Abstract
Forkhead box, class O (FOXO) transcription factors are major regulators of diverse cellular processes, including fuel metabolism, oxidative stress response and redox signalling, cell cycle progression and apoptosis. Their activities are controlled by multiple posttranslational modifications and nuclear‐cytoplasmic shuttling. Recently, post‐transcriptional regulation of FOXO synthesis has emerged as a new regulatory level of their functions. Accumulating evidence suggests that this post‐transcriptional mode of regulation of FOXO activity operates in response to stressful stimuli, including oxidative stress. Here, we give a brief overview on post‐transcriptional regulation of FOXO synthesis by microRNAs (miRNAs) and by RNA‐binding regulatory proteins, human antigen R (HuR) and quaking (QKI). Aberrant post‐transcriptional regulation of FOXOs is frequently connected with various disease states. We therefore discuss characteristic examples of FOXO regulation at the post‐transcriptional level under various physiological and pathophysiological conditions, including oxidative stress and cancer. The picture emerging from this summary points to a diversity of interactions between miRNAs/miRNA‐induced silencing complexes and RNA‐binding regulatory proteins. Better insight into these complexities of post‐transcriptional regulatory interactions will add to our understanding of the mechanisms of pathological processes and the role of FOXO proteins.
Linked Articles
This article is part of a themed section on Redox Biology and Oxidative Stress in Health and Disease. To view the other articles in this section visit http://onlinelibrary.wiley.com/doi/10.1111/bph.v174.12/issuetoc
Abbreviations
- 5‐FU
5‐fluorouracil
- AD
Alzheimer's disease
- Ago2
argonaute 2
- antagomiR/antimiR
synthetic miRNA inhibitor
- ARE
AU‐rich element
- Bim
Bcl‐2‐interacting mediator of cell death
- CAT‐1
cationic amino acid transporter‐1
- FasL
Fas ligand
- FOXO
forkhead box, class O
- GlcNAcylation
O‐linked‐D‐N‐acetylglucosamine addition
- HSC
haematopoietic stem cell
- HuR
human antigen R
- I/R
ischaemia/reperfusion
- LAT
linker for activation of T cells
- miRISC
miRNA‐induced silencing complex
- miRNA
microRNA
- MITF‐M
microphthalmia‐associated transcription factor‐M
- MnSOD
manganese superoxide dismutase
- oncomiR
miRNA associated with cancer
- PTEN
phosphatase and tensin homologue
- QKI
quaking
- QRE
QKI response element
- Sp1
specificity protein 1
- UTR
untranslated region
Tables of Links
| TARGETS | |
|---|---|
| Enzymes a | Other proteins b |
| Akt | Bim, Bcl‐2‐interacting mediator of cell death |
| p38 kinase | Transporters c |
| PTEN, phosphatase and tensin homologue | CAT‐1, cationic amino acid transporter 1, SLC7A1 |
| Sirtuin 1 |
These Tables list key protein targets and ligands in this article which are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Pawson et al., 2014) and are permanently archived in the Concise Guide to PHARMACOLOGY 2015/16 (a,b,cAlexander et al., 2015a, 2015b, 2015c).
Introduction
FOXOs and regulation of FOXO activity – posttranslational modification and beyond
Transcription factors of the forkhead box, class O (FOXO) family are major players in the regulation of metabolic processes and in the control of decisions on cellular life and death. FOXO targets include genes coding for proteins at crucial positions in fuel metabolism (such as the gluconeogenesis enzymes, glucose 6‐phosphatase or phosphoenolpyruvate carboxykinase; Barthel et al., 2005), in the regulation of cell cycle, apoptosis and autophagy (e.g. p27Kip, Fas ligand (FasL) and beclin1, respectively – see Eijkelenboom and Burgering, 2013) and in stress response and antioxidant defence [such as manganese superoxide dismutase (MnSOD), catalase and certain selenoproteins; Klotz et al., 2015].
There are four FOXO isoforms in humans, FOXO1, FOXO3, FOXO4 and FOXO6 – all of which are expressed in most human tissues and have overlapping target gene patterns. FOXO activity is regulated at several levels (Figure 1) – most acutely by posttranslational modification. FOXOs are substrates of various kinases (for a recent list of kinases and FOXO phosphorylation sites, see Klotz et al., 2015) and are acetylated, ubiquitinylated (Eijkelenboom and Burgering, 2013), GlcNAcylated (Housley et al., 2009) and even oxidized (Dansen et al., 2009). These modifications, in turn, affect FOXO activity, that is, DNA binding and transactivation activity, FOXO subcellular localization and/or FOXO stability/degradation as well as interaction with transcriptional coregulators.
Figure 1.
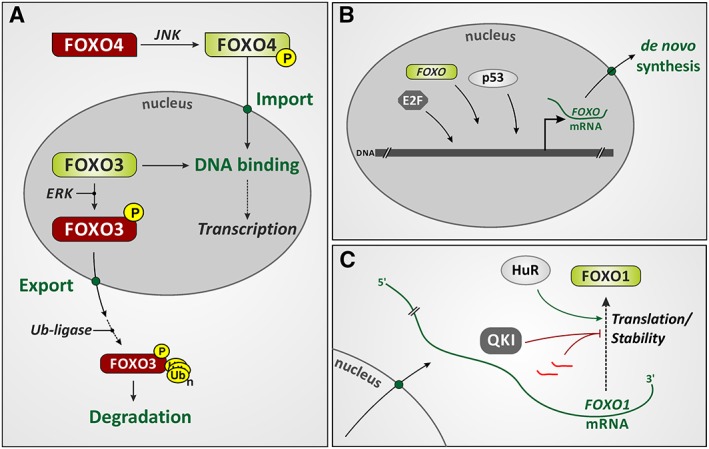
Levels of regulation of FOXO activity and expression. (A) Posttranslational modification of FOXOs affects their activity, subcellular localization and stability. The figure illustrates the effects of phosphorylation of two FOXO isoforms (FOXO3 and FOXO4) by two different kinases, c‐Jun‐N‐terminal kinases (JNK) and extracellular signal‐regulated kinases (ERK). The figure indicates consequences of FOXO phosphorylation: nuclear import and export, respectively, as well as increased or attenuated DNA‐binding activity (red: inactive; green: active FOXO form). A consequence of FOXO3 phosphorylation by ERK is nuclear exclusion and ubiquitination (leading to proteasomal degradation). For further detail regarding FOXO posttranslational modification, see Klotz et al., 2015. (B) Transcriptional regulation of FOXO expression. Transcription factors controlling FOXO expression include certain E2F isoforms, p53 and FOXOs (specifically, FOXO3 regulating FOXO1 expression; see text). (C) FOXO mRNA stability and translation are affected by miRNAs (bright red) and RNA‐binding proteins such as HuR and quaking (see text for details).
Thus, phosphorylation is a major regulator of FOXO nucleocytoplasmic shuttling but also affects DNA binding of FOXOs. For example, phosphorylation elicited by the serine/threonine kinase Akt can cause FOXO nuclear exclusion and inactivation. These appear to be two independent consequences of FOXO phosphorylation, as suggested by studies on FOXO6: while phosphorylation of FOXO isoforms 1, 3 or 4 by Akt (e.g. upon stimulation of cells with insulin, which results in Akt activation via phosphoinositide 3′‐kinase) results in their nuclear exclusion, this is not seen with FOXO6, which remains nuclear but nevertheless is inactivated (van der Heide et al., 2005).
Modification of lysines to yield acetylated or ubiquitinylated FOXOs has consequences for FOXO activity and its stability, respectively. FOXO acetylation appears to attenuate its DNA binding and transactivation activity (different from acetylation of histones, which would support FOXO‐dependent transcription), and it may render FOXOs susceptible towards phosphorylation and nuclear exclusion. However, it also stabilizes FOXOs by preventing ubiquitinylation of the same lysine residues and subsequent proteasomal degradation (Daitoku et al., 2011).
Oxidation of FOXO cysteines mediates interaction with coregulators and may contribute to stress‐induced changes in subcellular localization (Putker et al., 2015). Moreover, it affects FOXO acetylation and activity. For example, FOXO4 was demonstrated to form mixed disulfides with p300 acetyltransferase upon exposure of cells to hydrogen peroxide and under conditions that cause endogenous generation of ROS; this disulfide formation was required for H2O2‐induced FOXO4 acetylation and acetylation‐induced loss of FOXO activity to occur (Dansen et al., 2009).
Long‐term regulation of FOXO levels and activity occurs through transcriptional regulation. Major transcriptional regulators of FOXO expression include E2F, p53 and FOXO3 itself (see Klotz et al., 2015, for review).
More recently, post‐transcriptional regulation of FOXO expression under different (patho‐) physiological conditions has been of interest, focusing primarily on the effects of various microRNAs (miRNAs) on FOXO levels in various types of cancer. Because of their crucial role in the regulation and integration of metabolism, stress response, cell cycle progression and apoptosis (Furukawa‐Hibi et al., 2005; Eijkelenboom and Burgering, 2013; Klotz et al., 2015), FOXO proteins are generally regarded as tumour suppressors (Dansen and Burgering, 2008); hence, their translational repression in some types of tumours seems to be essential to allow the uncontrolled proliferation of transformed cells.
A high metabolic rate in cancer cells supports their rapid proliferation and often comes with elevated steady‐state levels of ROS as compared with untransformed cells, which is assumed – for example, through modulation of redox‐sensitive signalling cascades – to contribute to their metabolic adaptation, proliferation and survival (Schieber and Chandel, 2014; Lennicke et al., 2015). Moreover, redox signalling is involved in the regulation of quiescent and proliferative growth states. Intracellular ROS levels periodically change during the cell cycle, sometimes referred to as a ‘redox cycle within the cell cycle’ (Burhans and Heintz, 2009; Sarsour et al., 2014). Interestingly, the activity of MnSOD, encoded by SOD2, a FOXO target gene, fluctuates during the cell cycle as well, and its loss generally leads to aberrant proliferation whereas its high activity is linked to the quiescent state (reviewed by Sarsour et al., 2014).
We here present a brief overview of the literature on post‐transcriptional regulation of FOXO expression, followed by a more detailed discussion of selected examples on physiological and pathological regulation of FOXOs by miRNAs, including regulation under oxidative stress conditions and in cancer. Moreover, we will discuss post‐transcriptional regulation of FOXO expression by RNA‐binding proteins and possible interactions of these proteins with miRNAs and miRNA‐induced silencing complexes (miRISCs).
Post‐transcriptional regulation of FOXO expression: overview
FOXO activity is controlled by posttranslational modification. As the expression of modifying enzymes, such as the deacetylase sirtuin 1 (Abdelmohsen et al., 2007), may in turn be controlled at the level of mRNA stability, it can of course be argued that FOXO activity is also modulated by post‐transcriptional regulatory processes. This article, however, will focus on post‐transcriptional processes controlling FOXO expression by targeting FOXO mRNAs.
Many miRNAs have now been identified as regulators of FOXO expression, some of which will be dealt with in the following sections. Tables 1, 2 provide a list of miRNAs identified as regulating the formation of FOXO isoforms, along with information on cell/tissue type and/or biological processes in which FOXOs and these miRNAs were involved.
Table 1.
A list of miRNAs demonstrated to target FOXO mRNAa
| miRNA | Target mRNA (FOXO isoform) | Target cell/tissue or biological process (organismb) | Reference |
|---|---|---|---|
| miR‐9 | FOXO1 | Breast cancer cells (Hs) | Yang et al. (2014a) |
| FOXO1 | Neural stem cell differentiation (Mm) | Kim et al. (2015) | |
| FOXO1 and FOXO3 | Haematopoietic cells (Hs and Mm) | Senyuk et al. (2013) | |
| miR‐10b | FOXO3 | Glioblastoma multiforme (Hs) | Lin et al. (2012) |
| miR‐21 | FOXO1 | Glioblastoma cells (Hs) | Lei et al. (2014a) |
| FOXO1 | Diffuse large B‐cell lymphoma (Hs) | Go et al. (2015) | |
| FOXO1 | Pancreatic ductal adenocarcinoma (Hs) | Song et al. (2015) | |
| miR‐23a | FOXO3 | Cardiac hypertrophy (Mm) | Wang et al. (2012b) |
| miR‐23b | FOXO4 | Vascular smooth muscle cells (Hs and Rn) | Iaconetti et al. (2015) |
| miR‐27a | FOXO1 | Renal cell carcinoma (Hs) | Zhou et al. (2012) |
| FOXO1 | Liver cancer (Hs and Mm) | Sun et al. (2015a) | |
| FOXO3 | Glioblastoma (Hs) | Ge et al. (2013) | |
| miR‐29a | FOXO3 | Chondrogenic differentiation of mesenchymal stem cells (Hs and Mm) | Guerit et al. (2014) |
| miR‐30d | FOXO3 | Cardiomyocyte pyroptosis in diabetic cardiomyopathy (Rn and Hs) | Li et al. (2014d) |
| miR‐96 | FOXO1 | Bladder cancer (Hs) | Guo et al. (2012) |
| FOXO1 | Prostate cancer (Hs) | Fendler et al. (2013) | |
| FOXO1 | Prostate cancer (Hs) | Haflidadottir et al. (2013) | |
| FOXO1 and FOXO3 | HepG2 hepatoma cells (Hs) | Xu et al. (2013a) | |
| FOXO3 | Breast cancer (Hs) | Lin et al. (2010) | |
| FOXO3 | Idiopathic pulmonary fibrosis fibroblasts (Hs) | Nho et al. (2014) | |
| FOXO3 | Non‐small cell lung cancer (Hs) | Li et al. (2015a) | |
| FOXO3 | Prostate cancer (Hs) | Wang et al. (2015a) | |
| miR‐107 | FOXO1 | Gastric cancer cells (Hs) | Li et al. (2014b) |
| miR‐126 | FOXO3 | Vascular smooth muscle cells (Hs and Mm) | Zhou et al. (2013) |
| miR‐132 | FOXO1 | Gastric cancer cells (Hs) | Li et al. (2015b) |
| FOXO3 | Acute inflammatory lung injury (Mm) | Rao et al. (2015) | |
| miR‐132‐3p | FOXO1 | Alzheimer's disease (Hs) | Lau et al. (2013) |
| miR‐135a | FOXO1 | Spermatogonial stem cells in cryptochorid testes (Rn) | Moritoki et al. (2014) |
| FOXO1 | Bladder cancer (Hs) | Mao et al. (2015) | |
| FOXO1 | Malignant melanoma cells (Hs) | Ren et al. (2015) | |
| miR‐135b | FOXO1 | Anaplastic large cell lymphoma (Hs) | Matsuyama et al. (2011) |
| FOXO1 | Hepatocellular carcinoma (Hs and Mm) | Jung et al. (2014) | |
| FOXO1 | Osteosarcoma cells (Hs) | Pei et al. (2015) | |
| miR‐137 | FOXO1 | Hepatocellular carcinoma (Hs) | Tan et al. (2015) |
| miR‐139 | FOXO1 | Mouse hepatocytes (Mm) | Hasseine et al. (2009) |
| miR‐145 | FOXO1 | White adipose tissue (Mm) | Lin et al. (2014) |
| miR‐155 | FOXO3 | Breast cancer (Hs) | Kong et al. (2010) |
| FOXO3 | Hypoxic lung cancer cells (Hs) | Babar et al. (2011) | |
| FOXO3 | Multifunctional Treg cell line (Hs) | Yamamoto et al. (2011) | |
| FOXO3 | Vascular stem cell niche in bone marrow (Hs) | Spinetti et al. (2013) | |
| FOXO3 | Glioma (Hs) | Ling et al. (2013) | |
| FOXO3 | Ulcerative colitis (Hs) | Min et al. (2014) | |
| FOXO3 | Lymphoproliferation in linker for activation of T‐cells (LAT) mutant mice (Mm) | Rouquette‐Jazdanian et al. (2015) | |
| FOXO3 | Pancreatic cancer (Hs) | Wang et al. (2015c) | |
| miR‐182 | FOXO1 | Activated helper T‐lymphocytes (Mm and Hs) | Stittrich et al. (2010) |
| FOXO1 | Osteoblasts and skeletogenesis (Mm and Dr) | Kim et al. (2012) | |
| FOXO1 | Rejecting cardiac allografts and mononuclear cell infiltrate (Mm) | Wei et al. (2012) | |
| FOXO1 | Hydrogen peroxide‐induced apoptosis in neuroblastoma cells (Hs) | Gheysarzadeh and Yazdanparast (2015) | |
| FOXO1 and FOXO3 | Colon cancer – African and Caucasian Americans (Hs) | Li et al. (2014a) | |
| FOXO3 | Melanoma metastasis (Hs and Mm) | Segura et al. (2009) | |
| FOXO3 | Advanced ovarian carcinoma (Hs) | McMillen et al. (2012) | |
| FOXO3 | Skeletal muscle (Mm and Rn) | Hudson et al. (2014) | |
| FOXO3 | High‐grade serous ovarian carcinoma (Hs) | Levanon et al. (2014) | |
| FOXO3 | Ovarian cancer cells – orthotopic xenografts (Hs) | Xu et al. (2014a) | |
| FOXO3 | Lung cancer (Hs) | Yang et al. (2014b) | |
| miR‐183 | FOXO1 | Human‐specific miR‐183 target site (Hs) | McLoughlin et al. (2014) |
| FOXO1 | Non‐small cell lung cancer cells (Hs) | Zhang et al. (2015a) | |
| miR‐196a | FOXO1 | Cervical cancer (Hs) | Hou et al. (2014) |
| miR‐205 | FOXO3 | Lung squamous cell carcinoma (Hs) | Huang et al. (2014) |
| miR‐217 | FOXO3 | Angiogenesis of cytomegalovirus‐infected endothelial cells (Hs) | Zhang et al. (2013) |
| miR‐223 | FOXO1 | Colorectal cancer, cervical cancer and hepatoma cells (Hs) | Wu et al. (2012a) |
| FOXO1 and FOXO3 | Analysis of decoy oligonucleotides as modulators of miRNA activity in various cell lines (Hs) | Wu et al. (2013) | |
| miR‐370 | FOXO1 | Prostate cancer cells (Hs) | Wu et al. (2012b) |
| FOXO1 | Gastric carcinoma (Hs) | Fan et al. (2013) | |
| miR‐374a | FOXO1 | Osteosarcoma (Hs) | He et al. (2015) |
| miR‐421 | FOXO4 | Nasopharyngeal carcinoma (Hs) | Chen et al. (2013) |
| miR‐486 | FOXO1 | Cardiac and skeletal muscle (Mm and Rn) | Small et al. (2010) |
| FOXO1 | Muscle wasting in chronic kidney disease (Mm) | Xu et al. (2012) | |
| FOXO1 | Cardiac/skeletal muscle cells in breast cancer and mouse models (Hs and Mm) | Chen et al. (2014) | |
| miR‐498 | FOXO3 | Ovarian cancer cells (Hs) | Liu et al. (2015) |
| miR‐499‐5p | FOXO4 | Colorectal cancer (Hs) | Liu et al. (2011) |
| miR‐582‐5p | FOXO1 | Monocytes (Hs) | Liu et al. (2013) |
| miR‐664 | FOXO4 | Osteosarcoma cells (Hs) | Chen et al. (2015) |
| miR‐708 | FOXO3 | Childhood acute lymphoblastic leukaemia (Hs) | Han et al. (2011) |
| miR‐1269 | FOXO1 | Hepatocellular carcinoma (Hs) | Yang et al. (2014c) |
| miR‐1274a | FOXO4 | Gastric cancer (Hs) | Wang et al. (2015b) |
Only reports on FOXO regulation by a single miRNA are listed here. For reports on FOXO regulation by several miRNAs simultaneously, see Table 2.
Hs, human; Mm, mouse; Rn, rat; Dr, zebrafish.
Table 2.
A list of miRNAs demonstrated to target FOXO mRNA simultaneously
| miRNA | Target mRNA | Target cell/tissue or biological process (organisma) | Reference |
|---|---|---|---|
| miR‐9, miR‐27, miR‐96, miR‐153, miR‐182, miR‐183 and miR‐186 | FOXO1 | Endometrial cancer (Hs) | Myatt et al. (2010) |
| miR‐27a, miR‐96 and miR‐182 | FOXO1 | Breast cancer cells (Hs) | Guttilla and White (2009) |
| miR‐27a and miR‐24 | FOXO1 | Differentiation of embryonic stem cells (Mm) | Ma et al. (2015) |
| miR‐96, miR‐182 and miR‐183 | FOXO1 | Classical Hodgkin lymphoma (Hs) | Xie et al. (2012) |
| miR‐96, miR‐182 and miR‐183 | FOXO1 | Glioma (Hs) | Tang et al. (2013) |
| miR‐96, miR‐182 and miR‐183 | FOXO1 | Granulosa cells (Bt) | Gebremedhn et al. (2015) |
| miR‐96, miR‐182 and miR‐183 | FOXO1 | Hepatocellular carcinoma (Hs) | Leung et al. (2015) |
| miR‐96 and miR‐182 | FOXO3 | Ovarian cancer cells (Hs) | Xu et al. (2013b) |
| miR‐132 and miR‐212 | FOXO3 | Cardiac hypertrophy and cardiomyocyte autophagy (Mm and Rn) | Ucar et al. (2012) |
| miR‐132 and miR‐212 | FOXO3 | Alzheimer's disease (Hs, Mm and Rn) | Wong et al. (2013) |
| miR‐132 and miR‐212 | FOXO3 | Haematopoietic stem cell maintenance (Mm) | Mehta et al. (2015) |
| miR‐182 and miR‐223 | FOXO3 | Skeletal muscle – hormone replacement therapy (Hs and Mm) | Olivieri et al. (2014) |
Hs, human; Mm, mouse; Rn, rat; Bt, bovine.
Examples of such physiological and pathophysiological processes requiring post‐transcriptional control of FOXO expression include clonal expansion of antigen‐activated TH lymphocytes (Stittrich et al., 2010), maintenance of haematopoietic stem cells (HSCs) during ageing (Mehta et al., 2015), cardiac hypertrophy (Ucar et al., 2012) and certain aspects of neurodegeneration (Wong et al., 2013). A number of miRNAs directly targeting FOXO mRNAs are also implicated in tumour promotion, growth or metastasis: for example, miR‐182 targets FOXO3 mRNA in lung cancer (Yang et al., 2014b) and melanoma (Segura et al., 2009); the same FOXO3 mRNA is also targeted by miR‐155, promoting oxidative stress in pancreatic cancer (Wang et al., 2015c). Additionally, a polymorphism in a miR‐137‐binding site in the 3′‐untranslated region (UTR) of human FOXO1 mRNA has been described that decreases hereditary susceptibility to hepatocellular carcinoma (Tan et al., 2015).
The following sections will focus on selected miRNAs and their role in regulating FOXO expression, followed by examples of multiple miRNAs affecting FOXO levels through concerted action. A final section will then highlight mRNA‐binding proteins now known to control FOXO expression and their physiological relevance.
Selected miRNAs regulating FOXO expression and their (patho‐) physiological significance
miR‐182
miR‐182 was initially characterized as a member of an miRNA cluster (comprising the paralogues miR‐96, miR‐182 and miR‐183), which is highly expressed in sensory organ tissues, such as the retina and olfactory epithelium (Kloosterman et al., 2006; Weston et al., 2006; Xu et al., 2007). It is essential, together with miR‐183, for sustaining outer segments of cone photoreceptors of the eye (Busskamp et al., 2014) and contributes to the regulation of T‐cell differentiation and osteogenesis (Stittrich et al., 2010; Kim et al., 2012). Furthermore, miR‐182 is involved in the aetiology of multiple types of cancer and at various stages of cancerogenesis (see the following discussions, and Wei et al., 2015). According to current nomenclature, the publications on FOXO1 and FOXO3 and miR‐182 that are discussed in this section concern, more specifically, miR‐182‐5p (Kozomara and Griffiths‐Jones, 2014).
A common function of down‐regulation of FOXO expression by miR‐182 under physiological conditions is to allow cell cycle progression and cell proliferation. Conversely, down‐regulation of miR‐182 may be required for protection against excessive oxidative stress, inhibition of proliferation, induction of cell cycle arrest and/or apoptosis by FOXO.
For example, FOXO1 controls the resting state of helper T‐cells by inducing synthesis of the cell cycle inhibitor p27Kip1, which in turn blocks cell cycle progression (Peng, 2008). Upon activation by antigen, transition of helper T‐cells to a proliferative stage is achieved, which requires inactivation of FOXO1. FOXO1 is first inactivated by post‐translational modification (by phosphorylation), resulting in its exclusion from the nucleus. However, the initial T‐cell receptor‐dependent signal resulting in FOXO phosphorylation is transient and stops within a day after antigen stimulation (Stahl et al., 2002; Fabre et al., 2005; Peng, 2008; Stittrich et al., 2010). But what suppresses FOXO1 activity during the later phase of clonal expansion, when proliferation of helper T‐lymphocytes is promoted by IL‐2? Stittrich et al. (2010) showed that IL‐2/IL‐2 receptor‐dependent signalling results in stimulation of transcription factor STAT5 and in binding of activated STAT5 to the regulatory region of the miR‐182 gene, up‐regulating its expression up to 200‐fold. Mature miR‐182, in turn, decreases formation of FOXO1 protein by interacting with a specific site in the 3′‐UTR of its mRNA, hence enabling the replication of activated TH lymphocytes (Stittrich et al., 2010; reviewed in Haftmann et al., 2012).
It was previously demonstrated that oxidative stress may inhibit the stimulation of JAK‐STAT signalling by certain cytokines (Kaur et al., 2005; Di Bona et al., 2006; reviewed in Bourgeais et al., 2013). In line with this notion, Gheysarzadeh and Yazdanparast (2015) have shown recently that, under conditions of oxidative stress, the STAT5 ↑/miR‐182↑/FOXO1↓ axis can be altered to rapidly up‐regulate FOXO1 activity (STAT5↓/miR‐182↓/FOXO1↑). Exposure of human SK‐N‐MC neuroblastoma cells to 300 μM H2O2 led to inactivation of STAT5 and a 10‐fold down‐regulation of miR‐182, which resulted in a fourfold increase in FOXO1 protein levels (while FOXO1 mRNA levels did not change). This, in turn, led to an increased formation of the pro‐apoptotic FOXO1 target gene products, Bcl‐2‐associated X protein and Bcl‐2‐interacting mediator of cell death (Bim), and to activation of caspase‐3 (Gheysarzadeh and Yazdanparast, 2015).
The miR‐182/FOXO axis was also shown to control early versus late stages of lung cancer progression; here, human FOXO3 expression was attenuated by miR‐182 (Yang et al., 2014b). In their previous study, the authors had shown that transcription factor ‘specificity protein 1’ (Sp1) was highly expressed in early‐stage lung adenocarcinoma, which was important for cancer cell proliferation and tumour growth (Hsu et al., 2012). However, a decrease in Sp1 levels was observed in highly invasive and migratory late‐stage lung adenocarcinomas. Sp1 overexpression in these late‐stage cancers inhibited both migration and invasiveness of lung adenocarcinoma cells in vitro and their metastasis in vivo, while its knockdown enhanced these abilities (Hsu et al., 2012). The authors then identified the gene coding for miR‐182 as a target of Sp1 and demonstrated that expression of Sp1 inversely correlates with expression of FOXO3, the known target of miR‐182 (Segura et al., 2009). Hence, Sp1 was identified as another transcription factor stimulating the formation of miR‐182, which in turn repressed translation of a FOXO mRNA (Yang et al., 2014b) (Figure 2).
Figure 2.
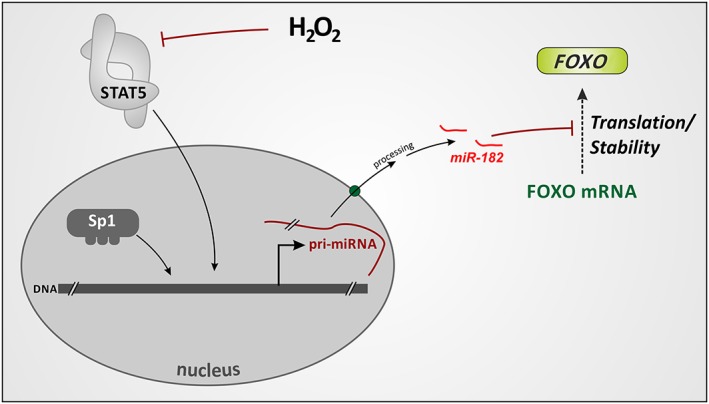
Regulation of FOXO synthesis through transcriptional control of microRNA formation. Transcription factors STAT5 or Sp1 stimulate miR‐182 generation. MiR‐182, in turn, blocks FOXO1 and FOXO3 mRNA translation or decreases its stability. Hydrogen peroxide was shown to attenuate STAT5 activation, thus up‐regulating FOXO levels through preventing miR‐182 production (see text).
Decreased levels of FOXO3 correlated with stimulated proliferation and tumour growth, possibly by down‐regulation of p27Kip1, whose gene is a FOXO3 target (Dijkers et al., 2000; Medema et al., 2000). Interestingly, Yang et al. (2014b) found that Sp1 also up‐regulates FOXO3 promoter activity – but any effect this might have on FOXO3 transcription was overcome by the miR‐182‐mediated translational repression in terms of the FOXO3 protein level. In the late tumour stages, Sp1 and miR‐182 were down‐regulated, and FOXO3 protein expression up‐regulated accordingly. Furthermore, the knockdown of miR‐182 led to an increased expression of the N‐cadherin gene and several other cancer metastasis‐related genes. It also led to the enhanced cell migration and invasion in vitro and to the higher capacity to metastasize in vivo (Yang et al., 2014b). In contrast, the simultaneous knockdown of miR‐182 and FOXO3 decreased invasive cell behaviour and expression of N‐cadherin, indicating an important role of FOXO3 in cell migration and metastasis during late‐stage lung adenocarcinomas. Peculiarly, miR‐182 appears to function in lung adenocarcinoma both as an oncomiR for growth and as a suppressor of metastasis. Further experiments are required to ascertain whether FOXO3, generally considered to act as a tumour suppressor (Paik et al., 2007; Dansen and Burgering, 2008; Kloet and Burgering, 2011), can also act as an oncogene during late stages of lung adenocarcinoma.
For melanoma cell lines, Segura et al. (2009) showed that overexpression of miR‐182 increases their capability to migrate (in vitro) and to metastasize (in vivo). Down‐regulation of miR‐182, on the other hand, hindered invasion and induced apoptosis. Moreover, the pro‐invasive effect of ectopically expressed miR‐182 was shown to be a consequence of translational repression of FOXO3 formation and of another transcription factor, microphthalmia‐associated transcription factor‐M (MITF‐M), by miR‐182. Overexpression of either of the two transcription factors along with miR‐182 blocked the pro‐invasive impact of miR‐182. In human tissue samples, the levels of miR‐182 were significantly higher in metastatic melanoma as compared with primary melanoma and nevi. In metastatic tissues, they also inversely correlated with the FOXO3 and MITF protein levels, as determined by immunohistochemistry (Segura et al., 2009).
The gene coding for miR‐182 is located in the 7q32.2 human chromosomal region, in a cluster with two other miRNAs, miR‐96 and miR‐183 (Landgraf et al., 2007; Xu et al., 2007). The broader region 7q31–34 also includes genes coding for the c‐MET and B‐RAF proto‐oncogenes and is often amplified in advanced human melanomas (Bastian et al., 1998; Lin et al., 2008). Indeed, copy number analysis performed by Segura et al. (2009) revealed seven cell lines with increased locus copy number out of 13 cell lines with elevated miR‐182. High miR‐182 levels in melanomas can thus be a consequence of locus amplification.
Taken together, miR‐182 expression can be differentially regulated in various cancer types in the course of cancer progression. Furthermore, many groups have shown, outside the FOXO context, that miR‐182 can act as an oncomiR or as a tumour suppressor, depending on the cell or tumour type. For example, the role of miR‐182 in cancer progression and metastasis has been well established in breast cancer (Lei et al., 2014b; Li et al., 2014c), hepatocellular carcinoma (Wang et al., 2012a; Du et al., 2015; Leung et al., 2015) and in primary (soft tissue) sarcomas (Sachdeva et al., 2014; Dodd et al., 2015). On the other hand, miR‐182 can suppress proliferation and tumourigenicity of renal cell carcinoma (Xu et al., 2014b), induce apoptosis in cervical cancer (Sun et al., 2015b) and act as tumour suppressor in glioblastoma (Kouri et al., 2015) and neuroblastoma (Rihani et al., 2015). Its down‐regulation has also been associated with the growth and invasion of osteosarcoma cells (Hu et al., 2015). It will be interesting to explore the expression and possible roles of FOXO mRNAs as miR‐182 targets in all these malignancies.
miR‐132/miR‐212
miR‐132 and miR‐212 are paralogous miRNAs encoded by a cluster on human chromosome 17. They play various roles in neuronal morphogenesis, maturation and function, while their deregulation is connected with neurological disorders. Additionally, accumulating evidence points to their involvement in the immune functions and inflammation (reviewed in Wanet et al., 2012). According to current nomenclature, the publications discussed in this section are concerned with, more specifically, miR‐132‐3p and miR‐212‐3p (Kozomara and Griffiths‐Jones, 2014).
FOXOs are known stress‐responsive regulators of the cellular generation of antioxidant proteins (Klotz et al., 2015). Likewise, they have an important role in the response of HSCs to physiological oxidative stress (Tothova et al., 2007). Among the FOXO isoforms, FOXO3 is required for HSC self‐renewal and for maintaining HSC quiescence and the HSC pool in aged mice (Miyamoto et al., 2007). Moreover, FOXO3 is vital to drive a protective autophagy programme in HSC, including old HSCs, upon metabolic stress (starvation) in order to support HSC survival (Warr et al., 2013). FOXO3 is also required for proper regulation of mitochondrial metabolism in HSCs (Rimmele et al., 2015).
Mehta et al. (2015) identified the miR‐132/miR‐212 cluster as being responsible for controlling FOXO3 expression in ageing HSCs, which in turn ensures balanced HSC maintenance, survival and function. The authors show that FOXO3 mRNA is directly targeted by miR‐132, and miR‐132 regulates HSC cycling and function through FOXO3. Notably, miR‐132 is up‐regulated in HSCs with age (Mehta et al., 2015). Taken together, the study demonstrates a molecular mechanism by which FOXO3 function is differentially regulated in the ageing organism. These findings are of particular interest, as several polymorphisms in the FOXO3 locus have been shown to be associated with exceptional human longevity (reviewed in Morris et al., 2015).
Recently, two groups have implicated miR‐132/miR‐212 and FOXO3 in two serious pathological conditions. Wong et al. (2013) found that both miRNAs are down‐regulated in temporal cortical areas and CA1 hippocampal neurons of human Alzheimer's disease (AD) brains. The authors identified phosphatase and tensin homologue (PTEN), FOXO3 and p300 as direct targets of miR‐132/212 in primary neurons and PC12 cells and showed that specific inhibition of miR‐132/212 with anti‐miRs (antagomirs) led to apoptosis of these cultured cells, which was counteracted by siRNA‐induced down‐regulation of PTEN, FOXO3 and p300. In contrast, overexpression of miR‐132 and miR‐212 rendered cultured hippocampal neurons less sensitive to hydrogen peroxide. Furthermore, mRNA and protein levels of PTEN, FOXO3 and p300 and of FOXO3 target genes coding for pro‐apoptotic factors were increased in AD brains. The authors suggest that a ‘miR‐132/miR‐212/PTEN/FOXO3’ axis exists that contributes to the neurodegeneration in AD (Wong et al., 2013).
In a second report, Ucar et al. (2012) uncovered the role of miR‐132/212 in cardiac hypertrophy and cardiomyocyte autophagy. Myocardial stress induces pathological hypertrophy (Barry and Townsend, 2010), and miR‐132/212 expression is up‐regulated by pro‐hypertrophic stimuli. Both miRNAs are necessary and sufficient for hypertrophic cardiomyocyte growth (Ucar et al., 2012). Overexpression of the two miRNAs leads to down‐regulation of FOXO3, which results in impairment of the autophagic response upon starvation. Intriguingly, antagomir injection in mice to block miR‐132 function prevents cardiac hypertrophy and heart failure (Ucar et al., 2012).
Simultaneous regulation of FOXO levels by several miRNAs
The real scenario within cells is likely to be more complicated, as commonly one mRNA can be simultaneously regulated by multiple miRNAs (Krek et al., 2005). For example, some studies showed that miR‐182 regulates FOXO1 expression in concert with several other miRNAs. Among these, miR‐27a and miR‐96 (which has the same seed region as miR‐182) were identified as regulating FOXO1 expression together with miR‐182 and to be highly expressed in MCF‐7 (miR‐27a/miR‐96/miR‐182) and MDA‐MB‐231 (miR‐27a/miR‐182 only) breast cancer cells, causing down‐regulation of FOXO1 protein levels and thus contributing to maintenance of a proliferative state while impairing apoptotic responses (Guttilla and White, 2009). In line with this, FOXO1 mRNA was significantly down‐regulated in breast tumours of various stages compared with normal breast tissue.
Interestingly, FOXO3 and FOXO4 mRNA levels were not significantly altered in these tumour samples (Guttilla and White, 2009), although FOXO3 mRNA has been shown to be the target of miR‐27a or of miR‐182 in multiple cell types (Table 1). One explanation for this discrepancy may be that the accessibility of a miRNA‐binding site in the respective 3′‐UTR may be affected by other 3′‐UTR interacting factors, depending on cell/tissue type and its developmental, physiological or pathological state.
For FOXO1 mRNA, two regions were identified in the 3′‐UTR (nucleotides 204–492 and 1907–2247 of the 3′‐UTR, which correspond to nucleotides 2557–2845 and 4260–4600, respectively, of the whole mRNA depicted in Figure 3), each containing one binding site for miR‐27a, miR‐96 and miR‐182. In functional assays using luciferase reporter constructs, a higher repression was observed with the construct containing both 3′‐UTR regions, as compared with the two constructs with one or the other 3′‐UTR region only, suggesting the combined action of multiple miRNA target sites in the FOXO1 3′‐UTR (Guttilla and White, 2009).
Figure 3.
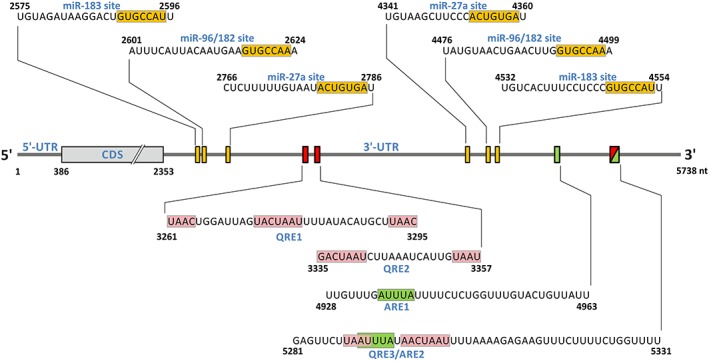
Position of microRNA, HuR and quaking target sites within the human FOXO1 3′‐UTR. Position of microRNA target sites is depicted as orange boxes on the RNA; the sequences interacting with the respective miRNA are shown above the RNA, with sequences pairing with the respective miRNA seed region boxed in orange. For miR‐96/182, the orange‐boxed sequence indicates pairing with the miR‐96 seed region, whereas pairing with the miR‐182 seed region would be 1 nt shorter at the 5′ end (Guttilla and White, 2009; McLoughlin et al., 2014). Quaking response elements (QRE1/2/3) (Yu et al., 2014) are shown as red boxes on the RNA and the actual sequences given underneath; the ‘core’ and ‘half site’ sequences corresponding to the bipartite consensus target RNA motif (Galarneau and Richard, 2005) are boxed in pink. Note that the ‘half site’ can be located downstream or upstream of the ‘core’ sequence. QRE1 appears to have two consensus‐corresponding ‘half sites’. HuR‐responsive AU‐rich elements (ARE1/2) are shown as green boxes on the RNA, and the corresponding sequences are shown underneath (Li et al., 2013), with the ARE consensus pentamer AUUUA boxed in green. Note that QRE3 and ARE2 overlap (see text for details). The scheme and sequences numberings are according to the human FOXO1 mRNA sequence entry in the National Center for Biotechnology Information Reference Sequence Database (O'Leary et al., 2016), NM_002015.3 (GI:133930787).
An array of seven miRNAs that cooperate in repressing FOXO1 protein levels, including miR‐27, miR‐96 and miR‐182, was identified in endometrial cancer (Myatt et al., 2010; Lam et al., 2012) (see also Table 2), the most common uterine cancer (Amant et al., 2005). FOXO1 is expressed in the normal human endometrium and up‐regulated during differentiation of endometrial stromal cells into decidual cells. Decidualization is induced by progesterone and cAMP signalling and accompanied by increased ROS levels and oxidative stress. FOXO1 has been shown to be an important regulator of this process (Christian et al., 2002; Kajihara et al., 2006; Labied et al., 2006; Takano et al., 2007).
FOXO1 protein levels were much lower in endometrial hyperplasia and carcinoma samples than in the normal endometrium (Myatt et al., 2010). Consistent with previous reports (Burney et al., 2007; Goto et al., 2008; Ward et al., 2008), FOXO1 mRNA levels were also lower in the cancer than in the normal tissue, although not markedly so – suggesting post‐transcriptional regulation of FOXO1 synthesis.
Using several miRNA target prediction programmes and quantitative RT‐PCR analysis, the authors then identified a collection of evolutionarily conserved miRNAs that were significantly up‐regulated in endometrial tumours. To verify FOXO1 targeting by these miRNAs, two endometrial cancer cell lines with profoundly different FOXO1 protein levels were used. As reported previously (Goto et al., 2008; Ward et al., 2008), HEC‐1B cells abundantly expressed FOXO1 protein, whereas its level was very low in the Ishikawa cell line. Although FOXO1 mRNA was substantially lower in Ishikawa cells, compared with the HEC‐1B cells, the difference in mRNA levels was clearly not reflected in the difference in protein levels, analogous to the picture observed in cancer tissue samples versus normal endometrium. Remarkably, most of the miRNAs that were up‐regulated in the endometrial tumours were also at very high levels in the Ishikawa cells but very low in HEC‐1B cells (Myatt et al., 2010).
In a series of miRNA overexpression and antagomir experiments, it was then demonstrated that a cooperative action of multiple miRNAs is essential to effectively repress the translation of FOXO1 in endometrial cancer cells (Myatt et al., 2010).
In Ishikawa cells, miRNA repression using antagomirs resulted in FOXO1 up‐regulation and induced cell cycle arrest and apoptosis. Simultaneous depletion of FOXO1 using siRNA targeting the FOXO1 mRNA coding region decreased cell death, confirming involvement of FOXO1 in this process (Myatt et al., 2010).
The additive effect of three of the miRNAs identified in these studies – miR‐96, miR‐182 and miR‐183 – on FOXO1 down‐regulation has also been reported in human glioma‐derived cell lines (Tang et al., 2013). This miRNA cluster is strongly up‐regulated in glioma tissue compared with the normal cerebrum. Further overexpression of the three miRNAs in glioma cells, individually or as a pool, led to a decreased FOXO1 expression and increased cell proliferation. Conversely, knockdown of these miRNAs resulted in up‐regulated FOXO1 expression and increased apoptosis. Interestingly, overexpression of miRNAs was correlated with lower ROS production (although FOXO proteins are usually connected with cellular antioxidant protein production), whereas their knockdown was accompanied by the rise of ROS levels (Tang et al., 2013). It is unclear, however, how and to what effect ROS levels are modulated by these procedures.
Regulation of FOXO expression by RNA‐binding proteins
Post‐transcriptional regulation of FOXO expression not only occurs through miRNA‐mediated modulation of mRNA stability or translation but is also regulated by RNA‐binding proteins, such as human antigen R (HuR) and quaking (QKI), which have recently been shown to control the levels of FOXO1 transcript and protein (Li et al., 2013; Guo et al., 2014; Yu et al., 2014).
HuR
HuR is an AU‐rich element (ARE)‐binding regulatory protein that interacts with a wide repertoire of mRNAs involved in cell survival, cell division, immune response and differentiation. It commonly acts as mRNA‐stabilizing or translation‐promoting factor (Srikantan et al., 2012). Abundantly present in the nucleus, it responds to diverse stressful stimuli by shuttling to the cytoplasm. These stimuli include conditions that induce oxidative stress, such as UV radiation (Wang et al., 2000; Fernau et al., 2010), exposure to arsenite (Bhattacharyya et al., 2006), tert‐butylhydroquinone (Song et al., 2005) or hydrogen peroxide (Wang et al., 2000; Tran et al., 2003)) or induce endoplasmic reticulum stress (thapsigargin; Bhattacharyya et al., 2006) or conditions of amino acid deprivation (Bhattacharyya et al., 2006). Exposure to these stimuli results in activation of p38‐MAPK, which directly or indirectly (e.g. via MAPK‐activated protein kinase‐2, MK2) triggers HuR phosphorylation, which in turn leads to binding of its target mRNAs and cytoplasmic accumulation. Furthermore, HuR itself was hypothesized to function as redox sensor. HuR attains its full activity as a homodimer, and a cysteine residue in the first of the three HuR RNA recognition motifs was suggested to contribute to homodimerization through formation of an intermolecular disulfide bond (Benoit et al., 2010). As a redox‐sensitive reactive cysteine, it would render HuR responsive towards oxidative stimuli, with an elevated HuR activity due to homodimerization (Benoit et al., 2010; Benoit and Auer, 2011).
Using photoactivatable ribonucleoside‐enhanced crosslinking and immunoprecipitation (PAR‐CliP, Hafner et al., 2010) and other high‐throughput targeting techniques, thousands of putative HuR‐binding sites were identified in the human transcriptome (Lebedeva et al., 2011; Mukherjee et al., 2011). The majority of binding sites were located in the 3′‐UTRs and, surprisingly, in intronic regions of pre‐mRNAs, indicating involvement of HuR in pre‐mRNA processing and regulation of mRNA stability. HuR‐binding sites were identified both in the 3′‐UTR and in the intronic regions of FOXO1 RNA (Lebedeva et al., 2011; Mukherjee et al., 2011), while only intronic HuR binding was found in FOXO3 pre‐mRNA (Mukherjee et al., 2011).
Li et al. (2013) characterized two AREs recognized by HuR in the 3′‐UTR of human FOXO1 mRNA. Overexpression of HuR in MDA‐MB‐231 breast cancer cells stabilized FOXO1 mRNA and elevated FOXO1 protein levels. Furthermore, stress‐induced up‐regulation of endogenous HuR levels using 5‐fluorouracil (5‐FU) also led to an increase in cellular FOXO1 levels. 5‐FU‐induced apoptosis was almost completely inhibited by simultaneous siRNA‐mediated knockdown of endogenous HuR and restored by overexpression of ectopic FOXO1 in the presence of HuR‐siRNA. Taken together, these data suggest a HuR‐mediated up‐regulation of FOXO1 upon exposure to a stressful stimulus, which in turn induces apoptosis in cancer cells (Figure 4).
Figure 4.
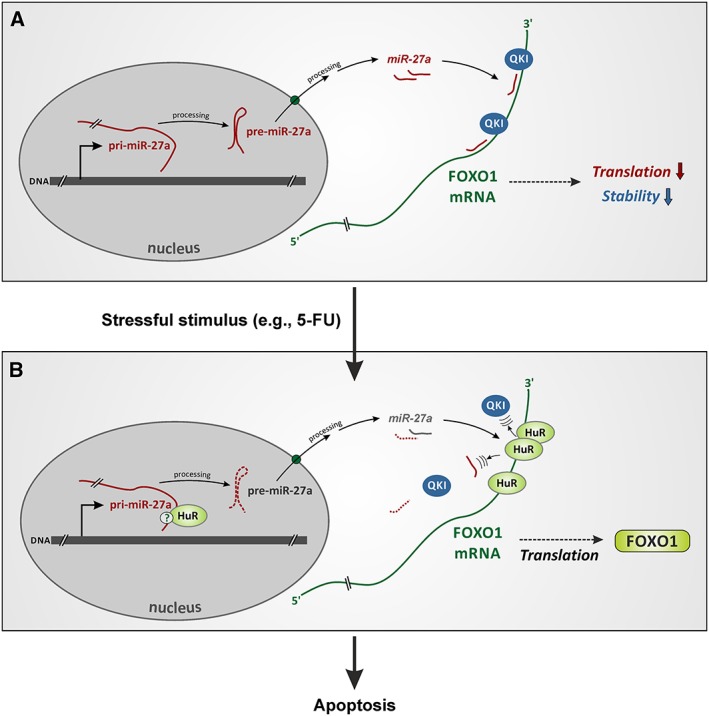
Hypothetical interactions between miRNAs, HuR and quaking in breast cancer cells upon stressful stimulus. (A) MiR‐27a and miR‐182 (not shown here), together with quaking (QKI), impair FOXO1 mRNA translation and decrease its stability (Guttilla and White, 2009; Yu et al., 2014). We hypothesize that QKI and miRNAs/miRISCs cooperate in this process. (B) Stressful stimuli, such as exposure to 5‐fluorouracil (5‐FU), up‐regulate HuR and cause its accumulation in the cytoplasm; simultaneously, QKI is down‐regulated. HuR binds FOXO1 mRNA, which leads to miRNA and possibly also QKI dissociation, resulting in mRNA stabilization and up‐regulation of its translation (Guttilla and White, 2009; Li et al., 2013; Yu et al., 2014). Notably, HuR may compete with QKI for one of their binding sites in this process (Figure 3). Additionally, a second synergistic level of miRNA regulation by HuR is proposed to take place. As HuR binds mature miR‐27a and/or, for example, its pri‐miRNA (Mukherjee et al., 2011), it may inhibit miR‐27a/RISC biogenesis. FOXO1 protein up‐regulation in turn induces apoptosis in breast cancer cells (see text for details and references).
Regarding FOXO1 up‐regulation by HuR, an interaction between HuR and miRNA(s) bound to FOXO1 3′‐UTR may contribute, analogous to findings by Bhattacharyya et al. (2006) who revealed that miRNA‐mediated translational repression can be reversed by HuR under a variety of stress conditions. They showed that the mRNA of cationic amino acid transporter‐1 (CAT‐1) and reporter constructs containing the CAT‐1 3′‐UTR were subjected to repression by miR‐122 in human Huh7 hepatocarcinoma cells. Upon conditions of amino acid deprivation, oxidative stress induced by arsenite, or endoplasmic reticulum stress, HuR binds an ARE located in the 3′‐UTR of CAT‐1 mRNA and relieves CAT‐1 from translational repression by miR‐122. Notably, this derepression occurs despite the fact that the aforementioned ARE is located at some distance from miR‐122‐binding sites (Bhattacharyya et al., 2006; Kundu et al., 2012). The ability of HuR to oligomerize along RNA was suggested as a potential underlying molecular mechanism, leading to a dissociation of miRNA and associated proteins from the corresponding 3′‐UTR site (Kundu et al., 2012).
In addition to the direct removal of miRNAs/miRISCs by HuR from the 3′‐UTR of a given mRNA, a second level of miRNA regulation by HuR has recently been described. HuR can bind primary transcripts of specific miRNA genes, thus inhibiting their processing and formation of mature miRNA (Lebedeva et al., 2011; Choudhury et al., 2013; Zhang et al., 2015b). For instance, HuR, in complex with Musashi homolog 2 protein, suppresses maturation of miR‐7 in non‐neural cells (Lebedeva et al., 2011; Choudhury et al., 2013). Moreover, maturation of miR‐199a is suppressed by HuR in hepatocellular carcinoma cells under hypoxic conditions, which contributes to the cancer cell switch to glycolytic metabolism (Zhang et al., 2015b).
Notably, transcriptome‐wide mapping of HuR‐binding sites in human cells identified miR‐27a as a HuR target (see Table S2 in Mukherjee et al., 2011, under cluster ID: G71353.1). It is currently unclear, whether HuR binds mature miR‐27a and/or any of its precursors, such as pri‐miRNA‐27a (as hypothesized in Figure 4). In any case, this finding raises a possibility that HuR employs two synergistic mechanisms to up‐regulate FOXO1 protein under stress conditions described previously: (i) dissociation of miR‐27a and miR‐182 from FOXO1 mRNA and (ii) inhibition of miR‐27a/RNA‐induced silencing complex (RISC) biogenesis (see Figure 4 for further details).
As outlined previously, FOXO1 expression is likely to be repressed by miR‐27a and miR‐182 in MDA‐MB‐231 breast cancer cells (Guttilla and White, 2009). Their target sites are located in two regions of the 3′‐UTR, with HuR‐binding AREs positioned further downstream from these regions (see Figure 3 for the details). We therefore hypothesize that stress induced by 5‐FU (Li et al., 2013) and oxidative stress inducing HuR activity may lead to HuR‐mediated dissociation of miR‐27a and miR‐182 from FOXO1 mRNA, resulting in its translational derepression (Figure 4).
Quaking
A second RNA‐binding protein involved in the post‐transcriptional regulation of FOXO1 is QKI (Guo et al., 2014; Yu et al., 2014), a regulator of mRNA splicing, transport, stability and translation. It is essential for embryogenesis and for myelination during postnatal development. It coordinates differentiation and cell cycle and functions as tumour suppressor (Artzt and Wu, 2010; Biedermann et al., 2010; Feng and Bankston, 2010). Galarneau and Richard (2005) defined a consensus sequence (NACUAAY‐N1‐20‐UAAY) for the QKI response element (QRE) and identified >1400 putative mRNA targets of QKI in the mouse using bioinformatics analysis, including FOXO1.
Yu et al. (2014) demonstrated interaction of QKI with three QREs located within the FOXO1 3′‐UTR. QKI was shown to decrease FOXO1 mRNA stability and to impair FOXO1 expression in breast cancer cells: siRNA‐mediated knockdown of QKI led to an up‐regulation of FOXO1 mRNA and protein, whereas QKI overexpression resulted in FOXO1 down‐regulation. Importantly, inhibition of proliferation by all‐trans‐retinoic acid or treatment with 5‐FU led to down‐regulation of QKI, up‐regulation of FOXO1 expression and up‐regulation of luciferase activity in the FOXO1 3′‐UTR assay.
Two recent studies demonstrate that the QKI↑/FOXO1↓ regulatory axis may play an important role in susceptibility to ischemic heart injury (Guo et al., 2011; Guo et al., 2014). QKI is highly expressed in rat cardiomyocytes; however, following ischaemia in vivo or simulated ischaemia in vitro, its predominant isoform QKI‐5 was strongly down‐regulated – even more so after reperfusion (ischaemia/reperfusion, I/R) (Guo et al., 2011). In line with the aforementioned QKI/FOXO1 antagonism, FOXO1 levels were strongly up‐regulated by ischaemia and I/R and further enhanced when siRNA‐mediated QKI knockdown preceded I/R. Elevated FOXO1 levels resulted in up‐regulation of Bim and FasL (known FOXO target gene products) and increased apoptosis (Guo et al., 2011). Interestingly, QKI knockdown alone, that is, without I/R, was not sufficient to induce FOXO1 in H9C2 cardiac myoblasts. FOXO1 is a target gene of transcription factor E2F‐1 (Nowak et al., 2007), and E2F‐1 itself has been shown to be up‐regulated by I/R and at the same time was necessary and sufficient to up‐regulate FOXO1 after I/R, both in vivo and in vitro (Angelis et al., 2011). In view of these data, we propose that FOXO1 is doubly regulated during I/R in the heart: I/R induces its transcription and abolishes its post‐transcriptional repression by QKI.
In a follow‐up report, Guo et al. (2014) extended their studies to the hearts of leptin‐deficient (ob/ob) mice; these mice exhibit type 2 diabetes‐like symptoms. They observed that QKI‐5 was down‐regulated in murine ob/ob hearts as compared with the wild‐type controls. Accordingly, FOXO1 was up‐regulated and strongly activated in the mutant in comparison with the wild‐type mice, as shown by its predominant localization in cellular nuclei. Interestingly, FOXO1 activation was linked to myocardial nitrosative stress and endoplasmic reticulum stress, further promoting myocardial I/R injury and apoptosis. Both siRNA‐mediated down‐regulation of FOXO1 and overexpression of ectopic QKI‐5 in ob/ob hearts in vivo attenuated stress and I/R injury. In agreement with the results obtained in breast cancer cells (Yu et al., 2014), QKI overexpression lowered FOXO1 mRNA stability in primary neonatal cardiomyocytes (Guo et al., 2014).
The molecular mechanism by which QKI destabilizes FOXO1 mRNA remains unknown. Could QKI cooperate with miRNAs/miRISCs that are bound to the FOXO1 3′‐UTR? Wang et al. (2010) showed that the QKI isoform QKI‐6 can interact with argonaute 2 (Ago2), the catalytic component of RISC, in a human glioblastoma cell line. Furthermore, as a result of an oxidative stress‐inducing treatment with arsenic oxide, QKI‐6 co‐localized with Ago2 and myelin basic protein mRNA in the cytoplasmic stress granules. Additionally, Chen et al., (2012) reported that QKI binds and stabilizes mature miR‐20a in doxorubicin‐treated astrocytes. It therefore seems likely that crosstalk between QKI and particular miRNA/miRISC may exist under certain conditions.
As discussed already, the 3′‐UTR of FOXO1 mRNA is bound both by miR‐27a, miR‐96 and miR‐182 (Guttilla and White, 2009) and by the QKI protein (Yu et al., 2014) in MCF‐7 breast cancer cells. Targeting of these miRNAs with antisense inhibitors had the same effect on endogenous FOXO1 expression as the knockdown of QKI, in both cases leading to up‐regulation of FOXO1 mRNA and protein levels (compare Guttilla and White, 2009; Yu et al., 2014). Future experiments will be required to ascertain whether the two effects are independent, that is, the simultaneous miRNA inhibition and QKI knockdown in one experiment would have an additive effect and lead to much stronger FOXO1 up‐regulation. Alternatively, the effect of miRNAs and QKI may be interconnected, and then a treatment targeting only one of the factors (e.g. targeting of miRNAs) would be expected to also block binding of the other (e.g. QKI) to the 3′‐UTR.
We compared the locations of AREs recognized by HuR within the 3′‐UTR of FOXO1 mRNA, as reported by Li et al. (2013), with the locations of QKI response elements (Galarneau and Richard, 2005; Yu et al., 2014). Interestingly, the second ARE overlaps with the third QRE in the 3′‐UTR (Figure 3). This binding site arrangement raises a possibility of mutually exclusive binding of the two regulatory proteins to the FOXO1 transcript. Moreover, 5‐FU treatment of MDA‐MB‐231 breast cancer cells led to FOXO1 up‐regulation in both reports, with concurrent up‐regulation of HuR (Li et al., 2013) and down‐regulation of QKI (Yu et al., 2014). Based on these data, it seems reasonable to propose that post‐transcriptional regulation of FOXO1 involves functional interplay between specific miRNAs, HuR and QKI. In such a way, FOXO1 protein formation could be up‐regulated post‐transcriptionally via these interactions in response to various stressful stimuli, including oxidative stress (Figure 4).
Conclusions and perspectives
Post‐transcriptional regulation emerges as a new level of control of FOXO functions. As demonstrated by numerous recent publications, a wide range of miRNAs can directly regulate FOXO transcripts. This occurs in a variety of cell types and tissues and under diverse physiological and pathological conditions, including cancer (Table 1). A vast majority of these reports addresses an involvement of only a single miRNA, in a given cell type and process, and shows its role in FOXO translational inhibition or mRNA destabilization.
However, an accumulating body of evidence (unrelated to the FOXO field) points to a considerably more complex nature of post‐transcriptional regulatory mechanisms and their diverse effects on mRNA translation and stability. For example, under certain physiological conditions, some miRNAs may (in addition to repressing translation of their target mRNAs) also contribute to the stimulation of target mRNA translation by recruiting activating regulatory proteins (Vasudevan and Steitz, 2007; Vasudevan et al., 2007; reviewed by Steitz and Vasudevan, 2009).
Similarly, HuR – described previously as a positive regulator of mRNA stability and translation by antagonizing the repressive effect of miRNAs – can also cooperate with specific miRNAs to destabilize and repress particular mRNAs: HuR and miRNA let‐7/RISC both bind the 3′‐UTR of c‐Myc mRNA and cooperate in its repression (Kim et al., 2009; reviewed in Meisner and Filipowicz, 2010; Srikantan et al., 2012). Intriguingly, by cooperating with let‐7/Ago2 to destabilize a different RNA, a long intergenic noncoding RNA (lincRNA‐p21), HuR can indirectly derepress the translation of JunB and CTNNB1 (β‐catenin) mRNAs. This is because under low HuR levels, lincRNA‐p21 accumulates in the cell and directly represses the translation of JunB and β‐catenin (Yoon et al., 2012). This example particularly nicely illustrates the complexity of post‐transcriptional regulatory interactions.
Like HuR, QKI can also act in an ‘opposite direction’. Larocque et al. (2005) showed in oligodendrocyte progenitor cells that QKI binds and stabilizes the mRNA coding for the cyclin‐dependent kinase inhibitor, p27Kip1, which in turn increases the level of p27Kip1 protein, induces cell cycle arrest and thus ensures proper differentiation into mature oligodendrocytes. This process is disturbed, in part, in the mouse mutant harbouring a mutation in the quaking (Qk) locus (the so‐called Quaking viable mice), leading to abnormal myelination in the central nervous system (Sidman et al., 1964; Ebersole et al., 1996). Of note, as the p27Kip1‐encoding gene is a target of FOXO1 (Dijkers et al., 2000; Medema et al., 2000; Machida et al., 2003), it will be interesting to see whether and how FOXO1 mRNA is regulated by QKI during oligodendrocyte progenitor cell differentiation.
In summary, the same regulatory molecules appear to be capable of exerting quite opposite effects, depending on target mRNA, cell type and conditions.
In addition to HuR and QKI, many other RNA‐binding proteins and interacting cofactors are being identified as post‐transcriptional regulators of gene expression, often acting under diverse conditions, such as during oxidative stress (Abdelmohsen et al., 2008) or in development (Kedde and Agami, 2008). As an illustration, the evolutionarily conserved RNA‐binding protein dead end 1 counteracts the activity of several miRNAs by precluding them from binding their target sites. This is essential for correct development of primordial germ cells in mammals and zebrafish (Kedde et al., 2007; Kedde and Agami, 2008) and was hypothesized to contribute to the regulation of cell–cell communication via gap junctions (Klotz, 2012). It is therefore very likely that many more regulatory proteins will turn out to control or modulate FOXO functions post‐transcriptionally, in concert with the growing battery of miRNAs (Table 1).
In view of the findings already discussed – both related and unrelated to FOXO – a number of questions arise concerning post‐transcriptional regulation of FOXO expression. For example, do miRNAs and QKI cooperate in down‐regulating FOXO1 protein? How does HuR abolish the inhibitory effect of miRNAs/QKI on FOXO1 expression? Does HuR suppress miR‐27a biogenesis? Can HuR and QKI exert an inhibitory (rather than stimulatory) and stimulatory (rather than attenuating) action, respectively, on FOXO expression under certain physiological conditions? Are there other RNA‐binding regulatory proteins that control or modulate FOXO expression post‐transcriptionally?
It currently appears that miRNA studies are advancing at a faster pace than research on regulatory proteins – probably due, in part, to the ease with which miRNAs can be experimentally manipulated. AntagomiRs (antimiRs, miRNA inhibitors) and miRNA mimics have become powerful standard tools. This is mirrored in recent attempts to introduce efficient and safe miRNA‐targeted and miRNA‐based therapeutic modalities (see Mendell and Olson, 2012; Piva et al., 2013; Garofalo et al., 2014). The molecular picture, however, will only be complete after we obtain a detailed knowledge of all interactions between mRNAs, miRNAs and RNA‐binding proteins.
Here, we have provided an overview on post‐transcriptional regulation of FOXO expression from the viewpoint of an emerging diversity of interactions between miRNAs and regulatory RNA‐binding proteins. Accumulating evidence suggests that this post‐transcriptional mode of regulation of FOXO activity is employed in reactions and adaptations to various stressful stimuli, including oxidative stress. Abnormal post‐transcriptional regulation of FOXO is often linked to various disease states. Future research into the complexities of post‐transcriptional regulatory interactions will bring new knowledge about the mechanisms of disease processes and open novel ways towards therapeutic intervention.
Conflict of interest
P.U. and L.O.K. have nothing to disclose.
Acknowledgements
The present work was supported by the European Cooperation in Science and Research (COST Action BM1203/EU‐ROS). We thank three anonymous reviewers for stimulating suggestions.
Urbánek, P. , and Klotz, L.‐O. (2017) Posttranscriptional regulation of FOXO expression: microRNAs and beyond. British Journal of Pharmacology, 174: 1514–1532. doi: 10.1111/bph.13471.
References
- Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M (2008). Posttranscriptional gene regulation by RNA‐binding proteins during oxidative stress: implications for cellular senescence. Biol Chem 389: 243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen K, Pullmann R Jr, Lal A, Kim HH, Galban S, Yang X et al. (2007). Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell 25: 543–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Fabbro D, Kelly E, Marrion N, Peters JA, Benson HE et al. (2015a). The Concise Guide to PHARMACOLOGY 2015/16: Enzymes. Br J Pharmacol 172: 6024–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Kelly E, Marrion N, Peters JA, Benson HE, Faccenda E et al. (2015b). The Concise Guide to PHARMACOLOGY 2015/16: Overview. Br J Pharmacol 172: 5729–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Kelly E, Marrion N, Peters JA, Benson HE, Faccenda E et al. (2015c). The Concise Guide to PHARMACOLOGY 2015/16: Transporters. Br J Pharmacol 172: 6110–6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amant F, Moerman P, Neven P, Timmerman D, Van Limbergen E, Vergote I (2005). Endometrial cancer. Lancet 366: 491–505. [DOI] [PubMed] [Google Scholar]
- Angelis E, Zhao P, Zhang R, Goldhaber JI, Maclellan WR (2011). The role of E2F‐1 and downstream target genes in mediating ischemia/reperfusion injury in vivo . J Mol Cell Cardiol 51: 919–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artzt K, Wu JI (2010). STAR trek: an introduction to STAR family proteins and review of quaking (QKI). Adv Exp Med Biol 693: 1–24. [PubMed] [Google Scholar]
- Babar IA, Czochor J, Steinmetz A, Weidhaas JB, Glazer PM, Slack FJ (2011). Inhibition of hypoxia‐induced miR‐155 radiosensitizes hypoxic lung cancer cells. Cancer Biol Ther 12: 908–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry SP, Townsend PA (2010). What causes a broken heart – molecular insights into heart failure. Int Rev Cell Mol Biol 284: 113–179. [DOI] [PubMed] [Google Scholar]
- Barthel A, Schmoll D, Unterman TG (2005). FoxO proteins in insulin action and metabolism. Trends Endocrinol Metab 16: 183–189. [DOI] [PubMed] [Google Scholar]
- Bastian BC, LeBoit PE, Hamm H, Brocker EB, Pinkel D (1998). Chromosomal gains and losses in primary cutaneous melanomas detected by comparative genomic hybridization. Cancer Res 58: 2170–2175. [PubMed] [Google Scholar]
- Benoit R, Auer M (2011). A direct way of redox sensing. RNA Biol 8: 18–23. [DOI] [PubMed] [Google Scholar]
- Benoit RM, Meisner NC, Kallen J, Graff P, Hemmig R, Cebe R et al. (2010). The X‐ray crystal structure of the first RNA recognition motif and site‐directed mutagenesis suggest a possible HuR redox sensing mechanism. J Mol Biol 397: 1231–1244. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W (2006). Relief of microRNA‐mediated translational repression in human cells subjected to stress. Cell 125: 1111–1124. [DOI] [PubMed] [Google Scholar]
- Biedermann B, Hotz HR, Ciosk R (2010). The quaking family of RNA‐binding proteins: coordinators of the cell cycle and differentiation. Cell Cycle 9: 1929–1933. [DOI] [PubMed] [Google Scholar]
- Bourgeais J, Gouilleux‐Gruart V, Gouilleux F (2013). Oxidative metabolism in cancer: a STAT affair? JAKSTAT 2: e25764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burhans WC, Heintz NH (2009). The cell cycle is a redox cycle: linking phase‐specific targets to cell fate. Free Radic Biol Med 47: 1282–1293. [DOI] [PubMed] [Google Scholar]
- Burney RO, Talbi S, Hamilton AE, Vo KC, Nyegaard M, Nezhat CR et al. (2007). Gene expression analysis of endometrium reveals progesterone resistance and candidate susceptibility genes in women with endometriosis. Endocrinology 148: 3814–3826. [DOI] [PubMed] [Google Scholar]
- Busskamp V, Krol J, Nelidova D, Daum J, Szikra T, Tsuda B et al. (2014). miRNAs 182 and 183 are necessary to maintain adult cone photoreceptor outer segments and visual function. Neuron 83: 586–600. [DOI] [PubMed] [Google Scholar]
- Chen AJ, Paik JH, Zhang H, Shukla SA, Mortensen R, Hu J et al. (2012). STAR RNA‐binding protein quaking suppresses cancer via stabilization of specific miRNA. Genes Dev 26: 1459–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Bao Y, Chen X, Yi J, Liu S, Fang Z et al. (2015). Mir‐664 promotes osteosarcoma cells proliferation via downregulating of FOXO4. Biomed Pharmacother 75: 1–7. [DOI] [PubMed] [Google Scholar]
- Chen D, Goswami CP, Burnett RM, Anjanappa M, Bhat‐Nakshatri P, Muller W et al. (2014). Cancer affects microRNA expression, release, and function in cardiac and skeletal muscle. Cancer Res 74: 4270–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Tang Y, Wang J, Yan Z, Xu R (2013). miR‐421 induces cell proliferation and apoptosis resistance in human nasopharyngeal carcinoma via downregulation of FOXO4. Biochem Biophys Res Commun 435: 745–750. [DOI] [PubMed] [Google Scholar]
- Choudhury NR, de Lima AF, de Andres‐Aguayo L, Graf T, Caceres JF, Rappsilber J et al. (2013). Tissue‐specific control of brain‐enriched miR‐7 biogenesis. Genes Dev 27: 24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Zhang X, Schneider‐Merck T, Unterman TG, Gellersen B, White JO et al. (2002). Cyclic AMP‐induced forkhead transcription factor, FKHR, cooperates with CCAAT/enhancer‐binding protein beta in differentiating human endometrial stromal cells. J Biol Chem 277: 20825–20832. [DOI] [PubMed] [Google Scholar]
- Daitoku H, Sakamaki J, Fukamizu A (2011). Regulation of FoxO transcription factors by acetylation and protein–protein interactions. Biochim Biophys Acta 1813: 1954–1960. [DOI] [PubMed] [Google Scholar]
- Dansen TB, Burgering BM (2008). Unravelling the tumor‐suppressive functions of FOXO proteins. Trends Cell Biol 18: 421–429. [DOI] [PubMed] [Google Scholar]
- Dansen TB, Smits LM, van Triest MH, de Keizer PL, van Leenen D, Koerkamp MG et al. (2009). Redox‐sensitive cysteines bridge p300/CBP‐mediated acetylation and FoxO4 activity. Nat Chem Biol 5: 664–672. [DOI] [PubMed] [Google Scholar]
- Di Bona D, Cippitelli M, Fionda C, Camma C, Licata A, Santoni A et al. (2006). Oxidative stress inhibits IFN‐alpha‐induced antiviral gene expression by blocking the JAK‐STAT pathway. J Hepatol 45: 271–279. [DOI] [PubMed] [Google Scholar]
- Dijkers PF, Medema RH, Pals C, Banerji L, Thomas NS, Lam EW et al. (2000). Forkhead transcription factor FKHR‐L1 modulates cytokine‐dependent transcriptional regulation of p27(KIP1). Mol Cell Biol 20: 9138–9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd RD, Sachdeva M, Mito JK, Eward WC, Brigman BE, Ma Y et al. (2015). Myogenic transcription factors regulate pro‐metastatic miR‐182. Oncogene. doi: 10.1038/onc.2015.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Weng X, Hu W, Lv Z, Xiao H, Ding C et al. (2015). Hypoxia‐inducible MiR‐182 promotes angiogenesis by targeting RASA1 in hepatocellular carcinoma. J Exp Clin Cancer Res 34: 67. doi: 10.1186/s13046-015-0182-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersole TA, Chen Q, Justice MJ, Artzt K (1996). The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins. Nat Genet 12: 260–265. [DOI] [PubMed] [Google Scholar]
- Eijkelenboom A, Burgering BM (2013). FOXOs: signalling integrators for homeostasis maintenance. Nat Rev Mol Cell Biol 14: 83–97. [DOI] [PubMed] [Google Scholar]
- Fabre S, Lang V, Harriague J, Jobart A, Unterman TG, Trautmann A et al. (2005). Stable activation of phosphatidylinositol 3‐kinase in the T cell immunological synapse stimulates Akt signaling to FoxO1 nuclear exclusion and cell growth control. J Immunol 174: 4161–4171. [DOI] [PubMed] [Google Scholar]
- Fan C, Liu S, Zhao Y, Han Y, Yang L, Tao G et al. (2013). Upregulation of miR‐370 contributes to the progression of gastric carcinoma via suppression of FOXO1. Biomed Pharmacother 67: 521–526. [DOI] [PubMed] [Google Scholar]
- Fendler A, Jung M, Stephan C, Erbersdobler A, Jung K, Yousef GM (2013). The antiapoptotic function of miR‐96 in prostate cancer by inhibition of FOXO1. PLoS One 8: e80807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Bankston A (2010). The star family member QKI and cell signaling. Adv Exp Med Biol 693: 25–36. [PubMed] [Google Scholar]
- Fernau NS, Fugmann D, Leyendecker M, Reimann K, Grether‐Beck S, Galban S et al. (2010). Role of HuR and p38MAPK in ultraviolet B‐induced post‐transcriptional regulation of COX‐2 expression in the human keratinocyte cell line HaCaT. J Biol Chem 285: 3896–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa‐Hibi Y, Kobayashi Y, Chen C, Motoyama N (2005). FOXO transcription factors in cell‐cycle regulation and the response to oxidative stress. Antioxid Redox Signal 7: 752–760. [DOI] [PubMed] [Google Scholar]
- Galarneau A, Richard S (2005). Target RNA motif and target mRNAs of the quaking STAR protein. Nat Struct Mol Biol 12: 691–698. [DOI] [PubMed] [Google Scholar]
- Garofalo M, Leva GD, Croce CM (2014). MicroRNAs as anti‐cancer therapy. Curr Pharm Des 20: 5328–5335. [DOI] [PubMed] [Google Scholar]
- Ge YF, Sun J, Jin CJ, Cao BQ, Jiang ZF, Shao JF (2013). AntagomiR‐27a targets FOXO3a in glioblastoma and suppresses U87 cell growth in vitro and in vivo . Asian Pac J Cancer Prev 14: 963–968. [DOI] [PubMed] [Google Scholar]
- Gebremedhn S, Salilew‐Wondim D, Ahmad I, Sahadevan S, Hossain MM, Hoelker M et al. (2015). MicroRNA expression profile in bovine granulosa cells of preovulatory dominant and subordinate follicles during the late follicular phase of the estrous cycle. PLoS One 10: e0125912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheysarzadeh A, Yazdanparast R (2015). STAT5 reactivation by catechin modulates H2O 2‐induced apoptosis through miR‐182/FOXO1 pathway in SK‐N‐MC cells. Cell Biochem Biophys 71: 649–656. [DOI] [PubMed] [Google Scholar]
- Go H, Jang JY, Kim PJ, Kim YG, Nam SJ, Paik JH et al. (2015). MicroRNA‐21 plays an oncogenic role by targeting FOXO1 and activating the PI3K/AKT pathway in diffuse large B‐cell lymphoma. Oncotargets 6: 15035–15049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto T, Takano M, Albergaria A, Briese J, Pomeranz KM, Cloke B et al. (2008). Mechanism and functional consequences of loss of FOXO1 expression in endometrioid endometrial cancer cells. Oncogene 27: 9–19. [DOI] [PubMed] [Google Scholar]
- Guerit D, Brondello JM, Chuchana P, Philipot D, Toupet K, Bony C et al. (2014). FOXO3A regulation by miRNA‐29a controls chondrogenic differentiation of mesenchymal stem cells and cartilage formation. Stem Cells Dev 23: 1195–1205. [DOI] [PubMed] [Google Scholar]
- Guo W, Jiang T, Lian C, Wang H, Zheng Q, Ma H (2014). QKI deficiency promotes FoxO1 mediated nitrosative stress and endoplasmic reticulum stress contributing to increased vulnerability to ischemic injury in diabetic heart. J Mol Cell Cardiol 75: 131–140. [DOI] [PubMed] [Google Scholar]
- Guo W, Shi X, Liu A, Yang G, Yu F, Zheng Q et al. (2011). RNA binding protein QKI inhibits the ischemia/reperfusion‐induced apoptosis in neonatal cardiomyocytes. Cell Physiol Biochem 28: 593–602. [DOI] [PubMed] [Google Scholar]
- Guo Y, Liu H, Zhang H, Shang C, Song Y (2012). miR‐96 regulates FOXO1‐mediated cell apoptosis in bladder cancer. Oncol Lett 4: 561–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttilla IK, White BA (2009). Coordinate regulation of FOXO1 by miR‐27a, miR‐96, and miR‐182 in breast cancer cells. J Biol Chem 284: 23204–23216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haflidadottir BS, Larne O, Martin M, Persson M, Edsjo A, Bjartell A et al. (2013). Upregulation of miR‐96 enhances cellular proliferation of prostate cancer cells through FOXO1. PLoS One 8: e72400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P et al. (2010). Transcriptome‐wide identification of RNA‐binding protein and microRNA target sites by PAR‐CLIP. Cell 141: 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haftmann C, Stittrich AB, Sgouroudis E, Matz M, Chang HD, Radbruch A et al. (2012). Lymphocyte signaling: regulation of FoxO transcription factors by microRNAs. Ann N Y Acad Sci 1247: 46–55. [DOI] [PubMed] [Google Scholar]
- Han BW, Feng DD, Li ZG, Luo XQ, Zhang H, Li XJ et al. (2011). A set of miRNAs that involve in the pathways of drug resistance and leukemic stem‐cell differentiation is associated with the risk of relapse and glucocorticoid response in childhood ALL. Hum Mol Genet 20: 4903–4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasseine LK, Hinault C, Lebrun P, Gautier N, Paul‐Bellon R, Van Obberghen E (2009). miR‐139 impacts FoxO1 action by decreasing FoxO1 protein in mouse hepatocytes. Biochem Biophys Res Commun 390: 1278–1282. [DOI] [PubMed] [Google Scholar]
- He W, Feng L, Xia D, Han N (2015). MiR‐374a promotes the proliferation of human osteosarcoma by downregulating FOXO1 expression. Int J Clin Exp Med 8: 3482–3489. [PMC free article] [PubMed] [Google Scholar]
- Hou T, Ou J, Zhao X, Huang X, Huang Y, Zhang Y (2014). MicroRNA‐196a promotes cervical cancer proliferation through the regulation of FOXO1 and p27Kip1. Br J Cancer 110: 1260–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housley MP, Udeshi ND, Rodgers JT, Shabanowitz J, Puigserver P, Hunt DF et al. (2009). A PGC‐1alpha‐O‐GlcNAc transferase complex regulates FoxO transcription factor activity in response to glucose. J Biol Chem 284: 5148–5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu TI, Wang MC, Chen SY, Yeh YM, Su WC, Chang WC et al. (2012). Sp1 expression regulates lung tumor progression. Oncogene 31: 3973–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Lv G, Zhou S, Zhou Y, Nie B, Duan H et al. (2015). The downregulation of MiR‐182 is associated with the growth and invasion of osteosarcoma cells through the regulation of TIAM1 expression. PLoS One 10: e0121175. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Huang W, Jin Y, Yuan Y, Bai C, Wu Y, Zhu H et al. (2014). Validation and target gene screening of hsa‐miR‐205 in lung squamous cell carcinoma. Chin Med J (Engl) 127: 272–278. [PubMed] [Google Scholar]
- Hudson MB, Rahnert JA, Zheng B, Woodworth‐Hobbs ME, Franch HA, Price SR (2014). miR‐182 attenuates atrophy‐related gene expression by targeting FoxO3 in skeletal muscle. Am J Physiol Cell Physiol 307: C314–C319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaconetti C, De Rosa S, Polimeni A, Sorrentino S, Gareri C, Carino A et al. (2015). Down‐regulation of miR‐23b induces phenotypic switching of vascular smooth muscle cells in vitro and in vivo . Cardiovasc Res 107: 522–533. [DOI] [PubMed] [Google Scholar]
- Jung HS, Seo YR, Yang YM, Koo JH, An J, Lee SJ et al. (2014). Galpha12gep oncogene inhibits FOXO1 in hepatocellular carcinoma as a consequence of miR‐135b and miR‐194 dysregulation. Cell Signal 26: 1456–1465. [DOI] [PubMed] [Google Scholar]
- Kajihara T, Jones M, Fusi L, Takano M, Feroze‐Zaidi F, Pirianov G et al. (2006). Differential expression of FOXO1 and FOXO3a confers resistance to oxidative cell death upon endometrial decidualization. Mol Endocrinol 20: 2444–2455. [DOI] [PubMed] [Google Scholar]
- Kaur N, Lu B, Monroe RK, Ward SM, Halvorsen SW (2005). Inducers of oxidative stress block ciliary neurotrophic factor activation of Jak/STAT signaling in neurons. J Neurochem 92: 1521–1530. [DOI] [PubMed] [Google Scholar]
- Kedde M, Agami R (2008). Interplay between microRNAs and RNA‐binding proteins determines developmental processes. Cell Cycle 7: 899–903. [DOI] [PubMed] [Google Scholar]
- Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C et al. (2007). RNA‐binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 131: 1273–1286. [DOI] [PubMed] [Google Scholar]
- Kim DY, Hwang I, Muller FL, Paik JH (2015). Functional regulation of FoxO1 in neural stem cell differentiation. Cell Death Differ 22: 2034–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M (2009). HuR recruits let‐7/RISC to repress c‐Myc expression. Genes Dev 23: 1743–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KM, Park SJ, Jung SH, Kim EJ, Jogeswar G, Ajita J et al. (2012). miR‐182 is a negative regulator of osteoblast proliferation, differentiation, and skeletogenesis through targeting FoxO1. J Bone Miner Res 27: 1669–1679. [DOI] [PubMed] [Google Scholar]
- Kloet DE, Burgering BM (2011). The PKB/FOXO switch in aging and cancer. Biochim Biophys Acta 1813: 1926–1937. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH (2006). In situ detection of miRNAs in animal embryos using LNA‐modified oligonucleotide probes. Nat Methods 3: 27–29. [DOI] [PubMed] [Google Scholar]
- Klotz LO (2012). Posttranscriptional regulation of connexin‐43 expression. Arch Biochem Biophys 524: 23–29. [DOI] [PubMed] [Google Scholar]
- Klotz LO, Sanchez‐Ramos C, Prieto‐Arroyo I, Urbanek P, Steinbrenner H, Monsalve M (2015). Redox regulation of FoxO transcription factors. Redox Biol 6: 51–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D et al. (2010). MicroRNA‐155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem 285: 17869–17879. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kouri FM, Hurley LA, Daniel WL, Day ES, Hua Y, Hao L et al. (2015). miR‐182 integrates apoptosis, growth, and differentiation programs in glioblastoma. Genes Dev 29: 732–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths‐Jones S (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ et al. (2005). Combinatorial microRNA target predictions. Nat Genet 37: 495–500. [DOI] [PubMed] [Google Scholar]
- Kundu P, Fabian MR, Sonenberg N, Bhattacharyya SN, Filipowicz W (2012). HuR protein attenuates miRNA‐mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res 40: 5088–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labied S, Kajihara T, Madureira PA, Fusi L, Jones MC, Higham JM et al. (2006). Progestins regulate the expression and activity of the forkhead transcription factor FOXO1 in differentiating human endometrium. Mol Endocrinol 20: 35–44. [DOI] [PubMed] [Google Scholar]
- Lam EW, Shah K, Brosens JJ (2012). The diversity of sex steroid action: the role of micro‐RNAs and FOXO transcription factors in cycling endometrium and cancer. J Endocrinol 212: 13–25. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A et al. (2007). A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larocque D, Galarneau A, Liu HN, Scott M, Almazan G, Richard S (2005). Protection of p27(Kip1) mRNA by quaking RNA binding proteins promotes oligodendrocyte differentiation. Nat Neurosci 8: 27–33. [DOI] [PubMed] [Google Scholar]
- Lau P, Bossers K, Janky R, Salta E, Frigerio CS, Barbash S et al. (2013). Alteration of the microRNA network during the progression of Alzheimer's disease. EMBO Mol Med 5: 1613–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M et al. (2011). Transcriptome‐wide analysis of regulatory interactions of the RNA‐binding protein HuR. Mol Cell 43: 340–352. [DOI] [PubMed] [Google Scholar]
- Lei BX, Liu ZH, Li ZJ, Li C, Deng YF (2014a). miR‐21 induces cell proliferation and suppresses the chemosensitivity in glioblastoma cells via downregulation of FOXO1. Int J Clin Exp Med 7: 2060–2066. [PMC free article] [PubMed] [Google Scholar]
- Lei R, Tang J, Zhuang X, Deng R, Li G, Yu J et al. (2014b). Suppression of MIM by microRNA‐182 activates RhoA and promotes breast cancer metastasis. Oncogene 33: 1287–1296. [DOI] [PubMed] [Google Scholar]
- Lennicke C, Rahn J, Lichtenfels R, Wessjohann LA, Seliger B (2015). Hydrogen peroxide – production, fate and role in redox signaling of tumor cells. Cell Commun Signal 13: 39. doi: 10.1186/s12964-015-0118-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung WK, He M, Chan AW, Law PT, Wong N (2015). Wnt/beta‐catenin activates MiR‐183/96/182 expression in hepatocellular carcinoma that promotes cell invasion. Cancer Lett 362: 97–105. [DOI] [PubMed] [Google Scholar]
- Levanon K, Sapoznik S, Bahar‐Shany K, Brand H, Shapira‐Frommer R, Korach J et al. (2014). FOXO3a loss is a frequent early event in high‐grade pelvic serous carcinogenesis. Oncogene 33: 4424–4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Ji P, Ouyang N, Zhang Y, Wang XY, Rubin DC et al. (2014a). Differential expression of miRNAs in colon cancer between African and Caucasian Americans: implications for cancer racial health disparities. Int J Oncol 45: 587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Liu B, Gao Y, Liu Y, Xu Y, Tong W et al. (2014b). Upregulation of microRNA‐107 induces proliferation in human gastric cancer cells by targeting the transcription factor FOXO1. FEBS Lett 588: 538–544. [DOI] [PubMed] [Google Scholar]
- Li J, Li P, Chen T, Gao G, Chen X, Du Y et al. (2015a). Expression of microRNA‐96 and its potential functions by targeting FOXO3 in non‐small cell lung cancer. Tumour Biol 36: 685–692. [DOI] [PubMed] [Google Scholar]
- Li P, Sheng C, Huang L, Zhang H, Cheng Z, Zhu Q (2014c). MiR‐183/‐96/‐182 cluster is up‐regulated in most breast cancers and increases cell proliferation and migration. Breast Cancer Res 16: 473. doi: 10.1186/s13058-014-0473-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhang J, Chen T, Yin P, Yang J, Cao Y (2015b). miR‐132 upregulation promotes gastric cancer cell growth through suppression of FoxO1 translation. Tumour Biol . [DOI] [PubMed] [Google Scholar]
- Li X, Du N, Zhang Q, Li J, Chen X, Liu X et al. (2014d). MicroRNA‐30d regulates cardiomyocyte pyroptosis by directly targeting foxo3a in diabetic cardiomyopathy. Cell Death Dis 5: e1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yu J, Du D, Fu S, Chen Y, Yu F et al. (2013). Involvement of post‐transcriptional regulation of FOXO1 by HuR in 5‐FU‐induced apoptosis in breast cancer cells. Oncol Lett 6: 156–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Dai T, Xiong H, Zhao X, Chen X, Yu C et al. (2010). Unregulated miR‐96 induces cell proliferation in human breast cancer by downregulating transcriptional factor FOXO3a. PLoS One 5: e15797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Teo S, Lam DH, Jeyaseelan K, Wang S (2012). MicroRNA‐10b pleiotropically regulates invasion, angiogenicity and apoptosis of tumor cells resembling mesenchymal subtype of glioblastoma multiforme. Cell Death Dis 3: e398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM et al. (2008). Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res 68: 664–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YY, Chou CF, Giovarelli M, Briata P, Gherzi R, Chen CY (2014). KSRP and MicroRNA 145 are negative regulators of lipolysis in white adipose tissue. Mol Cell Biol 34: 2339–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling N, Gu J, Lei Z, Li M, Zhao J, Zhang HT et al. (2013). microRNA‐155 regulates cell proliferation and invasion by targeting FOXO3a in glioma. Oncol Rep 30: 2111–2118. [DOI] [PubMed] [Google Scholar]
- Liu R, Liu F, Li L, Sun M, Chen K (2015). MiR‐498 regulated FOXO3 expression and inhibited the proliferation of human ovarian cancer cells. Biomed Pharmacother 72: 52–57. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang Z, Sun L, Chai N, Tang S, Jin J et al. (2011). MicroRNA‐499‐5p promotes cellular invasion and tumor metastasis in colorectal cancer by targeting FOXO4 and PDCD4. Carcinogenesis 32: 1798–1805. [DOI] [PubMed] [Google Scholar]
- Liu Y, Jiang J, Wang X, Zhai F, Cheng X (2013). miR‐582‐5p is upregulated in patients with active tuberculosis and inhibits apoptosis of monocytes by targeting FOXO1. PLoS One 8: e78381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Yao N, Liu G, Dong L, Liu Y, Zhang M et al. (2015). Functional screen reveals essential roles of miR‐27a/24 in differentiation of embryonic stem cells. EMBO J 34: 361–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida S, Spangenburg EE, Booth FW (2003). Forkhead transcription factor FoxO1 transduces insulin‐like growth factor's signal to p27Kip1 in primary skeletal muscle satellite cells. J Cell Physiol 196: 523–531. [DOI] [PubMed] [Google Scholar]
- Mao XP, Zhang LS, Huang B, Zhou SY, Liao J, Chen LW et al. (2015). Mir‐135a enhances cellular proliferation through post‐transcriptionally regulating PHLPP2 and FOXO1 in human bladder cancer. J Transl Med 13: 86. doi: 10.1186/s12967-015-0438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama H, Suzuki HI, Nishimori H, Noguchi M, Yao T, Komatsu N et al. (2011). miR‐135b mediates NPM‐ALK‐driven oncogenicity and renders IL‐17‐producing immunophenotype to anaplastic large cell lymphoma. Blood 118: 6881–6892. [DOI] [PubMed] [Google Scholar]
- McLoughlin HS, Wan J, Spengler RM, Xing Y, Davidson BL (2014). Human‐specific microRNA regulation of FOXO1: implications for microRNA recognition element evolution. Hum Mol Genet 23: 2593–2603. [DOI] [PubMed] [Google Scholar]
- McMillen BD, Aponte MM, Liu Z, Helenowski IB, Scholtens DM, Buttin BM et al. (2012). Expression analysis of MIR182 and its associated target genes in advanced ovarian carcinoma. Mod Pathol 25: 1644–1653. [DOI] [PubMed] [Google Scholar]
- Medema RH, Kops GJ, Bos JL, Burgering BM (2000). AFX‐like forkhead transcription factors mediate cell‐cycle regulation by Ras and PKB through p27kip1. Nature 404: 782–787. [DOI] [PubMed] [Google Scholar]
- Mehta A, Zhao JL, Sinha N, Marinov GK, Mann M, Kowalczyk MS et al. (2015). The microRNA‐132 and microRNA‐212 cluster regulates hematopoietic stem cell maintenance and survival with age by buffering FOXO3 expression. Immunity 42: 1021–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner NC, Filipowicz W (2010). Properties of the regulatory RNA‐binding protein HuR and its role in controlling miRNA repression. Adv Exp Med Biol 700: 106–123. [PubMed] [Google Scholar]
- Mendell JT, Olson EN (2012). MicroRNAs in stress signaling and human disease. Cell 148: 1172–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min M, Peng L, Yang Y, Guo M, Wang W, Sun G (2014). MicroRNA‐155 is involved in the pathogenesis of ulcerative colitis by targeting FOXO3a. Inflamm Bowel Dis 20: 652–659. [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S et al. (2007). Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell 1: 101–112. [DOI] [PubMed] [Google Scholar]
- Moritoki Y, Hayashi Y, Mizuno K, Kamisawa H, Nishio H, Kurokawa S et al. (2014). Expression profiling of microRNA in cryptorchid testes: miR‐135a contributes to the maintenance of spermatogonial stem cells by regulating FoxO1. J Urol 191: 1174–1180. [DOI] [PubMed] [Google Scholar]
- Morris BJ, Willcox DC, Donlon TA, Willcox BJ (2015). FOXO3: a major gene for human longevity – a mini‐review. Gerontology 61: 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M et al. (2011). Integrative regulatory mapping indicates that the RNA‐binding protein HuR couples pre‐mRNA processing and mRNA stability. Mol Cell 43: 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myatt SS, Wang J, Monteiro LJ, Christian M, Ho KK, Fusi L et al. (2010). Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Res 70: 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nho RS, Im J, Ho YY, Hergert P (2014). MicroRNA‐96 inhibits FoxO3a function in IPF fibroblasts on type I collagen matrix. Am J Physiol Lung Cell Mol Physiol 307: L632–L642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak K, Killmer K, Gessner C, Lutz W (2007). E2F‐1 regulates expression of FOXO1 and FOXO3a. Biochim Biophys Acta 1769: 244–252. [DOI] [PubMed] [Google Scholar]
- O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44: D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri F, Ahtiainen M, Lazzarini R, Pollanen E, Capri M, Lorenzi M et al. (2014). Hormone replacement therapy enhances IGF‐1 signaling in skeletal muscle by diminishing miR‐182 and miR‐223 expressions: a study on postmenopausal monozygotic twin pairs. Aging Cell 13: 850–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z et al. (2007). FoxOs are lineage‐restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 128: 309–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson AJ, Sharman JL, Benson HE, Faccenda E, Alexander SPH, Buneman OP et al. (2014). The IUPHAR/BPS Guide to PHARMACOLOGY: an expert‐driven knowledge base of drug targets and their ligands. Nucleic Acids Res 42 (Database Issue): D1098–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H, Jin Z, Chen S, Sun X, Yu J, Guo W (2015). MiR‐135b promotes proliferation and invasion of osteosarcoma cells via targeting FOXO1. Mol Cell Biochem 400: 245–252. [DOI] [PubMed] [Google Scholar]
- Peng SL (2008). Foxo in the immune system. Oncogene 27: 2337–2344. [DOI] [PubMed] [Google Scholar]
- Piva R, Spandidos DA, Gambari R (2013). From microRNA functions to microRNA therapeutics: novel targets and novel drugs in breast cancer research and treatment (Review). Int J Oncol 43: 985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putker M, Vos HR, van Dorenmalen K, de Ruiter H, Duran AG, Snel B et al. (2015). Evolutionary acquisition of cysteines determines FOXO paralog‐specific redox signaling. Antioxid Redox Signal 22: 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R, Nagarkatti P, Nagarkatti M (2015). Role of miRNA in the regulation of inflammatory genes in staphylococcal enterotoxin B‐induced acute inflammatory lung injury and mortality. Toxicol Sci 144: 284–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren JW, Li ZJ, Tu C (2015). MiR‐135 post‐transcriptionally regulates FOXO1 expression and promotes cell proliferation in human malignant melanoma cells. Int J Clin Exp Pathol 8: 6356–6366. [PMC free article] [PubMed] [Google Scholar]
- Rihani A, Van Goethem A, Ongenaert M, De Brouwer S, Volders PJ, Agarwal S et al. (2015). Genome wide expression profiling of p53 regulated miRNAs in neuroblastoma. Sci Rep 5: 9027. doi: 10.1038/srep09027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmele P, Liang R, Bigarella CL, Kocabas F, Xie J, Serasinghe MN et al. (2015). Mitochondrial metabolism in hematopoietic stem cells requires functional FOXO3. EMBO Rep 16: 1164–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquette‐Jazdanian AK, Kortum RL, Li W, Merrill RK, Nguyen PH, Samelson LE et al. (2015). miR‐155 controls lymphoproliferation in LAT mutant mice by restraining T‐cell apoptosis via SHIP‐1/mTOR and PAK1/FOXO3/BIM pathways. PLoS One 10: e0131823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva M, Mito JK, Lee CL, Zhang M, Li Z, Dodd RD et al. (2014). MicroRNA‐182 drives metastasis of primary sarcomas by targeting multiple genes. J Clin Invest 124: 4305–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarsour EH, Kalen AL, Goswami PC (2014). Manganese superoxide dismutase regulates a redox cycle within the cell cycle. Antioxid Redox Signal 20: 1618–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieber M, Chandel NS (2014). ROS function in redox signaling and oxidative stress. Curr Biol 24: R453–R462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez‐Diaz S et al. (2009). Aberrant miR‐182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia‐associated transcription factor. Proc Natl Acad Sci U S A 106: 1814–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senyuk V, Zhang Y, Liu Y, Ming M, Premanand K, Zhou L et al. (2013). Critical role of miR‐9 in myelopoiesis and EVI1‐induced leukemogenesis. Proc Natl Acad Sci U S A 110: 5594–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidman RL, Dickie MM, Appel SH (1964). Mutant mice (quaking and jimpy) with deficient myelination in the central nervous system. Science 144: 309–311. [DOI] [PubMed] [Google Scholar]
- Small EM, O'Rourke JR, Moresi V, Sutherland LB, McAnally J, Gerard RD et al. (2010). Regulation of PI3‐kinase/Akt signaling by muscle‐enriched microRNA‐486. Proc Natl Acad Sci U S A 107: 4218–4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song IS, Tatebe S, Dai W, Kuo MT (2005). Delayed mechanism for induction of gamma‐glutamylcysteine synthetase heavy subunit mRNA stability by oxidative stress involving p38 mitogen‐activated protein kinase signaling. J Biol Chem 280: 28230–28240. [DOI] [PubMed] [Google Scholar]
- Song W, Li Q, Wang L, Wang L (2015). Modulation of FoxO1 expression by miR‐21 to promote growth of pancreatic ductal adenocarcinoma. Cell Physiol Biochem 35: 184–190. [DOI] [PubMed] [Google Scholar]
- Spinetti G, Cordella D, Fortunato O, Sangalli E, Losa S, Gotti A et al. (2013). Global remodeling of the vascular stem cell niche in bone marrow of diabetic patients: implication of the microRNA‐155/FOXO3a signaling pathway. Circ Res 112: 510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantan S, Tominaga K, Gorospe M (2012). Functional interplay between RNA‐binding protein HuR and microRNAs. Curr Protein Pept Sci 13: 372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M, Dijkers PF, Kops GJ, Lens SM, Coffer PJ, Burgering BM et al. (2002). The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL‐2. J Immunol 168: 5024–5031. [DOI] [PubMed] [Google Scholar]
- Steitz JA, Vasudevan S (2009). miRNPs: versatile regulators of gene expression in vertebrate cells. Biochem Soc Trans 37: 931–935. [DOI] [PubMed] [Google Scholar]
- Stittrich AB, Haftmann C, Sgouroudis E, Kuhl AA, Hegazy AN, Panse I et al. (2010). The microRNA miR‐182 is induced by IL‐2 and promotes clonal expansion of activated helper T lymphocytes. Nat Immunol 11: 1057–1062. [DOI] [PubMed] [Google Scholar]
- Sun B, Li J, Shao D, Pan Y, Chen Y, Li S et al. (2015a). Adipose tissue‐secreted miR‐27a promotes liver cancer by targeting FOXO1 in obese individuals. Onco Targets Ther 8: 735–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Ji J, Huo G, Song Q, Zhang X (2015b). miR‐182 induces cervical cancer cell apoptosis through inhibiting the expression of DNMT3a. Int J Clin Exp Pathol 8: 4755–4763. [PMC free article] [PubMed] [Google Scholar]
- Takano M, Lu Z, Goto T, Fusi L, Higham J, Francis J et al. (2007). Transcriptional cross talk between the forkhead transcription factor forkhead box O1A and the progesterone receptor coordinates cell cycle regulation and differentiation in human endometrial stromal cells. Mol Endocrinol 21: 2334–2349. [DOI] [PubMed] [Google Scholar]
- Tan C, Liu S, Tan S, Zeng X, Yu H, Li A et al. (2015). Polymorphisms in microRNA target sites of forkhead box O genes are associated with hepatocellular carcinoma. PLoS One 10: e0119210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Bian Y, Tu C, Wang Z, Yu Z, Liu Q et al. (2013). The miR‐183/96/182 cluster regulates oxidative apoptosis and sensitizes cells to chemotherapy in gliomas. Curr Cancer Drug Targets 13: 221–231. [DOI] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE et al. (2007). FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128: 325–339. [DOI] [PubMed] [Google Scholar]
- Tran H, Maurer F, Nagamine Y (2003). Stabilization of urokinase and urokinase receptor mRNAs by HuR is linked to its cytoplasmic accumulation induced by activated mitogen‐activated protein kinase‐activated protein kinase 2. Mol Cell Biol 23: 7177–7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ucar A, Gupta SK, Fiedler J, Erikci E, Kardasinski M, Batkai S et al. (2012). The miRNA‐212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun 3: 1078. doi: 10.1038/ncomms2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heide LP, Jacobs FM, Burbach JP, Hoekman MF, Smidt MP (2005). FoxO6 transcriptional activity is regulated by Thr26 and Ser184, independent of nucleo‐cytoplasmic shuttling. Biochem J 391: 623–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Steitz JA (2007). AU‐rich‐element‐mediated upregulation of translation by FXR1 and argonaute 2. Cell 128: 1105–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA (2007). Switching from repression to activation: microRNAs can up‐regulate translation. Science 318: 1931–1934. [DOI] [PubMed] [Google Scholar]
- Wanet A, Tacheny A, Arnould T, Renard P (2012). miR‐212/132 expression and functions: within and beyond the neuronal compartment. Nucleic Acids Res 40: 4742–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BD, Ceniccola K, Yang Q, Andrawis R, Patel V, Ji Y et al. (2015a). Identification and functional validation of reciprocal microRNA–mRNA pairings in African American prostate cancer disparities. Clin Cancer Res 21: 4970–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GJ, Liu GH, Ye YW, Fu Y, Zhang XF (2015b). The role of microRNA‐1274a in the tumorigenesis of gastric cancer: accelerating cancer cell proliferation and migration via directly targeting FOXO4. Biochem Biophys Res Commun 459: 629–635. [DOI] [PubMed] [Google Scholar]
- Wang J, Li J, Shen J, Wang C, Yang L, Zhang X (2012a). MicroRNA‐182 downregulates metastasis suppressor 1 and contributes to metastasis of hepatocellular carcinoma. BMC Cancer 12: 227. doi: 10.1186/1471-2407-12-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Lin ZQ, Long B, Li JH, Zhou J, Li PF (2012b). Cardiac hypertrophy is positively regulated by MicroRNA miR‐23a. J Biol Chem 287: 589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zhu CF, Ma MZ, Chen G, Song M, Zeng ZL et al. (2015c). Micro‐RNA‐155 is induced by K‐Ras oncogenic signal and promotes ROS stress in pancreatic cancer. Oncotargets 6: 21148–21158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y et al. (2000). HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol 20: 760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lacroix G, Haines J, Doukhanine E, Almazan G, Richard S (2010). The QKI‐6 RNA binding protein localizes with the MBP mRNAs in stress granules of glial cells. PLoS One 5: e12824. doi: 10.1371/journal.pone.0012824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward EC, Hoekstra AV, Blok LJ, Hanifi‐Moghaddam P, Lurain JR, Singh DK et al. (2008). The regulation and function of the forkhead transcription factor, forkhead box O1, is dependent on the progesterone receptor in endometrial carcinoma. Endocrinology 149: 1942–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr MR, Binnewies M, Flach J, Reynaud D, Garg T, Malhotra R et al. (2013). FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature 494: 323–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Wang M, Qu X, Mah A, Xiong X, Harris AG et al. (2012). Differential expression of microRNAs during allograft rejection. Am J Transplant 12: 1113–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Lei R, Hu G (2015). Roles of miR‐182 in sensory organ development and cancer. Thorac Cancer 6: 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston MD, Pierce ML, Rocha‐Sanchez S, Beisel KW, Soukup GA (2006). MicroRNA gene expression in the mouse inner ear. Brain Res 1111: 95–104. [DOI] [PubMed] [Google Scholar]
- Wong HK, Veremeyko T, Patel N, Lemere CA, Walsh DM, Esau C et al. (2013). De‐repression of FOXO3a death axis by microRNA‐132 and ‐212 causes neuronal apoptosis in Alzheimer's disease. Hum Mol Genet 22: 3077–3092. [DOI] [PubMed] [Google Scholar]
- Wu L, Li H, Jia CY, Cheng W, Yu M, Peng M et al. (2012a). MicroRNA‐223 regulates FOXO1 expression and cell proliferation. FEBS Lett 586: 1038–1043. [DOI] [PubMed] [Google Scholar]
- Wu LH, Cai QQ, Dong YW, Wang R, He BM, Qi B et al. (2013). Decoy oligonucleotide rescues IGF1R expression from microRNA‐223 suppression. PLoS One 8: e82167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Sun H, Zeng W, He J, Mao X (2012b). Upregulation of mircoRNA‐370 induces proliferation in human prostate cancer cells by downregulating the transcription factor FOXO1. PLoS One 7: e45825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Ushmorov A, Leithauser F, Guan H, Steidl C, Farbinger J et al. (2012). FOXO1 is a tumor suppressor in classical Hodgkin lymphoma. Blood 119: 3503–3511. [DOI] [PubMed] [Google Scholar]
- Xu D, He X, Chang Y, Xu C, Jiang X, Sun S et al. (2013a). Inhibition of miR‐96 expression reduces cell proliferation and clonogenicity of HepG2 hepatoma cells. Oncol Rep 29: 653–661. [DOI] [PubMed] [Google Scholar]
- Xu J, Li R, Workeneh B, Dong Y, Wang X, Hu Z (2012). Transcription factor FoxO1, the dominant mediator of muscle wasting in chronic kidney disease, is inhibited by microRNA‐486. Kidney Int 82: 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D (2007). MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ‐specific miRNA cluster. J Biol Chem 282: 25053–25066. [DOI] [PubMed] [Google Scholar]
- Xu X, Ayub B, Liu Z, Serna VA, Qiang W, Liu Y et al. (2014a). Anti‐miR182 reduces ovarian cancer burden, invasion, and metastasis: an in vivo study in orthotopic xenografts of nude mice. Mol Cancer Ther 13: 1729–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Dong Z, Li Y, Yang Y, Yuan Z, Qu X et al. (2013b). The upregulation of signal transducer and activator of transcription 5‐dependent microRNA‐182 and microRNA‐96 promotes ovarian cancer cell proliferation by targeting forkhead box O3 upon leptin stimulation. Int J Biochem Cell Biol 45: 536–545. [DOI] [PubMed] [Google Scholar]
- Xu X, Wu J, Li S, Hu Z, Zhu Y, Liang Z et al. (2014b). Downregulation of microRNA‐182‐5p contributes to renal cell carcinoma proliferation via activating the AKT/FOXO3a signaling pathway. Mol Cancer 13: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Kondo E, Takeuchi M, Harashima A, Otani T, Tsuji‐Takayama K et al. (2011). miR‐155, a modulator of FOXO3a protein expression, is underexpressed and cannot be upregulated by stimulation of HOZOT, a line of multifunctional Treg. PLoS One 6: e16841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Li T, Gao C, Lv X, Liu K, Song H et al. (2014a). FOXO1 3′UTR functions as a ceRNA in repressing the metastases of breast cancer cells via regulating miRNA activity. FEBS Lett 588: 3218–3224. [DOI] [PubMed] [Google Scholar]
- Yang WB, Chen PH, Hsu TS, Fu TF, Su WC, Liaw H et al. (2014b). Sp1‐mediated microRNA‐182 expression regulates lung cancer progression. Oncotargets 5: 740–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XW, Shen GZ, Cao LQ, Jiang XF, Peng HP, Shen G et al. (2014c). MicroRNA‐1269 promotes proliferation in human hepatocellular carcinoma via downregulation of FOXO1. BMC Cancer 14: 909. doi: 10.1186/1471-2407-14-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S et al. (2012). LincRNA‐p21 suppresses target mRNA translation. Mol Cell 47: 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Jin L, Yang G, Ji L, Wang F, Lu Z (2014). Post‐transcriptional repression of FOXO1 by QKI results in low levels of FOXO1 expression in breast cancer cells. Oncol Rep 31: 1459–1465. [DOI] [PubMed] [Google Scholar]
- Zhang L, Quan H, Wang S, Li X, Che X (2015a). MiR‐183 promotes growth of non‐small cell lung cancer cells through FoxO1 inhibition. Tumour Biol 36: 8121–8126. [DOI] [PubMed] [Google Scholar]
- Zhang LF, Lou JT, Lu MH, Gao C, Zhao S, Li B et al. (2015b). Suppression of miR‐199a maturation by HuR is crucial for hypoxia‐induced glycolytic switch in hepatocellular carcinoma. EMBO J 34: 2671–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Liu L, Wang R, Tuo H, Guo Y, Yi L et al. (2013). MicroRNA‐217 promotes angiogenesis of human cytomegalovirus‐infected endothelial cells through downregulation of SIRT1 and FOXO3A. PLoS One 8: e83620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Li YS, Nguyen P, Wang KC, Weiss A, Kuo YC et al. (2013). Regulation of vascular smooth muscle cell turnover by endothelial cell‐secreted microRNA‐126: role of shear stress. Circ Res 113: 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Yin B, Liu Y, Hong Y, Zhang C, Fan J (2012). Mechanism and function of decreased FOXO1 in renal cell carcinoma. J Surg Oncol 105: 841–847. [DOI] [PubMed] [Google Scholar]


