Abstract
Colon cancer is one of the most prevalent cancer types in developed countries. Metastasis and drug resistance are two contributing factors to the high mortality rate. Accumulating evidence suggest that cancer stem-like cells (CSCs) represents as a major contributor to these malignant features. Here, we identified and isolated colon cancer stem-like cells using side-population (SP) method from human colon cancer cell lines. SP colon cells demonstrate cancer stem-like cell properties including enhanced sphere-forming ability and resistance towards fluorouracil (5-FU). The CSC properties were associated with the increased expression level of major oncogenic and stem cell markers including β-catenin, NF-kB, Akt/mTOR, KRAS and c-Myc. Trichostatin A (TSA), an antifungal antibiotic also a HDAC inhibitor, was found to function not only to decrease the expression of oncogenic markers but also the colon CSC properties. Importantly, TSA and 5-FU combined treatment synergistically suppressed colon cancer viability. Finally, in vivo results demonstrated that TSA alone and in combination with 5-FU effectively suppressed colon tumorigenesis. Collectively, this study provides preclinical evidence that TSA may function as a potential colon cancer therapeutic agent by targeting CSC and overcoming 5-FU resistance.
Keywords: Drug resistance, colon cancer, trichostatin A (TSA), cancer stem-like cells, side-population cells, 5-FU resistance
Introduction
Colon cancer is one of the most diagnosed malignancies in the developed countries with an estimated 9% of all cancer incidence [1]. The underlying causes for the development of colon cancer are multifactorial and complex. The contributing factors may include environmental risk factors, genetic risks and dietary habits [2]. Several frequently detected oncogenes such as KRAS, c-myc, β-catenin and other less frequent but powerful tumorigenic pathways have been shown to promote colon tumorigenesis [3,4].
Accumulating evidence suggest that the existence of a small subset of dormant cells termed cancer stem-like cells (CSCs) are involved in tumorigenic processes including initiation, progression, metastasis and drug resistance [5]. In both experimental and clinical settings, CSCs have been identified in a spectrum of cancer types, including colon cancer [6]. It is generally considered that resistance against current therapies observed clinically are, in part, due to the presence of CSCs and their enrichment post therapy. The CSCs are characterized to have an enhanced self-renewal potential to regenerate into different lineages of cancer cells within the tumor mass post cancer treatment. This key feature of CSCs drive continued tumorigenesis and contribute to the disease recurrence. Thus, the presence and the generation of CSCs is thought to be one of the major factors of treatment failure [7]. The discovery and development of an agent which can prevent the generation and/or eliminate CSCs could in theory prevent the aforementioned clinical challenges faced and significantly improve the prognosis of patients with colon cancer.
Tumorigenesis may be induced by different mechanisms, one of which is via epigenetic control. Accumulating evidence suggests that many tumor suppressors are silenced and oncogenes activated epigenetically in virtually all tumor types [8,9]; these epigenetically modifications in the cancer genome are directly linked to the overexpressed histone deacetylase (HDAC). Thus HDAC inhibitors (HDACi) may represent a promising class of compounds for the cancer therapy [10]. Trichostatin A (TSA) is a pan-HDACi and has been predominantly used in preclinical studies. The antitumor effect of TSA may be of multiple targets. Through its interactions with histone and nonhistone proteins, TSA has been shown to reactivate once-silenced tumor suppressors, inhibit cell cycle progression and promote apoptosis [11].
In this study, using side-population method we isolated cancer stem-like colon cancer cells with enhanced tumor sphere forming ability and drug resistance. In the search of anti-CSC agents, we initially identified TSA as a potential candidate via a bioinformatics tool. Subsequently, we demonstrated that TSA treatment not only suppressed the cell viability of colon cancer cells but also that of tumor spheres. In addition, the combination of TSA and 5-FU synergistically suppressed the generation of colon tumor spheres. The CSC-inhibitory effects mediated by TSA was associated with the downregulation of stemness and oncogenic markers such as Nanog, β-catenin, Kras and c-Myc. Our findings provide preclinical evidence and rationale for the potential usage of TSA in conjunction with 5-FU in drug-resistant colon cancer patients.
Materials and methods
Human colon cancer cell lines in vitro culture
The human colon cancer cell lines SW480, SW620, HCT116 and DLD-1 were purchased from the American Type Culture Collection (ATCC) (Manassas, VA, USA) and maintained and passaged as recommended by the ATCC. The culture media were supplemented with 1% penicillin/streptomycin and 10% fetal bovine serum (FBS) (Gibco). Cells were grown in a humidified incubator at 37°C containing 5% CO2.
Isolation of side-population (SP) cancer cells using Fluorescence-activated cell sorting (FACS)
The side-population colon cancer cells were determined and isolated using FACSAria™ III sorter (BD Biosciences, Taiwan). Verapamil (100 µM final concentration) was used as a negative control. Verapamil was added 15 min before Hoechst incubation. SP cells enriched with ABCG2 and with the ability to efflux Hoechst 33342 were identified and sorted for further analyses.
In vitro cell viability determination
Sulforhodamine B (SRB) dye (Sigma-Aldrich Chemie GmbH, Munich, Germany) was used to test cell viability. Cells were harvested and diluted with culture medium to a final DMSO concentration of <0.05%. Cells (SP, non-SP and parental) cells were seeded into 96 well plates at 3000 cells/well. The cells were incubated in the presence of 5-FU (ranging from 0 to 400 µM) or TSA (ranging from 0 to 2 µM) for 48 h. Subsequently, drug-treated cells were collected and fixed with 10% TCA. The plates were washed, dried and followed by staining with 0.4% (w/v) SRB dissolved in 1% (v/v) acetic acid. The bound SRB was solubilized in 20 mmol/L Tris base (100 μl/well) briefly. Finally, the optical density (OD) of the samples was measured using a microplate reader (Molecular Devices, Sunnyvale, CA) at 562 nm.
Western blot analysis
SW480 and SW620 human colon cancer cells (SP, non-SP, and drug-treated samples) after different treatments were subjected to SDS-PAGE and western blot analyses. In short, cells were lysed in cold lysis buffer (50 mM Tris-HCl pH 7.5, 2 mM EDTA, 2 mM EGTA, 10 mM b-glycerophosphate, 150 mM NaCl, 0.5% NP-40, 1 mM phenyl-methyl sulfonyl fluoride, 1 mM NaF, 1 mM DTT, 1% β-mercaptoethanol and protease inhibitor cocktail (EMD Biosciences). Cell lysates were centrifuged at 15,000 g and protein concentration was determined using commercial kit (Pierce, Rockford, IL) with bovine serum albumin served as standards. Protein samples (30 µg each) were dissolved in sample buffer, denatured and separated using 10% SDS-PAGE gels. The proteins were transferred onto nitrocellulose membranes and blocked in TBST (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.2% Tween-20 and skim milk) for 1 h. Membranes were sequentially treated with the primary and secondary antibodies, washed in TBST. The signals were detected using enhanced chemiluminescence (ECL-Plus, Amersham Pharmacia Biotech, Piscataway, NJ) and measured using BioSpectrum® Imaging System (Upland, CA).
Sphere forming assay
For the analysis of sphere forming ability, colon cancer cells were cultured under serum-deprived conditions and in Ultra-low Attachment Plates (Corning Incorporated). The culture conditions are modified from the original protocol described by Lo et al [12]. Colon cancer cells (103 cells/ml) were suspended and seeded in the tumorsphere medium containing: 20 ng/ml epidermal growth factor, 10 ng/ml basic fibroblast growth factor, 5 μg/ml insulin, 0.4% Bovine Serum Albumin. Approximately 3-5 days of incubation (depending on the cell type), tumor sphere numbers were counted under a phase-contrast microscope using the 40 × magnification lens. The ability of tumor formation was represented by the average number of spheres obtaining from counts from different views (at least 3 random fields).
Animal study
The animal study protocol was approved by the Animal Care and User Committee at Taipei Medical University (Affidavit of Approval of Animal Use Protocol # LAC-101-0026). Female NOD/SCID mice aged 4 to 6 weeks were purchased from the BioLASCO (Taipei, Taiwan). Mice were housed in specific pathogen-free conditions with an alternating 12-hour light: dark cycle. One week acclimation was allowed prior to experiments. Mice were subcutaneously inoculated with SW480 colon cancer cells (1 × 106 cells in 100 μL of PBS) subcutaneously. When tumors reached a palpable size, the tumor volumes were measured and recorded once a week with the following established formula: Tumor size = (L1 × L22)/2, where L1 is the long axis and L2 is the short axis of the tumor. The animals were randomly divided into 4 groups (N = 3 per group): vehicle control, TSA (0.5 mg/kg, i.p injection, twice a week); 5-FU (25 mg/kg, i.p injection, twice a week), and TSA + 5-FU (twice a week). All mice were allowed free access to a standard rodent LabDiet (Lab 5001, Purina Mills, St. Louis, MO) and water over the 8-week experimental period. The body weight of the mice was recorded weekly. After the experiment, the animals were humanely sacrificed by cervical dislocation and tumor samples harvested for further analysis.
Statistical analysis
All experiments were carried out at least three times. Two-tailed t tests were used to analyze the in vitro and in vivo data. The statistical analysis was performed using GraphPad Prism software where a p value <0.05 was considered as statistically significant and is indicated with an asterisk.
Results
Colon cancer side-population (SP) cells exhibited increased sphere-forming ability and expressed higher stemness markers
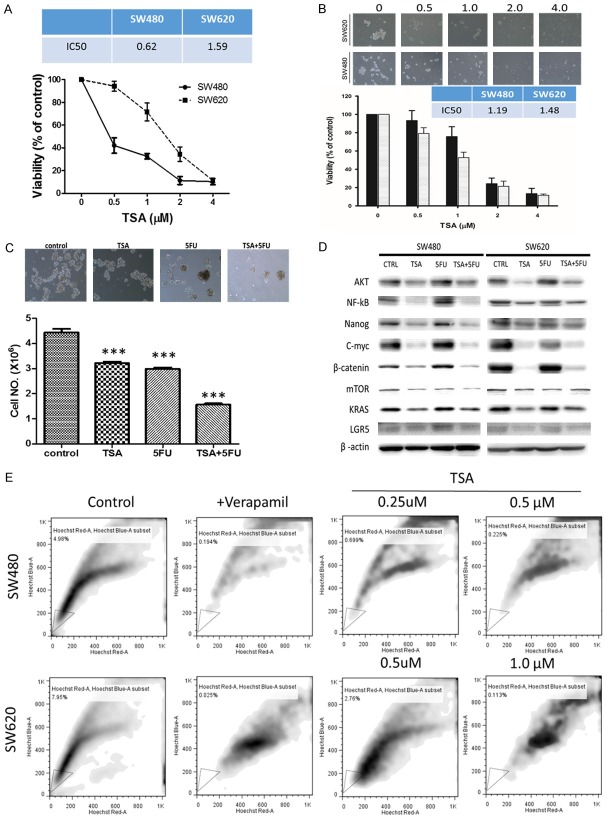
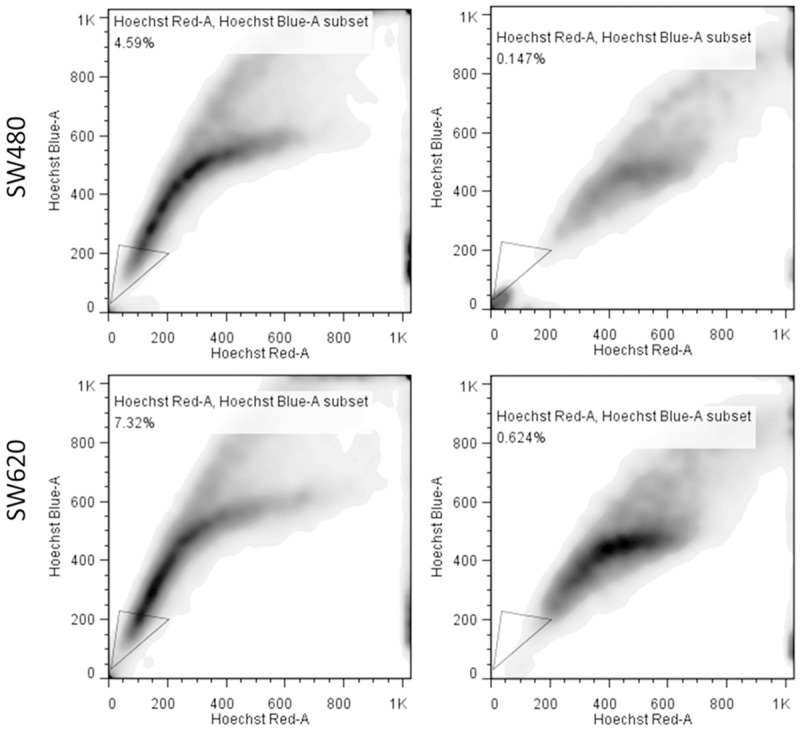
Using flow cytometry, we identified a small percentage of cancer cells exhibiting high ABCG activity, namely the side-population (SP) cells. In both SW480 and SW620 cell lines, using verapamil as the negative control, approximately 4.4% and 6.7% of SP cells were identified in SW480 and SW620 cells respectively under our experimental conditions (Figure 1). Subsequently, we isolated the SP cells and cultured them under serum-deprived culture conditions. We found that SP colon cancer cells generated a significantly higher number of colon spheres as compared to their non-SP counterparts (Figure 2A) and SW620 SP cells significantly formed more colon spheres than SW480 SP cells. We then tested these colon spheres against chemotherapeutic agent 5-FU and found that the spheres generated from SP cells showed a significantly increased resistance against 5-FU (Figure 2B). For instance, the IC50 values of the spheres generated from non-SP versus SP SW480 cells were approximately 10.52 and 42.80 µM respectively (a 4-fold increase) while 19.30 and 97.48 µM for SW620 non-SP and SP spheres respectively (Figure 2B). Western blots analysis showed that in the SP cells, the expression of stemness markers (Nanog, β-catenin), oncogenic molecules (KRAS, c-Myc, mTOR and NF-kB) was significantly higher comparing to their non-SP counterparts (Figure 2C).
Figure 1.
Identification of colon cancer stem-like cells. A representative flow cytometric analysis demonstrates the usage of side-population assay to identify and isolate cancer stem-like cell population in human colon cancer cell lines, SW480 and SW620. Experiments were performed in triplicates.
Figure 2.
Side-population (SP) colon cancer cells shows increased CSC properties. A. Representative micrographs of colon spheres of both SW480 and SW620 cells generated under serum free medium. SP cells clearly formed significantly more spheres as compared to non-SP counterparts. B. Colon spheres generated from SP cells were significantly more resistant against 5-FU treatment. The representative IC50 values of 5-FU in non-SP versus SP colon cancer cells are listed. C. Representative western blots depicting major oncogenic and/or CSC markers were elevated in SP cells as compared to their non-SP counterparts. These markers include Nanog, β-catein, c-Myc, and mTOR.
Identification of trichostatin A (TSA) as a potential inhibitor of colon tumorigenesis and colon sphere formation
We utilized the public search tool Connection Digest (Lincscloud, Broad Institute [13]) and found that trichostatin A (cmap_id: CGS001-3845) had a high connectivity score with KRAS expression in 9 cell lines and 11 microarray experiments. Since KRAS is one of the most mutated oncogenes detected in colon cancer [14-16], this in-silico result prompted us to examine the potential role of TSA as an antagonist of colon tumorigenesis. In addition, we analyzed a GEO profile where hepatocellular carcinoma was treated with TSA and the resultant mRNA level of both KRAS (profile ID: GDS2213/1559204_x_at) and LGR5 (profile ID: GDS2213/210393_at) was significantly reduced (data not shown) [17]. In our SRB assay, TSA treatment effectively inhibited the cell viability in both SW480 and SW620 cells with respective IC50 values of 0.62 and 1.59 µM (Figure 3A). The ability of generating colon spheres in both SW480 and SW620 cell lines were also significantly suppressed by the treatment of TSA (IC50 values 1.19 and 1.48 µM respectively, Figure 3B). Next, we tested the TSA and 5-FU combination on SW480 colon spheres and found that the combination of TSA (0.5 µM) and 5-FU (20 µM) had the highest inhibitory effect on the sphere forming ability as comparing to TSA alone or 5-FU alone (Figure 3C). Using western blots, we demonstrated that the TSA was able to suppress major oncogenic signaling pathways including c-Myc, mTOR/Akt, NF-kB, and KRAS (as predicted by Lincscloud) while also effectively in suppressing stemness markers such as Nanog, LGR5 and β-catenin (Figure 3D); interestingly, 5-FU appeared to be less effective than TSA according to our experimental results. The combination of TSA and 5-FU also significantly suppressed the expression of the aforementioned markers (Figure 3D). Furthermore, TSA treatment reduced the percentage of sidepopulation cells in both SW480 and SW620. Consistently, SW620 cells contained a relatively higher percentage of SP cells (approximately 7.93%) as compared to 4.79% in SW480 (Figure 3E); at the same TSA concentration (0.5 µM).
Figure 3.
TSA treatment effectively inhibited colon tumor cells and spheres. A. SRB assay results demonstrated that TSA effectively inhibited the cell viability of both SW480 and SW620 cells with the IC50 values below 2 µM. B. Colon sphere viability assay results indicated that TSA eliminated colon spheres from both SW480 and SW620 effectively with the IC50 values below 2 µM. C. The combination of TSA and 5-FU significantly reduced the cell viability of the tumor spheres. ***P<0.001 as compared to the control). D. Western blot analysis showed that TSA treatment suppressed oncogenic pathways including mTOR/Akt, NF-kB and stemness markers such as Nanog, LGR5 and β-catenin. The combination of TSA and 5-FU further suppressed the expression. E. TSA effectively suppressed the percentage of side-population in both SW480 and SW620 colon cancer cells.
TSA and 5-FU synergistically suppresses viability of colon cancer cells
Next, we examined the possibility of combining TSA and 5-FU in treating colon cancer cells. Different combinations of TSA (ranging from 0-0.1 µM) and 5-FU (from 0-30 µM) and incubation times (24-48 h) were used to determine the combination index for drug effect interactions. First, SW480 cells were incubated with TSA for 5 h followed by the addition of 5-FU and cultured for 48 h. This treatment regimen as demonstrated by Fa-CI plots, worked synergistically to suppress SW480 viability (Figure 4A, numbers in bold font). While SW620 responded towards a different TSA and 5-FU combinations where SW620 cells were first treated with TSA for 24 h, followed by the addition of 5-FU for another 24 h (Figure 4B). Thus, we demonstrated TSA and 5-FU functioned synergistically to suppress colon cancer cell viability but with different treatment conditions.
Figure 4.
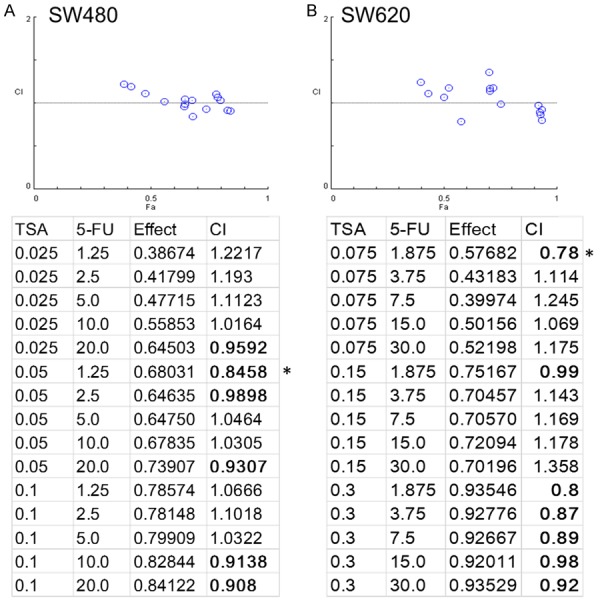
TSA and 5-FU combination analysis. SW480 and SW620 cells were subjected to different concentrations of TSA and 5-FU to test the effectiveness of the treatment. Definition: synergism (CI<1), additive effect (CI = 1), and antagonism (CI>1). The most potent combination was marked by an asterisk.
TSA inhibits colon tumorigenesis in vivo
After establishing that TSA treatment suppressed colon tumorigenesis in vitro, xenograft mouse model was used for in vivo validation. SW480 cells were inoculated subcutaneously into NOD/SCID mice to establish tumorigenesis. Tumor-bearing mice were then divided into four groups: vehicle control, TSA (0.5 mg/kg, 2 times/week), 5-FU (25 mg/kg, 2 times/week) and TSA plus 5-FU. Mice receiving TSA alone and 5-FU alone appeared to exhibit a similar tumor burden according to our regimen while the tumor burden appeared to be the lowest in the combination group (Figure 5A, 5B). The body weight in all groups did not seem to be significantly different as the body weight increased over time in all groups (Figure 5C), suggesting no apparent systematic toxicity in our treatment regimens.
Figure 5.
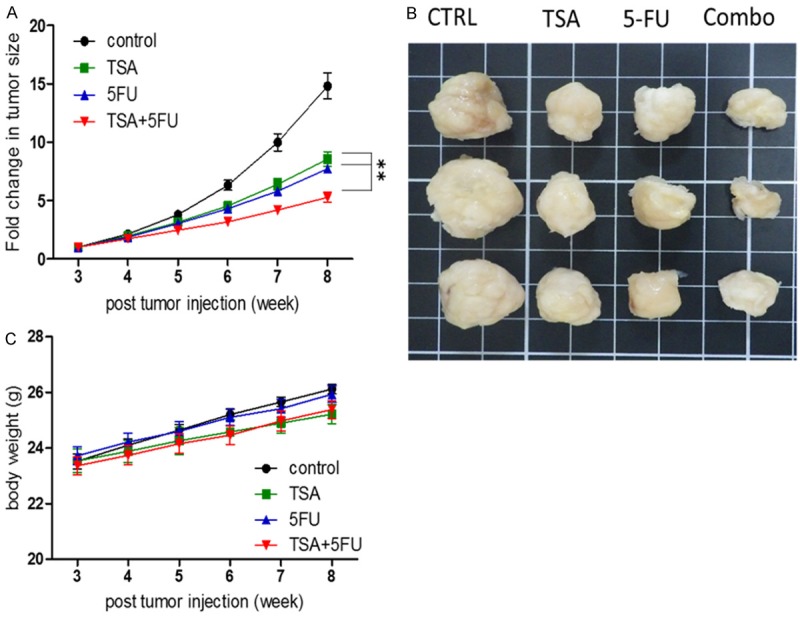
In vivo evaluation of TSA in suppressing colon tumorigenesis. A. SW480 subcutaneous xenograft mouse model was used to evaluate the tumor inhibitory effect of TSA and 5-FU. TSA alone significantly suppressed SW480 tumor growth (to a similar extent as 5-FU alone) as compared to those vehicle control mice. TSA and 5-FU combination showed the most significant inhibition to tumor growth. B. Photographs of tumor samples harvested from different treatment groups. The tumor size of the TSA + 5FU combination group was clearly the smallest followed by TSA and 5-FU alone groups. C. Body weight of the animals was monitored over the experiment and showed no significant difference among different treatment groups.
Discussion
One of the most clinically challenging task is to overcome drug resistance which is almost inevitable in the majority of patients with advanced colon cancer. For the past decade, both experimental and clinical evidence suggest the existence of cancer stem-like cells (CSCs) in virtually all cancer types and now considered as a major contributing factor to drug resistance, distance metastasis and disease recurrence [18]. Rationally, agents that can target and eliminate CSCs, may offer a more effective strategy for cancer treatment.
In this study, we used a generally accepted method termed side-population [19,20], to identify and isolate colon CSCs. Human colon cancer cell lines, SW480 and SW620 were found to contain approximately 4.4% and 6.7% SP cell population, respectively (Figure 1). Comparatively, SP colon cells showed an enhanced ability to form colon spheres than their non-SP counterparts, indicative of enhanced self-renewal ability (Figure 2A). More importantly, these colon spheres showed a significantly increased resistance towards 5-FU treatment. For instance, non-SP SW620 spheres had an IC50 value of 19.3 µM, but the value for SP SW620 spheres was approximately 5-fold higher (97.5 µM). In agreement, studies have indicated that tumor spheres derived from cell lines or patient samples serves a great model for exploring the molecular mechanisms associated with stem cell-like properties including drug resistance [5,21]. We showed that the expression of stemness genes, Nanog, β-catenin and LGR5 were elevated in colon spheres generated from the SP SW480 and SW620 cell lines. Both Nanog, β-catenin and LGR5 have all shown to participate in the maintenance of the undifferentiated state of colon stem cells and promote drug resistance in colon cancer [22-25]. Hence, we provide evidence for using tumor spheres generated from SP colon cancer cells as an in vitro tool for anti-CSC drug screening.
Clinically, 5-FU serves as one of the standard chemotherapeutic agents for treating colon cancer. However, tumor cells often develop resistance against it leading to a relatively low efficacy (approximately 40%). A recent study shows that 5-FU treatment may actually increase the CD133+ tumor population in human colon cancer patients with the increased resistance towards 5-FU [26]. Our observations are in agreement with the literature where the SP colon cancer cells showed an increased ability to form colon spheres and resist 5-FU treatment (Figure 2). These results supported our notion that SP-cancer cells serve an important in vitro tool for anti-CSC drug screening purpose.
Using a bioinformatics platform (Lincscloud App, Broad institute, USA), we identified that trichostatin A (TSA) as one of the top-ranking perturbagens negatively associated with the expression of KRAS across different cancer cell lines. Notably, KRAS is one of the most frequently observed mutated genes in colon cancer patients and play key roles in promoting colon tumorigenesis [16], generating colon cancer stem cells [27,28] and inducing drug resistance [29,30]. First, our in vitro test confirmed that TSA’s ability to suppress the viability of colon cancer cells with a significantly lower concentrations comparing to 5-FU (Figure 3). More importantly, KRAS expression was reduced by the treatment of TSA in concert of other key tumorigenic markers such as c-Myc, Akt/mTOR and stemness promoting markers, LGR5, Nango and β-catenin (Figure 3). This result validated our in silico analysis where TSA is predicted to suppress KRAS expression. Furthermore, colon cancer cells that are resistant to 5-FU damage have been shown to express Nanog and other stemness markers including LGR5 [31,32], suggesting their role in 5-FU resistance. More importantly, a recent report shows that the PI3K/Akt/mTOR pathway plays a key role in maintaining the stemness of colon cancer stem cells and inhibition of this pathway represents a potential venue to combat drug resistance and metastasis in colon cancer [33]. Similarly, suppressing PI3K/Akt/NF-κB signaling axis results in the significantly reduced the pluripotency in pancreatic stem cells [34]. Collectively, our data indicated that TSA exerts an overall inhibitory effect on signaling pathways associated with tumorigenesis and stemness-maintenance.
After establishing that TSA as a potential anti-CSC agent alone, we also evaluated the possibility of using TSA with 5-FU. Interestingly, we found that SW480 and SW620 responded differentially towards TSA and 5-FU combined therapy. For instance, SW480 cells were more sensitive to the regimen where cells were incubated with TSA for 5 hours first and followed by the 5-FU treatment for the total of 48 hours. The SW620 cells appeared to require a longer TSA incubation (24 hours) and less 5-FU (24 hours). This is interesting since SW480 and SW620 cells were derived from the same patient where SW480 was obtained from the primary tumor while SW620 from the lymph node metastasis. The observation where a longer TSA incubation was required for SW620 cells may be associated with the higher metastatic potential and gene-signature of SW620 over its non-metastatic counterpart SW480 cells [35]. More importantly, we found that when combining TSA and 5-FU, a significantly lower dosage was required to achieve synergy. This is important for future clinical consideration since 5-FU has been shown to pose serious adverse effects in a sub-population of patients [36,37]. By reducing the clinical dosage of 5-FU significantly may reduce and avoid 5-FU induced side effects. Together, our findings showed that TSA and 5-FU combined regimen synergistically suppressed colon cancer viability at significantly lower concentrations. Our findings supports the notion where precision medicine should be exercised to achieve the best possible efficacy.
In conclusion, we demonstrated that SP colon cancer cells as an important in vitro tool for anti-CSC drug screening where SP colon cancer cells demonstrate two key CSC-like properties namely increased colon sphere forming ability, and drug resistance. Equally important, we provided evidence that TSA identified in silico has the potential as an antagonist for colon cancer stem cells. TSA-treated SW480 and SW620 cells demonstrated a significantly decreased oncogenic markers (KRAS, NF-kB, Akt/mTOR) and CSC signature Nanog, LGR5 and β-catenin. Our in vivo experimental data showed that TSA alone suppressed colon tumorigenesis and to a more significant degree when it was combined with 5-FU. TSA may be repurposed for treating drug-resistant colon cancer with further clinical investigation in the future.
Acknowledgements
Mr. Oliver Yang and Mr. Ben Ho’s technical assistance is recognized and appreciated. This study is funded by Taipei Medical University and Taipei Medical University Hospital joint research fund (103TMU-SHH-09) to Tsu-Yi Chao and Alexander TH Wu; the Ministry of Science and Technology (MOST 105-2314-B-182A-133) and Chang-Gung Memorial Hospital Research Foundation (CMRPG2F0141~2 and CMRPG2D0091~3), Taiwan to Tse-Hung Huang.
Disclosure of conflict of interest
None.
References
- 1.Boyle P, Langman JS. ABC of colorectal cancer: epidemiology. BMJ. 2000;321:805–808. doi: 10.1136/bmj.321.7264.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. 2009;22:191–197. doi: 10.1055/s-0029-1242458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calvert PM, Frucht H. The genetics of colorectal cancer. Ann Intern Med. 2002;137:603–612. doi: 10.7326/0003-4819-137-7-200210010-00012. [DOI] [PubMed] [Google Scholar]
- 4.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 5.Dotse E, Bian Y. Isolation of colorectal cancer stem-like cells. Cytotechnology. 2016;68:609–19. doi: 10.1007/s10616-014-9806-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhan HX, Xu JW, Wu D, Zhang TP, Hu SY. Pancreatic cancer stem cells: new insight into a stubborn disease. Cancer Lett. 2015;357:429–437. doi: 10.1016/j.canlet.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Tanase CP, Neagu AI, Necula LG, Mambet C, Enciu AM, Calenic B, Cruceru ML, Albulescu R. Cancer stem cells: involvement in pancreatic cancer pathogenesis and perspectives on cancer therapeutics. World J Gastroenterol. 2014;20:10790–10801. doi: 10.3748/wjg.v20.i31.10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damaskos C, Valsami S, Kontos M, Spartalis E, Kalampokas T, Kalampokas E, Athanasiou A, Moris D, Daskalopoulou A, Davakis S, Tsourouflis G, Kontzoglou K, Perrea D, Nikiteas N, Dimitroulis D. Histone deacetylase inhibitors: An attractive therapeutic strategy against breast cancer. Anticancer Res. 2017;37:35–46. doi: 10.21873/anticanres.11286. [DOI] [PubMed] [Google Scholar]
- 9.Samanta S, Rajasingh S, Cao T, Dawn B, Rajasingh J. Epigenetic dysfunctional diseases and therapy for infection and inflammation. Biochim Biophys Acta. 2017;1863:518–528. doi: 10.1016/j.bbadis.2016.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walkinshaw DR, Yang XJ. Histone deacetylase inhibitors as novel anticancer therapeutics. Curr Oncol. 2008;15:237–243. doi: 10.3747/co.v15i5.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh BN, Zhou H, Li J, Tipton T, Wang B, Shao G, Gilbert EN, Li Q, Jiang SW. Preclinical studies on histone deacetylase inhibitors as therapeutic reagents for endometrial and ovarian cancers. Future Oncol. 2011;7:1415–1428. doi: 10.2217/fon.11.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lo PK, Kanojia D, Liu X, Singh UP, Berger FG, Wang Q, Chen H. CD49f and CD61 identify Her2/neu-induced mammary tumor-initiating cells that are potentially derived from luminal progenitors and maintained by the integrin-TGFbeta signaling. Oncogene. 2012;31:2614–2626. doi: 10.1038/onc.2011.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamb J. The connectivity map: a new tool for biomedical research. Nat Rev Cancer. 2007;7:54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- 14.Carethers JM, Jung BH. Genetics and genetic biomarkers in sporadic colorectal cancer. Gastroenterology. 2015;149:1177–1190. e1173. doi: 10.1053/j.gastro.2015.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Temraz S, Mukherji D, Shamseddine A. Dual inhibition of MEK and PI3K pathway in KRAS and BRAF mutated colorectal cancers. Int J Mol Sci. 2015;16:22976–22988. doi: 10.3390/ijms160922976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waring P, Tie J, Maru D, Karapetis CS. RAS mutations as predictive biomarkers in clinical management of metastatic colorectal cancer. Clin Colorectal Cancer. 2016;15:95–103. doi: 10.1016/j.clcc.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 17.Dannenberg LO, Edenberg HJ. Epigenetics of gene expression in human hepatoma cells: expression profiling the response to inhibition of DNA methylation and histone deacetylation. BMC Genomics. 2006;7:181–181. doi: 10.1186/1471-2164-7-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoian M, Stoica V, Radulian G. Stem cells and colorectal carcinogenesis. J Med Life. 2016;9:6–11. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Mathew G, Timm EA Jr, Sotomayor P, Godoy A, Montecinos VP, Smith GJ, Huss WJ. ABCG2-mediated DyeCycle Violet efflux defined side population in benign and malignant prostate. Cell Cycle. 2009;8:1053–1061. doi: 10.4161/cc.8.7.8043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petriz J. Flow cytometry of the side population (SP) Curr Protoc Cytom. 2013;9:Unit9.23. doi: 10.1002/0471142956.cy0923s39. [DOI] [PubMed] [Google Scholar]
- 21.Davies JM, Santaolalla R, Abreu MT. Use of cancer stem cells to investigate the pathogenesis of colitis-associated cancer. Inflamm Bowel Dis. 2016;22:976–983. doi: 10.1097/MIB.0000000000000756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han Y, Xue X, Jiang M, Guo X, Li P, Liu F, Yuan B, Shen Y, Guo X, Zhi Q, Zhao H. LGR5, a relevant marker of cancer stem cells, indicates a poor prognosis in colorectal cancer patients: a meta-analysis. Clin Res Hepatol Gastroenterol. 2015;39:267–273. doi: 10.1016/j.clinre.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 23.Lin YU, Wu T, Yao Q, Zi S, Cui L, Yang M, Li J. LGR5 promotes the proliferation of colorectal cancer cells via the Wnt/beta-catenin signaling pathway. Oncol Lett. 2015;9:2859–2863. doi: 10.3892/ol.2015.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osawa H, Takahashi H, Nishimura J, Ohta K, Haraguchi N, Hata T, Yamamoto H, Mizushima T, Takemasa I, Doki Y, Mori M. Full-length LGR5-positive cells have chemoresistant characteristics in colorectal cancer. Br J Cancer. 2016;114:1251–1260. doi: 10.1038/bjc.2016.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan Q, Meng L, Ye J, Wei X, Shang Y, Tian Y, He Y, Peng Z, Chen L, Chen W, Bian X, Wang R. Transcriptional repression of miR-200 family members by Nanog in colon cancer cells induces epithelial-mesenchymal transition (EMT) Cancer Lett. 2017;392:26–38. doi: 10.1016/j.canlet.2017.01.039. [DOI] [PubMed] [Google Scholar]
- 26.Paschall AV, Yang D, Lu C, Redd PS, Choi JH, Heaton CM, Lee JR, Nayak-Kapoor A, Liu K. CD133+CD24lo defines a 5-Fluorouracil-resistant colon cancer stem cell-like phenotype. Oncotarget. 2016;7:78698–78712. doi: 10.18632/oncotarget.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Rolle AF, Chiu TK, Zeng Z, Shia J, Weiser MR, Paty PB, Chiu VK. Oncogenic KRAS activates an embryonic stem cell-like program in human colon cancer initiation. Oncotarget. 2016;7:2159–2174. doi: 10.18632/oncotarget.6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong X, Zheng L, Shen J, Zhang D, Xiong M, Zhang Y, He X, Tanyi JL, Yang F, Montone KT, Chen X, Xu C, Xiang AP, Huang Q, Xu X, Zhang L. Suppression of MicroRNA 200 family expression by oncogenic KRAS activation promotes cell survival and epithelial-mesenchymal transition in KRAS-driven cancer. Mol Cell Biol. 2016;36:2742–2754. doi: 10.1128/MCB.00079-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong S, Kim S, Kim HY, Kang M, Jang HH, Lee WS. Targeting the PI3K signaling pathway in KRAS mutant colon cancer. Cancer Med. 2016;5:248–255. doi: 10.1002/cam4.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Tan R, Zhu X, Zhang Y, Tan Z, Su B, Li Y. Oncogenic K-ras confers SAHA resistance by up-regulating HDAC6 and c-myc expression. Oncotarget. 2016;7:10064–10072. doi: 10.18632/oncotarget.7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song N, Jia XS, Jia LL, Ma XB, Li F, Wang EH, Li X. Expression and role of Oct3/4, Nanog and Sox2 in regeneration of rat tracheal epithelium. Cell Prolif. 2010;43:49–55. doi: 10.1111/j.1365-2184.2009.00653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobayashi S, Yamada-Okabe H, Suzuki M, Natori O, Kato A, Matsubara K, Jau Chen Y, Yamazaki M, Funahashi S, Yoshida K, Hashimoto E, Watanabe Y, Mutoh H, Ashihara M, Kato C, Watanabe T, Yoshikubo T, Tamaoki N, Ochiya T, Kuroda M, Levine AJ, Yamazaki T. LGR5-positive colon cancer stem cells interconvert with drug-resistant LGR5-negative cells and are capable of tumor reconstitution. Stem Cells. 2012;30:2631–2644. doi: 10.1002/stem.1257. [DOI] [PubMed] [Google Scholar]
- 33.Chen J, Shao R, Li F, Monteiro M, Liu JP, Xu ZP, Gu W. PI3K/Akt/mTOR pathway dual inhibitor BEZ235 suppresses the stemness of colon cancer stem cells. Clin Exp Pharmacol Physiol. 2015;42:1317–1326. doi: 10.1111/1440-1681.12493. [DOI] [PubMed] [Google Scholar]
- 34.Erdogan S, Doganlar O, Doganlar ZB, Serttas R, Turkekul K, Dibirdik I, Bilir A. The flavonoid apigenin reduces prostate cancer CD44(+) stem cell survival and migration through PI3K/Akt/NF-kappaB signaling. Life Sci. 2016;162:77–86. doi: 10.1016/j.lfs.2016.08.019. [DOI] [PubMed] [Google Scholar]
- 35.Ayinde O, Wang Z, Griffin M. Tissue transglutaminase induces epithelial-mesenchymal-transition and the acquisition of stem cell like characteristics in colorectal cancer cells. Oncotarget. 2017;8:20025–20041. doi: 10.18632/oncotarget.15370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Offer SM, Diasio RB. Biomarkers of fluorouracil toxicity: insight from the PETACC-8 trial. JAMA Oncol. 2016 doi: 10.1001/jamaoncol.2015.5463. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 37.Saif MW, Diasio RB. Benefit of uridine triacetate (Vistogard) in rescuing severe 5-fluorouracil toxicity in patients with dihydropyrimidine dehydrogenase (DPYD) deficiency. Cancer Chemother Pharmacol. 2016;78:151–156. doi: 10.1007/s00280-016-3063-1. [DOI] [PubMed] [Google Scholar]