Abstract
RIZ1 has been considered as an important tumor suppressor gene. Our previous studies have already demonstrated that the expression of RIZ1 is closely related to the occurrence and development of meningioma. In addition, we also found that the expression of UbcH10 was related to the pathologic grade of meningioma which also affected the prognosis of these patients. However, we are lack of the understanding of the effect of UbcH10 on cell proliferation, cell apoptosis, cell cycle and other functions in meningioma cells. Besides, the regulation mechanism between RIZ1 and UbcH10 still remains unclear. In this study, we attempted to demonstrated that UbcH10 was a downstream target of RIZ1 and reported that UbcH10 silencing might negatively regulate cell proliferation, migration, invasion and promote apoptosis, which is similar to the cell phenotype that of over expressed RIZ1. Mechanistically, we proved that UbcH10 was a c-Myc target gene and that RIZ1 regulated UbcH10 expression in a c-Myc-dependent manner. For the first time, our study demonstrated that UbcH10 played a key role in the proliferation, metastasis and apoptosis of primary human malignant meningioma cells. In addition, the mechanism of RIZ1 regulating UbcH10 is also clear. Our study can also provide a potential target and new idea for the follow-up molecular intervention in clinical malignant meningiomas.
Keywords: c-Myc, meningioma, RIZ1, UbcH10
Introduction
Meningioma is one of the most common primary tumors of central nervous system (CNS), accounting for about 30% of intracranial primary tumors. In the light of 2016 World Health Organization (WHO) classification criteria, meningioma can be classified as benign, atypical or anaplastic (malignant) [1]. Compared with the benign ones, high-grade meningioma has worse prognosis and higher risk of recurrence. Despite advanced therapies for high-grade meningioma, including surgery and radiation, the prognosis of these patients have not been significantly improved [2,3]. Thus, other treatments, such as gene targeting therapy, have been rapidly developed, and finding new molecular targets for innovative approaches to reduce recurrence and improve survival is urgently needed [4].
Retinoblastoma protein-interacting zinc finger protein1 (RIZ1), which is considered to be an important tumor suppressor gene, is located on 1p36. Recent studies have indicated that the expression of RIZ1 is closely associated with the occurrence and development of glioma, breast cancer, hepatocellular carcinoma, colon cancer and other tumors [5-8]. In our previous studies, we found that the expression of RIZ1 protein was negatively correlated with the pathological grade of meningioma, and over-expression of RIZ1 in malignant meningioma cell lines can inhibit cell proliferation and activate G2/M checkpoint mechanism so as to promote its apoptosis [9]. Further study demonstrated that forced expression of RIZ1 could decrease the expression of c-Myc [10].
UbcH10, also named UBE2C, is an important member of ubiquitin-conjugating enzymes family, which exerts oncogenic function [11]. Our previous study found that the expression of UbcH10 protein in meningioma tissues was positively correlated with the pathological grade of meningiomas. Even more significantly, patients with higher UbcH10 expression had poor prognosis [12]. Bredel et al found that over-expression of c-Myc could promote the expression of UbcH10 [13]. Therefore, we hypothesized that RIZ1 may regulate the function of c-Myc and UbcH10 in meningiomas, and maintain the malignant proliferation of meningioma cells.
Materials and methods
Cell culture
Four primary meningioma cells (including M1: WHO grade I, M2: WHO grade II, M3: WHO grade II, M4: WHO grade III) were obtained from Changzheng Hospital, Shanghai, China [10]. All cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% fetal bovine serum (FBS). Cells were maintained at 37°C in a humidified atmosphere with 5% CO2. All of the procedures were reviewed and approved by Shanghai Changzheng Hospital Ethics Committees.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted using Trizol reagent (Invitrogen, USA) and was reverse-transcribed to cDNA using Prime Script RT Master Mix (Takara, Japan) according to the manufacturer’s instructions. The expression status of target genes and GAPDH were conducted by qRT-PCR using SYBR Premix Ex TaqTM II (Takara, Japan) on 7900HT Real-Time PCR system (Applied Biosystems, USA). Primers used were listed in Table 1. All of the samples were performed triplicate independently.
Table 1.
Primers sequences for quantitative PCR
| Primer | Primer sequence |
|---|---|
| RIZ1 | F: 5’-GGGACCTCGGAACTCAAC-3’ |
| R: 5’-TGTATCTGCCTGGGACTG-3’ | |
| c-Myc | F: 5’-AGGCTATTCTGCCCATTT-3’ |
| R: 5’-TCGTAGTCGAGGTCATAGTTC-3’ | |
| UbcH10 | F: 5’-GACCTGAGGTATAAGCTCTCGC-3’ |
| R: 5’-TTACCCTGGGTGTCCACGTT-3’ | |
| GAPDH | F: 5’-GGGAAACTGTGGCGTGAT-3’ |
| R: 5’-GAGTGGGTGTCGCTGTTGA-3’ |
Western blot
Primary meningioma cell extracts were prepared in RIPA Lysis Buffer (Beyotime, China). Protein concentrations were quantified by BCA assay (Thermo, USA). Protein (20 μg) were separated on SDS-PAGE and then transferred onto PVDF membranes (Millipore, USA). Besides, the membranes were blocked with 5% skim milk in TBST for 1 h and then incubated overnight at 4°C with primary antibody: RIZ1 (1:1000, Abcam, UK), c-Myc, UbcH10, Bax, Bcl-2, caspase3, GAPDH (1:1000, Proteintech, USA). After the membranes being washed, secondary antibodies (1:4000, Proteintech, USA) were used for 1 h at room temperature and chemiluminescence was detected through using luminol reagent (Santa Cruz, USA).
Samples collection and immunohistochemistry analysis
Shanghai Changzheng Hospital Ethics Committees reviewed and approved this study. All patients were performed meningioma resection at Changzheng Hospital, Shanghai, China, from June 2005 to June 2013. Before an operation, each patient was given informed written consent. Clinical tissue samples were obtained. The grade of pathology was determined by two pathology experts in our hospital based on the 2016 WHO classification of tumors of the CNS. Tissue samples treated by formalin fixed with paraffin embedding, specimens were then cut into 3-μm-thick sample sections. They were deparaffinized in xylene and rehydrated through graded alcohols. Microwave method was used for antigen retrieval. After the devitalization of endogenous peroxidase activity, the slides were blocked with 10% normal goat serum for 30 min to avoid nonspecific binding. Then the sections were incubated with RIZ1 antibody (Abcam, UK) and UbcH10 antibody (Proteintech, USA) at 4°C overnight. Next day, the sections were washed in TBST and then incubated with biotin-labeled secondary antibody (Proteintech, USA) for 30 min at 37°C. Then the sections were performed for visualization by the horseradish peroxidase streptavidin-biotin complex, and counterstained with hematoxylin. The results were visualized with 3,3’-diaminobenzidine (DAB). Two experienced pathology experts carried out the assessment of the slides independently. Percentage of positive cells was calculated by means of counting under high magnification (×400).
Transfection of siRNA
M1 (WHO I) and M4 (WHO III) cells were transiently transfected with siRNA targeting RIZ1 and UbcH10 respectively based on HiPerFect Transfection Reagent (QIAGEN, Germany). The sequence of RIZ1 siRNA was 5’-GAUCCAGAGAAGGGAAACUTT-3’, sequence of UbcH10 siRNA was: 5’-CCUGCAAGAAACCUACUCA-3’. A non-specific scramble siRNA sequence was used as negative control (NC), sequence was 5’-UUCUCCGAACGUGUCACGUTT-3’.
Cell proliferation assay
Transfected and non-transfected M4 (WHO III) cells were seeded at a density of 2×103 cells per well into 96-well plates. Cell proliferation was detected by using the Cell Proliferation Reagent WST-1 (Roche, Germany). At specified time points, 10 μl WST-1 was added per well and incubated for 4 h. Optical density (OD) was measured with micro-plate reader (KHB ST-360, China) at 450 nm wavelength. At least three wells were observed in each treatment group.
Cell cycle assay
Transfected and non-transfected M4 (WHO III) cells were harvested and fixed in 70% ice-cold ethanol centrifuged at 4°C for at least 18 h. After being washed with PBS, cells were incubated with PI/RNase Staining Buffer (BD Biosciences, USA) for 15 minutes at room temperature. The results were analyzed by a FACScan flow cytometry (BD Biosciences, USA).
Cell migration and invasion assay
Quantitative cell migration and invasion assays were carried out using 24-well Transwell and Matrigel Invasion Chamber (Corning, USA), respectively. Transfected and non-transfected M4 (WHO III) cells (4×104) were seeded into the upper well of the chambers with serum-free medium and medium containing 10% FBS in the lower chamber. After incubation at 37°C for 24 h, cells that migrated or invaded through the filters were fixed and stained. Three random fields were counted under the microscope.
Apoptosis assay
Transfected and non-transfected M4 (WHO III) cells were harvested and washed with cold PBS, resuspended in 1× binding buffer, mixed with 5 μL of PE Annexin V and 5 μL of 7-AAD (BD Biosciences, USA) according to manufacturer’s instructions, and then incubated at room temperature for 15 min in the dark. The results were analyzed by a FACScan flow cytometry (BD Biosciences, USA).
Plasmids
The over-expression vector of RIZ1 gene pCMV-RIZ1 was obtained from Changzheng Hospital, Shanghai, China [9]. The expressing plasmid which carried c-Myc gene pIRES2-c-Myc was purchased from Obio Technology Corp., Ltd., Shanghai, China.
Lentivirus infection
The pLKD-CMV-R&PR-U6-shRNA-c-Myc lentiviral vector was purchased from Obio Technology Corp., Ltd., Shanghai, China and polybrene was used to improve transfection efficiency. Based on medium containing puromycin, the stable expression clones were screened. The clones stably knocking down c-Myc were validated by qRT-PCR and western blotting. The sequence of shRNA targeting c-Myc (NM_002467) is 5’-GGCGAACACACAACGTCTT-3’. An empty lentiviral vector was used as a negative control.
Chromatin immunoprecipitation (ChIP) assay
Chip assays were performed through using PierceTMAgaroseChIP Kit (Thermo, USA) according to the manufacturer’s protocol. Anti-c-Myc (Abcam, UK) was used for immunoprecipitation. 3000 bases upstream of translation initiation codon ATG, which containing binding site of c-Myc in the promoter of UbcH10, were analyzed. A pair of primers were respectively designed every 300 bases, which were used to amplify specific UbcH10 promoter regions. Sequences of these primers were listed in Table 2. Besides, this assay was carried out in M3 (WHO II) and M4 (WHO III) cells.
Table 2.
Primers sequences for ChIP assay
| Primer | Primer sequence |
|---|---|
| Primer-1 (-3000/-2701 bp) | F: 5’-TTGAGGCAGCAACCCTTG-3’ |
| R: 5’-ATAAAATAAAAGGTAGCT-3’ | |
| Primer-2 (-2700/-2401 bp) | F: 5’-TTTACTACTCTATTTATGCC-3’ |
| R: 5’-ATTCCTTGAGTAATTTGT-3’ | |
| Primer-3 (-2400/-2101 bp) | F: 5’-ACACAATTATTCCTGACC-3’ |
| R: 5’-AAAGCCAGCAAAATATAT-3’ | |
| Primer-4 (-2100/-1801 bp) | F: 5’-CTTGCCATTTTGTTATAACC-3’ |
| R: 5’-GGGGATGCGGTTTACAAT-3’ | |
| Primer-5 (-1800/-1501 bp) | F: 5’-CGCTTCCACCTAGTAGCA-3’ |
| R: 5’-CTTCAGATCCAGGCATCC-3’ | |
| Primer-6 (-1500/-1201 bp) | F: 5’-CTAGAGGAATTGAAATCC-3’ |
| R: 5’-TTTACAAATATAGAACCA-3’ | |
| Primer-7 (-1200/-901 bp) | F: 5’-GTTATTGGGATAAGAAACAAT-3’ |
| R: 5’-CAACTCTATTTTTCATAT-3’ | |
| Primer-8 (-900/-601 bp) | F: 5’-GCTTTTATTGCTCCCATT-3’ |
| R: 5’-AGGATAGGGAGACCTGTC-3’ | |
| Primer-9 (-600/-301 bp) | F: 5’-GTTGCTCAGGCTGGTCTC-3’ |
| R: 5’-AGTGCCTCCAATCCTGCA-3’ | |
| Primer-10 (-300/-1 bp) | F: 5’-CCCTGCCCGAGGGAAATTGGATC-3’ |
| R: 5’-GCTTGGGTTTGATCCAGC-3’ |
Construction of UbcH10 reporter plasmid
The human UbcH10 upstream region (-2000/-1 bp relative to the translational codon ATG) was amplified by PCR and cloned into the pGL3-basic luciferase reporter vector. The plasmid pGL3-UbcH10 was used as a template for conducting experiments of site-directed mutagenesis. KOD-Plus-Neo (TOYOBO, Japan) was used for point mutation PCR to induce mutations into the two c-Myc binding sites. Site 1 located at -1329/-1324 bp and Site 2 located at -212/-207 bp were respectively mutated, which were designated as UbcH10-mutation1 and UbcH10-mutation2 (UbcH10-mutation1: -1329/-1324 bp, CACGTG→GTGGAC and UbcH10-mutation2: -212/-207 bp, CACGTG→GTGGAC). At the same time, the two sites were simultaneously mutated which was called UbcH10-mutation1&2. All the gene segments were verified based on sequence analysis. Besides, primers used for point mutation PCR were listed in Table 3.
Table 3.
Primers sequences for promoter constructs and mutagenesis
| Primer | Primer sequence |
|---|---|
| Mutation-1 | F: 5’-CCTCTTCCTGTGGACGCTGAGGCCACCGCTGGGAC-3’ |
| R: 5’-GTCCACAGGAAGAGGGCTTTGGAGGA-3’ | |
| Mutation-2 | F: 5’-GGTTCATCCGTGGACGACGTTTTCTTGGCCCTTTA-3’ |
| R: 5’-GTCCACGGATGAACCCCTCCTTCTGG-3’ |
Luciferase reporter assay
M4 (WHO III) cells were seeded in 96-well plates and pCMV-RIZ1 or pIRES2-c-Myc was co-transfected with luciferase reporter plasmids. Besides, a Renilla luciferase reporter gene vector was used as an internal control and Luciferase activity was measured through using Dual-Luciferase Reporter Assay System (Promega, USA) and normalized to Renilla luciferase activity.
Statistical analysis
All data are presented as the mean ± SD of three repeated experiments. Statistical analysis was performed using Student’s t-test and one-way ANOVA followed by conducting Dunnett’s t test. Spearman rank correlation test was used to research the relationship between RIZ1 and UbcH10. In addition, statistically significant difference was accepted at P values <0.05 and statistical analysis were also performed through using the software SPSS 19.0 and Graphpad Prism 5 was also used for data plotting.
Results
UbcH10 is a target of RIZ1 in meningioma
The expression of RIZ1 and UbcH10 in four primary meningioma cells was detected by western blotting. According to the results, it can be found that the expression of RIZ1 decreased with the increase of pathological grade. On the contrary, the UbcH10 expression was positively correlated with the malignancy grade (Figure 1A). Subsequently, we transfected pCMV-RIZ1 into M4 (WHO III) cells, and then decreased expression of UbcH10 was detected (Figure 1B). Conversely, downregulation of RIZ1 expression by siRNA in M1 (WHO I) cells resulted in increased expression of UbcH10 (Figure 1C). In addition, immunohistochemistry revealed high expression of RIZ1 in the cytoplasm and nucleus of benign meningiomas, whereas the staining was significantly reduced in meningiomas with the increase of pathological grade. The UbcH10 detected in cell nucleus was significantly lower in benign meningiomas than that in atypical/anaplastic meningiomas (Figure 1D). Besides, the research findings also demonstrated that RIZ1 expression had an obvious negative correlation with UbcH10 expression through using Spearman rank correlation (P<0.0001, Figure 1G). In summary, these data strongly suggest that UbcH10 is a downstream molecule of RIZ1.
Figure 1.
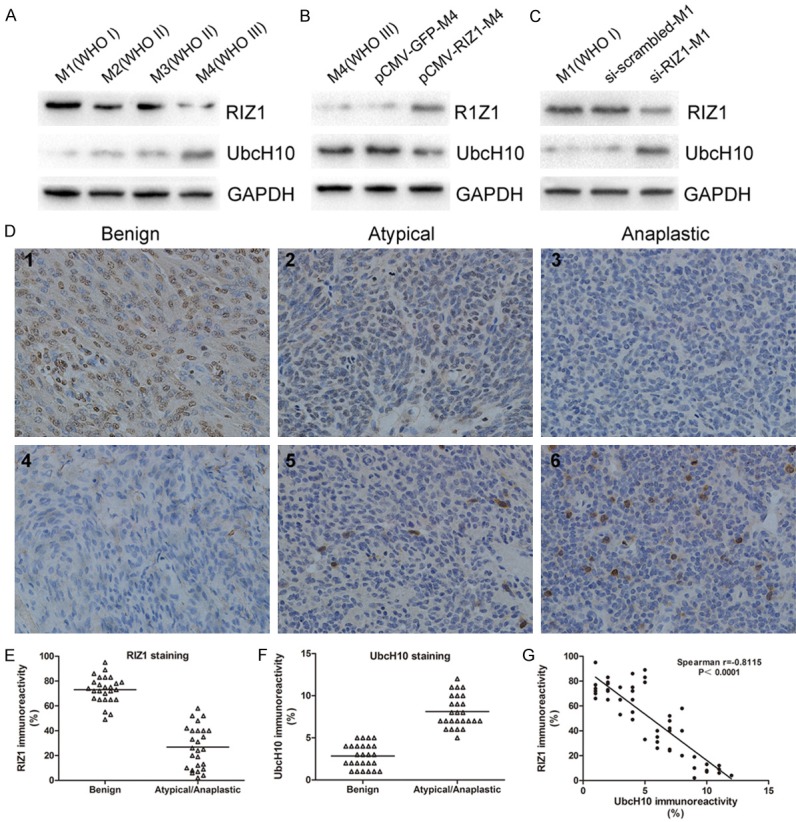
Statistical analysis of the correlation between the expression of RIZ1 and UbcH10 in meningiomas. A: Expression of RIZ1 decreased with the increase of pathological grade and the UbcH10 expression was positively correlated with the malignancy grade in primary meningioma cells. B: Overexpression of RIZ1 decreased the UbcH10 protein level in M4 (WHO III) cells. C: RIZ1 silencing increased the UbcH10 Protein level in M1 (WHO I) cells. D: High expression of RIZ1 in the cytoplasm and nucleus of benign meningiomas (1), whereas the staining was significantly reduced in meningiomas with the increase of pathological grade (2, 3). The UbcH10 detected in cell nucleus was significantly lower in benign meningiomas (4) than that in atypical/anaplastic meningiomas (5, 6) by immunohistochemistry. E, F: Quantitative analysis of RIZ1 and UbcH10 expression by immunohistochemistry in meningiomas of different grades. G: RIZ1 and UbcH10 expression showed a positive correlation in meningioma patient samples.
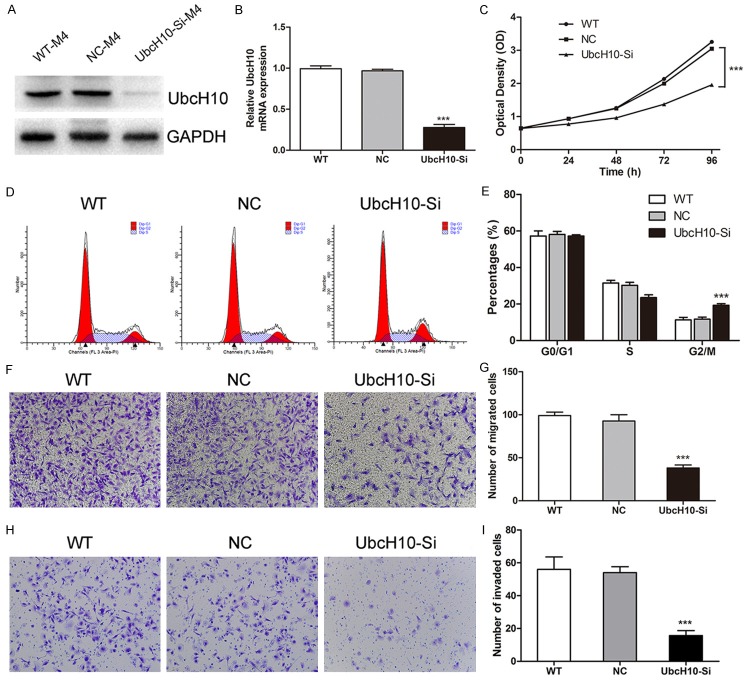
Knockdown of UbcH10 in primary meningioma cells
Based on previous western blotting (Figure 1A), we found than M4 (WHO III) cells had highest UbcH10 expression. As a result, we knocked down the expression of UbcH10 in M4 (WHO III) cells by siRNA transfection to investigate the effects of RNA interference on cell proliferation, cell cycle, cell migration, cell invasion, and cell apoptosis. By western blotting and qRT-PCR, the silence efficiency of siRNA was analyzed. As shown in Figure 2A and 2B, UbcH10 expression level was down-regulated in siRNA group compared with NC group. Then, we began to conduct the study of meningioma cell function. Firstly, we assessed the effect of siRNA on the proliferation through using WST-1 assay. Compared with NC group, cell proliferation was significantly inhibited in M4 (WHO III) cells with UbcH10 silenced (Figure 2C). In the next place, we analyzed the cell cycle distribution based on flow cytometry, indicating that the population of G2/M phase in M4 (WHO III) cells transfected with UbcH10 siRNA was significantly increased to 19.29±0.87%. However, in WT and NC groups, the proportion of the G2/M phase was still maintained at 11.29±1.37% and 11.68±1.13%, respectively (P<0.001, Figure 2D and 2E). Then, we investigated the effect of RNA interference on cell motility through transwell assays. The results proved that suppressing of UbcH10 expression brought out a strong inhibitory motility of M4 (WHO III) cells (Figure 2F and 2G). At the same time, we investigated the influence of UbcH10 siRNA on the invasion ability of meningiomas cells. In vitro cell invasion assay, the invasion ability of M4 (WHO III) cells was significantly decreased by UbcH10 siRNA interference compared with NC group (Figure 2H and 2I). Finally, we explored the influence of UbcH10 knockdown on apoptosis in M4 (WHO III) cells. The number of apoptotic cells was significantly increased by interference with UbcH10 siRNA at different time points post transfection (P<0.01, Figure 3A and 3B). In order to further study the mechanism of UbcH10 siRNA promoting cell apoptosis, we detected the expression level of apoptosis related proteins, Bax, Bcl-2 and caspase3. When transfected with UbcH10 siRNA 48 h, the expression of Bax and caspase3 were dramatically increased, whereas Bcl-2 expression was significantly decreased (Figure 3C). These results imply that UbcH10 is an important molecule influencing the occurrence and development of meningiomas.
Figure 2.
Silencing of UbcH10 inhibits cell proliferation, G2/M phase transition, migration and invasion in primary malignant meningioma cells. A, B: UbcH10 protein and mRNA level was down-regulated in siRNA group compared with NC group in M4 (WHO III) cells transfected with siRNA targeting UbcH10. WT: wild type cells; NC: scrambled siRNA transfected cells. C: siRNA targeting UbcH10 inhibited M4 (WHO III) cell growth as determined by WST-1 assay. D, E: Effect of siRNA-UbcH10 on cell cycle in M4 (WHO III) cells was detected through using flow cytometry, the population of G2/M phase was significantly increased after transfection. F-I: Silencing of UbcH10 inhibited cell migration and invasion in M4 (WHO III) cells. Data were based on at least 3 independent experiments, and shown as mean ± S.D. (*P<0.05, **P<0.01, ***P<0.001 as compared with NC).
Figure 3.
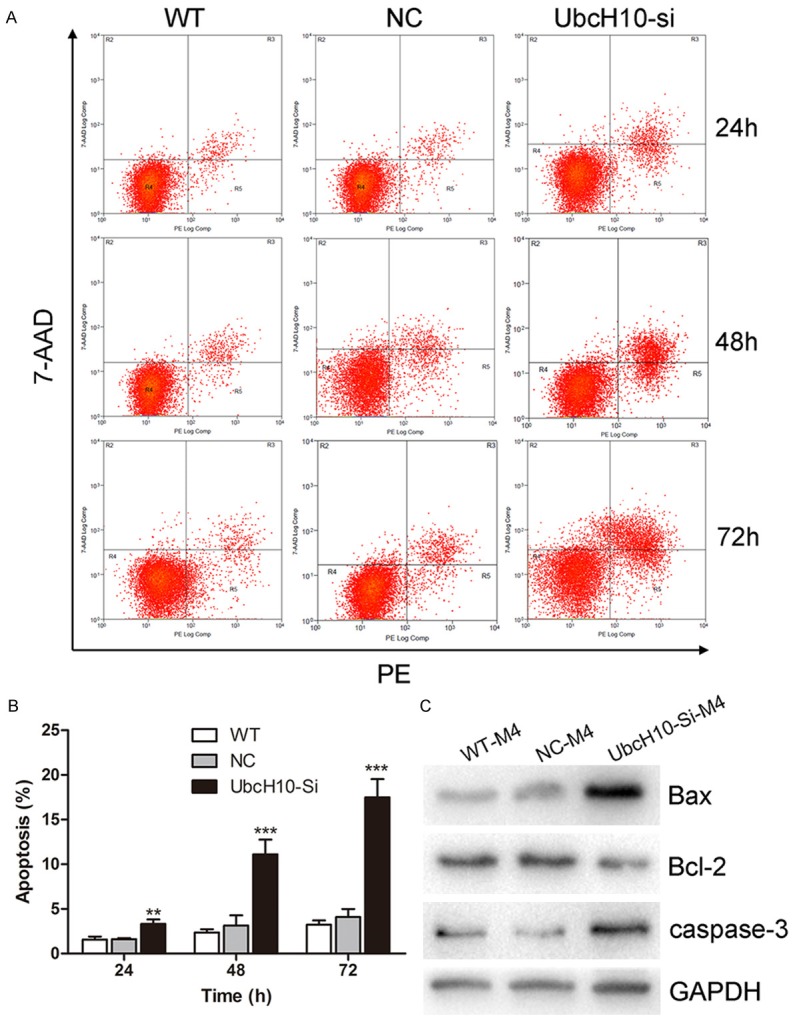
Effect of siRNA targeting UbcH10 on cell apoptosis in M4 (WHO III) cells. A: Induction of apoptosis was measured by PE/7-AAD staining at 24 h, 48 h, 72 h time point post transfection. B: Apoptotic cell population were significantly increased after UbcH10 siRNA transfection compared with NC group in M4 (WHO III) cells. C: Effect of UbcH10 siRNA on expression of Bax, Bcl-2, caspase3 protein in M4 (WHO III) cells, when transfected with UbcH10 siRNA 48 h, the expression of Bax and caspase3 were dramatically increased, whereas Bcl-2 expression was significantly decreased. Data were based on at least 3 independent experiments, and shown as mean ± S.D. (*P<0.05, **P<0.01, ***P<0.001 as compared with NC).
RIZ1 regulates UbcH10 in a c-Myc-dependent manner
To determine whether RIZ1 regulates UbcH10 through c-Myc, we first validated that c-Myc was able to bind to the UbcH10 promoter region through performing ChIP assays. Both in M3 (WHO II) and M4 (WHO III) cells, we detected two binding regions of c-Myc in the promoter region of UbcH10, which were located in -1500/-1201 bp and the -300/-1 bp (Figure 4A). These two regions contained E-box elements, consisting of the CACGTG nucleotide sequence and could be occupied by c-Myc Figure 4B). Site 1 was located at -1329/-1324 bp and Site 2 was found at -212/-207 bp. To confirm the function of the two c-Myc binding sites, we constructed the luciferase reporter plasmid pGL3-UbcH10, pGL3-UbcH10-mutation1, pGL3-UbcH10-mutation2 and pGL3-UbcH10-mutation1&2, as described in methods section. The luciferase reporter gene assay indicated that co-transfection with pGL3-UbcH10 and pIRES2-c-Myc increased UbcH10 promoter activity, while the mutation constructs pGL3-UbcH10-mutation1 and pGL3-UbcH10-mutation2 both inhibited the transcriptional activity compared with pGL3-UbcH10, decreased by about 50% respectively. Expectedly, co-transfected with pIRES2-c-Myc and pGL3-UbcH10-mutation1&2 did not make influence on the luciferase activity (Figure 4C). These results suggested that both of the two c-Myc binding sites had imposed a regulatory effect on the UbcH10 promoter.
Figure 4.
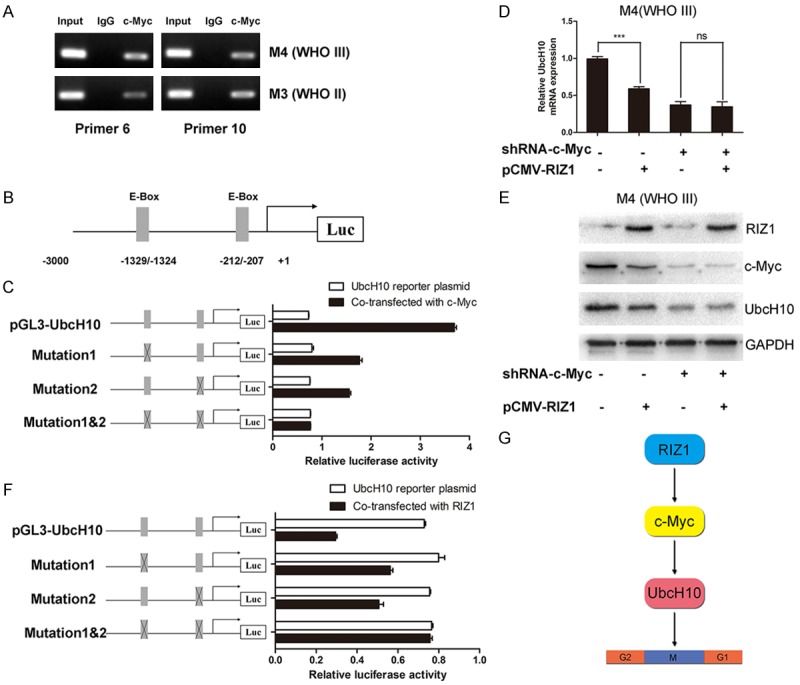
RIZ1 regulates UbcH10 expression via c-Myc. A: c-Myc occupies the E-box of the UbcH10 prompter region, as measured by ChIP assay in M3 (WHO II) and M4 (WHO III) cells, primer 6 and primer 10 showed DNA positive strand. B: c-Myc binding sites in the UbcH10 promoter were located in -1500/-1201 bp and -300/-1 bp. C: Luciferase reporter gene assay indicated that co-transfection with pGL3-UbcH10 and pIRES2-c-Myc increased UbcH10 promoter activity, while the mutation constructs pGL3-UbcH10-mutation1 and pGL3-UbcH10-mutation2 both inhibited the transcriptional activity compared with pGL3-UbcH10. Co-transfected with pIRES2-c-Myc and pGL3-UbcH10-mutation1&2 did not make influence on the luciferase activity. The sketches of wild-type and mutated reporter constructs of UbcH10 were showed on the leftside. D, E: Transfection of pCMV-RIZ1 into M4 (WHO III) cells induced a significant decrease in UbcH10 expression, however, almost no reduction of UbcH10 expression was observed after transfection of pCMV-RIZ1 into the c-Myc shRNA-transduced M4 (WHO III) cells, both at the mRNA level and protein level. F: Luciferase reporter gene assay revealed that co-transfected with RIZ1 and pGL3-UbcH10 decreased the luciferase activity, whereas it exerted no significant effect on UbcH10 promoter activity when the reporter plasmid was pGL3-UbcH10-mutation1&2. G: Proposed model of the mechanism of RIZ1 inhibits the occurrence and development of meningioma via the c-Myc/UbcH10 axis. (*P<0.05, **P<0.01, ***P<0.001 as compared with NC).
In order to verify whether RIZ1 regulated UbcH10 in a c-Myc dependent manner, we established the c-Myc stably knocked-down M4 (WHO III) cells by the transfection of lentivirus vector. Transfection of pCMV-RIZ1 into M4 (WHO III) cells induced a significant decrease in UbcH10 expression. Nevertheless, almost no reduction of UbcH10 expression was observed after transfection of pCMV-RIZ1 into the c-Myc shRNA-transduced M4 (WHO III) cells, both at the mRNA level and protein level (Figure 4D and 4E). In addition, luciferase reporter gene assay revealed that co-transfected with RIZ1 and pGL3-UbcH10 decreased the luciferase activity, whereas it exerted no significant effect on UbcH10 promoter activity when the reporter plasmid was pGL3-UbcH10-mutation1&2 (Figure 4F). Briefly, these results demonstrate that RIZ1 inhibits tumor progression in meningioma based on the c-Myc/UbcH10 axis (Figure 4G).
Discussion
As the common intracranial tumors, the main treatments of meningiomas include surgical operation and radiation treatment, etc. In recent years, with the progress of the treatment concept and method, the prognosis of patients and quality of life have been significantly improved. However, for the patients with atypical or anaplastic meningiomas, the rate of recurrence after completely resection in 5 years was still up to 38% and 78% respectively [3]. Similar to other tumors, the occurrence and malignant evolution of meningiomas were concerned with numerous factors. New treatments, such as gene targeting therapy, are expected to be one of the means to cure meningioma, yet there is still lack of ideal therapeutic targets. Therefore, it has become one of the hot spots in the field of neuroscience to search for the proteins that regulate the malignant biological behavior of meningiomas, as well as to clarify the pathological mechanism in the pathogenesis of meningiomas.
RIZ was confirmed to have a close relationship with various types of tumors. Our previous study demonstrated that the expression of RIZ1 was extremely low in malignant meningiomas, indicating RIZ1 played an important role in the progression of meningiomas [9]. In addition, UbcH10 is an important checkpoint proteins of G2/M phase, which is vital for chromosome segregation [14,15]. The abnormal expression of UbcH10 may lead to an unusually early spindle assembly checkpoint termination signal by degradation of Cyclin A, Cyclin B1 and other cell cycle proteins. It will result in an increase number of chromosome missegregation, which may be the molecular pathological mechanism of malignant meningioma proliferation [11,16,17]. Several recent studies have indicated that knockdown of UbcH10 expression in tumor cells leads to cell G2/M arrest, which is similar to the cell phenotype that of over expressed RIZ1 [18-20]. For a long time, the c-Myc gene has been known to be closely associated with the occurrence and development of many malignant tumors. Kazumoto et al found that the expression rate of c-Myc in meningioma was 63%, which is significantly higher than that in normal brain tissue [21]. In addition, Detta A et al also reported increased expression of c-Myc in meningiomas, which revealed that the high expression of c-Myc had an important role in meningiomas [22]. Most interestingly, as an important transcription factor, c-Myc can directly bind to the promoter region of multiple genes [23,24]. Previous reports indicated that c-Myc expression decreased after forced expression of RIZ1 and over-expression of c-Myc could promote the expression of UbcH10 [9,13]. Consequently, RIZ1 and c-Myc have the premise and foundation to regulate UbcH10. Combined with early studies, we inferred that RIZ1 may regulate the proliferation of meningioma cells by acting on the checkpoint protein UbcH10. In order to test the above hypothesis, we firstly investigated whether c-Myc could directly bind to the UbcH10 promoter. We found that there are two stable c-Myc binding sites in the promoter region of UbcH10, and c-Myc could increaseUbcH10 promoter activity and promote UbcH10 expression. Finally, we validated that RIZ1 regulated UbcH10 in a c-Myc dependent manner through constructing mutated reporter plasmids of UbcH10 promoter region.
To conclude, our study primarily demonstrated that UbcH10 played an important role in the proliferation, metastasis and apoptosis of primary primary human malignant meningioma cells. In addition, the mechanism of RIZ1 regulating UbcH10 is also clear. This study proved that RIZ1 is an essential molecule in the occurrence and development of meningiomas. In addition, our study also provides a potential target as well as new idea for the follow-up molecular intervention in clinical malignant meningiomas.
Acknowledgements
This work was supported by the National Natural Science Fund Project of China (No. 81372717).
Disclosure of conflict of interest
None.
References
- 1.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Soussain C, Ricard D, Fike JR, Mazeron JJ, Psimaras D, Delattre JY. CNS complications of radiotherapy and chemotherapy. Lancet. 2009;374:1639–1651. doi: 10.1016/S0140-6736(09)61299-X. [DOI] [PubMed] [Google Scholar]
- 3.Maier H, Ofner D, Hittmair A, Kitz K, Budka H. Classic, atypical, and anaplastic meningioma: three histopathological subtypes of clinical relevance. J Neurosurg. 1992;77:616–623. doi: 10.3171/jns.1992.77.4.0616. [DOI] [PubMed] [Google Scholar]
- 4.Wen PY, Quant E, Drappatz J, Beroukhim R, Norden AD. Medical therapies for meningiomas. J Neurooncol. 2010;99:365–378. doi: 10.1007/s11060-010-0349-8. [DOI] [PubMed] [Google Scholar]
- 5.Zhang C, Zhu Q, He H, Jiang L, Qiang Q, Hu L, Hu G, Jiang Y, Ding X, Lu Y. RIZ1: a potential tumor suppressor in glioma. BMC Cancer. 2015;15:990. doi: 10.1186/s12885-015-2023-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chadwick RB, Jiang GL, Bennington GA, Yuan B, Johnson CK, Stevens MW, Niemann TH, Peltomaki P, Huang S, de la Chapelle A. Candidate tumor suppressor RIZ is frequently involved in colorectal carcinogenesis. Proc Natl Acad Sci U S A. 2000;97:2662–2667. doi: 10.1073/pnas.040579497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He L, Yu JX, Liu L, Buyse IM, Wang MS, Yang QC, Nakagawara A, Brodeur GM, Shi YE, Huang S. RIZ1, but not the alternative RIZ2 product of the same gene, is underexpressed in breast cancer, and forced RIZ1 expression causes G2-M cell cycle arrest and/or apoptosis. Cancer Res. 1998;58:4238–4244. [PubMed] [Google Scholar]
- 8.Jiang G, Liu L, Buyse IM, Simon D, Huang S. Decreased RIZ1 expression but not RIZ2 in hepatoma and suppression of hepatoma tumorigenicity by RIZ1. Int J Cancer. 1999;83:541–546. doi: 10.1002/(sici)1097-0215(19991112)83:4<541::aid-ijc17>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 9.Liu ZY, Wang JY, Liu HH, Ma XM, Wang CL, Zhang XP, Tao YQ, Lu YC, Liao JC, Hu GH. Retinoblastoma protein-interacting zinc-finger gene 1 (RIZ1) dysregulation in human malignant meningiomas. Oncogene. 2013;32:1216–1222. doi: 10.1038/onc.2012.155. [DOI] [PubMed] [Google Scholar]
- 10.Ding MH, Wang Z, Jiang L, Fu HL, Gao J, Lin XB, Zhang CL, Liu ZY, Shi YF, Qiu GZ, Ma Y, Cui DX, Hu GH, Jin WL. The transducible TAT-RIZ1-PR protein exerts histone methyltransferase activity and tumor-suppressive functions in human malignant meningiomas. Biomaterials. 2015;56:165–178. doi: 10.1016/j.biomaterials.2015.03.058. [DOI] [PubMed] [Google Scholar]
- 11.van Ree JH, Jeganathan KB, Malureanu L, van Deursen JM. Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J Cell Biol. 2010;188:83–100. doi: 10.1083/jcb.200906147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang L, Wang T, Bao Y, Qian J, Wu XJ, Hu GH, Lu YC. A study of UbcH10 expression and its association with recurrence of meningiomas. J Surg Oncol. 2012;106:327–331. doi: 10.1002/jso.22141. [DOI] [PubMed] [Google Scholar]
- 13.Bredel M, Scholtens DM, Harsh GR, Bredel C, Chandler JP, Renfrow JJ, Yadav AK, Vogel H, Scheck AC, Tibshirani R, Sikic BI. A network model of a cooperative genetic landscape in brain tumors. JAMA. 2009;302:261–275. doi: 10.1001/jama.2009.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Gramont A, Ganier O, Cohen-Fix O. Before and after the spindle assembly checkpoint--an APC/C point of view. Cell Cycle. 2006;5:2168–2171. doi: 10.4161/cc.5.18.3219. [DOI] [PubMed] [Google Scholar]
- 15.Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124:89–103. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 16.Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- 17.Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 2004;432:588–595. doi: 10.1038/nature03023. [DOI] [PubMed] [Google Scholar]
- 18.Chen SM, Jiang CY, Wu JY, Liu B, Chen YJ, Hu CJ, Liu XX. RNA interference-mediated silencing of UBCH10 gene inhibits colorectal cancer cell growth in vitro and in vivo. Clin Exp Pharmacol Physiol. 2010;37:525–529. doi: 10.1111/j.1440-1681.2009.05348.x. [DOI] [PubMed] [Google Scholar]
- 19.Wang ST, Li DZ, Li JM, Fang J, Li HZ, Tong PJ, Liu FC. Lentivirus-mediated RNA interference targeting UbcH10 reduces cell growth and invasion of human osteosarcoma cells via inhibition of Ki-67 and matrix metalloproteinases. Oncol Lett. 2015;9:2171–2176. doi: 10.3892/ol.2015.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang L, Huang CG, Lu YC, Luo C, Hu GH, Liu HM, Chen JX, Han HX. Expression of ubiquitin-conjugating enzyme E2C/UbcH10 in astrocytic tumors. Brain Res. 2008;1201:161–166. doi: 10.1016/j.brainres.2008.01.037. [DOI] [PubMed] [Google Scholar]
- 21.Kazumoto K, Tamura M, Hoshino H, Yuasa Y. Enhanced expression of the sis and c-myc oncogenes in human meningiomas. J Neurosurg. 1990;72:786–791. doi: 10.3171/jns.1990.72.5.0786. [DOI] [PubMed] [Google Scholar]
- 22.Detta A, Kenny BG, Smith C, Logan A, Hitchcock E. Correlation of proto-oncogene expression and proliferation and meningiomas. Neurosurgery. 1993;33:1065–1074. doi: 10.1227/00006123-199312000-00015. [DOI] [PubMed] [Google Scholar]
- 23.Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res. 2009;15:6479–6483. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller DM, Thomas SD, Islam A, Muench D, Sedoris K. c-Myc and cancer metabolism. Clin Cancer Res. 2012;18:5546–5553. doi: 10.1158/1078-0432.CCR-12-0977. [DOI] [PMC free article] [PubMed] [Google Scholar]