ABSTRACT
Flavobacterium johnsoniae and many related bacteria secrete proteins across the outer membrane using the type IX secretion system (T9SS). Proteins secreted by T9SSs have amino-terminal signal peptides for export across the cytoplasmic membrane by the Sec system and carboxy-terminal domains (CTDs) targeting them for secretion across the outer membrane by the T9SS. Most but not all T9SS CTDs belong to the family TIGR04183 (type A CTDs). We functionally characterized diverse CTDs for secretion by the F. johnsoniae T9SS. Attachment of the CTDs from F. johnsoniae RemA, AmyB, and ChiA to the foreign superfolder green fluorescent protein (sfGFP) that had a signal peptide at the amino terminus resulted in secretion across the outer membrane. In each case, approximately 80 to 100 amino acids from the extreme carboxy termini were needed for efficient secretion. Several type A CTDs from distantly related members of the phylum Bacteroidetes functioned in F. johnsoniae, supporting the secretion of sfGFP by the F. johnsoniae T9SS. F. johnsoniae SprB requires the T9SS for secretion but lacks a type A CTD. It has a conserved C-terminal domain belonging to the family TIGR04131, which we refer to as a type B CTD. The CTD of SprB was required for its secretion, but attachment of C-terminal regions of SprB of up to 1,182 amino acids to sfGFP failed to result in secretion. Additional features outside the C-terminal region of SprB may be required for its secretion.
IMPORTANCE Type IX protein secretion systems (T9SSs) are common in but limited to members of the phylum Bacteroidetes. Most proteins that are secreted by T9SSs have conserved carboxy-terminal domains that belong to the protein domain family TIGR04183 (type A CTDs) or TIGR04131 (type B CTDs). Here, we identify features of T9SS CTDs of F. johnsoniae that are required for protein secretion and demonstrate that type A CTDs from distantly related members of the phylum function with the F. johnsoniae T9SS to secrete the foreign protein sfGFP. In contrast, type B CTDs failed to target sfGFP for secretion, suggesting a more complex association with the T9SS.
KEYWORDS: Bacteroidetes, protein secretion
INTRODUCTION
Many members of the phylum Bacteroidetes secrete proteins across the outer membrane using the type IX secretion system (T9SS) (1, 2). The components of the T9SS were first identified and characterized in Porphyromonas gingivalis and Flavobacterium johnsoniae (3, 4) and have more recently been studied in other members of the phylum Bacteroidetes (5–7). Components of the secretion system (Fig. 1) include GldK, GldL, GldM, GldN, SprA, SprE, SprT, PorU, and PorV (referred to as PorK, PorL, PorM, PorN, Sov, PorW, PorT, PorU, and PorV, respectively, in P. gingivalis). F. johnsoniae also has a paralog of GldN, GldO, and deletion of the genes encoding both proteins is required for complete loss of secretion (8). An additional component, PorZ, was also recently described in P. gingivalis (9). Most of the proteins listed above are unique to members of the phylum Bacteroidetes. Some of the components of the T9SS were first identified in F. johnsoniae as proteins that are required for gliding motility (10, 11), and these were later realized to function in protein secretion in both F. johnsoniae and in P. gingivalis (3, 12, 13). The F. johnsoniae cell surface motility adhesins SprB and RemA require the T9SS for their delivery across the outer membrane (3, 13, 14). Rapid movement of SprB and RemA along the cell surface is responsible for cell movement (15, 16). Mutations in the T9SS genes prevent proper assembly of the motility apparatus. Some components of the T9SS may also have more direct functions in motility (13, 17). Most but not all bacteria that have T9SSs exhibit gliding motility (1). P. gingivalis is one of the exceptions, as it has a T9SS but is nonmotile.
FIG 1.
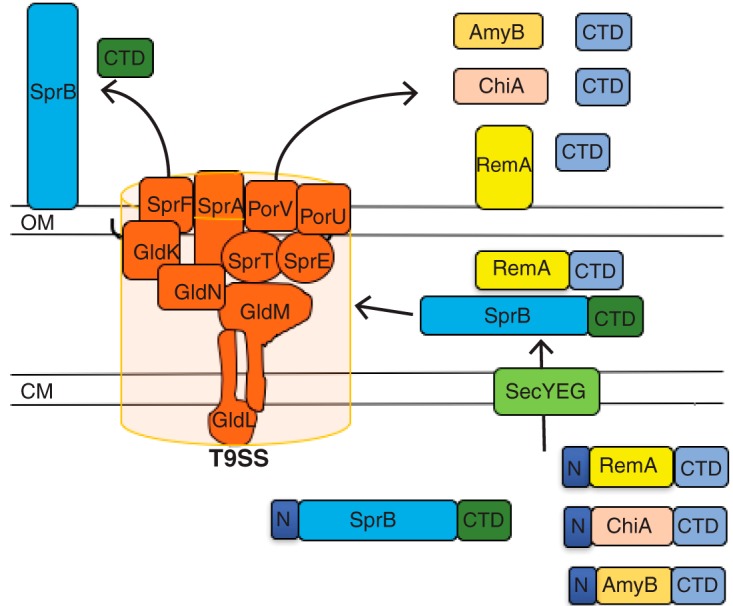
The F. johnsoniae type IX protein secretion system. Proteins required for secretion include the cytoplasmic membrane proteins GldL and GldM and the outer membrane-associated proteins GldK, GldN, SprA, SprE, and SprT. SprF is required for the secretion of SprB but not for the secretion of other proteins by the T9SS. Proteins secreted by T9SSs have type A (blue) or type B (green) C-terminal domains (CTDs) required for secretion. PorV appears to be required for the secretion of proteins that have type A CTDs but is not needed for the secretion of SprB or for the secretion of some other proteins that have type B CTDs. Black lines on the lipoproteins GldK and SprE indicate lipid tails. OM, outer membrane; CM, cytoplasmic membrane.
Proteins secreted by T9SSs have N-terminal signal peptides allowing export across the cytoplasmic membrane by the Sec system (2, 18, 19). They also typically have conserved C-terminal domains (CTDs) that are thought to target them to the T9SS for secretion across the outer membrane (2, 18–20). Once delivered across the outer membrane, some proteins are released in soluble form (18, 21), whereas others are modified and attached to the cell surface (2, 3, 13, 22). The CTDs are removed during or after secretion. In P. gingivalis, this is thought to involve PorU, which has been described as a C-terminal signal peptidase (23). The secretion process and attachment of secreted proteins to the cell surface are somewhat reminiscent of Gram-positive bacterial sortase-mediated secretion and cell surface localization, although the proteins involved are not related in sequence (22). T9SS CTDs typically fall into one of two protein domain families corresponding to TIGR04183 (here referred to as type A CTDs) or TIGR04131 (here referred to as type B CTDs) (18, 19). The conserved CTDs allow predictions to be made regarding the number of proteins secreted by the T9SSs of individual bacterial species. For example, F. johnsoniae and Cytophaga hutchinsonii are predicted to secrete 53 and 147 proteins, respectively, using their T9SSs (7, 13). Secretion of many of these proteins has been verified experimentally (2, 18). These predictions may underestimate the actual number of secreted proteins, since some proteins secreted by T9SSs, such as F. johnsoniae ChiA, have CTDs that are required for secretion but exhibit no obvious sequence similarity to either type A or type B CTDs (21).
T9SS CTDs have been functionally characterized for two P. gingivalis proteins, the gingipain protease RgpB, and hemin-binding protein 35 (HBP35) (20, 24–26). RgpB and HBP35 both have type A CTDs. HBP35 required the C-terminal 22 amino acids (aa) for secretion and cell surface attachment (25). Deletion of this region or modification of a conserved lysine within this region resulted in decreased secretion. Attachment of C-terminal regions to GFP allowed secretion of the foreign protein GFP by P. gingivalis. Analysis of CTDs from F. johnsoniae has been limited to a single protein, the soluble chitinase ChiA (21). As indicated above, ChiA requires the T9SS for secretion but lacks a recognizable conserved CTD. Nevertheless, attachment of the C-terminal 105 aa of ChiA to the foreign protein mCherry resulted in the secretion of mCherry from the cell (21). Here, we functionally examine the C-terminal regions of five diverse F. johnsoniae proteins that require the T9SS for secretion. These included two type A CTDs, two type B CTDs, and the ChiA CTD. We also demonstrate that some CTDs from distantly related members of the phylum Bacteroidetes can function with the F. johnsoniae T9SS.
RESULTS
Prevalence of T9SSs and T9SS-associated CTDs.
Genome analyses indicate that T9SSs are found in most but not all members of the phylum Bacteroidetes (see Table S3 in the supplemental material). Individual strains of 104 different species with complete genome sequences were analyzed. This included members of 65 genera. Members of 60 genera and 90 of the 104 species had each of the five core T9SS genes (gldK, gldL, gldM, gldN, and sprA) that have been assigned to TIGRFAM families. Although most members of the Bacteroidetes have T9SSs, a few do not. For example, most members of the well-studied genus Bacteroides lack T9SS genes (Table S3) and use other mechanisms to secrete proteins (1, 27, 28).
The 104 genomes of Bacteroidetes species were also analyzed for proteins with predicted T9SS CTDs belonging to TIGR04183 (type A CTDs) and TIGR04131 (type B CTDs). Of the 90 species and strains that had each of the T9SS genes, all had proteins with type A CTDs, and all but one (Arachidicoccus sp. strain BS20) also had proteins with type B CTDs. Most of these had many T9SS CTD-containing proteins. Fluviicola taffensis had the highest number of each, with 180 proteins with type A CTDs and 50 proteins with type B CTDs. The 14 Bacteroidetes species that lacked components of T9SSs were also examined, and none had predicted proteins with type A or type B CTDs. The results demonstrate a strong correlation between both types of CTDs and the T9SS.
Members of other phyla of bacteria were also analyzed, including 3,777 complete genome sequences (Table S4). None had orthologs of gldK, gldL, gldM, or gldN, but seven species had orthologs of sprA. These were members of the phyla Rhodothermaeota, Chlorobi, Fibrobacteres, Ignavibacteriae, and Gemmatimonadetes. None of the 3,777 non-Bacteroidetes bacterial genomes were predicted to encode proteins with type B CTDs, but 11 species had predicted proteins with type A CTDs. Interestingly, six of the seven species mentioned above that had SprA each had many proteins with type A CTDs (ranging from 18 to 147), suggesting a link between type A CTDs and the large outer membrane protein SprA. As indicated above, there were only five other bacterial species that had any predicted type A CTDs. Each of these had a single example. Three of the apparent CTDs from these five bacteria were not found at the C terminus, suggesting that they may have been false positives or may have functions unrelated to those of the Bacteroidetes CTDs. The high prevalence within the phylum Bacteroidetes of orthologs of GldK, GldL, GldM, and GldN and of proteins with type B CTDs, as well as their complete absence outside the phylum Bacteroidetes are indicative of the extremely low number of false positives obtained using the trusted cutoffs of the TIGRFAM assignments. The near absence of type A CTDs in bacteria lacking SprA also attests to the low level of false positives and indicates a strong correlation between the presence of the outer membrane protein SprA and proteins with type A CTDs.
Members of the archaea and eukarya were also examined for orthologs of T9SS components. Two hundred eighteen completed archaeal genome sequences and 36 completed eukaryal genomes were analyzed, but none had orthologs of gldK, gldL, gldM, gldN, or sprA, and none had predicted proteins with type A or type B CTDs. The eukaryotes examined included protists, fungi, plants (Arabidopsis thaliana and Zea mays) and animals (Caenorhabditis elegans, Danio rerio, and Homo sapiens).
Characteristics of F. johnsoniae type A and type B CTDs.
Multiple-sequence alignments of F. johnsoniae type A and type B CTDs were assembled to identify conserved features (Fig. S1 and S2). The conserved regions of type A and type B CTDs extend approximately 70 to 100 amino acids from the C termini of the proteins. F. johnsoniae type A CTDs have several highly conserved regions, including YPNP approximately 70 amino acids from the C terminus, G(I/L/V)Y approximately 20 amino acids from the C terminus, and (K/R)(I/L/V)(I/L/V)K followed immediately by the C-terminal residue. These sequences are also conserved in type A CTDs from many other Bacteroidetes (data not shown). F. johnsoniae type B CTDs are quite different in sequence, with F(T/S)PNGDGXND approximately 80 amino acids from the C terminus, IFXR(W/Y)G approximately 55 amino acids from the C terminus, and GX(L/F)X(L/I)X(R/K) at the C terminus.
Growth phase of cells affects function of the T9SS.
To examine the function of the T9SS, we fused the C-terminal 97 amino acids of RemA to the C terminus of the foreign superfolder green fluorescent protein (sfGFP), resulting in the fusion protein SPRemA-sfGFP-CTDRemA. The recombinant protein also carried the N-terminal signal peptide from RemA to facilitate export across the cytoplasmic membrane by the Sec system. Little sfGFP accumulated in the growth medium of wild-type cells growing exponentially in rich medium, whereas cells from stationary phase (approximately 8 h post-exponential phase) exhibited substantial secretion of SPRemA-sfGFP-CTDRemA (Fig. 2). The size of the protein in the culture fluid corresponded to that of sfGFP, suggesting that both the N-terminal signal peptide and the CTD had been removed. The presence of sfGFP in the spent medium was not the result of cell lysis, since analysis of total proteins by Coomassie blue staining revealed only trace amounts of protein in the culture fluid (Fig. S3). Further, the coexpression of SP-sfGFP and SP-mCherry-CTDRemA allowed secretion of mCherry but not leakage of sfGFP (Fig. S4), indicating that expression of excess CTD-containing protein from plasmid did not cause nonspecific T9SS-mediated leakage of periplasmic proteins. SPRemA-sfGFP-CTDRemA depended on the components of the T9SS for its secretion, since it was not secreted by cells of the ΔgldNO T9SS mutant (Fig. 2B). Mutations in genes encoding other core components of the T9SS (gldK, gldL, gldM, sprA, sprE, and sprT) also resulted in a lack of secretion (Fig. S5). The levels of components of the T9SS were examined in an attempt to explain the low level of secretion from exponentially growing cells. Exponentially growing cells had similar levels of GldK, GldL, GldM, GldN, and SprE, as did stationary-phase cells, but they had reduced levels of SprA (Fig. 3). SprA is important for secretion, and the low level present in cells growing exponentially in rich medium may explain the lack of secretion described above. Although lack of a suitable regulatable promoter prevented overexpression of SprA in exponential cells, increased levels of SprA in stationary-phase cells carrying sprA on a plasmid resulted in apparent increased levels of secretion of SP-sfGFP-CTDRemA (Fig. S6). Because of the lack of secretion during exponential growth, all subsequent analyses of secretion involved cells grown to approximately 8 h post-exponential phase.
FIG 2.
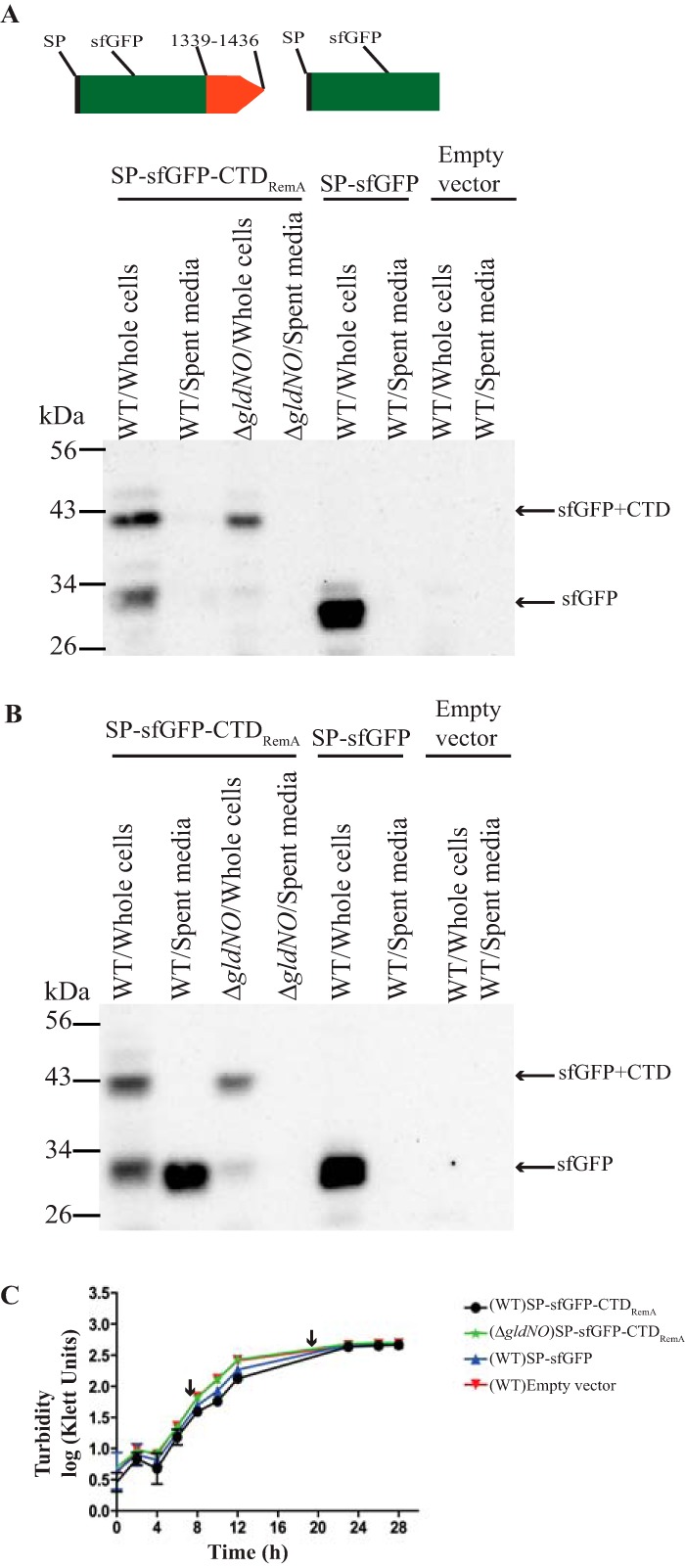
T9SS-mediated secretion of sfGFP fused to the CTD of RemA. Cultures of wild-type (WT) cells or of the T9SS ΔgldNO mutant were incubated in CYE at 25°C with shaking and harvested in exponential phase (10 h) and stationary phase (22 h). One-milliliter samples were centrifuged at 22,000 × g for 15 min. The culture supernatant (spent medium) and intact cells were analyzed for sfGFP by Western blotting. Cells carried either pCP23 (empty vector); pSK37, which expresses sfGFP with the N-terminal signal peptide from RemA (SP-sfGFP); or pSK30, which expresses SP-sfGFP fused to the 97-amino-acid CTD of RemA (SP-sfGFP-CTDRemA). Cartoons at the top indicate the plasmid-encoded proteins and apply to panels A and B, with 1339-1436 indicating the C-terminal 97 amino acids of RemA and SP indicating signal peptide. Whole-cell samples corresponded to 10 μg of protein per lane, and samples from spent medium corresponded to the volume of spent medium that contained 10 μg of cell protein before the cells were removed. Samples were separated by SDS-PAGE, and sfGFP was detected using antiserum against GFP. (A) Detection of sfGFP from whole cells and spent medium for samples harvested from the exponential phase of growth. (B) Detection of sfGFP from whole cells and spent medium for samples harvested from stationary phase. (C) Growth curves, with arrows indicating the points at which cells were harvested. Growth curves were determined in triplicate, and the error bars indicate standard deviation.
FIG 3.
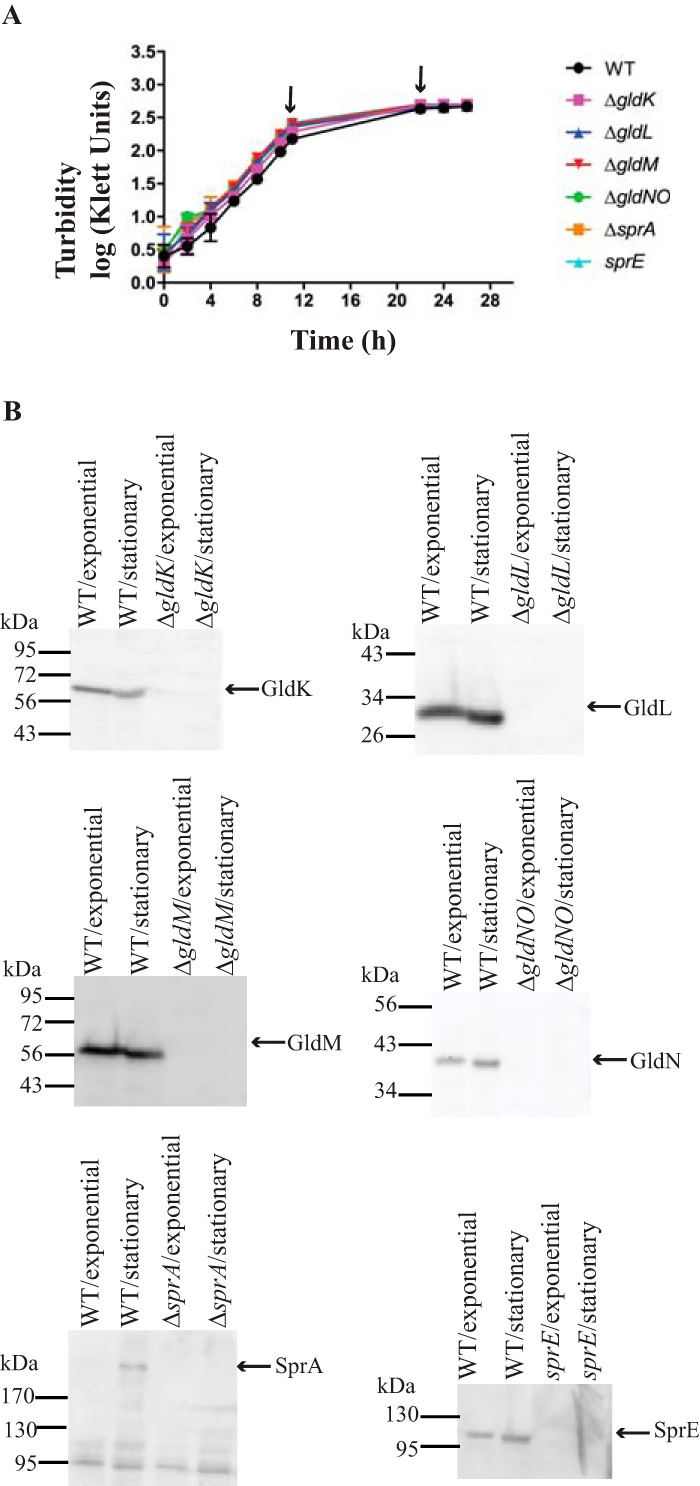
Levels of T9SS proteins in cells grown to exponential and stationary phases. (A) Cultures of wild-type cells or of cells of the T9SS ΔgldK, ΔgldL, ΔgldM, ΔgldNO, and ΔsprA mutants and of the sprE mutant FJ149 were incubated in CYE at 25°C with shaking and harvested in late-exponential phase (11 h) and stationary phase (22 h), as indicated by the arrows. Growth curves were determined in triplicate, and the error bars indicate standard deviation. (B) Cells were analyzed for T9SS proteins by Western blotting. Equal amounts (10 μg) of cell protein were loaded per lane and separated by SDS-PAGE, and antibodies were used to detect the respective proteins.
Identification of regions of the CTDs of F. johnsoniae RemA, AmyB, and ChiA that allow secretion of the foreign protein sfGFP.
To elucidate the features of type A CTDs that are important for secretion, we examined the F. johnsoniae proteins RemA and AmyB (Fjoh_1208). Different lengths of CTDRemA were attached to sfGFP to determine the minimal region needed for secretion. C-terminal regions of 97 and 87 amino acids facilitated the secretion of sfGFP, whereas a region spanning the final 62 amino acids of RemA did not (Fig. 4A). sfGFP carrying the 62-amino acid CTD appeared to be unstable, since decreased amounts of sfGFP were detected. In contrast, sfGFP carrying no CTD attachment was stable (Fig. 2). We do not know the reason for the instability of sfGFP carrying the final 62 amino acids of RemA. It might be due to improper folding of the fusion protein resulting in degradation by periplasmic proteases, although other explanations are possible.
FIG 4.
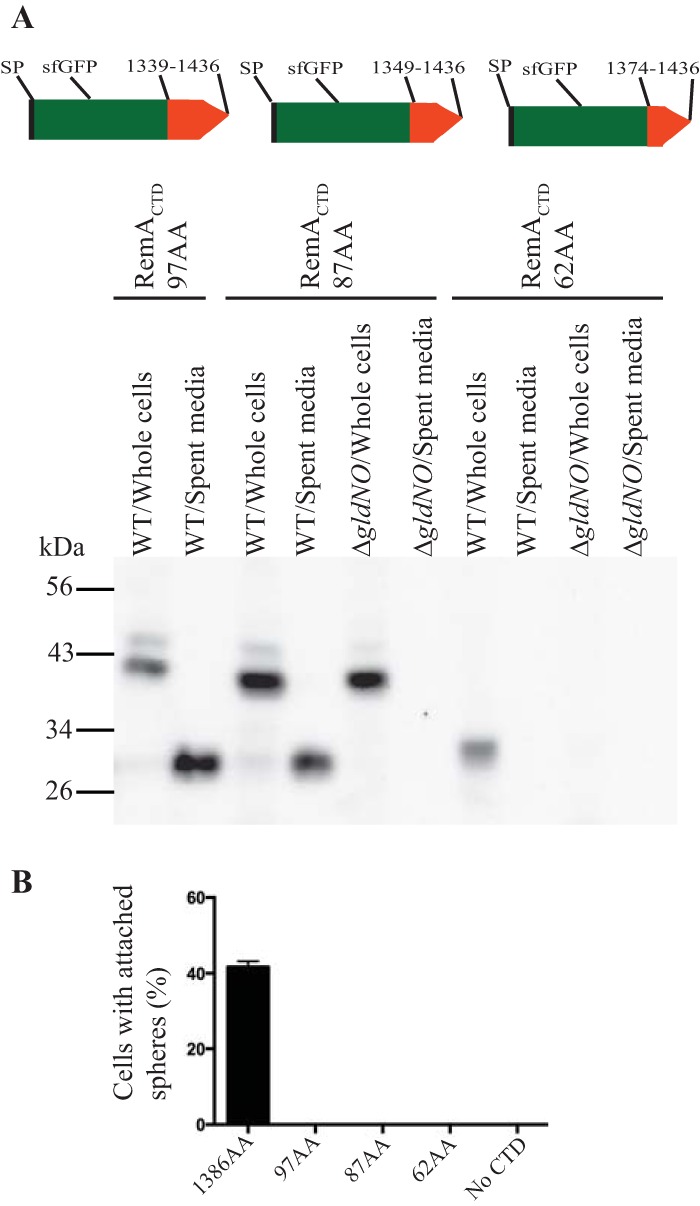
Determination of the region of RemA needed to allow secretion of sfGFP. (A) The minimum region necessary for sfGFP secretion was determined by examining cells carrying plasmids that expressed SPRemA-sfGFP-CTDRemA with CTD regions of 62 amino acids (pSK81), 87 amino acids (pSK71), and 97 amino acids (pSK30). Cartoons at the top indicate the plasmid-encoded proteins carrying CTDs extending from amino acids 1339 to 1436, 1349 to 1436, and 1374 to 1436 of RemA. Cell-free spent media and whole cells were examined for sfGFP by SDS-PAGE, followed by Western blotting using antiserum against sfGFP. Whole-cell samples corresponded to 10 μg of protein per lane, and samples from spent media corresponded to the volume of spent medium that contained 10 μg of cell protein before the cells were removed. (B) Attachment of sfGFP on the cell surface was determined by examining cells carrying plasmids that expressed SPRemA-sfGFP-CTDRemA with CTD regions of 62 amino acids, 87 amino acids, 97 amino acids, or 1,386 amino acids (pYT180). Anti-GFP antiserum and 0.5-μm-diameter protein G-coated polystyrene spheres were added to cells, as described in Materials and Methods. Samples were introduced into a tunnel slide, incubated for 3 min at 25°C, and examined using a phase-contrast microscope. Images were recorded for 30 s, and 100 randomly selected cells were examined for the presence of spheres that remained attached to the cells during this time. Error bars indicate standard deviations from three measurements.
In wild-type cells, RemA is found both on the cell surface and in soluble form in the spent medium (18). We examined cells expressing sfGFP fused to various regions of RemA to determine if sfGFP was targeted to the cell surface. This was done by incubating cells with protein G-coated latex spheres carrying antibodies against GFP and determining the percentage of cells to which spheres attached. Whereas full-length RemA fused to sfGFP (RemA C-terminal region of 1,386 amino acids) resulted in the attachment of spheres indicating surface localization, none of the other fusion proteins containing C-terminal regions of 97, 87, or 62 amino acids did (Fig. 4B and S7).
The constructs described above each had the N-terminal signal peptide from RemA. In order to determine if this region had any role in secretion by the T9SS beyond targeting to the Sec system for initial export across the cytoplasmic membrane, we replaced it with the N-terminal signal peptide from the predicted F. johnsoniae periplasmic cytochrome c, Fjoh_1634. The cytochrome c signal peptide allowed the secretion of SPFjoh_1634-sfGFP-CTDRemA (carrying 97 amino acids of the RemA CTD) by the T9SS (Fig. 5), suggesting that any cleavable N-terminal signal peptide that facilitates export across the cytoplasmic membrane may be sufficient to allow secretion across the outer membrane by the T9SS, provided that an appropriate CTD is present. Such secretion was not observed in the ΔgldNO mutant, indicating that the T9SS was required.
FIG 5.
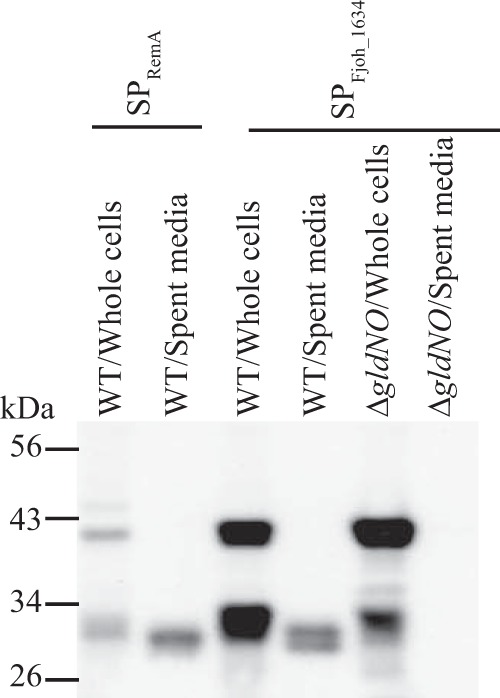
The N-terminal signal peptide from a periplasmic protein allows secretion of SP-sfGFP-CTDRemA. Cell-free spent medium and whole cells were analyzed for wild-type (WT) cells carrying fusion plasmid pSK30, which expresses SPRemA-sfGFP-CTDRemA, or carrying pSK84, which expresses SPFjoh_1634-sfGFP-CTDRemA. Both fusion proteins had 97-amino-acid C-terminal regions of RemA. Cells and spent media from cultures of the T9SS ΔgldNO mutant carrying pSK84 were also analyzed. Whole-cell samples corresponded to 10 μg of protein per lane, and samples from spent media corresponded to the volume of spent medium that contained 10 μg of cell protein before the cells were removed. Samples were separated by SDS-PAGE, and sfGFP was detected using antiserum against GFP.
The extreme C terminus of CTDRemA was explored in more detail. Attachment of amino acids 1339 to 1424 of RemA (97-aa CTD minus the C-terminal 12 aa) failed to support the secretion of SPRemA-sfGFP (Fig. 6A). The final 12 amino acids have a lysine residue that is highly conserved among type A CTDs, including those from F. johnsoniae (Fig. S1). Replacement of this lysine with alanine resulted in a failure to secrete SPRemA-sfGFP, suggesting that it may be important in secretion by the F. johnsoniae T9SS (Fig. 6B). Similar results have previously been reported for the distantly related bacteroidete P. gingivalis. Deletion of the C-terminal 13 amino acids or 2 amino acids of gingipain RgpB resulted in an accumulation of the truncated protein in the periplasm (20, 24), and conversion of the conserved lysine to alanine at amino acid 732 also resulted in decreased secretion (20). Amino acids near the C terminus appear to be important components of the Bacteroidetes T9SS secretion signal.
FIG 6.
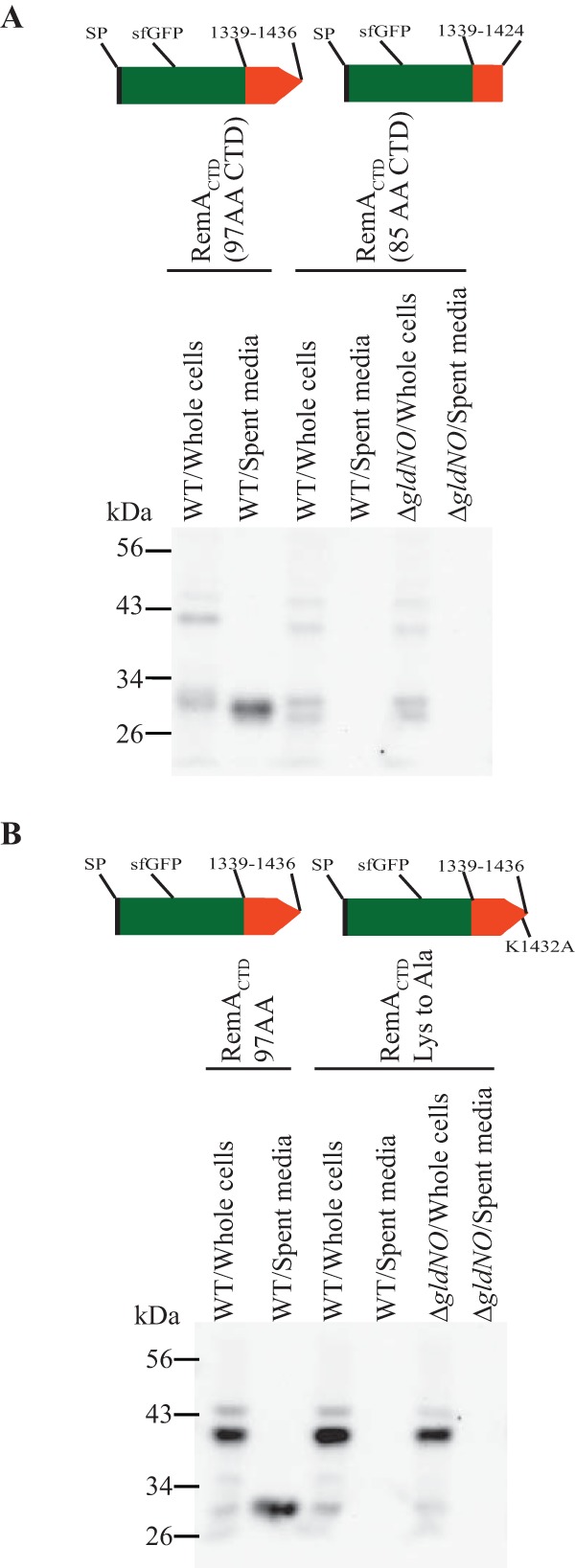
The C-terminal 12 amino acids of RemA are critical for secretion. Cells carrying pSK30, which expresses SPRemA-sfGFP fused to the C-terminal 97 amino acids of RemA (amino acids 1339 to 1436), pSK79, which expresses SPRemA-sfGFP fused to amino acids 1339 to 1424 of RemA but lacking the C-terminal 12 amino acids, and pSK91, which expresses SPRemA-sfGFP fused to the C-terminal 97 amino acids of RemA but with the conserved lysine 1432 (Fig. S1) replaced by alanine, were examined for sfGFP in intact cells and in cell-free spent medium by Western blotting. Cultures of wild-type (WT) and a T9SS mutant (ΔgldNO) carrying the plasmids were analyzed. Whole-cell samples corresponded to 10 μg of protein per lane, and samples from spent media corresponded to the volume of spent medium that contained 10 μg of cell protein before the cells were removed. (A) Effect of deletion of the C-terminal 12 amino acids of SPRemA-sfGFP-CTDRemA on secretion. (B) Effect on secretion of replacement of conserved lysine 1432 of CTDRemA with alanine.
PorU and PorV are involved in the secretion of some but not all proteins that are targeted to the T9SS (18, 23, 29). PorU has been suggested to function as a C-terminal signal peptidase, whereas the function of PorV is less certain. RemA requires PorV but not PorU for secretion by the T9SS (18). A porV deletion mutant failed to secrete SPRemA-sfGFP carrying the 97-amino-acid CTDRemA (Fig. 7). Cells of a porU deletion mutant secreted some SPRemA-sfGFP-CTDRemA but less than did wild-type cells. F. johnsoniae PorU is not essential for secretion but appears to allow more efficient secretion.
FIG 7.
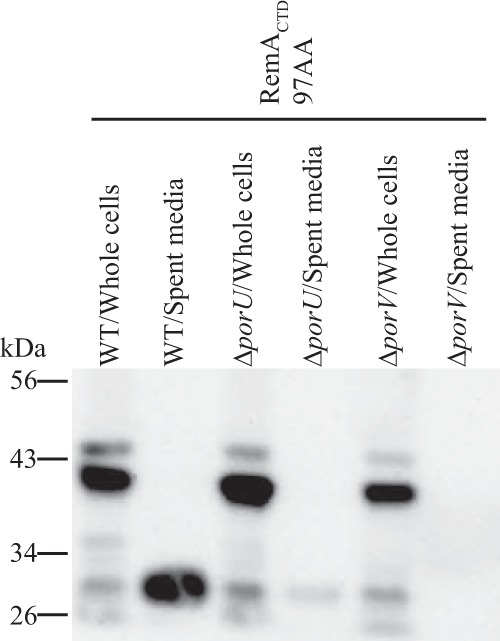
Effect of deletion of porU and porV on secretion of SPRemA-sfGFP-CTDRemA. Cell-free spent medium and whole cells were analyzed for cells of the wild type (WT), porU deletion mutant (ΔporU), and porV deletion mutant (ΔporV), each carrying pSK30. pSK30 expresses SPRemA-sfGFP-CTDRemA, where CTDRemA refers to the 97-amino-acid C-terminal region of RemA. Samples were separated by SDS-PAGE, and sfGFP was detected using antiserum against GFP. Whole-cell samples corresponded to 10 μg protein per lane, and samples from spent media corresponded to the volume of spent medium that contained 10 μg of cell protein before the cells were removed.
The results described above indicate that the C-terminal 87-amino-acid region of RemA is sufficient to target a foreign protein carrying an N-terminal signal peptide for secretion by the T9SS. Similar regions of the CTDs of F. johnsoniae AmyB (Fjoh_1208) and ChiA were examined for their ability to facilitate the secretion of SPRemA-sfGFP. The CTD of AmyB is a member of the protein domain family TIGR04183 (type A CTD) and is much closer to the consensus type A CTD sequence than is the CTD of RemA (Fig. S1). In contrast, the ChiA CTD is not similar in sequence to members of this or any other protein domain family (21). The 99-amino-acid CTDAmyB facilitated the efficient secretion of SPRemA-sfGFP, whereas the 73-amino-acid CTDAmyB did not (Fig. 8A). For ChiA, the 105-amino-acid CTDChiA and 79-amino-acid CTDChiA both facilitated the secretion of SPRemA-sfGFP by the T9SS (Fig. 8B). Attachment of the 79-amino-acid region of ChiA had a negative effect on the accumulation of sfGFP in whole cells. In contrast, a 62-amino-acid CTDChiA failed to support secretion but allowed the accumulation of sfGFP within the cells. The 79-amino-acid CTDChiA may have interacted with components of the T9SS in a way that destabilized the attached sfGFP, perhaps making it susceptible to proteolysis, although other explanations are possible.
FIG 8.
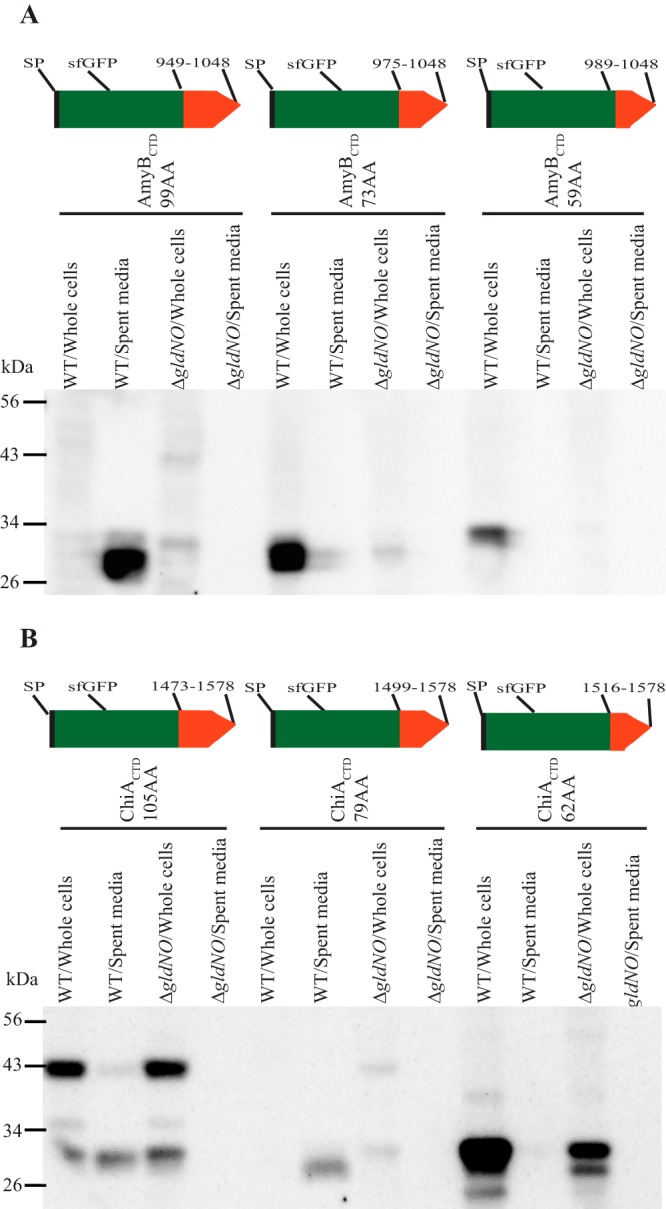
T9SS-mediated secretion of SPRemA-sfGFP-CTDAmyB and SPRemA-sfGFP-CTDChiA. Cell-free spent media and whole cells were analyzed for wild-type (WT) cells and for cells of the T9SS ΔgldNO mutant. Cells carried plasmids expressing either SPRemA-sfGFP-CTDAmyB (A) or SPRemA-sfGFP-CTDChiA (B), with CTD regions of various lengths. (A) pSK82, pSK85, and pSK86 produced SPRemA-sfGFP-CTDAmyB with CTD regions of 99, 73, and 59 amino acids, respectively, as indicated by the cartoons. (B) pCB3, pSK89, and pCB4 produced SPRemA-sfGFP-CTDChiA with CTD regions of 105, 79, and 62 amino acids, respectively, as indicated by the cartoons. Samples were separated by SDS-PAGE, and sfGFP was detected using antiserum against GFP. Whole-cell samples corresponded to 10 μg of protein per lane, and samples from spent media corresponded to the volume of spent medium that contained 10 μg of cell protein before the cells were removed.
Some type A CTDs from other Bacteroidetes facilitate secretion of sfGFP by the F. johnsoniae T9SS.
T9SSs and proteins with type A CTDs are common among members of the phylum Bacteroidetes (1). F. johnsoniae (class Flavobacteriia) has 40 proteins with type A CTDs, the marine bacterium Cellulophaga algicola DSM 14237 (class Flavobacteriia) has 13 proteins with type A CTDs, C. hutchinsonii ATCC 33406 (class Cytophagia) has 118 proteins with type A CTDs, and P. gingivalis ATCC 33277 (class Bacteroidia) has 17 proteins with type A CTDs (Table S3). We examined a single representative type A CTD from C. algicola, from C. hutchinsonii, and from P. gingivalis to determine if they support the secretion of SPRemA-sfGFP by the F. johnsoniae T9SS. Plasmids expressing sfGFP carrying the N-terminal RemA signal peptide and C-terminal regions from C. algicola Celal_2532 (AmyA), C. hutchinsonii Cel9B (30), and P. gingivalis RgpB (26) were introduced into F. johnsoniae. The CTD regions from C. algicola AmyA and C. hutchinsonii Cel9B functioned in F. johnsoniae, resulting in the secretion of sfGFP from wild-type cells (Fig. 9). They failed to support secretion in the ΔgldNO mutant, indicating that a functional T9SS was required. The results indicate that T9SS CTDs are not species specific. In contrast to the results described above, the CTD from P. gingivalis RgpB did not support the secretion of SPRemA-sfGFP (Fig. 9), suggesting that some foreign T9SS CTDs may not function properly with T9SSs from distantly related bacteria. The CTD of RgpB lacks the consensus YPNP sequence that is found in most F. johnsoniae type A CTD proteins at approximately 80 amino acids from the C terminus (Fig. S1), which may explain its inability to function with the F. johnsoniae T9SS.
FIG 9.
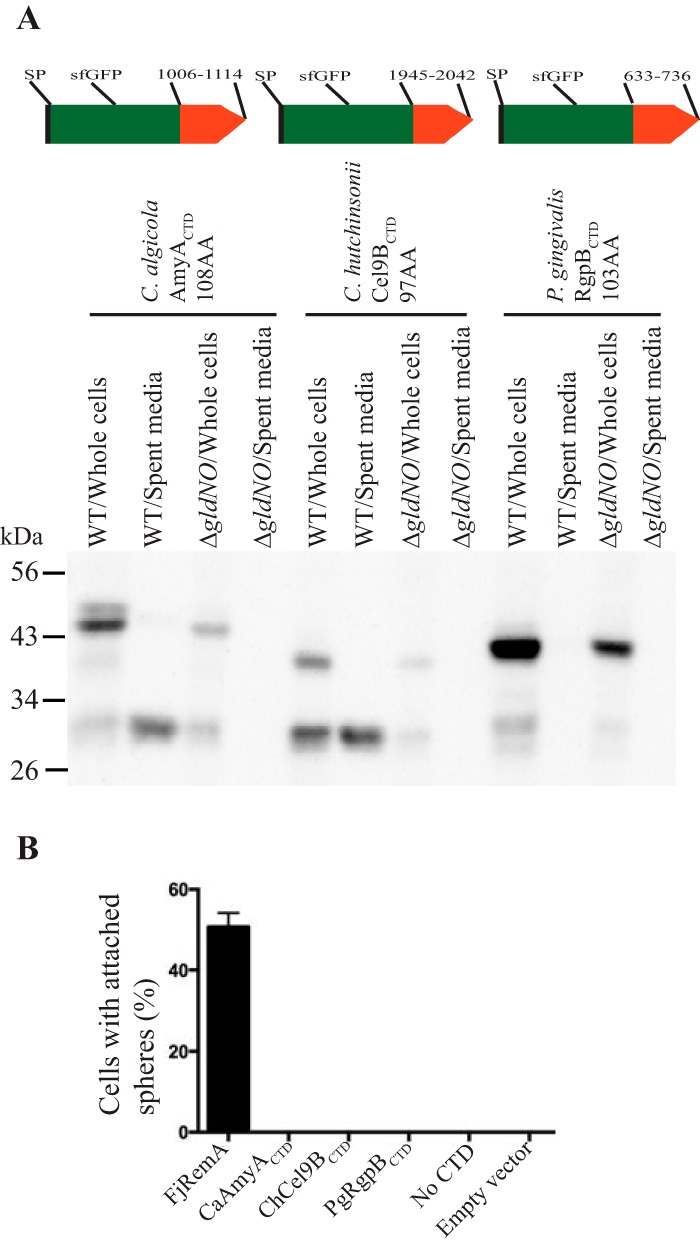
C-terminal regions from Cellulophaga algicola AmyA and Cytophaga hutchinsonii Cel9B target sfGFP for secretion by the F. johnsoniae T9SS. (A) Cell-free spent media and whole cells were analyzed for wild-type (WT) cells and for cells of the T9SS ΔgldNO mutant. Cells carried pSK65, pSK76, or pSK75 expressing SPRemA-sfGFP fused to the C-terminal region of C. algicola AmyA (108 amino acids), C. hutchinsonii Cel9B (97 amino acids), or P. gingivalis RgpB (103 amino acids), respectively. Samples were separated by SDS-PAGE, and sfGFP was detected using antiserum against GFP. Cell samples corresponded to 10 μg of protein per lane, and samples from spent media corresponded to the volume of spent medium that contained 10 μg of cell protein before the cells were removed. (B) To determine if any sfGFP was attached on the cell surface, anti-GFP antiserum and 0.5-μm-diameter protein G-coated polystyrene spheres were added to cells, as described in Materials and Methods. Samples were introduced into a tunnel slide, incubated for 3 min at 25°C, and examined using a phase-contrast microscope. Images were recorded for 30 s, and 100 randomly selected cells were examined for the presence of spheres that remained attached to the cells during this time. Error bars indicate standard deviations from three measurements. FjRemA is full-length RemA with sfGFP inserted immediately after the N-terminal signal peptide and was produced from pYT180. FjRemA was used as a positive control. “No CTD” refers to cells expressing SPRemA-sfGFP without a CTD, and “empty vector” refers to cells carrying pCP23.
The CTD of SprB is required for secretion but is not closely related to type A CTDs or to CTDChiA.
SprB has a type B CTD and requires the T9SS for its secretion to the cell surface (3, 8, 13). Previous results suggested that this region might be involved in secretion, because SprB and six other F. johnsoniae proteins that had type B CTDs were secreted by wild-type cells but not by cells of the T9SS ΔgldNO mutant (18). The genome analyses presented above also linked type B CTDs to the T9SS (Table S3). Finally, a role for CTDSprB in secretion is also suggested by the properties of mutant FJ117, which has a HimarEm2 transposon inserted 101 nucleotides upstream of the stop codon of sprB (16). FJ117 produces truncated SprB lacking the C-terminal 34 amino acids (Fig. 10A). The cell-associated truncated SprB protein was apparently not present on the cell surface (Fig. S8), suggesting that it was instead trapped inside the cells. sprF, which lies immediately downstream of and is cotranscribed with sprB, is required for SprB secretion (31). SprF protein was detected in wild-type cells and in cells of the FJ117 mutant (Fig. 10B), indicating that the failure to secrete SprB was not caused by a polar effect of the transposon on sprF. HimarEm2 has an internal promoter that was predicted to allow the expression of sprF (31), which explains the presence of SprF protein in the mutant. Since the defect in secretion of SprB was not caused by a lack of SprF, it is likely that the absence of the C-terminal 34 amino acids of SprB was responsible for the defect, lending further support for a role for this type B CTD in T9SS-mediated secretion.
FIG 10.
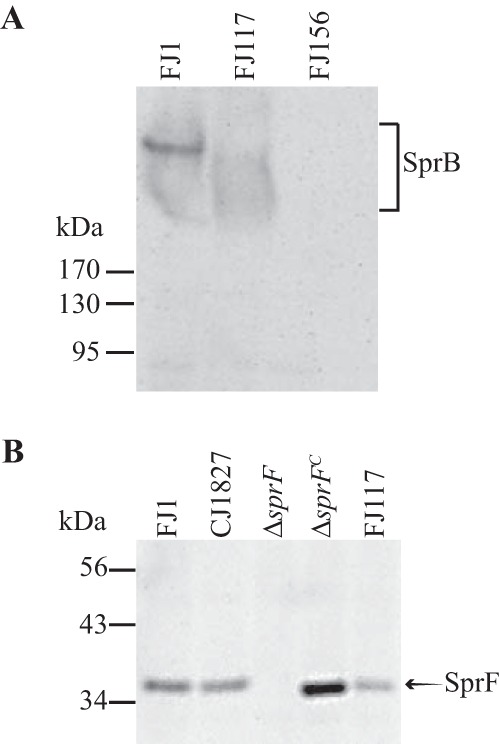
The C-terminal region of SprB is required for secretion. (A) Immunodetection of SprB from wild-type (WT) cells and from cells of the sprB transposon mutants FJ117 and FJ156 that should produce truncated SprB proteins lacking the C-terminal 34 and 5,880 amino acids, respectively. Total cell extracts were prepared by boiling in SDS loading buffer, and samples corresponding to 25 μg of protein per lane were analyzed for SprB by SDS-PAGE and Western blot analysis. (B) The transposon insertion in FJ117 is not polar on the downstream gene sprF. Cells were examined for SprF by SDS-PAGE and Western blot analysis using antiserum against SprF. Wild-type F. johnsoniae CJ1827 and FJ1, the ΔsprF mutant, the ΔsprF mutant complemented with pRR48 (ΔsprFc), and the transposon mutant FJ117 were examined. An equal amount (10 μg protein) of each sample was loaded in each lane.
C-terminal regions of SprB and of Fjoh_3952 fail to support secretion of sfGFP.
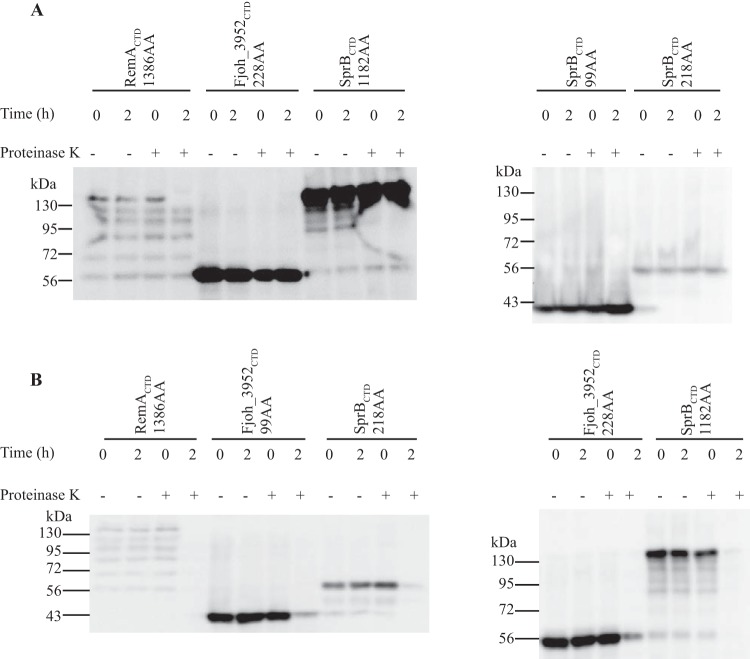
The highly conserved regions of type B CTDs extend approximately 80 to 100 amino acids from the C termini (Fig. S2). Plasmids were constructed that encode SPRemA-sfGFP with various lengths (99 to 1,182 amino acids) of the C-terminal region of SprB. None of these facilitated the secretion of sfGFP from wild-type cells (Fig. 11). sfGFP was not detected in the cell-free spent medium and was also not detected on intact cells using latex spheres coated with anti-sfGFP. In addition, the sfGFP fusion proteins in intact cells were not susceptible to proteinase K, further indicating that they were not secreted and attached on the cell surface (Fig. 12). We also examined the type B CTD of Fjoh_3952, which is secreted by wild-type cells but not by cells of a T9SS mutant (18). SPRemA-sfGFP carrying the C-terminal 228 aa of Fjoh_3952 failed to be secreted by wild-type cells (Fig. 11). The results indicate that whereas short (97 amino acids or less) type A CTDs were sufficient to target a foreign protein for secretion by the T9SS, even much longer regions of type B CTDs were not. Type B CTDs may require additional regions of the secreted protein to interact productively with the T9SS.
FIG 11.
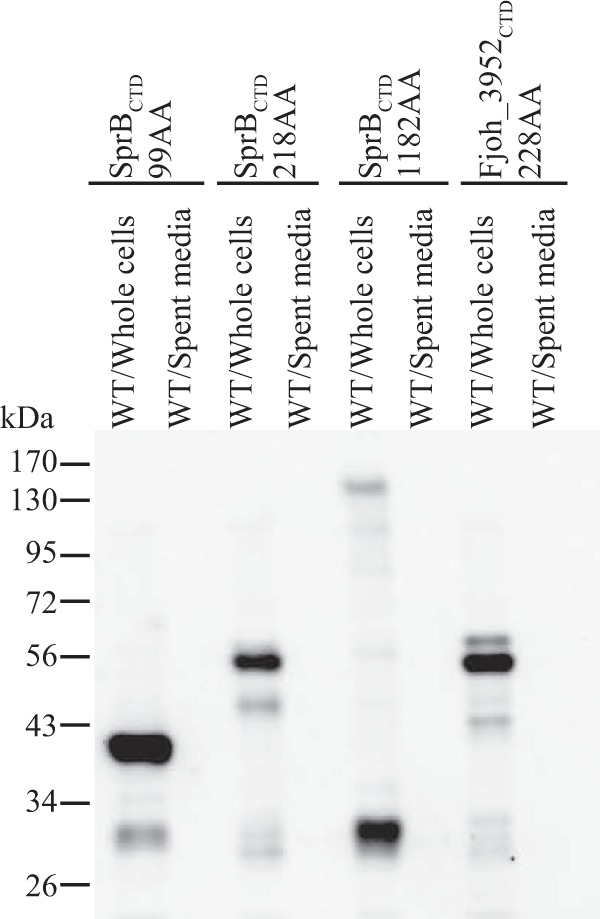
C-terminal regions of SprB fused to SPRemA-sfGFP failed to result in secretion of sfGFP. (A) Cell-free spent media and whole cells were analyzed for cultures of wild-type cells carrying fusion plasmids pSK93, pSK56, and pSK62, which express SPRemA-sfGFP with 99, 218, and 1,182-amino-acid C-terminal regions of SprB, respectively. Cells carrying fusion plasmid pSK58, which expresses SPRemA-sfGFP with the 228-amino-acid C-terminal region of the SprB-like protein Fjoh_3952, were also analyzed. Samples were separated by SDS-PAGE, and sfGFP was detected using antiserum against GFP. Whole-cell samples corresponded to 10 μg of protein per lane, and samples from spent media corresponded to the volume of spent medium that contained 10 μg of cell protein before the cells were removed.
FIG 12.
Proteinase K treatment to determine if SPRemA-sfGFP-CTDSprB localizes to the cell surface. Strains expressing sfGFP fusion proteins with SprBCTDs of 99 amino acids (pSK93), 218 amino acids (pSK56), and 1,182 amino acids (pSK62) and with Fjoh_3952CTD of 228 amino acids (pSK58) were analyzed. A strain expressing SPRemA-sfGFP-CTDRemA with 1,386 amino acids of RemACTD, which is cell surface localized, was used as a positive control. Proteinase K was added (+) at a final concentration of 1 mg/ml to intact cells (A) and to cell extracts prepared by French pressure cell treatment (B), and cells and extracts were incubated at 25°C. Samples were removed at 0 h and 2 h for immunoblot analyses. Samples were separated by SDS-PAGE, and sfGFP was detected using antiserum against GFP. Samples not exposed to proteinase K (−) were also included.
DISCUSSION
T9SSs are common in members of the phylum Bacteroidetes but have only recently been recognized and studied. Many secreted proteins of P. gingivalis have conserved CTDs (4, 20, 24), and it is now clear that this extends to the many other members of the phylum that have T9SSs (1, 2, 18). There is considerable diversity among these CTDs, but most belong to protein domain family TIGR04183 (type A CTDs) or TIGR04131 (type B CTDs). There have been only a few functional studies of the CTDs of secreted proteins, most of which were conducted on P. gingivalis (24, 26, 32). These studies focused on P. gingivalis RgpB and HBP35, which have type A CTDs. The only other functional studies involved F. johnsoniae ChiA, which has a unique CTD that is apparently unrelated to either type A or type B CTDs (21). The full spectrum of CTDs that facilitate secretion by T9SSs has not yet been thoroughly explored. Here, we examined the characteristics of several F. johnsoniae T9SS CTDs, including two type A CTDs, two type B CTDs, and the novel CTD of the chitinase ChiA.
The results confirm findings from P. gingivalis indicating that type A CTDs of <100 amino acids in length are sufficient to target foreign proteins for secretion by T9SSs. The lengths of the two F. johnsoniae type A CTDs required for efficient secretion of sfGFP were between 80 and 100 amino acids, and similar results were obtained for the ChiA CTD. Even shorter CTDs allowed the secretion of P. gingivalis RgpB and HBP35 (25). The results also confirm previous findings from P. gingivalis regarding the importance of the extreme C-terminal amino acids. Deletion of the final 12 amino acids of an otherwise functional CTDRemA eliminated secretion of the foreign protein sfGFP to which it was attached. Conversion of a conserved lysine within this terminal region to alanine had a similar effect. The similarity of these results to those reported for the P. gingivalis T9SS CTDs suggests that targeting of proteins for secretion by type A CTDs may be broadly similar throughout the phylum Bacteroidetes.
The components of T9SSs are conserved across the many members of the phylum Bacteroidetes that use this system (see Table S3 in the supplemental material). Recognizable type A CTDs are found in all members of the phylum that have the core components of the T9SS, but they are absent in the few species that lack T9SSs. The T9SS components from diverse members of the phylum, although conserved, also exhibit considerable divergence in sequence. For example, F. johnsoniae GldK, GldL, GldM, GldN, SprA, SprE, and SprT exhibit only 14 to 33% amino acid identity with their orthologs from C. hutchinsonii (Table S5). This is perhaps not surprising, since F. johnsoniae (class Flavobacteriia) and C. hutchinsonii (class Cytophagia) are only distantly related (33). In spite of the divergence in sequence between their T9SS components, the C. hutchinsonii Cel9B CTD functioned with the F. johnsoniae T9SS to allow secretion of the foreign protein sfGFP. While not all CTDs functioned in this way, the results indicate that individual CTDs can function with the secretion system from a distantly related member of the phylum. The structure of one CTD was recently solved (34). It is likely that the 3-dimensional structures of the CTDs and of the components of the T9SS, rather than the primary sequences, are most important in directing proteins for secretion by the T9SS. A determination of the structures of additional T9SS CTDs may help to determine if the diversity of primary sequences obscures a common fold that allows recognition of diverse CTDs by T9SSs. Similarly, determination of the structure of the secretion apparatus and its components may help determine how secreted proteins interact with this machine. Electron microscopic analyses recently revealed structural features of a complex composed of P. gingivalis PorK and PorN (orthologs of GldK and GldN), suggesting that they form a large ring on the periplasmic side of the outer membrane (35). Evidence for a cell envelope-spanning complex consisting of P. gingivalis PorK, PorL, PorM, PorN, and PorP was also recently presented (36). Further studies will undoubtedly reveal additional structural features of the T9SS. Regardless, it is clear that CTDs play a role in the secretion of proteins by the T9SS. The ability to predictably target proteins for secretion by the addition of a type A CTD allows engineering of F. johnsoniae or other members of the Bacteroidetes to secrete proteins of interest.
Given the ability of type A CTDs to target foreign proteins for secretion, the inability of type B CTDs to function similarly was somewhat surprising. All of the F. johnsoniae proteins with type B CTDs are large. SprB (6,497 aa in length) has a type B CTD that appears to be required for its secretion. The secretion of such large proteins may require additional secretion system proteins to be involved and additional regions of the secreted protein to interact with the secretion system. For example, sprF, which lies immediately downstream of sprB, is required for the secretion of SprB but not for secretion of RemA or ChiA (14, 21, 31). Most genes encoding secreted proteins with type B CTDs have sprF-like genes immediately downstream, and it has been suggested that these may be involved in the secretion of their cognate proteins (31). Additional experiments are needed to determine the features of type B CTD-containing proteins that are necessary for secretion by the T9SS. This may be challenging given the large sizes of these proteins.
Genome analyses indicate that T9SSs are found in most but not all members of the phylum Bacteroidetes (1) (Table S3). Our results suggest that the function of type A CTDs in secretion may be similar across diverse members of the phylum. The cooccurrence of type A CTDs and SprA across the entire phylum Bacteroidetes and extending to the phyla Rhodothermaeota, Chlorobi, Fibrobacteres, and Ignavibacteriae suggests the possibility that type A CTDs interact directly or indirectly with SprA. Our results also highlight the diversity of CTDs involved in secretion and suggest that type A CTDs and type B CTDs may interact differently with the T9SS to facilitate the secretion of specific sets of proteins.
Our results also identified the growth phase dependence of secretion. The F. johnsoniae T9SS appeared to function poorly in cells growing exponentially in rich medium, but it supported secretion in cells that were in the stationary phase of growth. The exponentially growing cells had little, if any, SprA protein, which may explain the secretion defect. The F. johnsoniae T9SS is needed for the secretion of numerous proteases, polysaccharide-digesting enzymes, and motility proteins. Polymer digestion and cell movement may only be needed when cells are nutrient limited, and the T9SS may be regulated so that it is fully expressed only under these conditions. Little is known regarding regulation of the genes encoding T9SS components. Further research is needed to explore this topic and to fully understand the structure and function of the T9SS apparatus in the many diverse members of the phylum Bacteroidetes that rely on these secretion machines.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
F. johnsoniae ATCC 17061T strain UW101 was the wild-type strain used in this study (37–39). F. johnsoniae strains were grown in Casitone-yeast extract (CYE) medium at 30°C, as previously described (40). Escherichia coli strains were grown in Luria-Bertani medium (LB) at 37°C (41). The strains and plasmids used in this study are listed in Table S1 in the supplemental material, and the primers are listed in Table S2. Antibiotics were used at the following concentrations when needed: ampicillin, 100 μg/ml; kanamycin, 30 μg/ml; erythromycin, 100 μg/ml; streptomycin, 100 μg/ml; and tetracycline, 20 μg/ml.
Generation of plasmids that express sfGFP with signal peptides at the N terminus and with regions of F. johnsoniae T9SS CTDs at the C terminus.
A plasmid expressing the N-terminal signal peptide of RemA fused to sfGFP, which was in turn fused to the C-terminal 97 amino acids of RemA (SPRemA-sfGFP-CTDRemA), was constructed as follows. A 511-bp fragment spanning the remA promoter, start codon, and N-terminal signal peptide-encoding region was amplified using Phusion DNA polymerase (New England BioLabs, Ipswich, MA) and primers 1269 (engineered KpnI site) and 1270 (engineered BamHI site). This fragment was inserted into the KpnI and BamHI sites of pCP23 to generate pYT40. A 711-bp region of sfGFP without a stop codon was amplified from pTB263 (Table S1) using primers 1389 (engineered BamHI site) and 1427 (engineered GGSGGGSG linker and XbaI site). This fragment was inserted into the BamHI and XbaI sites of pYT40, generating pYT179. To introduce the 97-amino-acid-CTD-encoding region of remA, 339 bp was amplified using primers 1488 (engineered XbaI site) and 1489 (engineered SphI site). The product was cloned into pYT179 to generate pSK30. A similar construct in a vector with a different antibiotic resistance marker (ermF) was obtained by amplifying the fragment encoding SPRemA-sfGFP-CTDRemA from pSK30 using primers 1269 and 1489 and inserting this into the SalI site of pMM105.A that had been made blunt using end-conversion mix from the Perfectly Blunt cloning kit (Novagen, Madison, WI), to generate pSK97. The orientation of the insert was confirmed by restriction enzyme digestion and by DNA sequencing.
A plasmid expressing full-length RemA with sfGFP inserted immediately after the N-terminal signal peptide was also constructed. For this purpose, the 4,383-bp region spanning all of remA, including the stop codon but lacking the region encoding the N-terminal signal peptide, was amplified using primers 1271 (engineered XbaI site) and 1272 (engineered XbaI site). This fragment was inserted into XbaI site of pYT179, and the orientation of insertion was confirmed by sequencing, generating pYT180. pSK37, expressing the RemA N-terminal signal peptide fused to sfGFP without CTDRemA, was constructed as a control. To construct this plasmid, the gene encoding sfGFP was amplified using primers 1389 (engineered BamHI site) and 1390 (engineered XbaI site and introduced stop codon) and introduced into BamHI- and XbaI-digested pYT40. A similar construct in a vector with a different antibiotic resistance marker (ermF) was obtained by amplifying the fragment encoding SPRemA-sfGFP from pSK37 using primers 1269 and 1390 and inserting this into the SalI site of pCP11 that had been made blunt using end-conversion mix from the Perfectly Blunt cloning kit to generate pSK96. The orientation of the insert was confirmed by restriction enzyme digestion and by DNA sequencing. Fragments encoding the CTDRemA regions of 87 amino acids and 62 amino acids were cloned in pYT179, generating plasmids pSK71 and pSK81, respectively. A 258-bp region encoding 85 amino acids near the C terminus of RemA but lacking the C-terminal 12 amino acids was also cloned into pYT179, generating plasmid pSK79. Similarly, a plasmid producing SPRemA-sfGFP fused to the C-terminal 97 amino acids of RemA, in which a conserved lysine of RemA was replaced with an alanine (K1432A), was constructed. For this purpose, a 300-bp fragment was amplified using primers 1488 (engineered XbaI site) and 1962 (engineered SphI site and engineered alanine codon). This fragment was inserted into pYT179, generating pSK91.
The RemA signal peptide was replaced by the cytochrome c (Fjoh_1634) signal peptide to determine if the signal peptide from a normally periplasmic protein would be sufficient to allow CTD-mediated secretion by the T9SS. A 396-bp region spanning the promoter and N-terminal signal sequence of Fjoh_1634 was amplified using primers 1946 (engineered KpnI site) and 1947 (engineered BamHI site). This fragment was inserted into the KpnI and BamHI sites of pSK30, generating plasmid pSK84.
Constructs expressing SPRemA-sfGFP fused to CTD-containing regions of F. johnsoniae ChiA, AmyB (Fjoh_1208), SprB, and the SprB-like protein Fjoh_3952 were also generated as follows. A 735-bp region of sfGFP was amplified from pTB263 using primers 1389 (engineered BamHI site) and 1427 (engineered XbaI site). This fragment was cloned into the BamHI and XbaI sites of pSSK52 (21), generating pCB3, which encodes sfGFP fused to the 105-amino-acid C-terminal region of ChiA. Similarly, fragments encoding CTDChiA regions of 79 amino acids and 62 amino acids were cloned in pCB3, generating plasmids pSK89 and pCB4, respectively. For F. johnsoniae AmyB, a 390-bp region encoding the C-terminal 99 amino acids was inserted into the XbaI and SphI sites of pYT179, generating plasmid pSK82. Similarly, regions encoding the C-terminal 73 amino acids and 59 amino acids were also inserted into the XbaI and SphI sites of pYT179, generating plasmids pSK85 and pSK86, respectively. For F. johnsoniae SprB, 300-bp, 657-bp, and 3,549-bp regions encoding the C-terminal 99, 218, and 1,182 amino acids were amplified and inserted into the XbaI and SphI sites of pYT179, generating plasmids pSK93, pSK56, and pSK62, respectively. For the F. johnsoniae SprB-like protein Fjoh_3952, a 687-bp region encoding the C-terminal 228 amino acids was inserted into the XbaI and SphI sites of pYT179, generating pSK58.
Generation of plasmids expressing SPRemA-sfGFP fused to regions of C. algicola, C. hutchinsonii, and P. gingivalis T9SS CTDs.
A 417-bp region encoding the C-terminal 108 amino acids of Celal_2532 (AmyA) was amplified from the C. algicola DSM 14237 genome using primers 1885 (engineered XbaI site) and 1886 (engineered SphI site). This fragment was inserted into pYT179, generating pSK65. A 294-bp region encoding the C-terminal 97 amino acids of CHU_1335 (Cel9B) was amplified from the C. hutchinsonii ATCC 33406 genome using primers 1925 (engineered XbaI site) and 1926 (engineered SphI site). This fragment was inserted into pYT179, generating pSK76. A 339-bp region encoding the C-terminal 103 amino acids of PGN_1466 (RgpB) was amplified from the P. gingivalis ATCC 33277 genome using primers 1923 (engineered XbaI site) and 1924 (engineered SphI site). This fragment was inserted into pYT179, generating pSK75.
Microscopic observation of binding of cells to protein G-coated polystyrene spheres to detect surface-localized SprB or sfGFP.
Cells were grown in motility medium (MM) (42) at 25°C without shaking. One microliter of purified anti-SprB (16) or anti-GFP (0.5 mg/ml; GenScript), 0.5-μm-diameter protein G-coated polystyrene spheres (1 μl of a 0.1% stock preparation; Spherotech, Inc., Libertyville, IL), and bovine serum albumin (BSA) (1 μl of 1% solution) were added to 7 μl of cells. The mixture was introduced into a tunnel slide, prepared as described previously (14), using Nichiban NW-5 double-sided tape (Nichiban Co., Tokyo, Japan) to hold a glass coverslip over a glass slide. Samples were incubated for 3 min and observed using an Olympus BH-2 phase-contrast microscope. Images were recorded using a Photometrics Cool-SNAPcf2 camera and analyzed using the Metamorph software. Images were recorded for 30 s, and 100 randomly selected cells were examined for the presence of spheres that remained attached to the cells during this time.
Analysis of secretion of SPRemA-sfGFP-CTDRemA during different stages of growth.
Two hundred microliters of F. johnsoniae cells (optical density at 600 nm [OD600], ∼1) carrying pSK30 expressing SPRemA-sfGFP-CTDRemA was inoculated in 50 ml of CYE containing tetracycline (10 μg/ml) in side-arm flasks and incubated at 25°C with shaking. In exponential phase (approximately 10 h) and stationary phase (approximately 22 h), 1-ml cultures were taken from the flasks and centrifuged at 22,000 × g for 15 min. In each case, the cells (pellet) and the spent medium (supernatant) were flash-frozen in dry-ice–ethanol baths and stored at −80°C until needed. The pellet and spent medium samples were thawed, cells were suspended in the original culture volume of phosphate-buffered saline consisting of 137 mM NaCl, 2.7 mM KCl, 10 mM Na2PO4, and 2 mM KH2PO4 (pH 7.4), and both samples were subjected to SDS-PAGE and Western blot analysis to detect sfGFP, as described below. Cells prepared in the same way were also examined for levels of the T9SS proteins GldK, GldL, GldM, GldN, SprA, and SprE using antisera specific for each.
Western blot analyses.
F. johnsoniae cells were grown to early stationary phase in CYE at 25°C with shaking. Cells were pelleted by centrifugation at 22,000 × g for 15 min, and the culture supernatant (spent medium) and cell pellet were separated. For whole-cell samples, the cells were suspended in the original culture volume of phosphate-buffered saline. Equal amounts of spent medium and whole cells were boiled in SDS-PAGE loading buffer for 10 min. Proteins were separated by SDS-PAGE, and Western blot analyses were performed as previously described (8). Equal amounts of each sample based on the starting material were loaded in each lane. For cell extracts, this corresponded to 10 μg of protein, whereas for spent medium, this corresponded to the equivalent volume of spent medium that contained 10 μg of cell protein before the cells were removed. For detection of sfGFP by Western blotting, anti-GFP antibodies (0.5 mg/ml) were used at a dilution of 1:3,000.
For the detection of full-length SprB, cells were mixed with SDS-PAGE loading buffer and incubated at 98°C for 10 min. Proteins (25 μg of protein was loaded per lane) were separated on 3 to 8% Criterion XT Tris-acetate acrylamide gels (Bio-Rad, Hercules, CA) and detected by Western blotting using antibodies against SprB (16) at a dilution of 1:2,500.
GldK, GldL, GldM, GldN, SprA, and SprE were detected by Western blotting, as previously described (8, 11, 13, 43). SprF was detected similarly, using polyclonal antibodies against SprF peptides that were produced by Biomatik Corporation (Cambridge, Ontario, Canada) at a dilution of 1:4,000. In each case, 10 μg of proteins was loaded per lane.
Proteinase K treatment of cells to determine the localization of sfGFP-SprB fusion protein.
Cells of F. johnsoniae were grown in CYE at 25°C with shaking. Cells were collected, washed, and suspended in 20 mM sodium phosphate-10 mM MgCl2 (pH 7.5) and diluted to an OD600 reading of 1.5. To examine sfGFP-CTDSprB, proteinase K was added to the intact cells to a final concentration of 1 mg/ml and incubated at 25°C with gentle inverting. At various times, 150 μl of cells was sampled, 10 mM phenylmethylsulfonyl fluoride was added, and the samples were boiled for 1 min to stop digestion. SDS-PAGE loading buffer was added and the samples were boiled for another 7 min. Equal volumes were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes, and proteins were detected with antiserum against GFP. As a positive control for the digestion of sfGFP fusion proteins by proteinase K, an identical cell sample was lysed using a French pressure cell, unbroken cells and debris were removed by centrifugation, and proteinase K was added as described above. Control samples that were not exposed to proteinase K were also included.
Genome analyses.
Genome sequences were analyzed for T9SS genes that encode proteins that belong to appropriate TIGRFAM multiple-sequence alignment families (44). This was accomplished using the Integrated Microbial Genomes (IMG version 4.0.1 [https://img.jgi.doe.gov/]) Function Profile Tool to examine the genomes for sequences predicted to encode orthologs of GldK (TIGR03525), GldL (TIGR03513), GldM (TIGR03517), GldN (TIGR03523), and SprA (TIGR04189). The genomes were also examined for genes encoding proteins with type A CTDs (TIGR04183) and type B CTDs (TIGR04131) in the same way. In each case, the trusted cutoffs assigned by The J. Craig Venter Institute (JCVI) were used to identify family members. These cutoffs allow identification of the vast majority of family members with vanishingly few false positives (44).
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by MCB-1516990 from the National Science Foundation.
The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00884-16.
REFERENCES
- 1.McBride MJ, Zhu Y. 2013. Gliding motility and Por secretion system genes are widespread among members of the phylum Bacteroidetes. J Bacteriol 195:270–278. doi: 10.1128/JB.01962-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Veith PD, Nor Muhammad NA, Dashper SG, Likic VA, Gorasia DG, Chen D, Byrne SJ, Catmull DV, Reynolds EC. 2013. Protein substrates of a novel secretion system are numerous in the Bacteroidetes phylum and have in common a cleavable C-terminal secretion signal, extensive post-translational modification and cell surface attachment. J Proteome Res 12:4449–4461. doi: 10.1021/pr400487b. [DOI] [PubMed] [Google Scholar]
- 3.Sato K, Naito M, Yukitake H, Hirakawa H, Shoji M, McBride MJ, Rhodes RG, Nakayama K. 2010. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc Natl Acad Sci U S A 107:276–281. doi: 10.1073/pnas.0912010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato K, Sakai E, Veith PD, Shoji M, Kikuchi Y, Yukitake H, Ohara N, Naito M, Okamoto K, Reynolds EC, Nakayama K. 2005. Identification of a new membrane-associated protein that influences transport/maturation of gingipains and adhesins of Porphyromonas gingivalis. J Biol Chem 280:8668–8677. doi: 10.1074/jbc.M413544200. [DOI] [PubMed] [Google Scholar]
- 5.Kita D, Shibata S, Kikuchi Y, Kokubu E, Nakayama K, Saito A, Ishihara K. 2016. Involvement of the type IX secretion system in Capnocytophaga ochracea gliding motility and biofilm formation. Appl Environ Microbiol 82:1756–1766. doi: 10.1128/AEM.03452-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomek MB, Neumann L, Nimeth I, Koerdt A, Andesner P, Messner P, Mach L, Potempa JS, Schaffer C. 2014. The S-layer proteins of Tannerella forsythia are secreted via a type IX secretion system that is decoupled from protein O-glycosylation. Mol Oral Microbiol 29:307–320. doi: 10.1111/omi.12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu Y, McBride MJ. 2014. Deletion of the Cytophaga hutchinsonii type IX secretion system gene sprP results in defects in gliding motility and cellulose utilization. Appl Microbiol Biotechnol 98:763–775. doi: 10.1007/s00253-013-5355-2. [DOI] [PubMed] [Google Scholar]
- 8.Rhodes RG, Samarasam MN, Shrivastava A, van Baaren JM, Pochiraju S, Bollampalli S, McBride MJ. 2010. Flavobacterium johnsoniae gldN and gldO are partially redundant genes required for gliding motility and surface localization of SprB. J Bacteriol 192:1201–1211. doi: 10.1128/JB.01495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lasica AM, Goulas T, Mizgalska D, Zhou X, de Diego I, Ksiazek M, Madej M, Guo Y, Guevara T, Nowak M, Potempa B, Goel A, Sztukowska M, Prabhakar AT, Bzowska M, Widziolek M, Thogersen IB, Enghild JJ, Simonian M, Kulczyk AW, Nguyen KA, Potempa J, Gomis-Ruth FX. 2016. Structural and functional probing of PorZ, an essential bacterial surface component of the type-IX secretion system of human oral-microbiomic Porphyromonas gingivalis. Sci Rep 6:37708. doi: 10.1038/srep37708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braun TF, Khubbar MK, Saffarini DA, McBride MJ. 2005. Flavobacterium johnsoniae gliding motility genes identified by mariner mutagenesis. J Bacteriol 187:6943–6952. doi: 10.1128/JB.187.20.6943-6952.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson SS, Glocka PP, Agarwal S, Grimm DP, McBride MJ. 2007. Flavobacterium johnsoniae SprA is a cell-surface protein involved in gliding motility. J Bacteriol 189:7145–7150. doi: 10.1128/JB.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saiki K, Konishi K. 2007. Identification of a Porphyromonas gingivalis novel protein Sov required for the secretion of gingipains. Microbiol Immunol 51:483–491. doi: 10.1111/j.1348-0421.2007.tb03936.x. [DOI] [PubMed] [Google Scholar]
- 13.Shrivastava A, Johnston JJ, van Baaren JM, McBride MJ. 2013. Flavobacterium johnsoniae GldK, GldL, GldM, and SprA are required for secretion of the cell-surface gliding motility adhesins SprB and RemA. J Bacteriol 195:3201–3212. doi: 10.1128/JB.00333-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shrivastava A, Rhodes RG, Pochiraju S, Nakane D, McBride MJ. 2012. Flavobacterium johnsoniae RemA is a mobile cell-surface lectin involved in gliding. J Bacteriol 194:3678–3688. doi: 10.1128/JB.00588-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakane D, Sato K, Wada H, McBride MJ, Nakayama K. 2013. Helical flow of surface protein required for bacterial gliding motility. Proc Natl Acad Sci U S A 110:11145–11150. doi: 10.1073/pnas.1219753110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson SS, Bollampalli S, McBride MJ. 2008. SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. J Bacteriol 190:2851–2857. doi: 10.1128/JB.01904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shrivastava A, Lele PP, Berg HC. 2015. A rotary motor drives Flavobacterium gliding. Curr Biol 25:338–341. doi: 10.1016/j.cub.2014.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kharade SS, McBride MJ. 2015. Flavobacterium johnsoniae PorV is required for secretion of a subset of proteins targeted to the type IX secretion system. J Bacteriol 197:147–158. doi: 10.1128/JB.02085-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato K, Yukitake H, Narita Y, Shoji M, Naito M, Nakayama K. 2013. Identification of Porphyromonas gingivalis proteins secreted by the Por secretion system. FEMS Microbiol Lett 338:68–76. doi: 10.1111/1574-6968.12028. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen K-A, Travis J, Potempa J. 2007. Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of Gram-negative bacteria? J Bacteriol 189:833–843. doi: 10.1128/JB.01530-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kharade SS, McBride MJ. 2014. Flavobacterium johnsoniae chitinase ChiA is required for chitin utilization and is secreted by the type IX secretion system. J Bacteriol 196:961–970. doi: 10.1128/JB.01170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorasia DG, Veith PD, Chen D, Seers CA, Mitchell HA, Chen YY, Glew MD, Dashper SG, Reynolds EC. 2015. Porphyromonas gingivalis type IX secretion substrates are cleaved and modified by a sortase-like mechanism. PLoS Pathog 11:e1005152. doi: 10.1371/journal.ppat.1005152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glew MD, Veith PD, Peng B, Chen YY, Gorasia DG, Yang Q, Slakeski N, Chen D, Moore C, Crawford S, Reynolds EC. 2012. PG0026 is the C-terminal signal peptidase of a novel secretion system of Porphyromonas gingivalis. J Biol Chem 287:24605–24617. doi: 10.1074/jbc.M112.369223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seers CA, Slakeski N, Veith PD, Nikolof T, Chen YY, Dashper SG, Reynolds EC. 2006. The RgpB C-terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C-terminal-domain family found in Porphyromonas gingivalis. J Bacteriol 188:6376–6386. doi: 10.1128/JB.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shoji M, Sato K, Yukitake H, Kondo Y, Narita Y, Kadowaki T, Naito M, Nakayama K. 2011. Por secretion system-dependent secretion and glycosylation of Porphyromonas gingivalis hemin-binding protein 35. PLoS One 6:e21372. doi: 10.1371/journal.pone.0021372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slakeski N, Seers CA, Ng K, Moore C, Cleal SM, Veith PD, Lo AW, Reynolds EC. 2011. C-terminal domain residues important for secretion and attachment of RgpB in Porphyromonas gingivalis. J Bacteriol 193:132–142. doi: 10.1128/JB.00773-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coyne MJ, Roelofs KG, Comstock LE. 2016. Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genomics 17:58. doi: 10.1186/s12864-016-2377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson MM, Anderson DE, Bernstein HD. 2015. Analysis of the outer membrane proteome and secretome of Bacteroides fragilis reveals a multiplicity of secretion mechanisms. PLoS One 10:e0117732. doi: 10.1371/journal.pone.0117732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishiguro I, Saiki K, Konishi K. 2009. PG27 is a novel membrane protein essential for a Porphyromonas gingivalis protease secretion system. FEMS Microbiol Lett 292:261–267. doi: 10.1111/j.1574-6968.2009.01489.x. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Y, Han L, Hefferon KL, Silvaggi NR, Wilson DB, McBride MJ. 2016. Periplasmic Cytophaga hutchinsonii endoglucanases are required for use of crystalline cellulose as the sole source of carbon and energy. Appl Environ Microbiol 82:4835–4845. doi: 10.1128/AEM.01298-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhodes RG, Nelson SS, Pochiraju S, McBride MJ. 2011. Flavobacterium johnsoniae sprB is part of an operon spanning the additional gliding motility genes sprC, sprD, and sprF. J Bacteriol 193:599–610. doi: 10.1128/JB.01203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shoji M, Ratnayake DB, Shi Y, Kadowaki T, Yamamoto K, Yoshimura F, Akamine A, Curtis MA, Nakayama K. 2002. Construction and characterization of a nonpigmented mutant of Porphyromonas gingivalis: cell surface polysaccharide as an anchorage for gingipains. Microbiology 148:1183–1191. doi: 10.1099/00221287-148-4-1183. [DOI] [PubMed] [Google Scholar]
- 33.Krieg NR, Ludwig W, Euzeby J, Whitman WB. 2011. Phylum XIV. Bacteroidetes phyl. nov., p 25–469. In Parte A, Krieg NR, Ludwig W, Whitman WB, Hedlund BP, Paster BJ, Staley JT, Ward N, Brown D (ed), Bergey's manual of systematic bacteriology, vol 4 Springer, New York, NY. [Google Scholar]
- 34.de Diego I, Ksiazek M, Mizgalska D, Koneru L, Golik P, Szmigielski B, Nowak M, Nowakowska Z, Potempa B, Houston JA, Enghild JJ, Thogersen IB, Gao J, Kwan AH, Trewhella J, Dubin G, Gomis-Ruth FX, Nguyen KA, Potempa J. 2016. The outer-membrane export signal of Porphyromonas gingivalis type IX secretion system (T9SS) is a conserved C-terminal beta-sandwich domain. Sci Rep 6:23123. doi: 10.1038/srep23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorasia DG, Veith PD, Hanssen EG, Glew MD, Sato K, Yukitake H, Nakayama K, Reynolds EC. 2016. Structural insights into the PorK and PorN components of the Porphyromonas gingivalis type IX secretion system. PLoS Pathog 12:e1005820. doi: 10.1371/journal.ppat.1005820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vincent MS, Canestrari MJ, Leone P, Stathopulos J, Ize B, Zoued A, Cambillau C, Kellenberger C, Roussel A, Cascales E. 2017. Characterization of the Porphyromonas gingivalis type IX secretion trans-envelope PorKLMNP core complex. J Biol Chem 292:3252–3261. doi: 10.1074/jbc.M116.765081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang LYE, Pate JL, Betzig RJ. 1984. Isolation and characterization of nonspreading mutants of the gliding bacterium Cytophaga johnsonae. J Bacteriol 159:26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McBride MJ, Braun TF. 2004. GldI is a lipoprotein that is required for Flavobacterium johnsoniae gliding motility and chitin utilization. J Bacteriol 186:2295–2302. doi: 10.1128/JB.186.8.2295-2302.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McBride MJ, Xie G, Martens EC, Lapidus A, Henrissat B, Rhodes RG, Goltsman E, Wang W, Xu J, Hunnicutt DW, Staroscik AM, Hoover TR, Cheng YQ, Stein JL. 2009. Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl Environ Microbiol 75:6864–6875. doi: 10.1128/AEM.01495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McBride MJ, Kempf MJ. 1996. Development of techniques for the genetic manipulation of the gliding bacterium Cytophaga johnsonae. J Bacteriol 178:583–590. doi: 10.1128/jb.178.3.583-590.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, NY. [Google Scholar]
- 42.Liu J, McBride MJ, Subramaniam S. 2007. Cell-surface filaments of the gliding bacterium Flavobacterium johnsoniae revealed by cryo-electron tomography. J Bacteriol 189:7503–7506. doi: 10.1128/JB.00957-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rhodes RG, Samarasam MN, Van Groll EJ, McBride MJ. 2011. Mutations in Flavobacterium johnsoniae sprE result in defects in gliding motility and protein secretion. J Bacteriol 193:5322–5327. doi: 10.1128/JB.05480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haft DH, Selengut JD, Richter RA, Harkins D, Basu MK, Beck E. 2013. TIGRFAMs and genome properties in 2013. Nucleic Acids Res 41:D387–D395. doi: 10.1093/nar/gks1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.