Abstract
Studies have shown that the abnormal expression of Fms related tyrosine kinase 1 (Flt1) is associated with multiple malignancies, yet its role in glioblastoma pathology remains to be elucidated. In this study, we investigated the role of Flt1 in regulating proliferation, migration and invasion of glioblastoma cells by establishing glioblastoma cell strains with constitutively silenced or elevated Flt1 expression. We demonstrate that ectopic expression of Flt1 promotes glioblastoma cells migration, invasion through cell scratching and Transwell assays. Further study has indicated that Flt1 knockdown prevents the spread of glioblastoma cells in vivo. Conversely, we also show that suppression of Flt1 expression inhibits migration and invasion of glioblastoma cells. Finally, our findings demonstrate that Flt1 promotes invasion and migration of glioblastoma cells through sonic hedgehog (SHH) signaling pathway. Our study suggests that galectin-1 represents a crucial regulator of glioblastoma cells metastasis. Thus, the detection and targeted treatment of Flt1-expressing cancer serves as a new therapeutic target for glioblastoma.
Keywords: Flt1, glioblastoma, metastasis, SHH
Introduction
Glioblastoma is one of the most malignant and aggressive tumors and has a very poor prognosis with a mean survival time of less than 2 years even with the recent development of surgery, radiation and temozolomide based intensive treatment [1]. One changeling in treating glioblastoma is substantial tumor cell and genetic heterogeneity leading to aberrant activation of multiple signaling pathways [2]. Several linked to survival; proliferation and metastasis of glioblastoma, such as the mTOR signaling, EGFR expression and STAT3 signaling have been reported. Meanwhile, glioblastoma is an aggressive cancer, which is often diagnosed at a late stage and glioblastoma cells are highly motile and invade the healthy brain tissue surrounding the primary tumor site [3]. Furthermore, invading glioblastoma cells are less sensitive to the current standard therapies including radiation and chemotherapy with DNA alkylating agents. Metastasis is a crucial problem in the clinical field, which must be overcome to extent the overall survival of glioblastoma patients. The mechanisms underlying migration and invasion of glioblastoma cells within the brain are complex and involve a sequence of events which includes: glioblastoma cells adhesion o the extracellular matrix (ECM) and remodeling and degradation of ECM, which is stimulated by multiple factors that are either secreted by tumor cells themselves or by the surrounding stroma, in order to create a “permissive” extracellular space [4]. Then, glioblastoma cells invasive into the modified tissue and form new metastases.
As with other malignancies, the growth and metastasis of glioblastoma is also reliant on the tumor angiogenic process. VEGF-A is one of the most important factors promoting tumor angiogenesis, and the activation of VEGF-A pathway requires the binding of the ligand (VEGF-A), to its receptor Flt1, thus generating downstream signals to stimulate the proliferation and progression of tumor cells [5]. Flt1 is a tyrosine kinase receptor, that binds VEGF-A with about 10-fold higher affinity than other kinase insert domain receptors (KDR), and it has been shown to promote tumor growth and metastasis [6]. VEGF-A and Flt1 are shown to be up-regulated in brain tumor vasculature, signifying the importance of angiogenesis in glioblastoma [7]. Suppressing expression of VEGF-A in tumoral cells by anti-sense RNA technology was shown to inhibit the growth of glioblastoma cells in nude mice. Together, these results suggest that the VEGF-A-Flt1 signaling pathway plays a central role in progression of glioblastoma cells [8]. Bevacizumab is a monoclonal antibody that binds to VEGF-A and neutralizes VEGF-A from binding to Flt1, thereby inhibiting the downstream growth signal that could induce angiogenesis [3]. Indeed, bevacizumab has been shown to be effective in combination with irinotecan for recurrent high grade glioma with a radiographic response noted in around 60% of patients, and a 6-month overall survival of 77% in patients treated with this regimen [9]. Other phase II trials have shown and confirmed that bevacizumab as a single agent or in combination with irinotecan are active and tolerable in patients with recurrent glioblastoma [10].
Despite these observations, virtually nothing is known about whether Flt1 and its associated factors may play a key role during human glioblastoma cells metastasis. In this work, we characterized the role of Flt1 in the proliferation of glioblastoma cells and found that Flt1 is widely expression in glioblastoma cell lines at both mRNA and protein levels. Interfering with Flt1 by shRNA knock-down impaired glioblastoma cells proliferation in vitro and tumor growth in vivo. In contrast, enforced Flt1 expression stimulated tumor growth. Furthermore, ectopic expression of Flt1 results in increased cellular migration and invasion, as well as metastasis in vivo. Conversely, Flt1 inhibition via shRNA knock-down results in significant reversion of these phenotypes. Finally, efficient Flt1 driven tumorigensis require the sonic hedgehog (SHH) signaling pathway. Combined, our results highlight a functional role for Flt1 in glioblastoma biology and uncover a potential a molecular target for glioblastoma metastasis.
Materials and methods
Cell lines and cell culture
Human glioblastoma cell lines (T98G, U373, U87 and LNT-229) were obtained from the Chinese Academy of Sciences Cell Bank of Type Culture Collection (CBTCCCAS, Shanghai, China). Cells were cultured in RPMI 1640 or DMEM medium supplemented with 10% fetal bovine serum (Wisent, Quebec, Canada), 100 μg/mL penicillin, and 100 μg/mL streptomycin in humidified 5% CO2 atmosphere at 37°C.
One solution cell proliferation assay
The cell viability was determined by CellTiter 96® AQueous One Solution cell proliferation assay (Promega, Madison, WI, USA). Briefly, cells were seeded in 96-well cell culture plates and treated with indicated agents. After incubation for indicated time period, 20 μL of One Solution reagent were added to each well and incubation was continued for additional 4 h. The absorbance was measured at 490 nm using Synergy™ HT Multi-Mode Microplate Reader (Bio-Tek, Winooski, VT, USA). The effect of Flt1 shRNA or overexpression on cell viability was assessed as the percent of cell viability compared with control cells (transfected with Scramble or vector), which were arbitrarily assigned 100% viability.
RNA interference and plasmid constructs
Human shRNA constructs against Flt1 and human SHH siRNA kit was purchased from Santa Cruz Biotech. The kit contains three predesigned duplexes targeting a specific gene of interest, and we used a pool of two target siRNAs to ensure work efficiency. Cells were transfected with plasmids and shRNAs or siRNAs by Lipofectamine 2000 reagent (Thermo Fischer Scientific). Human Flt1 complementary DNA (cDNA) or human SHH complementary DNA (cDNA) was amplified with PCR and cloned into the pcDNA3.1 (+) expression vector (Invitrogen) before being transfected into T98G or LNT-229 cells using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer’s instructions. Cells transfected with empty vector was used as control [11].
Cell migration assay
Briefly, cells were seeded in six-well plates and cultured at 37°C overnight to culture to a confluent monolayer. After starvation in serum-free medium for 24 h, a wound across the cell monolayer was created by scratching the monolayer with a 200 µl plastic tip. The six-well plates was then washed twice with phosphate-buffered saline (PBS) to remove cell debris and incubated with fresh complete medium. Photographic imaging was performed using the Olympus inverted microscopes at 0 h and 24 h after scratching respectively. The wound healing effect was determined by measuring the percentage of the remaining cell-free area compared with the area of the initial wound [12].
Invasion assay
The human glioblastoma cells invasion assay was carried out as previously described. Briefly, 2 × 106 cells suspended in serum-free medium (0.4 ml) were seeded onto the upper chamber with filters (pore size of 8-um), which coated with Matrigel. 0.6 ml medium with 20% FBS was applied to the lower chamber as a chemoattractant. After 18 h, the invaded cells on the bottom surface of the membrane were fixed with 4% formaldehyde, stained with 1% crystal violet, and counted in five randomly selected microscopic fields for each replicat were counted [13].
Flow cytometry analysis
Cells were detached by trypsin enzyme and washed with cold PBS. Precipitated cells were fixed by 500 μL cold 70% ethanol 10 min on ice. Cells was washed twice with PBS and re-suspended for anti-Flt1-APC (Biolegend) staining at 1 × 106 cells/100 uL in 1X PBS/1% BSA. Data were acquired using a FACS CALIBUR instrument (BD Biosciences) and analysis was performed by using Summit 5.0.0 (Dako, Agilent Technologies, CA, USA).
Immunofluorescence staining and cells immunocytochemistry
Human glioblastoma cells were fixed in 4% paraformaldehyde for 20 min and permeabilized in 0.1% Triton X-100 for 5 min. Flt1 was detected by staining with anti-Flt1 antibody (1 μg/ml) overnight at 4°C followed by incubating with Alexa 488-conjugated anti-mouse antibody (Molecular Probes) for 1 hr at room temperature. Cell nuclei were stained with DAPI for 3 min. T98G or LNT-229 cells were seeded on glass coverslips. After 24 h, they were fixed by pre-cold acetone, then rinsed three times with PBS. The cells were permeabilized in 0.1% Triton X-100 and incubated with 1% BSA/PBS to block nonspecific binding. Subsequently, the cells were immunostained by incubating with rabbit monoclonal antibody against Flt1 (diluted 1:1000, Epitomics) overnight at 4°C. After being washed with PBS, cells nuclei were counterstained with Mayer’s hematoxylin solution (Amber Scientific, Midvale, WA). Images were taken and analyzed using the ZEN 2011 imaging software on a Zeiss invert microscope (CarlZeiss, Hallbergnoos, Germany) under 200-fold magnification.
Colony formation assay
0.5 ml complete culture medium containing 1% agarose gel was added to a 25 cm2 cell culture plate in triplicates and allowed to harden. 2 × 103 T98G or LNT-229 cells were suspended in 1 ml culture medium containing 0.1% agarose gel and overlaid on the hardened bottom layer. 1 ml fresh medium was added to each well once a week for 4 weeks. The colonies were visualized by staining with 0.1% crystal violet for 4 hours at 37°C [14].
Microarray analysis
In order to screen for Flt1 target genes in human glioblastoma cells, microarray experimental procedures were carried out following the manufacturer’s protocols. Total RNA (1 μg) was extracted from parental LNT-229 and LNT-229 Flt1 knock-down cells and RNA was labeled by either Cy5 or Cy3 in transcription process. About 0.8 μg of Cy-labeled cRNA was cleavaged to an average size of about 50-100 nucleotides by incubation with fragmentation buffer (Agilent Technologies, CA) at 60°C for 30 min. Equal Cy-labeled cRNA was pooled and hybridized to SurePrint G3 Human Gene Expression 8 × 60 K v2 (Agilent Array®) at 65°C for 17 h. After washing and drying by nitrogen gun blowing, microarrays were scanned by an Agilent microarray scanner (Agilent Technologies, CA) at 535 nm for Cy3 and 625 nm for Cy5. Microarrays were scanned using an Agilent Scanner controlled by Agilent Scan Control 7.0 software. Data were extracted with Agilent Feature Extraction 9.1 software. Differentially expressed targets were identified by using processed signal intensities and p value log ratios as described elsewhere.
Western blot analysis
Whole-cell lysates were prepared with RIPA buffer containing protease and phosphatase inhibitors. Equal amounts of cell lysates (30 μg) were loaded on 8% SDS-PAGE and transferred onto PVDF membranes. After membranes were blocked, they were incubated with monoclonal antibody against Flt1 (1:1000, Signalway Antibody), SHH (1:1000, Cell Signaling Technology) and β-actin (1:1000, Bioworld Technology) followed by incubation with horseradish peroxidase-conjugated IgGs (1:10000, Bioworld Biotechnology). Target proteins were detected by the ECL system (Millipore, Braunschweig, Germany) and visualized with the ChemiDoc XRS system (Bio-Rad, Hercules, CA, USA) [15].
Quantitative real-time PCR (qPCR) analysis
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). First-strand cDNA was synthesized with 1 μg total RNA using a PrimeScript RT reagent kit (TakaraBio, Tokyo, Japan). qPCR was performed using IQTM SYBR Green supermix and the iQ5 real-time detection system (Bio-Rad Laboratories, Hercules, CA). The comparative cycle threshold (Ct) method was applied to quantify the expression levels through calculating the 2(-∆∆Ct) method. The primers used for PCR were as follows: β-actin: Forward Primer, 5’-AAGGAGCCCCACGAGAAAAAT-3’ and Reverse Primer, 5’-ACCGAACTTGCATTGATTCCAG-3’; Flt1: Forward Primer, 5’-TTTGCCTGAAATGGTGAGTAAGG-3’ and Reverse Primer, 5’-TGGTTTGCTTGAGCTGTGTTC-3’. cDNAs amplification and relative expression values were obtained from three independent experiments.
Subcutaneous xenograft models
All animal experiments were approved and conducted by the Institutional Animal Care and Treatment Committee of The First People’s Hospital of Huaian. SW1353 tumors were established by injecting T98G cells transduced with control vector or Flt1 plasmid and LNT-229 cells transfected with control scramble or shFlt1 into the dorsal area of 7-8 week old female athymic BALB/c nude mice. Tumor growth and body weight was measured every three days during the treatment. Tumor volumes were calculated using the formula as follow: volume (mm3) = 0.5 × length (mm) × width (mm)2.
In vivo tumor metastasis
BALB/c nude mice were purchased from Shanghai Slac Laboratory Animal Co. Ltd and maintained in SPF conditions. All animals were used in accordance with institutional guidelines and the current experiments were approved by the Use Committee for Animal Care of the First People s Hospital of Huaian. For glioblastoma cells metastasis assays, 1 × 107 T98G transduced with control vector or Flt1 plasmid and LNT-229 cells transfected with control scramble or shFlt1 were re-suspended in PBS and were injected into the tail vein of BALB/c nude mice. All the mice were killed by CO2 25 days after injection. The metastasis nodules in the lung tissues were stained with hematoxylin and eosin [16].
Statistical analysis
The data were presented as mean ± SD. Differences in the results of two groups were evaluated using either two-tailed Student’s t test or one-way ANOVA followed by post hoc Dunnett’s test. The differences with P < 0.05 were considered statistically significant.
Results
High level of tumor Flt1 expression was correlated with poor survival in glioblastoma patient
To investigate whether Flt1 and its associated factors are involved in human glioblastoma progression, we first examined their expression patterns in the publicly accessible Oncomine microarray database [17,18]. In two independent clinical data sets containing Flt1 information, Flt1 expression was markedly reduced in breast cancer tissues, especially in the invasive carcinoma, when compared with the matched normal tissues (Figure 1A). The prognostic value of the Flt1 genes in glioblastoma was analyzed using SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis (TCGA-Glioblastoma June 2016). Kaplan-Meier plotter analysis [19] in overall lung cancer showed correlation between overexpression of Flt1 and overall lower survival rates (Figure 1B). As up-regulation of Flt1 in human glioblastoma has been reported previously, we focused on Flt1 in this study. We examined Flt1 expression by qPCR and immunoblotting in four glioblastoma cell lines such as T98G, LNT-229, U373, and U87, which range from low- to high-level Flt1 expression. We found that Flt1 expression was relatively high in LNT-229 cell and low in T98G cell line (Figure 1C, 1D). FACS analysis after staining with anti-Flt1 antibody revealed the existence of LNT-229 and T98G cells expressing the receptor (Figure 1E). To future determine the expression of Flt1 in glioblastoma, immunohistochemistry (IHC) analysis was applied. The results showed that the expression of Flt1 appeared to be much higher in the LNT-229 cells and lower expression in T98G cells (Figure 1F).
Figure 1.
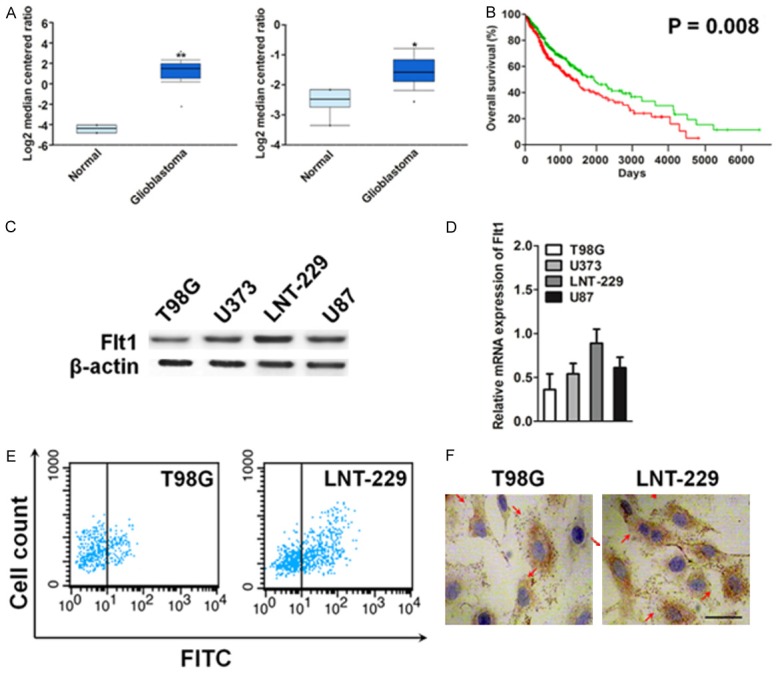
Flt1 is over-expressed in human glioblastoma. A. Box plots show increased levels of Flt1 in glioblastoma (right) compared with normal brain tissues in two microarray data sets. *P < 0.05, **P < 0.01, compared with normal brain tissues was determined by the Student’s t test. B. Kaplan-Meier survival curves for the glioblastoma patients. The overall survival times in the low (green, n = 330) and high Flt1 (red, n = 329) groups, with a hazard ratio of 1.41 (95% confidence interval (CI) 1.09-1.82) and P = 0.008. C. Western blotting analysis of the levels of Flt1 in various glioblastoma cell lines. β-actin was used as loading controls. D. qPCR analysis of Flt1 mRNA levels in various glioblastoma cell lines. PCR values were normalized to the levels of β-actin. Data were presented as the mean ± SD from three independent measurements. E. Representative FACS dot plots of LNT-229 and T98G cells stained with the anti-Flt1 antibody or with an isotype matched antibody (as a background control). Histograms reported the percentage of Flt1 positive cells as assessed by FACS. Mean ± SD of three independent experiments. F. Representative images of Flt1 expression in LNT-229 and T98G cells. Flt1 expression is notably higher in LNT-229 than T98G cells. Scale bars represent 50 μm.
Flt1 promoted the proliferation and migration of glioblastoma cells
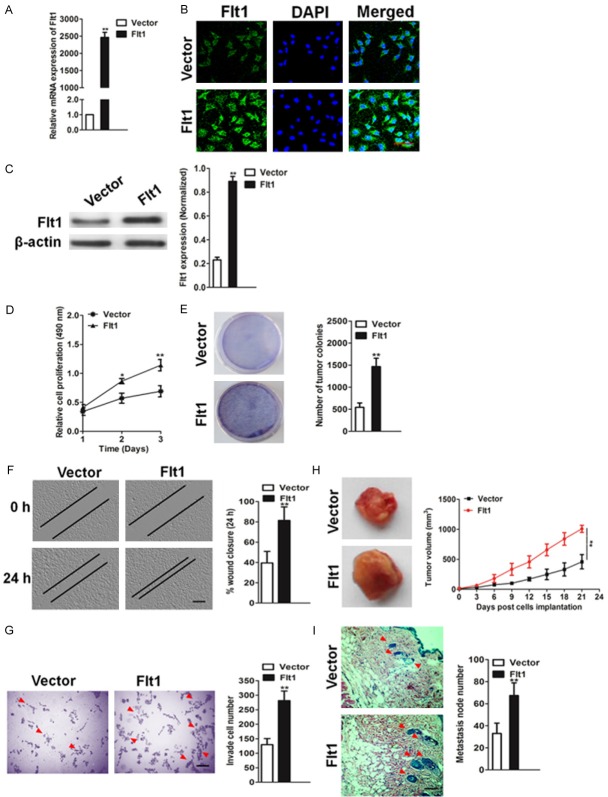
Given the clinical evidence of its association with poor over survival in glioblastoma patients, we assessed the precise role of Flt1 in glioblastoma cells biology. We stably expressed an empty vector and a vector expressing Flt1 plasmid in the T98G cell line, which expresses the lowest relative levels of Flt1 compared with a panels of glioblastoma cell lines. Functional assessment of the exogenous Flt1 was achieved using qPCR (Figure 2A) and immunofluorescence assay (Figure 2B). T98G cells exhibited strong up-regulation of Flt1 expression when infected with the Flt1 plasmid, which also confirmed by immunoblotting analysis (Figure 2C). Ectopic expression of Flt1 increased cellular proliferation (Figure 2D) and colony formation assay revealed that enhanced Flt1 expression increase tumor colonies (Figure 2E). In addition, wound healing and Transwell invasion assays showed that Flt1 over-expression significantly increased cellular mobility (Figure 2F) and T98G cells invasion (Figure 2G). Lastly, we examined the effects of Flt1 on T98G glioblastoma cells growth and dissemination in vivo, using preclinical nude mice models of glioblastoma. First, to assess the ability of Flt1 to promote survival and implantation of glioblastoma tumors within nude mice, Flt1 over-expressing or control cells were implanted subcutaneously into the posterior flank of nude mice (n = 6). Remarkably, Flt1 over-expressing cells formed solid tumor within the inoculation site (Figure 2H). Next, to assess the effect of Flt1 on metastasis, we utilized the xenograft model wherein the control or the Flt1 over-expressing cells were directly injected via tail-vein. Similar to the systems, mice injected with Flt1 expressing cells had a greater degree of metastasis foics compared with the vector control (Figure 2I). Taken together, our findings indicate that Flt1-overexpression contributes to glioblastoma cells progression in vivo.
Figure 2.
Enhanced expression of Flt1 increases glioblastoma cells migration and invasion. A. The expression of Flt1 in T98G glioblastoma cells transfected with the vector expressing Flt1 plasmid was evaluated by qPCR. B. T98G cells were immunostained with rabbit anti-Flt1 antibody (green) and DAPI (blue) for observation by laser confocal microscopy. Scale bars: 50 μm. C. Increased protein level of Flt1 was revealed by western blotting after transfection with the vector expressing Flt1 plasmid. Band intensities were measured by densitometry and normalized to β-actin expression. D. Up-regulation of Flt1 caused a significant growth promotion of T98G glioblastoma cells as revealed by proliferation assay. Values shown were the Mean absorbance ± SD for five wells from one experiment, and were representations of three independent experiments. E. Colonies are shown in purple post staining with crystal violet. F. Wound-healing assay to assess the effects of Flt1 on cellular motility by calculating per cent wound closure over time points shown. Scale bar, 100 μm. **P < 0.01 compared with vector. G. Transwell invasion assay of Flt1-expressing cells compared with control cell lines. Scale bar, 100 μm. **P < 0.01 compared with vector. H. Tumor growth kinetics (mean ± SD) of vector control T98G or Flt1 over-expressing cells in nude mice (n = 6 each) I. Representative images of the H&E-stained lung sections from mice injected with control or Flt1 over-expressing cells. Scale bar, 100 μm. Quantification of the number of metastatic nodules was shown in the bar graph below. n = 6 in each group. Data in this figure were presented as the mean ± SD, and **P < 0.01 was determined by the Student’s t test.
Down-regulation of Flt1 inhibits the aggressiveness of glioblastoma cells
shRNA specific to Flt1 was utilized to elucidate the effect of Flt1 inhibition on glioblastoma cells phenotypes. The cancer cell line LNT-229, which has relatively high expression levels of Flt1, was stably transduced with control shRNA (Scramble) or two independent Flt1-targeting sequences (shFlt1 #1 and shFlt1 #2). Cells transduced with shRNA Flt1 exhibited decreased the protein and mRNA of Flt1 when compared with cells transduced with scramble, which was confirmed by qPCR and western blot (Figure 3A and 3B). In addition, staining for corroborated that targeted inhibition of Flt1 resulted cells losing Flt1 expression (Figure 3C). We next examined whether specific silencing affect glioblastoma cells growth in vitro. One solution cell proliferation assay to analyze cells proliferation displayed that Flt1 depletion strongly inhibited the LNT-229 cells proliferation (Figure 3D). Similarly, the down-regulation of Flt1 in LNT-229 cells significantly decreased the colony formation ability both in numbers and mean size of soft agar colonies (Figure 3E). Transduction of LNT-229 cells with two distinct shRNAs targeting Flt1 inhibited LNT-229 glioblastoma cells growth in nude mice compared to that of vector controls (Figure 3F). To determine whether Flt1 knock-down also results in an decrease in LNT-229 cells metastasis in vivo, the LNT-229 control or shFlt1 cells were injected intravenously into the nude mice, and lung metastasis was examined 12 weeks later. Histological analyses revealed a significant decrease in the number of metastatic lesions produced by shFlt1 cells when compared with that produced by the control cells (Figure 3G). Finally, to determine the precise role of Flt1 in glioblastoma cancer development, the Transwell and wound healing assay were used to examine invasion and migration of LNT-229 cells. Functionally, Flt1 inhibition significantly reduced the number of cells invaded through the membrane compared with the control (Figure 3H). Likewise, wound closure ratio was markedly reduced (P < 0.01) in Flt1-shRNA transfected LNT-229 cells (Figure 3I).
Figure 3.
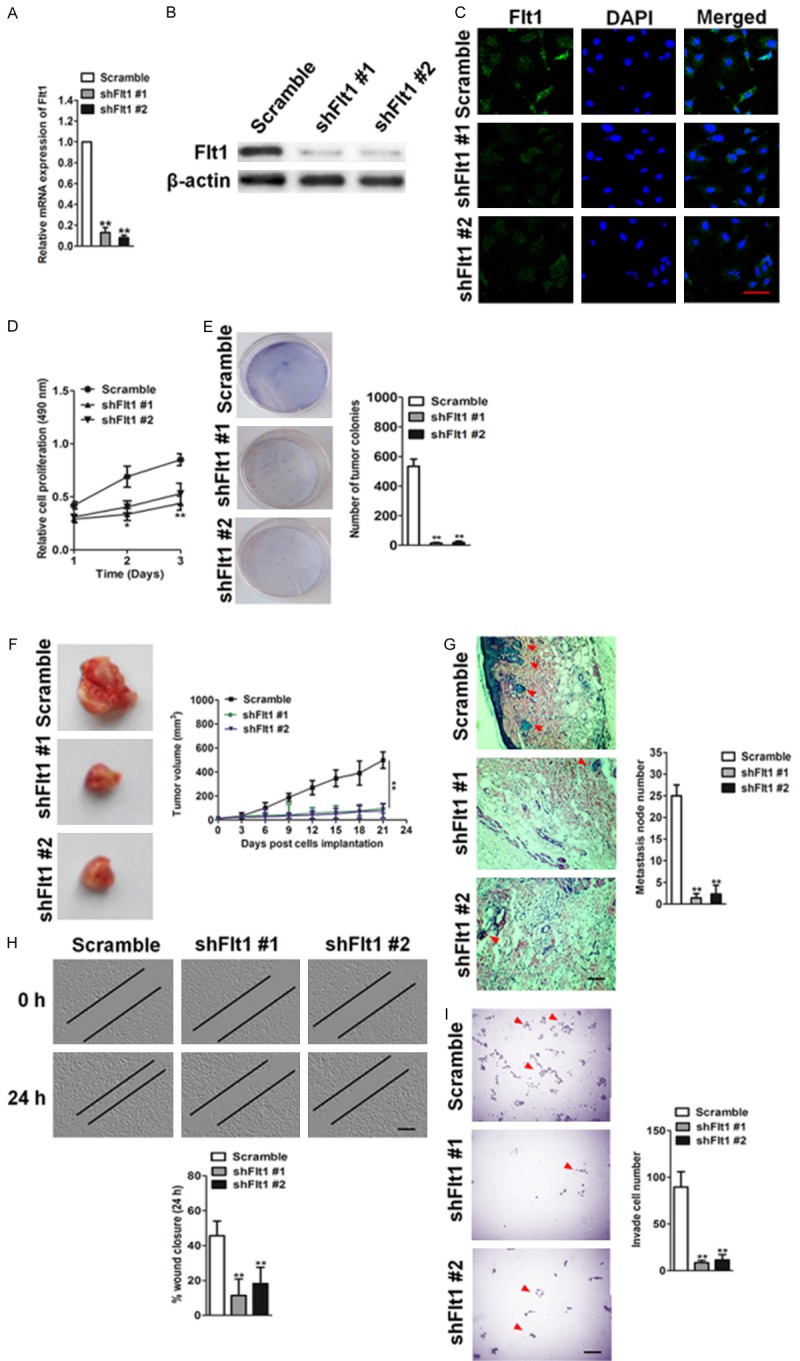
Silencing Flt1 inhibits malignant progression of glioblastoma cells LNT-229. A. Western blotting shows effective knock-down of Flt1 in LNT-229 cells stably expressing shFlt1 #1 or sFlt1 #2. B. qPCR analysis of Flt1 RNA level in LNT-229 cells stably expressing scramble control or shFlt1s. C. Effects of shRNA Flt1 on Flt1 expression as evaluated by immunofluorescence. Scale bars: 50 μM. D. Silencing Flt1 in LNT-229 cells results in decreased cell proliferation. Proliferation assay was performed to analyze LNT-229 cells growth after transfected Scramble or shRNA Flt1. **P < 0.01 compared with Scramble. E. Clonogenic assay to assess cellular survival in LNT-229 cells shown as fold change in number of colonies in control versus shFlt1 cells. Colonies are shown in purple post staining with crystal violet. F. Tumor growth kinetics (mean ± SD) of Flt1-shRNA versus control Scramble in nude mice (n = 6 each). G. Representative images of the H&E-stained lung sections from mice injected with control or shFlt1 cells. Scale bar, 100 μm. Quantification of the number of metastatic nodules is shown in the bar graph below. n = 6 in each group. Data in this figure are presented as the mean ± SD. H. Scratch-wound assays were performed to analyze the migration of LNT-229 cells. Data were representative of three independent experiments. Scale bars: 100 μM. I. Transwell invasion assay of silencing Flt1 LNT-229 cells compared with control cells. Scale bars: 100 μM.
The regulation of Flt1 on the sonic hedgehog (SHH) signaling pathway
To identify the cellular pathways regulated by Flt1 and defined specific targets driving the observed metastasis phenotype in glioblastoma cells, we performed microarray on cell lines with a combination of computational target prediction analysis using a threshold of minimum fold change > 5 or fold change < 5. Sonic hedgehog (SHH) was the most significantly ds-regulation among those genes altered by > 5 fold change or 5 < fold changes (Figure 4A). Immunoblot analysis was utilized to future define the expression of SHH protein level were reduced in knockdown LNT-229 cells (Figure 4B), whereas overexpression of Flt1 increased the amount of SHH in T98G cells (Figure 4C). To determine whether SHH inhibition can phenocopy the effects of Flt1 down-regulation on glioblastoma cells, we generated two independent cell clones expressing siRNA targeting SHH. Our data revealed that targeted knockdown of SHH results in a decreasing of migration (Figure 4D) and invasion (Figure 4E) phenotype.
Figure 4.
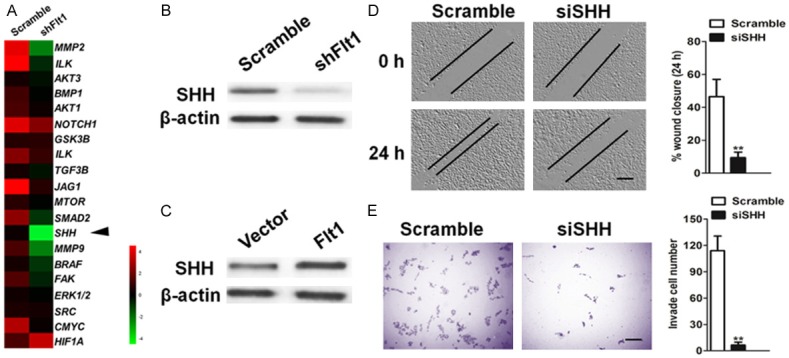
Flt1 regulates the sonic hedgehog (SHH) signaling pathway. A. Microarray-based gene expression and Venn diagram. The levels of SHH mRNA in Flt1 knockdown and Flt1 overexpressing glioblastoma cells. B. The protein level of SHH in Flt1-knockdown glioblastoma LNT-229 cells after transfection with the indicated shRNAs as determined by immunoblotting analysis. C. SHH protein expression level was measured by immunoblotting assay in T98G cells after transfection with the vector expressing Flt1 plasmid. D. Analysis of migration potential of the Scramble, siSHH cell lines by a wound healing assay (left) and the quantification of wound closure (right). Scale bars, 100 μM. E. Analysis of invasion potential of the Scramble, siSHH cell lines by a Transwell invasion assay (left) and the quantification of cell invasion (right). Scale bars, 100 μM.
SHH knockdown reverses Flt1-mediated cell migration and invasion
We, therefore, examined whether the oncogenic effects of Flt1 could be reversed by SHH silencing. We transfected Flt1 overexpressing T98G cells with SHH siRNA plasmid, and the efficacy of siRNA was validated by immunoblot analysis (Figure 5A). Knockdown of SHH decreases Flt1 overexpression-mediated cell migration and invasion (Figure 5B and 5C). To further investigate the involvement of SHH on Flt1-mediated cancer progression, we infected Flt1-knockdown LNT-229 cells with SHH cDNA (Figure 5C). Over-expression of SHH restores the migration and invasion abilities of Flt1-knockdown LNT-229 cells (Figure 5D). To investigate whether SHH and its associated factors are involved in human glioblastoma progression, we examined its expression patterns in the publicly accessible Oncomine microarray database. In two independent clinical data sets containing SHH information [20,21], SHH expression was markedly increased in primary glioblastoma tissues, when compared with the matched normal tissues (Figure 5E). We further analyzed the glioblastoma TCGA data set (TCGA-Glioblastoma June 2016) that contains information on the clinical outcomes of patients to investigate the correlation between SHH level and clinical characteristics. Statistical analysis revealed that up-regulation of SHH correlated with decreased overall survival (Figure 5F).
Figure 5.
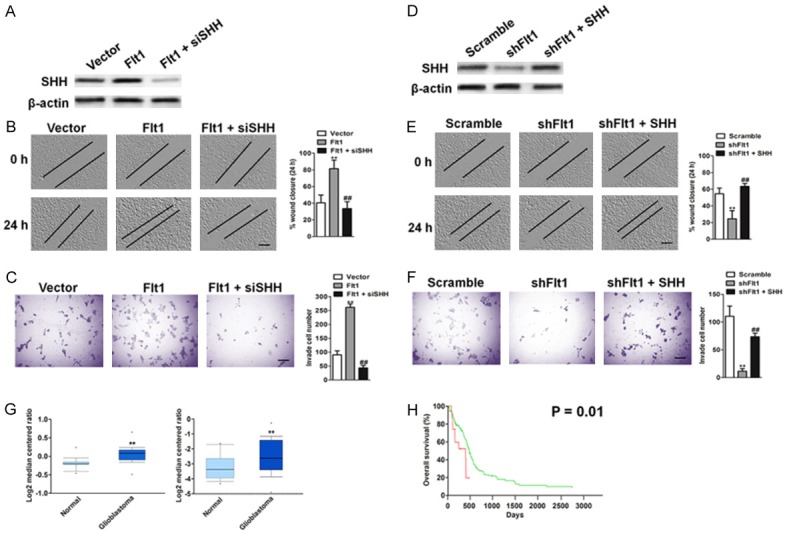
SHH knockdown decreases Flt1-mediated cell migration and invasion. (A) Flt1 overexpressing T98G cells were transfected with control shRNA or SHH siRNA. The protein level of SHH in indicated T98G cells were determined by immunoblotting analysis. Knockdown of SHH decreases cell migration (B) and invasion (C) in Flt1 overexpressing T98G cells. Scale bars, 100 μM. (D) Flt1 shRNA LNT-229 cells were transfected with control vector or SHH overexpressing plasmid. The protein level of SHH in indicated LNT-229 cells was determined by western blotting assay. (E) Ectopic expression of SHH reverses the phosphorylation of cell migration in Flt1-knockdown LNT-229 cells. Flt1-knockdown LNT-229 cells were transfected with SHH overexpressing plasmid. The migration ability of cells was quantified as described above. Scale bars, 100 μM. (F) Flt1-knockdown LNT-229 cells were transfected with SHH overexpressing plasmid. Ectopic expression of SHH reverses the phosphorylation of cell invasion in Flt1-knockdown LNT-229 cells. Scale bars, 100 μM. The invasiveness ability of cells was quantified as described above. Each value was the mean ± SD of three determinations. The asterisk indicated a significant difference among the indicated comparison groups, as analyzed by Student’s t-test (**P < 0.01, ##P < 0.01). (G) Box plots show increased levels of SHH in glioblastoma (right) compared with normal brain tissues in two microarray data sets. **P < 0.01, compared with normal brain tissues was determined by the Student’s t test. (H) Kaplan-Meier analysis of overall survival of glioblastoma patients stratified by the expression of SHH. The overall survival times in the low (green, n = 134) and high Flt1 (red, n = 14) groups, with a hazard ratio of 2.33 (95% confidence interval (CI) 1.19-4.59) and P = 0.01.
Discussion
The issues of tumor recurrence, drug resistance and enhanced cancer cells metastasis remain a challenge in the treatment and the clinical management of glioblastoma [22]. Despite a series of investigation were performed to better understanding of glioblastoma genetic alterations, the ability to translate these findings into effective treatments have been limited by the difficulty in identifying functionally relevant drivers of the disease. The processes of cancer metastasis are complex. In the first step of metastasis, cancer cells dissociate from a primary tumor by losing cell-cell contact and transforming into a mesenchymal phenotype, resulting in increased motility and invasive abilities. Hence, identifying molecular drivers of glioblastoma will be crucial to a better understanding of its biology and, ultimately, for the development of a therapeutic target in metastasis. Here, we identified that Flt1 can modulate sonic hedgehog (SHH) signaling affect further downstream events of tumor cell migration, invasion and dissemination in vivo. Ectopic expression of Flt1 in glioblastoma cells increases the capacity for migration and invasion. In contrast, in metastatic glioblastoma cells that express high levels of endogenous Flt1, transfection of specific shRNA against Flt1 decreases migration and invasion abilities. We also used glioblastoma cell lines to determine that Flt1 supports cell proliferation and tumor growth in vivo. Upon depletion of Flt1 using multiple shRNAs, glioblastoma cells showed impaired primary tumor growth in vivo, diminished tumor colony size and reduced cell proliferation in vitro. Conversely, cells with Flt1 over-expression accelerated the growth of cancer cells both in vitro and in vivo.
Flt1, as a kinase receptor in endothelial cells is also expressed by human epithelial cancer cells, such as those derived from human pancreatic, colorectal, and breast tumors, and melanomas [23]. Activation of Flt1 promotes epithelial-mesenchymal transition and an aggressive phenotype in specific cancer cells. Other recent studies support the crucial roles that Flt1 play in mediating epithelial-mesenchymal transition and consequent aggressive disease traits. In addition to its role in angiogenesis, Flt-1 might mediate a variety of hitherto unappreciated biological functions, such as liver regeneration, inflammatory process and cancer metastasis [24]. Recent studies have also reported that increased expression of Flt1 was observed in grade 4 diffusely infiltrating astrocytomas [25]. However, whether Flt1 is involved in glioblastoma metastatic potential has not yet been determined. Consequently, the role of Flt1 in mediating lung cancer tumor development has been investigated in the current study. Consequently, we show that the expressions of Flt1 significantly correlate with overall survival in glioblastoma patient.
Microrray analysis identified an unbiased list of downstream regulatory genes potentially responsible for glioblastoma cells phenotypes changes caused by Flt1 depletion or over-expression. Near the top of this list was SHH, a signaling molecule that promotes primary cell growth, and stimulates invasion and metastasis. Consistent with this, SHH level is significantly elevated in glioblastoma cells compared with normal brain tissue and its high expression associates poor survival of glioblastoma patients [26]. Furthermore, inhibition of SHH signaling by shRNA knock-down essentially eliminated the effect of SHH on cells migration and invasion. Several studies have demonstrated that sonic hedgehog (SHH) signaling pathway contribute to cancer cell plasticity through the regulation of cellular adhesion to the extracellular matrix, thereby increasing cell motility and invasion, and contribute to poor patient outcome. In this study, we have shown that ectopic expression of Flt1 increases the expression of SHH, whereas elimination of Flt1 markedly decreases SHH mRNA and protein level in glioblastoma cells. The functional role of SHH on Flt1 mediated cancer cells metastasis is also revealed by the inhibition of SHH decreasing the oncogenic potential of Flt1 mediated cells migration and invasion. Moreover, in Flt1 depleted cells, re-expression of SHH were able to rescue Flt1-mediated glioblastoma cells phenotypes.
Glioblastoma is the most aggressive form of brain cancer due to its highly invasive nature that impedes the surgical removal of all tumor cells, making relapse inevitable [27]. However, the mechanisms used by glioblastoma cells to metastasis into the surrounding tissue are still unclear. Our findings provide strong evidence that Flt1 promotes glioblastoma metastasis by using a loss-of-function model. Our present study provides an understanding of how enhanced expression of Flt1 can confer metastasis traits of glioblastoma cells through the modulation of SHH signaling pathway (Figure 6), and therefore, inhibition of Flt1 may serve as a potential target for the development of therapies against metastatic events.
Figure 6.
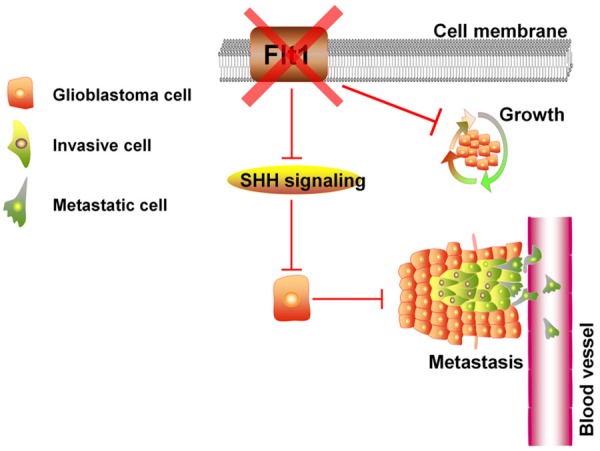
Proposed model by Flt1/SHH signaling promotes glioblastoma cells metastasis.
Acknowledgements
The study was supported by the Science and Technology Development Foundation of Huai’an to Dr. L.S. Ding (China, No. HAS2015018).
Disclosure of conflict of interest
None.
References
- 1.Cihoric N, Tsikkinis A, Minniti G, Lagerwaard FJ, Herrlinger U, Mathier E, Soldatovic I, Jeremic B, Ghadjar P, Elicin O, Lossl K, Aebersold DM, Belka C, Herrmann E, Niyazi M. Current status and perspectives of interventional clinical trials for glioblastoma-analysis of ClinicalTrials. gov. Radiat Oncol. 2017;12:1. doi: 10.1186/s13014-016-0740-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yi Y, Hsieh IY, Huang X, Li J, Zhao W. Glioblastoma stem-like cells: characteristics, microenvironment, and therapy. Front Pharmacol. 2016;7:477. doi: 10.3389/fphar.2016.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang SD, Leung KL, McCrudden CM, Kwok HF. The prognostic significance of combining VEGFA, FLT1 and KDR mRNA expressions in brain tumors. J Cancer. 2015;6:812–818. doi: 10.7150/jca.11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong R, Turlova E, Feng ZP, Rutka JT, Sun HS. Activation of TRPM7 by naltriben enhances migration and invasion of glioblastoma cells. Oncotarget. 2017 doi: 10.18632/oncotarget.14496. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kloepper J, Riedemann L, Amoozgar Z, Seano G, Susek K, Yu V, Dalvie N, Amelung RL, Datta M, Song JW, Askoxylakis V, Taylor JW, Lu-Emerson C, Batista A, Kirkpatrick ND, Jung K, Snuderl M, Muzikansky A, Stubenrauch KG, Krieter O, Wakimoto H, Xu L, Munn LL, Duda DG, Fukumura D, Batchelor TT, Jain RK. Ang-2/VEGF bispecific antibody reprograms macrophages and resident microglia to anti-tumor phenotype and prolongs glioblastoma survival. Proc Natl Acad Sci U S A. 2016;113:4476–4481. doi: 10.1073/pnas.1525360113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamerlik P, Lathia JD, Rasmussen R, Wu Q, Bartkova J, Lee M, Moudry P, Bartek J Jr, Fischer W, Lukas J, Rich JN, Bartek J. Autocrine VEGF-VEGFR2-Neuropilin-1 signaling promotes glioma stem-like cell viability and tumor growth. J Exp Med. 2012;209:507–520. doi: 10.1084/jem.20111424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu AR, Cai W, Veeravagu A, Mohamedali KA, Chen K, Kim S, Vogel H, Hou LC, Tse V, Rosenblum MG, Chen X. Multimodality molecular imaging of glioblastoma growth inhibition with vasculature-targeting fusion toxin VEGF121/rGel. J Nucl Med. 2007;48:445–454. [PubMed] [Google Scholar]
- 8.Cantarella G, Risuglia N, Dell’eva R, Lempereur L, Albini A, Pennisi G, Scoto GM, Noonan DN, Bernardini R. TRAIL inhibits angiogenesis stimulated by VEGF expression in human glioblastoma cells. Br J Cancer. 2006;94:1428–1435. doi: 10.1038/sj.bjc.6603092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gravina GL, Mancini A, Marampon F, Colapietro A, Delle Monache S, Sferra R, Vitale F, Richardson PJ, Patient L, Burbidge S, Festuccia C. The brain-penetrating CXCR4 antagonist, PRX177561, increases the antitumor effects of bevacizumab and sunitinib in preclinical models of human glioblastoma. J Hematol Oncol. 2017;10:5. doi: 10.1186/s13045-016-0377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du C, Ren J, Zhang R, Xin T, Li Z, Zhang Z, Xu X, Pang Q. Effect of bevacizumab plus temozolomide-radiotherapy for newly diagnosed glioblastoma with different MGMT methylation status: a meta-analysis of clinical trials. Med Sci Monit. 2016;22:3486–3492. doi: 10.12659/MSM.899224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Zhu L, Fang J, Ge Z, Li X. LRG1 modulates epithelial-mesenchymal transition and angiogenesis in colorectal cancer via HIF-1alpha activation. J Exp Clin Cancer Res. 2016;35:29. doi: 10.1186/s13046-016-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su X, Wang J, Chen W, Li Z, Fu X, Yang A. Overexpression of TRIM14 promotes tongue squamous cell carcinoma aggressiveness by activating the NF-kappaB signaling pathway. Oncotarget. 2016;7:9939–9950. doi: 10.18632/oncotarget.6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang HL, Jiang Y, Wang YH, Chen T, He HJ, Liu T, Yang T, Yang LW, Chen J, Song ZQ, Yao W, Wu B, Liu G. FBXO31 promotes cell proliferation, metastasis and invasion in lung cancer. Am J Cancer Res. 2015;5:1814–1822. [PMC free article] [PubMed] [Google Scholar]
- 14.Khotskaya YB, Goverdhan A, Shen J, Ponz-Sarvise M, Chang SS, Hsu MC, Wei Y, Xia W, Yu D, Hung MC. S6K1 promotes invasiveness of breast cancer cells in a model of metastasis of triple-negative breast cancer. Am J Transl Res. 2014;6:361–376. [PMC free article] [PubMed] [Google Scholar]
- 15.Feldmann G, Habbe N, Dhara S, Bisht S, Alvarez H, Fendrich V, Beaty R, Mullendore M, Karikari C, Bardeesy N, Ouellette MM, Yu W, Maitra A. Hedgehog inhibition prolongs survival in a genetically engineered mouse model of pancreatic cancer. Gut. 2008;57:1420–1430. doi: 10.1136/gut.2007.148189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao T, Jiang W, Wang X, Wang H, Zheng C, Li Y, Sun Y, Huang C, Han ZB, Yang S, Jia Z, Xie K, Ren H, Hao J. ESE3 inhibits pancreatic cancer metastasis by upregulating E-cadherin. Cancer Res. 2017;77:874–885. doi: 10.1158/0008-5472.CAN-16-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bredel M, Bredel C, Juric D, Harsh GR, Vogel H, Recht LD, Sikic BI. Functional network analysis reveals extended gliomagenesis pathway maps and three novel MYC-interacting genes in human gliomas. Cancer Res. 2005;65:8679–8689. doi: 10.1158/0008-5472.CAN-05-1204. [DOI] [PubMed] [Google Scholar]
- 18.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, Park JK, Fine HA. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 19.Tian J, Hachim MY, Hachim IY, Dai M, Lo C, Raffa FA, Ali S, Lebrun JJ. Cyclooxygenase-2 regulates TGFbeta-induced cancer stemness in triple-negative breast cancer. Sci Rep. 2017;7:40258. doi: 10.1038/srep40258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, Du J, Kau T, Thomas RK, Shah K, Soto H, Perner S, Prensner J, Debiasi RM, Demichelis F, Hatton C, Rubin MA, Garraway LA, Nelson SF, Liau L, Mischel PS, Cloughesy TF, Meyerson M, Golub TA, Lander ES, Mellinghoff IK, Sellers WR. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey R, Rosenblum M, Mikkelsen T, Fine HA. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Xu H, Hu Y, Qiu W. Potential mechanisms of microRNA-129-5p in inhibiting cell processes including viability, proliferation, migration and invasiveness of glioblastoma cells U87 through targeting FNDC3B. Biomed Pharmacother. 2017;87:405–411. doi: 10.1016/j.biopha.2016.12.100. [DOI] [PubMed] [Google Scholar]
- 23.Takano S, Ishikawa E, Matsuda M, Sakamoto N, Akutsu H, Yamamoto T, Matsumura A. The anti-angiogenic role of soluble-form VEGF receptor in malignant gliomas. Int J Oncol. 2017;50:515–524. doi: 10.3892/ijo.2016.3810. [DOI] [PubMed] [Google Scholar]
- 24.Bhattacharya R, Ye XC, Wang R, Ling X, McManus M, Fan F, Boulbes D, Ellis LM. Intracrine VEGF signaling mediates the activity of prosurvival pathways in human colorectal cancer cells. Cancer Res. 2016;76:3014–3024. doi: 10.1158/0008-5472.CAN-15-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dimitropoulos K, Giannopoulou E, Argyriou AA, Zolota V, Petsas T, Tsiata E, Kalofonos HP. The effects of anti-VEGFR and anti-EGFR agents on glioma cell migration through implication of growth factors with integrins. Anticancer Res. 2010;30:4987–4992. [PubMed] [Google Scholar]
- 26.Cherepanov SA, Cherepanova KI, Grinenko NF, Antonova OM, Chekhonin VP. Effect of hedgehog signaling pathway activation on proliferation of high-grade gliomas. Bull Exp Biol Med. 2016;161:674–678. doi: 10.1007/s10517-016-3483-2. [DOI] [PubMed] [Google Scholar]
- 27.Feng X, Yao J, Gao X, Jing Y, Kang T, Jiang D, Jiang T, Feng J, Zhu Q, Jiang X, Chen J. Multi-targeting peptide-functionalized nanoparticles recognized vasculogenic mimicry, tumor neovasculature, and glioma cells for enhanced anti-glioma therapy. ACS Appl Mater Interfaces. 2015;7:27885–27899. doi: 10.1021/acsami.5b09934. [DOI] [PubMed] [Google Scholar]