Abstract
The high degree of conservation in microRNA from Caenorhabditis elegans to humans has enabled relatively rapid implementation of findings in model systems to the clinic. The convergence of the capacity for genomic screening being implemented in the prevailing precision medicine initiative and the capabilities of microRNA to address these changes holds significant promise. However, prostate, ovarian and breast cancers are heterogeneous and face issues of evolving therapeutic resistance. The transforming growth factor-beta (TGFβ) signaling axis plays an important role in the progression of these cancers by regulating microRNAs. Reciprocally, microRNAs regulate TGFβ actions during cancer progression. One must consider the expression of miRNA in the tumor microenvironment a source of biomarkers of disease progression and a viable target for therapeutic targeting. The differential expression pattern of microRNAs in health and disease, therapeutic response and resistance has resulted in its application as robust biomarkers. With two microRNA mimetics in ongoing restorative clinical trials, the paradigm for future clinical studies rests on the current observational trials to validate microRNA markers of disease progression. Some of today’s biomarkers can be translated to the next generation of microRNA-based therapies.
Keywords: biomarker, TGF-β, clinical trials, microRNA, microenvironment
Introduction
Precision medicine refers to leveraging larger scale somatic or genomic data in the informing medical decision-making. Although the principles of precision medicine are extending into proteomics and metabolomics, a vast amount of patient data currently exists in the form of genetic material. The genome expresses more non-coding RNA than the code for proteins. First recognized in 1993 in Caenorhabditis elegans, the noncoding RNAs, let-7 and lin-4 induced mRNA degradation (Lee et al. 1993). The mature miR let-7 sequence, conserved from C. elegans to humans, is downregulated in multiple cancers, which highlights the significance of the 3’UTR region during gene regulation (Pasquinelli 2000, Vella et al. 2004). MicroRNA (miR), most studied constituent of the non-coding RNAs, was termed to describe small (~22 nt) RNA regulators that influence gene functions (Lagos-Quintana et al. 2001). About 30–50% of all protein-coding genes are possibly regulated by miRs in health and deregulated in disease. However, the exquisite tissue and developmental stage specificity of miR expression mean that the regulation of miRs themselves can determine disease progression (Lagos-Quintana et al. 2001, Ruvkun 2001, Calin et al. 2002). Calin and coworkers found that specific miR downregulation was associated with the incidences of B cell chronic lymphocytic leukemia (Calin et al. 2002). Now what was once recognized as the passive circulation of miR (found in all body fluids) is emerging as signaling molecules between cells and robust biomarkers of cancer progression and therapy response.
Gene regulation by miRs and reciprocal regulation of miRs have now been studied for over 15 years and extensively reviewed (Vidigal & Ventura 2015). Briefly, miRs are transcribed as pri-miRNAs that are cleaved by the Drosha/DGCR8 complex in the nucleus (Han et al. 2004). The resulting hairpin-shaped pre-miRNA is further processed by Dicer in the cytoplasm (Zhang et al. 2002). The mature miRs employ the RNA-induced silencing complex (RISC) in the two mechanisms of gene regulation through mRNA degradation and translation inhibition (Fabian & Sonenberg 2012). The core of complex includes an argonaute protein for miR targeting and GW182 protein to serve as the scaffold on the mRNA poly-A tail (Song et al. 2004, Hutvagner & Simard 2008). The degree of pairing between the miR and the target mRNA seems to be a determinant of the mechanism of regulation. Direct complementarity will induce target mRNA cleavage by argonaute and be immediately repressive. In the indirect or imperfect complementarity scenario, miRs bind to the 3′ UTR prior to the poly-A tail to cause deadenylation of the mRNA. Loss of the poly-A tail leads to mRNA degradation. Imperfect complementation can additionally induce translational repression by blocking initiation of causing ribosomal drop-off. While the perfect pairing may be a more efficient repressor, there is a possibility that partial complementation may have a greater long-term efficacy: an issue considered in therapeutic design. Although miRs can bind the ORF or 5′UTR to enhance translation (Orom et al. 2008, Lee et al. 2009), the 3′ UTR has been the target for miRs in the clinic. The regulation of the miR expression by hormones and cytokines will be further detailed here with a focus on the tumor and tumor associated microenvironment cell types.
The clinical application of miRs has rapidly matured from aspirational to now exploiting its diagnostic and therapeutic potential. We refer the reader to the comprehensive reviews describing the mechanisms of miR action (Wagner et al. 2014, Fendler et al. 2016, Masliah-Planchon et al. 2016). The direct role of miRs in cancer was made by the seminal publication of mutations in miR processing enzymes, DICER and DROSHA leading to ovarian cancer (Merritt et al. 2008). Now there are two observational clinical trials for ovarian cancer patients to determine the effects on miR expression in patients (ClinicalTrials.gov identifier: Nbib1970696 and Nbib1572467). The pleotropic functions of miRs associated with transforming growth factor-beta/bone morphogenic protein (TGF-β/BMP) signaling will be used as an example as many of the miRs in current clinical utility seem to converge on this pervasive pathway. To appreciate the progress in the field, we need to start with one of the first mammalian miR identified, let-7. Its notoriety stems from KRas being one of its many targets. Although KRas is the most mutated gene in cancer, its activity is often elevated in the absence of mutations. Many miRs have been described to stimulate the RAS-MAPK pathway (miR-31, miR-143, miR-4689) (Hiraki et al. 2015, Chen et al. 2016, Edmonds et al. 2016). Regardless, in breast and ovarian cancer, let-7 is frequently downregulated, leading to increased KRas expression (Johnson et al. 2005, Dai et al. 2015). Since both of these cancers are commonly associated with activating Ras mutations, the loss of let-7 is even more significant in potentiating tumor progression. Although prostate cancer is not one of the cancers associated with Ras mutations through its tumor evolution, it is also associated with let-7 downregulation (Albino et al. 2016). The let-7 example points to one of the many similarities the three cancers have. However, the rationale for active clinical trials using let-7 mimetics has a broader basis, and that is the fact that let-7 is a potent down regulator of HMGB1, c-myc, IL-6 and cyclin D2. Thus, therapeutic targeting of a single miR can potentially address a multitude of genetic and epigenetic changes that need to be addressed when dealing with the heterogeneous disease of each ovarian, prostate or breast cancer patient. However, as with any ‘dirty drug’, antagonizing or mimicking a single miR with the many gene targets will result in unwanted side effects. Although the path from bench-to-bedside for miRs is not very different from any other oncology therapeutic or biomarker, the stark difference in miR mechanism-of-action make it important to understand this topic in the era of precision medicine. The mandate of the Precision Medicine Initiative (NIH) is to ultimately have data-driven care for patients. Cancer genomic and RNAseq analysis of patients is the most developed unbiased high throughput means of patient screening. Accordingly, miRs are prominent biomarkers currently being validated in the clinic today to support diagnosis, prognosis or therapeutic efficacy. Further, miRs are beginning to reach the clinic as therapeutics themselves to address some of the changes revealed by large-scale genomic analysis. However, it is important to remember miRs are generated by the cancer epithelia as well as its microenvironment. And in most cases, systemic administration of miR-based therapy will impact both cancer cells and its microenvironment. Thus, for miR studies to successfully translate to the patient, many cell types need to be considered (Sood et al. 2006).
Endocrine-related cancers of breast, prostate and ovaries will be the focus of this review. We discuss the mechanisms that regulate miRs during cancer progression. The clinical applications used to target miR functions increase precision, especially when standard therapies are less effective. Therefore, the urgency to understand the mechanisms by which miRs function during endocrine-related cancers is reflected in the studies focused on miR signaling in the tumor microenvironment.
Rationale and pitfalls in targeting TGF-β/BMP signaling
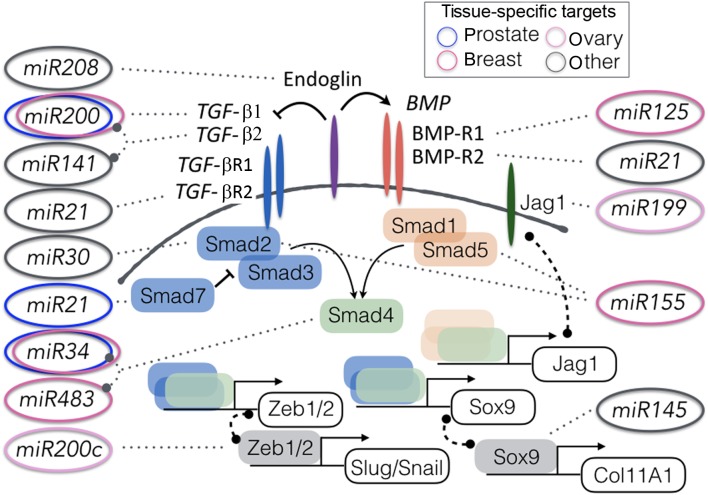
AS TGF-β signaling is a master regulator of tumorigenesis, the tumor microenvironment, and metastatic progression, this pathway impacts many miRs with significant clinical potential. The broad range of pathways influenced by TGF-β signaling is reflected in nearly 40 members comprising the TGF-β gene family. These members include the three TGF-β ligands, growth differentiation factors (GDFs), inhibin, activin, nodal, endoglin, lefty and BMP ligands (Chang et al. 2002). The complexities of stromal–epithelial interactions involving TGF-β signaling manifest itself as a tumor suppressor in tumorigenesis, yet demonstrate tumor promoting activity in metastatic progression (Bhowmick et al. 2003, 2004, Taylor et al. 2011, Lebrun 2012, Smith & Kang 2013, Neuzillet et al. 2015). The cross-talk between TGF-β signaling and miR functions is based on an autoregulatory feedback loop observed (Butz et al. 2011). The miR profiles of pituitary, prostate and breast cancer directly correlate with the disruptive functions of TGF-β. Several miRs, such as miR-21, miR-34a, and the miR-200 family cluster target TGF-β signaling in prostate, breast and thyroid cancer for their functions during tumor progression and promotion (Braun et al. 2010, Shen et al. 2012, Chen et al. 2016) (Fig. 1). Interestingly, miR-21 can also in turn promote TGF-β1-induced epithelial to mesenchymal transdifferentiation (Li et al. 2016). Conversely, miR-200 expression down regulates known targets, including TGF-β2, that are particularly up regulated in many cancers (Benlhabib et al. 2015). These few miRs serve just as examples of the cross-regulation with the expression with TGF-β ligands.
Figure 1.
Modulation of the TGF-β/BMP signaling pathways by miRs. Validated targets in prostate, ovarian, and breast cancer are indicated by the corresponding colored ovals. miR targets identified in other tissues are indicated in grey ovals, as miR targets in one tissue may not be effective in another tissue. Only relevant miRs discussed in the paper are shown here and by no means depict all the TGF-β/BMP-associated miRs.
Small molecule and neutralizing antibody-based antagonism of the TGF-β signaling axis is being pursued for multiple cancers. However, with the pleotropic roles of TGF-β it was not surprising that targeting the TGF-β ligands or receptors was associated with some unwanted side effects (Kulkarni et al. 1993, Anderton et al. 2011). It is possible that the specificity of a miR to a particular TGF-β ligand isoform or downstream effectors may limit some of these unwanted effects. Of these effectors, Smad2, Smad3 and Smad4 activate downstream transcription, but can be inhibited by STRAP and Smad7 activation endogenously. In addition, p38MAP kinase, AKT and RhoA are downstream of TGF-β in a cell-specific manner, with distinct and overlapping functions with Smad signaling (Bakin et al. 2000, Bhowmick et al. 2001a,b, 2003). Smad2 and Smad3 are targeted by miR-155 and miR-200a, respectively (Kong et al. 2008, Park et al. 2008, Louafi et al. 2010, Ahn et al. 2012). Smad4, however, is targeted by miR-130a, miR-182, miR-205 and miR-483 (Hao et al. 2011, Geraldo et al. 2012, Egawa et al. 2016). Identification of miRs that target TGF-β downstream effectors presents an advantage of specificity. However, since Smad4 is a shared effector of much of the broader TGF-β family of ligands it may share some of the same side effects faced with the TGF-β receptor kinase inhibitors. The function of such miRs, if used in a therapeutic function, would ultimately be dictated by the pleotropic nature of the Smad proteins. For example, Smads regulate transcriptional repressors (i.e., Snail, ZEB1/2) associated with EMT, binding to proteins required to produce miRs (i.e., p68) and influencing microenvironment components, such as cancer-associated fibroblasts (CAF).
The role of TGF-β during the biogenesis and maturation of miRs has been analyzed extensively in several disease states. Both TGF-β and BMP signaling regulate vascular integrity, acting through the vascular smooth muscle cells. Inhibition of either TGF-β or BMP signaling can disrupt vascular smooth muscle cell contractile phenotype. TGF-β/BMP-induced miR-21 had an essential role in the expression of smooth muscle specific genes CNN1 (calponin) and ACTA2 (α-smooth muscle actin) (Davis et al. 2008). The study found that TGF-β/BMP signaling regulated miR-21 expression by potentiating Smad1/5 or Smad2/3 to p68, a component of the DROSHA miR processing complex (Davis et al. 2008). This indicated that TGF-β/BMP signaling can regulate miR biosynthesis and not just transcription. It could mean that many miRs, besides miR-21, may be regulated in this fashion. On its own, this does not necessarily affect miR-based therapeutic design, but it would suggest that miRs that target the Smad proteins might affect the biosynthesis of other miRs. Due to the broad target range of any individual miR, it would require that all such Smad-directed miR be tested for effects on other miRs.
miRs impact epithelial–mesenchymal transdifferentiation
Epithelial–mesenchymal transition (EMT) is a multi-step process that regulates changes in cell morphology during embryogenesis, tissue development and tumor development (Kalluri & Weinberg 2009, Smith & Bhowmick 2016). One of the pro-tumorigenic roles of TGF-β signaling includes the potentiation of EMT (Oft et al. 1996, Bakin et al. 2000, Bhowmick et al. 2001a,b, 2003). The miRs associated with EMT have some overlap with those discussed regulated by TGF-β. They are important to note due to their relevance to tumor aggressiveness and therapeutic resistance. Snail1 and Slug are transcriptional repressors of epithelial markers (E-cadherin, zona occludin) and upregulate the expression of mesenchymal genes (vimentin, N-cadherin) (Thiery et al. 2009). miR-9 was found to downregulate Snail resulting in E-cadherin upregulation in melanoma to ultimately decrease growth and metastasis in vivo (Liu et al. 2012). Recently, miR-9 and miR-200c was found to regulate PDGFR-mediated endothelial differentiation of triple negative breast cancer (D’Ippolito et al. 2016). miR-9 inhibition or miR-200c restoration, delivered peritumorally to MDA-MB-231 xenografts, decreased the number of vascular lacunae, associated with breast tumor growth and dissemination (D’Ippolito et al. 2016). Another target of miR-200 is Zeb1/Zeb2 in the maintenance of cellular plasticity, a major hallmark of tumor cell morphology (Brabletz & Brabletz 2010). Interestingly, Zeb1 and Zeb2 bind to the E- and Z-box enhancer sequences of miR-200f promoter regions. In further cross-talk within the EMT regulators, Snail1 assists in the transcriptional repression of miR-200f, to enhance EMT (Diaz-Lopez et al. 2015). Importantly, miR-16 and miR-200 family members silences TGF-β signaling and blocks EMT (Brabletz & Brabletz 2010, Wang et al. 2014b, Tang et al. 2016). Individuals have the potential to influence several mRNA targets to regulate cancer progression (Davis-Dusenbery and Hata 2010). However, in the study of their endogenous expression in cancer, families of miRs can be expressed concomitantly to have broad effects. When inhibiting a miR, therapeutic efficacy may be limited as other miR family members can compensate for the targeted miR. However, the use of specific miR mimetics would not be plagued by this issue. In a specific example, ectopic expression of miR-200 members significantly reduced anaplastic thyroid carcinoma cell invasion in a ZEB1/ZEB2-dependent manner (Fig. 1) (Braun et al. 2010). As anaplastic thyroid carcinoma is a highly metastatic disease with poor prognosis, these findings suggest that a similar strategy for ovarian and triple negative breast cancer could be promising. It is important not to automatically assume that the regulation of a particular miR or its targets will be conserved from one tissue to another. However, findings in one tissue can suggest its testing in another tissue of interest. miR-10b is one such example where it was originally found to regulate EMT in a TGF-β dependent manner in breast cancer (Ma et al. 2007). Later, in independent mouse models, miR-10b was found to inhibit breast and pancreatic tumor metastasis (Moriarty et al. 2010, Ouyang et al. 2014). Although in both reports EMT was attributed to be rational for the role of miR-10b on metastatic potential, the targets identified were entirely different. Since the targets were not cross-validated in the two tissues, there is no way to know if the common phenotypic and physiologic readout of miR action was a result of the same target or targets. Despites the caveats on the specificity of miR targets, the role of miRs on EMT and metastatic progression are well supported.
Addressing the tumor microenvironment with miRs
The concept of tumors being composed of disconnected ‘rogue’ masses of malignant cells has long been abandoned. We now understand that tumors function like organs, and the components that make up tumors and the surrounding environment plays intimate roles in cancer progression. The interactions between miR-21 and Smad7, for example, have been shown to predict the formation of reactive skin cancer-associated fibroblasts (CAF) (Li et al. 2013b). As a target of miR-21, Smad7 translation is inhibited leading to an increase in cells with the CAF phenotype. However, when miR-21 is blocked or Smad7 was overexpressed, TGF-β1-induced CAF formation was inhibited. The CAF and tumor epithelia of many cancer types rely on TGF-β signaling to mediate communication and increase tumor growth and potentiate therapy resistance (Coulouarn et al. 2008, Vonlaufen et al. 2008, Duner et al. 2010, Coulouarn & Clément 2014). However, due to the importance of the spatiotemporal nature of miR expression and action (Sood et al. 2006), we divided the discussion of specific miRs to the CAF associated with the three particular cancer types and immune cells in the next sub-sections.
Regulation of miRs within the prostate tumor microenvironment
The prostate luminal and basal epithelia are encircled by smooth muscle cells that differentiate into activated fibroblastic cells during cancer progression. The tumor inductive nature of CAF was first coined in the context of prostate cancer (Olumi et al. 1999). CAFs are a major component of the tumor stroma with undoubted cancer-promoting effects. Of particular interest, TGF-β signaling influences CAF ability to mediate oncogenic transformation of normal epithelial cells (Bhowmick et al. 2004). The heterogeneous CAF populations can communicate within the stromal compartment in depositing certain ECM components and express cytokines/growth factors to facilitate cancer invasion and even instruct carcinoma cells to the site of metastatic invasion (Kiskowski et al. 2011, Li et al. 2013b). In prostate cancer, the CAF often lose TGF-β receptor type II expression due to promoter methylation (Banerjee et al. 2014). However, we have found that there is a juxtaposition of TGF-β-responsive and -nonresponsive CAF in the prostate cancer microenvironment (Kiskowski et al. 2011). The loss of TGF-β responsiveness in the CAF can lead to the accumulation of DNA damage and expression of multiple growth factors (Coppe et al. 2008, Banerjee et al. 2014). P53-induced miR-34a was found to restore DNA integrity (He et al. 2007, Yamakuchi et al. 2008). Interestingly, miR-34a expression in CAF has not been closely studied with respect to stromal DNA damage accumulation and its activation. Doldi and coworkers suggested that in prostate cancer the CAF could be activated in one of two ways: by IL-6 or TGF-β (Doldi et al. 2015). Both these cytokines can be sourced from the adjacent cancer epithelia in vivo. In this study, miR-133b emerged as a common mediator of CAF activation, as determined by the expression of markers such as, ACTA2, FAP, S100A4 and COL4A2 (Doldi et al. 2015). The expression of miR-409 by CAF was associated with prostatic tumor epithelial expansion and EMT (Josson et al. 2015). The surrounding prostate epithelia seemed to take up the miR-409, leading to increased tumor migration and invasion. Tumor growth continued due to miR-409-directed inhibition of RSU1 and STAG2 tumor suppressors. miR-15 and miR-16 are downregulated in prostate cancer CAF in the majority of 23 patients (Musumeci et al. 2011). These expression patterns inversely correlated with upregulation of FGF2 and FGFR1, supportive of the tumor inductive nature of CAF. In addition to these miRs that potentially serve as biomarkers for prostate cancer progression, Shen and coworkers found that plasma miR-21, miR-20a, miR-145 and miR-221 could risk stratify patients for tumor relapse (n = 82) (Shen et al. 2012). Patients with high or intermediate risk had particularly elevated expression of miR-21 (P = 0.047) and miR-145 (P = 0.011) compared to lower risk scores (Shen et al. 2012). Although the source of circulating miRs may be from either the cancer or the microenvironment, the capacity to affect the gene expression on cells that do not express the miR itself is eye-opening when it comes to expression profiling tumors, as is the practice of precision medicine. It also means that therapeutic targeting of miRs needs to consider both the tumor epithelia and its microenvironment.
Regulation of miRs within the ovarian tumor microenvironment
All three major cell types of the ovaries (epithelial, germ and stromal cells) can develop malignancies. Although the epithelial tumors are the most common, the contribution of CAF in this disease is evident by shear bulk and prevalent cross-talk. The fibrotic scarring and development of fibroblast populations closely related to the epithelial tumor cells seem to correlate with miR functions. It has been difficult to stratify serous ovarian cancer. The pathologic similarities of this common ovarian cancer type do not reveal the level of chemoresistance of these tumors. However, a set of ECM and ECM remodeling proteins (10-gene signature: AEBP1, COL11A1, COL5A1, COL6A2, LOX, POSTN, SNAI2, THBS2, TIMP3 and VCAN) were identified as associated with poor overall survival (Cheon et al. 2014). The signature genes are TGF-β regulated and their involvement with miR functions is broad. MMPs, TIMPs and Smads have a defined interplay with miRs in cancer metastasis. Examining miR silencing of Smad3 by miR-200a, for example, resulted in downregulation of a different set of ECM proteins (i.e., collagen I, collagen IV, fibronectin, PAI-1 and α-smooth muscle actin) (Wang et al. 2011). Careful interrogation of the ovarian cancer-associated signature resolved COL11A1 to be a determinant of lethal disease progression (Cheon et al. 2014). COL11A1expression is regulated by a TGFβ target transcription factor, SOX9. SOX9 is a direct target of miR-145 and is found to regulate COL11A1 expression (Yang et al. 2011). As the primary tumor microenvironment can predispose the cancer epithelia for metastasis, the metastatic site itself clearly has an influence on the adaptability of the tumor cells (Bhowmick 2012). One way the secondary site can be made adaptive to the tumor cells is the transport of cancer-derived miRs via exosomes. A seminal example of this phenomenon was described for pancreatic cancer metastasis to the liver (Hoshino et al. 2015). As another highly metastatic tumor type, ovarian cancer frequently spreads to the omentum adipose fat. In one example, the omental fat cells and associated CAF were found to express exosomes containing miR-21 (Au Yeung et al. 2016). miR-21 is induced by many cytokines including TGF-β and is associated with collective migration (Dean et al. 2015). The ovarian cancer epithelia were found to accept these miR-21 containing exosomes to support chemoresistance by directly silencing its target, APAF1 (Au Yeung et al. 2016). Metastatic breast and prostate cancer cells also overexpress miR-21. miRs have been identified in exosomes and microvesicles derived from several tissues, plasma, saliva and urine to support a means of cell to cell transfer and the viability of the use of exosomes as biomarker capsules. Thus, it is fitting that therapeutic microvesicles can be generated to transfer miR cargo to counter metastatic progression.
Regulation of miRs within the breast tumor microenvironment
miRs identified in CAF can regulate the expression of paracrine factors. Dysregulated miRs in breast CAF included upregulation of miR-221, miR-31, and miR-221 with the downregulation of miR205, miR-200b, miR-200c, miR-141, miR-101, miR-342, let-7g and miR-26b affecting all aspects of cell differentiation and paracrine regulation (Zhao et al. 2012). Although in the epithelia miR-205 and miR-200 family members (miR-200c, miR-200b and miR-141) are associated with EMT progression, in fibroblastic cells they clearly have a different function. In the breast CAF, miR-200 family members seem to contributed to ECM remodeling through the expression of fibronectin and lysyl oxidase to potentiate cancer epithelial invasion (Tang et al. 2016). In the absence of somatic mutations in the CAF population, epigenetic changes in the form of DNA methylation and miR expression can dictate the fate of adjacent epithelia. However, the redundancy of miR-200 downregulation in both the epithelial and stromal compartments is highly supportive of using miR mimetic of this species.
We have discussed examples where the miR itself can act in a paracrine manner. Another example of this for breast cancer was identified in exosomes expressed by CAF recently. Upregulation of miR-133b in CAF was found to travel via gap junction proteins into breast cancer epithelia, where it has anti-apoptotic and pro-growth molecule with oncomiRs properties (Katakowski et al. 2010, Xin et al. 2012). Interestingly, miR-133b transfer could be limited by the use a gap junction inhibitor (Xin et al. 2012), revealing a novel way of countering the miRs identified in primary tumors to prevent future metastatic progression. Incidentally, miR-133b was also identified to be a poor prognostic indicator in prostate cancer, in causing further aggressive behavior (Li et al. 2014). For prostate cancer, miR-133b was of epithelial origin. But the same miR-133b expressed by breast cancer CAF suggests that an individual miR can have a role in the cancer and its microenvironment regardless of its source.
miR regulation in immune system signaling
The introduction of checkpoint inhibitors in the armamentarium against cancer has caused us to reevaluate our understanding of T cell biology, in terms of clinical applications. Now, companion biomarkers and combination therapy candidates are a focus in many labs. miRs can play an important role in both these fronts. As TGF-β is a known immune-suppressor, it is not surprising that it has a central role in adaptive and innate immune function. The Smad2 silencing by miR-155 can promote B-cell activation as well as enable CD4 T cells mature to TH17 and TH1 cells (Mehta & Baltimore 2016). miR-155 is also critical in cells of the myeloid lineage. miR-155 expression is upregulated by NF-κB signaling, downstream of toll-like receptor activity to drive dendritic cells and macrophage differentiation (O’Connell et al. 2007). Smad4-mediated stimulation of granulocyte differentiation by TGF-β is tempered by miR-130a expression (Hager et al. 2011). The miR-34a is a member of miR-34 family that target TGF-β/Smad4 signaling in T-regulatory (Treg) cell tumor recruitment. Yang and coworkers demonstrated that miR-34a downregulation in liver tissues results in CCL22 expression for the recruitment of Treg-mediated escape from immune surveillance (Yang et al. 2012). Numerous publications on miR-34a suggesting its role as a tumor suppressor, demonstrate a reduction in several cancer types including breast, ovarian and prostate cancer (Corney et al. 2010, Peurala et al. 2011, Benassi et al. 2012). Analysis of miR-34 in human epithelial ovarian cancer showed that there was a 100% decrease in miR-34, and a 72% decrease in miR-34b*/c in the context of p53 mutation (Corney et al. 2010). Wild type p53 correlated with a 93% decrease in miR-34 in ovarian cancer cells. Clinically, stage III and stage IV tumors were analyzed where reduced miR-34 was significant (P = 0.0029, P = 0.0171, respectively). Interestingly, miR-34a downregulation is frequently regulated by promoter methylation in cancer cells. These counter acting epigenetic regulations impact the expression of endogenous oncogenes (e.g., MYC) as well as exogenous immune infiltration (Benassi et al. 2012, Yang et al. 2012). The confounding epigenetic mechanisms need to be considered before long-term clinical use of miR-based therapeutics is in practice.
Clinical applications of miRs in ovarian, prostate and breast cancer
miRs are currently part of many trials examining genomic signatures to contend with the issue of patient selection to identify companion biomarkers for conventional therapeutics. As miRs are increasingly examined in patients, the importance of not making generalizations of a miR being oncogenic or tumor suppressor is clear. Further sources of variability in miR detection in bodily fluids can be associated with factors such as diet, age and circadian rhythms. Yet, in multiple independent studies miR-200 family was upregulated in metastatic breast and prostate cancers (Lin et al. 2014, Taylor et al. 2016). However, miR-200 family members have been reported to be both up- as well as downregulated in ovarian cancer (Park et al. 2008, Kan et al. 2012). Notwithstanding, miRs are attractive due to their stability and conservation among species (Chen et al. 2008). miRs gain their extraordinary stability from their small size (20–24 nt) being less exposed to degradation (even during FFPE sample processing), encapsulated in protective exosomes or microvessicles and most commonly its association with RNA binding proteins. Exosome-free miRs associated with argonaute proteins make up much of what is detectable in circulation (Turchinovich et al. 2011). It is not just a coincidence that miRs have diagnostic value for cancer patients, but rather that miRs are frequently located at fragile chromosomal sites associated with cancer hotspots (Calin & Croce 2006). Accordingly, screening both free and exosome-associated miRs by high-throughput sequencing platforms have revealed changes in breast, prostate and ovarian cancer patients.
RNA-based therapies have been tested for twenty years, with antisense oligonucleotides having the longest track record in the clinic. As of 2016, eight of those therapies had reached phase III clinical trials. Short interfering RNAs (siRNAs) have been used in the clinic, but none have reached phase III yet. However, there are only two current trials using miRs in interventional trials. In these phase I studies, miR mimetics have been used to restore miR-34 (MRX34) and miR-16 (TargomiRs) activity. Some of the roles of miR-34 have already been discussed, as one of its targets is Smad4. As previously described, miR-16 inhibits TGF-β-induced EMT by silencing p-FAK and p-Akt expression, disrupting NF-κB and Slug transcriptional activity (Wang et al. 2014b). Although it is not possible to assess the clinical efficacy of the miR-based therapeutics at this stage, these two drugs were well tolerated with minimal inflammatory side effects. Regardless, it should be noted that the FDA has not approved an RNA-based drug to date. The reasons for the low success rate are not entirely clear, but a critical hurdle of therapeutic delivery seems to have improved. The lessons learned through the long history of antisense RNA-therapy are evident, as both miR trials use nanoparticle-based formulations to enhance drug availability. In the near term, we may also see further improvements in tissue targeting and sustained release of miRs or anti-miRs encapsulated in nanoparticles conjugated to antibodies or peptides. However, with about a 7% likelihood of FDA approval of any oncology drug, many of the challenges are not unique to RNA-based therapy (Hay et al. 2014). The need for understanding tumor heterogeneity, mechanisms of host–tumor interaction, and patient selection are only a few obvious issues that we still need to address.
miRs in prostate cancer patient care
Prostate cancer is the most common non-cutaneous cancer in men and, next to lung cancer, is the second leading cancer killer in men. Improvements in survival from the disease have been due to advances in first line interventions (surgery and radiation) as well as newer antagonists to the androgen signaling axis. Yet, metastatic prostate cancer has no cure as resistance to androgen antagonists are inevitable, and taxane therapy was approved by the FDA for providing an increase in three months of overall survival (Petrylak et al. 2004, Tannock et al. 2004). Thus, markers for patient selection and sensitizing agents for current therapies are a focus in the field. Currently, there are a number of clinical trials that aim to analyze the miRs in prostate cancer and treat the malignancies by using miRs, detailed in Table 1. As an example, the Medical College of Wisconsin conducted a study to identify exosomal miRs from peripheral blood of prostate cancer patients with systemic disease to predict response to androgen deprivation therapy. In the study, they gathered blood sample at the time of treatment, 3 months post-treatment and upon disease progression to determine changes on disease response (ClinicalTrials.gov Identifier: Nbib2366494). The study further aims to compare exosomal RNA levels between groups of patients who have relapsed as opposed to those in remission within 2 years of treatment. In a phase II study, expression levels of circulating miRs are being validated as a biomarker for sensitivity to abiraterone acetate (androgen synthesis inhibitor) and to predict metastatic prostate cancer progression (ClinicalTrials.gov identifier: Nbib1503229). These observational and interventional study examples would help in the process of decision-making for patient care. One could envision such a set of markers complementing imaging techniques in identifying metastatic status. Most prostatic metastasis is invisible to current imaging techniques in the early stages. If stable miRs could predict such an eventuality, it could suggest the use of chemotherapy, such as a taxane, at a time when it could appreciably alter overall survival.
Table 1.
Prostate cancer clinical trials involving miRs.
| Trial reference | Study type | Institution | Trial title |
|---|---|---|---|
| NCT2366494 | Observational | Medical College of Wisconsin | MicoRNAs to predict response to androgen deprivation therapy |
| NCT1503229 | Interventional | University of Washington | Abiraterone acetate in treating patients with metastatic hormone-resistant prostate cancer |
| NCT1220427 | Observational | Wuerzburg University Hospital | microRNA expression profiles in high risk prostate cancer |
| NCT2471469 | Observational | Radboud University | Personalizing enzalutamide therapy by understanding the relation between tumor mRNAs, miRNAs and treatment response |
| NCT2391051 | Interventional | University of Erlangen-Nürnberg Medical School | Focal brachytherapy in patients with selected ‘low-risk’ prostate cancer – a phase-II-trial |
Another set of observational trials attempts to identify therapeutic efficacy by measuring dynamic changes in the expression of specific miRs in circulation in response to androgen deprivation therapy. A current study underway at Radboud University (Nijmegen, Netherlands) explores the possibility of dosing enzalutamide therapy (androgen receptor antagonist) based on reduction in a panel of select miRs to assess treatment response (ClinicalTrials.gov identifier: Nbib2471469). A phase II trial conducted by University of Erlangen-Nürnberg Medical School (Nürnberg, Germany) explores the feasibility and toxicity of focal brachytherapy in 50 patients with low-risk prostate cancer (ClinicalTrials.gov identifier: Nbib2391051). The secondary outcome of this study is to correlate miR-375 and miR-141 to visible efficacy of the radiation therapy. This secondary endpoint is supported by a number of previous clinical studies including, one where miR-21, miR-141 and miR-221 was detected in the plasma of a prostate cancer cohort of 51 patients (locally advanced or metastatic) had higher expression compared to 20 healthy controls (Yaman Agaoglu et al. 2011). It turned out that miR-21 helped distinguish between healthy and prostate cancer patients, but miR-141 (miR-200 family member) enabled distinction between localized and metastatic subjects. A related study screening 667 abundant miRs in the serum of prostate cancer patients identified miR-375 and miR-141 to be closely associated with disease progression (ClinicalTrials.gov identifier: Nbib2391051) (Brase et al. 2011). The study therefore proposed that circulating miR-375 and miR-141 can be used as a non-invasive biomarker for tumor progression (Brase et al. 2011). The near-term ramifications of determining treatment efficacy is to be able to change therapies to one that may be effective for the patient and not subject him to the side effects associated with an ineffective drug. However, it may also suggest an opportunity to provide a combination therapy of a particular miR mimetic with a conventional intervention. For those patients that do not manifest a drop in miR-141 with radiation, an anti-miR-141 could be supplemented with radiation. As the use of miRs for prostate cancer therapeutics is at a nascent stage, results from such observational trials could lead back to the bench to test potential complementary therapeutic approaches pre-clinically.
The first-in-man miR-based therapeutic trial, MXR34 (Nbib1829971), delivered in a liposomal nanoparticles was in part based on the important work by Liu and coworkers showing that systematic delivery of miR-34a reduced prostate tumor and metastasis in mice (Table 1) (Liu et al. 2011). As CD44 was validated as a target for miR-34a in the publication, it was attributed to preventing prostate cancer metastasis and regeneration (Liu et al. 2011). CD44 silencing in LAPC4 cells prevented lung metastasis and expansion of orthotopic tumors (Liu et al. 2011). Since that study, we have a better appreciation of the pro-metastatic properties of hyaluranon (a CD44 ligand) (Hiraga et al. 2013). However, considering other known targets of miR-34a, such as Notch, CDK4 and CyclinD1, it is difficult to attribute a miR’s systemic effects in vivo on any one target. As a direct target of p53, miR-34 targeting can potentially have effects on any number of cancer types. But, the highest levels of miR-34a are in the ovary, prostate and testes. Thus, its downregulation in the respective cancers in those organs would likely benefit the greatest from a miR-34 mimetic like MXR34. However, as with all miRs, the targets are highly dependent on the cell type. For the tested prostate cells, CD44 has emerged as the reproducible target of miR-34a. However, it is important to note that the current MXR34 study in unresettable liver cancer patients where delivery system and potential off-target effects will be revealed. Liver cancer was the chosen disease for the MRX34 study since the liposomes that contain the double-stranded RNA payload tend to accumulate in liver tissues. For prostate targeting, the lyposomes maybe functionalized with PSMA (prostate specific membrane antigen) or other such agent. MXR34 tries to address the two primary obstacles in gene therapy, tissue-specific delivery and cellular uptake.
miRs in ovarian cancer patient care
The most lethal form of gynecologic malignancy is ovarian cancer, with a dismal 44% five-year survival rate. Over 200,000 women around the world are diagnosed with ovarian cancer each year and cancer antigen 125 (CA-125) can only detect late stage disease with little predictive value as to therapy response or relapse. Unfortunately, since three-fourths of the patients present with advanced disease, surgical cytoreduction and platinum/taxane-based chemotherapy are among the first lines of intervention. Benson and coworkers first reported that the concentration of circulating miRs is altered after administration of carboplatin and decitabine chemotherapy regime in platinum resistant recurrent ovarian cancer patients, in a phase II clinical trial of 18 subjects (Benson et al. 2015). A tumor suppressive role for miR-148b-5p has been suggested in multiple solid tumors (Song et al. 2011, Zhao et al. 2013). The phase II trial showed reduced circulating miR-148b-5p was associated with longer progression free survival on chemotherapy (Benson et al. 2015). This study suggested that chemotherapy induced miRs concentration changes could be used as a prognostic biomarker of therapeutic response in ovarian cancer patients. A similar decision enabling miR was described for prostate cancer. However, for a disease that can progress rapidly, miR-448-5p could be especially important to limit ovarian cancer recurrence (Kim et al. 2010).
EphA2 is a well-recognized marker and evolving therapeutic target for multiple cancers including ovarian and breast cancer. EphA2 is part of a receptor family associated with epithelial–endothelial interaction via juxtacrine signaling by Eph ligands. Interestingly, miR-520d-3p with EphA2 is an independent prognostic marker for serous ovarian cancer (Nishimura et al. 2013). Many mechanisms of EphA2 targeting involving peptide- and antibody-conjugates of chemotherapy as well as CAR-T cell therapy have had preclinical promise (Kiewlich et al. 2006, Chow et al. 2013, Wang et al. 2013). SiRNA-mediated silencing of EphA2 has reduced ovarian tumor growth in mouse models (Landen et al. 2005). More recently, miR-520d-3p mimetic was found to synergize with EphA2 siRNA to reduce ovarian tumor size and invasive capacity (Nishimura et al. 2013). Both complementary therapeutic platforms depend on AGO2-RISC complex to downregulate EphA2 expression.
TargomiRs is the second of the two miR mimetics currently in clinical trials (Australian New Zealand Trial # ACTRN12614001248651). It is being tested for safety in late stage pleural mesothelioma and non-small cell lung cancer subjects. TargomiRs are EGF receptor-targeted antibody conjugated to minicells containing a miR-16 mimic. The mimic is a double-stranded RNA molecule in a bacteria-based nanoparticle (minicell). As previously mentioned, miR-16 is implicated as a tumor suppressor for many cancers. However, there are compelling preclinical reports in which the ovarian cancer models were treated with cisplatin alone or in combination with miR-15a and miR-16 mimetics (Dwivedi et al. 2016). There was a significant inhibition of tumor growth of the chemo-resistant ovarian cancer cells by the addition of the miR mimetics that targeted BMI1 and the cisplatin transporter, ATP7B (Dwivedi et al. 2016). The study demonstrated that in vivo nano-liposomal delivery of miR-15a and miR-16 decrease tumor growth in preclinical chemo-resistant orthotopic ovarian cancer mouse model in support of combination therapies. In another study, restoration of miR-199b-5p increased sensitivity to cisplatin induced cytotoxicity in vitro and in vivo ovarian cancer models via inactivation of Jag1 mediated Notch signaling pathway (Liu et al. 2014b). A combination therapy possibility with miRs and taxane-based chemotherapy was also suggested. With the reports miR-200c role in ZEB1/ZEB2-mediated EMT progression, Cittelly and coworkers found that restoration of miR-200c induced chemo- and ankoikis sensitivity (Cittelly et al. 2012). Furthermore, miR-220c restoration also helped reduce tumor growth alone and in combination with paclitaxel (Cittelly et al. 2012). The point of this study was to test if the recovery of miR-200c can improve response to chemotherapy. Ovarian cancer has a high rate of relapse within 18 months of current therapeutic interventions with no targeted therapies approved. To identify patients that will benefit from current lines of chemotherapy, an interventional trial profiling serum miRs from patients prior to chemotherapy treatment is being performed (ClinicalTrials.gov identifier: Nbib1391351, Table 2). For those that may not fare as well on chemotherapy, miRs may be used to overcome drug resistant variants that emerge in ovarian cancer progression.
Table 2.
Ovarian cancer clinical trials involving miRs.
| Trial reference | Study type | Sponsor | Trial title |
|---|---|---|---|
| NCT2758652 | Observational | Tampere University Hospital | Molecular mechanisms leading to chemo-resistance in epithelial ovarian cancer |
| NCT1391351 | Interventional | Centre Francois Baclesse | Search for predictors of therapeutic response in patients with carcinoma of the ovary, the fallopian tube or peritoneal serous-type advanced |
| NCT1572467 | Observational | Children’s Oncology Group | DICER1 mutations and miRNA in ovarian and testicular sex cord stromal tumors of childhood |
| NCT1970696 | Observational | Children’s Hospitals and Clinics of Minnesota | International ovarian and testicular stromal tumor registry |
| NCT1879436 | Observational | Meir Medical Center | The effect of human placental explants and pregnant women sera on cancer cells |
miRs in breast cancer patient care
Breast cancer is the second leading killer of women, with only lung cancer resulting in more cancer deaths. Yet, more than 3.1 million women with a history of breast cancer are alive in the US with the potential need for further means of intervention. The estrogen receptor positive subtype is a critical discriminator, accounting for nearly 75% of all cases of breast cancer. However, non-hormone responsive (triple negative breast cancer) results account for the poor prognosis, often treated with a cytotoxic chemotherapy or/and targeted therapy (with exception of HER2 antagonists). As with prostate cancer, hormone responsive breast cancer can evolve to a hormone-nonresponsive disease in heterogeneous tumors. As with other cancers discussed, there is a lack of reliable biomarkers for patient selection and therapeutic efficacy where miRs can play an important role. Heneghan and coworkers demonstrated that among the elevated breast cancer-associated miRs present in circulation, miR-195 can help distinguish from other malignancies (melanoma, renal, prostate and colon cancer) and from non-malignant subjects (Heneghan et al. 2010). All the cancer patients generally expressed let-7a, miR-10b and miR-155, but miR-195 provided strong specificity and sensitivity with other covariates.
With MXR34 being tested and the breadth of supporting literature for its application, many patients including those with breast cancer may benefit following pharmacokinetics/maximum tolerated dose (PK/MTD) are established in the phase I trial. In a recent report, anti-tumorigenic effect of miR-34a administration in triple negative breast cancer models inhibited proliferation, invasion and metastasis (Adams et al. 2016). Moreover, the delivery of miR-34a replacement therapy retarded the tumor growth of subcutaneously and orthotopically xenografted tumors (Adams et al. 2016). In a seminal publication where breast cancer patients were treated with the chemotherapeutic 5-flurouracil (5-FU), miR-34a was found to be downregulated (Li et al. 2013a). Accordingly, a miR-34a mimetic was given with chemotherapeutic 5-flurouracil (5-FU) to reveal a more efficient anti-tumor effect than either single agent treatment (Li et al. 2013a). These studies support the promise of miR-34a (MXR34) as a potential therapeutic agent for breast and other cancer patients.
Majority of the breast cancer clinical trials are observational in terms of profiling circulating miR expression, like the one by Cancer Trials Ireland which aims to identify miR in response to neoadjuvant and adjuvant chemotherapy (ClinicalTrials.gov identifier: Nbib1722851, Table 3). Similarly, another clinical study investigates the circulating miRs as markers of hormone resistance and sensitivity in patients with metastatic invasive breast cancer or locally advanced breast cancer (ClinicalTrials.gov identifier: Nbib1612871, Table 3). It is worth noting the paradigm set by identifying miRs as a surrogate of side effect of certain chemotherapies. Doxorubicin (Adriamycin, Rubex), used for many cancers including breast cancer, is a DNA damaging agent that can cause cardiac damage. Years of work on miRs in the cardiology field has revealed miR-208 to be elevated in cases of cardiac hypertrophy and heart failure (Callis et al. 2009, Montgomery et al. 2011, Oliveira-Carvalho & Bocchi 2016). miR-208 expression is associated with fibrosis and EMT progression as well as specifically targeting the BMP co-receptor, endoglin (Fig. 1) (Liu et al. 2014a, Wang et al. 2014a). If the clinical trial demonstrates miR-208 as a robust marker for cardiac damage, there will be a pre-clinical basis to consider the use of an antimiR-208 as a viable cardio-protective for chemotherapy patients (Callis et al. 2009, Montgomery et al. 2011, Tony et al. 2015). Such an application of a miR-based therapy would follow the logic of a preclinical study where Xue and coworkers identified miR-621 elevation associated with paclitaxel and carboplatin (PTX/CBP) sensitivity (Xue et al. 2016). They subsequently administered miR-621 mimetic to silence the FBXO11, a poor prognostic indicator, in sensitizing breast tumors to PTX/CBP (Xue et al. 2016). As most of the miR-associated trials listed in Tables 1, 2 and 3 are observational, in identifying changes in miR expression, this paradigm of targeting the deregulated miRs for therapy could follow. With appropriate pre-clinical validation, these same miRs may be targeted by a miR mimetic or anti-miR. However, as with any such line of reasoning, not all good biomarkers are consequential to disease etiology and conversely, miRs that are central to disease progression or therapeutic resistance may not serve to be robust biomarkers.
Table 3.
Breast cancer clinical trials involving miRs.
| Trial reference | Study type | Sponsor | Trial title |
|---|---|---|---|
| NCT1722851 | Observational | Cancer Trials Ireland | Circulating miRNAs: novel breast cancer biomarkers and their use for guiding and monitoring response to chemotherapy |
| NCT2127073 | Interventional | Sheldon Feldman | Pilot study of oxytocin and microRNA identification in NAF, serum, and tissue in women with breast cancer |
| NCT1957332 | Observational | University Medical Center Groningen | Imaging patients for cancer drug selection – metastatic breast cancer (IMPACT-MBC) |
| NCT1598285 | Observational | Spanish Breast Cancer Research Group | A combined GWAS and miRNA for the Identification of bevacizumab response predictors in metastatic breast cancer |
| NCT1231386 | Observational | City of Hope Medical Center | miRNAs profiling of breast cancer in patients undergoing neoadjuvant or adjuvant treatment for locally advanced and inflammatory breast cancer |
| NCT1612871 | Interventional | Institut Claudius Regaud | Circulating miRNAs as biomarkers of hormone sensitivity in breast cancer? Pilot study |
| NCT2656589 | Observational | Sun Yat-Sen Memorial Hospital of Sun Yat-Sen University | A perspective study of the predictive value of microRNA in patients with HER2 positive advanced stage breast cancer who were treated with herceptin |
| NCT2065908 | Observational | West Pomeranian Cancer Center | Circulating microRNAs as a novel biomarker of early cardiotoxicity in breast cancer patients treated with anthracyclines |
| NCT581750 | Observational | Memorial Sloan Kettering Cancer Center | Molecular genetic basis of invasive breast cancer risk associated with lobular carcinoma in situ |
| NCT1965522 | Interventional | Juravinski Cancer Center | Anti-proliferative effects of vitamin D and melatonin in breast cancer (MELO-D) |
| NCT2288806 | Interventional | Hamilton Health Sciences Corporation | Melatonin and vitamin d in breast cancer (MELO-D) |
| NCT2103140 | Interventional | Georgetown University | An exercise randomized controlled trial targeting African-American women with metabolic syndrome and high risk for breast cancer |
| NCT1907438 | Observational | Hadassah Medical Organization | Identification of the transformation potential of normal estrogen exposed BRCA1 (breast cancer susceptibility gene 1) and BRCA2 (breast cancer susceptibility gene 2) heterozygous epithelial breast cells due to irradiation |
| NCT773695 | Interventional | Hoffmann-La Roche | A multicenter, randomized, ph II clinical trial to evaluate the effect of avastin in combination with neoadj treatment regimens on the molecular and metabolic characteristics and changes in the primary tumors with ref to the obtained responses in patients with large primary HER2 Neg breast cancers |
| NCT1724450 | Interventional | University of Sao Paulo | Carvedilol effect in preventing chemotherapy – induced cardiotoxicity. a randomized double blind study |
| NCT2437318 | Interventional | Novartis Pharmaceuticals | A phase III randomized double-blind, placebo controlled study of alpelisib in combination with fulvestrant for men and postmenopausal women with hormone receptor positive, HER2-negative advanced breast cancer which progressed on or after aromatase inhibitor treatment |
| NCT1879436 | Observational | Meir Medical Center | The effect of human placental explants and pregnant women sera on cancer cells |
| NCT2678650 | Interventional | Capital Medical University | MicroRNA mediates volatile anesthetics preconditioning induced artery protection |
Summary and perspective
Many examples of miR-based biomarkers and potential therapeutics in prostate, ovarian and breast cancers have been presented. These examples serve to highlight the mechanism of action and ongoing clinical trials for the diseases. In the post-genomic age a trove of patient/tumor-specific data can potentially guide and prioritize the clinical findings. However, the challenge in the field is the ability to predict miR target sites with high confidence in a cell type of interest. miRs typically repress target gene expression, but the reciprocal effect on targets by miRs is less clear. Although miR-based therapeutics is in its infancy it faces many of the same limitations as other targeted therapies. As the miR field progresses the focus may be less on a specific target, but a genomically defined phenotype. Conceptually, miRs could complement CRISPR/Cas9 in engineering networks. As we better characterize the heterogeneous nature of a patient’s tumors and consider genetic/epigenetic drift associated with disease progression and therapeutic response, miR-based therapies become more attractive due to their stability, sequence specificity, and relative ease of synthesis.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of this review.
Funding
This work was supported by the Veterans Administration Merit Award (grant number bib1BX001040) and National Cancer Institute at the National Institutes of Health (grant number R01CA108646).
Acknowledgments
The authors thank Joan D Ramos for editorial help.
References
- Adams BD, Wali VB, Cheng CJ, Inukai S, Booth CJ, Agarwal S, Rimm DL, Gyorffy B, Santarpia L, Pusztai L, et al. 2016. miR-34a silences c-SRC to attenuate tumor growth in triple-negative breast cancer. Cancer Research 76 927–939. ( 10.1158/0008-5472.CAN-15-2321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SM, Cha JY, Kim J, Kim D, Trang HT, Kim YM, Cho YH, Park D, Hong S. 2012. Smad3 regulates E-cadherin via miRNA-200 pathway. Oncogene 31 3051–3059. ( 10.1038/onc.2011.484) [DOI] [PubMed] [Google Scholar]
- Albino D, Civenni G, Dallavalle C, Roos M, Jahns H, Curti L, Rossi S, Pinton S, D’Ambrosio G, Sessa F, et al. 2016. Activation of the Lin28/let-7 axis by loss of ESE3/EHF promotes a tumorigenic and stem-like phenotype in prostate cancer. Cancer Research 76 3629–3643. ( 10.1158/0008-5472.CAN-15-2665) [DOI] [PubMed] [Google Scholar]
- Anderton MJ, Mellor HR, Bell A, Sadler C, Pass M, Powell S, Steele SJ, Roberts RR, Heier A. 2011. Induction of heart valve lesions by small-molecule ALK5 inhibitors. Toxicologic Pathology 39 916–924. ( 10.1177/0192623311416259) [DOI] [PubMed] [Google Scholar]
- Au Yeung CL, Co NN, Tsuruga T, Yeung TL, Kwan SY, Leung CS, Li Y, Lu ES, Kwan K, Wong KK, et al. 2016. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nature Communications 7 11150 ( 10.1038/ncomms11150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakin AV, Tomlinson AK, Bhowmick NA, Moses HL, Arteaga CL. 2000. Phosphatidylinositol 3-kinase function is required for transforming growth factor beta-mediated epithelial to mesenchymal transition and cell migration. Journal of Biological Chemistry 275 36803–36810. ( 10.1074/jbc.M005912200) [DOI] [PubMed] [Google Scholar]
- Banerjee J, Mishra R, Li X, Jackson RS, 2nd, Sharma A, Bhowmick NA. 2014. A reciprocal role of prostate cancer on stromal DNA damage. Oncogene 33 4924–4931. ( 10.1038/onc.2013.431) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benassi B, Flavin R, Marchionni L, Zanata S, Pan Y, Chowdhury D, Marani M, Strano S, Muti P, Blandino G, et al. 2012. MYC is activated by USP2a-mediated modulation of microRNAs in prostate cancer. Cancer Discovery 2 236–247. ( 10.1158/2159-8290.CD-11-0219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlhabib H, Guo W, Pierce BM, Mendelson CR. 2015. The miR-200 family and its targets regulate type II cell differentiation in human fetal lung. Journal of Biological Chemistry 290 22409–22422. ( 10.1074/jbc.M114.636068) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson EA, Skaar TC, Liu Y, Nephew KP, Matei D. 2015. Carboplatin with decitabine therapy, in recurrent platinum resistant ovarian cancer, alters circulating miRNAs concentrations: a pilot study. PLoS ONE 10 e0141279 ( 10.1371/journal.pone.0141279) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick NA. 2012. Metastatic ability: adapting to a tissue site unseen. Cancer Cell 22 563–564. ( 10.1016/j.ccr.2012.10.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick NA, Ghiassi M, Bakin A, Aakre M, Lundquist CA, Engel ME, Arteaga CL, Moses HL. 2001a. Transforming growth factor-beta1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Molecular Biology of Cell 12 27–36. ( 10.1091/mbc.12.1.27) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick NA, Zent R, Ghiassi M, McDonnell M, Moses HL. 2001b. Integrin beta 1 signaling is necessary for transforming growth factor-beta activation of p38MAPK and epithelial plasticity. Journal of Biological Chemistry 276 46707–46713. ( 10.1074/jbc.M106176200) [DOI] [PubMed] [Google Scholar]
- Bhowmick NA, Ghiassi M, Aakre M, Brown K, Singh V, Moses HL. 2003. TGF-beta-induced RhoA and p160ROCK activation is involved in the inhibition of Cdc25A with resultant cell-cycle arrest. PNAS 100 15548–15553. ( 10.1073/pnas.2536483100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick NA, Chytil A, Plieth D, Gorska AE, Dumont N, Shappell S, Washington MK, Neilson EG, Moses HL. 2004. TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science 303 848–851. ( 10.1126/science.1090922) [DOI] [PubMed] [Google Scholar]
- Brabletz S, Brabletz T. 2010. The ZEB/miR-200 feedback loop – a motor of cellular plasticity in development and cancer? EMBO Reports 11 670–677. ( 10.1038/embor.2010.117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brase JC, Johannes M, Schlomm T, Falth M, Haese A, Steuber T, Beissbarth T, Kuner R, Sultmann H. 2011. Circulating miRNAs are correlated with tumor progression in prostate cancer. International Journal of Cancer 128 608–616. ( 10.1002/ijc.25376) [DOI] [PubMed] [Google Scholar]
- Braun J, Hoang-Vu C, Dralle H, Huttelmaier S. 2010. Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene 29 4237–4244. ( 10.1038/onc.2010.169) [DOI] [PubMed] [Google Scholar]
- Butz H, Liko I, Czirjak S, Igaz P, Korbonits M, Racz K, Patocs A. 2011. MicroRNA profile indicates downregulation of the TGFbeta pathway in sporadic non-functioning pituitary adenomas. Pituitary 14 112–124. ( 10.1007/s11102-010-0268-x) [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. 2006. MicroRNA signatures in human cancers. Nature Reviews Cancer 6 857–866. ( 10.1038/nrc1997) [DOI] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. 2002. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. PNAS 99 15524–15529. ( 10.1073/pnas.242606799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, et al. 2009. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. Journal of Clinical Investigation 119 2772–2786. ( 10.1172/JCI36154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Brown CW, Matzuk MM. 2002. Genetic analysis of the mammalian transforming growth factor-beta superfamily. Endocrine Reviews 23 787–823. ( 10.1210/er.2002-0003) [DOI] [PubMed] [Google Scholar]
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al. 2008. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Research 18 997–1006. ( 10.1038/cr.2008.282) [DOI] [PubMed] [Google Scholar]
- Chen W, Zhou S, Mao L, Zhang H, Sun D, Zhang J, Li J, Tang JH. 2016. Crosstalk between TGF-beta signaling and miRNAs in breast cancer metastasis. Tumor Biology 37 10011–10019. ( 10.1007/s13277-016-5060-8) [DOI] [PubMed] [Google Scholar]
- Cheon DJ, Tong Y, Sim MS, Dering J, Berel D, Cui X, Lester J, Beach JA, Tighiouart M, Walts AE, et al. 2014. A collagen-remodeling gene signature regulated by TGF-beta signaling is associated with metastasis and poor survival in serous ovarian cancer. Clinical Cancer Research 20 711–723. ( 10.1158/1078-0432.CCR-13-1256) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow KK, Naik S, Kakarla S, Brawley VS, Shaffer DR, Yi Z, Rainusso N, Wu MF, Liu H, Kew Y, et al. 2013. T cells redirected to EphA2 for the immunotherapy of glioblastoma. Molecular Therapy 21 629–637. ( 10.1038/mt.2012.210) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cittelly DM, Dimitrova I, Howe EN, Cochrane DR, Jean A, Spoelstra NS, Post MD, Lu X, Broaddus RR, Spillman MA, et al. 2012. Restoration of miR-200c to ovarian cancer reduces tumor burden and increases sensitivity to paclitaxel. Molecular Cancer Therapeutics 11 2556–2565. ( 10.1158/1535-7163.MCT-12-0463) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J. 2008. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biology 6 2853–2868. ( 10.1371/journal.pbio.0060301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corney DC, Hwang CI, Matoso A, Vogt M, Flesken-Nikitin A, Godwin AK, Kamat AA, Sood AK, Ellenson LH, Hermeking H, et al. 2010. Frequent downregulation of miR-34 family in human ovarian cancers. Clinical Cancer Research 16 1119–1128. ( 10.1158/1078-0432.CCR-09-2642) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulouarn C, Clément B. 2014. Stellate cells and the development of liver cancer: therapeutic potential of targeting the stroma. Journal of Hepatology 60 1306–1309. ( 10.1016/j.jhep.2014.02.003) [DOI] [PubMed] [Google Scholar]
- Coulouarn C, Factor VM, Thorgeirsson SS. 2008. Transforming growth factor-beta gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology 47 2059–2067. ( 10.1002/hep.22283) [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ippolito E, Plantamura I, Bongiovanni L, Casalini P, Baroni S, Piovan C, Orlandi R, Gualeni A, Gloghini A, Rossini A, et al. 2016. MiR-9 and miR-200 regulate PDGFR-mediated endothelial differentiation of tumor cells in triple negative breast cancer. Cancer Research 76 5562–5572. ( 10.1158/0008-5472.CAN-16-0140) [DOI] [PubMed] [Google Scholar]
- Dai X, Jiang Y, Tan C. 2015. Let-7 sensitizes KRAS mutant tumor cells to chemotherapy. PLoS ONE 10 e0126653 ( 10.1371/journal.pone.0126653) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A. 2008. SMAD proteins control DROSHA-mediated microRNA maturation. Nature 454 56–61. ( 10.1038/nature07086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis-Dusenbery BN, Hata A. 2010. Mechanisms of control of microRNA biogenesis. Journal of Biochemistry 148 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean ZS, Riahi R, Wong PK. 2015. Spatiotemporal dynamics of microRNA during epithelial collective cell migration. Biomaterials 37 156–163. ( 10.1016/j.biomaterials.2014.10.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Lopez A, Diaz-Martin J, Moreno-Bueno G, Cuevas EP, Santos V, Olmeda D, Portillo F, Palacios J, Cano A. 2015. Zeb1 and Snail1 engage miR-200f transcriptional and epigenetic regulation during EMT. International Journal of Cancer 136 E62–E73. ( 10.1002/ijc.29177) [DOI] [PubMed] [Google Scholar]
- Doldi V, Callari M, Giannoni E, D’Aiuto F, Maffezzini M, Valdagni R, Chiarugi P, Gandellini P, Zaffaroni N, et al. 2015. Integrated gene and miRNA expression analysis of prostate cancer associated fibroblasts supports a prominent role for interleukin-6 in fibroblast activation. Oncotarget 6 31441–31460. ( 10.18632/oncotarget.5056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duner S, Lindman J, Ansari D, Gundewar C, Andersson R. 2010. Pancreatic cancer: the role of pancreatic stellate cells in tumor progression. Pancreatology 10 673–681. ( 10.1159/000320711) [DOI] [PubMed] [Google Scholar]
- Dwivedi SK, Mustafi SB, Mangala LS, Jiang D, Pradeep S, Rodriguez-Aguayo C, Ling H, Ivan C, Mukherjee P, Calin GA, et al. 2016. Therapeutic evaluation of microRNA-15a and microRNA-16 in ovarian cancer. Oncotarget 7 15093–15104. ( 10.18632/oncotarget.7618) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds MD, Boyd KL, Moyo T, Mitra R, Duszynski R, Arrate MP, Chen X, Zhao Z, Blackwell TS, Andl T, et al. 2016. MicroRNA-31 initiates lung tumorigenesis and promotes mutant KRAS-driven lung cancer. Journal of Clinical Investigation 126 349–364. ( 10.1172/JCI82720) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa H, Jingushi K, Hirono T, Ueda Y, Kitae K, Nakata W, Fujita K, Uemura M, Nonomura N, Tsujikawa K. 2016. The miR-130 family promotes cell migration and invasion in bladder cancer through FAK and Akt phosphorylation by regulating PTEN. Scientific Reports 6 20574 ( 10.1038/srep20574) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N. 2012. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nature Structural and Molecular Biology 19 586–593. ( 10.1038/nsmb.2296) [DOI] [PubMed] [Google Scholar]
- Fendler A, Stephan C, Yousef GM, Kristiansen G, Jung K. 2016. The translational potential of microRNAs as biofluid markers of urological tumours. Nature Reviews Urology 13 734–752. ( 10.1038/nrurol.2016.193) [DOI] [PubMed] [Google Scholar]
- Geraldo MV, Yamashita AS, Kimura ET. 2012. MicroRNA miR-146b-5p regulates signal transduction of TGF-beta by repressing SMAD4 in thyroid cancer. Oncogene 31 1910–1922. ( 10.1038/onc.2011.381) [DOI] [PubMed] [Google Scholar]
- Hager M, Pedersen CC, Larsen MT, Andersen MK, Hother C, Gronbaek K, Jarmer H, Borregaard N, Cowland JB. 2011. MicroRNA-130a-mediated down-regulation of Smad4 contributes to reduced sensitivity to TGF-beta1 stimulation in granulocytic precursors. Blood 118 6649–6659. ( 10.1182/blood-2011-03-339978) [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. 2004. The Drosha-DGCR8 complex in primary microRNA processing. Genes and Development 18 3016–3027. ( 10.1101/gad.1262504) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J, Zhang S, Zhou Y, Hu X, Shao C. 2011. MicroRNA 483–3p suppresses the expression of DPC4/Smad4 in pancreatic cancer. FEBS Letters 585 207–213. ( 10.1016/j.febslet.2010.11.039) [DOI] [PubMed] [Google Scholar]
- Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J. 2014. Clinical development success rates for investigational drugs. Nature Biotechnology 32 40–51. ( 10.1038/nbt.2786) [DOI] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. 2007. A microRNA component of the p53 tumour suppressor network. Nature 4471130–1134. ( 10.1038/mtna.2015.5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneghan HM, Miller N, Kelly R, Newell J, Kerin MJ. 2010. Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease. Oncologist 15 673–682. ( 10.1634/theoncologist.2010-0103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga T, Ito S, Nakamura H. 2013. Cancer stem-like cell marker CD44 promotes bone metastases by enhancing tumorigenicity, cell motility, and hyaluronan production. Cancer Research 73 4112–4122. ( 10.1158/0008-5472.CAN-12-3801) [DOI] [PubMed] [Google Scholar]
- Hiraki M, Nishimura J, Takahashi H, Wu X, Takahashi Y, Miyo M, Nishida N, Uemura M, Hata T, Takemasa I, et al. 2015. Concurrent targeting of KRAS and AKT by MiR-4689 is a novel treatment against mutant KRAS colorectal cancer. Molecular Therapy: Nucleic Acids 4 e231 ( 10.1038/mtna.2015.5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, et al. 2015. Tumour exosome integrins determine organotropic metastasis. Nature 527 329–335. ( 10.1038/nature15756) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. 2008. Argonaute proteins: key players in RNA silencing. Nature Reviews Molecular Cell Biology 9 22–32. ( 10.1038/nrm2321) [DOI] [PubMed] [Google Scholar]
- Lebrun J-J. 2012. The dual role of TGF-β in human cancer: from tumor suppression to cancer metastasis. ISRN Molecular Biology 2012 381428 ( 10.5402/2012/381428) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. 2005. RAS is regulated by the let-7 microRNA family. Cell 120 635–647. ( 10.1016/j.cell.2005.01.014) [DOI] [PubMed] [Google Scholar]
- Josson S, Gururajan M, Sung SY, Hu P, Shao C, Zhau HE, Liu C, Lichterman J, Duan P, Li Q, et al. 2015. Stromal fibroblast-derived miR-409 promotes epithelial-to-mesenchymal transition and prostate tumorigenesis. Oncogene 34 2690–2699. ( 10.1038/onc.2014.212) [DOI] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. 2009. The basics of epithelial-mesenchymal transition. Journal of Clinical Investigation 119 1420–1428. ( 10.1172/JCI39104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan CW, Hahn MA, Gard GB, Maidens J, Huh JY, Marsh DJ, Howell VM. 2012. Elevated levels of circulating microRNA-200 family members correlate with serous epithelial ovarian cancer. BMC Cancer 12 627 ( 10.1186/1471-2407-12-627) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katakowski M, Buller B, Wang X, Rogers T, Chopp M. 2010. Functional microRNA is transferred between glioma cells. Cancer Research 70 8259–8263. ( 10.1158/0008-5472.CAN-10-0604) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiewlich D, Zhang J, Gross C, Xia W, Larsen B, Cobb RR, Biroc S, Gu JM, Sato T, Light DR, et al. 2006. Anti-EphA2 antibodies decrease EphA2 protein levels in murine CT26 colorectal and human MDA-231 breast tumors but do not inhibit tumor growth. Neoplasia 8 18–30. ( 10.1593/neo.05544) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Kim YK, Kwon Y, Heo JH, Kang H, Kim G, An HJ. 2010. Deregulation of miR-519a, 153, and 485-5p and its clinicopathological relevance in ovarian epithelial tumours. Histopathology 57 734–743. ( 10.1111/j.1365-2559.2010.03686.x) [DOI] [PubMed] [Google Scholar]
- Kiskowski MA, Jackson RS, 2nd, Banerjee J, Li X, Kang M, Iturregui JM, Franco OE, Hayward SW, Bhowmick NA. 2011. Role for stromal heterogeneity in prostate tumorigenesis. Cancer Research 71 3459–3470. ( 10.1158/0008-5472.CAN-10-2999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, Cheng JQ. 2008. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Molecular and Cellular Biology 28 6773–6784. ( 10.1128/MCB.00941-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni AB, Huh CG, Becker D, Geiser A, Lyght M, Flanders KC, Roberts AB, Sporn MB, Ward JM, Karlsson S. 1993. Transforming growth factor beta 1 null mutation in mice causes excessive inflammatory response and early death. PNAS 90 770–774. ( 10.1073/pnas.90.2.770) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294 853–858. ( 10.1126/science.1064921) [DOI] [PubMed] [Google Scholar]
- Landen CN, Jr, Chavez-Reyes A, Bucana C, Schmandt R, Deavers MT, Lopez-Berestein G, Sood AK. 2005. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Research 65 6910–6918. ( 10.1158/0008-5472.CAN-05-0530) [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75 843–854. ( 10.1016/0092-8674(93)90529-Y) [DOI] [PubMed] [Google Scholar]
- Lee I, Ajay SS, Yook JI, Kim HS, Hong SH, Kim NH, Dhanasekaran SM, Chinnaiyan AM, Athey BD. 2009. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Research 19 1175–1183. ( 10.1101/gr.089367.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Yuan L, Luo J, Gao J, Guo J, Xie X. 2013a. MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clinical and Experimental Medicine 13 109–117. ( 10.1007/s10238-012-0186-5) [DOI] [PubMed] [Google Scholar]
- Li Q, Zhang D, Wang Y, Sun P, Hou X, Larner J, Xiong W, Mi J. 2013b. MiR-21/Smad 7 signaling determines TGF-beta1-induced CAF formation. Scientific Reports 3 2038 ( 10.1038/srep02038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wan X, Chen H, Yang S, Liu Y, Mo W, Meng D, Du W, Huang Y, Wu H, et al. 2014. Identification of miR-133b and RB1CC1 as independent predictors for biochemical recurrence and potential therapeutic targets for prostate cancer. Clinical Cancer Research 20 2312–2325. ( 10.1158/1078-0432.CCR-13-1588) [DOI] [PubMed] [Google Scholar]
- Li C, Song L, Zhang Z, Bai XX, Cui MF, Ma LJ. 2016. MicroRNA-21 promotes TGF-beta1-induced epithelial-mesenchymal transition in gastric cancer through up-regulating PTEN expression. Oncotarget 7 66989–67003. ( 10.18632/oncotarget.11888) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HM, Castillo L, Mahon KL, Chiam K, Lee BY, Nguyen Q, Boyer MJ, Stockler MR, Pavlakis N, Marx G, et al. 2014. Circulating microRNAs are associated with docetaxel chemotherapy outcome in castration-resistant prostate cancer. British Journal of Cancer 110 2462–2471. ( 10.1038/bjc.2014.181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, et al. 2011. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nature Medicine 17 211–215. ( 10.1038/nm.2284) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Kumar SM, Lu H, Liu A, Yang R, Pushparajan A, Guo W, Xu X. 2012. MicroRNA-9 up-regulates E-cadherin through inhibition of NF-kappaB1-Snail1 pathway in melanoma. Journal of Pathology 226 61–72. ( 10.1002/path.2964) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Shao C, Jin G, Liu R, Hao J, Song B, Ouyang L, Hu X. 2014a. miR-208-induced epithelial to mesenchymal transition of pancreatic cancer cells promotes cell metastasis and invasion. Cell Biochemistry and Biophysics 69 341–346. ( 10.1007/s12013-013-9805-3) [DOI] [PubMed] [Google Scholar]
- Liu MX, Siu MK, Liu SS, Yam JW, Ngan HY, Chan DW. 2014b. Epigenetic silencing of microRNA-199b-5p is associated with acquired chemoresistance via activation of JAG1-Notch1 signaling in ovarian cancer. Oncotarget 5 944–958. ( 10.18632/oncotarget.1458) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louafi F, Martinez-Nunez RT, Sanchez-Elsner T. 2010. MicroRNA-155 targets SMAD2 and modulates the response of macrophages to transforming growth factor-{beta}. Journal of Biological Chemistry 285 41328–41336. ( 10.1074/jbc.M110.146852) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA. 2007. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449 682–688. ( 10.1038/nature06174) [DOI] [PubMed] [Google Scholar]
- Masliah-Planchon J, Garinet S, Pasmant E. 2016. RAS-MAPK pathway epigenetic activation in cancer: miRNAs in action. Oncotarget 7 38892–38907. ( 10.18632/oncotarget.6476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Baltimore D. 2016. MicroRNAs as regulatory elements in immune system logic. Nature Reviews Immunology 16 279–294. ( 10.1038/nri.2016.40) [DOI] [PubMed] [Google Scholar]
- Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick AM, et al. 2008. Dicer, Drosha, and outcomes in patients with ovarian cancer. New England Journal of Medicine 359 2641–2650. ( 10.1056/NEJMoa0803785) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery RL, Hullinger TG, Semus HM, Dickinson BA, Seto AG, Lynch JM, Stack C, Latimer PA, Olson EN, van Rooij E. 2011. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation 124 1537–1547. ( 10.1161/CIRCULATIONAHA.111.030932) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty CH, Pursell B, Mercurio AM. 2010. miR-10b targets Tiam1: implications for Rac activation and carcinoma migration. Journal of Biological Chemistry 285 20541–20546. ( 10.1074/jbc.M110.121012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musumeci M, Coppola V, Addario A, Patrizii M, Maugeri-Sacca M, Memeo L, Colarossi C, Francescangeli F, Biffoni M, Collura D, et al. 2011. Control of tumor and microenvironment cross-talk by miR-15a and miR-16 in prostate cancer. Oncogene 30 4231–4242. ( 10.1038/onc.2011.140) [DOI] [PubMed] [Google Scholar]
- Neuzillet C, Tijeras-Raballand A, Cohen R, Cros J, Faivre S, Raymond E, de Gramont A. 2015. Targeting the TGF-β pathway for cancer therapy. Pharmacology and Therapeutics 147 22–31. ( 10.1016/j.pharmthera.2014.11.001) [DOI] [PubMed] [Google Scholar]
- Nishimura M, Jung EJ, Shah MY, Lu C, Spizzo R, Shimizu M, Han HD, Ivan C, Rossi S, Zhang X, et al. 2013. Therapeutic synergy between microRNA and siRNA in ovarian cancer treatment. Cancer Discovery 3 1302–1315. ( 10.1158/2159-8290.CD-13-0159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. 2007. MicroRNA-155 is induced during the macrophage inflammatory response. PNAS 104 1604–1609. ( 10.1073/pnas.0610731104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oft M, Peli J, Rudaz C, Schwarz H, Beug H, Reichmann E. 1996. TGF-beta1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes and Development 10 2462–2477. ( 10.1101/gad.10.19.2462) [DOI] [PubMed] [Google Scholar]
- Oliveira-Carvalho V, Bocchi EA. 2016. Comment on ‘MicroRNA-208a silencing attenuates doxorubicin induced myocyte apoptosis and cardiac dysfunction’. Oxidative Medicine and Cellular Longevity 2016 article ID 7939626. ( 10.1155/2016/7939626) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tlsty TD, Cunha GR. 1999. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Research 59 5002–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Nielsen FC, Lund AH. 2008. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Molecular Cell 30 460–471. ( 10.1016/j.molcel.2008.05.001) [DOI] [PubMed] [Google Scholar]
- Ouyang H, Gore J, Deitz S, Korc M. 2014. microRNA-10b enhances pancreatic cancer cell invasion by suppressing TIP30 expression and promoting EGF and TGF-beta actions. Oncogene 33 4664–4674. ( 10.1038/onc.2013.405) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME. 2008. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes and Development 22 894–907. ( 10.1101/gad.1640608) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart B, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, et al. 2000. Conservation across animal phylogeny of the sequence and temporal regulation of the 21 nucleotide C. elegans let-7 heterochronic regulatory RNA Nature 408 86–89. ( 10.1038/35040556) [DOI] [PubMed] [Google Scholar]
- Petrylak DP, Tangen CM, Hussain MH, Lara PN, Jr, Jones JA, Taplin ME, Burch PA, Berry D, Moinpour C, Kohli M, et al. 2004. Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. New England Journal of Medicine 351 1513–1520. ( 10.1056/NEJMoa041318) [DOI] [PubMed] [Google Scholar]
- Peurala H, Greco D, Heikkinen T, Kaur S, Bartkova J, Jamshidi M, Aittomaki K, Heikkila P, Bartek J, Blomqvist C, et al. 2011. MiR-34a expression has an effect for lower risk of metastasis and associates with expression patterns predicting clinical outcome in breast cancer. PLoS ONE 6 e26122 ( 10.1371/journal.pone.0026122) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G. 2001. Molecular biology. Glimpses of a tiny RNA world. Science 294 797–799. ( 10.1126/science.1066315) [DOI] [PubMed] [Google Scholar]
- Shen J, Hruby GW, McKiernan JM, Gurvich I, Lipsky MJ, Benson MC, Santella RM. 2012. Dysregulation of circulating microRNAs and prediction of aggressive prostate cancer. Prostate 72 1469–1477. ( 10.1002/pros.22499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BN, Bhowmick NA. 2016. Role of EMT in metastasis and therapy resistance. Journal of Clinical Medicine Research 5 E17 ( 10.3390/jcm5020017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HE, Kang Y. 2013. The metastasis-promoting roles of tumor-associated immune cells. Journal of Molecular Medicine 91 411–429. ( 10.1007/s00109-013-1021-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. 2004. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305 1434–1437. ( 10.1126/science.1102514) [DOI] [PubMed] [Google Scholar]
- Song YX, Yue ZY, Wang ZN, Xu YY, Luo Y, Xu HM, Zhang X, Jiang L, Xing CZ, Zhang Y. 2011. MicroRNA-148b is frequently down-regulated in gastric cancer and acts as a tumor suppressor by inhibiting cell proliferation. Molecular Cancer 10 1. ( 10.1186/1476-4598-10-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. 2006. Cell-type-specific signatures of microRNAs on target mRNA expression. PNAS 103 2746–2751. ( 10.1073/pnas.0511045103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Hou Y, Yang G, Wang X, Tang S, Du YE, Yang L, Yu T, Zhang H, Zhou M, et al. 2016. Stromal miR-200s contribute to breast cancer cell invasion through CAF activation and ECM remodeling. Cell Death and Differentiation 23 132–145. ( 10.1038/cdd.2015.78) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock IF, de Wit R, Berry WR, Horti J, Pluzanska A, Chi KN, Oudard S, Theodore C, James ND, Turesson I, et al. 2004. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. New England Journal of Medicine 351 1502–1512. ( 10.1056/NEJMoa040720) [DOI] [PubMed] [Google Scholar]
- Taylor MO, Lee Y-H, Schiemann WP. 2011. Role of TGF-β and the tumor microenvironment during mammary tumorigenesis. Gene Expression 15 117–132. ( 10.3727/105221611X13176664479322) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RA, Chang CF, Goods BA, Hammond MD, Grory BM, Ai Y, Steinschneider AF, Renfroe SC, Askenase MH, McCullough LD, et al. 2016. TGF-beta1 modulates microglial phenotype and promotes recovery after intracerebral hemorrhage. Journal of Clinical Investigation 127 280–292. ( 10.1172/JCI88647) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. 2009. Epithelial-mesenchymal transitions in development and disease. Cell 139 871–890. ( 10.1016/j.cell.2009.11.007) [DOI] [PubMed] [Google Scholar]
- Tony H, Yu K, Qiutang Z. 2015. MicroRNA-208a silencing attenuates doxorubicin induced myocyte apoptosis and cardiac dysfunction. Oxidative Medicine and Cellular Longevity 2015 article ID 597032. ( 10.1155/2015/597032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchinovich A, Weiz L, Langheinz A, Burwinkel B. 2011. Characterization of extracellular circulating microRNA. Nucleic Acids Research 39 7223–7233. ( 10.1093/nar/gkr254) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella MC, Reinert K, Slack FJ. 2004. Architecture of a validated microRNA::target interaction. Trends in Cell Biology 11 1619–1623. ( 10.1016/j.chembiol.2004.09.010) [DOI] [PubMed] [Google Scholar]
- Vidigal JA, Ventura A. 2015. The biological functions of miRNAs: lessons from in vivo studies. Trends in Cell Biology 25 137–147. ( 10.1016/j.tcb.2014.11.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonlaufen A, Phillips P, Xu Z, Goldstein D, Pirola RC, Wilson JS, Apte MV. 2008. Pancreatic stellate cells and pancreatic cancer cells: an unholy alliance. Cancer Research 68 7707–7710. ( 10.1158/0008-5472.CAN-08-1132) [DOI] [PubMed] [Google Scholar]
- Wagner S, Ngezahayo A, Murua Escobar H, Nolte I. 2014. Role of miRNA let-7 and its major targets in prostate cancer. BioMed Research International 2014 article ID 376326. ( 10.1155/2014/376326) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Koh P, Winbanks C, Coughlan MT, McClelland A, Watson A, Jandeleit-Dahm K, Burns WC, Thomas MC, Cooper ME, et al. 2011. miR-200a prevents renal fibrogenesis through repression of TGF-beta2 expression. Diabetes 60 280–287. ( 10.2337/db10-0892) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Noberini R, Stebbins JL, Das S, Zhang Z, Wu B, Mitra S, Billet S, Fernandez A, Bhowmick NA, et al. 2013. Targeted delivery of paclitaxel to EphA2-expressing cancer cells. Clinical Cancer Research 19 128–137. ( 10.1158/1078-0432.CCR-12-2654) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BW, Wu GJ, Cheng WP, Shyu KG. 2014a. MicroRNA-208a increases myocardial fibrosis via endoglin in volume overloading heart. PLoS ONE 9 e84188 ( 10.1371/journal.pone.0084188) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li X, Zhu Y, Yang P. 2014b. MicroRNA-16 suppresses epithelial-mesenchymal transitionrelated gene expression in human glioma. Molecular Medicine Reports 10 3310–3314. ( 10.3892/mmr.2014.2583) [DOI] [PubMed] [Google Scholar]
- Xin H, Li Y, Buller B, Katakowski M, Zhang Y, Wang X, Shang X, Zhang ZG, Chopp M. 2012. Exosome-mediated transfer of miR-133b from multipotent mesenchymal stromal cells to neural cells contributes to neurite outgrowth. Stem Cells 30 1556–1564. ( 10.1002/stem.1129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Chi Y, Chen Y, Huang S, Ye X, Niu J, Wang W, Pfeffer LM, Shao ZM, Wu ZH, et al. 2016. MiRNA-621 sensitizes breast cancer to chemotherapy by suppressing FBXO11 and enhancing p53 activity. Oncogene 35 448–458. ( 10.1038/onc.2015.96) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M, Ferlito M, Lowenstein CJ. 2008. miR-34a repression of SIRT1 regulates apoptosis. PNAS 105 13421–13426. ( 10.1073/pnas.0801613105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaman Agaoglu F, Kovancilar M, Dizdar Y, Darendeliler E, Holdenrieder S, Dalay N, Gezer U. 2011. Investigation of miR-21, miR-141, and miR-221 in blood circulation of patients with prostate cancer. Tumor Biology 32 583–588. ( 10.1007/s13277-011-0154-9) [DOI] [PubMed] [Google Scholar]
- Yang B, Guo H, Zhang Y, Chen L, Ying D, Dong S. 2011. MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS ONE 6 e21679 ( 10.1371/journal.pone.0021679) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Li QJ, Feng Y, Zhang Y, Markowitz GJ, Ning S, Deng Y, Zhao J, Jiang S, Yuan Y, et al. 2012. TGF-beta-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell 22 291–303. ( 10.1016/j.ccr.2012.07.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. 2002. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO Journal 21 5875–5885. ( 10.1093/emboj/cdf582) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Sun Y, Hou Y, Peng Q, Wang L, Luo H, Tang X, Zeng Z, Liu M. 2012. MiRNA expression analysis of cancer-associated fibroblasts and normal fibroblasts in breast cancer. International Journal of Biochemistry and Cell Biology 44 2051–2059. ( 10.1016/j.biocel.2012.08.005) [DOI] [PubMed] [Google Scholar]
- Zhao G, Zhang JG, Liu Y, Qin Q, Wang B, Tian K, Liu L, Li X, Niu Y, Deng SC, et al. 2013. miR-148b functions as a tumor suppressor in pancreatic cancer by targeting AMPKalpha1. Molecular Cancer Therapeutics 12 83–93. ( 10.1158/1535-7163.MCT-12-0534-T) [DOI] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a