ABSTRACT
Although mycoplasmas have small genomes, many of them, including the HIV-associated opportunist Mycoplasma penetrans, construct a polar attachment organelle (AO) that is used for both adherence to host cells and gliding motility. However, the irregular phylogenetic distribution of similar structures within the mycoplasmas, as well as compositional and ultrastructural differences among these AOs, suggests that AOs have arisen several times through convergent evolution. We investigated the ultrastructure and protein composition of the cytoskeleton-like material of the M. penetrans AO with several forms of microscopy and biochemical analysis, to determine whether the M. penetrans AO was constructed at the molecular level on principles similar to those of other mycoplasmas, such as Mycoplasma pneumoniae and Mycoplasma mobile. We found that the M. penetrans AO interior was generally dissimilar from that of other mycoplasmas, in that it exhibited considerable heterogeneity in size and shape, suggesting a gel-like nature. In contrast, several of the 12 potential protein components identified by mass spectrometry of M. penetrans detergent-insoluble proteins shared certain distinctive biochemical characteristics with M. pneumoniae AO proteins, although not with M. mobile proteins. We conclude that convergence between M. penetrans and M. pneumoniae AOs extends to the molecular level, leading to the possibility that the less organized material in both M. pneumoniae and M. penetrans is the substance principally responsible for the organization and function of the AO.
IMPORTANCE Mycoplasma penetrans is a bacterium that infects HIV-positive patients and may contribute to the progression of AIDS. It attaches to host cells through a structure called an AO, but it is not clear how it builds this structure. Our research is significant not only because it identifies the novel protein components that make up the material within the AO that give it its structure but also because we find that the M. penetrans AO is organized unlike AOs from other mycoplasmas, suggesting that similar structures have evolved multiple times. From this work, we derive some basic principles by which mycoplasmas, and potentially all organisms, build structures at the subcellular level.
KEYWORDS: Mycoplasma, cytoskeleton, electron microscopy, evolution, fractionation, mass spectrometry, transcriptomics
INTRODUCTION
Mycoplasmas, which are genomically reduced, cell wall-lacking bacteria that live parasitically or commensally in nature but can be cultured axenically, occupy a unique space within the field of bacterial cell polarization that makes them potentially informative concerning mechanisms by which polarization is achieved, as well as the evolutionary origins of such mechanisms. Some mycoplasmas have a differentiated prosthecal tip structure, known as an attachment organelle (AO), at one pole (reviewed in reference 1). The AO houses adhesin proteins for interactions with host cells and surfaces, as well as machinery that carries out unidirectional gliding motility, and appears to be physically associated with the chromosome in some cases (reviewed in reference 2). The interior of the AO contains a complex of structural proteins that serves as a cytoskeletal scaffold for AO assembly (reviewed in reference 2). In the sense that the AO is constructed at a cell pole and is involved in functions associated with polarity, this structure parallels other polar structures of bacteria, such as the Caulobacter crescentus stalk (2).
The majority of information regarding the components, assembly, and substructures of mycoplasma AOs comes from studies of two different species, Mycoplasma pneumoniae and Mycoplasma mobile. These two species are members of two distantly related phylogenetic clusters within the genus (3). Characterization of the structures and proteins that make up the interior of the AO of each of these species has shown them to be very different both ultrastructurally and compositionally, with no AO protein homologs shared between these organisms (4; reviewed in reference 1), suggesting that, like the C. crescentus stalk, these structures have evolved independently and recently, compared with other members of their respective phyla.
Mycoplasma penetrans is another species that has a polar AO, representing a third mycoplasma phylogenetic cluster (5–7). M. penetrans is best known for its association with AIDS patients, in whom it is commonly found in the urogenital tract, but it has also been isolated from an HIV-negative patient with antiphospholipid syndrome (5, 6, 8). The major focus of studies of M. penetrans has been its putative role as an AIDS cofactor (9–12). We showed previously that the M. penetrans AO contains material distinct in structure from that of M. pneumoniae and M. mobile (7). Furthermore, M. penetrans lacks any homologs of the AO structural proteins of M. pneumoniae or M. mobile (4, 13, 14). Special difficulty in working with this organism stems from its genetic intractability, leaving biochemical and cell biological routes as the best options for its study.
The ability to establish cellular polarization and to localize specific biomolecules to non-uniformly distributed subcellular positions is critical for many bacteria. Many fundamental processes, such as DNA segregation, cell division, and placement of specialized structures, rely on morphological polarity as a cue for appropriate spatial organization. The cytoskeleton, which is composed of a network of polymeric proteins, plays an integral role in maintaining the morphological and functional integrity of bacterial cells by establishing polarity (reviewed in reference 15). In addition to the widely phylogenetically distributed MreB and FtsZ, which form dynamic filaments that contribute to the nearly universal bacterial processes of peptidoglycan synthesis and cell division (reviewed in reference 16), there is increasing evidence of a diverse array of other cytoskeletal elements, with narrow phylogenetic distribution, that facilitate more recently evolved functions (reviewed in reference 17). In contrast to the highly dynamic MreB and FtsZ, many of these proteins form static structures. The α-helical coiled-coil-rich, intermediate filament-like protein crescentin from C. crescentus is associated with the establishment of cell shape (18, 19). Many bacterial scaffold proteins contain an abundance of coiled-coil domains, which are important for oligomerization and close protein-protein interactions and are referred to as coiled-coil-rich proteins (CCRPs) (reviewed in reference 20). CCRPs serve an array of functions, such as maintaining cell morphology and enabling cell motility, often by participating in polar localization of proteins (reviewed in reference 19). The widely distributed bactofilins are characterized by the conserved DUF583 domain and form stable nucleotide-independent filaments that serve as spatial landmarks (21, 22). Other bacterial cytoskeletal proteins include DivIVA and PopZ, which assemble into 2- or 3-dimensional arrays at extant or nascent cell poles and interact with proteins involved in pole-associated processes (reviewed in reference 19). It is essential to understand processes that rely on bacterial cell polarization, and this understanding can be achieved by establishing an inventory of cytoskeletal proteins across bacterial phylogeny and determining their characteristics and how these properties enable them to carry out their functions.
We sought to identify the proteins of the interior of the M. penetrans AO and to examine their properties in relation to other proteins of mycoplasma AOs. Because mycoplasma AOs seem to have evolved in a convergent manner, we hypothesized that the proteins involved in the structure and assembly of the M. penetrans AO would be distinct from those of other mycoplasmas, and we predicted that their composition would reveal important information about the properties of the AO. To test these ideas, we investigated the protein content of the detergent-insoluble material of M. penetrans, which constitutes the AO interior, and characterized the structure of this material. We found that, although many of the proteins have some properties resembling those of the corresponding proteins of M. pneumoniae, the M. penetrans AO is quite distinct from its counterparts in other mycoplasmas, consistent with convergent evolution of AOs and of the mechanisms of polarization of bacterial cells. We also examined changes in gene expression of M. penetrans upon incubation with human cervical epithelioid HeLa cells, to test whether AO and other genes were subject to transcriptional regulation upon encountering the host, and we found that they were not.
(This work was performed in partial fulfillment of the doctoral dissertation requirements of S. L. Distelhorst.)
RESULTS
Internal structure of M. penetrans AO.
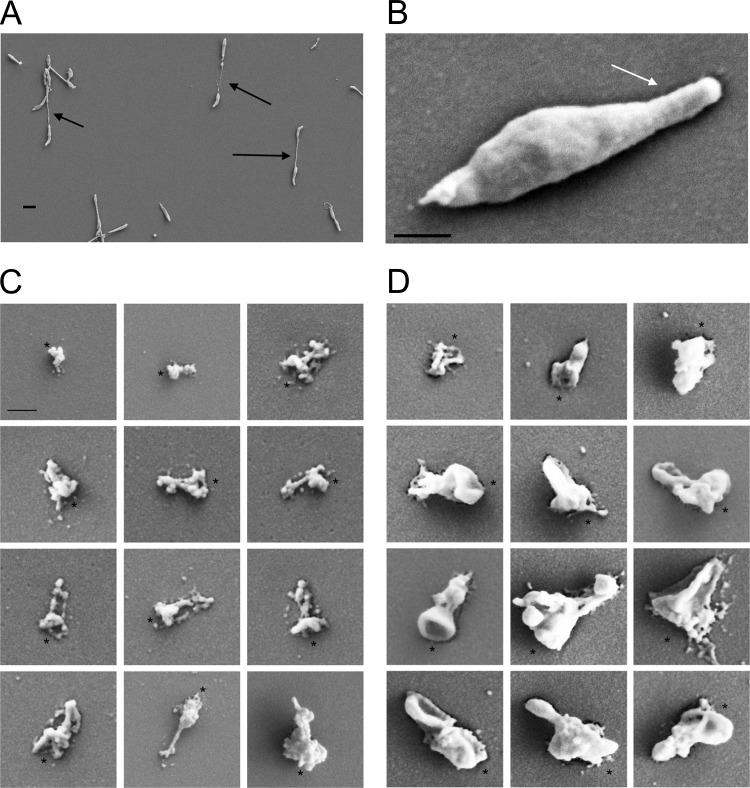
As described previously, M. penetrans cells are elongated (Fig. 1B), with a polar tip structure that functions as an AO. The AO confers attachment and gliding motility, which contributes to cell division; dividing cells often take the form of two cell bodies linked by a membranous connecting filament (7) (Fig. 1A). M. penetrans cells attach to host cells by means of a pole, whose interior is distinctly differentiated from the cytoplasm of the rest of the cell despite being adjacent to it, as established previously by transmission electron microscopy (TEM) (5, 6). Treatment of M. penetrans cells attached to plastic with the nonionic detergent Triton X-100 results in solubilization of a large amount of cellular material, leaving behind discrete detergent-insoluble objects with a range of dimensions, as visualized by scanning electron microscopy (SEM) (Fig. 1C) (7). Unlike the AO structures obtained similarly from species of the M. pneumoniae cluster (23), treatment with DNase had no effect on the appearance of these objects (data not shown), indicating that they did not contain significant amounts of DNA.
FIG 1.
SEM images of M. penetrans whole cells and detergent-insoluble structures. (A) Field of M. penetrans cells. Arrows, connecting filaments. Scale bar, 1 μm. (B) Individual M. penetrans cell. Arrow, AO. Scale bar, 200 nm. (C and D) Detergent-insoluble structures from M. penetrans cells extracted with Triton X-100 (C) or Tween 20 (D). Asterisks, wide ends of structures. Scale bar, 200 nm.
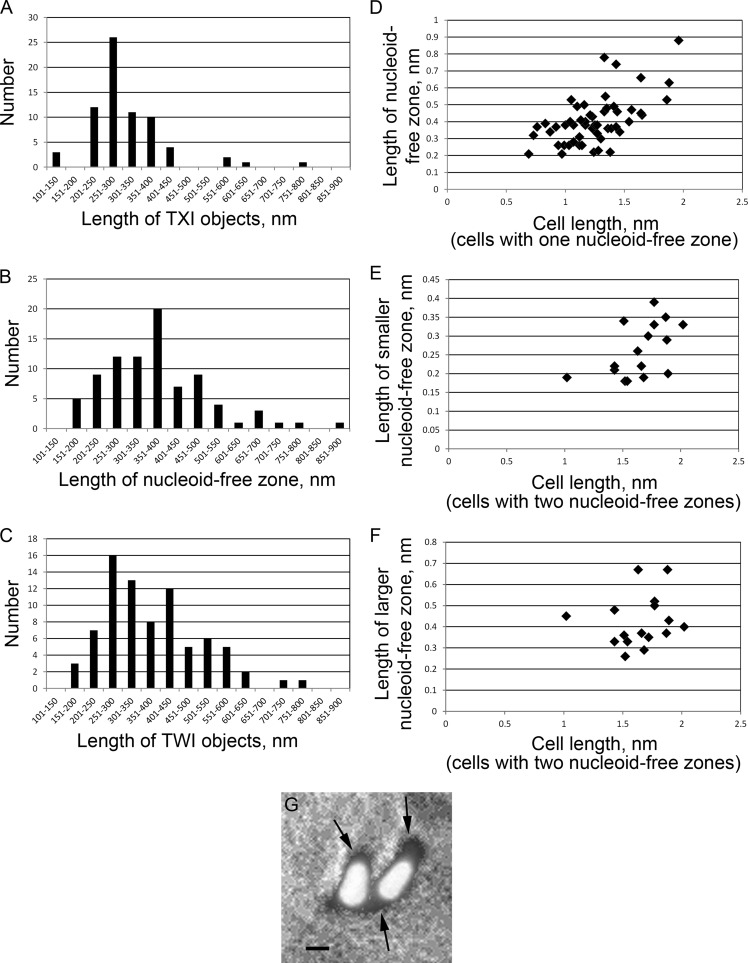
Close examination of these Triton X-100-insoluble (TXI) structures revealed that, despite considerable heterogeneity in size and shape, they generally consisted of a wider, irregular, ball-like object from which emanated one or more rod-like filaments, which were often periodically punctuated with other material along the filaments. The structures had an average length of 320 ± 100 nm (n = 70) (Fig. 2A). This range of lengths was significantly ∼15% smaller than that of the polar nucleoid-free zones of M. penetrans cells (n = 84; P < 0.001) (Fig. 2), as revealed by overlaying phase-contrast images and images of the DNA stained with 4′,6-diamidino-2-phenylindole (DAPI) (Fig. 2G) (7). The mean length of the nucleoid-free zones was 380 ± 140 nm. In cells with single nucleoid-free zones, there was a positive correlation between the length of the cell and the length of the nucleoid-free zone (n = 52; P < 0.0001) (Fig. 2D), suggesting that growth of this structure is coupled to cell growth. In cells with two such zones at each pole, the length of the smaller one was correlated with cell length (n = 16; P = 0.04) (Fig. 2E), whereas that of the larger one was not (n = 16; P = 0.52) (Fig. 2F), consistent with the larger one being a preexisting structure whose net growth had slowed or stopped.
FIG 2.
AO-associated objects and their lengths. (A) Distribution of the lengths of TXI objects observed by SEM. (B) Distribution of the lengths of nucleoid-free zones observed by light and fluorescence microscopy. (C) Distribution of the lengths of TWI objects observed by SEM. (D) Correspondence of the lengths of nucleoid-free zones to the fractions of the cell length occupied by the nucleoid-free zones in 52 cells with a single nucleoid-free zone. (E and F) Correspondence of the lengths of nucleoid-free zones to the fractions of the cell length occupied by the nucleoid-free zones in 16 cells with two nucleoid-free zones, a shorter one (E) and a longer one (F). (G) Example of nucleoid-free zones. Black, cell outline viewed by phase-contrast microscopy; white, nucleoid stained with DAPI. Arrows, nucleoid-free zones, measured along the long axis of the cell for panel B. Scale bar, 500 nm.
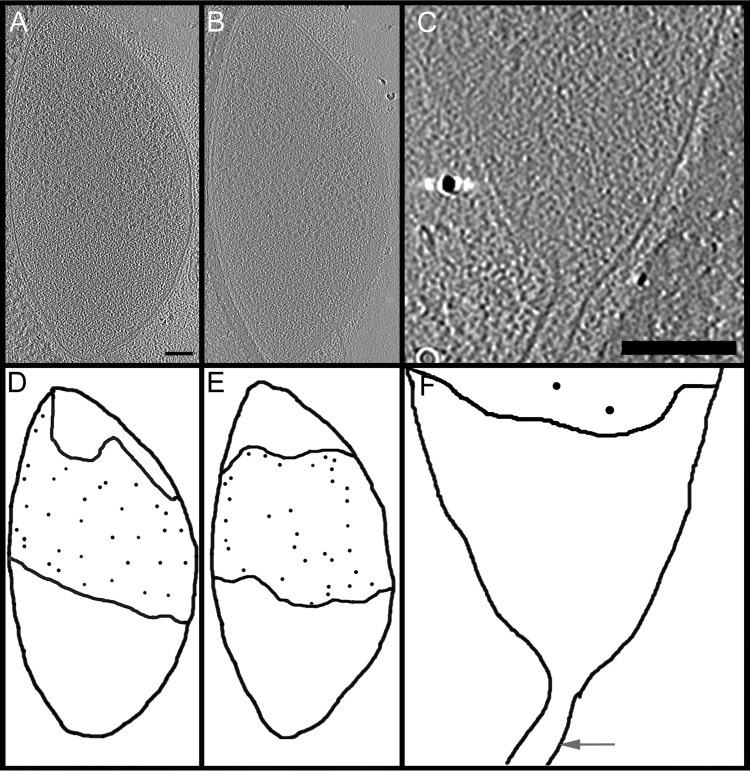
To determine whether another nonionic detergent might yield similar structures, we used SEM to examine the structures that remained following extraction of cells grown under identical conditions with the chemically distinct Tween 20 (Fig. 1D). These objects were generally similar to those obtained using Triton X-100, although they were larger and had a generally smoother appearance. Indeed, their mean length of 380 ± 130 nm was identical to that of the nucleoid-free zones (n = 79; P > 0.8) (Fig. 2C). The variability in the sizes and shapes of the detergent-insoluble objects was recapitulated in electron cryotomography (ECT) images, which showed ribosome-containing zones in the central area of the cell that were distinct from ribosome-free zones at both cell poles. These ribosome-free zones had irregular shapes and boundaries (Fig. 3A and B), often extending into the filament connecting cells undergoing division (Fig. 3C) (7). Given the similarities in size distributions, previous TEM images of similar material at M. penetrans poles (5, 6, 24), and the limited amount of space in an M. penetrans cell for large objects, we concluded that the polar nucleoid-free zones visualized with DAPI staining, the polar ribosome-free zones visualized with ECT, and the DNA-lacking, detergent-insoluble objects visualized with SEM are the same objects, with Triton X-100 solubilizing some component that Tween 20 does not. Interestingly, whereas the poles of whole M. penetrans cells, as visualized with SEM (Fig. 1B) or light microscopy (Fig. 2G), were distinctly narrower than the cell bodies, there was no abrupt narrowing at the poles in cells imaged by ECT, with the cells exhibiting a more ovoid shape (Fig. 3).
FIG 3.
Internal organization of M. penetrans observed by ECT. (A to C) Sections of different whole cells. (D to F) Corresponding schematics. In panels A to C, ∼20-nm granules are ribosomes (reviewed in reference 70). The insertion site of a connecting filament was observed at one cell pole (C and F). Black lines, cell membranes and boundaries of ribosome-free zones; circles, ribosomes (positions are schematic and do not represent actual ribosomes); arrow in panel F, location of connecting filament. Scale bars, 100 nm.
Identification and characterization of detergent-insoluble proteins.
M. penetrans lacks homologs of the AO cytoskeletal proteins of other mycoplasmas (14). To determine which proteins make up the TXI and Tween 20-insoluble (TWI) material, we examined the protein profiles of whole-cell lysates, TXI proteins, and TWI proteins using SDS-PAGE. The detergent-insoluble material was generated by extracting attached M. penetrans cells in flasks and scraping the material that remained attached to the flasks, ensuring that it was identical to the material that was visualized by SEM, which consisted almost entirely of structures similar in shape but heterogeneous in size, as described above (Fig. 1C and D). Several proteins were prominent in the TXI and TWI fractions, and some were enriched, compared to the whole-cell lysate (Fig. 4). Consistent with the similar appearances of the structures, the profile of the TWI proteins was similar to that of the TXI proteins, with the principal exception being one prominent TWI band migrating at ∼40 kDa that was absent in the TXI fraction (Fig. 4). In general, the profiles featured mostly the same proteins but often in different relative amounts.
FIG 4.
SDS-PAGE of the M. penetrans whole-cell lysate and TWI and TXI proteins. A total of 22 μg each of protein from the whole-cell lysate (lane 2), the TWI fraction (lane 3), and the TXI fraction (lane 4) was separated by SDS-PAGE for comparison and for excision of candidate bands chosen for identification. Lane 1 contains protein standards, with sizes indicated to the left. The 66-kDa band in lane 4 was present only in some preparations. Numbers in the gel indicate specific bands chosen for MALDI-TOF mass spectrometry and correspond to protein identifications in Table 1.
We selected eight bands from SDS-polyacrylamide gels that appeared to be enriched in both detergent-insoluble fractions, compared to the whole-cell lysate, as well as the one band that was present in the TWI fraction but not the TXI fraction, for identification using matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (Fig. 4). The 12 proteins that were identified were associated with a variety of functional characteristics, including four with well-established biochemical roles, three putative lipoproteins, and five of unknown function (Table 1). The four with characterized biochemical roles were lactate dehydrogenase, ribosomal protein S3, pyruvate dehydrogenase subunit E2, and OppF, an oligopeptide ABC transporter ATP-binding protein. Two of the three putative lipoproteins were identified as P35 and P42, both members of the P35 lipoprotein family; P42 was the protein that was present only in the TWI fraction. The third lipoprotein was a homolog of Mycoplasma genitalium G37 MG309. Of the five identified proteins that were uncharacterized, four were encoded by genes found very close to one another in the genome, MYPE1530, MYPE1550, MYPE1560, and MYPE1570. Together with MYPE4000, they shared no specific sequence homology with other proteins except in a close relative of M. penetrans, namely, Mycoplasma iowae (25); however, the first four were labeled as predicted cytoskeletal proteins in the original annotation of the M. penetrans genome, on the basis of predicted extensive α-helical coiled-coil structures, reminiscent of Mycoplasma pneumoniae AO protein HMW2 (14), which is predicted to constitute a major structural element within the M. pneumoniae AO cytoskeleton (26–28).
TABLE 1.
TXI and TWI proteins identified by MALDI-TOF
| Protein | Band no. in Fig. 4 | Functional annotation | Size (kDa) |
|---|---|---|---|
| MYPE1530 | 3 | NAa | 122 |
| MYPE1550 | 1 | NA | 386 |
| MYPE1560 | 4 | NA | 99 |
| MYPE1570 | 4 | NA | 93 |
| MYPE4000 | 2 | NA | 99 |
| MYPE5100 | 6 | Pyruvate dehydrogenase subunit E2, PdhC | 51 |
| MYPE6630 | 8 | P42 lipoprotein (P35 family) | 42 |
| MYPE6810 | 9 | P35 lipoprotein (P35 family) | 38 |
| MYPE6960 | 3 | Lipoprotein | 138 |
| MYPE8750 | 5 | Oligopeptide ABC transporter ATP-binding protein, OppF | 92 |
| MYPE9640 | 9 | Lactate dehydrogenase, Ldh | 34 |
| MYPE10090 | 7 | Ribosomal subunit S3, RpsC | 48 |
NA, not applicable.
The absence of sequence homology of these five proteins to the proteins of the AO cytoskeleton of M. pneumoniae (28) or the analogous structure in M. mobile (29), together with the disparity in the detergent-insoluble structures between M. penetrans and mycoplasmas from other phylogenetic clusters, is consistent with a model in which attachment organelles arose independently multiple times during the course of mycoplasma evolution (7, 30). We examined whether there were general features shared in common among the cytoskeletal proteins of M. penetrans, M. pneumoniae, and M. mobile. Interestingly, the five putative M. penetrans cytoskeletal proteins have molecular masses of ≥93 kDa, isoelectric points of ∼4.5, and predicted α-helical coiled-coil regions (extensive in all except MYPE4000). Many of the proteins of the M. pneumoniae attachment organelle, but not of the tip structure of M. mobile, share these features (Table 2).
TABLE 2.
Comparison of AO protein features
| Species and protein | Coiled coil | Molecular mass of >70 kDa | Extreme pI (<5 or >9) | Total no. of features |
|---|---|---|---|---|
| M. pneumoniae | ||||
| HMW1 (MPN447) | X | X | X | 3 |
| HMW2 (MPN310) | X | X | X | 3 |
| HMW3 (MPN453) | X | X | 2 | |
| P24 (MPN312) | 0 | |||
| P41 (MPN311) | X | X | 2 | |
| P65 (MPN309) | X | X | 2 | |
| P200 (MPN567) | X | X | 2 | |
| TopJ (MPN119) | X | X | 2 | |
| M. mobile | ||||
| MMOB0150 | 0 | |||
| MMOB1620 | 0 | |||
| MMOB1630 | X | X | 2 | |
| MMOB1640 | 0 | |||
| MMOB1650 | X | 1 | ||
| MMOB1660 | 0 | |||
| MMOB1670 | 0 | |||
| MMOB4530 | X | 1 | ||
| MMOB4860 | X | 1 | ||
| MMOB5430 | X | 1 | ||
| M. penetrans | ||||
| MYPE1530 | X | X | X | 3 |
| MYPE1550 | X | X | X | 3 |
| MYPE1560 | X | X | X | 3 |
| MYPE1570 | X | X | X | 3 |
| MYPE4000 | X | X | X | 3 |
| MYPE5100 | 0 | |||
| MYPE6630 | 0 | |||
| MYPE6810 | 0 | |||
| MYPE6960 | X | X | 2 | |
| MYPE8750 | X | X | X | 3 |
| MYPE9640 | 0 | |||
| MYPE10090 | 0 |
Cotranscription of putative cytoskeletal genes.
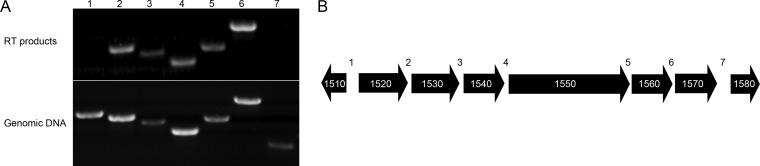
Examination of the genomic organization of the genes encoding proteins MYPE1530, MYPE1550, MYPE1560, and MYPE1570, as well as predicted structural and compositional similarities among the predicted protein products, suggested that, along with the genes encoding the structurally and compositionally similar proteins MYPE1520 and MYPE1540, they might form a transcriptional unit. We performed reverse transcription (RT)-PCR to test whether these genes, each separated by no more than 30 bp, were cotranscribed. We used a primer complementary to a region near the 3′ end of MYPE1570, which is positioned downstream of the other genes in question, to create cDNA. From this cDNA, we amplified regions spanning the junctions between the six candidate genes (MYPE1520 to MYPE1570) as well as MYPE1510, which is ∼180 bp away from MYPE1520 and on the opposite strand. We also made cDNA using a primer complementary to MYPE1580, which is also ∼180 bp away, and we attempted to amplify cDNA linking MYPE1570 and MYPE1580. There were RT-PCR products for all of the junctions between MYPE1520 and MYPE1570 but no products for the MYPE1510-MYPE1520 junction (Fig. 5A). Amplification of the junction between MYPE1570 and MYPE1580 sometimes yielded a faint, poorly reproducible product, suggesting partial read-through (data not shown). These results suggest that MYPE1520 to MYPE1570 constitute an operon (Fig. 5B).
FIG 5.
RT-PCR analysis of the putative cytoskeletal operon of M. penetrans. (A) Agarose gel containing PCR products from RT-generated transcripts that span the gene junctions numbered in panel B. Top, products amplified from reverse-transcribed RNA using a primer complementary to a region at the 3′ end of mype1570; bottom, products amplified from genomic DNA used as a positive control. (B) Schematic of genes encoding MYPE1520 to MYPE1570, showing the genomic orientation and relative sizes, including the 5′ and 3′ flanking genes. Numbers 1 to 7 refer to the positions of the primer pairs used to span gene junctions for RT-PCR.
Transcriptomic analysis of M. penetrans grown with HeLa cells.
Because the major function of the AO is adherence to host cells, which is expected to occur at the initial stages of infection, we hypothesized that the genes involved in AO formation and function, including those encoding the proteins identified in the TXI fraction, would be upregulated during the early stages of infection. The interactions between M. penetrans and HeLa cells, including the events that occur during invasion, have been documented (31–34). To identify genes that undergo differential expression upon interaction with host cells, we performed transcriptome sequencing (RNA-Seq) with M. penetrans cells grown alone and grown in the presence of HeLa cells, 6 h after infection (representing a time point at which cytadherence and invasion had occurred [33]), and we compared global gene expression profiles between the two conditions. Surprisingly, there was almost no statistically significant difference in gene expression between the two conditions, including the AO genes, with the exception of three genes that were upregulated and one gene that was downregulated 2-fold (see Data Set S1 in the supplemental material). The three upregulated genes were MYPE1035, MYPE3985, and MYPE8260, all of which encode small hypothetical proteins of less than 70 amino acids. The single downregulated gene was MYPE20130, which codes for a tRNA for arginine and whose corresponding codon is predicted to be commonly used; its expression was 3-fold lower in mycoplasmas incubated with HeLa cells.
DISCUSSION
Convergent evolution of attachment organelles.
The overall organization of the AO-associated structures of M. penetrans was quite distinct from those of M. pneumoniae (35, 36), Mycoplasma mobile (29), and Mycoplasma insons (30), which represent three other phylogenetic lineages or clusters within the genus. Despite superficial similarities in the morphology and function of the AOs of M. pneumoniae and M. mobile, dissimilarities in both the organization and the composition of their internal structures are well documented (37). In contrast, the structures and compositions of AO cytoskeletal structures are shared among the close relatives of M. pneumoniae in the M. pneumoniae cluster (23). These differences have been taken to suggest that the AOs of M. pneumoniae and M. mobile arose through convergent evolution (2), which is not altogether surprising, considering that the two species are separated by one of the deepest phylogenetic divisions within the genus, with M. pneumoniae being in the pneumoniae group and M. mobile in the hominis group (3). However, the subsequent finding that Mycoplasma insons (a species in the pneumoniae group that is not a close relative of M. pneumoniae) is unidirectionally motile but lacks a distinct tip structure and instead contains cytoskeletal elements throughout its entire rod-shaped body (30) raised the possibility that even within the pneumoniae group there has been independent evolution of polar attachment and motility. If so, then these structures have an even more recent evolutionary origin. Alternatively, the M. insons organization could represent a variation of the M. pneumoniae organization whose homology is not obvious, pending further studies of this organism. M. penetrans is also in the pneumoniae group but within a different cluster than either M. pneumoniae or M. insons (38). The AO of this species, whose motility is driven by a fundamentally different mechanism than that of M. mobile (39), provided an opportunity to test whether there were detectable conserved elements of AO organization within the pneumoniae group.
The absence of homology between M. penetrans and M. pneumoniae AO proteins strongly suggests that these parallel systems evolved independently. M. mobile, with no apparent equivalent of a ribosome-free space (29) and with its cytoskeletal components lacking the hallmarks of the M. pneumoniae and M. penetrans AO proteins identified in the present work, appears to have evolved a different approach to generating a protrusive and therefore superficially similar structure. M. insons achieves polarization of adherence and motility without a protrusive structure (30), representing a completely different model for polarization.
Properties of M. penetrans AO interior.
As described previously (7), there can be little doubt that the detergent-insoluble, DNA-free objects observed by SEM, whose abundance on coverslips is routinely parallel to that of unextracted M. penetrans cells grown from the same inoculum (data not shown), occupy the space in the nucleoid-free zones of M. penetrans cells and represent the material observed at the cell poles in thin sections and also observed here by ECT. Indeed, the material underlying the cell poles in ECT images, which closely resemble the material at the cell poles observed in TEM images of thin sections (5, 6, 24), are strikingly reminiscent of ribosome-free PopZ-containing structures at the poles of C. crescentus cells when PopZ is overproduced (40). Our deeper study of these objects has revealed that the organization of the TXI and TWI structures of the M. penetrans AO is completely unlike that of the M. pneumoniae structures. Whereas the dimensions of the M. pneumoniae structures appear to be tightly constrained (23), the dimensions are highly variable in M. penetrans, as visualized in a number of ways. SEM images of these variable objects revealed some commonalities, however, including a wider portion and a narrower portion, the latter often containing one or more flexible rod-like elements. This heterogeneity, which is distinct from the great regularity of the M. pneumoniae AO structures, suggests the possibility of growth by accretion of material onto, or absorption of material into, a preexisting structure. ECT images suggest that the two poles contain the same differentiated material, and this material even extends into the filaments connecting dividing M. penetrans cells. These facts are consistent with a model in which nucleation and biogenesis of the AO underlying structure are linked to the cell cycle, perhaps with new material being deposited into spaces in the cytoplasm vacated during chromosome condensation and segregation. The broader size distribution of the nucleoid-free zones and the detergent-insoluble structures suggests that growth of the M. penetrans AO, though capped, is not capped at a uniform size.
In addition to the heterogeneity in size, the pleomorphy of the detergent-insoluble objects is striking. We noted that, whereas our SEM images of whole M. penetrans cells and previously described TEM images of thin sections (5–7, 24) showed that the region of the cell at the pole is constricted along the long axis, this was not the case in ECT images. One possible interpretation of this discrepancy is that the material within the M. penetrans AO is highly hydrated and therefore subject to shrinkage during the dehydration steps associated with standard SEM and TEM processing techniques, whereas ECT lacks a dehydration step. Conceivably, this hydration results from attraction of water molecules to the highly negatively charged proteins present in the structure. If the structural elements are principally organized parallel to the long axis, then dehydration would result predominantly in a decrease in width rather than length. Another possibility is that, because the cell bodies visualized by ECT are not attached firmly to surfaces but instead are present in the holes of the carbon-coated grids, they are in a relaxed conformation. In either case, we predict that the material within the AO is flexible like a gel.
M. penetrans AO composition and gene expression.
Of the proteins we identified in the TXI and similar TWI fractions of M. penetrans by MALDI-TOF mass spectrometry, four have characterized biochemical roles. Whether any or all of them are part of the internal structure of the AO is unclear. Pyruvate dehydrogenase subunit E2 normally functions as part of a large structure that could be insoluble in nonionic detergents; however, none of the other subunits of pyruvate dehydrogenase was identified, suggesting a possible alternative or additional role for this protein, which also functions as a surface adhesin in other mycoplasmas (41). OppF is normally a peripheral membrane component of the oligopeptide ABC transporter (42), including in mycoplasmas (43). The substrate-binding protein component of this transporter, OppA, doubles as an adhesin in Mycoplasma hominis (44, 45), raising the possibility that M. penetrans OppF anchors the material underlying the AO to OppA, acting as an adhesin. However, none of the proteins of this complex other than OppF was identified in the M. penetrans detergent-insoluble fractions.
Surprisingly, three lipoproteins were identified in the detergent-insoluble fraction. Two of these lipoproteins are members of the P35 lipoprotein family, which comprises 44 total M. penetrans genes (14). The P35 lipoprotein and its paralogs, which are distributed across the surface of M. penetrans cells, are immunodominant (46–49). The expression of genes encoding these proteins undergoes extensive antigenic variation, which is thought to be a major factor for immune evasion (50). The presence of two of these P35 homologs in the detergent-insoluble fractions may suggest that they indirectly interact with the AO interior, although it is also possible, given their extensive distribution across the cell surface, that they aggregate or form multiple interactions with each other that prevent their solubilization in nonionic detergents. Indeed, the difference in appearance between the TWI and TXI fractions might be entirely attributable to the presence of P42 in the former. The third lipoprotein identified, MYPE6960, has no known function, and its distribution and presence at the cell surface of M. penetrans cells is unknown. However, this protein is a homolog of MG309, a protein in M. genitalium that binds and activates Toll-like receptors (51), making MYPE6960 a particularly interesting candidate for further study with respect to pathogenesis. Conceivably, this lipoprotein plays a role in the adherence of M. penetrans to host cells and is associated with AO proteins.
The remaining five proteins that were identified in this study had no predicted function or conserved domains, making them ideal candidates for principal structural elements of the AO. The genes encoding four of these proteins are located very close together in the genome of M. penetrans and were found to be transcribed as a polycistronic message with two other genes. Three of these, MYPE1550, MYPE1560, and MYPE1570, were among the most abundant proteins in the detergent-insoluble fractions. All six of these proteins, as well as MYPE4000, share an unusual set of characteristics, including predicted α-helical coiled-coil regions, molecular masses of >90 kDa, and pI values of approximately 4.5 (Table 2). Interestingly, these characteristics are shared with many of the M. pneumoniae AO cytoskeletal proteins (Table 2) despite the lack of sequence homology. It is highly unlikely, given that the M. penetrans proteins lack characteristics of M. pneumoniae AO proteins such as proline-rich domains and sequence elements such as the EAGR box, that the M. pneumoniae and M. penetrans AO proteins share a common ancestry. The shared properties are intriguing, however, when the functional analogy of the structures that they build is considered. These shared properties suggest that one solution to forming a polar cytoskeletal structure, arrived at independently in M. pneumoniae and M. penetrans, involves the assembly of large, highly charged proteins that interact with each other over an extensive surface area via α-helical coiled coils.
Although another group reported changes in the expression of a specific set of phase-variable genes in response to infection of a host (52), this study is the first, to our knowledge, to report on the expression of an entire mycoplasma transcriptome upon exposure to host cells. Particularly striking was the absence of major differences in the transcript profiles of M. penetrans cells grown independently and grown in the presence of HeLa cells for 6 h, which is enough time for interactions to occur between the two cell types, including invasion (31, 33). We anticipate that, after 6 h in an environment filled with signals from host cells, M. penetrans cells will have had ample opportunity to adjust their gene expression profiles to prepare for interactions, whether or not all M. penetrans cells have adhered to and invaded the HeLa cells. Therefore, even if, as a consequence of the use of HeLa cells at 70 to 80% confluence, only ∼75% of M. penetrans cells interacted with host cells, we would expect to see significant changes in the overall gene expression pattern if a change were occurring. The absence of transcriptional regulation is a formal but unlikely possibility, as mycoplasmas, despite having limited machinery for transcriptional regulation, nonetheless exhibit it (53–55). A more likely explanation is that M. penetrans, which naturally occurs as an obligate commensal or parasite like all mycoplasmas, has no evolutionary adaptation to living in the absence of host cells and therefore is always in a mode in which it is deployed for interaction with the host. Relatedly, it is possible that the fetal bovine serum endows SP-4 broth with all of the relevant factors provided by HeLa cells.
Implications for mechanisms of AO function.
Unlike the M. penetrans AO, which consists entirely of a poorly organized matrix, the corresponding M. pneumoniae feature is visually dominated by a dense, highly organized structure flanked by a ribosome-excluding zone in contact with the cell membrane (35). The structure, which has been called the electron-dense core because of its prominent staining with metal-based dyes (56), has been the focus of considerable study (reviewed by Balish [2]). Because the ribosome-excluding zone is directly adjacent to the core, even in cells in which the core is detached from the membrane (35, 57, 58), it is likely formed by some fine substance linked to the core, probably regions of the same proteins that make up the core itself. Indeed, the failure to identify any non-core-associated proteins in the M. pneumoniae AO (58) supports this model. In contrast to M. pneumoniae, the M. penetrans AO consists solely of material whose lack of obvious organization is more reminiscent of the lucent area of M. pneumoniae than the core. Because both AOs, one with a core and one without, function in adherence and motility, it is possible that the core itself is dispensable for these functions in M. pneumoniae, as suggested previously (23, 59), although this assertion is in opposition to some models (35, 58). Therefore, we propose that the less organized material, constituting the ribosome-excluding zone in M. pneumoniae and perhaps the entire AO interior in M. penetrans, is the principal organizer of the AO, driving the concentration and organization of adhesins at the AO in both species and leaving motility to be carried out directly by adhesins with motor properties, as described for the more distantly related M. mobile (37).
Regarding M. pneumoniae, if it is the lucent material that organizes the AO, then the core may be dispensable for adherence and motility. Superficially, this statement appears to be counter to observations (reviewed in reference 60). However, if the lucent and core regions of the M. pneumoniae AO are actually composed of different regions of the same proteins, then mutants in which core proteins are lost would also be defective in the functions associated with the lucent region. Indeed, if this model is correct, then M. pneumoniae mutants affected only in the lucent area and not in the core have never been studied. The physical association of DNA with the core in M. pneumoniae relatives (23) raises the possibility that the M. pneumoniae core itself is primarily involved in chromosome segregation. In M. pneumoniae, a newly formed AO is displaced from the preexisting pole to the pole created during binary fission during cell division (61), possibly carrying the newly replicated chromosome with it to the daughter cell (2). Because M. penetrans cells appear to acquire a mass of AO material opposite the pole of the functioning AO, and not adjacent to it, during the process of division and because DNA is not associated with the M. penetrans detergent-insoluble material, it is reasonable to propose that the M. penetrans AO is not directly involved in chromosome segregation.
MATERIALS AND METHODS
Bacterial culture.
Hyperadherent Mycoplasma penetrans strain HP88 cells (39) were grown to the mid-log phase at 37°C in plastic tissue culture flasks (Thermo Scientific BioLite) containing SP-4 broth (62).
Detergent extraction and protein analysis.
Cultures of M. penetrans were decanted and washed twice with warm potassium-free phosphate-buffered saline (PBS) (145 mM NaCl, 2.83 mM NaH2PO4·H2O, 7.20 mM NaHPO4·7H2O [pH 7.2]). For detergent extraction, Triton X-100 or Tween 20 in 20 mM Tris-HCl-150 mM NaCl (pH 7.2) was added to the cultures to a final concentration of 1% in 25 ml PBS. Detergent extracts were then incubated for 30 min at 37°C, scraped, and centrifuged for 20 min at 17,050 × g. Whole-cell lysates were treated in the same way without the addition of detergent. Following centrifugation, the pellets were washed three times and resuspended in PBS. Protein concentrations were determined using a bicinchoninic acid protein assay kit (Pierce). Equal amounts of protein of each fraction were subjected to 9% SDS-PAGE (63) and stained with Coomassie brilliant blue or Sypro Ruby (Molecular Probes). Detergent-extracted proteins were separated by SDS-PAGE, and select bands, chosen on the basis of being abundant or enriched, compared to the total-cell lysate, were excised, reduced and alkylated, digested with trypsin, and analyzed by MALDI-TOF mass spectrometry (AB Sciex 4800 MALDI-TOF/TOF analyzer) at the University of Cincinnati Proteomics Laboratory, as described previously (64). MultiCoil (65) was used to test predicted amino acid sequences for the presence of coiled-coil domains; the presence of regions with >50% probability of forming α-helical coiled coils was recorded as positive.
Scanning electron microscopy.
Cells were prepared for SEM as described previously (66). Briefly, cells were grown on glass coverslips for 6 to 24 h at 37°C in SP-4 medium supplemented with 3% gelatin. For examination of TXI structures or TWI structures, detergent in 20 mM Tris-HCl (pH 7.5)-150 mM NaCl was added to cells at a final concentration of 1%, and cells were incubated for 30 min at 37°C. The samples were fixed for 30 min at room temperature in 1.5% glutaraldehyde-1% paraformaldehyde-0.1 M sodium cacodylate (pH 7.2). Following fixation, the coverslips were washed four times in 0.1 M sodium cacodylate (pH 7.2) and dehydrated through a series of ethanol washes from 25% to 100%. The coverslips were then critical point dried, gold sputter coated, and viewed on a Zeiss Supra 35 FEG-VP scanning electron microscope at the Miami University Center for Advanced Microscopy and Imaging.
Electron cryotomography.
M. penetrans was grown at 37°C on Quantifoil Au-finder grids (Quantifoil Micro Tools GmbH) in SP-4 broth to the mid-log phase, which was indicated by a color change in the medium from red to orange. The electron microscopy grids with attached M. penetrans cells were removed from the medium, loaded onto tweezers, and washed with fresh medium. Colloidal gold (10 nm) was added to the grids, and the grids were blotted (Whatman grade 40 filter paper) prior to being plunge-frozen in liquid ethane on a gravity plunger freezer. Grids were loaded into a 300-kV FEI Polara G2 electro-cryotransmission electron microscope equipped with a field emission gun, a lens-coupled Gatan UltraCam (4,000- by 4,000-pixel resolution), and a Gatan imaging filter (GIF). Samples were maintained at liquid nitrogen temperature as tilt series of whole cells were captured. The tilt series were recorded from −60° to +60° at 1° increments, with a 10-μm defocus, using Leginon (67). All tilt series were collected through the GIF around zero-loss energy with a slit width of 20 keV, for a cumulative dose of 180 e−/Å2 for each tilt series.
Measurements and analysis.
To measure the lengths of nucleoid-free zones, M. penetrans cells were grown overnight on coverslips, fixed as described previously (7), and mounted on slides with Vectashield containing DAPI (Vector Laboratories). Fields were imaged using a 100× objective, as described previously (7). Phase-contrast and DAPI fluorescence images were overlaid, and the lengths of 84 polar nucleoid-free zones were measured using SPOT software. To measure the lengths of TXI and TWI objects, SEM images of the objects from multiple fields, captured as described above, were measured along their long axes. Objects that appeared to be clusters of individual objects were excluded. For TXI objects, 70 were measured; for TWI objects, 79 were measured. Statistical comparisons of lengths were performed by one-way analysis of variance (ANOVA), after a Shannon-Wilk test was used to establish that the distributions of lengths were not normal. Statistical comparisons of the lengths of the cells and the nucleoid-free zones were performed using least-squares regression analysis.
RNA extraction.
RNA was harvested as described previously (52). M. penetrans cells were harvested by centrifugation at 17,400 × g for 20 min, followed by three washes in PBS. The cell pellet was resuspended in 1 ml of TRI Reagent solution (Ambion). Total RNA was then purified using the Direct-zol RNA MiniPrep kit (Zymo Research), according to the manufacturer's instructions. The remaining DNA was removed from the sample by treatment with DNase I twice using the DNA-free kit (Applied Biosystems), according to the manufacturer's instructions. Quantitative PCR was performed to ensure that the RNA was free of DNA. The quality of the RNA was then examined using an Agilent 2100 bioanalyzer (Agilent Technologies).
RNA sequencing.
M. penetrans cells were grown for 6 h at 37°C in SP-4 broth, either alone or together with HeLa cells grown to 70 to 80% confluence, at a multiplicity of infection (MOI) of 100. Three biological replicates were used for each of the conditions. For each biological replicate, RNA was extracted as described above and divided into three technical replicates. Purified bacterial RNA was enriched by performing poly(A) depletion using the NEBNext poly(A) mRNA magnetic isolation module (New England BioLabs) on all samples, as described previously (52). The RNA was then depleted of both prokaryotic and eukaryotic rRNA by using the Ribo-Zero Gold rRNA removal kit (Illumina), as directed by the manufacturer's instructions, and was cleaned and concentrated by using the Zymo RNA Clean and Concentrator-25 kit (Zymo Research), as described previously (52). The RNA was eluted in 25 μl RNase-free water, examined for quality using an Agilent 2100 Bioanalyzer (Agilent Technologies), and used for cDNA synthesis.
cDNA libraries were created using the Illumina TruSeq stranded mRNA library preparation kit (Illumina), following the manufacturer's protocol, as described previously (52). The cDNA libraries were examined for quality using the Agilent 2100 Bioanalyzer and then were quantified using a library quantification kit (Kapa Biosystems). Libraries were normalized to 10 nM, pooled, diluted, denatured, and loaded onto a MiSeq reagent kit v3 cartridge, as directed by the manufacturer (Illumina). The samples were sequenced on a MiSeq system using a 75-bp paired-end approach.
Data from the RNA-Seq experiment were analyzed as described previously for Mycoplasma gallisepticum data (52). Briefly, the fastq files were assembled, mapped, and analyzed for differential gene expression using Rockhopper (http://cs.wellesley.edu/~btjaden/Rockhopper) (68), with the M. penetrans HF-2 genome as the reference genome. Parameters were as described previously (52). For comparison of gene expression values, the data were first normalized by determining the ratio of reads per kilobase per million (RPKM) for mycoplasmas grown with HeLa cells to those for mycoplasmas grown axenically and then the fold change was calculated by taking the log2 transformation of the RPKM difference between the two biological samples.
Reverse transcription-coupled PCR.
The RT reaction was carried out as described previously for Mycoplasma iowae (69). Briefly, cDNA synthesis was performed with 100 ng of total RNA from M. penetrans and random hexamers, using the Verso cDNA synthesis kit (Thermo Scientific), according to the manufacturer's instructions. One microliter of cDNA was then used as the template in a 50-μl PCR mixture with Taq polymerase (New England BioLabs), using EasyStart PCR tubes (Molecular Bioproducts), according to the manufacturer's instructions. The primers used in each reaction were designed to span the adjacent gene junctions (Table 3). Reactions were amplified by incubation at 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 53°C for 1 min, and 68°C for 1 min, with a final extension at 68°C for 5 min. The PCR products were subjected to gel electrophoresis. Reactions with genomic DNA and without the addition of reverse transcriptase or template were performed as positive and negative controls, respectively.
TABLE 3.
Primers used for RT-PCR
| Primer designation | Sequence (5′ to 3′) | Target | Product size (bp) |
|---|---|---|---|
| 1510end(up) | AATGAGCAGTAAGTGCTAGC | MYPE1510-MYPE1520 | 603 |
| 1520begin(down) | GTCTTAGGAATTTGAACAGGTG | ||
| 1520end(up) | GGAAAAAGAAGCTGTCACAG | MYPE1520-MYPE1530 | 566 |
| 1530begin(down) | GGTTGATTGTCAGCAGAACC | ||
| 1530end(up) | TTCCAGCTCCTGCATATGAC | MYPE1530-MYPE1540 | 527 |
| 1540begin(down) | GTCAGTTCCCTGAAATTGTTC | ||
| 1540end(up) | CTAAATTGTTCCAATATTGAATAATCGC | MYPE1540-MYPE1550 | 454 |
| 1550begin(down) | ATAGATGAGATTCTTTATCAAAGTTCTC | ||
| 1550end(up) | ATCAAACCCAAGCGATTCTG | MYPE1550-MYPE1560 | 541 |
| 1560begin(down) | TCAACATCGTCTTGTCTTGG | ||
| 1560end(up) | TGCTCCTAGTAGTGAGTTGC | MYPE1560-MYPE1570 | 696 |
| 1570begin(down) | CTTCTCTAGCTTGAACTTGG | ||
| 1570end(up) | TTTGGAAGACCAGCAGGAAG | MYPE1570-MYPE1580 | 380 |
| 1580begin(down) | GTGTTGCCAGGTCTAAGATC | ||
| 1580RT(down) | CATAGCCGTTTGAACAGTGT | NAa | NA |
NA, not applicable.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (grant R15 AI073994 to M.F.B.).
We thank A. Kiss (Miami University Center for Bioinformatics and Functional Genomics) and K. Pflaum (University of Connecticut) for help with RNA-Seq. We thank R. J. Hickey (Miami University), W. Ambrosius (Wake Forest University), and members of the Balish laboratory for insightful discussions.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00069-17.
REFERENCES
- 1.Balish MF. 2006. Subcellular structures of mycoplasmas. Front Biosci 11:2017–2027. doi: 10.2741/1943. [DOI] [PubMed] [Google Scholar]
- 2.Balish MF. 2014. Mycoplasma pneumoniae, an underutilized model for bacterial cell biology. J Bacteriol 196:3675–3682. doi: 10.1128/JB.01865-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johannson KE, Pettersson B. 2002. Taxonomy of Mollicutes, p 1–29. In Razin S, Herrmann R (ed), Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum Publishers, New York, NY. [Google Scholar]
- 4.Jaffe JD, Stange-Thomann N, Smith C, DeCaprio D, Fisher S, Butler J, Calvo S, Elkins T, FitzGerald MG, Hafez N, Kodira CD, Major J, Wang S, Wilkinson J, Nicol R, Nusbaum C, Birren B, Berg HC, Church GM. 2004. The complete genome and proteome of Mycoplasma mobile. Genome Res 14:1447–1461. doi: 10.1101/gr.2674004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lo SC, Hayes MM, Wang RY, Pierce PF, Kotani H, Shih JW. 1991. Newly discovered mycoplasma isolated from patients infected with HIV. Lancet 338:1415–1418. doi: 10.1016/0140-6736(91)92721-D. [DOI] [PubMed] [Google Scholar]
- 6.Lo SC, Hayes MM, Tully JG, Wang RY, Kotani H, Pierce PF, Rose DL, Shih JW. 1992. Mycoplasma penetrans sp. nov., from the urogenital tract of patients with AIDS. Int J Syst Bacteriol 42:357–364. doi: 10.1099/00207713-42-3-357. [DOI] [PubMed] [Google Scholar]
- 7.Jurkovic DA, Newman JT, Balish MF. 2012. Conserved terminal organelle morphology and function in Mycoplasma penetrans and Mycoplasma iowae. J Bacteriol 194:2877–2883. doi: 10.1128/JB.00060-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yáñez A, Cedillo L, Neyrolles O, Alonso E, Prévost MC, Rojas J, Watson HL, Blanchard A, Cassell GH. 1999. Mycoplasma penetrans bacteremia and primary antiphospholipid syndrome. Emerg Infect Dis 5:164–167. doi: 10.3201/eid0501.990122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nir-Paz R, Israel S, Honigman A, Kahane I. 1995. Mycoplasmas regulate HIV-LTR-dependent gene expression. FEMS Microbiol Lett 128:63–68. doi: 10.1111/j.1574-6968.1995.tb07501.x. [DOI] [PubMed] [Google Scholar]
- 10.Sasaki Y, Blanchard A, Watson HL, Garcia S, Dulioust A, Montagnier L, Gougeon ML. 1995. In vitro influence of Mycoplasma penetrans on activation of peripheral T lymphocytes from healthy donors or human immunodeficiency virus-infected individuals. Infect Immun 63:4277–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iyama K, Ono S, Kuwano K, Ohishi M, Shigematsu H, Arai S. 1996. Induction of tumor necrosis factor alpha (TNFα) and enhancement of HIV-1 replication in the J22HL60 cell line by Mycoplasma penetrans. Microbiol Immunol 40:907–914. doi: 10.1111/j.1348-0421.1996.tb01159.x. [DOI] [PubMed] [Google Scholar]
- 12.Shimizu T, Kida Y, Kuwano K. 2004. Lipid-associated membrane proteins of Mycoplasma fermentans and M. penetrans activate human immunodeficiency virus long-terminal repeats through Toll-like receptors. Immunology 113:121–129. doi: 10.1111/j.1365-2567.2004.01937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li BC, Herrmann R. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res 24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasaki Y, Ishikawa J, Yamashita A, Oshima K, Kenri T, Furuya K, Yoshino C, Horino A, Shiba T, Sasaki T, Hattori M. 2002. The complete genomic sequence of Mycoplasma penetrans, an intracellular bacterial pathogen in humans. Nucleic Acids Res 30:5293–5300. doi: 10.1093/nar/gkf667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho H. 2015. The role of cytoskeletal elements in shaping bacterial cells. J Microbiol Biotechnol 25:307–316. doi: 10.4014/jmb.1409.09047. [DOI] [PubMed] [Google Scholar]
- 16.Margolin W. 2009. Sculpting the bacterial cell. Curr Biol 19:R812–R822. doi: 10.1016/j.cub.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pilhofer M, Jensen GJ. 2013. The bacterial cytoskeleton: more than twisted filaments. Curr Opin Cell Biol 25:125–133. doi: 10.1016/j.ceb.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ausmees N, Kuhn JR, Jacobs-Wagner C. 2003. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell 115:705–713. doi: 10.1016/S0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- 19.Lin L, Thanbichler M. 2013. Nucleotide-independent cytoskeletal scaffolds in bacteria. Cytoskeleton 70:409–423. doi: 10.1002/cm.21126. [DOI] [PubMed] [Google Scholar]
- 20.Burkhard P, Stetefeld J, Strelkov SV. 2001. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol 11:82–88. doi: 10.1016/S0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- 21.Kühn J, Briegel A, Mörschel E, Kahnt J, Leser K, Wick S, Jensen GJ, Thanbichler M. 2010. Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus. EMBO J 29:327–339. doi: 10.1038/emboj.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koch MK, McHugh CA, Hoiczyk E. 2011. BacM, an N-terminally processed bactofilin of Myxococcus xanthus, is crucial for proper cell shape. Mol Microbiol 80:1031–1051. doi: 10.1111/j.1365-2958.2011.07629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatchel JM, Balish MF. 2008. Attachment organelle ultrastructure correlates with phylogeny, not gliding motility properties, in Mycoplasma pneumoniae relatives. Microbiology 154:286–295. doi: 10.1099/mic.0.2007/012765-0. [DOI] [PubMed] [Google Scholar]
- 24.Neyrolles O, Brenner C, Prevost MC, Fontaine T, Montagnier L, Blanchard A. 1998. Identification of two glycosylated components of Mycoplasma penetrans: a surface-exposed capsular polysaccharide and a glycolipid fraction. Microbiology 144:1247–1255. doi: 10.1099/00221287-144-5-1247. [DOI] [PubMed] [Google Scholar]
- 25.Pritchard RE, Prassinos AJ, Osborne JD, Raviv Z, Balish MF. 2014. Reduction of hydrogen peroxide accumulation and toxicity by a catalase from Mycoplasma iowae. PLoS One 9:e105188. doi: 10.1371/journal.pone.0105188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balish MF, Ross SM, Fisseha M, Krause DC. 2003. Deletion analysis identifies key functional domains of the cytadherence-associated protein HMW2 of Mycoplasma pneumoniae. Mol Microbiol 50:1507–1516. doi: 10.1046/j.1365-2958.2003.03807.x. [DOI] [PubMed] [Google Scholar]
- 27.Bose SR, Balish MF, Krause DC. 2009. Mycoplasma pneumoniae cytoskeletal protein HMW2 and the architecture of the terminal organelle. J Bacteriol 191:6741–6748. doi: 10.1128/JB.01486-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakane D, Kenri T, Matsuo L, Miyata M. 2015. Systematic structural analyses of attachment organelle in Mycoplasma pneumoniae. PLoS Pathog 11:e1005299. doi: 10.1371/journal.ppat.1005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakane D, Miyata M. 2007. Cytoskeletal “jellyfish” structure of Mycoplasma mobile. Proc Natl Acad Sci U S A 104:19518–19523. doi: 10.1073/pnas.0704280104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Relich RF, Friedberg AJ, Balish MF. 2009. Novel cellular organization in a gliding mycoplasma, Mycoplasma insons. J Bacteriol 191:5312–5314. doi: 10.1128/JB.00474-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andreev J, Borovsky Z, Rosenshine I, Rottem S. 1995. Invasion of HeLa cells by Mycoplasma penetrans and the induction of tyrosine phosphorylation of a 145-kDa host cell protein. FEMS Microbiol Lett 132:189–194. doi: 10.1111/j.1574-6968.1995.tb07832.x. [DOI] [PubMed] [Google Scholar]
- 32.Borovsky Z, Tarshis M, Zhang P, Rottem S. 1998. Protein kinase C activation and vacuolation in HeLa cells invaded by Mycoplasma penetrans. J Med Microbiol 47:915–922. doi: 10.1099/00222615-47-10-915. [DOI] [PubMed] [Google Scholar]
- 33.Tarshis M, Yavlovich A, Katzenel A, Ginsburg I, Rottem S. 2004. Intracellular location and survival of Mycoplasma penetrans within HeLa cells. Curr Microbiol 49:136–140. doi: 10.1007/s00284-004-4298-3. [DOI] [PubMed] [Google Scholar]
- 34.Zeiman E, Tarshis M, Rottem S. 2008. Mycoplasma penetrans under nutritional stress: influence on lipid and lipoprotein profiles and on the binding to and invasion of HeLa cells. FEMS Microbiol Lett 287:243–249. doi: 10.1111/j.1574-6968.2008.01322.x. [DOI] [PubMed] [Google Scholar]
- 35.Henderson GP, Jensen GJ. 2006. Three-dimensional structure of Mycoplasma pneumoniae's attachment organelle and a model for its role in gliding motility. Mol Microbiol 60:376–385. doi: 10.1111/j.1365-2958.2006.05113.x. [DOI] [PubMed] [Google Scholar]
- 36.Seybert A, Herrmann R, Frangakis AS. 2006. Structural analysis of Mycoplasma pneumoniae by cryo-electron tomography. J Struct Biol 156:342–354. doi: 10.1016/j.jsb.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 37.Miyata M. 2010. Unique centipede mechanism of Mycoplasma gliding. Annu Rev Microbiol 64:519–537. doi: 10.1146/annurev.micro.112408.134116. [DOI] [PubMed] [Google Scholar]
- 38.Brown DR, May M, Bradbury JM, Balish MF, Calcutt MJ, Glass JI, Tasker S, Messick JB, Johansson KE, Neimark H. 2010. Genus I. Mycoplasma Nowak 1929, 1349 nom. Cons. Jud. Comm. Opin 22, 1958, 166AL, p 575–613. In Krieg NR, Staley JT, Brown DR, Hedlund BP, Paster BJ, Ward NL, Ludwig W, Whitman WB (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 4 Springer, New York, NY. [Google Scholar]
- 39.Jurkovic DA, Hughes MR, Balish MF. 2013. Analysis of energy sources for Mycoplasma penetrans gliding motility. FEMS Microbiol Lett 338:39–45. doi: 10.1111/1574-6968.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. 2008. A self-associating protein critical for chromosome attachment, division, and polar organization in Caulobacter. Cell 134:956–968. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gründel A, Friedrich K, Pfeiffer M, Jacobs E, Dumke R. 2015. Subunits of the pyruvate dehydrogenase cluster of Mycoplasma pneumoniae are surface-displayed proteins that bind and activate human plasminogen. PLoS One 10:e0126600. doi: 10.1371/journal.pone.0126600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Higgins CF. 2001. ABC transporters: physiology, structure and mechanism: an overview. Res Microbiol 152:205–210. doi: 10.1016/S0923-2508(01)01193-7. [DOI] [PubMed] [Google Scholar]
- 43.Hopfe M, Henrich B. 2004. OppA, the substrate-binding subunit of the oligopeptide permease, is the major ecto-ATPase of Mycoplasma hominis. J Bacteriol 186:1021–1928. doi: 10.1128/JB.186.4.1021-1028.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henrich B, Feldmann RC, Hadding U. 1993. Cytoadhesins of Mycoplasma hominis. Infect Immun 61:2945–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henrich B, Hopfe M, Kitzerow A, Hadding U. 1999. The adherence-associated lipoprotein P100, encoded by an opp operon structure, functions as the oligopeptide-binding domain OppA of a putative oligopeptide transport system in Mycoplasma hominis. J Bacteriol 181:4873–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang RY, Shih JW, Grandinetti T, Pierce PF, Hayes MM, Wear DJ, Alter HJ, Lo SC. 1992. High frequency of antibodies to Mycoplasma penetrans in HIV-infected patients. Lancet 340:1312–1316. doi: 10.1016/0140-6736(92)92493-Y. [DOI] [PubMed] [Google Scholar]
- 47.Neyrolles O, Chambaud I, Ferris S, Prevost MC, Sasaki T, Montagnier L, Blanchard A. 1999. Phase variations of the Mycoplasma penetrans main surface lipoprotein increase antigenic diversity. Infect Immun 67:1569–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neyrolles O, Eliane JP, Ferris S, da Cunha RA, Prevost MC, Bahraoui E, Blanchard A. 1999. Antigenic characterization and cytolocalization of P35, the major Mycoplasma penetrans antigen. Microbiology 145:343–355. [DOI] [PubMed] [Google Scholar]
- 49.Röske K, Blanchard A, Chambaud I, Citti C, Helbig JH, Prevost MC, Rosengarten R, Jacobs E. 2001. Phase variation among major surface antigens of Mycoplasma penetrans. Infect Immun 69:7642–7651. doi: 10.1128/IAI.69.12.7642-7651.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horino A, Sasaki Y, Sasaki T, Kenri T. 2003. Multiple promoter inversions generate surface antigenic variation in Mycoplasma penetrans. J Bacteriol 185:231–242. doi: 10.1128/JB.185.1.231-242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGowin CL, Ma L, Martin DH, Pyles RB. 2009. Mycoplasma genitalium-encoded MG309 activates NF-κB via Toll-like receptors 2 and 6 to elicit proinflammatory cytokine secretion from human genital epithelial cells. Infect Immun 77:1175–1181. doi: 10.1128/IAI.00845-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pflaum K, Tulman ER, Beaudet J, Liao X, Geary SJ. 2015. Global changes in Mycoplasma gallisepticum phase-variable lipoprotein gene vlhA expression during in vivo infection of the natural chicken host. Infect Immun 84:351–355. doi: 10.1128/IAI.01092-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Güell M, van Noort V, Yus E, Chen WH, Leigh-Bell J, Michalodimitrakis K, Yamada T, Arumugam M, Doerks T, Kühner S, Rode M, Suyama M, Schmidt S, Gavin AC, Bork P, Serrano L. 2009. Transcriptome complexity in a genome-reduced bacterium. Science 326:1268–1271. doi: 10.1126/science.1176951. [DOI] [PubMed] [Google Scholar]
- 54.Dorman CJ. 2011. Regulation of transcription by DNA supercoiling in Mycoplasma genitalium: global control in the smallest known self-replicating genome. Mol Microbiol 81:302–304. doi: 10.1111/j.1365-2958.2011.07718.x. [DOI] [PubMed] [Google Scholar]
- 55.Torres-Puig S, Broto A, Querol E, Piñol J, Pich OQ. 2015. A novel sigma factor reveals a unique regulon controlling cell-specific recombination in Mycoplasma genitalium. Nucleic Acids Res 43:4923–4936. doi: 10.1093/nar/gkv422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Biberfeld G, Biberfeld P. 1970. Ultrastructural features of Mycoplasma pneumoniae. J Bacteriol 102:855–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cloward JM, Krause DC. 2011. Loss of co-chaperone TopJ impacts adhesin P1 presentation and terminal organelle maturation in Mycoplasma pneumoniae. Mol Microbiol 81:528–539. doi: 10.1111/j.1365-2958.2011.07712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kawamoto A, Matsuo L, Yamamoto H, Namba K, Miyata M. 2016. Periodicity in attachment organelle revealed by electron cryotomography suggests conformational changes in gliding mechanism of Mycoplasma pneumoniae. mBio 7:e00243-16. doi: 10.1128/mBio.00243-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.García-Morales L, González-González L, Querol E, Piñol J. 2016. A minimized motile machinery for Mycoplasma genitalium. Mol Microbiol 100:125–138. doi: 10.1111/mmi.13305. [DOI] [PubMed] [Google Scholar]
- 60.Krause DC, Balish MF. 2004. Cellular engineering in a minimal microbe: structure and assembly of the terminal organelle of Mycoplasma pneumoniae. Mol Microbiol 51:917–924. doi: 10.1046/j.1365-2958.2003.03899.x. [DOI] [PubMed] [Google Scholar]
- 61.Hasselbring BM, Jordan JL, Krause RW, Krause DC. 2006. Terminal organelle development in the cell wall-less bacterium Mycoplasma pneumoniae. Proc Natl Acad Sci U S A 103:16478–16483. doi: 10.1073/pnas.0608051103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tully JG, Rose DL, Whitcomb RF, Wenzel RP. 1979. Enhanced isolation of Mycoplasma pneumoniae from throat washings with a newly-modified culture medium. J Infect Dis 139:478–482. doi: 10.1093/infdis/139.4.478. [DOI] [PubMed] [Google Scholar]
- 63.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 64.Eismann T, Huber N, Shin T, Kuboki S, Galloway E, Wyder M, Edwards MJ, Greis KD, Shertzer HG, Fisher AB, Lentsch AB. 2009. Peroxiredoxin-6 protects against mitochondrial dysfunction and liver injury during ischemia-reperfusion in mice. Am J Physiol Gastrointest Liver Physiol 296:G266–G274. doi: 10.1152/ajpgi.90583.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wolf E, Kim PS, Berger B. 1997. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci 6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hatchel JM, Balish RS, Duley ML, Balish MF. 2006. Ultrastructure and gliding motility of Mycoplasma amphoriforme, a possible human respiratory pathogen. Microbiology 152:2181–2189. doi: 10.1099/mic.0.28905-0. [DOI] [PubMed] [Google Scholar]
- 67.Suloway C, Shi J, Cheng A, Pulokas J, Carragher B, Potter CS, Zheng SQ, Agard DA, Jensen GJ. 2009. Fully automated, sequential tilt-series acquisition with Leginon. J Struct Biol 167:11–18. doi: 10.1016/j.jsb.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CJ, Tjaden B. 2013. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res 41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pritchard RE, Balish MF. 2015. Mycoplasma iowae: relationships among oxygen, virulence, and protection from oxidative stress. Vet Res 46:36. doi: 10.1186/s13567-015-0170-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jensen GJ, Briegel A. 2007. How electron cryotomography is opening a new window onto prokaryotic ultrastructure. Curr Opin Struct Biol 17:260–267. doi: 10.1016/j.sbi.2007.03.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.