ABSTRACT
The rare actinomycete Actinoplanes missouriensis forms sporangia, including hundreds of flagellated spores that start swimming as zoospores after their release. Under conditions suitable for vegetative growth, zoospores stop swimming and germinate. A comparative proteome analysis between zoospores and germinating cells identified 15 proteins that were produced in larger amounts in germinating cells. They include an orthologue of BldD (herein named AmBldD [BldD of A. missouriensis]), which is a transcriptional regulator involved in morphological development and secondary metabolism in Streptomyces. AmBldD was detected in mycelia during vegetative growth but was barely detected in mycelia during the sporangium-forming phase, in spite of the constant transcription of AmbldD throughout growth. An AmbldD mutant started to form sporangia much earlier than the wild-type strain, and the resulting sporangia were morphologically abnormal. Recombinant AmBldD bound a palindromic sequence, the AmBldD box, located upstream from AmbldD. 3′,5′-Cyclic di-GMP significantly enhanced the in vitro DNA-binding ability of AmBldD. A chromatin immunoprecipitation-sequencing analysis and an in silico search for AmBldD boxes revealed that AmBldD bound 346 genomic loci that contained the 19-bp inverted repeat 5′-NN(G/A)TNACN(C/G)N(G/C)NGTNA(C/T)NN-3′ as the consensus AmBldD-binding sequence. The transcriptional analysis of 27 selected AmBldD target gene candidates indicated that AmBldD should repress 12 of the 27 genes, including bldM, ssgB, whiD, ddbA, and wblA orthologues. These genes are involved in morphological development in Streptomyces coelicolor A3(2). Thus, AmBldD is a global transcriptional regulator that seems to repress the transcription of tens of genes during vegetative growth, some of which are likely to be required for sporangium formation.
IMPORTANCE The rare actinomycete Actinoplanes missouriensis undergoes complex morphological differentiation, including sporangium formation. However, almost no molecular biological studies have been conducted on this bacterium. BldD is a key global regulator involved in the morphological development of streptomycetes. BldD orthologues are highly conserved among sporulating actinomycetes, but no BldD orthologues, except one in Saccharopolyspora erythraea, have been studied outside the streptomycetes. Here, it was revealed that the BldD orthologue AmBldD is essential for normal developmental processes in A. missouriensis. The AmBldD regulon seems to be different from the BldD regulon in Streptomyces coelicolor A3(2), but they share four genes that are involved in morphological differentiation in S. coelicolor A3(2).
KEYWORDS: gene regulation, rare actinomycete, regulon, sporangium formation, transcriptional factor
INTRODUCTION
Actinomycetes are Gram-positive, mainly soil-inhabiting bacteria. Many genera of actinomycetes show filamentous growth and produce spores, and therefore, actinomycetes are generally characterized by a complex morphological development. For example, members of the genus Streptomyces, the most extensively studied actinomycetes, typically grow into a highly branched substrate mycelium and then produce aerial hyphae arising from the substrate mycelium. After apical growth of the aerial hyphae stops, septa are synchronously formed at regular intervals along the aerial hyphae to form unigenomic spore compartments. In contrast, members of the genus Actinoplanes form a substrate mycelium from a germinating spore and subsequently produce terminal sporangia of various shapes growing from the vegetative mycelia through a short sporangiophore (1). Terminal sporangia include spores that can swim by means of flagella, termed zoospores after their release (2, 3). They show chemotactic properties in response to a wide variety of compounds, such as aromatic compounds, sugars, and amino acids. Under conditions suitable for vegetative growth, zoospores stop swimming and begin to germinate (4, 5). Although Actinoplanes bacteria are of great interest due to their complex morphological differentiation, most of the molecular mechanisms that underlie their life cycles remain to be elucidated.
Actinoplanes missouriensis is a well-characterized species of the genus, and the complete genome sequence of A. missouriensis 431T (NBRC 102363T), the species' type strain, has already been reported (6). This strain forms globose or subglobose terminal sporangia that include a few hundred spherical spores each on the sporangium-forming HAT (humic acid and trace element) agar. On this agar medium, small immature sporangium-like structures are observed after 2 to 3 days of cultivation. Then, sporangia that are mature enough to release spores are formed after being incubated for 5 to 7 days. In contrast, A. missouriensis grows into a substrate mycelium and sporangium formation is not observed in PYM (peptone-yeast extract-MgSO4) liquid medium and on YBNM (yeast extract-beef extract-NZ amine-maltose monohydrate) agar, both of which are nutrient-rich media. On HAT agar, spores are released from sporangia upon contact with water and an as-yet-unknown substance(s) in soil extracts through the process termed dehiscence. Alternatively, sporangium dehiscence and spore release can be induced by pouring 25 mM NH4HCO3 solution on the sporangia that formed on the HAT agar. Released spores start swimming as zoospores at an astonishingly high speed and show chemotaxis toward substances that include sugars, amino acids, aromatic compounds, and inorganic ions (4, 5). When zoospores reach the proper environment for vegetative growth, they stop swimming and begin to germinate to form a substrate mycelium.
In Streptomyces bacteria, bld genes are required for aerial hypha formation, and mutations in the bld loci lead to a “bald” phenotype (7, 8). In the regulatory cascade controlling morphological development, BldD serves to repress the expression of genes required for aerial hypha formation and sporulation (9). BldD proteins are highly conserved among the sporulating actinomycetes (having 77 to 99% amino acid identity) (10), and BldD in Streptomyces coelicolor A3(2) (ScBldD) and its orthologue in Streptomyces venezuelae (SvBldD) have been well studied. ScBldD directly controls at least 167 genes, and 25% of its regulon (42 genes) encodes regulatory proteins (11). Over 80% of the ScBldD-binding regions contain a well-conserved 15-bp palindromic sequence, 5′-NTNACNC(A/T)GNGTNAN-3′, which functions as the ScBldD box. The ScBldD regulon contains many genes responsible for morphological differentiation, such as adpA (also known as bldH), bldA, bldC, bldM, bldN, ssgA, ssgB, whiB, whiD, whiG, ftsZ, and smeA-sffA, in addition to bldD itself (11). Because the mutations in the whi genes, such as whiB and whiD, lead to a “white” phenotype, these genes are required for sporulation but not for aerial mycelium formation (7). SsgA and SsgB recruit FtsZ to the sites of sporulation septation (12, 13), and the smeA-sffA operon is involved in chromosomal segregation into spores (14). Furthermore, 3′,5′-cyclic di-GMP (c-di-GMP) is the effector of SvBldD, and the c-di-GMP tetramer enables SvBldD to dimerize and specifically bind a BldD box (10). BldD orthologues are highly conserved among sporulating actinomycetes, including, for example, the genera Saccharopolyspora, Salinospora, and Frankia. However, outside the streptomycetes, only one BldD orthologue has been characterized, the BldD orthologue in Saccharopolyspora erythraea, which regulates the synthesis of erythromycin and morphological differentiation (15).
Here, we describe the functional analysis of the BldD orthologue in A. missouriensis (AmBldD), revealing that AmBldD is a global transcriptional regulator involved in the morphological development of this bacterium. To our knowledge, this is the first reported molecular biology study of sporangium formation in Actinoplanes bacteria.
RESULTS
Comparative proteome analysis identified AMIS62190, the BldD orthologue in A. missouriensis, as a germinating-cell-specific protein.
To know an aspect of the physiological differences between a swimming zoospore and a cell that starts mycelial growth, we conducted a comparative proteome analysis by 2-dimensional polyacrylamide gel electrophoresis (2D-PAGE) using cellular proteins prepared from zoospores and germinating cells. We were able to make the zoospores germinate synchronously by shaking them in the nutrient-rich PYM liquid medium at 30°C (see Fig. S2 in the supplemental material), which enabled us to prepare germinating cells for this analysis. The protein samples were separated by 2D-PAGE and stained with Coomassie brilliant blue (CBB). To minimize false positives, experiments were performed in triplicate with three independent biological replicates, and representative pictures are shown in Fig. 1. We successfully detected 18 spots with protein amounts that were at least three times larger in germinating cells than in zoospores. As a result, 15 germinating-cell-specific proteins were identified by peptide mass fingerprinting. They included proteins involved in vegetative growth, such as ribosomal protein L25 and enzymes for primary metabolism and chaperones, supporting the notion that the germinating cells initiate vegetative growth (Table 1). We also detected 30 spots that were at least three times larger in zoospores than in germinating cells and identified a total of 27 zoospore-specific proteins. They included proteins required for chemotaxis and flagellar synthesis, in addition to several functionally unknown proteins (see Table S2 in the supplemental material).
FIG 1.
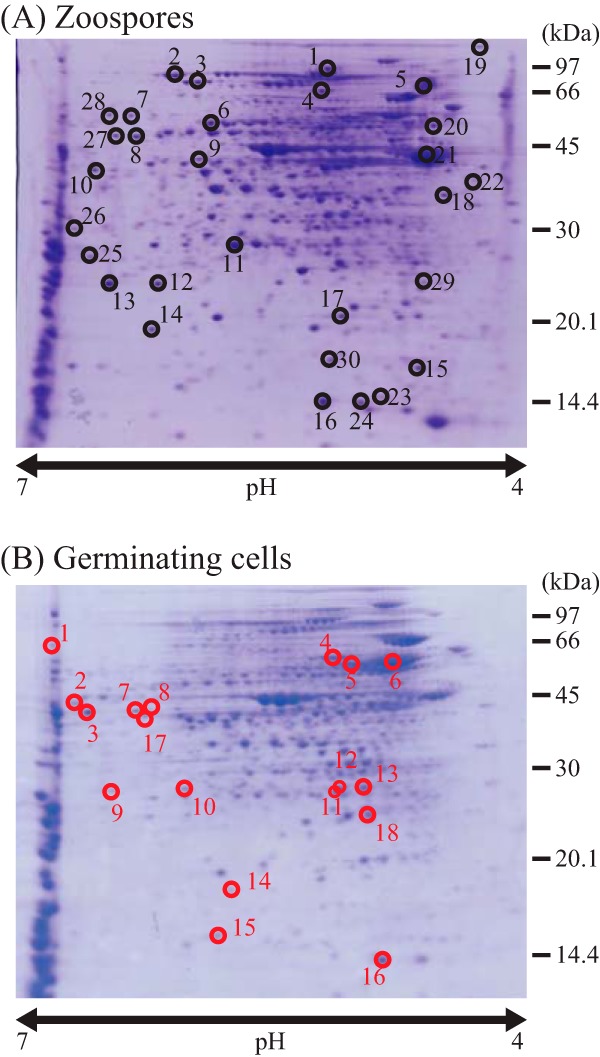
Comparative proteome analysis between zoospores and germinating cells. 2D-PAGE was performed using cellular proteins extracted from zoospores (A) and germinating cells (B). Germinating cells were prepared by shaking zoospores in PYM liquid medium at 30°C for 8 h. The gels were stained with Coomassie brilliant blue (CBB). Circles indicate zoospore-specific (A) and germinating cell-specific (B) protein spots, respectively, and the black and red numbers correspond to protein spot identifications in Table S2 in the supplemental material and Table 1, respectively.
TABLE 1.
Germinating-cell-specific proteins detected in 2D-PAGE
| Spot no.a | ORF no.b | Gene productc |
|---|---|---|
| 1 | AMIS2010 | Putative DEAD/DEAH box helicase |
| 2 | AMIS47630 | Unknown |
| 3 | AMIS77290 | Putative ABC transporter ATP-binding protein |
| 4 | AMIS6760 | Putative chaperonin GroEL |
| 5 | AMIS16260 | Putative FeS assembly protein |
| 6 | AMIS77060 | Putative chaperonin GroEL |
| 7, 8 | AMIS72620 | Putative ornithine aminotransferase |
| 9, 10 | AMIS71950 | Putative F-type proton-transporting ATPase delta chain |
| 11 | AMIS16160 | Putative 6-phosphogluconolactonase |
| 12, 13 | AMIS71020 | Unknown |
| 14 | AMIS77530 | Unknown |
| 15 | AMIS62190 | Putative DNA-binding protein (AmBldD) |
| 16 | AMIS6770 | Putative chaperonin GroES |
| 17 | AMIS1270 | Putative Glu/Leu/Phe/Val dehydrogenase |
| 18 | AMIS75010 | Putative ribosomal protein L25 |
Spot numbers are shown in Fig. 1B.
A total of 15 proteins from the 18 spots were identified by peptide mass fingerprinting. AMIS71950, AMIS72620, and AMIS71020 were identified from two spots each.
Possible functions and classifications of the identified proteins are shown.
Among the proteins identified, we focused on AMIS62190, which was predicted to function as a DNA-binding protein (Table 1). AMIS62190 shows 67.5% amino acid identity with ScBldD (Fig. 2A), and the organization of the genes near AMIS62190 and ScbldD is highly conserved (Fig. 2B). Therefore, we name AMIS62190 AmBldD herein. The helix-turn-helix DNA-binding motifs of ScBldD and AmBldD are highly conserved (Fig. 2A), suggesting that AmBldD binds sequences similar to that of the ScBldD box. Considering the function of BldD in Streptomyces, we hypothesized that AmBldD plays a pivotal role in the morphological differentiation of A. missouriensis rather than in the motility of zoospores and the germination process.
FIG 2.
In silico analysis of AmbldD and ScbldD. (A) Amino acid sequence alignment between AmBldD and ScBldD. The helix-turn-helix (HTH) motifs are highly conserved (87% identity). Asterisks show the nine residues that are required for the binding of c-di-GMP and dimerization in SvBldD. Motifs 1 and 2 were expected to interact directly with c-di-GMP (10). (B) Gene organization of AmbldD and ScbldD. Arrows indicate the locations of the open reading frames, including their lengths and directions. Gene identifiers are shown above or below the arrows. Amino acid identities are also shown between the arrows. A double diagonal line indicates that a gene is not shown.
AmBldD is produced in actively growing cells, but its protein level decreases during sporangium formation.
AmbldD expression in the wild-type (wt) strain was examined by Western blot analysis using a newly generated polyclonal anti-AmBldD antibody (Fig. 3A; see also Fig. S3 in the supplemental material). AmBldD was strongly detected in the cell lysates of vegetative hyphae in PYM liquid medium (Fig. 3A, lane V; see also Fig. S3, lane VP) and YBNM agar (see Fig. S3, lane VY) and in germinating cells (Fig. 3A, lane G). AmBldD was also weakly detected in the cell lysate of zoospores (Fig. 3A, lane Z). However, AmBldD was hardly detected in the cell lysate extracted from mixtures of substrate hyphae and sporangia on HAT agar (Fig. 3A, lanes S3 and S6). Thus, AmBldD is actively produced during vegetative growth immediately after germination, but its protein level decreases significantly in the process of sporangium formation.
FIG 3.
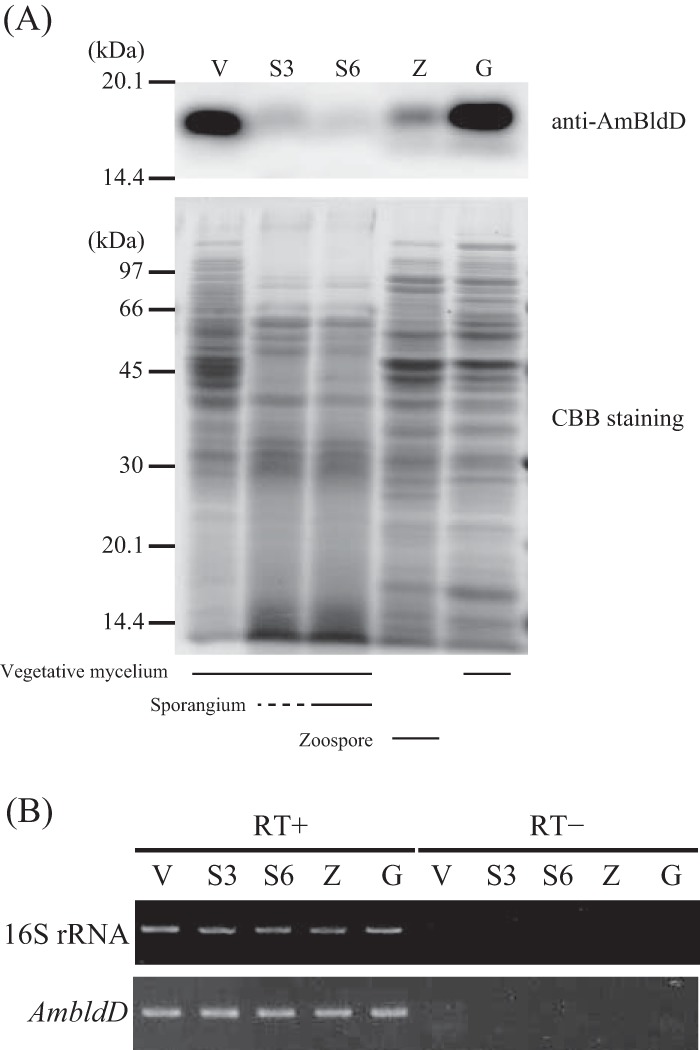
Expression of AmbldD under various culture conditions. (A) Protein levels of AmBldD. Protein concentrations were determined by the Bradford assay, and equal amounts of proteins were used for SDS-PAGE. AmBldD was detected using the anti-AmBldD polyclonal antibody (top). A CBB-stained gel is shown below. (B) Transcript levels of AmbldD. Transcript amounts were examined by a semiquantitative RT-PCR analysis. 16S rRNA was used as an internal standard to check the purity and amount of RNA used. RT+, with reverse transcriptase; RT-, without reverse transcriptase (negative control to show there was no contamination by genomic DNA in the RNA samples). Protein and RNA samples were prepared from vegetative mycelia grown in PYM liquid medium at 30°C for 48 h (V), mixtures of substrate mycelium and (immature) sporangia grown on HAT agar at 30°C for 3 days (S3) and 6 days (S6), zoospores released from sporangia grown on HAT agar at 30°C for 6 days (Z), and germinating cells prepared by shaking zoospores in PYM liquid medium at 30°C for 4 h (G).
We also analyzed the transcript levels of AmbldD by semiquantitative reverse transcription-PCR (semi-qRT-PCR) analysis (Fig. 3B). Because the transcripts of ScbldD are most abundant in the vegetative growth (16) and because AmBldD was hardly detected in a mixture of substrate mycelium and sporangia (Fig. 3A), we hypothesized that AmbldD was not transcribed in cells during sporangium formation. Contrary to our expectation, however, AmbldD transcripts were detected at almost identical levels in vegetative hyphae, mixtures of vegetative hyphae and sporangia, zoospores, and germinating cells (Fig. 3B). Previous transcriptome sequencing (RNA-Seq) analyses in our laboratory (29) also showed that AmbldD is actively transcribed in 1-, 3-, 6- and 40-day cultures on HAT agar (see Fig. S4 in the supplemental material). AmbldD has a typical σ70-dependent promoter sequence, TTGACC-N17-TACCGT, in the appropriate region upstream from the transcription start point determined by the 5′ rapid amplification of cDNA ends (5′-RACE) experiment (see below).
AmBldD plays a pivotal role in sporangium formation.
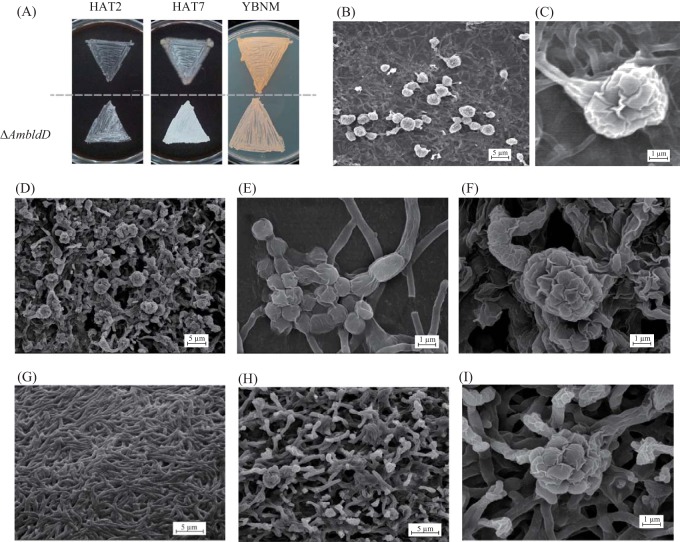
To examine the function of AmBldD, we generated an AmbldD null (ΔAmbldD) mutant strain (see Fig. S1 in the supplemental material). On HAT agar, the ΔAmbldD mycelium turned white in appearance earlier than that of the wt strain, suggesting that sporangium formation was enhanced in the mutant (Fig. 4A, left and middle). On YBNM agar, vegetative hyphae of the ΔAmbldD mutant were unable to penetrate sufficiently into the agar medium and upheaved into the air (Fig. 4A, right). To examine sporangium formation in detail, we observed the mycelia of the wt and ΔAmbldD strains by scanning electron microscopy (SEM). Both strains were grown on HAT agar for 7 days or on YBNM agar for 4 days prior to imaging. On HAT agar, the wt strain produced normal globose sporangia with short sporangiophores (Fig. 4B and C). In contrast, the ΔAmbldD mutant produced many abnormal sporangia with distorted shapes (Fig. 4D), including even “bare” spores, which were not surrounded by sporangial walls (Fig. 4E). Some sporangia of the mutant seemed to be formed directly on substrate hyphae without sporangiophores (Fig. 4F). On YBNM agar, the wt strain grew only as substrate mycelium (Fig. 4G), but the ΔAmbldD mutant formed morphologically abnormal hyphae whose tips turned into sporangial-wall-like rugose structures (Fig. 4H and I). Moreover, the ΔAmbldD mutant produced some immature sporangium-like structures upon vegetative mycelia (Fig. 4I). Thus, sporangium formation is accelerated and does not proceed appropriately in the ΔAmbldD mutant, indicating that AmBldD strictly regulates the initiation and proper progress of sporangium formation in A. missouriensis. An accelerated sporulation phenotype was also reported in the SvbldD-disrupted mutant of S. venezuelae (10).
FIG 4.
Phenotypic observation of the ΔAmbldD mutant strain. (A) Macroscopic observations of the colonies formed on agar media. The wt and ΔAmbldD strains were cultivated on HAT agar at 30°C for 2 days (HAT2) and 7 days (HAT7) and on YBNM agar at 30°C for 2 days (YBNM). (B to I) Observations by SEM. (B) The wt strain cultivated on HAT agar at 30°C for 7 days. (C) An enlarged view of a sporangium of the wt strain on HAT agar. (D) The ΔAmbldD mutant cultivated on HAT agar at 30°C for 7 days. (E, F) Enlarged views of a sporangium (F) and “bare” spores (E) of the ΔAmbldD mutant on HAT agar. (G) The wt strain grown on YBNM agar at 30°C for 4 days. (H) The ΔAmbldD mutant strain cultivated on YBNM agar at 30°C for 4 days. (I) An enlarged view of a sporangium-like structure of the ΔAmbldD mutant on YBNM agar.
Because of the extremely low transformation efficiency of the ΔAmbldD mutant, we could not carry out a gene complementation experiment. However, the same phenotypes were also observed in another two ΔAmbldD mutants that were generated by independent gene disruption experiments, supporting the hypothesis that the phenotypic changes were due to the AmbldD gene disruption (data not shown). Additionally, the growth of the ΔAmbldD mutant was much slower than that of the wt strain in submerged cultures. In fact, the wet cell weight of the ΔAmbldD mutant remained very low even after a 66-h cultivation in PYM liquid medium (Fig. 5). The significant growth defect of the bldD mutant was also reported in S. coelicolor A3(2) (11). The effects of the AmbldD disruption on the vegetative growth of hyphae are considered in Discussion.
FIG 5.
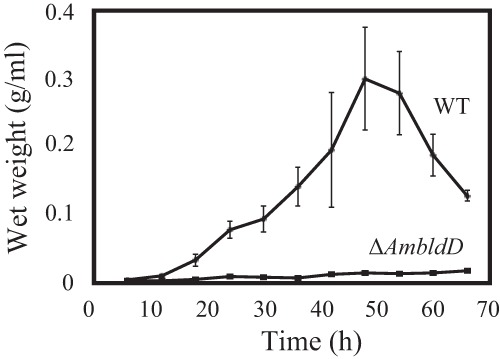
Growth curves of the wt and ΔAmbldD strains. Both strains were cultivated at 30°C in PYM liquid medium. The data are the mean results from five biological replicates ± standard deviations.
AmBldD binds to its own promoter.
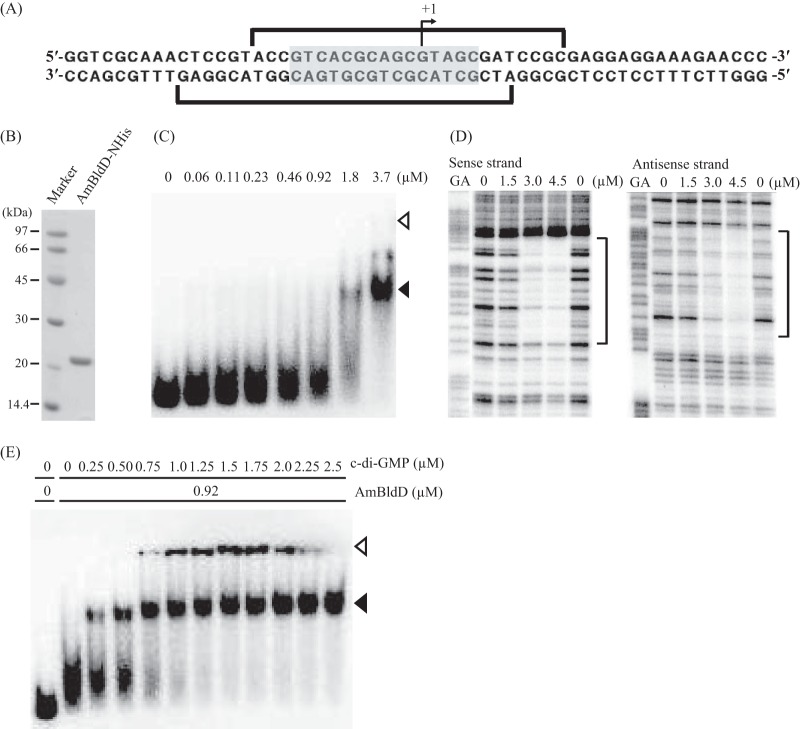
We determined the transcriptional start point of AmbldD to be 56 nucleotides upstream from the translational start codon by the 5′-RACE experiment (Fig. 6A). Because the N-terminal DNA-binding domains of AmBldD and ScBldD share a high sequence identity (Fig. 2A), we expected AmBldD to bind sequences similar to the ScBldD box. Such a palindromic sequence, 5′-GTCACGCAGCGTAGC-3′, was found around the transcriptional start point of AmbldD (from position −10 to +5, referring to the transcriptional start point as +1) (Fig. 6A). To examine whether AmBldD bound this sequence, we purified a recombinant AmBldD protein with a polyhistidine tag at its N-terminal end (AmBldD-NHis) using Escherichia coli as the host. Purified AmBldD-NHis produced a single 21.2-kDa band on SDS-PAGE (Fig. 6B). We prepared a 32P-labeled DNA probe that contained the ScBldD box-like sequence and used it for an electrophoretic mobility shift assay (EMSA). The probe produced a retarded signal in the presence of AmBldD-NHis, indicating that the protein bound this DNA fragment as expected (Fig. 6C). Then, the exact location of the AmBldD-NHis-binding site was determined by DNase I footprinting. AmBldD-NHis protected a sequence from position −13 to +12 on the sense strand and from −19 to +8 on the antisense strand (Fig. 6D). Thus, we confirmed that AmBldD-NHis binds the ScBldD box-like palindromic sequence (Fig. 6A).
FIG 6.
Binding of AmBldD-NHis to the AmbldD promoter region. (A) The nucleotide sequence around the transcriptional start point of AmbldD. The bent arrow indicates the transcriptional start point determined by the 5′-RACE experiment. A palindromic sequence similar to the ScBldD box is shaded. Brackets show the AmBldD-NHis-binding sequence determined in the experiment whose results are shown in panel D. (B) Recombinant AmBldD-NHis protein. The AmBldD-NHis protein was produced in E. coli and purified to homogeneity using affinity chromatography. The purified protein was analyzed by SDS-PAGE and visualized with CBB. (C) Binding of AmBldD-NHis to the AmbldD promoter-containing region. A DNA fragment of 224 bp (nucleotide positions from −197 to +27 with respect to the start codon of BldD) was 32P labeled and used as a probe in EMSAs. The amounts of AmBldD-NHis used are shown above the lanes. The positions of the wells and the AmBldD-NHis-bound probe are shown by open and closed triangles, respectively. (D) Determination of the AmBldD-NHis-binding site by a DNase I protection assay. The DNase I digests were run with the same probes that had been chemically cleaved (GA lanes). The amounts of AmBldD-NHis used are shown above the lanes. Lanes without AmBldD-NHis are control lanes. The protected nucleotides are indicated by the brackets. (E) Enhancement of the AmBldD-NHis DNA-binding activity by c-di-GMP. The DNA probe used in the experiment whose results are shown in panel C was incubated with a fixed concentration of AmBldD-NHis and increasing concentrations of c-di-GMP in the EMSA. The amounts of AmBldD-NHis and c-di-GMP used are shown above the lanes. The positions of the wells and the AmBldD-NHis-bound probe are shown by open and closed triangles, respectively.
Recently, a tetrameric form of c-di-GMP was reported to bind SvBldD to mediate its dimerization, allowing the protein to acquire a high affinity for its binding sequences. Because all eight of the amino acid residues important for the c-di-GMP binding of SvBldD (10) are conserved in AmBldD (Fig. 2A), we assumed that c-di-GMP bound AmBldD and enhanced its DNA-binding ability. To test this hypothesis, another EMSA was performed. Although the fixed concentration of AmBldD-NHis used in this assay (0.92 μM) was insufficient to elicit a complete retardation of the DNA (Fig. 6C), the addition of increasing concentrations of c-di-GMP (0.25 to 2.5 μM) clearly enhanced the binding of AmBldD-NHis to the target DNA sequence (Fig. 6E). Thus, c-di-GMP activates the DNA-binding ability of AmBldD in a manner similar to that for SvBldD (10). In the EMSA (Fig. 6C and 7C) and DNase I footprinting (Fig. 6D), AmBldD-NHis bound to the DNA fragments without the addition of c-di-GMP. We think that a portion of the recombinant AmBldD-NHis protein had become the active form, i.e., the dimer with c-di-GMP, in the host E. coli cells.
FIG 7.
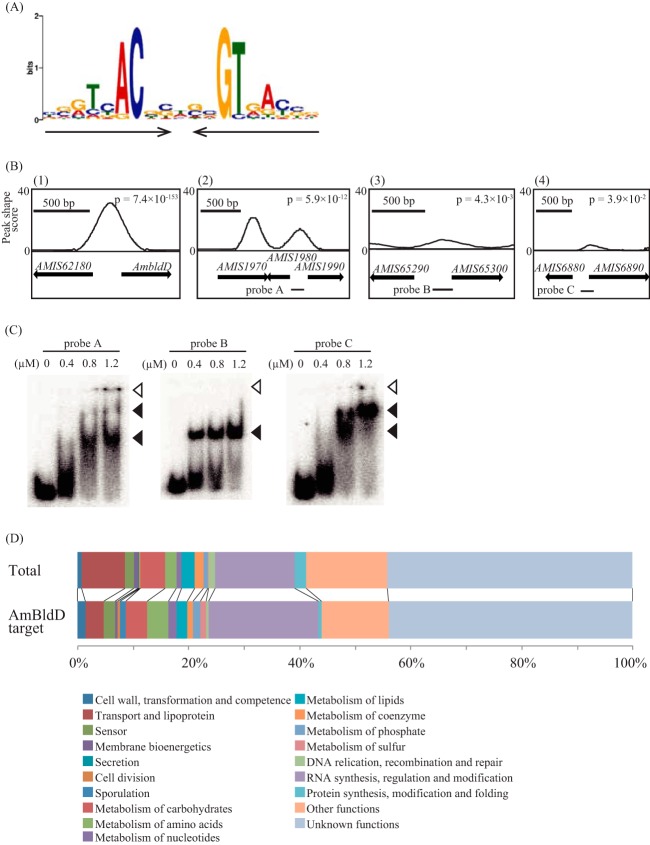
ChIP-Seq analysis and target genes of AmBldD. (A) Sequence logo of the consensus AmBldD-binding site. The conserved sequence motif was identified using the MEME algorithm by analyzing the genomic regions that were enriched in the AmBldD ChIP-Seq experiment. Black arrows indicate the palindromic sequence. (B) ChIP-Seq data for selected regions. Peaks indicate the AmBldD-binding sites. The horizontal axis shows the genomic locations, and the vertical axis shows the read count at each position. Arrows indicate the locations of the open reading frames, including their lengths and directions. The gene name or identifier (AMIS numbers) is shown above the arrows. Bars below arrows (probes A, B, and C) indicate the locations of the DNA probes used for the EMSAs whose results are shown in panel C. (C) Confirmation of AmBldD binding to the three promoter regions of putative target genes. EMSAs were performed using the purified AmBldD-NHis protein and the 32P-labeled probes A, B, and C. The concentrations of AmBldD-NHis used are shown above the lanes. Open and closed triangles indicate the wells and AmBldD-NHis-bound DNAs, respectively. (D) Classifications of A. missouriensis genes (top) and AmBldD target gene candidates (bottom) based on the NITE annotation database.
AmBldD binds to more than 346 sites on the A. missouriensis genome in vivo.
ScBldD directly regulates approximately 30 genes involved in morphological development and antibiotic production in S. coelicolor A3(2) (11). Therefore, we postulated that AmBldD also has many target genes, and we investigated the genome-wide distribution of AmBldD-binding sites using a chromatin immunoprecipitation sequencing (ChIP-Seq) analysis with the anti-AmBldD polyclonal antibody. Because AmBldD was detected strongly in the vegetative hyphae grown in PYM liquid medium at 30°C for 2 days (Fig. 3A), this culture was used for the ChIP-Seq analysis. A total of 163 peaks of AmBldD binding were detected throughout the genome when we used a strict threshold, a P value of <0.0005. A computational search of these enriched DNA sequences using the MEME algorithm established a 19-bp palindromic sequence, 5′-NN(G/A)TNACN(C/G)N(G/C)NGTNA(C/T)NN-3′, as the consensus AmBldD-binding sequence, namely, the AmBldD box, which is very similar to the ScBldD box (Fig. 7A; see also Table S3 in the supplemental material). All of the 163 peak regions contained at least one AmBldD box. When we loosened the threshold to a P value of <0.05 in the ChIP-Seq analysis, 437 peaks of AmBldD binding were detected. However, 91 (21.8%) of the 437 peak regions did not contain an AmBldD box, indicating that they are not genuine AmBldD-binding sites; these false-positive peaks may be caused by the cross-reaction of the anti-AmBldD antibody with DNA-binding proteins other than AmBldD. In this way, the loosened threshold increased the risk of false-positive signals, but the number of probable AmBldD-binding sites was increased to 346 by this analysis (see Table S4). Some representative results are shown in Fig. 7B; peaks in the peak shape score profile, which was calculated from the mapping data of deep-sequenced ChIP and control read libraries, indicate regions bound by AmBldD in vivo. Consistent with the EMSA (Fig. 6C), the AmbldD promoter region was enriched [Fig. 7B(1)]. Similarly, by EMSA, we confirmed the binding of AmBldD to DNA fragments containing the upstream regions of AMIS1980 (encoding a DNA-binding protein), AMIS65300 (ssgB), and AMIS6890 (bldM) (Fig. 7C).
Of the 346 probable AmBldD-binding sites, 161 (46.5%) were located at the possible regulatory region of an open reading frame (ORF) or two divergent ORFs (from position −200 to +50, referring to the transcriptional start point predicted by RNA-Seq analyses as +1), which indicated that AmBldD might regulate the expression of 207 ORFs that exist downstream from the AmBldD boxes (see Table S5 in the supplemental material). Here, to avoid prediction uncertainty, we only considered the first ORFs in some putative operons that might be regulated by AmBldD. Then, we classified these 207 ORFs by their putative functions based on the NITE annotation database (Fig. 7D; see also Table S5). Forty-one genes (19.8%) belong to the category of “RNA synthesis, regulation and modification.” Because the proportion of genes in this category out of all the A. missouriensis genes is 14.2%, the genes of this category appear to be meaningfully enriched in the AmBldD target gene candidates (Fig. 7D). The AmBldD target gene candidates classified into “RNA synthesis, regulation and modification” include 31 transcriptional regulator genes and 2 RNA polymerase sigma factor genes. Interestingly, the A. missouriensis genome has only two genes that were classified into the “sporulation” category by the NITE annotation database, and these two genes, AMIS65300 [ssgB; encoding a small acidic protein involved in the septation of sporulating hyphae in S. coelicolor A3(2)] and AMIS24370 (encoding a putative spore coat protein), are also possible AmBldD target genes.
Transcriptional analysis of candidates for AmBldD target genes.
As described above, 161 probable AmBldD-binding sites were found in possible regulatory regions of ORFs, but more than half of the probable AmBldD-binding sites were located on regions that were apparently not involved in the transcriptional regulation of any ORFs. Therefore, we hypothesized that AmBldD would not necessarily function as a transcriptional regulator even if it bound DNA. Accordingly, we examined a portion of the candidate AmBldD target genes for transcriptional regulation by AmBldD. Because we were interested in the genes involved in morphological development and its regulation, we focused on 134 possible AmBldD target genes that were classified into the “sporulation” (2 genes), “RNA synthesis, regulation and modification” (41 genes), and “unknown function” (91 genes) categories. Then, we used the amino acid sequence conservation among Actinoplanes as the first selection filter, based on the idea that genes important for morphological development should be conserved in the genus. Forty-one of the 134 possible AmBldD target genes are highly (more than 70% amino acid sequence identity) conserved among the following four Actinoplanes species: A. missouriensis, Actinoplanes sp. strain SE50/110, Actinoplanes sp. strain N902-109, and Actinoplanes friuliensis (see Table S4 in the supplemental material) (18, 33). Next, we used our previous RNA-Seq analysis data as the second selection filter, because performing a transcriptional analysis of weakly transcribed genes is difficult. Twenty-seven of the 41 selected genes were sufficiently transcribed (>200 reads per kilobase per million [RPKM]) during vegetative growth (day 1) and/or sporangium formation (day 3) (see Table S5).
We quantified the transcription levels of the 27 genes in the wt and ΔAmbldD strains using qRT-PCR (see Fig. S5 in the supplemental material). In this analysis, we used mRNAs extracted from the mycelia grown on YBNM agar at 30°C for 2 days, because the ΔAmbldD mutant showed abnormal growth and began to produce immature sporangium-like structures in this culture. The expression ratio of each gene (in the ΔAmbldD strain versus in the wt strain) is shown in Table 2. The transcription levels of 16 genes were significantly changed in the ΔAmbldD strain, with 12 and 4 genes being up- and downregulated, respectively, in the ΔAmbldD strain (Table 2). Unexpectedly, no significant correlation was observed between the expression ratios and the positions of the AmBldD-binding sites with respect to the transcriptional start point among these 27 genes. However, six of the seven genes that had an AmBldD-binding site with a central nucleotide located between positions −41 and +10 (referring to the transcriptional start point as +1) appeared to be repressed by AmBldD. This result is consistent with the predicted function of BldD as a transcriptional repressor. One exception was the AmbldD gene, which has an AmBldD-binding site at position −3, but its transcription was hardly affected by the disruption of AmbldD. We confirmed that transcription of AmbldD in the ΔAmbldD mutant was also not enhanced under other culture conditions, i.e., in PYM liquid medium or on HAT agar (see Fig. S6). These results indicated that AmbldD is not repressed by AmBldD. This is in contrast to the self-repression of the ScbldD gene by its gene product in S. coelicolor A3(2) (see Discussion) (11, 19). Additionally, three of the four genes that appeared to be activated by AmBldD have their respective AmBldD-binding sites at positions −53, −59, and −74, which are appropriate positions for being bound by a typical transcriptional activator. As suggested elsewhere (11, 15, 20), it is possible that BldD and its orthologues also function as transcriptional activators for some genes.
TABLE 2.
Comparative transcriptional analysis of the 27 selected possible AmBldD target genes in the wild-type and ΔAmbldD strains cultivated on YBNM agar for 48 h
| Gene | ΔAmbldD/wt expression ratioa | Position of AmBldD boxb | Orthologuec | Gene productd |
|---|---|---|---|---|
| AMIS6890 | 118.6* | +10 | bldM | LuxR family transcriptional regulator |
| AMIS65300 | 32.7* | −35** | ssgB | SsgA family protein |
| AMIS9230 | 24.2* | −64 | — | Hypothetical protein |
| AMIS6880 | 14.0* | −172** and −150** | whiD | WhiB family transcriptional regulator |
| AMIS15330 | 7.1* | −30 | ddbA | TraR/DksA family transcriptional regulator |
| AMIS78290 | 5.8* | −41** | wblA | WhiB family transcriptional regulator |
| AMIS14670 | 5.6* | −30 | — | Hypothetical protein |
| AMIS16220 | 5.4* | −164 | SCO1933 | Hypothetical protein |
| AMIS75470 | 3.8* | −17 | — | Thioredoxine |
| AMIS72380 | 2.8* | −17 | — | Hypothetical protein |
| AMIS77530 | 2.5* | −52 | SCO4232 | Hypothetical protein |
| AMIS78180 | 2.1 | −87 | — | Transcriptional regulator with a cyclic nucleotide-binding domain |
| AMIS1980 | 1.9* | +25 | — | DNA-binding protein |
| AMIS68580 | 1.8 | −63 | — | Hypothetical protein |
| AMIS14490 | 1.7 | −50 | — | Transcriptional regulator with a cyclic nucleotide-binding domain |
| AMIS41990 | 1.7 | −113 | — | Hypothetical protein |
| AMIS77890 | 1.6 | −67 | — | Hypothetical protein |
| AMIS78890 | 1.4 | −44 | — | MerR family transcriptional regulator |
| AMIS9410 | 1.2 | −38 | — | Hypothetical protein of Mov34/MPN/PAD-1 family |
| AMIS64740 | 1.1 | −76 | — | Hypothetical protein |
| AMIS67900 | 1.1 | +29 | sigU | RNA polymerase ECF subfamily sigma factor |
| AMIS9450 | 1.1 | −134 | — | Hypothetical protein |
| AmbldD downstream | −1.3 | −3** | bldD | DNA-binding protein |
| AmbldD upstream | −1.5 | −3** | bldD | DNA-binding protein |
| AMIS78900 | −2.1* | +26 | — | Hypothetical protein |
| AMIS80980 | −2.4* | −74 | — | PadR-like family transcriptional regulator |
| AMIS72910 | −2.8* | −59 | wblE | WhiB family transcriptional regulator |
| AMIS80970 | −2.8* | −53 | — | Hypothetical protein |
The expression ratio of each gene (in the ΔAmbldD strain versus in the wt strain) determined by qRT-PCR (see Fig. S5 in the supplemental material) is shown. The AmbldD transcript was analyzed by two primer sets; one is for the 5′ region of AmbldD (AmbldD upstream), and the other is for the 3′ region (AmbldD downstream). See Fig. S1C for the amplification regions. *, value was statistically significant by the t test at a 1% level.
The position of the central nucleotide of each AmBldD box is shown. The transcriptional start points determined by 5′-RACE (for values with double asterisks) and predicted by previous RNA-Seq analyses were set as +1. The RNA samples for the qRT-PCR experiment were used for the 5′-RACE experiment. The RNA-Seq analyses were performed using RNA samples prepared from the cells cultivated on HAT agar for 1, 3, 6, and 40 days (29).
Orthologues in S. coelicolor A3(2). When the organization of each gene locus is conserved in S. coelicolor A3(2), the gene is regarded as an orthologue of the corresponding S. coelicolor A3(2) gene. Dashes indicate no orthologue in S. coelicolor A3(2). The probable ScBldD target genes are indicated by underlining.
The predicted function of each gene product is shown.
The gene product was detected as a zoospore-specific protein in our study (Fig. 1A; see also Table S2 in the supplemental material).
DISCUSSION
BldD is located at the top of the regulatory hierarchy that controls morphological differentiation in Streptomyces species, serving as the transcriptional repressor of sporulation genes (8). In this study, we showed that the BldD orthologue AmBldD plays an important role in the morphological differentiation of A. missouriensis. AmBldD is actively produced during vegetative growth (Fig. 3A) and presumably represses the transcription of the genes responsible for sporangium formation to control the timing of the transition from vegetative growth to sporangium formation. Although the morphological development of Actinoplanes species is quite different from that of Streptomyces species, our study clearly showed that BldD orthologues are pivotal determinants of the initiation of sporulation in both genera.
In the cells grown on HAT agar and in swimming zoospores, the AmBldD protein levels were very low (Fig. 3A), which is consistent with the results of the comparative proteome analysis, where the AmBldD protein was detected in germinating cells but not in swimming zoospores (Fig. 1). However, AmbldD transcripts were detected at almost the same levels at all of the time points examined (Fig. 3B). How is the amount of AmBldD protein controlled at low levels in sporangium-forming cells and zoospores despite the constant transcription of AmbldD? A possibility is that the AmBldD protein levels are regulated by proteolysis. In this regard, it is interesting to note that the ATP-dependent Clp protease plays an essential role in aerial mycelium formation in several Streptomyces bacteria (21) and that the C-terminal part of the recombinant ScBldD protein was degraded when incubated with cytoplasmic extracts at the aerial mycelium and spore-forming stages, suggesting that morphological development is modulated by the proteolytic cleavage of ScBldD in S. coelicolor A3(2) (22). Therefore, there may be a developmental-phase-specific proteolysis of AmBldD in A. missouriensis. Another possibility is that the translation of the AmbldD transcripts is repressed in sporangium-forming cells and zoospores. Thus, we hypothesize that the expression of AmBldD is regulated at the translational or posttranslational level, not at the transcriptional level. Constant AmbldD transcription in the presence or absence of AmBldD also indicates that AmBldD is not involved in the self-regulation of the AmbldD gene. This notion is also supported by the observation that the transcription of AmbldD was not affected by the deletion of AmbldD in the mycelia grown on YBNM agar at 30°C for 2 days, in which AmBldD should repress the genes required for the onset of morphological development. Therefore, we believe that the transcription of AmbldD is not autoregulated by AmBldD, although we showed that AmBldD bound to the region containing the AmbldD transcriptional start point both in vivo and in vitro. We speculate that the binding of AmBldD to this region is not strong enough to interfere with the transcription of AmbldD by RNA polymerase.
The growth of the vegetative hyphae of the ΔAmbldD mutant was impaired, especially under nutrient-rich conditions. Its vegetative hyphae failed to penetrate sufficiently into the YBNM agar medium and upheaved into the air (Fig. 4A, right). Additionally, it grew slowly and yielded a much smaller amount of biomass in PYM liquid medium (Fig. 5). On YBNM agar, sporangium formation underwent a disorderly initiation in the ΔAmbldD mutant, and many hyphal tips turned into sporangial-wall-like rugose structures (Fig. 4H and I). We speculated that these abnormal hyphal tip structures would also form in PYM liquid medium and that they might hamper quick mycelial growth achieved by the branching and tip extension.
It was reported that c-di-GMP is the effector of SvBldD and that the SvBldD–(c-di-GMP) complex, not SvBldD, controls the transcription of the BldD regulons (10). In accordance with this report, the DNA-binding ability of AmBldD was significantly enhanced by the addition of c-di-GMP (Fig. 6E). Therefore, the intracellular concentration of c-di-GMP seems to be a critical determinant of the entry into morphological development in A. missouriensis. In the transition from vegetative growth to sporangium formation, the intracellular c-di-GMP concentration is presumed to decrease to liberate the transcriptional repression of the developmental genes by AmBldD. The GGDEF domain is known to act as a diguanylate cyclase to synthesize c-di-GMP. In contrast, the EAL domain is responsible for the conversion of c-di-GMP into 5′-phosphoguanylyl-(3′-5′)-guanosine (5′-pGpG). Interestingly, there are at least 55 genes that encode proteins with the GGDEF domain on the A. missouriensis genome. Of these, 27 genes encode proteins with both the GGDEF and EAL domains (https://www.ncbi.nlm.nih.gov/Complete_Genomes/SigCensus/GGDEFactino2014.html#512565). The presence of many GGDEF domain-containing proteins in A. missouriensis suggests the importance of c-di-GMP in its life cycle, including not only morphological development but also dormancy and awakening of spores and motility of zoospores. We presume that other regulatory proteins than AmBldD that use c-di-GMP as the effector should be present in A. missouriensis.
In a previous study, ScBldD targets were identified by transcriptome and ChIP with microarray technology (ChIP-chip) analyses using a DNA microarray, which revealed that many genes involved in morphological development are under the control of ScBldD (11). Here, we identified 207 genes as AmBldD target gene candidates by ChIP-Seq analysis. Of these 207 genes, 27 were selected and their transcription was examined in the wt and ΔAmbldD strains, which indicated that AmBldD could repress 12 of the 27 genes. Of the 12 genes, 7 were considered to be orthologous to the corresponding S. coelicolor A3(2) genes. These genes were bldM, ssgB, whiD, ddbA, and wblA, all of which are involved in the morphological development of S. coelicolor A3(2) (23–25), and SCO1933 and SCO4232, both of which encode hypothetical proteins. Furthermore, the regulation of bldM, ssgB, whiD, and wblA by ScBldD has also been proposed (11). Thus, some genes of the BldD regulon seem to overlap between A. missouriensis and S. coelicolor A3(2). Meanwhile, another in silico analysis suggests that a large part of the BldD regulon differs between A. missouriensis and S. coelicolor A3(2), as described below. According to the OrtholugeDB, 3,249 orthologue (or homologue) pairs were found between A. missouriensis and S. coelicolor A3(2), and S. coelicolor A3(2) has 92 homologues of the 207 AmBldD target gene candidates. Of the genes containing predicted ScBldD-binding sites (11), only 15 of the 92 genes were found as predicted ScBldD target genes, suggesting that the AmBldD regulon is largely different from the ScBldD regulon. We are afraid that the 207 AmBldD target gene candidates include a considerable number of mistakenly selected genes that are not actual AmBldD targets. However, of the 207 genes, 52 genes (including 6 genes whose transcription was upregulated in the ΔAmbldD mutant) have an AmBldD-binding site at an appropriate position (between −41 and +10) for being repressed by AmBldD. Therefore, we hypothesize that AmBldD is a global transcriptional repressor, just like BldD in Streptomyces. We did not perform a comparative transcriptome analysis between the wt and ΔAmbldD strains in this study because a huge difference between their transcriptomes was assumed due to the abnormal morphological differentiation and very low hyphal growth rate in solid and liquid cultures, respectively, of the ΔAmbldD mutant. However, further detailed analyses, including such a comparative transcriptome analysis and examinations of the effects of AmBldD-binding site mutations on the transcription of each target gene (especially for the genes that have AmBldD boxes unusually far upstream from the promoters), are required to understand the entire AmBldD regulon more precisely. We expect that new genes involved in the morphological differentiation of A. missouriensis will be found through further analyses of the AmBldD regulon.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
A. missouriensis 431T (NBRC 102363T) was obtained from the National Institute of Technology and Evaluation (NITE, Chiba, Japan). A. missouriensis was grown on YBNM (0.1% yeast extract [Difco], 0.2% meat extract [Kyokuto], 0.2% NZ amine [Wako], 1% maltose monohydrate [Wako], pH 7.0) or HAT (0.1% sucrose, 0.01% Casamino Acids [Difco], 0.05% K2HPO4, 0.2% humic acid, 1% trace element solution, pH 7.5) agar at 30°C for solid culture and in PYM liquid medium (0.5% Bacto peptone [Difco], 0.3% yeast extract [Difco], 0.1% MgSO4 · 7H2O, pH 7.0) at 30°C for liquid culture. The agar media contained 2% agar. The trace element solution contained 0.004% ZnCl2, 0.02% FeCl3 · 6H2O, 0.001% CuCl2 · 2H2O, 0.001% MnCl2 · 4H2O, 0.001% Na2B4O7 · 10H2O, and 0.001% (NH4)6Mo7O24 · 4H2O. HAT agar was used for sporangium formation. GYMC agar medium (0.4% glucose, 0.4% yeast extract [Difco], 1% malt extract [Difco], 0.2% CaCO3, pH 7.2) was used for transformation by conjugation with Escherichia coli ET12567 (pUZ8002). E. coli ET12567 (pUZ8002) was obtained from the John Innes Centre (Norwich, United Kingdom). E. coli JM109 and vectors pUC19 and pColdI were purchased from TaKaRa Biochemicals (Shiga, Japan). pBlueScript II SK(+) was purchased from Agilent Technologies (CA, USA). E. coli BL21(DE3) was purchased from Merck Millipore (Darmstadt, Germany). The media and growth conditions for E. coli were described by Maniatis et al. (26). Apramycin (50 μg/ml), spectinomycin (50 μg/ml), and ampicillin (50 μg/ml) were added when necessary.
Preparation of protein samples from zoospores and germinating cells for 2D-PAGE.
A. missouriensis was cultured on HAT agar at 30°C for 7 days for sporangium formation. To collect zoospores, 25 mM NH4HCO3 solution (10 ml) was poured on sporangia formed on the HAT agar, followed by an incubation at 30°C for 1 h. Next, the solution was collected and centrifuged at 14,000 × g for 5 min to harvest zoospores. To induce germination, the harvested zoospores were suspended in PYM liquid medium (3 ml) and shaken at 30°C for 8 h. Next, the medium was centrifuged at 14,000 × g for 5 min to harvest germinating cells. To prepare proteins, the zoospores or germinating cells were suspended in 300 μl of lysis buffer (50 mM Tris-HCl [pH 8.0], 2 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM dithiothreitol [DTT]) and disrupted by sonication. After removal of cell debris, the lysate was centrifuged at 20,630 × g at 4°C for 30 min. Then, 4.2 ml of extraction buffer (0.7 M sucrose, 0.5 M Tris-HCl [pH 8.0], 50 mM EDTA, 0.1 M KCl, 40 mM DTT) and 4.2 ml of TE (10 mM Tris, 1 mM EDTA)-phenol (pH 8.0) were added to the supernatant, and the mixture was shaken at 4°C for 30 min, followed by centrifugation at 20,630 × g at 4°C for 30 min. An equal volume of TE-phenol (pH 8.0) was added to the supernatant, and the mixture was shaken at 4°C for 30 min, followed by centrifugation at 20,630 × g at 4°C for 30 min. Then, 5 times the volume of 100 mM ammonium acetate in ice-cold methanol was added to the supernatant, and the mixture was kept at −20°C for 2 h. Next, the mixture was centrifuged at 5,000 × g at 4°C for 30 min, and the precipitate was washed twice with methanol and once with 80% ice-cold acetone, air-dried briefly on ice, and suspended in 600 μl of Tris buffer or urea buffer (8 M urea, 40 mg/ml CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 100 mM PMSF, 40 mM DTT). The proteins in this solution were purified using the 2-D Clean-Up kit (GE Healthcare, Buckinghamshire, United Kingdom), and the precipitate was suspended in 8 M urea solution.
2D-PAGE and protein identification.
Equal volumes of swelling buffer (8 M urea, 4% CHAPS, 65 mM DTT, 0.5% immobilized pH gradient [IPG] buffer [pH 4 to 7], 0.001% bromophenol blue [BPB]) were added to the protein solutions, and the mixtures were applied to Immobiline drystrips (pH 4 to 7, 13 cm; GE Healthcare, Buckinghamshire, United Kingdom). First-dimension gel electrophoresis was performed according to the manufacturer's instructions as follows: at 500 V for 2 h, at 700 V for 1 h, at 1,000 V for 1 h, at 2,000 V for 1 h, at 3,000 V for 1 h, and at 3,500 V overnight. The first-dimension gels were mildly soaked with equilibration buffer (0.5 M Tris-HCl [pH 6.8], 6 M urea, 10% glycerol, 1% sodium dodecyl sulfate [SDS], 2% BPB, 25 mg/ml DTT) and applied to second-dimension electrophoresis using 12.5% SDS-PAGE gels. Then, the gels were stained with Coomassie brilliant blue (CBB) and scanned using the GT-8700 imaging scanner (Epson, Nagano, Japan). The images were analyzed using PDQuest software (Bio-Rad, CA). Protein spots were detected automatically, followed by a manual edit. Spots were also matched between different gels automatically, followed by a manual edit. Protein spots whose intensities were at least 3-fold higher in germinating cells than in zoospores were identified by the methods described in Akanuma et al. (27). Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) peptide mass fingerprint data were compared with the A. missouriensis sequence database (DDBJ accession number AP012319) using the MASCOT search engine (Matrix Science, London, United Kingdom), and the proteins were identified using the probability-based MOWSE score algorithm. The search parameters were as follows: mass accuracy, 300 ppm; missed cleavage, none; fixed modification, carbamidomethylation of Cys residues; and variable modification, oxidation of Met residues. A MOWSE score of >52 guarantees identification of the protein with >95% probability. All proteins identified in this study gave MOWSE scores of >52.
Purification of the recombinant AmBldD-NHis protein.
A 500-bp DNA fragment containing the AmbldD coding sequence was amplified by PCR using primers pColdI-BldD.f and pColdI-BldD.r. This fragment was digested with EcoRI and XbaI and cloned between the EcoRI and XbaI sites of pColdI, generating pColdI-AmBldD-NHis. The plasmid was introduced into E. coli BLR(DE3). The transformant was cultivated in LB (200 ml) at 37°C for 2.5 h and at 15°C for 30 min. Then, isopropyl β-d-1-thiogalactopyranoside was added to the culture with a final concentration of 1 mM and the culture was cultivated at 15°C for another 24 h. Cells were collected by centrifugation at 3,000 × g for 10 min and suspended in 5 ml of lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole), followed by disruption by sonication. After removal of cell debris, the cell extract was centrifuged at 12,000 × g for 30 min and the His-tagged AmBldD protein was purified using Ni-nitrilotriacetic acid (NTA) Superflow resin (Qiagen, Tokyo, Japan) according to the manufacturer's instructions. The AmBldD-NHis protein was eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, 20% glycerol). The quality of the purified protein was assessed by SDS-PAGE using a 12.5% acrylamide gel.
Preparation of cells for Western blot and RT-PCR analyses.
A. missouriensis vegetative hyphae were prepared by cultivation in PYM liquid medium at 30°C for 48 h. Mixtures of substrate hyphae and sporangia were harvested using a spatula from colonies grown on HAT agar at 30°C for 3 or 6 days. To prepare zoospores, 25 mM NH4HCO3 solution (10 ml) was poured on sporangia formed on the HAT agar to induce sporangium dehiscence. After incubation at 30°C for 1 h, released zoospores were collected by centrifugation at 21,880 × g for 10 min. To prepare germinating cells, zoospores were suspended in PYM liquid medium (3 ml) and incubated at 30°C for 4 h. Then, the germinating cells were collected by centrifugation at 21,880 × g for 10 min.
Western blotting.
Cells were disrupted by rubbing with a mortar and pestle, and the cell lysate was mixed with lysis buffer (50 mM Tris-HCl, 150 mM NaCl, pH 8.0). After the debris was eliminated by centrifugation, the protein concentrations of the supernatants were determined using Bradford reagent (Bio-Rad). Then, 2 μg of each protein sample was separated by SDS-PAGE using two replicative 12.5% acrylamide gels. One of the gels was stained with CBB, and another was used for protein transfer to the Immobilon-P polyvinylidene difluoride (PVDF) membrane (Millipore). Anti-AmBldD polyclonal antibody produced in rabbits using purified AmBldD-NHis protein as the antigen (Eurofins Genomics, Tokyo, Japan) was used to probe AmBldD. Horseradish peroxidase-coupled anti-rabbit IgG (SeraCare, Milford, MA) was used as the secondary antibody and detected by chemiluminescence using Clarity Western enhanced chemiluminescence (ECL) substrate (Bio-Rad) and the ImageQuant LAS 4000 system (GE Healthcare).
RNA extraction, semi-qRT-PCR, and qRT-PCR.
Cells were disrupted by rubbing with a mortar and pestle, and the lysate was mixed with the lysis/binding solution in the RNAqueous total RNA isolation kit (Ambion, TX). After the debris was removed by centrifugation, total RNAs were extracted according to the manufacturer's instructions. The total RNAs were treated with DNase I to eliminate contaminating genomic DNA and purified by phenol-chloroform extraction and ethanol precipitation. Then, each RNA sample (0.5 μg) was used for reverse transcription (RT) reactions using the ThermoScript RT-PCR system for first-strand cDNA synthesis (Life Technologies) according to the manufacturer's instructions. Following the treatment with RNase H, the synthesized cDNA libraries were used as templates for PCR. For semiquantitative RT-PCR (semi-qRT-PCR), a part of the 16S rRNA gene was amplified using primers sRT.16SrRNA.f and sRT.16SrRNA.r under the following conditions: 10 s at 98°C, followed by 20 cycles of 30 s at 55°C and 1 min at 72°C. For a part of the AmbldD gene, primers sRT.bldD.f and sRT.bldD.r were used, and the PCR cycling conditions were as follows: 10 s at 98°C, followed by 30 cycles of 30 s at 60°C and 30 s at 72°C. All qRT-PCRs were performed with the TaKaRa SYBR premix Ex Taq II reaction mixture (TaKaRa Biochemicals) using the SmartCycler II real-time PCR system (Cepheid, CA) under the following conditions: 5 min at 95°C, followed by 40 cycles of 5 s at 95°C and 20 s at 60°C. The 16S rRNA gene was used as an internal standard. Primer sets are shown in Table S1 in the supplemental material. All reactions were performed in triplicate using RNA samples extracted from three independent cultures, and the data were normalized using the average for the internal standard.
Construction of the AmbldD deletion mutant.
For construction of a ΔAmbldD mutant, the upstream and downstream regions of AmbldD were amplified by PCR using primers ΔbldD.up.f plus ΔbldD.up.r, and ΔbldD.down.f plus ΔbldD.down.r, respectively. The amplified fragments were digested with EcoRI and NheI (for the upstream region) and NheI and HindIII (for the downstream region), and cloned together between the EcoRI and HindIII sites of pK19mobsacB (28), whose kanamycin resistance gene had been replaced with the apramycin resistance gene aac(3)IV by standard DNA manipulation, generating pK19mobsacB-ΔAmbldD. Plasmid pK19mobsacB-ΔAmbldD was introduced into A. missouriensis by conjugation as described previously (29). Apramycin-resistant colonies resulting from a single-crossover recombination were isolated. One of them was cultivated in PYM liquid medium at 30°C for 36 h. Then, mycelia were suspended in 0.75% NaCl solution and spread onto Czapek dox agar medium (BD, NJ) containing 10% sucrose. After incubation at 30°C for 5 to 7 days, the sucrose-resistant colonies were inoculated onto YBNM agar with or without apramycin to confirm that they were sensitive to apramycin. The apramycin-sensitive and sucrose-resistant colonies resulting from the second crossover recombination event were isolated as candidates for the AmbldD disruptant. The disruption of AmbldD was confirmed by Southern hybridization with the probe amplified by PCR using primers bldD.southern.f and bldD.southern.r against BamHI-digested chromosomal DNA (Fig. S1 in the supplemental material).
Measurement of the A. missouriensis growth.
The wild-type (wt) and ΔAmbldD strains were grown on YBNM agar at 30°C for 3 days, and 0.01 g of the harvested cells of each was inoculated into five 500-ml shaking (Sakaguchi) flasks, each containing 150 ml PYM liquid medium for both strains. The flasks were reciprocally shaken at 120 rpm at 30°C. Every 6 h, a portion of culture medium (1 ml) was harvested from each flask and the wet cell weight was measured.
SEM.
The wt and ΔAmbldD strains were grown at 30°C on YBNM agar for 4 days or on HAT agar for 7 days. Scanning electron microscopy (SEM) was performed with the S-4800 scanning electron microscope (Hitachi, Tokyo, Japan) as described previously (17).
5′-RACE.
The transcriptional start point of AmbldD was determined with the 5′-full RACE (rapid amplification of cDNA ends) core set (TaKaRa Biochemicals). Using the total RNA extracted from the wt strain as a template, cDNA was synthesized with the 5′-phosphorylated primer bldD-5′RACE-RT. Template RNA was degraded with RNase H, and the cDNA was ligated into circularization or concatemer formation according to the manufacturer's instructions. Next, the region containing the transcriptional start point of AmbldD was amplified by a nested PCR using primer sets bldD-5′RACE-A1 plus bldD-5′RACE-S1 and bldD-5′RACE-A2 plus bldD-5′RACE-S2. The PCR product was digested with EcoRI and HindIII and cloned into pUC19. The insert DNA from four different clones was sequenced, and the identical cDNA end was regarded as a transcriptional start point. Similarly, the transcription start points of AMIS6880 (whiD), AMIS65300 (ssgB), and AMIS78290 (wblA) were determined using each RT primer and primer sets for nested PCR (see Table S1 in the supplemental material). The PCR products were cloned into pJET 1.2 and sequenced.
EMSAs.
DNA fragments (94 to 210 bp) including AmBldD-binding regions were prepared by PCR for electrophoretic mobility shift assays (EMSAs). The primer sets used were primers EMSA.bldD.f (positions −197 to −177 with respect to the start codon of AmBldD) plus EMSA.bldD.r (positions +8 to +27) for the AmbldD probe, EMSA.A.f plus EMSA.A.r for probe A, EMSA.B.f plus EMSA.B.r for probe B, and EMSA.C.f plus EMSA.C.r for probe C. The PCR products were 32P labeled at the 5′ ends using [γ-32P]ATP (PerkinElmer, Waltham, MA) and T4 polynucleotide kinase (TaKaRa Biochemicals). Recombinant AmBldD-NHis protein was incubated with the labeled probes at 25°C for 15 min in a buffer [25 mM NaH2PO4, 7.5% glycerol, 100 ng/μl bovine serum albumin, 25 ng/μl poly(dI-dC)], and the mixtures were applied to 6% PAGE at 34 mA for 2 h. When necessary, c-di-GMP (0.25 to 2.5 μM) was added to the reaction mixtures.
DNase I footprinting.
The probe for the sense strand was prepared by PCR using primers sense.f (positions −146 to −127 with respect to the start codon of AmbldD), 32P labeled at the 5′end, and sense.r (positions −17 to +3). The probe for the antisense strand was prepared with primers anti-sense.f (positions −146 to −127), 32P labeled at the 5′ end, and anti-sense.r (positions −17 to +3). The 50-μl reaction mixtures contained 32P-labeled DNA probe, 0.58 to 4.72 μg of AmBldD-NHis, 25 mM HEPES-KOH (pH 7.9), 0.5 mM EDTA-NaOH (pH 8.0), 50 mM KCl, and 10% glycerol. After incubation at 25°C for 30 min, DNase I was added at a final concentration of 20 μg/ml, and the mixtures were further incubated for 1 min. The DNase I digestion was stopped by adding 200 μl of stop solution (100 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1% sodium N-lauroylsarcosinate, 10 mM EDTA-NaOH [pH 8.0], 25 μg/ml of salmon sperm DNA) and 300 μl of phenol-chloroform (1:1). After ethanol precipitation, the DNA probe was washed with 80% ethanol, dissolved in 6 μl of a formamide-containing dye mixture, and separated using 6% PAGE. The exact region protected was determined with the Maxam-Gilbert sequencing ladder (A+G reactions) (30).
In vivo cross-linking and ChIP.
In vivo cross-linking and chromatin immunoprecipitation (ChIP) were performed essentially by the methods described in Higo et al. (31). For in vivo cross-linking of AmBldD and DNA in cells grown in liquid culture, the wt strain was grown in PYM liquid medium (100 ml) at 30°C for 40 h. The culture was centrifuged at 7,600 × g for 5 min to harvest the mycelium. The mycelium was treated with formaldehyde (1% final concentration) for 20 min at room temperature. Then, formaldehyde was quenched by adding glycine to a final concentration of 250 mM. Finally, the cells were washed with Tris-buffered saline (pH 7.5) three times and stored at −80°C until use. Cross-linked cells in 1 ml of ChIP buffer (50 mM HEPES-KOH, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 0.1% sodium deoxycholate, pH 7.5) were sonicated on ice to give an average DNA fragment size of 300 to 600 bp. After eliminating cell debris, the solution was centrifuged at 12,000 × g for 30 min and the supernatant was collected and diluted with ChIP buffer to 3 to 5 mg protein/ml. At the same time, 50 μl of Dynabeads M-280 sheep anti-rabbit IgG (Life Technologies, MA), which was washed twice with phosphate-buffered saline containing 5 mg/ml bovine serum albumin (PBS-BSA), was mixed with anti-AmBldD polyclonal antibody at 4°C for 4 h, washed twice with 1 ml of PBS-BSA, and resuspended with PBS-BSA. The anti-AmBldD antibody-conjugated beads were added to 1 ml of the diluted supernatant, followed by an incubation at 4°C for 18 h with gentle rotation. The beads were then washed once with ChIP buffer, once with ChIP high-salt buffer (ChIP buffer containing 300 mM NaCl), twice with ChIP wash buffer (100 mM Tris-HCl, 250 mM LiCl, 1 mM EDTA, and 0.5% sodium deoxycholate, pH 8.0), and twice with TE (10 mM Tris and 1 mM EDTA, pH 8.0). Proteins bound to the beads were eluted with 100 μl of the elution buffer (50 mM Tris-HCl, 10 mM EDTA, and 1% SDS, pH 8.0). After the cross-linking between AmBldD and DNA was destroyed by heating at 65°C overnight, 300 μl of TE was added to the reverse-cross-linked sample. The sample was then treated with RNase A (a final concentration of 0.4 mg/ml) at 37°C for 2 h and with proteinase K (a final concentration of 0.2 mg/ml) at 55°C for 2 h. Finally, DNA fragments were purified by ethanol precipitation and by using the QIAquick PCR purification kit (Qiagen) (ChIP DNA). DNA was also purified from the diluted supernatant before the addition of the Dynabeads by the same method (input DNA).
ChIP-Seq and in silico analysis.
DNA libraries were prepared from input and ChIP DNAs (10 ng each) using the NEBNext ChIP-Seq library prep master mix set for Illumina (NEB, Hertfordshire, United Kingdom) according to the manufacturer's instructions. The libraries were pooled at equimolar concentrations and deep sequenced using the Illumina MiSeq sequencer to generate 150-bp single-end reads. In silico analyses were performed with CLC Genomics Workbench version 8.5 (CLC Bio, Aarhus, Denmark). Low-quality bases (10 bp at the 5′ end and 3 bp at the 3′ end) were trimmed, and low-quality reads were discarded based on the following parameters: (i) removal of low-quality sequence, limit of 0.05; (ii) removal of 5′-terminal nucleotides, 10; (iii) removal of 3′-terminal nucleotides, 3; and (iv) removal of shorter reads than minimum number of nucleotides, 130. Undiscarded and trimmed reads were mapped to the reference genome sequence of A. missouriensis using the default parameters of the software. The mapping data of the ChIP DNA library were compared to those of the input DNA library with 0.0005 and 0.05 of the maximum P value used for peak calling to identify peak enrichments. The statistically significant peaks were visualized using the software, and 163 (P < 0.0005) and 437 (P < 0.05) peaks were produced. MEME (version 4.10.0) (32) was used with the default settings, except that “look for palindrome only = yes” was used to define the AmBldD-binding sequence. As a result, 163 and 346 sequences similar to the consensus AmBldD-binding sequence were found in peak regions selected using 0.0005 and 0.05 of the maximum P value, respectively. We predicted the transcriptional start points near the probable AmBldD-binding sites and found 161 sites whose center residues were located in the possible regulatory region of an ORF or two divergent ORFs (from nucleotides −200 to +50, referring to the transcriptional start point predicted by RNA-Seq analyses as +1). We identified these 207 ORFs as AmBldD target genes. Based on the NITE annotation database (http://www.bio.nite.go.jp/dogan/category_search/view/AM2), the AmBldD target genes were classified into 19 functions.
Accession number(s).
Sequencing data were deposited to the DDBJ Sequence Read Archive (http://trace.ddbj.nig.ac.jp/dra/index.html) under the accession numbers SAMD00056702 and SAMD00056703.
Supplementary Material
ACKNOWLEDGMENTS
We thank Genki Akanuma for his technical help in 2D-PAGE and protein identification.
This research was supported in part by a funding program for next-generation world-leading researchers from the Bureau of Science, Technology, and Innovation Policy, Cabinet Office, Government of Japan, and a Grant-in-Aid for Scientific Research (A) and a Grant-in-Aid for Young Scientists (B) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00840-16.
For a commentary on this article, see https://doi.org/10.1128/JB.00070-17.
REFERENCES
- 1.Couch JN. 1963. Some new genera and species of the Actinoplanaceae. J Elisha Mitchell Sci Soc Chapel Hill N C 79:53–70. [Google Scholar]
- 2.Uchida K, Jang MS, Ohnishi Y, Horinouchi S, Hayakawa M, Fujita N, Aizawa SI. 2011. Characterization of Actinoplanes missouriensis spore flagella. Appl Environ Microbiol 77:2559–2562. doi: 10.1128/AEM.02061-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higgins ML. 1967. Release of sporangiospores by a strain of Actinoplanes. J Bacteriol 94:495–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayakawa M, Tamura T, Nonomura H. 1991. Selective isolation of Actinoplanes and Dactylosporangium from soil by using γ-collidine as the chemoattractant. J Ferment Bioeng 72:426–432. doi: 10.1016/0922-338X(91)90049-M. [DOI] [Google Scholar]
- 5.Arora DK. 1986. Chemotaxis of Actinoplanes missouriensis zoospores to fungal conidia, chlamydospores and sclerotia. Microbiology 132:1657–1663. doi: 10.1099/00221287-132-6-1657. [DOI] [Google Scholar]
- 6.Yamamura H, Ohnishi Y, Ishikawa J, Ichikawa N, Ikeda H, Sekine M, Harada T, Horinouchi S, Otoguro M, Tamura T, Suzuki K, Hoshino Y, Arisawa A, Nakagawa Y, Fujita N, Hayakawa M. 2012. Complete genome sequence of the motile actinomycete Actinoplanes missouriensis 431(T) (= NBRC 102363(T)). Stand Genomic Sci 7:294–303. doi: 10.4056/sigs.3226659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandra G, Chater KF. 2014. Developmental biology of Streptomyces from the perspective of 100 actinobacterial genome sequences. FEMS Microbiol Rev 38:345–379. doi: 10.1111/1574-6976.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bush MJ, Tschowri N, Schlimpert S, Flärdh K, Buttner MJ. 2015. c-di-GMP signalling and the regulation of developmental transitions in streptomycetes. Nat Rev Microbiol 13:749–760. doi: 10.1038/nrmicro3546. [DOI] [PubMed] [Google Scholar]
- 9.Elliot MA, Bibb MJ, Buttner MJ, Leskiw BK. 2001. BldD is a direct regulator of key developmental genes in Streptomyces coelicolor A3(2). Mol Microbiol 40:257–269. doi: 10.1046/j.1365-2958.2001.02387.x. [DOI] [PubMed] [Google Scholar]
- 10.Tschowri N, Schumacher MA, Schlimpert S, Chinnam NB, Findlay KC, Brennan RG, Buttner MJ. 2014. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 158:1136–1147. doi: 10.1016/j.cell.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.den Hengst CD, Tran NT, Bibb MJ, Chandra G, Leskiw BK, Buttner MJ. 2010. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol Microbiol 78:361–379. doi: 10.1111/j.1365-2958.2010.07338.x. [DOI] [PubMed] [Google Scholar]
- 12.Noens EE, Mersinias V, Traag BA, Smith CP, Koerten HK, van Wezel GP. 2005. SsgA-like proteins determine the fate of peptidoglycan during sporulation of Streptomyces coelicolor. Mol Microbiol 58:929–944. doi: 10.1111/j.1365-2958.2005.04883.x. [DOI] [PubMed] [Google Scholar]
- 13.Willemse J, Borst JW, de Waal E, Bisseling T, van Wezel GP. 2011. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev 25:89–99. doi: 10.1101/gad.600211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ausmees N, Wahlstedt H, Bagchi S, Elliot MA, Buttner MJ, Flärdh K. 2007. SmeA, a small membrane protein with multiple functions in Streptomyces sporulation including targeting of a SpoIIIE/FtsK-like protein to cell division septa. Mol Microbiol 65:1458–1473. doi: 10.1111/j.1365-2958.2007.05877.x. [DOI] [PubMed] [Google Scholar]
- 15.Chng C, Lum AM, Vroom JA, Kao CM. 2008. A key developmental regulator controls the synthesis of the antibiotic erythromycin in Saccharopolyspora erythraea. Proc Natl Acad Sci U S A 105:11346–11351. doi: 10.1073/pnas.0803622105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliot M, Damji F, Passantino R, Chater K, Leskiw B. 1998. The bldD gene of Streptomyces coelicolor A3(2): a regulatory gene involved in morphogenesis and antibiotic production. J Bacteriol 180:1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamazaki H, Ohnishi Y, Horinouchi S. 2000. An A-factor-dependent extracytoplasmic function sigma factor (sigmaAdsA) that is essential for morphological development in Streptomyces griseus. J Bacteriol 182:4596–4605. doi: 10.1128/JB.182.16.4596-4605.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwientek P, Szczepanowski R, Rückert C, Kalinowski J, Klein A, Selber K, Wehmeier UF, Stoye J, Pühler A. 2012. The complete genome sequence of the acarbose producer Actinoplanes sp. SE50/110. BMC Genomics 13:112. doi: 10.1186/1471-2164-13-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliot M, Leskiw B. 1999. The BldD protein from Streptomyces coelicolor is a DNA-binding protein. J Bacteriol 181:6832–6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu H, Mao Y, Chen M, Pan H, Huang X, Ren M, Wu H, Li J, Xu Z, Yuan H, Geng M, Weaver DT, Zhang L, Zhang B. 2015. Capturing the target genes of BldD in Saccharopolyspora erythraea using improved genomic SELEX method. Appl Microbiol Biotechnol 99:2683–2692. doi: 10.1007/s00253-014-6255-9. [DOI] [PubMed] [Google Scholar]
- 21.de Crécy-Lagard V, Servant-Moisson P, Viala J, Grandvalet C, Mazodier P. 1999. Alternation of the synthesis of the Clp ATP-dependent protease affects morphological and physiological differentiation in Streptomyces. Mol Microbiol 32:505–517. doi: 10.1046/j.1365-2958.1999.01364.x. [DOI] [PubMed] [Google Scholar]
- 22.Lee CJ, Won HS, Kim JM, Lee BJ, Kang SO. 2007. Molecular domain organization of BldD, an essential transcriptional regulator for developmental process of Streptomyces coelicolor A3(2). Proteins 68:344–352. doi: 10.1002/prot.21338. [DOI] [PubMed] [Google Scholar]
- 23.Romero-Rodríguez A, Robledo-Casados I, Sánchez S. 2015. An overview on transcriptional regulators in Streptomyces. Biochim Biophys Acta 1849:1017–1039. doi: 10.1016/j.bbagrm.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 24.Aldridge M, Facey P, Francis L, Bayliss S, Del Sol R, Dyson P. 2013. A novel bifunctional histone protein in Streptomyces: a candidate for structural coupling between DNA conformation and transcription during development and stress? Nucleic Acid Res 41:4813–4824. doi: 10.1093/nar/gkt180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fowler-Goldsworthy K, Gust B, Mouz S, Chandra G, Findlay KC, Charter KF. 2011. The actinobacteria-specific gene wblA controls major developmental transitions in Streptomyces coelicolor A3(2). Microbiology 157:1312–1328. doi: 10.1099/mic.0.047555-0. [DOI] [PubMed] [Google Scholar]
- 26.Maniatis T, Fritsch EF, Sambrook J. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- 27.Akanuma G, Hara H, Ohnishi Y, Horinouchi S. 2009. Dynamic changes in the extracellular proteome caused by absence of a pleiotropic regulator AdpA in Streptomyces griseus. Mol Microbiol 73:898–912. doi: 10.1111/j.1365-2958.2009.06814.x. [DOI] [PubMed] [Google Scholar]
- 28.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 29.Jang MS, Mouri Y, Uchida K, Aizawa SI, Hayakawa M, Fujita N, Tezuka T, Ohnishi Y. 2016. Genetic and transcriptional analysis of the flagellar gene cluster in Actinoplanes missouriensis. J Bacteriol 198:2219–2227. doi: 10.1128/JB.00306-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maxam AM, Gilbert W. 1980. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol 65:499–560. doi: 10.1016/S0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 31.Higo A, Hara H, Horinouchi S, Ohnishi Y. 2012. Genome-wide distribution of AdpA, a global regulator for secondary metabolism and morphological differentiation in Streptomyces, revealed the extent and complexity of the AdpA regulatory network. DNA Res 19:259–273. doi: 10.1093/dnares/dss010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey TL, Elkan C. 1995. The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol 3:21–29. [PubMed] [Google Scholar]
- 33.Rückert C, Szczepanowski R, Albersmeier A, Goesmann A, Fischer N, Steinkämper A, Pühler A, Biener R, Schwartz D, Kalinowski J. 2014. Complete genome sequence of the actinobacterium Actinoplanes friuliensis HAG 010964, producer of the lipopeptide antibiotic friulimycin. J Biotechnol 178:41–42. doi: 10.1016/j.jbiotec.2014.03.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.