Abstract
Tumour heterogeneity poses a significant challenge for treatment stratification. The goals of this study were to quantify heterogeneity in hepatocellular carcinoma (HCC) using multiparametric magnetic resonance imaging (mpMRI), and to report preliminary data correlating quantitative MRI parameters with advanced histopathology and gene expression in a patient subset. Thirty-two HCC patients with 39 HCC lesions underwent mpMRI including diffusion-weighted imaging (DWI), blood-oxygenation-level-dependent (BOLD), tissue-oxygenation-level-dependent (TOLD) and dynamic contrast-enhanced (DCE)-MRI. Histogram characteristics [central tendency (mean, median) and heterogeneity (standard deviation, kurtosis, skewness) MRI parameters] in HCC and liver parenchyma were compared using Wilcoxon signed-rank tests. Histogram data was correlated between MRI methods in all patients and with histopathology and gene expression in 14 patients. HCCs exhibited significantly higher intra-tissue heterogeneity vs. liver with all MRI methods (P < 0.030). Although central tendency parameters showed significant correlations between MRI methods and with each of histopathology and gene expression, heterogeneity parameters exhibited additional complementary correlations between BOLD and DCE-MRI and with histopathologic hypoxia marker HIF1α and gene expression of Wnt target GLUL, pharmacological target FGFR4, stemness markers EPCAM and KRT19 and immune checkpoint PDCD1. Histogram analysis combining central tendency and heterogeneity mpMRI features is promising for non-invasive HCC characterization on the imaging, histologic and genomics levels.
Introduction
Quantitative functional multiparametric magnetic resonance imaging (mpMRI) of cancer allows for non-invasive assessment of several tumour characteristics, such as cellularity, perfusion and oxygenation, which can be used for tumour characterization and for assessing treatment response1. Apparent diffusion coefficient (ADC) quantification using diffusion-weighted imaging (DWI) has shown to correlate with tumour cellularity2. Tissue oxygenation can be indirectly assessed using blood oxygenation level-dependent (BOLD) and tissue oxygenation level-dependent (TOLD) MRI3. Tissue perfusion and flow can be quantitatively measured using dynamic contrast-enhanced MRI (DCE-MRI)4.
Many studies that employ mpMRI to assess/predict tumour response use central tendency parameters, such as mean or median, over entire regions of interest (ROIs) to determine longitudinal changes in the tumour tissue after treatment5. However, such analysis may not represent the exact tumour status, given the intrinsic heterogeneous tumour composition5. Heterogeneity analysis of tumour MRI measurements may provide accurate markers of tumour heterogeneity at the genetic, cellular and molecular levels5 and thereby allow for a better understanding of tumour characteristics that may affect treatment decisions.
Hepatocellular carcinoma (HCC) lesions are known to exhibit substantial intra- and inter-tumour heterogeneity, due to a large variety in etiological and genetic backgrounds and the long-time development of the disease6. Tumour heterogeneity poses a significant challenge for treatment stratification in HCC. While pathological and genetic heterogeneity in HCC lesions have been described7, imaging reports on HCC heterogeneity are extremely limited, with only one study reporting visual assessment of HCC heterogeneity on contrast-enhanced MRI8, with no study employing quantitative imaging measurements of HCC heterogeneity.
Tumour imaging phenotypes, including histogram features, may potentially correlate with the underlying genotype. Thus, non-invasive imaging, including computed tomography (CT) and MRI, can potentially be used as a surrogate for genomics and transcriptomics (radiogenomics)9–11. Radiogenomics could potentially be used for the prediction of certain molecular gene signatures12. A major advantage of imaging versus genomics or histopathological analysis is that imaging captures the entire tumour non-invasively12. In addition, combined analysis of imaging, histopathology and genomics data may improve characterization of tumours, because next to mutual information, the separate assessments may also provide additional, independent information on tumour properties, which may help predict outcomes12.
Immunotherapy has recently revolutionized treatment in cancer13, 14. The clinical experience with immunotherapy in HCC is growing, with recent and ongoing phase I trials15, 16. The success of such treatment heavily depends on tumour expression of immunotherapy targets, such as immune checkpoints. Identification of imaging features that correlate with gene expression of immunotherapy targets potentially allows for non-invasive prediction of immunotherapy outcome.
The objectives of our study were to: 1) quantify intra- and inter-tumour heterogeneity of HCC lesions using mpMRI, consisting of DCE-MRI, BOLD-MRI, TOLD-MRI and DWI, and 2) report our preliminary results correlating the quantitative MRI parameters with advanced histopathology and gene signatures and expression levels in a subset of patients.
Patients and Methods
Patients
This single-centre prospective study was compliant with the Health Insurance Portability and Accountability Act and approved by the institutional review board at the Icahn School of Medicine at Mount Sinai. Written informed consent was obtained from all subjects. From June 2013 to June 2016, 41 consecutive patients with HCC were enrolled. HCC was diagnosed based on routine imaging by two radiologists in consensus [observer 1 (MW) and observer 2 (CB), with 5 and 6 years of experience in body MRI, respectively], according to the Organ Procurement and Transplantation Network (OPTN) criteria17. Of the 41 initial patients, 9 patients were excluded because of severe motion in the DCE-MRI acquisition (n = 5), absence of contrast injection (n = 2) or low quality of BOLD-MRI data (n = 2). Thus, 32 patients were finally included (M/F 26/6, mean age 59 y, range 30–71 y), of which 31 had chronic liver disease [chronic hepatitis C (n = 18), chronic hepatitis B (n = 9), non alcoholic steatohepatitis (n = 2), cryptogenic cirrhosis (n = 1) and alcoholic cirrhosis (n = 1)]. For one patient without history of liver disease, HCC was confirmed at histopathology. Three patients underwent locoregional therapy with either transarterial chemoembolization (TACE) combined with radiofrequency ablation (n = 1), TACE only (n = 1) or radioembolization (n = 1) before MRI (range 111–178 days). Data of 23 of the included patients has been published in at least one of our previous publications3, 18, 19. The aim of the previous studies was mainly to quantify each of the individual MRI methods in HCC lesions and to assess test-retest repeatability. Assessment of tumour heterogeneity was beyond the scope of those studies.
MRI acquisition
The MRI acquisition was performed on 1.5 T (Siemens Aera, Siemens Healthineers, Erlangen, Germany; n = 19) or 3.0 T [Siemens Skyra (n = 5) or Siemens BioGraph mMR (n = 8)] systems. Each of the systems is equipped with a 32-channel spine and flexible body array coil for signal reception. Subjects were asked to fast for 6 hours to eliminate post-prandial effects on portal venous blood flow20. In addition to axial DWI, BOLD-MRI, TOLD-MRI and DCE-MRI, the MRI acquisition consisted of axial and coronal T2-weighted imaging, axial dual-echo chemical shift imaging, T1-weighted imaging before and at a delayed phase after contrast injection. The acquisition parameters for DWI, BOLD-MRI, TOLD-MRI and DCE-MRI are listed in Table 1. For BOLD-MRI and TOLD-MRI, R2* and R1 acquisitions were performed before and at the end of a respiratory oxygen challenge of 10–15 minutes at a flow rate of 15 l/min3. DCE-MRI acquisition consisted of 100 dynamic acquisitions. Eight seconds after the start of the acquisition, a half dose (0.05 mmol/kg) of gadobenate dimeglumine (Multihance, Bracco Diagnostics Inc) followed by a 25 ml saline flush was administered intravenously at a rate of 3 ml/s19. Half dose of the contrast agent was used to avoid saturation of the signal intensity in the DCE-MRI acquisition, because of the high T1 relaxivity of the contrast agent.
Table 1.
Acquisition parameters of the multiparametric MRI protocol.
| DWI | BOLD | TOLD | DCE-MRI | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Sequence type | Single-shot spin-echo EPI | 2D MGRE | 2D MGRE | 2D MGRE | 2D MGRE | 3D VFA | 2D IR-LL | 2D-IR-LL | 3D FLASH |
| Acquisition plane | Axial | Axial | Axial | Axial | Axial | Axial | Axial | Axial | Axial |
| MR system | All | Skyra (n = 5) | Aera (n = 10) | Aera (n = 9) | mMR (n = 8) | Skyra (n = 5) | Aera (n = 19) | mMR (n = 8) | All |
| TE (ms) | 74–81 | 2.5, 4.9, 7.4, 9.8, 12.3, 14.8, 17.2 | 4.8, 9.5, 14.3, 19.1, 23.9 | 1.7, 2.9, 4.3, 5.6, 6.9, 10.0, 15.0, 20.0, 25.0, 30.0, 35.0, 40.0 | 1.1, 2.4, 3.8, 5.2, 6.6, 8.0, 10.0, 12.0, 15.0, 20.0, 25.0, 30.0 | 1.2 | 1.0 | 1.2 | 1.0 |
| TR (ms) | 4500 (Skyra); one respiration (Aera/mMR); | 165 | 242 | 319 | 249 | 10 | 2.3 | 35.1 | 2.7 |
| FA (°) | 90 | 35 | 35 | 20 | 18 | 1, 10, 19 | 8 | 10 | 9.5–11.5 |
| TI (ms) | — | — | — | — | — | — | 42–1577 (32 TI’s) | 80–1445 (40 TI’s) | — |
| b-values (s/mm2) | 0, 15, 30, 45, 60, 75, 90, 105, 120, 135, 150, 175, 200, 400, 600, 800 | — | — | — | — | — | — | — | — |
| FOV (mm2) | 380 × 255 | 340 × 255 | 340 × 255 | 340 × 255 | 340 × 255 | 350 × 260 | 420 × 290 | 380 × 280 | 380 × 280 |
| Matrix | 320 × 240 | 512 × 384 | 384 × 288 | 384 × 288 | 384 × 288 | 384 × 288 | 128 × 88 | 384 × 288 | 384 × 288 |
| Slice thickness (mm) | 7–8 | 7 | 7 | 7 | 7 | 5 | 8 | 8 | 4–5 |
| Number of slices | 20–30 | 4–5 | 4–5 | 4–5 | 4–5 | 36 | 1–2 | 1 | 40 |
| Acceleration factor | 2 | 2 | 2 | 2 | 2 | 2 | — | — | 4 |
| Acquisition time (min:s) | 8:00 | 0:15 | 0:15 | 0:15 | 0:15 | 0:14 | 0:18 | 0:10 | 0:02 per dynamic |
EPI = echo planar imaging, FA = flip angle, FLASH = fast low angle shot, FOV = field-of-view, IR-LL = inversion recovery Look-Locker, MGRE = multi-echo gradient echo, TE = echo time, TI = inversion time, TR = repetition time, VFA = variable flip angle.
Image analysis
Image analysis was performed on MATLAB platform (version R2015a, MathWorks, Natick, MA, USA), unless specified otherwise. The analysis was performed by two observers in consensus [observer 1 and observer 3 (SH), an MRI physicist with 6 years of experience].
DWI analysis
DWI acquisition was performed with 16 b-values for the purpose of the estimation of intravoxel incoherent motion (IVIM) diffusion parameters21, 22. However, for this particular study, we only used 3 b-values (60, 400 and 800 s/mm2) to determine the apparent diffusion coefficient (ADC), which was estimated as the slope of a linear fit of the logarithmic signal intensity (SI) curves at these particular b-values.
BOLD/TOLD analysis
R2* maps were generated by fitting of a mono-exponential decay model to the pixel signal values at all acquired echoes3. T1 maps were calculated using the Look-Locker recovery equation23 or the Ernst equation for a spoiled gradient recalled echo (SPGR) sequence for the variable flip angle measurements24. The pixel T1 values were inverted to obtain R1 values. In addition, ΔR1 and ΔR2* maps were generated by subtraction of the baseline R1 and R2* measurements from the measurements after oxygen (ΔR1/2* = R1/2* post O2 − R1/2* pre O2)3.
DCE-MRI analysis
Prior to pharmacokinetic analysis, motion correction was applied on the DCE-MR images with 3D rigid registration using open-source image analysis software (Firevoxel, CAI2R, New York University, New York, NY, USA). Free-hand 3D regions-of-interest (ROIs) were drawn in the portal vein on the registered images and in the abdominal aorta at the level of the celiac trunk on the unregistered images. Pixel dynamic SI curves were converted to dynamic contrast agent concentration ([CA]) curves by using the signal equation for a SPGR sequence, pre-contrast R1 values as determined from the baseline TOLD measurements (registered to the DCE-MRI measurements, see below) and the contrast agent’s relaxivity25. For the portal vein and aorta, pre-contrast R1 values were based on literature26. A haematocrit value of 0.45 was used for conversion from blood [CA] to plasma [CA]. Contrast agent relaxivity values of 8.1 mM−1 s−1 and 6.3 mM−1 s−1 were used at 1.5 T and 3.0 T, respectively27.
Pharmacokinetic parameter maps were generated by fitting of the dual-input signal compartment model to the dynamic [CA] curves in each pixel28. The following parameters were estimated in tumours and liver parenchyma: arterial flow (Fa), portal flow (Fp), total flow (Ft = Fa + Fp), arterial fraction (ART = Fa/Ft), mean transit time (MTT) and the distribution volume fraction of the contrast agent through the tissue (DV).
Coregistration of mpMRI data
Although the orientation of all sequences was in the axial plane, there were acquisition differences related to different matrix size, field of view, offset and subject motion. Therefore, a coregistration algorithm (MATLAB function fitgeotrans) was used to register the mpMRI data. Landmark points were placed on both the raw DCE-MR images and the corresponding BOLD, TOLD or ADC parameter maps. Subsequently, nonrigid coregistration was performed, visually checked and repeated if necessary. The coregistration was only performed in slices that contained HCC lesion(s) and that were common to all acquisitions.
Region of interest (ROI) analysis
ROIs in the HCC lesions and liver parenchyma were placed by observer 1 on the registered DCE-MR images. For the HCC ROIs, the entire lesion was included, both viable and, if present, necrotic regions. Subsequently, the ROIs were copied and pasted on the BOLD, TOLD and DWI parameter maps and adjusted if necessary to avoid inclusion of artefacts. Histogram quantification included determination of central tendency (mean and median) and heterogeneity [standard deviation (SD), kurtosis and skewness] parameters. Kurtosis determines the peakedness of the distribution. A higher kurtosis has been associated with increased heterogeneity29, because distributions with high peaks often have fat tails of outlier values. A skewness value deviating from zero indicates that there is tail toward either the left or right side of the distribution, which is also indicative of heterogeneity.
Histopathology
Fifteen patients underwent partial hepatectomy after the MRI examination (time range between MRI and resection 0–40 days) and two patients had a biopsy of their index HCC lesion (57 days before and 15 days after the MRI exam). Histological grade30 was available for the HCC lesions in these 17 patients. For one patient that underwent resection, no sections were available since the resection was performed in an outside hospital. For each HCC lesion in the 14 remaining resected liver samples, two five-μm-thick formalin-fixed paraffin-embedded sections were available for advanced histopathological staining. The Multiplexed Immunohistochemical Consecutive Staining on Single Slide (MICSSS)31 technique was used to sequentially stain endothelial cells (CD31 monoclonal antibody, clone JC70), macrophages (CD68, clone KP1) and T-cells (CD3, clone 2GV6). For detection of hypoxia, a separate HCC section stained for hypoxia-inducible factor 1-alpha (HIF1α) activity was used for each lesion32. The stained HCC sections were digitally imaged at 20x magnification using a whole-slide scanner (Olympus, Tokyo, Japan). A threshold-based segmentation method was implemented in MATLAB to detect stained pixels (Fig. 1 ). For the segmentation, the red, green and blue intensity values from the images were retrieved and scaled from 0 to 255. Since the stained pixels generally had a dark red appearance, a pixel was considered stained if (1) the red intensity was higher than 1.1 times the green channel and (2) the average signal intensity was lower than 120. These thresholds were empirically chosen based on inspection of all histological images. From these segmentations, stained tumour fractions were derived.
Figure 1.
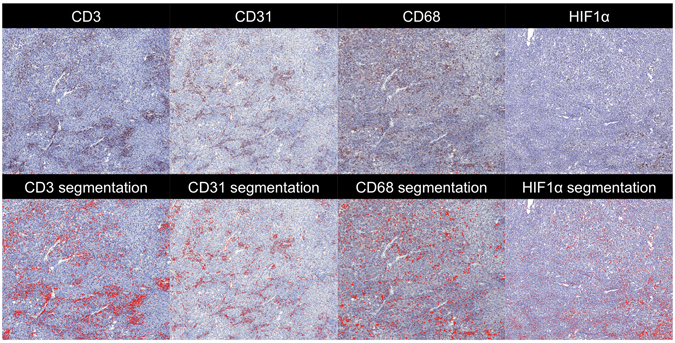
Histopathology processing of HCC sections obtained from a 70-year-old male patient with NASH cirrhosis and HCC. Zoomed microscopy images (×20) of an HCC section stained using Multiplexed Immunohistochemical Consecutive Staining on Single Slide (MICSSS) for CD3, CD31 and CD68 and a separate section stained for HIF1α are shown in the top row. Results of the automatic segmentation algorithm are shown in the bottom row, in which pixels identified as stained are coloured in red. CD3 = cluster of differentiation 3, CD31 = cluster of differentiation 31, CD68 = cluster of differentiation 68, HIF1α = hypoxia-inducible factor 1-alpha.
Gene expression analysis
Histologically distinct components of the 14 resected HCC tumours were macro-dissected using H&E staining of serial tissue sections as reference as previously described33. Isolated total RNA samples were profiled to determine transcriptomic HCC subtypes S1, S2 and S334 with digital transcript counting technology using the nearest template prediction algorithm as previously described33. In addition, the following HCC marker genes were simultaneously profiled: liver specific Wnt target overexpressed in S3 subtype (GLUL)35, stemness markers (EPCAM, KRT19)36, 37, early HCC markers (BIRC5, HSP70, LYVE1, EZH2)38, 39, a pharmacological target currently under clinical testing (FGFR4)40, potentially targetable angiogenesis marker (VEGFA)41 and targetable immune checkpoints (CFfigD274, PDCD1, CTLA4)42.
Statistical analysis
All values are presented as mean ± SD. For all correlation analyses, the Spearman correlation test was used. For all tests, a P-value lower than 0.05 was considered significant. For the analyses of differences and correlations between techniques, TOLD parameters, except ΔR1, were not compared with the DCE-MRI parameters, because the baseline R1 measurements were used as input for the DCE-MRI modelling.
Intra-tumour heterogeneity
Differences in kurtosis and skewness values between parameters of different MRI techniques were tested for significance using a Friedman test followed by post-hoc Bonferroni-corrected Wilcoxon signed rank tests. Mean, median and SD values were not compared between the techniques, because they directly depend on the actual MRI parameter values, which are inherently different between the MRI techniques. Liver and HCC histogram parameter values were compared using Wilcoxon signed-rank tests. For patients with multiple HCC lesions, the histogram features of the different lesions were averaged for comparison with liver parenchyma.
Inter-tumour heterogeneity
The absolute coefficient of variation (CV = 100*SD/absolute mean) between all MRI parameter features across lesions was measured. Absolute CV was calculated because of the presence of negative skewness values.
Correlation between MRI methods
The correlation of the mean, median, SD, kurtosis and skewness measures between the parameters of different MRI methods was assessed. The correlation analysis between mean and median parameters was performed both on all lesions as well as separately for lesions measured at 1.5 T and 3.0 T. To assess the effect of field strength on the correlations, differences between the correlations at both field strengths were tested for significance using a Fisher transformation test.
Correlation with histopathology and gene expression levels
The MRI histogram features in the tumour ROIs were correlated with histopathology stained tumour fractions and gene expression levels. In addition, differences in MRI parameters between HCC grades and molecular subclasses were assessed for statistical significance using Kruskal-Wallis tests.
Results
Forty-four HCCs were identified in 32 patients. Five lesions were excluded because they were not covered by TOLD/BOLD measurements (n = 4) or were completely necrotic due to previous treatment (n = 1). Two other treated lesions were included since they exhibited limited necrosis (<30%). The final 39 included lesions had an average size of 4.4 ± 3.3 cm (range 1.5–13 cm). The number of analysed lesions per patient was as follows: 1 (n = 27, 84%), 2 (n = 4, 13%) or 4 (n = 1, 3%). The distribution of histological grade for the lesions with pathologic validation was as follows: well differentiated (n = 2), moderately differentiated (n = 9), poorly differentiated (n = 5) and undifferentiated (n = 1).
Intra-tumour heterogeneity
Average mean, median, SD, kurtosis and skewness values for all assessed parameters in the liver parenchyma and HCC lesions are listed in Table 2. HCC lesions exhibited more intra-tissue heterogeneity compared to liver parenchyma (Fig. 2), with significantly higher SD (Fa, Fp, Ft, MTT, DV and R1 pre O2; P < 0.020), kurtosis (Fp and ADC; P < 0.030) and skewness (Fp, ART, R2* post O2 and R1 pre O2; P < 0.012). Kurtosis and skewness in HCC lesions were significantly different between MRI parameters of different techniques (P < 0.001). Specifically, significantly higher kurtosis was observed for Fp, R1 post O2 and ΔR1 compared to ADC (P < 0.045). A significantly higher skewness was observed for Fp vs. ΔR2* (P = 0.020), while ART showed a significantly lower skewness compared to R2* post O2, ΔR2*, ΔR1 and ADC (P < 0.030). Leptokurtosis, i.e. a kurtosis value higher than 3, which is indicative of intra-tumour heterogeneity, was observed in all lesions for at least one MRI parameter. In 9 lesions (23%) leptokurtosis was seen in all MRI methods (DCE-MRI, BOLD, TOLD and DWI), while the remaining lesions showed leptokurtosis in a subset of MRI methods [DCE-MRI, BOLD and TOLD in 18 lesions (46%); DCE-MRI, TOLD and DWI in 4 lesions (10%); DCE-MRI and TOLD in 4 lesions (10%); BOLD and TOLD in 1 lesion (3%); TOLD and DWI in 1 lesion (3%); DCE-MRI only in 2 lesions (5%)].
Table 2.
Central tendency (mean and median) and heterogeneity (SD, kurtosis, skewness) MRI parameters in 39 HCC lesions and liver parenchyma in 32 patients.
| Mean | Median | SD | Kurtosis | Skewness | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Liver | HCC | P * | Liver | HCC | P * | Liver | HCC | P * | Liver | HCC | P * | Liver | HCC | P * | |
| Fa | 49 ± 45 | 247 ± 240 | <0.001 | 47 ± 45 | 241 ± 250 | <0.001 | 16.3 ± 13.0 | 83.0 ± 66.7 | <0.001 | 4.3 ± 1.8 | 5.5 ± 6.0 | 0.754 | 0.76 ± 0.66 | 0.44 ± 1.34 | 0.232 |
| Fp | 145 ± 123 | 112 ± 156 | 0.048 | 146 ± 126 | 103 ± 166 | 0.010 | 39.9 ± 32.5 | 78.6 ± 78.6 | 0.020 | 6.3 ± 14.0 | 64.1 ± 183.6 | 0.005 | 0.54 ± 1.57 | 4.00 ± 6.41 | <0.001 |
| Ft | 193 ± 122 | 358 ± 337 | 0.023 | 193 ± 125 | 342 ± 350 | 0.092 | 43.7 ± 36.2 | 144.3 ± 126.3 | <0.001 | 4.5 ± 3.1 | 17.1 ± 54.9 | 0.969 | 0.62 ± 0.76 | 1.14 ± 3.11 | 0.984 |
| ART | 33.9 ± 27.9 | 78.9 ± 21.5 | <0.001 | 33.0 ± 29.1 | 80.3 ± 24.2 | <0.001 | 9.7 ± 5.1 | 13.1 ± 9.0 | 0.055 | 7.6 ± 10.5 | 30.1 ± 61.8 | 0.256 | 0.68 ± 1.71 | −2.49 ± 4.37 | <0.001 |
| MTT | 20.3 ± 12.3 | 21.2 ± 15.8 | 0.724 | 19.7 ± 12.1 | 18.8 ± 16.3 | 0.667 | 4.2 ± 2.9 | 10.9 ± 7.3 | <0.001 | 5.2 ± 4.8 | 9.1 ± 17.4 | 0.938 | 0.90 ± 0.87 | 1.54 ± 2.06 | 0.337 |
| DV | 37.4 ± 22.7 | 34.6 ± 24.7 | 0.695 | 36.8 ± 22.8 | 34.0 ± 26.0 | 0.710 | 4.9 ± 3.4 | 9.9 ± 5.7 | <0.001 | 3.7 ± 1.7 | 5.5 ± 10.8 | 0.240 | 0.58 ± 0.61 | 0.70 ± 1.59 | 0.468 |
| R2* pre O2 | 55.3 ± 34.4 | 34.5 ± 15.1 | <0.001 | 54.7 ± 34.8 | 33.1 ± 15.0 | <0.001 | 7.6 ± 7.5 | 7.6 ± 5.2 | 0.681 | 4.0 ± 2.9 | 5.2 ± 4.3 | 0.433 | 0.35 ± 0.89 | 0.92 ± 1.09 | 0.055 |
| R2* post O2 | 51.8 ± 30.8 | 33.7 ± 14.5 | <0.001 | 51.5 ± 31.4 | 32.7 ± 14.6 | <0.001 | 8.3 ± 10.7 | 8.0 ± 4.7 | 0.347 | 3.5 ± 1.8 | 4.0 ± 2.7 | 0.984 | 0.15 ± 0.64 | 0.72 ± 0.81 | 0.011 |
| ΔR2* | −3.47 ± 10.0 | −0.78 ± 8.74 | 0.281 | −3.12 ± 8.8 | −0.86 ± 7.48 | 0.264 | 8.8 ± 9.5 | 8.6 ± 5.2 | 0.638 | 3.9 ± 2.0 | 4.7 ± ± 3.6 | 0.830 | −0.18 ± 0.72 | −0.16 ± 0.94 | 0.799 |
| R1 pre O2 | 1.76 ± 0.47 | 1.67 ± 0.94 | 0.052 | 1.76 ± 0.46 | 1.58 ± 0.73 | 0.033 | 0.12 ± 0.14 | 0.75 ± 3.31 | 0.008 | 3.7 ± 2.2 | 8.1 ± 17.7 | 0.531 | −0.12 ± 0.76 | 0.72 ± 2.07 | 0.012 |
| R1 post O2 | 1.87 ± 0.73 | 1.90 ± 1.79 | 0.027 | 1.77 ± 0.63 | 1.54 ± 0.61 | 0.026 | 2.26 ± 7.73 | 1.44 ± 5.48 | 0.422 | 71.4 ± 257.6 | 16.5 ± 34.5 | 0.327 | 2.13 ± 8.07 | 1.41 ± 2.76 | 0.153 |
| ΔR1 | 0.11 ± 0.45 | 0.23 ± 1.15 | 0.176 | 0.01 ± 0.24 | −0.05 ± 0.25 | 0.829 | 2.26 ± 7.73 | 1.06 ± 3.46 | 0.196 | 72.2 ± 257.4 | 17.6 ± 44.6 | 0.814 | 2.14 ± 8.08 | 1.50 ± 3.03 | 0.112 |
| ADC | 1.31 ± 0.58 | 1.43 ± 0.68 | <0.001 | 1.31 ± 0.58 | 1.41 ± 0.70 | <0.001 | 0.2 ± 0.1 | 0.3 ± 0.2 | 0.142 | 3.8 ± 2.2 | 4.0 ± 6.4 | 0.030 | 0.10 ± 0.79 | 0.33 ± 0.90 | 0.337 |
*P-values from Wilcoxon signed rank tests. Significant P-values are bolded. ADC = apparent diffusion coefficient (10−3 mm2/s), ART = arterial fraction (%), DV = distribution volume (%), Fa = arterial flow (ml/100 g/min), Fp = portal flow (ml/100 g/min), Ft = total flow (ml/100 g/min), MTT = mean transit time (s), R1 = longitudinal relaxation rate (s−1), R2* = transverse relaxation rate (s−1).
Figure 2.

54 year-old male patient with cirrhosis secondary to chronic hepatitis B virus infection and HCC. (a) Representative magnified parametric maps of a large (8.3 cm) HCC. Location of the tumour within the liver is indicated by the white arrow on the T2-weighted image (bottom row, right). A distinct region in the anterior portion of the tumour of high arterial flow (Fa) and low R2* was observed, reflective of high tumour perfusion and normoxia (grey arrow in Fa and R2* pre O2 maps). The posterior portion of the tumour displays low Fa and high R2*, suggestive of poor perfusion and hypoxia (white arrow in Fa and R2* pre O2 maps). (b) Histograms of Fa, R2* pre O2, R1 pre O2 and ADC in the same lesion. The extensive heterogeneity observed in the parameter maps of Fa and R2* pre O2 is also reflected in the histograms, as illustrated by the fat tails and pronounced skewness, indicated by arrows. The R1 pre O2 histogram also exhibited skewness (black arrow). ADC = apparent diffusion coefficient, ART = arterial fraction, DV = distribution volume, Fa = arterial flow, Fp = portal flow, Ft = total flow, MTT = mean transit time, R1 = longitudinal relaxation rate, R2* = transverse relaxation rate.
Inter-tumour heterogeneity
CV values of MRI parameters across tumours are shown in Fig. 3. CV values were markedly higher for kurtosis and skewness vs. mean and median values, indicative of higher inter-tumour variation for heterogeneity parameters. SD overall showed similar CV values as the mean and median values, except for a substantial higher CV observed for SD of R1 pre and post O2.
Figure 3.
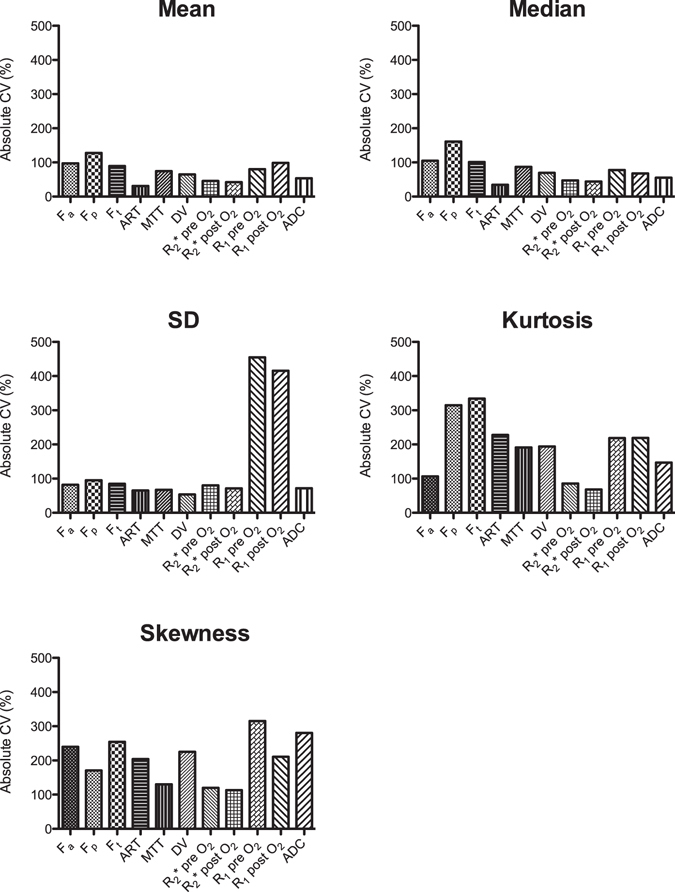
Inter-tumour variability in MRI parameters. Absolute coefficients of variation (CV) of mean, median, SD, kurtosis and skewness parameter values across 39 HCC tumours (in 32 patients). Kurtosis and skewness showed a substantially higher inter-tumour variability compared to mean and median values. ADC = apparent diffusion coefficient, ART = arterial fraction, DV = distribution volume, Fa = arterial flow, Fp = portal flow, Ft = total flow, MTT = mean transit time, R1 = longitudinal relaxation rate, R2* = transverse relaxation rate.
Correlation between MRI methods
Figure 4 shows heatmaps of significant correlations found between the MRI parameter values in tumours. Numerical values of the correlations are listed in Supplementary Table S1. For the central tendency parameters, significant correlations were found between ADC values and parameters from all other sequences (DCE-MRI, BOLD and TOLD), between DCE-MRI and ΔR1 and between BOLD and TOLD values. However, no significant correlations were found between BOLD and DCE-MRI central tendency parameter values; while these parameters showed significant correlation with respect to their MRI heterogeneity features, which indicates that these parameters exhibited similar intra-tumour distribution, as illustrated in Fig. 2. Differences in correlations between mean and median MRI parameters at both field strengths were not significant (P-value range mean 0.119–0.949, median 0.098–0.981).
Figure 4.
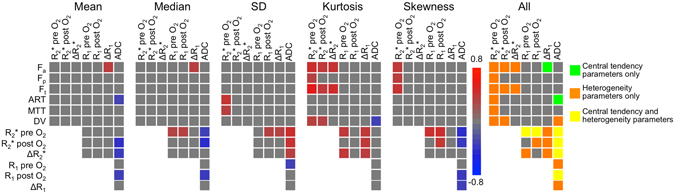
Heatmaps of correlations between mean, median, SD, kurtosis and skewness of MRI parameters in 39 HCC lesions (in 32 patients). Significant correlations (P < 0.05) are coloured according to the scale bar. The correlation between R1 pre and post O2 and DCE-MRI parameters was not assessed, because the baseline R1 measurements were used as input for the DCE-MRI modelling. A combined heatmap of all significant correlations between MRI features is shown on the right, illustrating additional information provided by heterogeneity parameters (SD, kurtosis and skewness). Significant correlations between DCE-MRI and BOLD were for example only seen for heterogeneity parameters and not for central tendency parameters (mean and median). ADC = apparent diffusion coefficient, ART = arterial fraction, DV = distribution volume, Fa = arterial flow, Fp = portal flow, Ft = total flow, MTT = mean transit time, R1 = longitudinal relaxation rate, R2* = transverse relaxation rate.
Correlation with histopathology
None of the MRI parameters showed a significant difference between histological grades (Kruskal-Wallis P-value range 0.085–0.410). Significant correlations between MRI parameters and histologically stained tumour fractions in 14 lesions (14 patients) are displayed in heatmaps in Fig. 5 (numerical values in Supplementary Table S2). CD3, CD31 and CD68 tumour fractions correlated moderately to strongly with both central tendency and heterogeneity parameter values. With regards of central tendency parameters, CD3 stained tumour fractions showed moderate significant negative correlations with mean and median R1 pre O2 (P < 0.029). CD31 staining significantly negatively correlated with mean and median R2* pre and post O2 (P < 0.009). CD68 stained tumour fractions correlated negatively with mean and median R1 pre O2 and median R1 post O2 and positively with mean and median ADC (P < 0.050). HIF1α tumour fractions did not correlate with any of the central tendency parameters, but did show significant positive correlation with SD of R2* pre and post O2 and skewness of R1 pre O2 (P < 0.048).
Figure 5.
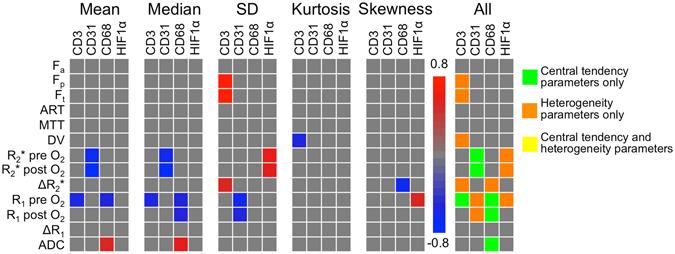
Heatmaps of correlations between MRI parameters and histopathology in 14 HCC lesions in 14 patients. Significant correlations (P < 0.05) are coloured according to the scale bar. A combined heatmap of all significant correlations between MRI features and histopathology is shown on the right, illustrating additional information provided by heterogeneity parameters (SD, kurtosis and skewness). For example, HIF1α tumour fractions correlated with heterogeneity parameters, but not with central tendency parameters (mean and median). ADC = apparent diffusion coefficient, ART = arterial fraction, CD3 = cluster of differentiation 3, CD31 = cluster of differentiation 31, CD68 = cluster of differentiation 68, DV = distribution volume, Fa = arterial flow, Fp = portal flow, Ft = total flow, HIF1α = hypoxia-inducible factor 1-alpha, MTT = mean transit time, R1 = longitudinal relaxation rate, R2* = transverse relaxation rate.
Gene expression analysis
The distribution of HCC molecular subclasses was as follows: S1 (n = 6, 43%), S2 (n = 3, 21%), S3 (n = 5, 36%). No significant differences between MRI parameters in the different subclasses were observed (P-value range 0.160–0.970). MRI central tendency and heterogeneity features were complementary in terms of correlations with gene expression levels, as illustrated in Fig. 6 (numerical correlation values in Supplementary Table S3). The expression level of CTLA4 only correlated with central tendency parameters, while expression of GLUL, FGFR4, tumour stemness markers KRT19 and EPCAM and immune checkpoint PDCD1 showed significant correlations with MRI heterogeneity parameters only. The expression levels of early HCC markers BIRC5, HSP70, LYVE and EZH2, angiogenesis marker VEGFA and immune checkpoint CD274 significantly correlated with both central tendency and heterogeneity parameters.
Figure 6.
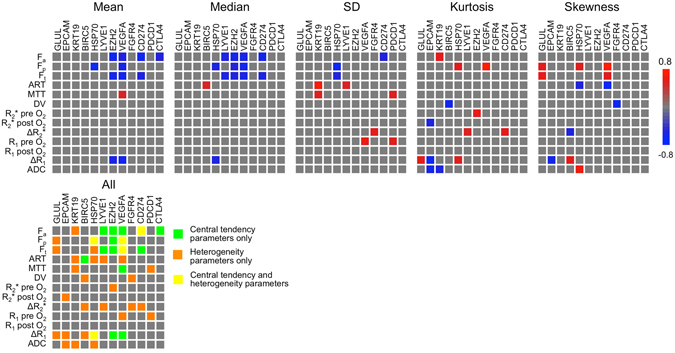
Heatmaps of correlations between MRI parameters and gene expression levels in 14 HCC lesions in 14 patients. Significant correlations (P < 0.05) are coloured according to the scale bar. A combined heatmap of all MRI features is shown at the figure bottom, illustrating additional information provided by heterogeneity parameters (SD, kurtosis and skewness) compared to central tendency parameters (mean and median) with respect to correlations between MRI and gene expression levels. ADC = apparent diffusion coefficient, ART = arterial fraction, BIRC5 = Baculoviral IAP repeat containing 5, CD274 = cluster of differentiation 274, CTLA4 = cytotoxic T-lymphocyte-associated protein 4, DV = distribution volume, EPCAM = epithelial cell adhesion molecule, EZH2 = enhancer of zeste homolog 2, Fa = arterial flow, FGFR4 = fibroblast growth factor receptor 4, Fp = portal flow, Ft = total flow, GLUL = glutamate-ammonia ligase, HSP70 = 70 kilodalton heat shock protein, KRT19 = keratin 19; LYVE1 = lymphatic vessel endothelial hyaluronan receptor 1, MTT = mean transit time, R1 = longitudinal relaxation rate, R2* = transverse relaxation rate, VEGFA = vascular endothelial growth factor A.
Discussion
In this study we quantified intra- and inter-tumour heterogeneity of HCC lesions using quantitative mpMRI and correlated the mpMRI parameters with histopathological and gene expression analysis in a subset of patients.
Intra-tumour heterogeneity, assessed by the presence of leptokurtosis in the MRI parameters, was observed in all lesions. DCE-MRI and TOLD parameters showed a significantly higher kurtosis compared to ADC, which suggests that intra-tumour heterogeneity in HCC is mainly caused by spatial variation in presence of (functional) vasculature/oxygenation rather than intra-tumoural differences in cellularity.
A higher inter-tumour variation was found for the heterogeneity parameters compared to the mean and median parameter values. This finding suggests that the proposed heterogeneity analysis could potentially allow for a more extensive differentiation between tumours than using the more traditional analysis of central tendency parameter values in ROIs.
With respect to inter-sequence correlations, we found that DCE-MRI and BOLD MRI central tendency parameters did not correlate, while heterogeneity features of these two sequences showed a significant correlation, indicating that DCE-MRI and BOLD-MRI parameters exhibited a similar intra-tumour distribution. Baudelet et al. found that DCE-MRI and BOLD MRI were also largely complementary for the characterization of vasculature in a preclinical fibrosarcoma model43. A possible explanation for the lack of a clear correlation between the techniques could be due to the fact that BOLD-MRI has intrinsic low sensitivity in tumour blood vessels with low haematocrit, as opposed to DCE-MRI, in which contrast agent influx is not affected by haematocrit43.
Significant correlations were found between BOLD, TOLD and DWI central tendency parameters and histopathology. The negative correlations between R1 pre O2 and CD3- and CD68-stained tumour fractions and the positive correlation between ADC and CD68 tumour fractions may be explained by a possible increase in interstitial fluid associated with inflammation44. The negative correlation between R2* (pre and post O2) and CD31 may be due to the fact that tumours with higher vascularity will most probably be less hypoxic and therefore exhibit a lower R2*.
Strong significant correlations were observed between gene expression levels and several MRI parameters. Poor tumour perfusion (low flow/ART and high MTT) was associated with high expression of HCC markers, VEGFA and immune checkpoints CD274 and CTLA4. The relation with HCC markers may be related to tumour progression, which is associated with decreased arterial flow, secondary to increased interstitial pressure and resulting closure of arterial capillaries45, and higher expression of HCC markers, including HSP70 and LYVE1 46, 47. The negative correlation between perfusion and VEGFA expression could potentially be explained by the fact that decreased perfusion may be associated with hypoxia, which triggers expression of VEGFA 48. However, counterintuitively, VEGFA expression was not associated with mean or median BOLD parameters and negatively associated with ΔR1, which suggests absence of correlation between MRI-measured hypoxia and expression of VEGFA. The underlying physiology of the negative correlations between tumour perfusion and CTLA4 and CD274 expression is unclear and needs to be verified in a larger study. The expression of immune checkpoints has shown to be positively associated with tumour progression in lung cancer49, but no data has been reported on this association in HCC. Interestingly, significant correlations with expression of several genes, including stemness markers and immunotherapy target PDCD1, were only observed with heterogeneity parameters, which further illustrates the complementary properties of central tendency and heterogeneity parameters in terms of tumour characterization.
In general, limited data is available on radiogenomics in HCC. There are a few reports that compare CT imaging traits with HCC genomics analysis10, 11. However, these studies have assessed only qualitative metrics, which are limited by inter- and intra-observer variability. The quantitative assessment of heterogeneity histogram features of functional mpMRI in the current study is probably less prone to inter-observer variability. Nevertheless, the proposed heterogeneity features can potentially be confounded by image noise, which introduces non-biological heterogeneity, and by inaccuracies in parameter estimation of single pixels50. However, we found that the HCC lesions exhibited more intra-tissue heterogeneity than liver parenchyma, which indicates that the observed intra-tumour heterogeneity in the HCC lesions is likely biological in nature.
There are a few published reports on the use of imaging to predict and assess immunotherapy outcome51. Jajamovich et al. found a significant correlation between ADC and a gene signature related to dendritic cell maturation in glioblastoma, which can be used to stratify patients with immunogenic tumours for immunotherapy52. Mayerhoefer et al. found that DWI is a promising technique for the evaluation of immunotherapy response in lymphoma patients53. The potential value of DWI for immunotherapy was also reported by Qin et al., who found that the decrease or stabilization of the lesion volume on ADC maps, after an initial increase after immunotherapy, was predictive of therapeutic benefit in glioblastoma54. Interestingly, we did not find significant correlations between ADC and any of the immunotherapy targets. The role of DWI for the prediction of immunotherapy outcome in HCC needs to be established in future studies. There is no published data assessing HCC response to immunotherapy, given the recent introduction of this therapy in HCC. More studies are needed to determine the exact value of mpMRI for the prediction of immunotherapy efficacy in HCC and other cancer types.
While mpMRI proved promising for the non-invasive assessment of histopathological and genomics properties of HCC lesions in our study, the separate imaging, histopathological and genomics measurements also provided independent information on tumour properties. For example, none of the mpMRI features could distinguish between different histopathological grades. We therefore do not believe that mpMRI could fully replace histopathologic assessment in HCC. The mpMRI analysis could rather be used to improve the characterization of HCC lesions in biopsied or resected samples, by providing mutual and additional information on tumour properties on a whole-tumour level12. Nevertheless, in the majority of patients HCC is diagnosed based on imaging alone, without histologic confirmation55. Treatment stratification in those patients without biopsy could ultimately benefit from knowledge of the correlation of imaging parameters with histopathological and genomics properties of HCC lesions in a training set of resected HCC samples.
The findings described in our study may have substantial clinical implications for characterization and treatment stratification of HCC. Tumours with high intra-tumour heterogeneity generally have a poorer prognosis, which may be secondary to a more aggressive biology or treatment resistance56. The observed correlations between MRI features and the expression of potential therapeutic markers for targeted molecular or immunotherapy of HCC57 suggest that the proposed mpMRI histogram analysis could be used to noninvasively predict treatment outcome at baseline. Knowledge of the expression of therapeutic targets using non-invasive imaging could aid in the stratification of patient-tailored, personalized treatment. A future study should assess the proposed mpMRI heterogeneity analysis as a predictor of response to immunotherapy. In such study, it could also be assessed which (combination of) MRI features have the highest diagnostic performance for prediction of treatment response.
Our study has several limitations. Assessment of intra-tumour heterogeneity was not feasible for the histopathology and genomics analysis, because only a small part of the lesion was available for these analyses. Another limitation was the relatively small sample size of HCC lesions for histopathology and genomics. Moreover, there was variability between MRI field strengths on which the patients were scanned, which could affect MRI parameter estimation. Nevertheless, the correlations between MRI parameters were not significantly different between both field strengths. Finally, the correlation of the mpMRI features with clinical parameters such as survival and treatment response was not assessed in this initial study.
In conclusion, our results show that central tendency and heterogeneity features of mpMRI data of HCC tumours are complementary in terms of correlations between sequences, with histopathology and with gene expression levels. The proposed histogram analysis is therefore promising for non-invasive HCC characterization on the imaging, immunohistochemical and genomics level.
Electronic supplementary material
Acknowledgements
MW: Fondation ARC Contract grant number: SAE20140601302. SL: RSNA Research Seed Grant #RSD1608. SG: Clinical Strategy Teams Grants from the Cancer Research Institute; NIH Grant P01 CA1 90174. MM: NIH Grants R01 CA173861, U19 AI 128949, U24 AI 118644, U19 AI 117873. YH: NIH Grant R01 DK099558, EU Grant ERC-2014-AdG-671231 HEPCIR, Irma T. Hirschl Trust, and U.S. Department of Defense W81XWH-16-1-0363. BT: NIH Grants U01 CA172320, 1R01DK087877.
Author Contributions
Study design: S.H., M.W., B.T. Development of methodology: S.H., M.W., O.B., R.R., S.G., M.M., Y.H., B.T., Acquisition of data: S.H., M.W., O.B., C.B., R.R., N.C., M.I.F., H.Z., Y.H. Analysis of data: S.H., M.W., C.B., Y.H. Writing and review of manuscript: S.H., M.W., O.B., C.B., S.L., R.R., S.G., Y.H., B.T.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-02706-z
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Abramson RG, et al. MR Imaging Biomarkers in Oncology Clinical Trials. Magn. Reson. Imaging Clin. N. Am. 2016;24:11–29. doi: 10.1016/j.mric.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo Y, et al. Differentiation of clinically benign and malignant breast lesions using diffusion-weighted imaging. J. Magn. Reson. Imaging. 2002;16:172–178. doi: 10.1002/jmri.10140. [DOI] [PubMed] [Google Scholar]
- 3.Bane O, et al. Feasibility and reproducibility of BOLD and TOLD measurements in the liver with oxygen and carbogen gas challenge in healthy volunteers and patients with hepatocellular carcinoma. J. Magn. Reson. Imaging. 2016;43:866–876. doi: 10.1002/jmri.25051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yankeelov TE, Gore JC. Dynamic Contrast Enhanced Magnetic Resonance Imaging in Oncology: Theory, Data Acquisition, Analysis, and Examples. Curr. Med. Imaging Rev. 2009;3:91–107. doi: 10.2174/157340507780619179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Just N. Improving tumour heterogeneity MRI assessment with histograms. Br. J. Cancer. 2014;111:2205–2213. doi: 10.1038/bjc.2014.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anfuso B, El-Khobar KE, Sukowati CH, Tiribelli C. The multiple origin of cancer stem cells in hepatocellular carcinoma. Clin. Res. Hepatol. Gastroenterol. 2015;39(Suppl 1):S92–97. doi: 10.1016/j.clinre.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 7.Friemel J, et al. Intratumor heterogeneity in hepatocellular carcinoma. Clin. Cancer Res. 2015;21:1951–1961. doi: 10.1158/1078-0432.CCR-14-0122. [DOI] [PubMed] [Google Scholar]
- 8.Fujita N, et al. Hepatocellular carcinoma: clinical significance of signal heterogeneity in the hepatobiliary phase of gadoxetic acid-enhanced MR imaging. Eur. Radiol. 2015;25:211–220. doi: 10.1007/s00330-014-3349-9. [DOI] [PubMed] [Google Scholar]
- 9.Rutman AM, Kuo MD. Radiogenomics: creating a link between molecular diagnostics and diagnostic imaging. Eur. J. Radiol. 2009;70:232–241. doi: 10.1016/j.ejrad.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 10.Kuo MD, Gollub J, Sirlin CB, Ooi C, Chen X. Radiogenomic analysis to identify imaging phenotypes associated with drug response gene expression programs in hepatocellular carcinoma. J. Vasc. Interv. Radiol. 2007;18:821–831. doi: 10.1016/j.jvir.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 11.Segal E, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat. Biotechnol. 2007;25:675–680. doi: 10.1038/nbt1306. [DOI] [PubMed] [Google Scholar]
- 12.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 15.Calabro L, et al. Tremelimumab for patients with chemotherapy-resistant advanced malignant mesothelioma: an open-label, single-arm, phase 2 trial. Lancet Oncol. 2013;14:1104–1111. doi: 10.1016/S1470-2045(13)70381-4. [DOI] [PubMed] [Google Scholar]
- 16.Sangro B, et al. A clinical trial of CTLA-4 blockade with tremelimumab in patients with hepatocellular carcinoma and chronic hepatitis C. J Hepatol. 2013;59:81–88. doi: 10.1016/j.jhep.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 17.OPTN policies, policy 9: Allocation of Livers and Liver-Intestines; 9.3.F Candidates with Hepatocellular Carcinoma. (2017).
- 18.Hectors SJ, et al. Intravoxel incoherent motion diffusion-weighted imaging of hepatocellular carcinoma: Is there a correlation with flow and perfusion metrics obtained with dynamic contrast-enhanced MRI? J. Magn. Reson. Imaging. 2016;44:856–864. doi: 10.1002/jmri.25194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jajamovich GH, et al. DCE-MRI of hepatocellular carcinoma: perfusion quantification with Tofts model versus shutter-speed model–initial experience. MAGMA. 2016;29:49–58. doi: 10.1007/s10334-015-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jajamovich GH, Dyvorne H, Donnerhack C, Taouli B. Quantitative liver MRI combining phase contrast imaging, elastography, and DWI: assessment of reproducibility and postprandial effect at 3.0 T. PLoS One. 2014;9:e97355. doi: 10.1371/journal.pone.0097355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dyvorne H, Jajamovich G, Kakite S, Kuehn B, Taouli B. Intravoxel incoherent motion diffusion imaging of the liver: optimal b-value subsampling and impact on parameter precision and reproducibility. Eur. J. Radiol. 2014;83:2109–2113. doi: 10.1016/j.ejrad.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dyvorne HA, et al. Diffusion-weighted imaging of the liver with multiple b values: effect of diffusion gradient polarity and breathing acquisition on image quality and intravoxel incoherent motion parameters–a pilot study. Radiology. 2013;266:920–929. doi: 10.1148/radiol.12120686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deichmann R, Haase A. Quantification of T1 Values by Snapshot-Flash Nmr Imaging. J. Magn. Reson. 1992;96:608–612. [Google Scholar]
- 24.Haase A. Snapshot FLASH MRI. Applications to T1, T2, and chemical-shift imaging. Magn. Reson. Med. 1990;13:77–89. doi: 10.1002/mrm.1910130109. [DOI] [PubMed] [Google Scholar]
- 25.Aronhime S, et al. DCE-MRI of the liver: effect of linear and nonlinear conversions on hepatic perfusion quantification and reproducibility. J. Magn. Reson. Imaging. 2014;40:90–98. doi: 10.1002/jmri.24341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, et al. In vivo blood T(1) measurements at 1.5 T, 3 T, and 7 T. Magn. Reson. Med. 2013;70:1082–1086. doi: 10.1002/mrm.24550. [DOI] [PubMed] [Google Scholar]
- 27.Pintaske J, et al. Relaxivity of Gadopentetate Dimeglumine (Magnevist), Gadobutrol (Gadovist), and Gadobenate Dimeglumine (MultiHance) in human blood plasma at 0.2, 1.5, and 3 Tesla. Invest. Radiol. 2006;41:213–221. doi: 10.1097/01.rli.0000197668.44926.f7. [DOI] [PubMed] [Google Scholar]
- 28.Materne R, et al. Assessment of hepatic perfusion parameters with dynamic MRI. Magn. Reson. Med. 2002;47:135–142. doi: 10.1002/mrm.10045. [DOI] [PubMed] [Google Scholar]
- 29.Yoon, S. H. et al. Tumor Heterogeneity in Lung Cancer: Assessment with Dynamic Contrast-enhanced MR Imaging. Radiology, 151367, doi:10.1148/radiol.2016151367 (2016). [DOI] [PubMed]
- 30.Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7:462–503. doi: 10.1002/1097-0142(195405)7:3<462::AID-CNCR2820070308>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 31.Remark R, et al. In-depth tissue profiling using multiplexed immunohistochemical consecutive staining on single slide. Science Immunology. 2016 doi: 10.1126/sciimmunol.aaf6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ziello JE, Jovin IS, Huang Y. Hypoxia-Inducible Factor (HIF)-1 regulatory pathway and its potential for therapeutic intervention in malignancy and ischemia. Yale J. Biol. Med. 2007;80:51–60. [PMC free article] [PubMed] [Google Scholar]
- 33.Tan PS, et al. Clinicopathological indices to predict hepatocellular carcinoma molecular classification. Liver Int. 2016;36:108–118. doi: 10.1111/liv.12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoshida Y, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goossens N, Sun X, Hoshida Y. Molecular classification of hepatocellular carcinoma: potential therapeutic implications. Hepat Oncol. 2015;2:371–379. doi: 10.2217/hep.15.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villanueva A, et al. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology. 2011;140:1501–1512 e1502. doi: 10.1053/j.gastro.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamashita T, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- 38.Llovet JM, et al. A molecular signature to discriminate dysplastic nodules from early hepatocellular carcinoma in HCV cirrhosis. Gastroenterology. 2006;131:1758–1767. doi: 10.1053/j.gastro.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 39.Di Tommaso L, et al. Diagnostic value of HSP70, glypican 3, and glutamine synthetase in hepatocellular nodules in cirrhosis. Hepatology. 2007;45:725–734. doi: 10.1002/hep.21531. [DOI] [PubMed] [Google Scholar]
- 40.Hagel M, et al. First Selective Small Molecule Inhibitor of FGFR4 for the Treatment of Hepatocellular Carcinomas with an Activated FGFR4 Signaling Pathway. Cancer Discov. 2015;5:424–437. doi: 10.1158/2159-8290.CD-14-1029. [DOI] [PubMed] [Google Scholar]
- 41.Horwitz E, et al. Human and mouse VEGFA-amplified hepatocellular carcinomas are highly sensitive to sorafenib treatment. Cancer Discov. 2014;4:730–743. doi: 10.1158/2159-8290.CD-13-0782. [DOI] [PubMed] [Google Scholar]
- 42.Harding JJ, El Dika I, Abou-Alfa GK. Immunotherapy in hepatocellular carcinoma: Primed to make a difference? Cancer. 2016;122:367–377. doi: 10.1002/cncr.29769. [DOI] [PubMed] [Google Scholar]
- 43.Baudelet C, Cron GO, Gallez B. Determination of the maturity and functionality of tumor vasculature by MRI: correlation between BOLD-MRI and DCE-MRI using P792 in experimental fibrosarcoma tumors. Magn. Reson. Med. 2006;56:1041–1049. doi: 10.1002/mrm.21047. [DOI] [PubMed] [Google Scholar]
- 44.Baronzio G, Parmar G, Baronzio M, Kiselevsky M. Tumor interstitial fluid: proteomic determination as a possible source of biomarkers. Cancer Genomics Proteomics. 2014;11:225–237. [PubMed] [Google Scholar]
- 45.Choi JY, Lee JM, Sirlin CB. CT and MR imaging diagnosis and staging of hepatocellular carcinoma: part I. Development, growth, and spread: key pathologic and imaging aspects. Radiology. 2014;272:635–654. doi: 10.1148/radiol.14132361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joo M, Chi JG, Lee H. Expressions of HSP70 and HSP27 in hepatocellular carcinoma. J. Korean Med. Sci. 2005;20:829–834. doi: 10.3346/jkms.2005.20.5.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kitagawa K, Nakajima G, Kuramochi H, Ariizumi SI, Yamamoto M. Lymphatic vessel endothelial hyaluronan receptor-1 is a novel prognostic indicator for human hepatocellular carcinoma. Mol Clin Oncol. 2013;1:1039–1048. doi: 10.3892/mco.2013.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Cox SR, Morita T, Kourembanas S. Hypoxia regulates vascular endothelial growth factor gene expression in endothelial cells. Identification of a 5′ enhancer. Circ. Res. 1995;77:638–643. doi: 10.1161/01.RES.77.3.638. [DOI] [PubMed] [Google Scholar]
- 49.Thommen DS, et al. Progression of Lung Cancer Is Associated with Increased Dysfunction of T Cells Defined by Coexpression of Multiple Inhibitory Receptors. Cancer Immunol Res. 2015;3:1344–1355. doi: 10.1158/2326-6066.CIR-15-0097. [DOI] [PubMed] [Google Scholar]
- 50.Asselin MC, O’Connor JP, Boellaard R, Thacker NA, Jackson A. Quantifying heterogeneity in human tumours using MRI and PET. Eur. J. Cancer. 2012;48:447–455. doi: 10.1016/j.ejca.2011.12.025. [DOI] [PubMed] [Google Scholar]
- 51.Garcia-Figueiras R, Padhani AR, Baleato-Gonzalez S. Therapy Monitoring with Functional and Molecular MR Imaging. Magn. Reson. Imaging Clin. N. Am. 2016;24:261–288. doi: 10.1016/j.mric.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 52.Jajamovich GH, Valiathan CR, Cristescu R, Somayajula S. Integrative analysis of diffusion-weighted MRI and genomic data to inform treatment of glioblastoma. J. Neurooncol. 2016;129:289–300. doi: 10.1007/s11060-016-2174-1. [DOI] [PubMed] [Google Scholar]
- 53.Mayerhoefer ME, et al. Evaluation of Diffusion-Weighted Magnetic Resonance Imaging for Follow-up and Treatment Response Assessment of Lymphoma: Results of an 18F-FDG-PET/CT-Controlled Prospective Study in 64 Patients. Clin. Cancer Res. 2015;21:2506–2513. doi: 10.1158/1078-0432.CCR-14-2454. [DOI] [PubMed] [Google Scholar]
- 54.Qin L, et al. Advanced MRI assessment to predict benefit of anti-programmed cell death 1 protein immunotherapy response in patients with recurrent glioblastoma. Neuroradiology. 2017;59:135–145. doi: 10.1007/s00234-016-1769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McEvoy SH, et al. Hepatocellular carcinoma: illustrated guide to systematic radiologic diagnosis and staging according to guidelines of the American Association for the Study of Liver Diseases. Radiographics. 2013;33:1653–1668. doi: 10.1148/rg.336125104. [DOI] [PubMed] [Google Scholar]
- 56.Davnall F, et al. Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging. 2012;3:573–589. doi: 10.1007/s13244-012-0196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Connell LC, Harding JJ, Abou-Alfa GK. Advanced Hepatocellular Cancer: the Current State of Future Research. Curr. Treat. Options Oncol. 2016;17:43. doi: 10.1007/s11864-016-0415-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


