Abstract
Breast cancer is the most common cancer in women worldwide. Triple-negative breast cancer is one of the most aggressive types of breast cancer as it has the worst clinical outcome for patients. microRNAs are a type of small non-coding RNA and play an important role in breast cancer. The purpose of this study was to explore the potential function and mechanism of miR-143-3p in triple-negative breast cancer (TNBC). MTT and colony formation assays, the effect of miR-143-3p modulation on MDA-MB-231 cell proliferation, revealed that increased miR-143-3p expression inhibited the proliferation of MDA-MB-231 TNBC cells. Moreover, miR-143-3p overexpression inhibited the movement of MDA-MB-231 TNBC cells in wound healing and transwell assays. To identify a potential miR-143-3p target, we investigated the effect of miR-143-3p modulation on LIMK1 expression level. Increased miR-143-3p expression caused a reduction in LIMK1 mRNA and protein, suggesting that LIMK1 is a target of miR-143-3p. In addition, dual-luciferase reporter assays showed that LIMK1 is a target gene of miR-143-3p. Flow cytometry analysis indicated that miR-143-3p arrested MDA-MB-231 TNBC cells at the G0/G1 phase. The TCGA (The Cancer Genome Atlas) database demonstrated that miR-143-3p was down-regulated in breast cancer tissues compared with normal breast tissues. These data demonstrated that miR-143-3p functioned as a suppressor gene in TNBC and that miR-143 targeted therapy may be a new strategy for TNBC treatment.
Keywords: miR-143-3p, LIMK1, triple-negative breast cancer (TNBC), progression, cell cycle
Introduction
Breast cancer currently represents approximately one-quarter of new cancer cases and is the most common malignant cancer in women worldwide [1]. Multiple molecular pathway are dysregulated, resulting in different strategies regarding prognosis, metastasis, treatment sensitivity and patient survival [2]. Breast cancer can be divided into different types based on molecular characterization of ER, PR, ki-67, and HER2. These molecular subtypes include Lumina A, Lumina B, Her-2 overexpression, basal-like tumors, and ‘normal-like’ breast tumors [3]. In addition, ‘triple negative breast cancer’ (TNBC) is defined as a tumor that is negative for ER, PR, and HER2 [3]. Because TNBC does not have any specific hormonal biomarker, effective treatment of this type of BC remains a challenge, and it lacks effective therapies to improve patient survival outcomes [4]. Numerous studies have demonstrated that TNBC exhibits a higher histological grade [5-7]. The incidence of metastases to visceral organs in TNBC patients is considerably increased compared with other BC patient types during the first 5 years after diagnosis [7]. Therefore, determining the mechanism of triple-negative breast cancer tumorigenesis is very important.
LIM kinase-1 (LIMK1) and LIM kinase-2 (LIMK2) belong to a small subfamily with a unique combination of 2 N-terminal LIM motifs and a C-terminal protein kinase domain. LIMK1 is a serine/threonine kinase that regulates actin polymerization via phosphorylation and inactivation of the actin binding factor cofilin (CFL1) [8]. The actin cytoskeleton is a very important regulator in cell biological activities [9]. Cell movement and cell cycle regulation can be regulated through phosphorylation and inactivation of CFL1 [9,10]. Activated LIMK1 can phosphorylate CFL1 and is therefore involved in the regulation of biological processes in cancer tumorigenesis and metastasis [11]. LIMK1 is highly expressed in breast cancer tissues, and over-expression of LIMK1 increased proliferation and promoted breast cancer metastasis to liver and lung [12].
As a type of small non-coding RNA of approximately 22 nucleotides, microRNAs (miRNAs) negatively regulate the expression of target genes. Although the miRNA group is composed of a few members compared with the enormous number of genes, one miRNA can interact with multiple target gene mRNAs. In addition, one target gene can be regulated by several miRNAs simultaneously [13]. The cellular processes regulated by miRNAs are highly tissue-specific and are tightly controlled during developmental stages, exerting a regulatory effect on a myriad of cellular physiological functions, including cell development, differentiation, proliferation and apoptosis [14]. In BC, several miRNAs function as courier messengers involved in BC tumorigenesis and metastasis [15].
Several studies reported that the expression of miR-143-3p was reduced in breast cancer tissues [16-19] and that over-expression of miR-143-3p suppresses proliferation by targeting ERBB3 [20] and CD44 [19]. Whether miR-143-3p has other target genes to function as a suppressor remains unknown. Here, in our study, we found that miR-143-3p suppresses the LIMK1/CFL1 pathway to suppress the proliferation and movement of TNBC. This information could help us better understand the mechanism of breast cancer and could be useful for target therapy for breast cancer.
Methods and materials
Cell culture and transfection
Triple-negative breast cancer cells MDA-MB-231 and HCC 1937 cells [21] were purchased from Chinese Scientific Institute (Shanghai, China). HEK 293T cell was a gift from the department of laboratory, Shanghai Tenth Hospital. All cells were cultured with Dulbecco’s modified Eagle’s medium (DMEM; Gibco, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, USA) and penicillin-streptomycin (Enpromise, China) in incubator at 37°C in 5% CO2.
The miR-143-3p mimics and negative-control mimics were purchased from IBS Biology Company (Ibsbio, Shanghai, China). Before mimics were transfected into cells, the cells were cultured in six-well plates (Corning, USA) at a concentration of 3*105 cells/well. When 40-50% cell density was achieved, the mimics were transfected with lipofectamine 2000 (lipo-2000, Invitrogen, USA) according to the manufacturer’s instruction. The concentration of mimics was 100 nm/l, and the ratio of mimics to lipo-2000 was 1.25:1 (volume). The miR-143-3p mimic sequence was sense: 5’-UGAGAUGAAGCACUGUAGCUC-3’ and anti-sense: 5’-GCUACAGUGCUUCAUCUCAUU-3’. The negative-control (NC) mimics sequence was sense: 5’-UCACAACCUCCUAGAAAGAGUAGA-3’ and anti-sense: 5’-UACUCUUUCUAGGAGGUUGUGAUU-3’.
Cell proliferation assays (clone formation assay and 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphe nyltetrazolium bromide (MTT) assay)
MDA-MB-231 breast cancer cells were first seeded in six-well plate at 50,000/well. After 40-50% confluence was achieved, the cells were treated with 100 nM miR-143-3p and NC mimics. Then, the treated cells were suspended in a single-cell suspension and then seeded in twelve-well plates (Corning, USA) at 600 cells/well. The medium was changed every three days. After two weeks, the clone-formation plate was washed by PBS twice and then treated with 4% polyformaldehyde for 10 minutes at room temperature (r.t.). MDA-MB-BC cells were then stained with 0.1% Crystal violet for 10 minutes at r.t. and then washed with DDH2O thrice. Pictures were obtained as soon as the plate dried.
For the proliferation assay, the mimic-treated MDA-MB-231 BC cells were seeded in ninety-six well plates (Corning, USA) at 3000 cells/well. The proliferation rate was measured by 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphe nyltetrazolium bromide (MTT) assay at 24, 48, and 72 h according to the manufacturer’s instructions. The absorbance at 490 nm of each sample was measured by a microplate spectrophotometer (BioTek, Vermont USA).
Cell migration and invasion assay
Wound healing and transwell assays were used to evaluate cell migration and invasion of mimic-treated MDA-MB-231 BC cells. MDA-MB-231 cells were treated by miR-143-3p or NC mimics in six-well plates.
For the wound healing assay, when cells were 90% confluent, a scratch was generated using a 200-μl-pipette tip. Pictures were obtained at 0 and 24 h in the same position to compare cell movement. Cell migration and invasion ability were also evaluated by transwell assay (with Matrigel for invasion and without Matrigel for migration). The transwell cups were placed in 24-well plate (Corning, USA). After treatment with miR-143-3p and NC mimics, 5*104 cells/well was placed in the upper chamber with 200 μl of DMEM with 0.1% BSA (Sigma, USA). The lower chamber was filled with 600 μl of 10% FBS DMEM. The cells were cultured for 12 to 16 h. Then, the outer surface of the cup was washed thrice, fixed with 4% polyformaldehyde, and stained with 0.1% Crystal violet for 10 minutes r.t. Pictures were obtained as soon as the cups dried.
Quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
MDA-MB-231 and HCC-1937 breast cancer cells were treated with miR-143-3p and NC mimics in six-well plates. After a 36-h incubation, TRIzol reagent (Invitrogen, USA) was used to isolate total RNA according to the manufacturer’s instructions, and the RT-PCR kit (Takara, Shiga, Japan) was used to generate the cDNA. QRT-PCR was performed on a 7900HT Fast RT-PCR instrument (Applied Biosystems, Singapore). The amplification procedure followed a previous method: 5 min at 95°C followed by 40 cycles at 95°C for 30 s and 65°C for 45 s. Expression of mRNA was assessed by evaluating threshold cycle (CT) values. The CT values of LIMK1 were normalized with GAPDH expression. The primer sequences were as follows: LIMK1-F: 5’-CAAGGGACTGGTTATGGTGGC-3’; LIMK1-R: 5’-CCCCGTCACCGATAAAGGTC-3’; GAPDH-F: 5’-CATGAGAAGTATGACAACAGCCT-3’; GAPDH-R: 5’-AGTCCTTCCACGATACCAAAGT-3’. The qRT-PCR results were analyzed with the method 2-ΔΔt [22].
Western blot analysis
After 72 h of treatment of MDA-MB-231 and HCC-1937 breast cancer cells, protein was collected with Ripa (Beyotime Biotechnology, Shanghai, China), and the concentration was measured with BCA Protein Assay Kit (Beyotime biotechnology, Shanghai, China) according to the manufacturer’s instructions.
Protein samples (60 μg) were denatured with 6× sodium dodecyl sulfate (SDS) loading buffer (100 mmol/L Tri-HCl pH 6.8, 4% SDS, 0.2% bromophenol blue, 20% glycerin, 200 mmol/L β-mercaptoethanol) at 95°C for 5 min. Protein samples were separated by 10% SDS polyacrylamide gel electrophoresis and transferred onto 0.45-μm nitrocellulose membranes (Beyotime biotechnology, Shanghai, China). Following 60 min of blocking with 5% fat-free milk, membranes were incubated with the primary antibody overnight at 4°C. Blots were washed and incubated for 1 h with the anti-rabbit/mouse secondary antibody (1:2000, Santa, USA). After three washes with PBST, immunoreactive protein bands were detected with an Odyssey Scanning system (Li-Cor, USA). The following primary antibodies were used:
PCNA (1:2000, CST, USA), MMP2 (1:1000, Arigo, Taiwan, China), MMP9 (1:1000, Arigo, Taiwan, China), LIMK1 (1:500, Arigo, Taiwan, China), CFL1 (1:500, Arigo, Taiwan, China), Phos-CFL1 (1:500, Arigo, Taiwan, China), CDK2 (1:2000, Bioworld, Shanghai, China), CDK4 (1:2000, Abcam, USA), CDK6 (1:2000, Abcam, USA), Cyclin D1 (1:10000, Abcam, USA), Cyclin E1 (1:2000, Abcam, USA), and β-actin (1:2000, Santa, USA).
Cell cycle assay
MDA-MB-231 breast cancer cells were treated with miR-143-3p and NC mimics. After 36 h, the cells were collected, washed and suspended with PBS, and fixed in cold 70% ethanol overnight at 4°C overnight. After a 30-min digestion in RNase (0.1 g/l), a total of 300 μL (0.05 g/l) propidium iodide (PI) staining solution was added to each sample, which was then incubated for 30 min at room temperature in the dark. Cell cycles were then analyzed by flow cytometry.
Dual-luciferase reporter assay
HEK 293T cells were seeded in 48-well plates and cultured until the cells reached 70% confluence. psiCHECK-2/LIMK1 3’-UTR and psiCHECK-2/LIMK1 3’-UTR mutant reporter plasmids were purchased from Ibsbio (Shanghai, China). HEK 293T cells were transiently co-transfected with 0.2 µg psiCHECK-2/LIMK1 3’-UTR or psiCHECK-2/LIMK1 3’-UTR mutant reporter plasmids and together with 100 nmol/l miR-143-3p or miR-NC using LipofectamineTM per the manufacturer’s instructions. After 72 h, firefly and renilla luciferase activities were measured using a Dual Luciferase Assay (Beyotime biotechnology, Shanghai, China). Firefly luciferase values were normalized to Renilla, and the ratio of firefly and renilla luciferase was presented.
Statistical analysis
Data from at least three separate experiments are presented as the mean ± standard (mean ± SD). The two tailed t-test and student t test were used for comparisons between groups. Differences were considered significant for p-values less than 0.05. GraphPad Prism version 6.0 (GraphPad, San Diego, CA, USA) was used for all statistical analyses.
Results
miR-143-3p suppresses breast cancer cell proliferation
MDA-MB-231 breast cancer cell proliferation was measured by clone formation and MTT assays. As shown in Figure 1A, clone formation was decreased by over-expression of miR-143-3p. MTT assays showed that miR-143-3p has a significant inhibitory effect compared with the NC group (Figure 1B). PCNA protein levels, which represent the level of proliferation, were also decreased by over-expression miR-143-3p in MDA-MB-231 cells and further confirmed in HCC-1937 cells (Figure 1C and 1D). The data suggested that miR-143-3p suppresses breast cancer proliferation.
Figure 1.
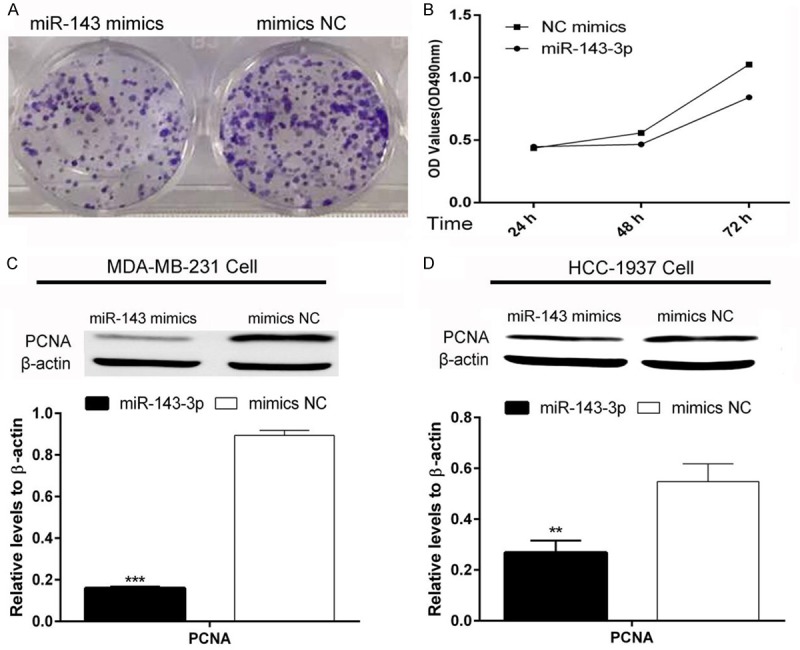
miR-143-3p suppressed the proliferation of TNBC cells. A and B: Clone formation assay and MTT assay showed that miR-143-3p inhibited MDA-MB-231 TNBC cell proliferation (student t-test and ANOVA test, *P<0.05); C and D: miR-143-3p inhibited PCNA protein expression in MDA-MB-231 and HCC-1937 TNBC cells (student t-test, **P<0.01;***P<0.001).
miR-143-3p suppresses migration and invasion of breast cancer cells
MDA-MB-231 breast cancer cell migration and invasion was measured using the wound healing test and transwell assay. As shown in Figure 2A, the scratch in the NC mimics group healed more rapidly compared with the miR-143-3p-treated group at 24 h, suggesting that miR-143-3p could suppress the movement ability of MDA-MB-231 breast cancer cells. The transwell assay also confirmed the result (Figure 2B). We then detected MMP2 and MMP9 protein levels as these proteins are involved in migration and invasion. As shown in Figure 2C and 2D, MMP2 and MMP9 expression was significantly decreased in MDA-MB-231 and HCC-1937 breast cancer cells. These data suggested that miR-143-3p suppresses the migration and invasion of breast cancer cells.
Figure 2.
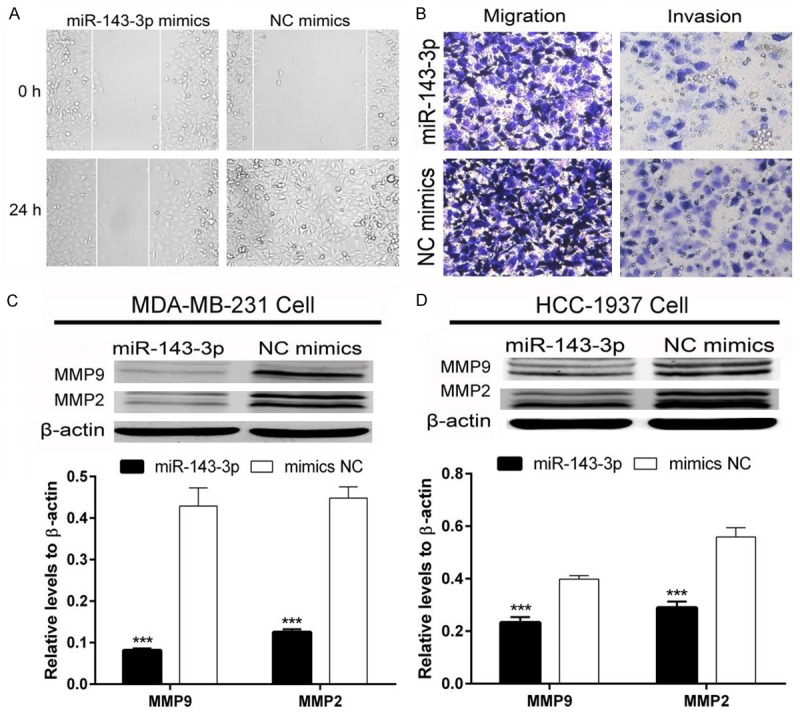
miR-143-3p suppressed TNBC cell migration and invasion. A and B: Wound healing and transwell assays showed that miR-143-3p suppressed MDA-MB-231 TNBC cell motility; C and D: miR-143-3p inhibited MMP2 and MMP9 protein expression in MDA-MB-231 and HCC-1937 TNBC cells (student t-test, ***P<0.001).
miR-143-3p suppresses LIMK1 expression
Several databases, such as TargetScan, PicTar and miRwalk, indicated that LIMK1 may be a potential target of miR-143-3p. We constructed two types of plasmids that contained the wild-type and mutant binding site of miR-143-3p with LIMK1 (Figure 3A). As shown in Figure 3B, miR-143-3p significantly reduces luciferase activity in the wild-type group compared with the NC group. The data suggested that LIMK1 is a target gene of miR-143-3p. As shown in Figure 4A and 4B, miR-143-3p significantly inhibited LIMK1 mRNA levels. miR-143-3p also inhibited LIMK1 protein levels and furthermore inhibited the CFL1 expression in MDA-MB-231 and HCC-1937 breast cancer cells (Figure 4C and 4D). These data indicated that miR-143-3p targets the LIMK1 gene, inhibits its expression at both the mRNA and protein level and furthermore affects CFL1 expression.
Figure 3.
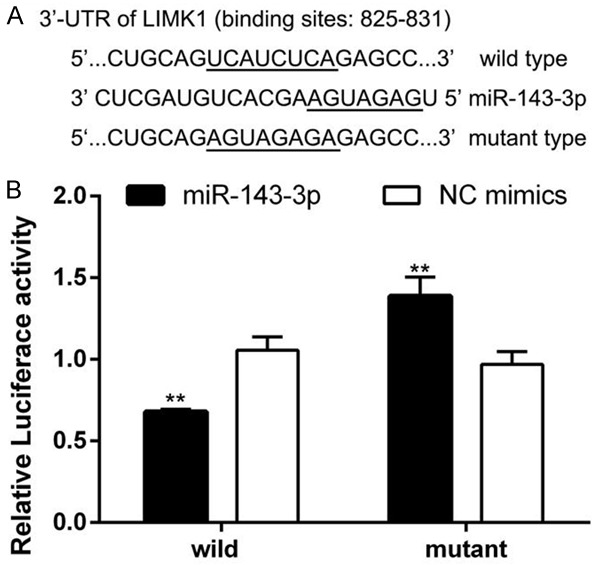
LIMK1 is a target gene of miR-143-3p. A: Wild-type and mutant binding site of miR-143-3p with LIMK1; B: Dual-luciferase reporter assay revealed that miR-143-3p reduced the luciferase activity of LIMK1, indicating that LIMK1 is a target gene of miR-143-3p (student’s t-test, **P<0.01).
Figure 4.
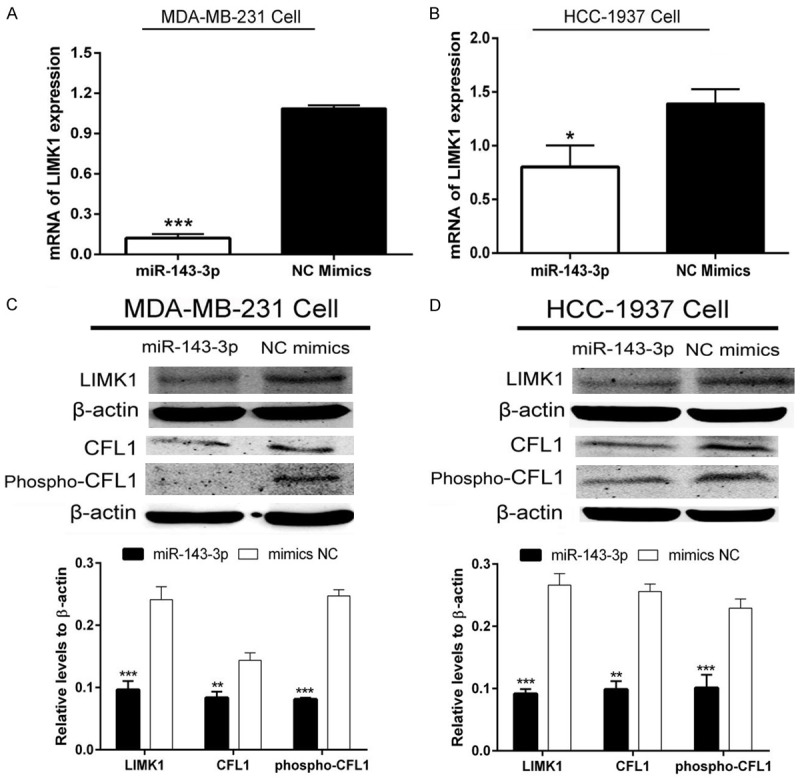
miR-143-3p inhibited the expression of the LIMK1/CFL1 signaling pathway. A and B: miR-143-3p reduced LIMK1 mRNA levels in MDA-MB-231 and HCC-1937 TNBC cells (student t-test, *P<0.05, ***P<0.001); C and D: miR-143-3p decreased LIMK1 protein levels and suppressed phosphorylation of CFL1 (student t-test, **P<0.01, ***P<0.001).
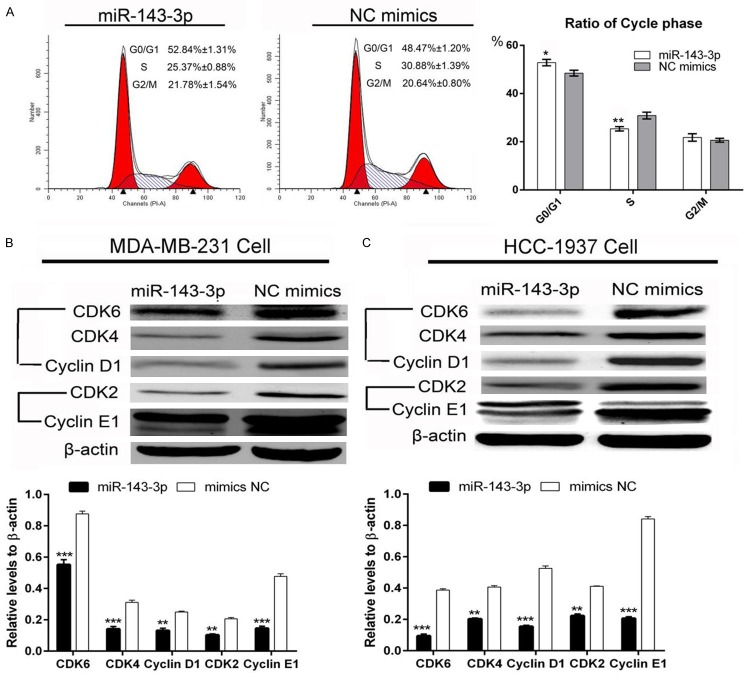
miR-143-3p arrested MDA-MB-231 cells at the G0/G1 cell phase
After treatment with miR-143-3p and NC mimics, cells were collected for flow cytometry analysis. Flow cytometry analysis results showed that miR-143-3p arrests MDA-MB-231 cells at the G0/G1 cell phase. In addition, the proportion of S phase cells decreased compared with the NC group (Figure 5A). Furthermore, we explored potential proteins that could affect the cell cycle. Expression of CDK4, 6/Cyclin D1 and CDK2/Cyclin E1 were also reduced by miR-143-3p in MDA-MB-231 and HCC-1937 breast cancer cells (Figure 5B and 5C). These data indicated that miR-143-3p arrest cells in the G0/G1 cell phase.
Figure 5.
Overexpression of miR-143-3p arrested MDA-MB-231 TNBC cells in the G0/G1 cell phase. A: The flow cytometry analysis indicated that miR-143-3p arrested MDA-MB-231 TNBC cells in the G0/G1 phase (student t-test, *P<0.05, **P<0.01); B and C: miR-143-3p suppressed the protein levels of potential regulated pathway components: CDK4, 6/Cyclin D1 and CDK2/Cyclin E1 (student t-test, **P<0.01, ***P<0.001).
miR-143-3p expression is decreased in breast cancer tissues
Furthermore, miRNA-seq data of miR-143-3P were retrieved from TCGA. In the TCGA database, we found that miR-143-3p expression was significantly decreased in breast cancer tissues compared with normal breast tissues (Figure 6).
Figure 6.
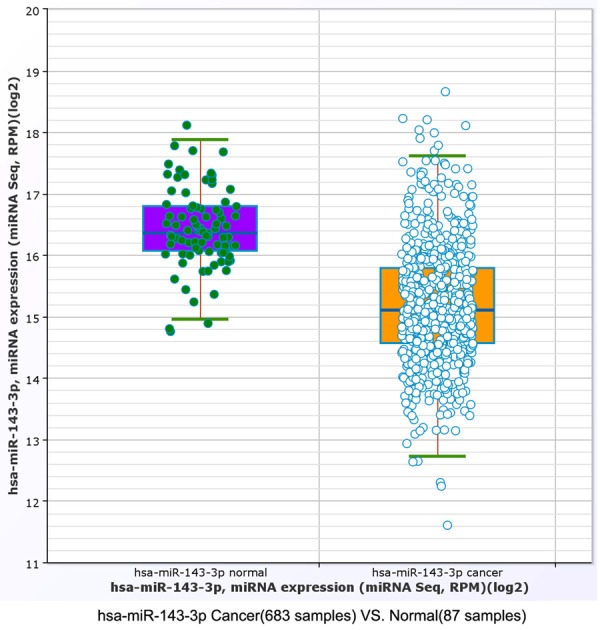
The TCGA database demonstrated that miR-143-3p expression was down-regulated in breast cancer tissues compared with normal breast tissues (student t-test, P=1.77636e-15; fold change =0.423152).
Discussion
According to several studies, TNBC accounts for approximately 15% of all breast cancer cases, and TNBC typically corresponds to a more aggressive clinical phenotype. The metastasis rate is 35% after 6-year follow-up [23]. To date, no specific therapy is available for TNBC, which typically exhibited a worse outcome than other types of breast cancers [24].
As one type of important epigenetic regulator, miRNAs play a key role in tumorigenesis and progression of BC [15]. The development of miRNA-based therapies has been one of the proposed strategies for BC treatment [25]. Indeed, several miRNAs has been reported to be involved in BC tumorigenesis and metastasis [15]. The inhibition of oncogenic miRNAs could be an innovative therapeutic approach for BC treatment [17,26,27].
Several studies reported reduced miR-143-3p expression in BC tissues [16-19]. In this study, we found that over-expression of miR-143-3p could significantly suppressed the proliferation of MDA-MB-231 BC cells. Given that PCNA protein typically reflects the trend of proliferation and is used as a clinical and experimental marker, we analyzed PCNA protein expression levels. In both MDA-MB-231 and HCC-1937 TNBC cancer cells, PCNA was significantly reduced upon treatment with the miR-143-3p mimic. In addition, we explored whether miR-143-3p inhibits TNBC cell migration and invasion. Wound healing and transwell assays showed that over-expression of miR-143-3p suppresses cell motility. In MDA-MB-231 and HCC-1937 cells, over-expression of miR-143-3p mimic caused a decrease in MMP2 and MMP9 protein expression. Our results confirm that miR-143-3p could be considered a tumor suppressor miRNA for TNBC, which is consistent with other findings [19,20,28]. Based on TargetScanPicTar and miRwalk database predictions, we found that LIMK1 may be a potential target gene for miR-143-3p. LIMK1 has only one known physiological substrate, CFL1, a small protein that is required for tumor cell movement [29]. During mitosis and/or meiosis, the LIMKs-CFL1 pathway may function as an important regulator of actin cytoskeletal rearrangements, chromosome segregation, and cytokinesis (REF). In addition, this pathway is involved in cell cycle regulation [9].
The plasmids containing the 3’-UTR of human LIMK1 with wild-type binding sites and/or mutant binding sites for miR-143-3p were bought from the IBS Biology Company (Shanghai, China). The plasmids were co-transferred into HEK293T cells together with mimic miR-143-3p. The luciferase activity assay showed that LIMK1 is a direct target gene of miR-143-3p. In the literature, it was reported that miR-143-3p targeted LIMK1 to inhibit the progression of lung cancer and prostate cancer [30-32]. In our study, we found that miR-143-3p inhibits the expression of LIMK1 at both the mRNA and protein level. Furthermore, miR-143-3p could inhibit the expression and phosphorylation of CFL1, which is the only known substrate of LIMK1. Flow cytometry analysis showed that miR-143-3p overexpression arrests MDA-MB-231 cells at the G0/G1 phase. We analyzed potential proteins that regulate this process in both MDA-MB-231 and HCC-1937 TNBC cells. The results showed that the expression of the CDK4, 6/Cyclin D1 complex and the CDK2/Cyclin E1 complex were suppressed by over-expression of miR-143-3p compared with the NC group. Our findings suggest that miR-143-3p inhibits TNBC cell proliferation through the control of CDK4, 6/Cyclin D1 and CDK2/Cyclin E and decreases the invasion ability of TNBC cells through the LIMK1/CFL1 pathway.
In conclusion, our findings suggest that miR-143-3p could act as a tumor suppressor miRNA by reducing the progression of TNBC cancer cells through partial regulation of the LIMK1/CFL1 pathway. Further work must be performed to clarify the role of this miRNA in in vivo animal models of TNBC.
Acknowledgements
This work was supported by grant no. 201640097 from the Shanghai Municipal Health Bureau of Shanghai, China and grant no. 82172240 from the National Natural Science Foundation of China (to L. Fang).
Disclosure of conflict of interest
None.
References
- 1.Cardoso F, Harbeck N, Barrios CH, Bergh J, Cortes J, El Saghir N, Francis PA, Hudis CA, Ohno S, Partridge AH, Sledge GW, Smith IE, Gelmon KA. Research needs in breast cancer. Ann Oncol. 2017;28:208–217. doi: 10.1093/annonc/mdw571. [DOI] [PubMed] [Google Scholar]
- 2.Santos JC, Ribeiro ML, Sarian LO, Ortega MM, Derchain SF. Exosomes-mediate microRNAs transfer in breast cancer chemoresistance regulation. Am J Cancer Res. 2016;6:2129–2139. [PMC free article] [PubMed] [Google Scholar]
- 3.Senkus E, Kyriakides S, Ohno S, Penault-Llorca F, Poortmans P, Rutgers E, Zackrisson S, Cardoso F ESMO Guidelines Committee. Primary breast cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26(Suppl 5):v8–30. [Google Scholar]
- 4.Feng T, Cao W, Shen W, Zhang L, Gu X, Guo Y, Tsai HI, Liu X, Li J, Zhang J, Li S, Wu F, Liu Y. Arctigenin inhibits STAT3 and exhibits anticancer potential in human triple-negative breast cancer therapy. Oncotarget. 2017;8:329–344. doi: 10.18632/oncotarget.13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reddy GM, Suresh PK, Pai RR. Clinicopathological features of triple negative breast carcinoma. J Clin Diagn Res. 2017;11:EC05–EC08. doi: 10.7860/JCDR/2017/21452.9187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maka VV, Pancha H, Shukla SN, Talati SS, Shah PM, Patel KM, Anand AS, Shah SA, Patel AA, Parikh S. Clinical characteristics and prognostic analysis of triple-negative breast cancer: single institute experience. Gulf J Oncolog. 2016;1:38–44. [PubMed] [Google Scholar]
- 7.Qiu J, Xue X, Hu C, Xu H, Kou D, Li R, Li M. Comparison of clinicopathological features and prognosis in triple-negative and non-triple negative breast cancer. J Cancer. 2016;7:167–173. doi: 10.7150/jca.10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mardilovich K, Baugh M, Crighton D, Kowalczyk D, Gabrielsen M, Munro J, Croft DR, Lourenco F, James D, Kalna G, McGarry L, Rath O, Shanks E, Garnett MJ, McDermott U, Brookfield J, Charles M, Hammonds T, Olson MF. LIM kinase inhibitors disrupt mitotic microtubule organization and impair tumor cell proliferation. Oncotarget. 2015;6:38469–38486. doi: 10.18632/oncotarget.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meng Y, Takahashi H, Meng J, Zhang Y, Lu G, Asrar S, Nakamura T, Jia Z. Regulation of ADF/cofilin phosphorylation and synaptic function by LIM-kinase. Neuropharmacology. 2004;47:746–754. doi: 10.1016/j.neuropharm.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 10.Su B, Su J, Zeng Y, Liu F, Xia H, Ma YH, Zhou ZG, Zhang S, Yang BM, Wu YH, Zeng X, Ai XH, Ling H, Jiang H, Su Q. Diallyl disulfide suppresses epithelial-mesenchymal transition, invasion and proliferation by downregulation of LIMK1 in gastric cancer. Oncotarget. 2016;7:10498–10512. doi: 10.18632/oncotarget.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.You T, Gao W, Wei J, Jin X, Zhao Z, Wang C, Li Y. Overexpression of LIMK1 promotes tumor growth and metastasis in gastric cancer. Biomed Pharmacother. 2015;69:96–101. doi: 10.1016/j.biopha.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 12.Bagheri-Yarmand R, Mazumdar A, Sahin AA, Kumar R. LIM kinase 1 increases tumor metastasis of human breast cancer cells via regulation of the urokinase-type plasminogen activator system. Int J Cancer. 2006;118:2703–2710. doi: 10.1002/ijc.21650. [DOI] [PubMed] [Google Scholar]
- 13.Peng F, Xiong L, Tang H, Peng C, Chen J. Regulation of epithelial-mesenchymal transition through microRNAs: clinical and biological significance of microRNAs in breast cancer. Tumour Biol. 2016;37:14463–14477. doi: 10.1007/s13277-016-5334-1. [DOI] [PubMed] [Google Scholar]
- 14.Casey MC, Sweeney KJ, Brown JA, Kerin MJ. Exploring circulating micro-RNA in the neoadjuvant treatment of breast cancer. Int J Cancer. 2016;139:12–22. doi: 10.1002/ijc.29985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jahagirdar D, Purohit S, Jain A, Sharma NK. Export of microRNAs: a bridge between breast carcinoma and their neighboring cells. Front Oncol. 2016;6:147. doi: 10.3389/fonc.2016.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng T, Zhang X, Wang Y, Yu X. Predicting associations between microRNAs and target genes in breast cancer by bioinformatics analyses. Oncol Lett. 2016;12:1067–1073. doi: 10.3892/ol.2016.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertoli G, Cava C, Castiglioni I. The potential of miRNAs for diagnosis, treatment and monitoring of breast cancer. Scand J Clin Lab Invest Suppl. 2016;245:S34–39. doi: 10.1080/00365513.2016.1208444. [DOI] [PubMed] [Google Scholar]
- 18.Sevinc ED, Cecener G, Ak S, Tunca B, Egeli U, Gokgoz S, Tolunay S, Tasdelen I. Expression and clinical significance of miRNAs that may be associated with the FHIT gene in breast cancer. Gene. 2016;590:278–284. doi: 10.1016/j.gene.2016.05.033. [DOI] [PubMed] [Google Scholar]
- 19.Yang Z, Chen D, Nie J, Zhou S, Wang J, Tang Q, Yang X. MicroRNA143 targets CD44 to inhibit breast cancer progression and stem cell-like properties. Mol Med Rep. 2016;13:5193–5199. doi: 10.3892/mmr.2016.5194. [DOI] [PubMed] [Google Scholar]
- 20.Yan X, Chen X, Liang H, Deng T, Chen W, Zhang S, Liu M, Gao X, Liu Y, Zhao C, Wang X, Wang N, Li J, Liu R, Zen K, Zhang CY, Liu B, Ba Y. miR-143 and miR-145 synergistically regulate ERBB3 to suppress cell proliferation and invasion in breast cancer. Mol Cancer. 2014;13:220. doi: 10.1186/1476-4598-13-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chavez KJ, Garimella SV, Lipkowitz S. Triple negative breast cancer cell lines: one tool in the search for better treatment of triple negative breast cancer. Breast Dis. 2010;32:35–48. doi: 10.3233/BD-2010-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Kong X, Li J, Luo Q, Li X, Shen L, Chen L, Fang L. miR-96 promotes tumor proliferation and invasion by targeting RECK in breast cancer. Oncol Rep. 2014;31:1357–1363. doi: 10.3892/or.2013.2934. [DOI] [PubMed] [Google Scholar]
- 23.Chen J, Shin VY, Siu MT, Ho JC, Cheuk I, Kwong A. miR-199a-5p confers tumor-suppressive role in triple-negative breast cancer. BMC Cancer. 2016;16:887. doi: 10.1186/s12885-016-2916-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masili-Oku SM, Bacchi CE, Fernandes FS, Filassi JR, Baracat EC, Carvalho FM. The apocrine profile of triple-negative breast carcinomas in patients aged 45 years or younger: favorable but rare features. Rev Bras Ginecol Obstet. 2016;38:512–517. doi: 10.1055/s-0036-1593854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gambari R, Brognara E, Spandidos DA, Fabbri E. Targeting oncomiRNAs and mimicking tumor suppressor miRNAs: nuew trends in the development of miRNA therapeutic strategies in oncology (Review) Int J Oncol. 2016;49:5–32. doi: 10.3892/ijo.2016.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5:1122–1143. doi: 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cava C, Bertoli G, Castiglioni I. Integrating genetics and epigenetics in breast cancer: biological insights, experimental, computational methods and therapeutic potential. BMC Syst Biol. 2015;9:62. doi: 10.1186/s12918-015-0211-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhai L, Ma C, Li W, Yang S, Liu Z. miR-143 suppresses epithelial-mesenchymal transition and inhibits tumor growth of breast cancer through down-regulation of ERK5. Mol Carcinog. 2016;55:1990–2000. doi: 10.1002/mc.22445. [DOI] [PubMed] [Google Scholar]
- 29.Cai S, Chen R, Li X, Cai Y, Ye Z, Li S, Li J, Huang H, Peng S, Wang J, Tao Y, Huang H, Wen X, Mo J, Deng Z, Wang J, Zhang Y, Gao X, Wen X. Downregulation of microRNA-23a suppresses prostate cancer metastasis by targeting the PAK6-LIMK1 signaling pathway. Oncotarget. 2015;6:3904–3917. doi: 10.18632/oncotarget.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertoli G, Cava C, Castiglioni I. MicroRNAs as biomarkers for diagnosis, prognosis and theranostics in prostate cancer. Int J Mol Sci. 2016;17:421. doi: 10.3390/ijms17030421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia H, Sun S, Wang B, Wang T, Liang C, Li G, Huang C, Qi D, Chu X. miR-143 inhibits NSCLC cell growth and metastasis by targeting Limk1. Int J Mol Sci. 2014;15:11973–11983. doi: 10.3390/ijms150711973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ngalame NN, Makia NL, Waalkes MP, Tokar EJ. Mitigation of arsenic-induced acquired cancer phenotype in prostate cancer stem cells by miR-143 restoration. Toxicol Appl Pharmacol. 2016;312:11–18. doi: 10.1016/j.taap.2015.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]