/198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shekhani MT, Jayanthy AS, Maddodi N, et al. Cancer stem cells and tumor transdifferentiation: implications for novel therapeutic strategies. American journal of stem cells. 2013;2(1):52–61. [PMC free article] [PubMed] [Google Scholar]
- 3.Somasundaram R, Villanueva J, Herlyn M. Intratumoral heterogeneity as a therapy resistance mechanism: role of melanoma subpopulations. Adv Pharmacol. 2012;65:335–359. doi: 10.1016/B978-0-12-397927-8.00011-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wicha MS. Cancer stem cells and metastasis: lethal seeds. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12(19):5606–5607. doi: 10.1158/1078-0432.CCR-06-1537. [DOI] [PubMed] [Google Scholar]
- 5.Lai CY, Schwartz BE, Hsu MY. CD133+ melanoma subpopulations contribute to perivascular niche morphogenesis and tumorigenicity through vasculogenic mimicry. Cancer research. 2012;72(19):5111–5118. doi: 10.1158/0008-5472.CAN-12-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roesch A, Fukunaga-Kalabis M, Schmidt EC, et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141(4):583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shmelkov SV, Butler JM, Hooper AT, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133− metastatic colon cancer cells initiate tumors. The Journal of clinical investigation. 2008;118(6):2111–2120. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park DM, Rich JN. Biology of glioma cancer stem cells. Molecules and cells. 2009;28(1):7–12. doi: 10.1007/s10059-009-0111-2. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Laterra J. Cancer stem cells: distinct entities or dynamically regulated phenotypes? Cancer research. 2012;72(3):576–580. doi: 10.1158/0008-5472.CAN-11-3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar SM, Liu S, Lu H, et al. Acquired cancer stem cell phenotypes through Oct4-mediated dedifferentiation. Oncogene. 2012;31(47):4898–4911. doi: 10.1038/onc.2011.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Li A, Glas M, et al. c-Met signaling induces a reprogramming network and supports the glioblastoma stem-like phenotype. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(24):9951–9956. doi: 10.1073/pnas.1016912108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barcellos-Hoff MH, Newcomb EW, Zagzag D, et al. Therapeutic targets in malignant glioblastoma microenvironment. Seminars in radiation oncology. 2009;19(3):163–170. doi: 10.1016/j.semradonc.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denysenko T, Gennero L, Roos MA, et al. Glioblastoma cancer stem cells: heterogeneity, microenvironment and related therapeutic strategies. Cell biochemistry and function. 2010;28(5):343–351. doi: 10.1002/cbf.1666. [DOI] [PubMed] [Google Scholar]
- 14.Reardon DA, Wen PY. Therapeutic advances in the treatment of glioblastoma: rationale and potential role of targeted agents. The oncologist. 2006;11(2):152–164. doi: 10.1634/theoncologist.11-2-152. [DOI] [PubMed] [Google Scholar]
- 15.Clarke MF, Becker MW. Stem cells: the real culprits in cancer? Scientific American. 2006;295(1):52–59. doi: 10.1038/scientificamerican0706-52. [DOI] [PubMed] [Google Scholar]
- 16.Herlyn M, Villanueva J. Sorting through the many opportunities for melanoma therapy. Pigment cell & melanoma research. 2011;24(5):975–977. doi: 10.1111/j.1755-148X.2011.00906.x. [DOI] [PubMed] [Google Scholar]
- 17.Schatton T, Frank MH. Cancer stem cells and human malignant melanoma. Pigment cell & melanoma research. 2008;21(1):39–55. doi: 10.1111/j.1755-148X.2007.00427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang D, Nguyen TK, Leishear K, et al. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer research. 2005;65(20):9328–9337. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- 19.Paulis YW, Soetekouw PM, Verheul HM, et al. Signalling pathways in vasculogenic mimicry. Biochimica et biophysica acta. 2010;1806(1):18–28. doi: 10.1016/j.bbcan.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Hendrix MJ, Seftor EA, Hess AR, et al. Vasculogenic mimicry and tumour-cell plasticity: lessons from melanoma. Nature reviews Cancer. 2003;3(6):411–421. doi: 10.1038/nrc1092. [DOI] [PubMed] [Google Scholar]
- 21.Folberg R, Hendrix MJ, Maniotis AJ. Vasculogenic mimicry and tumor angiogenesis. The American journal of pathology. 2000;156(2):361–381. doi: 10.1016/S0002-9440(10)64739-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Schaft DW, Seftor RE, Seftor EA, et al. Effects of angiogenesis inhibitors on vascular network formation by human endothelial and melanoma cells. Journal of the National Cancer Institute. 2004;96(19):1473–1477. doi: 10.1093/jnci/djh267. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Fukunaga-Kalabis M, Li L, et al. Developmental pathways activated in melanocytes and melanoma. Archives of biochemistry and biophysics. 2014;563:13–21. doi: 10.1016/j.abb.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hardy KM, Kirschmann DA, Seftor EA, et al. Regulation of the embryonic morphogen Nodal by Notch4 facilitates manifestation of the aggressive melanoma phenotype. Cancer research. 2010;70(24):10340–10350. doi: 10.1158/0008-5472.CAN-10-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin JZ, Stennett L, Bacon P, et al. p53-independent NOXA induction overcomes apoptotic resistance of malignant melanomas. Molecular cancer therapeutics. 2004;3(8):895–902. [PubMed] [Google Scholar]
- 26.Nickoloff BJ, Hendrix MJ, Pollock PM, et al. Notch and NOXA-related pathways in melanoma cells. The journal of investigative dermatology Symposium proceedings/the Society for Investigative Dermatology, Inc [and] European Society for Dermatological Research. 2005;10(2):95–104. doi: 10.1111/j.1087-0024.2005.200404.x. [DOI] [PubMed] [Google Scholar]
- 27.Howard JD, Moriarty WF, Park J, et al. Notch signaling mediates melanoma-endothelial cell communication and melanoma cell migration. Pigment cell & melanoma research. 2013;26(5):697–707. doi: 10.1111/pcmr.12131. [DOI] [PubMed] [Google Scholar]
- 28.Hsu MY, Elder DE, Herlyn M. The Wistar melanoma (WM) cell lines. In: Masters J, Palsson B, editors. Human cell culture. I. Nowell, MA: Kluwer; 1999. pp. 259–274. [Google Scholar]
- 29.Schnegg CI, Yang MH, Ghosh SK, et al. Induction of Vasculogenic Mimicry Overrides VEGF-A Silencing and Enriches Stem-like Cancer Cells in Melanoma. Cancer research. 2015;75(8):1682–1690. doi: 10.1158/0008-5472.CAN-14-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mak AB, Schnegg C, Lai CY, et al. CD133-targeted niche-dependent therapy in cancer: a multipronged approach. The American journal of pathology. 2014;184(5):1256–1262. doi: 10.1016/j.ajpath.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138(17):3593–3612. doi: 10.1242/dev.063610. [DOI] [PubMed] [Google Scholar]
- 32.Huynh C, Poliseno L, Segura MF, et al. The novel gamma secretase inhibitor RO4929097 reduces the tumor initiating potential of melanoma. PloS one. 2011;6(9):e25264. doi: 10.1371/journal.pone.0025264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arasada RR, Amann JM, Rahman MA, et al. EGFR blockade enriches for lung cancer stem-like cells through Notch3-dependent signaling. Cancer research. 2014;74(19):5572–5584. doi: 10.1158/0008-5472.CAN-13-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pasto A, Serafin V, Pilotto G, et al. NOTCH3 signaling regulates MUSASHI-1 expression in metastatic colorectal cancer cells. Cancer research. 2014;74(7):2106–2118. doi: 10.1158/0008-5472.CAN-13-2022. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Q, Lu C, Fang T, et al. Notch3 functions as a regulator of cell self-renewal by interacting with the beta-catenin pathway in hepatocellular carcinoma. Oncotarget. 2015;6(6):3669–3679. doi: 10.18632/oncotarget.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sansone P, Ceccarelli C, Berishaj M, et al. Self-renewal of CD133(hi) cells by IL6/Notch3 signalling regulates endocrine resistance in metastatic breast cancer. Nature communications. 2016;7:10442. doi: 10.1038/ncomms10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yen WC, Fischer MM, Axelrod F, et al. Targeting Notch signaling with a Notch2/Notch3 antagonist (tarextumab) inhibits tumor growth and decreases tumor-initiating cell frequency. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21(9):2084–2095. doi: 10.1158/1078-0432.CCR-14-2808. [DOI] [PubMed] [Google Scholar]
- 38.Nickoloff BJ, Osborne BA, Miele L. Notch signaling as a therapeutic target in cancer: a new approach to the development of cell fate modifying agents. Oncogene. 2003;22(42):6598–6608. doi: 10.1038/sj.onc.1206758. [DOI] [PubMed] [Google Scholar]
- 39.Liu ZJ, Xiao M, Balint K, et al. Notch1 signaling promotes primary melanoma progression by activating mitogen-activated protein kinase/phosphatidylinositol 3-kinase-Akt pathways and up-regulating N-cadherin expression. Cancer research. 2006;66(8):4182–4190. doi: 10.1158/0008-5472.CAN-05-3589. [DOI] [PubMed] [Google Scholar]
- 40.Balint K, Xiao M, Pinnix CC, et al. Activation of Notch1 signaling is required for beta-catenin-mediated human primary melanoma progression. The Journal of clinical investigation. 2005;115(11):3166–3176. doi: 10.1172/JCI25001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stine MJ, Wang CJ, Moriarty WF, et al. Integration of genotypic and phenotypic screening reveals molecular mediators of melanoma-stromal interaction. Cancer research. 2011;71(7):2433–2444. doi: 10.1158/0008-5472.CAN-10-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vartanian A, Gatsina G, Grigorieva I, et al. The involvement of Notch signaling in melanoma vasculogenic mimicry. Clinical and experimental medicine. 2013;13(3):201–209. doi: 10.1007/s10238-012-0190-9. [DOI] [PubMed] [Google Scholar]
- 43.Cui H, Kong Y, Xu M, et al. Notch3 functions as a tumor suppressor by controlling cellular senescence. Cancer research. 2013;73(11):3451–3459. doi: 10.1158/0008-5472.CAN-12-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J, Fukunaga-Kalabis M, Li L, et al. Developmental pathways activated in melanocytes and melanoma. Archives of biochemistry and biophysics. 2014 doi: 10.1016/j.abb.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lobry C, Oh P, Mansour MR, et al. Notch signaling: switching an oncogene to a tumor suppressor. Blood. 2014;123(16):2451–2459. doi: 10.1182/blood-2013-08-355818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Danza G, Di Serio C, Rosati F, et al. Notch signaling modulates hypoxia-induced neuroendocrine differentiation of human prostate cancer cells. Molecular cancer research : MCR. 2012;10(2):230–238. doi: 10.1158/1541-7786.MCR-11-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng Q, Li S, Chepeha DB, et al. Crosstalk between tumor and endothelial cells promotes tumor angiogenesis by MAPK activation of Notch signaling. Cancer cell. 2005;8(1):13–23. doi: 10.1016/j.ccr.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 48.Indraccolo S. Insights into the regulation of tumor dormancy by angiogenesis in experimental tumors. Advances in experimental medicine and biology. 2013;734:37–52. doi: 10.1007/978-1-4614-1445-2_3. [DOI] [PubMed] [Google Scholar]
- 49.Indraccolo S, Minuzzo S, Masiero M, et al. Cross-talk between tumor and endothelial cells involving the Notch3-Dll4 interaction marks escape from tumor dormancy. Cancer research. 2009;69(4):1314–1323. doi: 10.1158/0008-5472.CAN-08-2791. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Z, Wang H, Ikeda S, et al. Notch3 in human breast cancer cell lines regulates osteoblast-cancer cell interactions and osteolytic bone metastasis. The American journal of pathology. 2010;177(3):1459–1469. doi: 10.2353/ajpath.2010.090476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee SM, Moon J, Redman BG, et al. Phase 2 study of RO4929097, a gamma-secretase inhibitor, in metastatic melanoma: SWOG 0933. Cancer. 2015;121(3):432–440. doi: 10.1002/cncr.29055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nair JS, Sheikh T, Ho AL, et al. PTEN regulates sensitivity of melanoma cells to RO4929097, the gamma-secretase inhibitor. Anticancer Res. 2013;33(4):1307–1316. [PubMed] [Google Scholar]
- 53.Saito N, Fu J, Zheng S, et al. A high Notch pathway activation predicts response to gamma secretase inhibitors in proneural subtype of glioma tumor-initiating cells. Stem Cells. 2014;32(1):301–312. doi: 10.1002/stem.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morgan SL, Wyant GA, Dinulescu DM. “Take it up a NOTCH”: novel strategies for cancer therapy. Cell Cycle. 2013;12(2):191–192. doi: 10.4161/cc.23375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kang H, Jeong JY, Song JY, et al. Notch3-specific inhibition using siRNA knockdown or GSI sensitizes paclitaxel-resistant ovarian cancer cells. Molecular carcinogenesis. 2015 doi: 10.1002/mc.22363. [DOI] [PubMed] [Google Scholar]
- 56.Park JT, Chen X, Trope CG, et al. Notch3 overexpression is related to the recurrence of ovarian cancer and confers resistance to carboplatin. The American journal of pathology. 2010;177(3):1087–1094. doi: 10.2353/ajpath.2010.100316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McAuliffe SM, Morgan SL, Wyant GA, et al. Targeting Notch, a key pathway for ovarian cancer stem cells, sensitizes tumors to platinum therapy. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(43):E2939–2948. doi: 10.1073/pnas.1206400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Man CH, Wei-Man Lun S, Wai-Ying Hui J, et al. Inhibition of NOTCH3 signalling significantly enhances sensitivity to cisplatin in EBV-associated nasopharyngeal carcinoma. The Journal of pathology. 2012;226(3):471–481. doi: 10.1002/path.2997. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.