Abstract
MiRNA have been found to play a role in a plethora of cellular processes of cancer cells such as cell apoptosis, cell proliferation, invasion, migration metabolism and stem cell differentiation. Dysregulation of miR-146b-5p has been documented in a variety of human malignancies. However, the biological functions and molecular mechanisms of miR-146b-5p in ovarian cancer remain unknown. In this study, our results show that miR-146b-5p was unregulated in colorectal cancer (CRC) tissues compared with the adjacent non-cancerous tissues. Ectopic overexpression of miR-146b-5p in CRC promoted cell growth, invasion and glycolysis, while knockdown of miR-146b-5p inhibited the growth, invasion and glycolysis of CRC cells. The oncogenic effect of miR-146b-5p is also confirmed in vivo. Mechanically, miR-146b-5p targets the 3’-UTR of pyruvate dehydrogenase B (PDHB) and exerts oncogenic effect. Overexpression of PDHB abolished the oncogenic effects of miR-146b-5p on the growth, invasion and glycolysis of CRC cells. Taken together, our results show that miR-146b-5p is an oncogenic miRNA in CRC which exerts its effect by directly targeting PDHB.
Keywords: Colorectal cancer, miR-146b-5p, PDHB, glycolysis
Introduction
As the second most common type of human malignancies in women and third in men, colorectal cancer (CRC) accounts for 1.2 million new cases and results in about 700,000 dead cases every year [1]. Particularly, the incidence of colorectal cancer is increasing in China, which may be attributed to the adoption to modern life style featured by increased fat intake and reduced physical activity [2]. Although aggressive therapeutic strategies including surgery, radiotherapy and chemotherapy have significantly improve the prognosis of patients with CRC [3], almost 50% patients with CRC will develop repetition, leading to the high mortality of CRC [4]. Up to now, numerous factors has been investigated for their prognostic value and potential as a therapeutic target.
miRNAs are short, endogenous RNAs with a length of 22nt that demonstrate complementarity with 3’-untranslated region (3’-UTR) of their target messenger RNAs (mRNAs). The regulation of gene expression by miRNAs could be carried out through either posttranscriptional regulation of gene expression leading to target mRNA degradation or repression of its translation with consequent decrease in the particular protein levels or even by upregulation of the targets [5]. By modulating different target genes, miRNA have been found to play a role in a plethora of cellular processes of cancer cells such as cell apoptosis, cell proliferation, invasion, migration metabolism and stem cell differentiation [6-8]. Dysregulation of miR-146b-5p has been documented in a variety of human malignancies, including gastric cancer [9], thyroid carcinoma [10], osteosarcoma [11] and glioma [12]. However, the prognostic significance and role of miR-146b-5p in CRC remain unclear.
About 90 years ago, the Nobel laureate Otto Warburg proposed that cancer cells utilized the glycolytic phenotype (aerobic glycolysis) as energy resources rather than performing oxidative phosphorylation (OXPHOS). The process that glucose is converted into lactate for energy production in cancer cells is therefore named “the Warburg’s effect”, which supports energy production, macromolecular synthesis, and redox balance essential for tumor growth [13]. Therefore, targeting the glycolysis process can provide a novel insight in understanding cancer biology. Localized in the mitochondria, pyruvate dehydrogenase B (PDHB) is the enzyme that catalyzes the glucose-derived pyruvate to the acetyl-CoA and plays important roles in oxidative phosphorylation [14]. However, the mechanism by which PDHB is regulated remains poorly understood. In this study, the expression pattern of miR-146b-5p is assessed and the prognostic significance of miR-146b-5p is investigated. Furthermore, the effect of miR-146b-5p on CRC cell growth, metastatic behavior and bioenergetics is examined and the underlying molecular mechanisms are explored.
Materials and methods
Clinical samples
A total of 49 patients who were diagnosed with CRC and received radical resection surgery from May 2010 and March 2011 were enrolled in this study. This study was approved by the ethics committee of Henan Province People’s Hospital and written consent forms were obtained from all patients. CRC tissue and matched non-tumor tissue samples were collected and immediately frozen in liquid nitrogen until use. The diagnosis and histological classification of samples were blindly determined by two independent pathologists. Quantitative RT-PCR (qRT-PCR) was performed to determine the level of miR-146b-5p in tissue samples. Automated capillary western blot (WES) [15] was conducted to determine the level of PDHB in tissue samples.
Cell cultures
Human CRC cell lines LOVO, HT29, SW620, HCT116 and SW480 were purchased from ATCC (Shanghai, China). Normal human intestinal epithelial cell line (HIEC) was purchased from Cell Bank of the Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). All cells were maintained in DMEM medium (Invitrogen, Carlsbad, CA) containing 1% penicillin/streptomycin (Thermo Fisher Scientific, Waltham, MA) and 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA) in a humidified incubator at 37°C with 5% CO2.
Construct of miR-146b-5p mimic or anti-miR-146b-5p and cell transfection
The lentiviral constructs of miR-146b-5p mimic (miR-146b-5p), miR-146b-5p control (miR-con), anti-miR-146b-5p, and anti-miR control (anti-miR-con) were constructed by Genepharma (Shanghai, China). CRC cells were infected with the constructed lentivirus following standard protocol.
Quantitative real-time PCR (qRT-PCR)
TRIzol Reagent (Life Technology, Carlsbad, CA) was used to isolate total RNA from cultured CRC cells. The isolated RNA was used to cDNA synthesis using the reverse transcription kit (Promega, Madison, WI), following the manufacturer’s instructions. Then the expression level of PDHB mRNA was determined by RT-PCR using Power SYBR Green PCR Master Mix (Carlsbad, CA) with GAPDH was used as an internal control. The forward and reversed primer sequences synthesized by Sangon (Shanghai, China) as the following: forward primer, 5’-GATGAGAAGGTATTTCTGCT-3’, and reverse primer, 5’-GAGAAATTGAAGGTCATAAA-3’. The 2-ΔΔCt method was used to analyse the relative expression of target genes. To determin the levek of miR-146b-5p in cells and tissues, miRVANA Kit (Ambion, Carlsbad, MA) was utilized to isolate miRNA. A small RNA specific stem-loop RT primer was used for reverse transcription of cDNA. MiR-146b-5p level was determined by RT-PCR according to the TaqMan MicroRNA Assay (Thermo Fisher Scientific, Waltham, MA). Small nuclear RNA U6 was taken as control and the data were expressed as Log 2 fold change in respective miR/U6 snRNA levels.
Western blot
Western blotting was performed following standard protocols. Proteins were detected with specific primary antibodies. Goat anti-rabbit IgG-HRP used as the second antibodies were purchased from Beyotime (Shanghai, China). The relative levels of protein expression were normalized against β-actin, as internal control.
Bioinformatics and luciferase reporter assay
The molecular target of miR-146b-5p was predicted using the computational algorithm softwares, TargetScan [16] and microRNA.org [17]. The 3’-untranslated region (UTR) of PDHB was identified as a potential target gene. To verify whether PDHB is a direct target of miR-146b-5p n this case, a luciferase reporter was constructed by amplifying a wild-type 3’UTR segment of PDHB that contains putative miR-146b-5p binding sites and cloning into the XhoI and NotI sites downstream of the luciferase reporter gene in pGL3 vector (Promega, Madison, WI, USA). The primers used to amply the 3’-UTR of PDHB was designed and synthesized by Sangong (Shanghai, China). The mutagenesis of the 3’UTR segment of PDHB was performed as previously described [18]. Both wild-type and mutant luciferase reporter constructs were confirmed by sequencing. CRC cells were transiently co-transfected with 0.2 μg of pGL3-PDHB-3’UTR or pGL3-PDHB-3’UTR Mut, 0.02 μg of pRL-TK-Renilla luciferase reporter plasmids (Promega, Madison, WI) containing the Renilla-luciferase for normalization, and with 5 pmol of miR-146b-5p mimic or negative control. 48 hours after the transfection, the luciferase activity was examined using a Dual-luciferase assay system (Promega, Madison, WI).
Re-expression of in CRC cells
PDHB overexpressing vector was constructed as previously described using a plasmid vector pGCsi-H1 [19] and the transfection with expressing construct was performed using Lipofectamine 3000 reagent (Invitrogen, Grand Island, NY) according the manufacturer’s instructions. Cells transfected with empty vector were used as controls. 48 hours after transfection, the cells were rinsed before resuspended in fresh culture media and the overexpression was verified by western blot analysis.
Cell proliferation assay
Cell Counting Kit-8 (Beyotime, Shanghai, China) was used to assess the cell proliferation. Briefly, a total of 1×105 cells were plated in culture plates. Following an incubation of indicated time, the viable cells were examined by measuring absorbance at 450 nm (Tecan Group Ltd, Männedorf, Switzerland).
Colony formation
Cells suspended in DMEM agarose medium were seeded in each well of a 6-well plate over a bottom layer of solidified DMEM agarose medium. Cultures were maintained for 14 days without fresh medium feeding at 37°C in a humidified atmosphere of 95% air and 5.0% CO2. Then cell colonies with over 50 cells were enumerated and stained with violet crystal before being photographed using a digital camera (Olympus, Tokyo, Japan).
Flow cytometry
Following treatment, CRC cells were harvested and stained with Annexin V-PE and propidium iodide using an Apoptosis kit (BD Pharmingen, Franklin Lakes, NJ) according to the manufacturer’s instructions. Then the apoptotic percentage of treated cells was determined by a flow cytometer (Beckman Coulter Inc., Miami, FL).
Transwell invasion assay
24-well Transwells coated with Matrigel (8-μm pore size; BD Biosciences, San Jose, CA) were used for cell invasion assays. Equal numbers (1×105) of cells were plated on separate wells. Cells were cultured overnight in serum-free medium before trypsinization and re-suspended at a density of 2×105 cells/ml in DMEM containing 1% FBS. The cells were loaded to the upper chamber, with MEM containing 10% FBS as chemoattractant in the lower chamber. The medium containing 1% FBS in the lower chamber was used as a control. The Matrigel and the cells remaining in the upper chamber were removed by cotton swabs following 24-hour incubation. The cells in the lower surface of the membrane were stained with hematoxylin after the cells were fixed with formaldehyde solution. The cells in at least five random microscopic fields (×200) were counted and photographed.
Determination of the glucose consumption and lactate production
The cells were seeded onto six-well plate for 6 hours before the culture medium was replaced with fresh complete medium and incubated for additional 48 hours. The media were then collected to examine the glucose consumption and lactate production. A glucose assay kit (Sigma, St Louis, MI) was utilized to assess glucose level and glucose consumption was calculated by deducting the measured glucose concentration in the media from the original glucose concentration. A lactate assay kit (Sigma, St Louis, MI) was used to determine the lactate levels in culture medium.
In vivo xenograft model
The protocol of animal experiments were reviewed and approved by Medical Ethics Committee of Henan Province People’s Hospital. For tumor growth assay, BALB/c nude mice of four-week-old were used for the CRC xenograft models (n=6 per group). 1×107 HT29 cells transfected with control vector or miR-429 mimic construct were suspended in 100 μl medium was injected subcutaneously into the lower left flank regions of mice model. The tumour volume was measured every 3 days and the mice were sacrificed after 30 days.
For the lung metastasis assays, BALB/C-nu/nu nude mice of six-week-old were used and 1×106 LOVO cells transfected with control vector or miR-146b-5p mimic construct were injected into the lateral tail vein of mice model (n=3 per group). The mice were sacrificed 6 weeks later and the metastasis modules in lung were counted.
Statistical analysis
Values were presented as the mean ± SD. The comparison of miR-429 levels in tumor and normal tissue were performed using student’s t test. Statistical comparisons between cell lines were performed by one-way ANOVA followed by Dunnett’s t-test. The overall survival of patients was assessed by Kaplan-Meier survival analysis. GraphPad Prism software (GraphPad Software Inc., La Jolla, CA) was used to analyze experimental data and a P value less than 0.05 was considered to be statistically significant.
Results
Aberrant upregulation of miR-146b-5p is observed in CRC tissue and CRC cell lines
The level of miR-146b-5p was determined by qRT-PCR analysis. Our results showed that the expression of miR-146b-5p was significantly upregulated in CRC tissues compared to adjacent non-tumor tissue (Figure 1A). Moreover, when the expression of miR-146b-5p was compared in tissues collected from patients of different TNM stage, it was found that higher miR-146b-5p expression was associated with advanced stage of CRC (Figure 1B). Next, we compared the expression of miR-146b-5p in CRC cell lines with that of HIEC cells. As show in Figure 1C, miR-146b-5p level was significantly higher. Collectively, our findings showed that miR-146b-5p was highly expressed in CRC, which suggested the involvement of miR-146b-5p in CRC development and progression.
Figure 1.
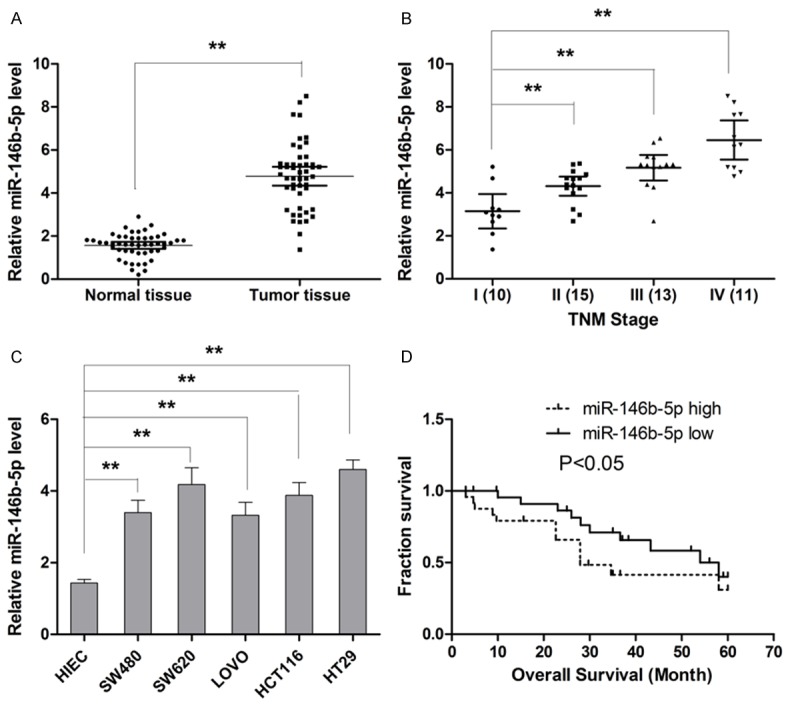
MiR-146b-5p is abnormally higher expressed in CRC tissues samples and cell lines, and predicts the overall survival of patients with CRC. A. The expression of miR-146b-5p is significantly higher in CRC tissue relative to non-cancerous tissue. B. Expression of miR-146b-5p increases with advancement of cancer stage. C. The expression of miR-146b-5p is upregulated in CRC cell lines compared with HIEC. D. Higher expression of miR-146b-5p correlates with shorter survival of patients with CRC. **P<0.01.
High miR-146b-5p level is associated with advanced tumor stage and poor prognosis
Based on our beforementioned results that higher miR-146b-5p expression was associated with advanced stage of CRC, CRC tissues expressing miR-146b-5p at levels less than the median expression level were allocated to the low group while and those samples with expression above the median value were allocated to the high expression group to analyze the correlation between miR-146b-5p expression and clinicopathological characteristics of patients. The analysis results showed that miR-146b-5p expression was positive associated with tumor stage progression and lymph node metastasis (Table 1). However, the other clinicopathologcial features including gender, tumor size, tumor location and histological grade were not found to correlate with miR-146b-5p level in tumor tissues. Then a Kaplan-Meier survival curve was plotted to establish the association between miR-146b-5p expression and survival time of patients. As show in Figure 1D, our results showed that higher miR-146b-5p expression significantly correlated with shorter median survival time (27.9 months for high miR-146b-5p group vs. 58.0 months for low miR-146b-5p group). To further evaluate the prognostic value of miR-146b-5p, both univariable and multivariable analysis were performed to assess the prognostic value of miR-146b-5p. As shown in Table 2, the overall survival of patients significantly correlated with miR-146b-5p expression, absence of lymph node metastasis and TNM stage and miR-146b-5p can be considered as an independent prognostic marker for overall survival of patients with CRC.
Table 1.
The expression of miR-146b-5p correlates with clinical features of patients with CRC
| Parameters | miR-146b-5p level | P value | |
|---|---|---|---|
|
| |||
| High (24) | Low (25) | ||
| Age | 0.5672 | ||
| ≤60 | 13 | 16 | |
| >60 | 11 | 9 | |
| Gender | 0.7688 | ||
| Male | 15 | 17 | |
| Female | 9 | 8 | |
| Tumor size | 0.5512 | ||
| ≤5 cm | 17 | 15 | |
| >5 cm | 7 | 10 | |
| Histological grade | 0.5607 | ||
| Well, moderate | 14 | 17 | |
| Poor | 10 | 8 | |
| TNM stage | 0.0421* | ||
| I and II | 6 | 14 | |
| III and IV | 18 | 11 | |
| Site | 0.2321 | ||
| Colon | 14 | 19 | |
| Rectum | 10 | 6 | |
| Lymph node metastasis | 0.0042** | ||
| Absence | 7 | 18 | |
| Presence | 17 | 7 | |
P<0.05;
P<0.01.
Table 2.
MiR-146b-5p is an independent prognostic biomarker for patients with CRC
| Parameters | Univariable analysis | Multivariable analysis | ||
|---|---|---|---|---|
|
| ||||
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Gender (Male or Female) | 0.987 (0.423-2.521) | 0.452 | - | - |
| Age (≤60/>60) | 1.206 (0.751-2.950) | 0.687 | - | - |
| Histological grade (Well, moderate/Poor) | 1.219 (0.681-3.295) | 0.435 | - | - |
| Tumor size (≤5 cm/>5 cm) | 1.644 (1.013-3.937) | 0.235 | - | - |
| Tumor site (colon/rectum) | 1.109 (0.685-1.562) | 0.335 | ||
| Lymph node metastasis (Absence/Presence) | 5.952 (1.984-9.687) | 0.004** | 3.952 (2.219-8.157) | 0.028* |
| TNM stage (I, II/III, IV) | 3.568 (1.441-9.672) | 0.016* | 3.454 (1.975-8.567) | 0.007** |
| miR-146b-5p expression (high/low) | 2.354 (0.919-7.654) | 0.008** | 1.954 (0.987-3.914) | 0.017* |
P<0.05;
P<0.01.
MiR-146b-5p promotes proliferation, invasion and glycolysis of CRC cells
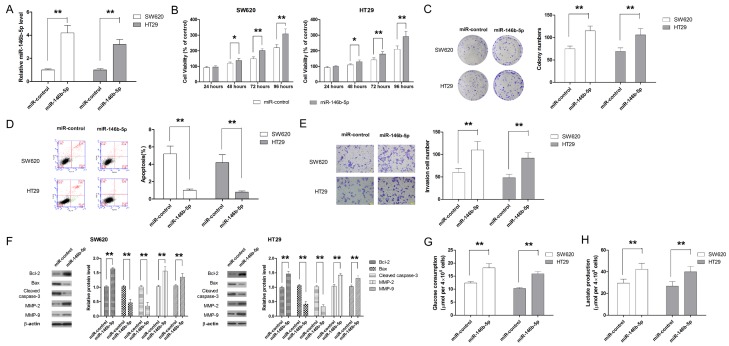
Two CRC cell lines, SW620 and HT29, were used as model cell line to investigate the effect of miR-146b-5p on cell proliferation, invasiveness and glycolysis. CRC cell lines were transfected with miR-146b-5p mimic to induce ectopic overexpression of miR-146b-5p while CRC cells transfected with negative control miRNA was taken as controls. As shown in Figure 2A, miR-146-5p mimic successful increased the expression of miR-146b-5p in SW620 and HT29 cells by about 4 and 3 folds, respectively. The effect of miR-146b-5p on cell proliferation was assessed by CCK-8 assay. Our results showed that miR-146b-5p was able to significantly increase the cell viability compared with control cells (Figure 2B). In addition, colony formation assay also confirmed the pro-proliferative effect of miR-146b-5p in CRC cells (Figure 2C). Flow cytometric assay demonstrated that miR-146b-5p significantly decreased the apoptotic cell population (Figure 2D). The effect of miR-146b-5p on cell apoptosis was also backed up by the findings that miR-146b-5p significantly increased the Bcl-2/Bax ratio and the level of cleaved caspase-3 in both SW620 and HT29 cells (Figure 2E). Next, the Transwell assay was performed to examine the effect of miR-146b-5p on cell invasion. As shown in Figure 2E, CRC cells transfected with miR-146b-5p mimic exhibited significantly higher invasiveness. In addition, miR-146b-5p mimic was also able to decrease the levels of MMP-2 and MMP-9, two key molecules involved in cell invasion (Figure 2F). The effect of miR-146b-5p on glucose metabolism of cells was also examined. As shown in Figure 2G and 2H, increase in glucose uptake and lactate production was observed when ectopic overexpression of miR-146b-5p was introduced.
Figure 2.
MiR-146b-5p promotes cell growth, inhibits apoptosis, enhances invasion and glycolysis in CRC cells. A. MiR-146b-5p mimic significantly enhances expression level of miR-146b-5p. B. Ectopic overexpression of miR-146b-5p promotes CRC cell growth, as demonstrated by CCK-8 assay. C. Ectopic overexpression of miR-146b-5p enhances long term cell proliferation, as colonigenic assay. D. Overexpression of miR-146b-5p is associated with less apoptosis in CRC cells. E. MiR-146b-5p enhances in cell invasion. F. MiR-146b-5p mimic transfection significantly changes the expression levels of marker molecules for apoptosis and invasion. G. MiR-146b-5p overexpression enhances the glucose consumption of CRC cells. H. MiR-146b-5p overexpression promotes the production of lactate. **P<0.01.
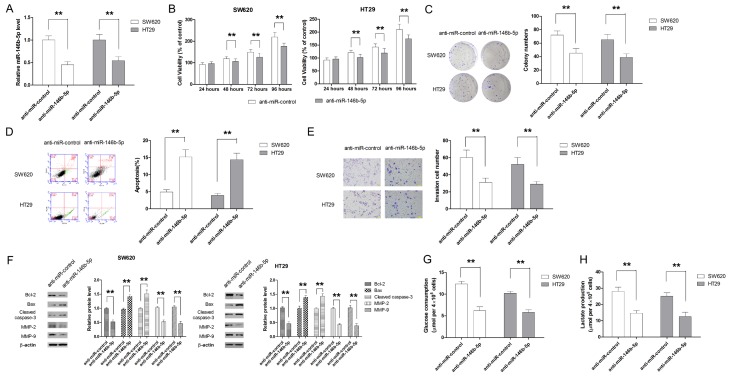
Transfection with anti-miR-146b-5p dampened growth, invasion and glycolysis of CRC cells
To further investigate the role of miR-146b-5p in CRC, anti-miR-146b-5p was used to suppress the endogenous expression of miR-146b-5p (Figure 3A). Then a series of biological experiments were conducted to examine the effect of anti-miR-146b-5p on growth, invasion and glycolysis of CRC cells. As shown in Figure 3B and 3C, both CCK-8 and colony formation assays showed that anti-miR-146b-5p significantly inhibited the growth of SW620 and HT29 cells. In addition, transfection with anti-miR-146b-5p significantly induced apoptosis and suppressed cell invasion (Figure 3D-F). Moreover, anti-miR-146b-5p significantly impaired the glucose consumption and lactate production of CRC cells (Figure 3G and 3H).
Figure 3.
Anti-miR-146b-5p inhibited cell growth, induces apoptosis, suppresses invasion and glycolysis of CRC cells. A. Knockdown of miR-146b-5p significantly decreases the expression of miR-146b-5p in CRC cells. B. Knockdown of miR-146b-5p inhibits cell growth, as demonstrated by CCK-8 assay. C. Knockdown of miR-146b-5p suppresses the long-term proliferation of CRC cells, as demonstrated by colonigenic assay. D. Anti-miR-146b-5p promotes cell apoptosis. E. Anti-miR-146b-5p is associated with significantly less invasion of CRC cells. F. Anti-miR-146b-5p regulated the expression of marker molecules involved in apoptosis and invasion. G. Anti-miR-146b-5p transfection leads to decrease in glucose consumption. H. Knockdown of miR-146b-5p results in less production of lactate. **P<0.01.
MiR-146b-5p promotes tumor growth and metastasis in vivo
Since our before mentioned results suggested that miR-146b-5p functioned an oncogene in vitro, mice models were utilized to examine the in vivo effect of miR-146b-5p. Consistent with the in vitro results, in vivo experiments also showed that HT29 cells transfected with miR-146b-5p correlated with significantly enhanced tumor growth (Figure 4A). HE staining and immunohistochemical assay also showed that miR-146b-5p group presented with more cell mitosis along with higher expression of Ki-67 and PCNA as well as lower level of activated caspase-3 (Figure 4B). Moreover, lung metastasis experiments also showed that miR-146b-5p overexpression was associated with significantly more metastasis nodules (Figure 4C and 4D). Taken together, our findings provided strong evidence that miR-146b-5p promoted tumor growth and metastasis in CRC.
Figure 4.
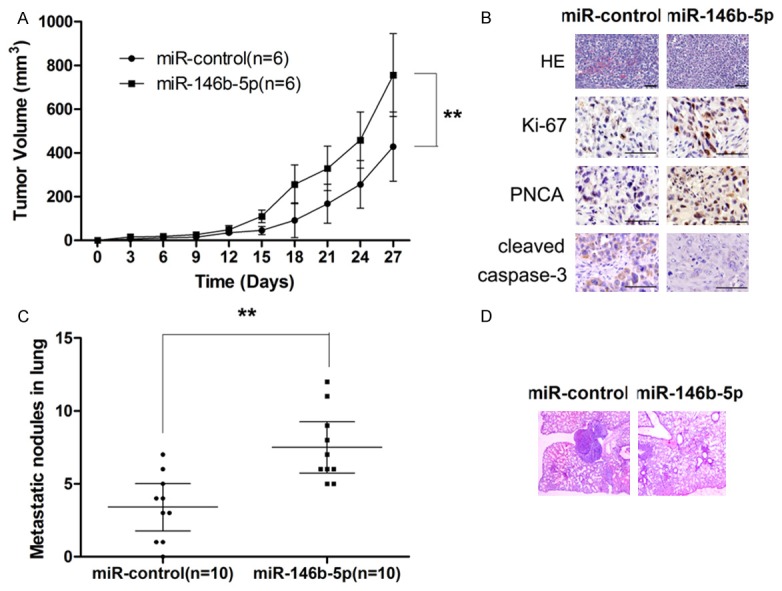
MiR-146b-5p promotes tumor growth and metastasis in vivo. A. MiR-146b-5p overexpression correlates significantly enhancement in tumor growth. B. MiR-146b-5p overexpression shows higher proliferation and less apoptosis, as demonstrated by HE staining and IHE examination of the xenograft tumors. C. MiR-146b-5p overexpression promotes pulmonary metastasis of CRC. D. MiR-146b-5p overexpression correlates with significantly more metastatic nodules in lung, as demonstrated by HE staining.
PDHB is a direct target of miR-146b-5p in CRC
To clarify the molecular mechanisms for the effect of miR-146b-5p, candidate target genes of miR-146b-5p was searched in bioinformatics database (MicroCosm and Targetscan). Give that miR-146b-5p could enhance the glycolysis of CRC cells, we postulated that PDHB is a direct target of miR-146b-5p in CRC. To confirm our postulation, a luciferase reporter was constructed according to the sequence shown in Figure 5A. As shown in Figure 5B, luciferase activity of the reporter containing wild-type 3’-UTR of PDHB was significantly decreased in cells transfected with miR-146b-5p mimic while no significant change in luciferase activity of the reporter containing mutated 3’-UTR was observed, providing direct evidence that PDHB was directly targeted by miR-146b-5p. To further study the mechanism by which miR-146b-5p regulated the expression of PDHB, the mRNA and protein expression in both SW620 and HT29 cells was examined following transfection with miR-146b-5p mimic. As shown in Figure 5C and 5D, miR-146b-5p did not significantly change the level of PDHB mRNA while significantly repressed the protein expression of PDHB, suggesting that miR-146b-5p mediated the post-transcriptional regulation of PDHB expression. The expression of mRNA and protein level of PDHB was also examined in tumors collected from xenograft mice models. Although no significant difference in PDHB mRNA levels was observed between miR-control and miR-146b-5p overexpressing group, PDHB protein level in tumor tissues from miR-146b-5p overexpressing group was significantly lower compared with control group (Figure 5E and 5F). Moreover, the correlation between PDHB expression and miR-146b-5p level in patient tissue samples was also examined, which also supported that miR-146b-5p regulated the expression of PDHB post-transcriptionally (Figure 5G and 5H).
Figure 5.
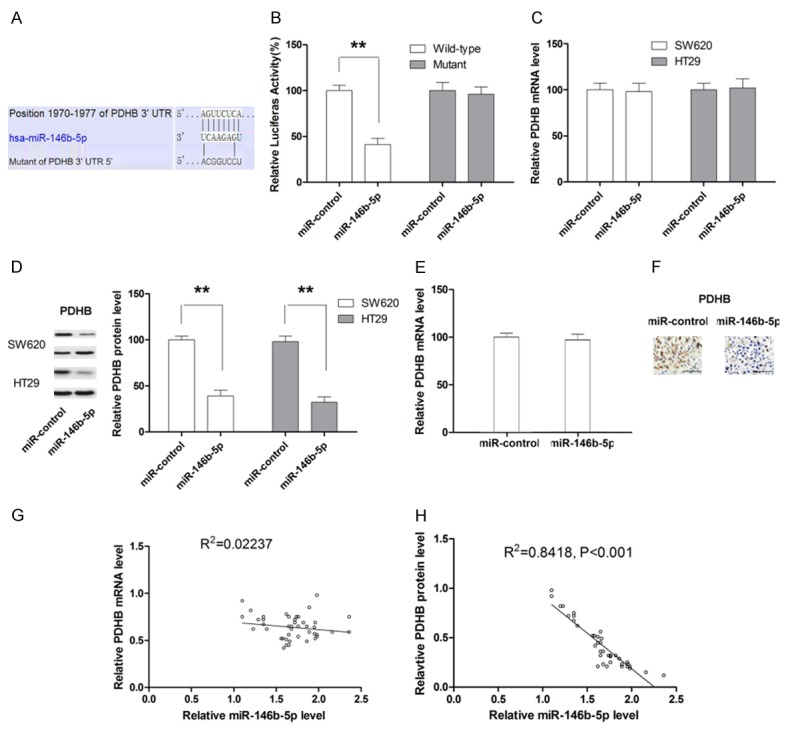
MiR-146b-5p regulates the expression of PDHB through post-transcriptional mechanism. A. Schematic illustration of the putative binding sites between wild-type or mutant 3’-UTR of PDHB and miR-146b-5p. B. Luciferase activity is decreased by miR-146b-5p mimic. C. MiR-146b-5p does not change the mRNA level of PDHB in CRC cell lines. D. MiR-146b-5p significantly changes the protein level of PDHB in CRC cell lines. E. The expression of PDHB mRNA in tumor xenograft is not affected by miR-146b-5p overexpression. F. The expression of PDHB protein in tumor xenograft is significantly repressed by miR-146b-5p overexpression. G. The expression level of miR-146b-5p is not associated with mRNA expression of PDHB in CRC specimen. H. The expression level of miR-146b-5p is negatively associated with mRNA expression of PDHB in CRC specimen. **P<0.01.
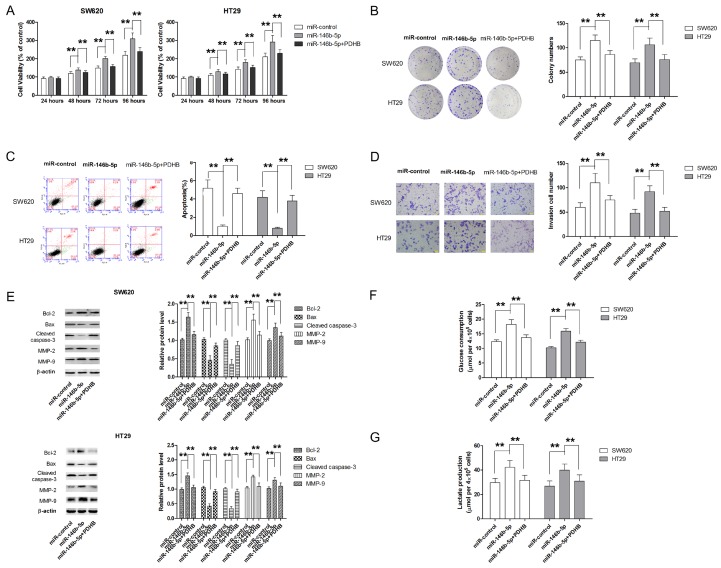
Restoration of PDHB reverses the promoting effect of miR-146b-5p on cell growth, invasion and glycolysis
To further appreciate the role of PDHB in the oncogenic effect of miR-146b-5p, CRC cells were cotransfected with miR-146b-5p mimic and PDHB overexpressing plasmid. As shown in Figure 6A, PDHB overexpression significantly attenuated the promoting effect of miR-146b-5p on cell proliferation in both SW620 and HT29 cells. Similar results were also obtained from colony formation assay (Figure 6B). Meanwhile, the apoptosis inhibition by miR-146b-5p mimic was also markedly abrogated by PDHB overexpression (Figure 6C and 6E). Transwell assay also revealed that ectopic overexpression of PDHB was able to partially reverse miR-146b-5p mimic-induced invasion in both SW620 and HT29 cells (Figure 6D and 6E). Furthermore, PDHB also significantly attenuated miR-146b-5p-caused enhancement in glucose metabolism (Figure 6F and 6G). Collectively, our findings demonstrated that miR-146b-5p exerted oncogenic effect, at least in part, by directly repressing PDHB expression.
Figure 6.
Restoration of PDHB attenuates the oncogenic effect of miR-146b-5p in CRC cells. A. Ectopic PDHB overexpression attenuated the promoting effect of miR-146b-5p on cell growth of CRC cells. B. Ectopic PDHB overexpression attenuated the long term pro-proliferative effect of miR-146b-5p in CRC cells. C. Ectopic PDHB overexpression results in significantly higher apoptotic rates in CRC cells. D. Ectopic PDHB overexpression leads to significantly inhibition in cell invasion. E. PDHB overexpression attenuates miR-146b-5p-induced changes in expression of molecular markers for apoptosis and invasion. F. PDHB overexpression reverses miR-146b-5p-induced increase in glucose consumption. G. PDHB overexpression reverses miR-146b-5p-induced increase in lactate production. **P<0.01.
Discussion
Accumulating evidence from preclinical and clinical studies has showed that miRNAs play in the development of human malignancies has been evidenced in [20]. The expression of miR-146b-5p has been found in almost all human organs and the dysregulation of miR-146b-5p has been reported in a variety of human malignancies [21]. The role of miR-146b-5p as oncogene was first identified in papillary thyroid carcinoma, which showed that over-expression of miR-146b-5p is associated with aggressiveness of papillary thyroid carcinoma [22]. In lung cancer, miR-146b-5p has also been reported to be negatively associated with prognosis [23]. A recent study by Yoon et al also suggested that aberrant upregulation of miR-146b-5p is a candidate biomarker predicting poor prognosis of gastric cancer patients [9]. However, conflicting results that support the role of miR-146b-5p as tumor suppressor have also been reported. A very recent study by Correia et al has reported that miR-146b-5p alters the motility, migration and invasion capacities of T-cell acute lymphoblastic leukaemia cell lines in vitro and in vivo, highlighting the involvement of miR-146b-5p in hematological malignancies [24]. Their findings were in line with a number of studies with solid tumors, including osteosarcoma (via AUF1 regulation) [25], breast cancer (via NF-κB regulation) [26,27], glioma (via MMP16 and EGFR regulation) [28,29], and pancreatic cancer (via MMP16 regulation) [30], all of which suggesting that miR-146b-5p serves a tumor suppressor. A previous study in 2012 reported dysregulation of miR-146b-5p in patients with CRC [31]. Nevertheless, the prognostic significance of miR-146b-5p in CRC has never been investigated. In this study, our findings showed that aberrantly high expression of miR-146b-5p correlated with advanced TNM stage and lymph mode metastasis. CRC patients with high miR-146b-5p expression had worse prognoses than did patients with a low miR-146b-5p expression. Moreover, we used Cox regression analysis to evaluate some clinical and pathological factors, which showed low miR-146b-5p expression is an independent indicator of poor patient prognosis.
Energy metabolism Reprogramming has been considered as a feature of cancer cells [32]. Even in the presence of oxygen, cancer cells can reprogram their glucose metabolism, and thus the ATP generation shifts from oxidative phosphorylation to glycolysis, leading to a state termed “aerobic glycolysis”, which is known as “Warburg effect” [13]. Different from normal cells, tumor cells meet their energy demand mainly through aerobic glycolysis by converting most incoming glucose to lactate rather than metabolizing it through oxidative phosphorylation. Given that ATP generated is far less for per unit of glucose consumed in glycolysis than oxidative phosphorylation, tumor cells demand abnormally high rate of glucose uptake to meet their increased energy and biosynthesis. Given the crucial role of metabolism in various biological activities of cancer cell, modulating the metabolism has been considered a promising therapeutic approach in cancer therapy. Up to date, a number of miRNAs have been found to play a role in regulating the bioenergetics of cancer cells. For instance, miR-195-5p [33] and miR-143 [34] have been found to suppress the glucose uptake and glycolysis processing by repressing the expression of glucose transporter 3 (GLUT3) and hexokinase 2, respectively. On other hand, miR-203 [35] and miR-26a [36] has been found to enhance glycolysis in cancer cells. Therefore, manipulating the expression of miRNAs, such as using oligonucleotide complementary or antisense oligonucleotides, to suppressing cell metabolism reprogramming might highlight a novel therapeutic strategy for cancer control [37].
In this study, our findings showed that ectopic overexpression of miR-146b-5p significantly increased the glucose uptake and lactate production in CRC cells while transfection with anti-miR-146b-5p correlated with significantly decreased glucose utilization and lactate production, which suggested that miR-146b-5p was able to enhance glycolysis to meet the need of energy and biosynthesis in CRC cells.
More importantly, our findings in present study provided molecular basis for the modulating effect of miR-146b-5p on glucose metabolism of CRC cells. Our results showed that miR-146b-5p repressed the expression of PDHB by direct targeting the conserved miR-146b-5p recognition motif of the 3’UTR of PDHB mRNA. PDHB, localizing in the mitochondrial, catalyzes the glucose derived pyruvate to the acetyl-CoA [14] and thus regulate the key step of glycolysis to the citric acid cycle in glucose metabolism. Aberrantly downregulation of PDHB has been reported in gastric cancer and cardiac cancer [38,39]. In gastric cancer, the expression of PDHB is found to be an independent prognostic marker and correlates with favorable clinical outcomes [39]. In our study, we also found that repressing PDHB by miR-146b-5p was associated with tumor growth and metastasis, supporting the tumor suppressor role of PDHB. On the contrary, Tang et al documented that PDH functioned as oncogene in the nasopharyngeal carcinoma cells to promote cell proliferation and migration [19]. It seems PDHB might exert distinct effects in different human malignancies.
In summary, our findings in current study provide experimental evidence that miR-146b-5p correlates with clinicopathological characteristics and survival of CRC patients. In addition, miR-146b-5p modulates cell growth, invasion and glycolysis directly targeting PDHB.
Disclosure of conflict of interest
None.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Byeon JS, Yang SK, Kim TI, Kim WH, Lau JY, Leung WK, Fujita R, Makharia GK, Abdullah M, Hilmi I, Sollano J, Yeoh KG, Wu DC, Chen MH, Kongkam P, Sung JJ Asia Pacific Working Group for Colorectal Cancer. Colorectal neoplasm in asymptomatic Asians: a prospective multinational multicenter colonoscopy survey. Gastrointest Endosc. 2007;65:1015–1022. doi: 10.1016/j.gie.2006.12.065. [DOI] [PubMed] [Google Scholar]
- 3.Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, Starling N. Colorectal cancer. Lancet. 2010;375:1030–1047. doi: 10.1016/S0140-6736(10)60353-4. [DOI] [PubMed] [Google Scholar]
- 4.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 5.Fabbri M, Croce CM, Calin GA. MicroRNAs. Cancer J. 2008;14:1–6. doi: 10.1097/PPO.0b013e318164145e. [DOI] [PubMed] [Google Scholar]
- 6.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 7.le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafre SA, Farace MG, Agami R. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon SO, Kim EK, Lee M, Jung WY, Lee H, Kang Y, Jang YJ, Hong SW, Choi SH, Yang WI. NOVA1 inhibition by miR-146b-5p in the remnant tissue microenvironment defines occult residual disease after gastric cancer removal. Oncotarget. 2016;7:2475–2495. doi: 10.18632/oncotarget.6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng X, Wu B, Xiao K, Kang J, Xie J, Zhang X, Fan Y. MiR-146b-5p promotes metastasis and induces epithelial-mesenchymal transition in thyroid cancer by targeting ZNRF3. Cell Physiol Biochem. 2015;35:71–82. doi: 10.1159/000369676. [DOI] [PubMed] [Google Scholar]
- 11.Xu E, Zhao J, Ma J, Wang C, Zhang C, Jiang H, Cheng J, Gao R, Zhou X. miR-146b-5p promotes invasion and metastasis contributing to chemoresistance in osteosarcoma by targeting zinc and ring finger 3. Oncol Rep. 2016;35:275–283. doi: 10.3892/or.2015.4393. [DOI] [PubMed] [Google Scholar]
- 12.Liu J, Xu J, Li H, Sun C, Yu L, Li Y, Shi C, Zhou X, Bian X, Ping Y, Wen Y, Zhao S, Xu H, Ren L, An T, Wang Q, Yu S. miR-146b-5p functions as a tumor suppressor by targeting TRAF6 and predicts the prognosis of human gliomas. Oncotarget. 2015;6:29129–29142. doi: 10.18632/oncotarget.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 14.Saunier E, Benelli C, Bortoli S. The pyruvate dehydrogenase complex in cancer: an old metabolic gatekeeper regulated by new pathways and pharmacological agents. Int J Cancer. 2016;138:809–817. doi: 10.1002/ijc.29564. [DOI] [PubMed] [Google Scholar]
- 15.Ridnour LA, Cheng RY, Weiss JM, Kaur S, Soto-Pantoja DR, Basudhar D, Heinecke JL, Stewart CA, DeGraff W, Sowers AL, Thetford A, Kesarwala AH, Roberts DD, Young HA, Mitchell JB, Trinchieri G, Wiltrout RH, Wink DA. NOS inhibition modulates immune polarization and improves radiation-induced tumor growth delay. Cancer Res. 2015;75:2788–2799. doi: 10.1158/0008-5472.CAN-14-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 17.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng B, Wang B, Fang J, Zhu X, Cao Z, Lin Q, Zhou L, Sun X. MiRNA-203 suppresses cell proliferation, migration and invasion in colorectal cancer via targeting of EIF5A2. Sci Rep. 2016;6:28301. doi: 10.1038/srep28301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang H, Luo X, Li J, Zhou Y, Li Y, Song L, Zhang X, Chen T. Pyruvate dehydrogenase B promoted the growth and migration of the nasopharyngeal carcinoma cells. Tumour Biol. 2016;37:10563–10569. doi: 10.1007/s13277-016-4922-4. [DOI] [PubMed] [Google Scholar]
- 20.Chi Y, Zhou D. MicroRNAs in colorectal carcinoma--from pathogenesis to therapy. J Exp Clin Cancer Res. 2016;35:43. doi: 10.1186/s13046-016-0320-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu SD, Chu CH, Tsou AP, Chen SJ, Chen HC, Hsu PW, Wong YH, Chen YH, Chen GH, Huang HD. miRNAMap 2.0: genomic maps of microRNAs in metazoan genomes. Nucleic Acids Res. 2008;36:D165–169. doi: 10.1093/nar/gkm1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geraldo MV, Fuziwara CS, Friguglieti CU, Costa RB, Kulcsar MA, Yamashita AS, Kimura ET. MicroRNAs miR-146-5p and let-7f as prognostic tools for aggressive papillary thyroid carcinoma: a case report. Arq Bras Endocrinol Metabol. 2012;56:552–557. doi: 10.1590/s0004-27302012000800015. [DOI] [PubMed] [Google Scholar]
- 23.Patnaik SK, Kannisto E, Mallick R, Yendamuri S. Overexpression of the lung cancer-prognostic miR-146b microRNAs has a minimal and negative effect on the malignant phenotype of A549 lung cancer cells. PLoS One. 2011;6:e22379. doi: 10.1371/journal.pone.0022379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Correia NC, Fragoso R, Carvalho T, Enguita FJ, Barata JT. MiR-146b negatively regulates migration and delays progression of T-cell acute lymphoblastic leukemia. Sci Rep. 2016;6:31894. doi: 10.1038/srep31894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Khalaf HH, Aboussekhra A. MicroRNA-141 and microRNA-146b-5p inhibit the prometastatic mesenchymal characteristics through the RNA-binding protein AUF1 targeting the transcription factor ZEB1 and the protein kinase AKT. J Biol Chem. 2014;289:31433–31447. doi: 10.1074/jbc.M114.593004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009;69:1279–1283. doi: 10.1158/0008-5472.CAN-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiang M, Birkbak NJ, Vafaizadeh V, Walker SR, Yeh JE, Liu S, Kroll Y, Boldin M, Taganov K, Groner B, Richardson AL, Frank DA. STAT3 induction of miR-146b forms a feedback loop to inhibit the NF-kappaB to IL-6 signaling axis and STAT3-driven cancer phenotypes. Sci Signal. 2014;7:ra11. doi: 10.1126/scisignal.2004497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia H, Qi Y, Ng SS, Chen X, Li D, Chen S, Ge R, Jiang S, Li G, Chen Y, He ML, Kung HF, Lai L, Lin MC. microRNA-146b inhibits glioma cell migration and invasion by targeting MMPs. Brain Res. 2009;1269:158–165. doi: 10.1016/j.brainres.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 29.Katakowski M, Zheng X, Jiang F, Rogers T, Szalad A, Chopp M. MiR-146b-5p suppresses EGFR expression and reduces in vitro migration and invasion of glioma. Cancer Invest. 2010;28:1024–1030. doi: 10.3109/07357907.2010.512596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin F, Wang X, Jie Z, Hong X, Li X, Wang M, Yu Y. Inhibitory effects of miR-146b-5p on cell migration and invasion of pancreatic cancer by targeting MMP16. J Huazhong Univ Sci Technolog Med Sci. 2011;31:509–514. doi: 10.1007/s11596-011-0481-5. [DOI] [PubMed] [Google Scholar]
- 31.Kanaan Z, Rai SN, Eichenberger MR, Barnes C, Dworkin AM, Weller C, Cohen E, Roberts H, Keskey B, Petras RE, Crawford NP, Galandiuk S. Differential microRNA expression tracks neoplastic progression in inflammatory bowel disease-associated colorectal cancer. Hum Mutat. 2012;33:551–560. doi: 10.1002/humu.22021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 33.Fei X, Qi M, Wu B, Song Y, Wang Y, Li T. MicroRNA-195-5p suppresses glucose uptake and proliferation of human bladder cancer T24 cells by regulating GLUT3 expression. FEBS Lett. 2012;586:392–397. doi: 10.1016/j.febslet.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 34.Peschiaroli A, Giacobbe A, Formosa A, Markert EK, Bongiorno-Borbone L, Levine AJ, Candi E, D’Alessandro A, Zolla L, Finazzi Agro A, Melino G. miR-143 regulates hexokinase 2 expression in cancer cells. Oncogene. 2013;32:797–802. doi: 10.1038/onc.2012.100. [DOI] [PubMed] [Google Scholar]
- 35.Xiaohong Z, Lichun F, Na X, Kejian Z, Xiaolan X, Shaosheng W. MiR-203 promotes the growth and migration of ovarian cancer cells by enhancing glycolytic pathway. Tumour Biol. 2016;37:14989–14997. doi: 10.1007/s13277-016-5415-1. [DOI] [PubMed] [Google Scholar]
- 36.Chen B, Liu Y, Jin X, Lu W, Liu J, Xia Z, Yuan Q, Zhao X, Xu N, Liang S. MicroRNA-26a regulates glucose metabolism by direct targeting PDHX in colorectal cancer cells. BMC Cancer. 2014;14:443. doi: 10.1186/1471-2407-14-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pereira DM, Rodrigues PM, Borralho PM, Rodrigues CM. Delivering the promise of miRNA cancer therapeutics. Drug Discov Today. 2013;18:282–289. doi: 10.1016/j.drudis.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 38.Goh WQ, Ow GS, Kuznetsov VA, Chong S, Lim YP. DLAT subunit of the pyruvate dehydrogenase complex is upregulated in gastric cancer-implications in cancer therapy. Am J Transl Res. 2015;7:1140–1151. [PMC free article] [PubMed] [Google Scholar]
- 39.Sun XR, Sun Z, Zhu Z, Guan HX, Li CY, Zhang JY, Zhang YN, Zhou H, Zhang HJ, Xu HM, Sun MJ. Expression of pyruvate dehydrogenase is an independent prognostic marker in gastric cancer. World J Gastroenterol. 2015;21:5336–5344. doi: 10.3748/wjg.v21.i17.5336. [DOI] [PMC free article] [PubMed] [Google Scholar]