Highlights
-
•
Smallest floR plasmid in the Pasteurellaceae.
-
•
Unique arrangement with complete strA, but partial strB and sul2 sequences.
-
•
Loss of mobilisation genes indicated by partial mobC; no other mob genes.
-
•
Not transferrable by conjugation or natural transformation.
Keywords: Antibiotic resistance, Florfenicol, Respiratory tract, Pasteurellaceae
Abstract
A small (3.9 kb) plasmid (p518), conferring resistance to florfenicol (MIC >8 μg/mL) and chloramphenicol (MIC >8 μg/mL) was isolated from an Actinobacillus pleuropneumoniae clinical isolate from Southeastern Brazil. To date, this is the smallest florfenicol resistance plasmid isolated from a member of the Pasteurellaceae. The complete nucleotide of this plasmid revealed a unique gene arrangement compared to previously reported florfenicol resistance plasmids found in other members of the Pasteurellaceae. In addition to the floR gene and a lysR gene, common to various florfenicol resistance plasmids, p518 also encodes strA and a partial strB sequence. An origin of replication (oriV) similar to that in the broad host range plasmid, pLS88, was identified in p518, and transformation into Escherichia coli MFDpir confirmed the ability to replicate in other species. Mobilisation genes appear to have been lost, with only a partial mobC sequence remaining, and attempts to transfer p518 from a conjugal donor strain (E. coli MFDpir) were not successful, suggesting this plasmid is not mobilisable. Similarly, attempts to transfer p518 into a competent A. pleuropneumoniae strain, MIDG2331, by natural transformation were also not successful. These results suggest that p518 may be only transferred by vertical descent.
1. Introduction
Porcine pleuropneumonia is a severe respiratory disease causing extensive economic losses in the swine industry worldwide, and Actinobacillus pleuropneumoniae, a member of the Pasteurellaceae family, is the main etiologic agent of this disease (Gottschalk, 2012). Reductions in morbidity and mortality can be achieved to some degree using good husbandry practices and vaccination, however subclinical and acute disease may still occur (Gottschalk, 2015). Treatment with antimicrobial agents is necessary to limit the spread and severity of disease in the case of clinically active infection.
In many countries, the most commonly used antimicrobial agents for treating porcine respiratory disease are tetracyclines, macrolides/lincosamides, aminoglycosides, beta-lactams, and trimethoprim/sulphonamides (Guardabassi et al., 2008). There is currently no information regarding relative amounts of different antimicrobial agents used for veterinary treatments in Brazil (Bokma et al., 2014), however florfenicol, an antimicrobial agent used exclusively in veterinary medicine, has only been used in Brazil since 1996 (SINDAN, 2013). Levels of resistance to florfenicol are still very low for A. pleuropneumoniae in most countries (Dayao et al., 2014, Garch et al., 2016, Vanni et al., 2012), except in Korea where extensive use of this antimicrobial agent has led to resistance in 34% of isolates tested (Yoo et al., 2014). A. pleuropneumoniae isolates resistant to florfenicol have been shown to harbour the floR gene encoding a chloramphenicol/florfenicol efflux pump, FloR (Kucerova et al., 2011, Yoo et al., 2014, Bossé et al., 2015). The prevalence of florfenicol resistance amongst A. pleuropneumoniae isolates in Brazil has yet to be determined, however we recently identified a clinical isolate, MV518, from Southeastern Brazil that carries the floR gene, as identified by Resfinder analysis (Zankari et al., 2012) of the draft genome sequence (Pereira et al., 2015). Here we describe the isolation and complete nucleotide sequence of a 3.9 kb plasmid, p518, from A. pleuropneumoiae MV518.
2. Materials and methods
2.1. Bacterial strain and antimicrobial susceptibility testing
A. pleuropneumoniae MV518, a serovar 8 clinical isolate, was kindly provided by Microvet (Minas Gerais). Minimum inhibitory concentrations (MICs) for florfenicol and chloramphenicol were determined by the broth microdilution susceptibility assay, according to the CLSI VET01-A4 guidance (CLSI, 2013).
2.2. Sequence analysis and plasmid isolation
The whole genome sequence of MV518 (accession number JSVZ00000000), previously reported (Pereira et al., 2015), was analysed using ResFinder (Zankari et al., 2012) for identification of resistance genes, using a threshold of 98% identity and minimum length of 60%. Further sequence analysis was performed using BLASTn and BLASTx (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Plasmid sequence alignments were performed using ClustalW (Larkin et al., 2007).
Plasmid DNA was extracted from MV518 using the Qiaprep extraction kit (Qiagen). Inverse PCR was performed, using 1U of Platinum® Taq DNA polymerase (Invitrogen) according to the manufacturer’s instructions, with outward facing primers (strA_I_F TAACGCCGAAGAGAACTGGG and strA_I_R AAGTTGCTGCCCCATTGACG) designed to amplify the plasmid sequence found between the 5′ and 3′ ends of the strA gene (see Fig. 1). The resulting amplicon was purified with the QIAquick PCR Purification Kit (Qiagen) and completely sequenced using a primer walking strategy. The 3937 bp sequence of p518 was manually annotated and deposited in Genbank under the accession number KT355773.
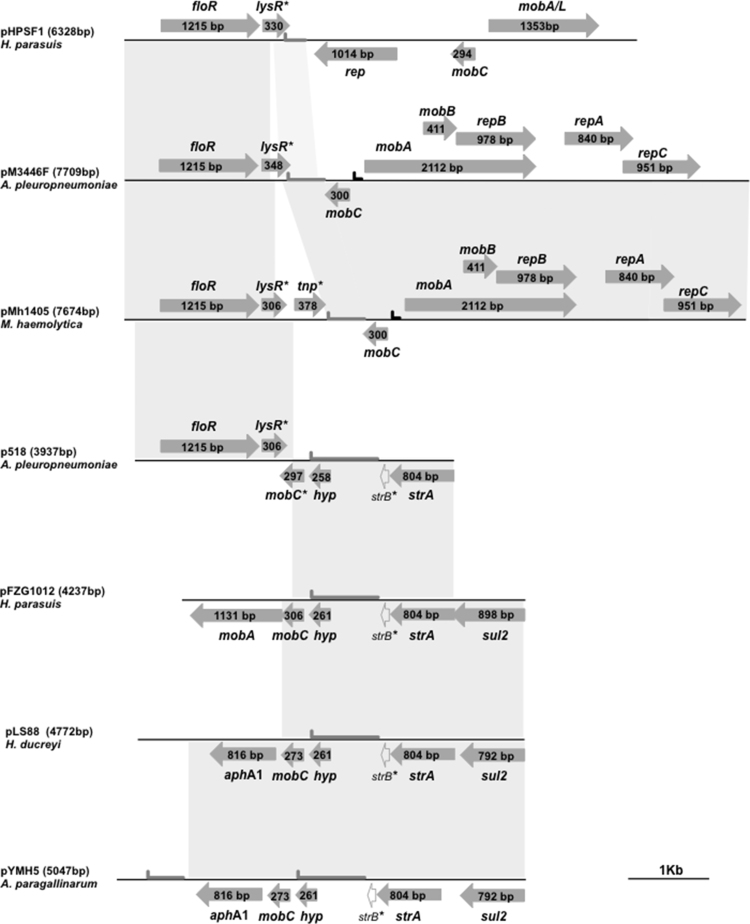
Fig. 1.
Schematic comparison of p518, a florfenicol resistance plasmid from A. pleuropneumoniae strain MV518, with other Pasteurellaceae plasmids. Open reading frames are indicated by arrows, with arrowheads showing direction of transcription (floR: florfenicol resistance; lysR*: partial lysR transcriptional regulator; tnp*: truncated transposase gene; mob, mobA, mobB, mobC, mobA/L: plasmid mobilisation; mobC*: partial mobC gene; repA, repB, repC: plasmid replication; sul2: sulphonamide resistance; strA: streptomycin resistance; strB*: partial strB streptomycin resistance; hyp: hypothetical gene). The predicted origins of replication (oriV;  ) and transfer (oriT;
) and transfer (oriT;  ) are shown. Grey blocks between sequences indicate ≥ 98% nucleotide sequence identity. A distance scale in kb is shown.
) are shown. Grey blocks between sequences indicate ≥ 98% nucleotide sequence identity. A distance scale in kb is shown.
2.3. Transformation and conjugation
Initially, p518 was transformed into E. coli MFDpir (Ferrieres et al., 2010) by heat shock, with selection on Luria-Bertani agar plates containing florfenicol (10 μg/mL) and 0.3 mM diaminopimelic acid (DAP: required for growth of the MFDpir strain). The presence of p518 in transformants was confirmed by PCR amplification of a floR gene sequence using primers floR1 (5′-GCGATATTCATTACTTTGGC-3′) and floR2 (5′-TAGGATGAAGGTGAGGAATG-3′), and plasmid size was confirmed by linearisation with SacI. Subsequently, mobilisation of p518 from E. coli MFDpir into the plasmid-free, florfenicol sensitive A. pleuropneumoniae strain MIDG2331 (Bossé et al., 2016), was assessed in three separate experiments, using a previously described conjugation protocol (Bossé et al., 2009). For determination of plasmid uptake by natural transformation, three separate experiments compared the ability of either 1 μg of uncut purified plasmid p518, or 1 μg of control genomic DNA (carrying a dfrA14 trimethoprim resistance gene inserted in the chromosome), to transform MIDG2331, as previously described (Bossé et al., 2014), with selection of transformants on BHI agar supplemented with 0.01% NAD and either 5 μg/mL florfenicol, or 10 μg/mL trimethoprim, as appropriate.
3. Results and discussion
Analysis of the draft genome of MV518 using Resfinder revealed a contig (Accession number JSVZ00000000, contig 40) containing the floR and strA resistance genes. When tested by the broth microdilution susceptibility assay (CLSI, 2013), strain MV518 had an MIC >8 μg/mL for both florfenicol and chloramphenicol, consistent with floR encoding a chloramphenicol/florfenicol efflux pump (Braibant et al., 2005).
A 3.9 kb plasmid, p518, was extracted from MV518 and the complete nucleotide sequence determined. Sequence analysis revealed that p518 appears to have arisen by recombination of a region encoding the 1215 bp floR gene and a 306 bp ORF showing similarity to predicted lysR genes, with sequences from a plasmid encoding an 804 bp strA gene followed by a truncated (98 bp) strB sequence (Fig. 1). Although the sequence containing the floR and lysR genes is common to numerous floR plasmids, including those found in members of the Pasteurellaceae such as pM3446F (accession number KP696484) from A. pleuropneumoniae (Bossé et al., 2015), and pHPSF1 (accession number KR262062) from Haemophilus parasuis (Li et al., 2015), the highest similarity (99% identity) over the first 1933 bp of p518 was seen with the Mannheimia haemolytica plasmid, pMh1405 (accession number AB621552) (Katsuda et al., 2012). The remaining sequence of p518 shows greatest identity (2003 of 2004 bp) to part of a 4237 bp plasmid from H. parasuis, strain FZG1012 (accession number HQ015158), which carries the resistance genes sul2 and strA, as well as mobilisation genes mobC and mobA (Liu et al., 2011). This 2 kb sequence is also highly similar to regions of pYMH5, a 5047 bp plasmid from Avibacterium paragallinarum (Hsu et al., 2007), and pLS88 (accession number L23118), a 4772 bp plasmid from Haemophilus ducreyi (Dixon et al., 1994), having 2002 or 1994 identical bases, respectively (Fig. 1). Although only 14 bases of the sul2 gene (3′ end), and 141 bases of the mobC gene (coding for the first 47 AAs), can be identified in the p518 sequence, the strA gene, as well as the broad-host-range origin of replication (oriV), are complete (Fig. 1). We confirmed the ability of p518 to replicate in another species by transforming it into E. coli MFDpir (Ferrieres et al., 2010).
Although a 297 bp ORF was identified which shares 100% identity over the first 142 bp with the mobC gene found in the other Pasteurellaceae sul2/strA plasmids described above, the remaining 155 bp do not match known mobC genes. It is likely that the recombination between the floR/lysR containing sequence and the strA containing plasmid resulted in an alternate stop codon for this ORF in p518, and it is not known if it encodes a functional protein. Similarly, the sequences of the lysR genes in the various plasmids shown in Fig. 1 all share a common 5′ end, but differ at their 3′ ends (suggesting different stop codons acquired following recombination), and it is not known if any of these genes encode a functional regulatory protein of the LysR family.
We tested the ability of p518 to be mobilised from the DAP-dependent E. coli donor strain, MFDpir, which encodes all of the genes required for generating conjugal transfer machinery (Ferrieres et al., 2010), into a plasmid-free recipient strain of A. pleuropneumoniae, MIDG2331 (Bossé et al., 2016), however we were unable to detect transconjugants containing p518. It appears that p518 is not capable of being transferred via conjugation, which is consistent with the lack of mobilisation genes in this plasmid.
As isolates of A. pleuropneumoniae, including MIDG2331, are known to be competent for natural transformation (Bossé et al., 2014), we investigated the possibility that p518 could be transferred horizontally by this mechanism. The sequence of this plasmid contains seven of the nine bases of the uptake sequence (ACAAGCGGT; bases in p518 are underlined) known to be required for efficient transformation of A. pleuropneumoniae (Redfield et al., 2006). A transformation frequency of ×10−5 was achieved using the control genomic DNA, whereas the p518 plasmid yielded no detectable transformants.
In summary, to the best of our knowledge, p518 is the smallest florfenicol resistance plasmid so far isolated from a member of the Pasteurellaceae, and this is the first report of florfenicol resistance in A. pleuropneumoniae in South America. Plasmid p518 shows a novel gene organization, sharing regions of high sequence identity with other Pasteurellaceae plasmids, suggesting that the floR gene may have been exchanged between different plasmids in closely related pathogens (and/or commensals) that share that same host environment. Despite the fact that we were not able to demonstrate horizontal transfer of p518 by either mobilisation or natural transformation, it is apparent from the conservation of the floR/lysR region in this and numerous other plasmids, that this is a highly recombinogenic sequence, and p518 could therefore act as a donor for spread of the floR gene into other, more mobile, plasmids. An antimicrobial susceptibility survey of A. pleuropneumoniae isolates in Brazil is required to determine the prevalence of resistance to florfenicol and other important antimicrobial agents, and continued surveillance to monitor spread of resistance is essential.
Conflict of interest
We declare that we have no conflict of interest.
Funding
This work was supported by Fundação de Amparo à Pesquisa do Estado de Minas Gerais – FAPEMIG (APQ-02732-15); Conselho Nacional de Desenvolvimento Científico e Tecnológico – CNPq; Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – CAPES/PROEX, and the Biotechnology and Biological Sciences Research Council (BB/K021109/1, BB/G018553, and BB/M023052/1). Funding for JB was also provided by CONFAP - the UK Academies Fellowship (FAPEMIG – APQ-00689-16).
Acknowledgements
The authors wish to thank Microvet – Microbiologia Veterinária Especial (Minas Gerais, Brazil) for providing the isolate, MV518, and Fábio Assad Féres Rodrigues for assistance with the antimicrobial susceptibility testing.
Contributor Information
Janine T. Bossé, Email: j.bosse@imperial.ac.uk.
Denise Mara Soares Bazzolli, Email: dbazzolli@ufv.br.
References
- Bokma M., Bondt N., Neijenhuis F., Mevius D., Ruiter S. Wageningen UR Livestock Research Report 714; 2014. Antibiotic Use in Brazilian Broiler and Pig Production: an Indication and Forecast of Trends.http://edepot.wur.nl/297414 [Google Scholar]
- Bossé J.T., Durham A.L., Rycroft A.N., Kroll J.S., Langford P.R. New plasmid tools for genetic analysis of Actinobacillus pleuropneumoniae and other Pasteurellaceae. Appl. Environ. Microbiol. 2009;75:6124–6131. doi: 10.1128/AEM.00809-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossé J.T., Soares-Bazzolli D.M., Li Y., Wren B.W., Tucker A.W., Maskell D.J., Rycroft A.N., Langford P.R., BRaDP1T consortium The generation of successive unmarked mutations and chromosomal insertion of heterologous genes in Actinobacillus pleuropneumoniae using natural transformation. PLoS One. 2014;9:e111252. doi: 10.1371/journal.pone.0111252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossé J.T., Li Y., Atherton T.G., Walker S., Williamson S.M., Rogers J., Chaudhuri R.R., Weinert L.A., Holden M.T.G., Maskell D.J., Tucker A.W., Wren B.W., Rycroft A.N., Langford P.R., BRaDP1T consortium Characterisation of a mobilisable plasmid conferring florfenicol and chloramphenicol resistance in Actinobacillus pleuropneumoniae. Vet. Microbiol. 2015;178:279–282. doi: 10.1016/j.vetmic.2015.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossé J.T., Chaudhuri, RR, Li Y., Leanse L.G., Fernandez Crespo R., Coupland P., Holden M.T.G., Bazzolli D.M., Maskell D.J., Tucker A.W., Wren B.W., Rycroft A.N., Langford P.R., BRaDP1T consortium Complete genome sequence of MIDG2331, a genetically tractable serovar 8 clinical isolate of Actinobacillus pleuropneumoniae. Genome Announc. 2016;4:15–e01667. doi: 10.1128/genomeA.01667-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braibant M., Chevalier J., Chaslus-Dancla E., Pagès J.M., Cloeckaert A. Structural and functional study of the phenicol-specific efflux pump FloR belonging to the major facilitator superfamily. Antimicrob. Agents Chemother. 2005;49:2965–2971. doi: 10.1128/AAC.49.7.2965-2971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals - Fourth Edition: Approved Standard VET01-A4. CLSI, Wayne, PA, USA, 2013.
- Dayao D.A.E., Gibson J.S., Blackall P.J., Turni C. Antimicrobial resistance in bacteria associated with porcine respiratory disease in Australia. Vet. Microbiol. 2014;171:232–235. doi: 10.1016/j.vetmic.2014.03.014. [DOI] [PubMed] [Google Scholar]
- Dixon L.G., Albritton W.L., Willson P.J. An analysis of the complete nucleotide sequence of the Haemophilus ducreyi broad-host-range plasmid pLS88. Plasmid. 1994;32:228–232. doi: 10.1006/plas.1994.1060. [DOI] [PubMed] [Google Scholar]
- Ferrieres L., Hemery G., Nham T., Guerout A.M., Mazel D., Beloin C., Ghigo J.M. Silent mischief: bacteriophage Mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. J. Bacteriol. 2010;192:6418–6427. doi: 10.1128/JB.00621-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garch El F., de Jong A., Simjee S., Moyaert H., Klein U., Ludwig C., Marion H., Haag-Diergarten S., Richard-Mazet A., Thomas V., Siegwart E. Monitoring of antimicrobial susceptibility of respiratory tract pathogens isolated from diseased cattle and pigs across Europe, 2009–2012: VetPath results. Vet. Microbiol. 2016 doi: 10.1016/j.vetmic.2016.04.009. [DOI] [PubMed] [Google Scholar]
- Gottschalk M. Actinobacillosis. In: Karriker A., Ramirez K., Stevenson G., Zimmerman J., editors. Diseases of Swine. 10th edn. Wiley Hoboken; NJ: 2012. pp. 653–669. [Google Scholar]
- Gottschalk M. The challenge of detecting herds sub-clinically infected with Actinobacillus pleuropneumoniae. Vet. J. 2015;206:30–38. doi: 10.1016/j.tvjl.2015.06.016. [DOI] [PubMed] [Google Scholar]
- Guardabassi L., Jensen L.B., Kruse H., editors. Blackwell Publishing, Ltd; Oxford, UK: 2008. Guide to Antimicrobial Use in Animals. [Google Scholar]
- Hsu Y.-M., Shieh H.K., Chen W.-H., Sun T.-Y., Shiang J.-H. Antimicrobial susceptibility, plasmid profiles and haemocin activities of Avibacterium paragallinarum strains. Vet. Microbiol. 2007;124:209–218. doi: 10.1016/j.vetmic.2007.04.024. [DOI] [PubMed] [Google Scholar]
- Katsuda K., Kohmoto M., Mikami O., Tamamura Y., Uchida I. Plasmid-mediated florfenicol resistance in Mannheimia haemolytica isolated from cattle. Vet. Microbiol. 2012;155:444–447. doi: 10.1016/j.vetmic.2011.09.033. [DOI] [PubMed] [Google Scholar]
- Kucerova Z., Hradecka H., Nechvatalova K., Nedbalcova K. Antimicrobial susceptibility of Actinobacillus pleuropneumoniae isolates from clinical outbreaks of porcine respiratory diseases. Vet. Microbiol. 2011;150:203–206. doi: 10.1016/j.vetmic.2011.01.016. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and clustal x version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li B., Zhang Y., Wei J., Shao D., Liu K., Shi Y., Qiu Y., Ma Z. Characterization of a novel small plasmid carrying the florfenicol resistance gene floR in Haemophilus parasuis. J. Antimicrob. Chemother. 2015;70:3159–3161. doi: 10.1093/jac/dkv230. [DOI] [PubMed] [Google Scholar]
- Liu Y., He Q., Chen P., Chen H. Multiresistance in Haemophilus parasuis is mediated by small plasmids. Proceedings of the 5th Asian Pig Veterinary Society Congress; Pattaya, Thailand; 2011. [Google Scholar]
- Pereira M.F., Rossi C.C., de Carvalho F.M., de Almeida L.G.P., Souza R.C., de Vasconcelos A.T.R., Bazzolli D.M.S. Draft genome sequences of six Actinobacillus pleuropneumoniae serotype 8 Brazilian clinical isolates: insight into new applications. Genome Announc. 2015;3:e01585–14. doi: 10.1128/genomeA.01585-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfield R.J., Findlay W.A., Bossé J.T., Kroll J.S., Cameron A.D., Nash J.H. Evolution of competence and DNA uptake specificity in the Pasteurellaceae. BMC Evol. Biol. 2006;6:82. doi: 10.1186/1471-2148-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindicato Nacional da Indústria de Produtos para a Saúde Animal (SINDAN) Compêndio De Produtos Veterinários. Editora MedVet; São Paulo, Brasil: 2013. 1ª ed. [Google Scholar]
- Vanni M., Merenda M., Barigazzi G., Garbarino C., Luppi A., Tognetti R., Intorre L. Antimicrobial resistance of Actinobacillus pleuropneumoniae isolated from swine. Vet. Microbiol. 2012;156:172–177. doi: 10.1016/j.vetmic.2011.10.022. [DOI] [PubMed] [Google Scholar]
- Yoo A.N., Cha S.B., Shin M.K., Won H.K., Kim E.H., Choi H.W., Yoo H.S. Serotypes and antimicrobial resistance patterns of the recent Korean Actinobacillus pleuropneumoniae isolates. Vet. Rec. 2014;174:223. doi: 10.1136/vr.101863. [DOI] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Voldby Larsen M. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]