Abstract
Long non-coding RNAs (lncRNAs) have been recently reported to be dysregulated and play a critical role in the progression of thyroid cancer. Here, we found that the lncRNA n340790 was highly expressed in human thyroid cancer tissues and was strongly correlated with the clinical characteristics of patients. There was a good prognostic value of n340790 for thyroid cancer. In vitro overexpression of n340790 promoted the development of thyroid cancer, while silencing n340790 inhibited this process. Additionally, n340790 accelerated the growth of thyroid cancer tumor in vivo. Furthermore, we discovered that n340790 could act as an endogenous sponge by directly binding to miR-1254 and downregulating miR-1254 expression. In addition, miR-1254 could inhibit the stimulatory effect of n340790 on the growth and invasion of thyroid cancer cells. In conclusion, n340790 promoted the development process of malignant thyroid cancer by regulating miR-1254, and targeting n340790 may be a promising strategy as a thyroid cancer therapy.
Keywords: LncRNA-n340790, miR-1254, thyroid cancer, growth, invasion
Introduction
Thyroid cancer is the most common endocrine malignancy of the thyroid with a 1.7% incidence rate of total cancer diagnoses [1]. In addition, the number of cases has been steadily increasing in the last few decades [2,3]. For example, the incidence rate has increased more than 2-fold in the United States in the last 3 decades [4]. Meanwhile, a study in China reported a thyroid cancer rate of 0.368 per 100,000 people [5]. As one of the most malignant tumors, thyroid cancers are usually related to specific genetic abnormalities and environmental factors [6]. Therefore, early diagnosis and treatment are urgent. Currently, there is a lack of effective biomarkers, and the diagnostic technology for thyroid cancer is limited. The molecular and functional mechanisms of thyroid cancer also require further research.
Studies have predicted that only 1-2% of genes encode proteins in the human genome [7], while most other mammalian genomes do not encode proteins and comprise a large number of non-coding RNAs (ncRNAs) [8]. lncRNAs are the main transcribed noncoding RNA, and the types and numbers of lncRNAs among species are large [9]. Many studies have found that lncRNAs participate in regulating gene expression [9,10], chromatin structure [11], epigenetic control [12], splicing [13], and nuclear transport [9]. Other studies have also indicated that lncRNAs are associated with the development and progression of cancer [14-16]. However, the role of the lncRNA n340790 in thyroid cancer development, invasion and metastasis remains unknown.
Previous studies have indicated that lncRNAs can serve as a competing endogenous RNA (ceRNA) to regulate miRNAs [17,18]. ceRNAs play a post-transcriptional regulatory role in miRNA molecule distribution within cells. MicroRNAs (miRNAs) are non-coding RNA molecules approximately 20 nucleotides in length that specifically regulate gene and protein production by targeting the 3’ untranslated region (UTR) of mRNAs [19,20]. They are involved in the regulation of a wide variety of biological processes [21]. Certain miRNAs may also serve as either tumor suppressor genes or oncogenes during the development and progression of tumors. The regulatory mechanism and function of most miRNAs remain unknown for thyroid cancer. In this study, we explored n340790, which acted as a ceRNA for miR-1254 and is involved in thyroid cancer progression.
Materials and methods
Clinical specimens
A total of 85 clinical samples were collected from patients who were diagnosed with thyroid cancer and had undergone thyroidectomy at the Second Hospital of Hebei Medical University between 2013 and 2015. None of them had received radiotherapy or chemotherapy before surgical resection. The histological diagnosis of thyroid cancer was evaluated based on criteria established by the World Health Organization (WHO). The study was approved by the ethics committee of the Second Hospital of Hebei Medical University. All patients provided written informed consent before enrollment. Thyroid cancer tissues and corresponding adjacent normal tissues were immediately stored at -80°C.
Cell culture
A human thyroid follicular epithelial cell line (Nthy-ori 3-1), a papillary TC cell line with a BRAF mutation (B-CPAP), and a thyroid squamous cell carcinoma cell line (SW579) were obtained from American Type Culture Collection (ATCC, Manassas, VA); HEK293T, follicular Hürthle cells (FTC-133), a medullary thyroid cancer (MTC) cell line, a papillary with RET/PTC rearrangement (TPC-1) cell line, and poorly differentiated thyroid cancer (PDTC) cells were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The Nthy-ori 3-1, FTC-133, B-CPAP, and SW579 cell lines were cultured in RPMI 1640 medium (Invitrogen). MTC, TPC-1, and PDTC cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen). All the media were supplemented with 10% fetal bovine serum (FBS) (Sigma Aldrich), and 100 U/mL penicillin/streptomycin (Invitrogen, Carlsbad, CA). Cells were incubated at 37°C in an appropriate incubator containing 5% CO2.
Lentiviral vector construction, production and transfection
Human n340790 cDNAs were amplified by PCR, with them RNA from FTC-133 cells serving as a template. A lentiviral vector expressing enhanced green fluorescent protein (EGFP) was used as a control. The objective genes were cloned into the lentiviral vector and then into pcDNA3.1 (Invitrogen). Meanwhile, the shn340790 sequences were designed, with shLUC used as a control. DNA fragments of shRNA was synthesized and cloned into the U6 promoter-containing pBluescript SK (+) plasmid (pU6). The U6-shRNA was cloned into a lentiviral vector [22,23]. The lentiviral vectors and associated packaging vectors (pMD2.G, pMDL-G/P-RRE, pRSV-REV) were co-transfected into HEK293T cells for 48 hrs. Lentiviruses were produced, harvested, and purified. FTC-133 and PDTC cells (1 × 104 cells/well) were seeded into a 24-well plate. FTC-133 cells were transfected with either n340790 and control, and PDTC cells were transfected with either shn340790 or control using 8 μg/mL polybrene (Sigma). G418 (800 μg/ml, Sigma) was used to screen for stably expressing cells.
Quantitative real-time reverse transcription PCR (qRT-PCR)
Total RNA was isolated using TRIzol reagent (Invitrogen, CA, USA) according to the manufacturer’s introductions. ARevertAid First Strand cDNA Synthesis kit (Thermo Fisher) was used to synthesize cDNA. Quantitative PCR was performed on the cDNA using a SYBR-Green PCR Master Mix kit (Takara). The primer sequences for GAPDH were 5’-CCTCGTCTCATAGACAAGATGGT-3’ (forward primer) and 5’-GGGTAGAGTCATACTGGAACATG-3’ (reverse primer), the primer sequences for n340790 were 5’-GCAGGACTGAGACGTTGAGA-3’ (forward primer) and 5’-GGCCTCCGAAAGTGTGATTA-3’ (reverse primer) [24], and the primer sequences for U6 were 5’-CTCGCTTCGGCAGCACA-3’ (forward primer) and 5’-AACGCTTCACGAATTTGCGT-3’ (reverse primer). GAPDH and U6 were used as internal control.
Western blot assay
Cells were lysed by a radio immunoprecipitation assay (RIPA) buffer (Thermo Scientific, Rockford, IL, USA). Protein lysates were separated on a 10% gel using SDS/PAGE and transferred onto a polyvinylidene fluoride membrane (PVDF, Millipore, Billerica, MA). The PVDF membranes were blocked and incubated with a rabbit anti-GAPDH antibody (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA, USA) or a rabbit anti-NF-κB antibody (1:1000; Cell Signaling Technology, Beverly, MA) at 4°C overnight. The following day, the membranes were incubated with HRP-conjugated anti-IgG. Signals were detected using an ECL system (Amersham Pharmacia, Piscataway, NJ, USA).
Luciferase assay
Cells (5 × 104 cells/well) were cultured in 24-well plates and co-transfected with miR-1254, either a wild-type or mutant n340790 fragment, and a Renillaplasmid (RL-SV40) using Lipofectamine 3000 (Invitrogen). The pRL-CMV Renillaplasmid acted as an internal control. After 48 hrs, the luciferase activities were measured using a dual-luciferase reporter assay system (Promega, Madison WI, USA) according to the manufacturer’s instructions.
Proliferation assay
Cell proliferation was measured by a 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide (MTT) assay. Cells at a concentration of 3 × 103 per well were seeded into a 96-well plate and incubated for 1, 2, 3, 4, or 5 days. Afterwards, 10 μl of MTT solution (5 mg/ml) was added to each well. After 4 hrs, 200 μl of dimethyl sulfoxide (DMSO) was used to dissolve the precipitates. The absorbance value was recorded at 490 nm using an ELx800 Reader (Bio-Tek Instruments Inc., Winooski, VT, USA). For the colony forming assay, cells were seeded in a 6-well plate and incubated in complete medium (culture medium was changed every three days) at 37°C in a 5% CO2 incubator. After 14 days, the cells were fixed with methanol and stained with 0.1% crystal violet. All visible colonies were counted.
Flow cytometric analysis
To analyze cell cycle progression, transduced FTC-133 and PDTC cells were obtained, fixed, and incubated with RNase A (0.25 mg/ml) followed by treatment with propidium iodide (KeyGen, Nanjing, China). Cell cycle was analyzed using a FACS Caliburflow cytometer (BD Biosciences, San Jose, CA, USA). For the cell apoptosis assay, cells (1 × 106 cells/mL) were digested, centrifuged, washed, resuspended with 1 × binding buffer, and double stained with an annexin V-FITC and phosphatidylethanolamine (PE) staining kit (BD Biosciences). Cell apoptosis was analyzed using a FACS Calibur (BD Biosciences) and FlowJo software (Tree Star Corp, Ashland, OR).
Scratch assay
Transduced FTC-133 and PDTC cells were seeded in a 6-well plate, incubated in complete medium for 12 hrs at 37°C in a 5% CO2 incubator, and then scratched with a pipette tip. After replacing the supernatant with complete medium, the cells were then incubated at 37°C for 48 hrs. Photographs were obtained every 6 hrs.
Invasion assays
The assay was performed in 24-well Transwell chambers (Corning, Painted Post, NY, USA). Matrigel (BD Biosciences, San Diego, CA, USA) was precoated onto the upper chambers. Then, 200 μl of transduced FTC-133 and PDTC cells (5 × 105) in serum-free medium were seeded in the upper chamber, and complete medium was added to the lower chamber. After 24 hrs, the cells that invaded through the transwell were fixed, stained in a 0.1% crystal violet solution, photographed and counted.
Tumor formation in nude mice
Animal experiments were approved by the Institutional Committee for Animal Research and performed in accordance with the national guidelines for the care and use of laboratory animals. FTC-133 cells were transfected with either n340790 or control plasmids, and PDTC cells were transfected with either shn340790 or control plasmids. Cells (1 × 107 cells in 100 μl) were subcutaneously injected into 6-week-old BALB/c athymic nude mice that were purchased from the Shanghai LAC Laboratory Animal Co. Ltd. After 40 days, photographs of the excised tumors were obtained. The tumor volume was also measured at 10, 15, 20, 25, 30, 35, and 40 days. The excised tumors were used for additional assays such as qRT-PCR and Western blot analysis.
Statistical analysis
Differences between groups were analyzed using Student’s t-test. An ROC analysis was performed, and the area under the curve was calculated using SPSS 21.0 software (SPSS Inc., Chicago, IL, USA). P < 0.05 was considered statistically significant. The data are expressed as the mean ± standard deviation (SD), and all experiments were performed in triplicate.
Results
The lncRNA n340790 was upregulated in human thyroid cancer tissues
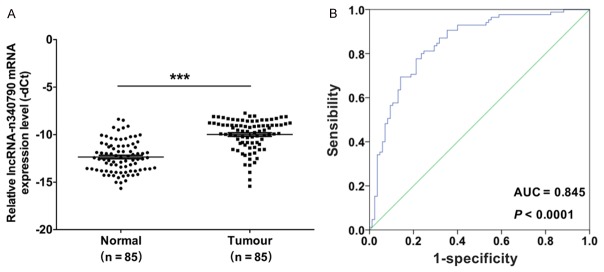
To study whether n340790 was involved in either the tumorigenesis or development of human thyroid cancer, the expression level of n340790 was examined by qRT-PCR in 85 pairs of thyroid cancer tissues (Tumor) and corresponding adjacent normal tissues (Normal). We found that n340790 expression was significantly upregulated in cancer tissues compared with the corresponding adjacent normal tissues (Figure 1A). Second, we investigated the relationships between n340790 expression and the clinical characteristics of 85 paired tissues from patients with thyroid cancer, and we discovered that the expression levels of n340790 were significantly associated with invasion, metastasis, and TNM stage. However, n340790 was not associated with sex or age (Table 1). Third, the receiver operating characteristic (ROC) curve was used to predict the prognostic value of n340790 in thyroid cancer, and the results indicated that the area under the curve (AUC) of n340790 was 0.845 (P < 0.0001, Figure 1B), suggesting that n340790 expression could serve as a molecular marker for thyroid cancer. Therefore, these data indicated an important role for n340790 in thyroid cancer.
Figure 1.
The lncRNA n340790 is upregulated in human thyroid cancer tissues. A. The mRNA expression levels of n340790 were examined by qRT-PCR and normalized to the GAPDH levels in 85 pairs of thyroid cancer tissues (Tumor) and corresponding adjacent normal tissues (Normal), (***P < 0.001). B. The receiver operating characteristic (ROC) curve predicted the prognosis of n340790 in thyroid cancer, as the area under the ROC curves of n340790 was 0.845 (P < 0.0001).
Table 1.
Correlation analysis between n340790 expression and the clinicopathological characteristics of patients with thyroid cancer
| Characteristics | No. of patients | Mean ± SD | P value |
|---|---|---|---|
| Total no. of patients | 85 | ||
| Age (year) | |||
| < 60 | 41 (48.2%) | 10.31 ± 1.32 | 0.414 |
| ≤ 60 | 44 (51.8%) | 10.09 ± 1.15 | |
| Gender | |||
| Male | 35 (41.2%) | 10.47 ± 1.62 | 0.077 |
| Female | 50 (58.8%) | 9.93 ± 1.16 | |
| Invasion | |||
| T0-T2 | 54 (63.5%) | 10.54 ± 1.26 | 0.032* |
| T3-T4 | 31 (36.5%) | 9.74 ± 2.13 | |
| Lymphatic metastasis | |||
| N0 | 59 (69.4%) | 11.14 ± 1.46 | < 0.0001*** |
| N1-N3 | 26 (30.6%) | 8.97 ± 2.02 | |
| Distal metastasis | |||
| M0 | 67 (78.8%) | 11.07 ± 2.47 | 0.014* |
| M1 | 18 (21.2%) | 9.48 ± 2.17 | |
| TNM stage | |||
| 0 & I | 61 (71.8%) | 10.94 ± 2.15 | < 0.0001*** |
| II & III & IV | 24 (28.2%) | 8.59 ± 1.57 |
Indicated statistical significance (P < 0.05);
Indicated statistical significance (P < 0.001).
n340790 promotes thyroid cancer cell proliferation in vitro
Based on the above results, we performed qRT-PCR to analyze n340790 expression inhuman thyroid follicular epithelial cells (Nthy-ori 3-1) and various thyroid cancer cell lines (FTC-133, MTC, TPC-1, B-CPAP, SW579, and PDTC). We found that the expression level of n340790 was dramatically higher in 6 thyroid cancer cell lines than in Nthy-ori 3-1 cells (Figure 2A). Based on the expression level of n340790 in these thyroid cancer cell lines, we chose FTC-133 and PDTC for all subsequent experiments. FTC-133 cells were transduced with either n340790 or control lentivirus. The relative n340790 expression level was determined by qRT-PCR. The overexpression efficiency was obtained in FTC-133 cells stably transduced with the n340790 plasmid (P < 0.001, Figure 2B). PDTC cells were transduced with either a shn340790 (RNA interference to knockdown of n340790) or control lentivirus. The expression level of n340790 was measured by qRT-PCR. The knockdown efficiency was obtained in PDTC cells stably transduced with the shn340790 plasmid (P < 0.001, Figure 2C). To further study the function of n340790 in thyroid cancer, MTT assays were used to detect the proliferative ability of the transfected FTC-133 cells, and we found that n340790 promoted FTC-133 cell proliferation (P < 0.001, Figure 2D). MTT assays were also performed to measure the proliferative ability of the transduced PDTC cells, and we found that silencing n340790 inhibited the proliferative ability of PDTC cells (P < 0.001, Figure 2E). In addition, the proliferative ability was further measured by colony formation assays in transduced FTC-133 and PDTC cells. These results also indicated that n340790 promoted cell proliferation (Figure 2F), while silencing n340790 inhibited cell proliferation (Figure 2G).
Figure 2.

n340790 promotes thyroid cancer cell proliferation in vitro. A. qRT-PCR analysis of n340790 expression in human thyroid follicular epithelial cells (Nthy-ori 3-1) and thyroid cancer cell lines (MTC, FTC-133, TPC-1, B-CPAP, SW579, and PDTC) (*P < 0.05, **P < 0.01, ***P < 0.001). B. FTC-133 cells were transduced with either n340790 or control lentivirus, and the relative n340790 expression levels were determined by qRT-PCR with the comparative delta-delta Ct method (2-ΔΔCt) (***P < 0.001). C. PDTC cells were transduced with either shn340790 or control lentivirus, and the mRNA expression levels of n340790 were measured by qRT-PCR (***P < 0.001). D. The MTT assays revealed the cell growth curves of the transduced FTC-133 cells (***P < 0.001). E. The MTT assays revealed the cell growth curves of the transduced PDTC cells (***P < 0.001, **P < 0.01). F. The proliferative ability of transduced FTC-133 cells was measured using colony formation assays. G. The proliferative ability of transduced PDTC cells was measured using colony formation assays.
n340790 accelerates cell cycle progression, migration and invasion as well as suppresses apoptosis of thyroid cancer cells in vitro
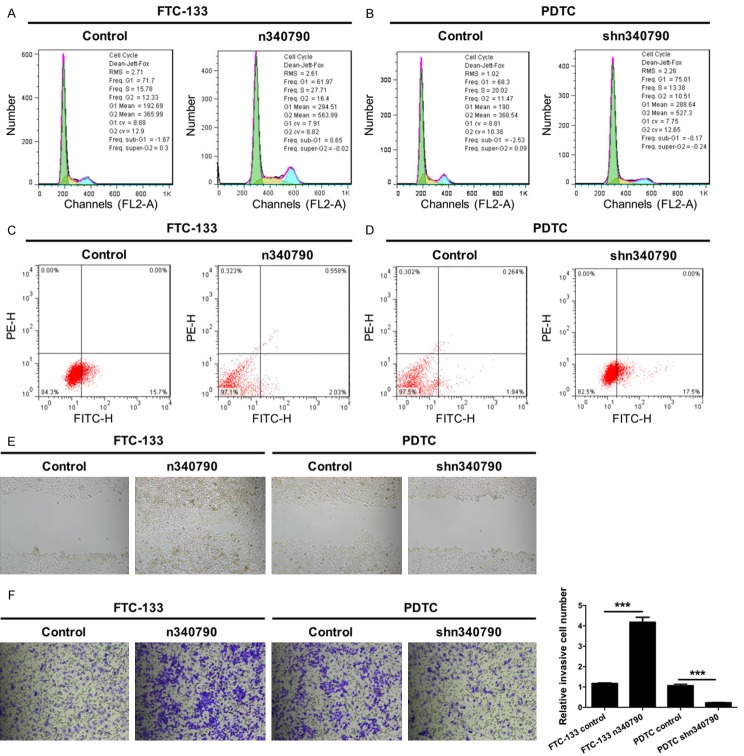
We further detected cell cycle distribution of the transduced FTC-133 and PDTC cells using flow cytometry. The results indicated that there was a significant decrease in the number of FTC-133 cells transduced with n340790 in G1phase (61.97% of total cells) compared with control cells (71.70% of total cells) and a significant increase in the number of FTC-133 cells transduced with n340790 in Sphase in (27.71% of total cells) compared with control cells (15.78% of total cells), suggesting that n340790 induced cell cycle progression in FTC-133 cells (Figure 3A). Likewise, there was a significant increase in the number of PDTC cells transduced with shn340790 in G1phase (75.01% of total cells) relative to the control cells (68.30% of total cells) and a significant decrease in the number of PDTC cells transfected with shn340790 in Sphase (13.38% of total cells) relative to the control cells (20.02% of total cells), indicating that silencing n340790 induced cell cycle arrest in PDTC cells (Figure 3B). Furthermore, the cell apoptosis assay was determined by flow cytometry analysis using annexin V-FITC and PE staining. The results revealed that n340790 inhibited cell apoptosis in FTC-133 cells (Figure 3C), while silencing n340790 accelerated cell apoptosis in PDTC cells (Figure 3D).
Figure 3.
n340790 accelerates cell cycle progression, migration and invasion as well as suppresses apoptosis of thyroid cancer cells in vitro. A. Cell cycle progression of FTC-133 cells transduced with n340790 or control lentivirus was examined by flow cytometry. B. Cell cycle progression of PDTC cells transduced with shn340790 or control lentivirus was detected by flow cytometry. C. Cell apoptosis was determined by flow cytometry analysis using annexin V-FITC and PE staining, and inhibited cell apoptosis was observed in FTC-133 cells transduced with n340790. D. Cell apoptosis was measured by flow cytometry analysis using annexin V-FITC and PE staining, and increased cell apoptosis was observed in PDTC cells transduced with shn340790. E. FTC-133 cells were transduced with n340790 or control lentivirus, and PDTC cells were transduced with shn340790 or control lentivirus. Cell migration ability was measured using the wound healing assay. F. Transwell assays were performed to detect the invasive ability of the cells. Magnification 200 ×, Scale bars = 10 μm, (***P < 0.001).
To further determine whether n340790 could promote the migration and invasion of thyroid cancer cells.FTC-133 cells were transduced with either n340790 or control lentivirus, and PDTC cells were transduced with either shn340790 or control lentivirus. The cell migratory ability was measured by the wound healing assay, and the results showed that n340790 promoted cell migration, whereas silencing n340790 inhibited cell migration (Figure 3E). In addition, Transwell assays were performed to detect the invasive ability, and the results indicated that n340790 promoted cell invasion, but silencing n340790 inhibited cell invasion (P < 0.001, Figure 3F).
n340790 promotes the proliferative and invasive abilities of thyroid cancer cells by targeting miR-1254
As mentioned above, n340790 was highly expressed in both thyroid cancer tissues and thyroid cancer cell lines (Figures 1A, 2A). We next investigated the role of miR-1254 in thyroid cancer by examining a panel of 85 matched sets of thyroid cancer tissues and adjacent normal tissues. The results indicated that miR-1254 expression was significantly downregulated in thyroid cancer tissues compared with that in the corresponding adjacent normal tissues (P < 0.0001, Figure 4A). We then analyzed the relationships between miR-1254 expression and clinicopathological characteristics and found that the expression level of miR-1254 was significantly associated with invasion, metastasis, and TNM stage (Table 2). The ROC curve of miR-1254 predicted the prognosis in thyroid cancer, and the results indicated that the area under the curve (AUC) of miR-1254 was 0.658 (P < 0.0001, Figure 4B), suggesting that miR-1254 expression may serve as a biomarker for thyroid cancer. Moreover, we found that there was a negative correlation between n340790 and miR-1254 expression levels in thyroid cancer tissues (R2 = 0.2962, P < 0.001, Figure 4C). Therefore, these data indicated an important role for miR-1254 in thyroid cancer.
Figure 4.

n340790 promotes the proliferative and invasive abilities of thyroid cancer cells by targeting miR-1254. (A) The expression levels of miR-1254 were examined by qRT-PCR and normalized to U6 expression in 85 pairs of thyroid cancer tissues (Tumor) and corresponding adjacent normal tissues (Normal), (***P < 0.001). (B) The receiver operating characteristic (ROC) curve predicted the prognosis of miR-1254 in thyroid cancer, and the area under the ROC curves of miR-1254 was 0.658 (P < 0.0001). (C) The correlation between n340790 and miR-1254 in thyroid cancer tissues was analyzed using Pearson’s correlation algorithm (R2 = 0.2962, P < 0.001). (D) Putative miR-1254 target sites in the n340790 sequence and the mutated sites in the n340790 sequence are shown. The relative fluorescence value was detected by the luciferase reporter gene assay in FTC-133 cells co-transfected with either wild-type or mutant n340790 and either control or miR-1254 (***P < 0.001). (E) The following combinations of transfections were conducted with FTC-133 cells: n340790; n340790 and miR-1254 mock; n340790 and miR-1254 mimics; and n340790 and miR-1254 inhibitors. The following combinations of transfections were conducted with PDTC cells: control; shn340790; shn340790 miR-1254 mock; shn340790 and miR-1254 mimics; and shn340790 and miR-1254 inhibitors. The qRT-PCR assay was performed to detect the expression levels of miR-1254 (***P < 0.001). (F) The MTT assay was used to measure the proliferative abilities of FTC-133 and PDTC cells treated as described in (E). (G) The Transwell assay was performed to analyze the invasive abilities of FTC-133 and PDTC cells treated as described in (E) (**P < 0.01, ***P < 0.001).
Table 2.
Correlation analysis between miR-1254 expression and the clinicopathological characteristics of patients with thyroid cancer
| Characteristics | No. of patients | Mean ± SD | P value |
|---|---|---|---|
| Total no. of patients | 85 | ||
| Age (year) | |||
| < 60 | 41 (48.2%) | 10.01 ± 1.94 | 0.289 |
| ≤ 60 | 44 (51.8%) | 9.59 ± 1.69 | |
| Gender | |||
| Male | 35 (41.2%) | 9.89 ± 1.38 | 0.317 |
| Female | 50 (58.8%) | 9.53 ± 1.77 | |
| Invasion | |||
| T0-T2 | 54 (63.5%) | 9.45 ± 1.26 | 0.005** |
| T3-T4 | 31 (36.5%) | 10.37 ± 1.68 | |
| Lymphatic metastasis | |||
| N0 | 26 (30.6%) | 10.14 ± 1.74 | 0.006** |
| N1-N3 | 59 (69.4%) | 9.25 ± 1.13 | |
| Distal metastasis | |||
| M0 | 18 (21.2%) | 10.87 ± 1.75 | 0.016* |
| M1 | 67 (78.8%) | 9.48 ± 2.21 | |
| TNM stage | |||
| 0 & I | 24 (28.2%) | 10.74 ± 2.07 | 0.012* |
| II & III & IV | 61 (71.8%) | 9.64 ± 1.64 |
Indicated statistical significance (P < 0.05);
Indicated statistical significance (P < 0.01).
Based on the above results, we hypothesized that the decrease inmiR-1254 levels might be related to n340790. TargetScan (www.targetscan.org), miRanda (www.microrna.org) and miR Base (www.mirbase.org) inquiries were performed to conduct a computational analysis for n340790 with complementary sites of miR-1254 in their 3’-UTR, and we found putative miR-1254 target sites in 3’-UTRs of n340790. We cloned the wild-type or mutant n340790 into a pGL3-basic luciferase reporter vector. FTC-133 cells were co-transfected with either wild-type or mutant n340790 and either control or miR-1254. The luciferase reporter gene assay results indicated that n340790 interacted with miR-1254 at the specific binding sites (P < 0.001, Figure 4D). To further study the role of miR-1254 on human thyroid cancer, we overexpressed n340790 in FTC-133 cells and observed decreased expression levels of miR-1254, while miR-1254 mimics could rescue this expression. Additionally, miR-1254 inhibitors could further inhibit miR-1254 expression. We also silenced n340790 in PDTC cells and found that these cells exhibited increased expression levels of miR-1254, while miR-1254 mimics could rescue the loss of expression. In addition, miR-1254 inhibitors also suppressed the miR-1254 expression levels (P < 0.001, Figure 4E). The MTT results showed that n340790 increased the proliferative abilities of FTC-133 cells, while miR-1254 mimics could rescue this phonotype. Silencing n340790 decreased the proliferative abilities of PDTC cells, butmiR-1254 inhibitors could rescue this phonotype (Figure 4F). Likewise, the results of the invasion assays revealed similar tendencies (P < 0.01, P < 0.001, Figure 4G). Therefore, these data suggest that n340790 promotes the proliferative and invasive abilities of thyroid cancer cells by regulating miR-1254.
n340790 promotes tumor growth in vivo
To study the effect of n340790 on proliferation in vivo, nude mice were injected with FTC-133 cells stably expressed n340790 or PDTC cells stably expressing shn340790y. Photographs of the excised tumors were obtained at 40 days (Figure 5A). There was a dramatic increase in the tumor volume in the n340790 group compared with the corresponding control group (P < 0.001, Figure 5B) as well as a dramatic decrease in the tumor volume in the shn340790 group compared with the corresponding control group (P < 0.001, Figure 5C). At 40 days, the tumors were excised, and the relative expression levels of miR-1254 were detected by qRT-PCR whereas the protein expression levels of NF-κB were measured by Western blotting. The results indicated that the expression levels of miR-1254 were significantly decreased in mice injected with FTC-133 cells transduced with n340790 compared with the respective control group (P < 0.001, Figure 5D) and were significantly increased in mice injected with PDTC cells transduced with shn340790 compared with the respective control group (P < 0.001, Figure 5E). In addition, we found that n340790 upregulated NF-κB expression, while silencing n340790 downregulated NF-κB expression in nude mice (Figure 5F). Our results are summarized in Figure 6.
Figure 5.
n340790 promotes the growth of tumors in vivo. A. Nude mice were injected with stably transduced FTC-133 or PDTC cells, and then photographs of excised tumors were obtained. B. The mean tumor volume was measured on the indicated days, and the data show that n340790 increased the tumor volume (*P < 0.05, **P < 0.01, ***P < 0.001). C. The mean tumor volume was measured on the indicated days, and silencing n340790 expression resulted in a decrease in the tumor volume (**P < 0.01, ***P < 0.001). D. Nude mice were injected with FTC-133 cells transduced with either n340790 or control lentivirus, the tumors were excised after 40 days, and the relative expression levels of miR-1254 were detected by qRT-PCR (***P < 0.001). E. Nude mice were injected with PDTC cells transduced with either shn340790 or control lentivirus, and the tumors were excised after 40 days, after which the relative expression levels of miR-1254 were detected by qRT-PCR (***P < 0.001). F. Western blotting was performed to detect NF-κB expression in vivo. GAPDH was used as a reference protein.
Figure 6.

A schematic model of n340790 in thyroid cancer. The lncRNA n340790 can regulate miR-1254 in tumor cells and cause upregulation of NF-κB, which induces an increase in thyroid cancer cell growth, invasion and migration.
Discussion
lncRNAs play an important role in regulating gene expression [25,26]. Many studies have shown that lncRNAs are involved in cell cycle progression [27], cell differentiation [27], apoptosis [28], and chromatin remodeling [29]. Other reports have indicated that dysregulated expression of lncRNAs was involved in the development of various cancers, including lung [30], liver [31], and breast cancers [32]. In our study, the results showed that n340790 was highly expressed in human thyroid cancer tissues. In addition, n340790 was significantly associated with invasion, metastasis, and TNM stage. n340790 also exhibited a good prognostic value for thyroid cancer. Furthermore, we found that n340790 could regulate cell cycle progression, proliferation, migration, and invasion as well as inhibit thyroid cancer cell apoptosis in vitro. We also observed that n340790 regulated the growth of thyroid cancer cells in vivo. Therefore, we suggested that n340790 may be a potential oncogene for thyroid cancer.
miRNAs are a type of non-coding RNA approximately 20 nucleotides in length that regulate gene expression by affecting mRNA transcripts [33]. Studies have revealed that miRNAs play important roles in the development of human cancers [34,35]. They are involved in various biological processes, such as proliferation, differentiation, apoptosis, migration and invasion [36,37]. They can also be considered as new biomarkers for human cancer therapy. In our study, the results revealed that human thyroid cancer tissues had low expression levels of miR-1254. In addition, we also found that miR-1254 was significantly associated with invasion, metastasis, and TNM stage. Therefore, we speculated that miR-1254 may be a potential target for treating thyroid cancer.
Recent studies have demonstrated that lncRNAs act as competitive endogenous RNAs (ceRNAs) that affect miRNA expression and modulate the derepression of miRNA targets via post-transcriptional regulation [38,39]. Alarge number of studies also indicated that lncRNAs, such as ceRNAs, were involved in the development of various diseases [40,41]. For example, the lncRNA H19, which is a sponge for miRNA, promoted colorectal cancer development [42]. In our study, we found that there was a negative correlation between n340790 and miR-1254 expression levels in thyroid cancer tissues, and we hypothesized that n340790 served as an endogenous sponge by directly binding to miR-1254 and downregulating miR-1254 expression. Furthermore, we also investigated whether miR-1254 could rescue the carcinogenesis effect of n340790 on thyroid cancer cells. Based on the results, we concluded that n340790 promoted thyroid cancer development by regulating miR-1254.
The transcription factor NF-κB is a nuclear factor that can interact with the enhancer element of the immunoglobulin kappa light-chain inactivated B cells [43]. Currently, many studies have indicated that NF-κB plays vital roles in the development of tumors, including proliferation, apoptosis, and metastasis [44]. In our study, the results revealed that n340790 increased the protein expression of NF-κB, while silencing n340790 decreased the protein expression of NF-κB, suggesting that n340790 can upregulate NF-κB expression in thyroid cancer. This may be another molecular mechanism of n340790 in promoting thyroid cancer tumorigenesis. In addition, further studies will be performed to explore how n340790 upregulates NF-κB expression (i.e., direct interaction or an indirect mechanism), as illustrated in Figure 6.
In summary, we found that n340790 was highly expressed, miR-1254 exhibited low levels of expression in human thyroid cancer tissues, and both of these molecules were good prognostic factors for thyroid cancer. n340790 and miR-1254 were simultaneously and strongly correlated with invasion, metastasis, and TNM stage of thyroid cancer. n340790 plays vital roles in the development of thyroid cancer both in vitro and in vivo. Furthermore, n340790 could act as an endogenous sponge by directly binding to miR-1254 and decreasing miR-1254 expression. In addition, miR-1254 could further suppress the stimulatory effect of n340790 on the growth and invasion of thyroid cancer cells. Therefore, n340790 is involved in the development of malignant thyroid cancer by regulating miR-1254 (Figure 6).
Acknowledgements
We thank Professor Xiaolu Wang (The Second Hospital of Hebei Medical University) for his technical help.
Disclosure of conflict of interest
None.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Carlberg M, Hedendahl L, Ahonen M, Koppel T, Hardell L. Increasing incidence of thyroid cancer in the Nordic countries with main focus on Swedish data. BMC Cancer. 2016;16:221–228. doi: 10.1186/s12885-016-2429-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang YX, Wang W. Increasing incidence of thyroid cancer in Shanghai, China, 1983-2007. Asia Pac J Public Health. 2015;27:Np223–Np229. doi: 10.1177/1010539512436874. [DOI] [PubMed] [Google Scholar]
- 4.Chen AY, Jemal A, Ward EM. Increasing incidence of differentiated thyroid cancer in the United States, 1988-2005. Cancer. 2009;115:3801–3807. doi: 10.1002/cncr.24416. [DOI] [PubMed] [Google Scholar]
- 5.La Vecchia C, Malvezzi M, Bosetti C, Garavello W, Bertuccio P, Levi F, Negri E. Thyroid cancer mortality and incidence: A global overview. Int J Cancer. 2015;136:2187–2195. doi: 10.1002/ijc.29251. [DOI] [PubMed] [Google Scholar]
- 6.Li XY, Abdel-Mageed AB, Mondal D, Kandil E. The nuclear factor Kappa-B signaling pathway as a therapeutic target against thyroid cancers. Thyroid. 2013;23:209–218. doi: 10.1089/thy.2012.0237. [DOI] [PubMed] [Google Scholar]
- 7.Djebali S, Davis CA, Merkel A, Guigo R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furuno M, Pang KC, Ninomiya N, Fukuda S, Frith MC, Bult C, Kai C, Kawai J, Carninci P, Hayashizaki Y, Mattick JS, Suzuki H. Clusters of internally primed transcripts reveal novel long noncoding RNAs. PLoS Genet. 2006;2:e37. doi: 10.1371/journal.pgen.0020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang JY, Liu XF, Wu HC, Ni PH, Gu ZD, Qiao YX, Chen N, Sun FY, Fan QS. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse x chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 13.Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, Bonilla F, de Herreros AG. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K, Kossai M, MacDonald TY, Fontugne J, Erho N, Vergara IA, Ghadessi M, Davicioni E, Jenkins RB, Palanisamy N, Chen Z, Nakagawa S, Hirose T, Bander NH, Beltran H, Fox AH, Elemento O, Rubin MA. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rincic M, Iourov IY, Liehr T. Thoughts about SLC16A2, TSIX and XIST gene like sites in the human genome and a potential role in cellular chromosome counting. Mol Cytogenet. 2016;9:56. doi: 10.1186/s13039-016-0271-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012;8:2289–2294. doi: 10.1039/c2mb25070e. [DOI] [PubMed] [Google Scholar]
- 17.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA Hypothesis: the rosetta stone of a Hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarthy N. Regulatory RNA layer by layer. Nat Rev Genet. 2011;12:804–804. doi: 10.1038/nrg3108. [DOI] [PubMed] [Google Scholar]
- 19.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ambros V. The evolution of our thinking about microRNAs. Nat Med. 2008;14:1036–1040. doi: 10.1038/nm1008-1036. [DOI] [PubMed] [Google Scholar]
- 21.Tang GL, Yan J, Gu YY, Qiao MM, Fan RW, Mao YP, Tang XQ. Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods. 2012;58:118–125. doi: 10.1016/j.ymeth.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Lin MC, Yao H, Wang H, Zhang AQ, Yu J, Hui CK, Lau GK, He ML, Sung J, Kung HF. Lentivirus-mediated RNA interference targeting enhancer of zeste homolog 2 inhibits hepatocellular carcinoma growth through downregulation of stathmin. Hepatology. 2007;46:200–208. doi: 10.1002/hep.21668. [DOI] [PubMed] [Google Scholar]
- 23.Jiang L, Lai YK, Zhang J, Wang H, Lin MC, He ML, Kung HF. Targeting S100P inhibits colon cancer growth and metastasis by lentivirus-mediated RNA interference and proteomic analysis. Mol Med. 2011;17:709–716. doi: 10.2119/molmed.2011.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lan XB, Zhang H, Wang ZH, Dong WW, Sun W, Shao L, Zhang T, Zhang DL. Genome-wide analysis of long noncoding RNA expression profile in papillary thyroid carcinoma. Gene. 2015;569:109–117. doi: 10.1016/j.gene.2015.05.046. [DOI] [PubMed] [Google Scholar]
- 25.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Li D, Zhang W, Guo M, Zhan Q. Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. EMBO J. 2012;31:4415–4427. doi: 10.1038/emboj.2012.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lakhotia SC. Long non-coding RNAs coordinate cellular responses to stress. Wiley Interdiscip Rev RNA. 2012;3:779–796. doi: 10.1002/wrna.1135. [DOI] [PubMed] [Google Scholar]
- 29.Saxena A, Carninci P. Long non-coding RNA modifies chromatin. Bioessays. 2011;33:830–839. doi: 10.1002/bies.201100084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enfield KS, Pikor LA, Martinez VD, Lam WL. Mechanistic roles of noncoding RNAs in lung cancer biology and their clinical implications. Genet Res Int. 2012;2012:737416. doi: 10.1155/2012/737416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 32.Piao Hl, Ma L. Non-coding RNAs as regulators of mammary development and breast cancer. J Mammary Gland Biol Neoplasia. 2012;17:33–42. doi: 10.1007/s10911-012-9245-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayes EL, Lewis-Wambi JS. Mechanisms of endocrine resistance in breast cancer: an overview of the proposed roles of noncoding RNA. Breast Cancer Res. 2015;17:40. doi: 10.1186/s13058-015-0542-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNA cancer regulation. Springer; 2013. MicroRNAs in human cancer; pp. 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Leva G, Croce CM. The role of microRNAs in the tumorigenesis of ovarian cancer. Front Oncol. 2013;3:153. doi: 10.3389/fonc.2013.00153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Muluhngwi P, Klinge CM. Roles for miRNAs in endocrine resistance in breast cancer. Endocr Relat Cancer. 2015;22:R279–R300. doi: 10.1530/ERC-15-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Filipska M, Skrzypski M, Bigda JJ, Jassem J. Biological role of prognostic microRNAs (MiRNAs) in squamous lung cancer cell lines. J Thorac Oncol. 2015;10:S391–S391. [Google Scholar]
- 38.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long Noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu LG, Liu CC, Yi JS, Zhang HF, Min W, Bennett AM, Gregory RI, Ding Y, Huang YQ. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3:1113–1121. doi: 10.1158/2159-8290.CD-13-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX, Zhang L, Xiao LJ, Wan D, Zhang JF. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–22525. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46:705–716. [PubMed] [Google Scholar]
- 44.Hoesel B, Schmid JA. The complexity of NF-kappaB signaling in inflammation and cancer. Mol Cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]