ABSTRACT
Many bacteria move through liquids and across surfaces by using flagella—filaments propelled by a membrane-embedded rotary motor. Much is known about the flagellum: its basic structure, the function of its individual motor components, and the regulation of its synthesis. However, we are only beginning to identify the dynamics of flagellar proteins and to understand how the motor structurally adapts to environmental stimuli. In this review, we discuss the external and cellular factors that influence the dynamics of stator complexes (the ion-conducting channels of the flagellar motor). We focus on recent discoveries suggesting that stator dynamics are a means for controlling flagellar function in response to different environments.
KEYWORDS: flagella, stators, MotAB, swarming, swimming, c-di-GMP
INTRODUCTION
The flagellar motor is an intricate machine that drives the rotation of the flagellum by converting ion motive force to mechanical energy (1). Torque is generated via association between two main motor components: membrane-embedded stator complexes and the cytoplasmic rotor (Fig. 1). On the basis of work with the model organism Escherichia coli, each stator complex is composed of protein subunits, MotA and MotB, assembled in a 4MotA-2MotB stoichiometry (2). MotB has been shown to interact with the P-ring protein FlgI (3) and is also thought to associate with peptidoglycan through its peptidoglycan-binding motif. The interactions made by the periplasmic domain of MotB likely serve to anchor stator complexes around the rotor, where the stators act as ion channels. It is thought that the ions passing through these channels are bound by a conserved aspartic acid residue in the transmembrane segment of MotB, causing conformational changes in MotA (4). These conformational changes are coupled to rotation by specific electrostatic interactions between MotA and the rotor protein FliG (5, 6). Work with Vibrio alginolyticus suggests that FliG contributes to both rotation and stator assembly (7).
FIG 1.
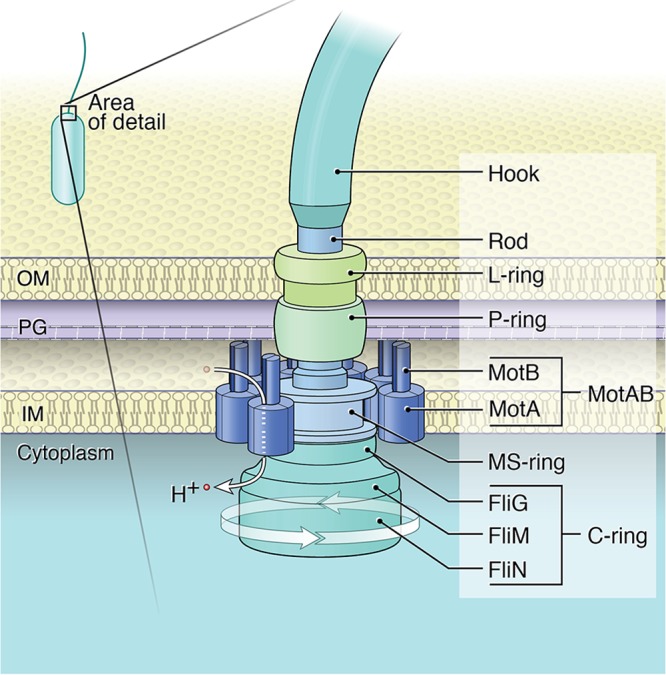
Flagellar motor structure. Stator complexes assemble in a ring in the inner membrane. Each stator unit is formed by four MotA and two MotB proteins, and the MotB protein anchors the complex to peptidoglycan. Each stator unit forms two channels through which ions can flow (H+ indicates protons in this example, with arrow indicating the direction of ion flow). Electrostatic interactions between the MotA and FliG proteins in the C ring generate the torque necessary to power rotation of the flagellum. OM, outer membrane; PG, peptidoglycan; IM, inner membrane. The arrow at the base of the motor indicates a motor complex capable of rotating the flagellum. Copyright, William Scavone; used with permission.
While stators are named for their “stationary” role as the nonrotating motor component, the composition of stators surrounding the motor is highly dynamic (Fig. 2). Stator dynamics were first demonstrated in “resurrection experiments.” These experiments showed that stators could incorporate themselves into and restore rotation to paralyzed flagellar motors, with each successive stator incorporation resulting in a stepwise increase in motor speed (8, 9). Importantly, stochastic fluctuations in torque were also observed in these experiments, in which torque would periodically decrease in steps (8). Specifically, 11 distinct rotational speeds were observed in E. coli, indicating that a maximum of at least 11 stator complexes can engage in the motor at any one time in this species (10). Stator turnover was directly observed and measured by total internal reflection fluorescence microscopy combined with fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching. These experiments demonstrated that green fluorescent protein (GFP)-labeled MotB proteins remain in the motor for an average of only 30 s and exchange with a pool of stators on reserve in the inner membrane (11).
FIG 2.
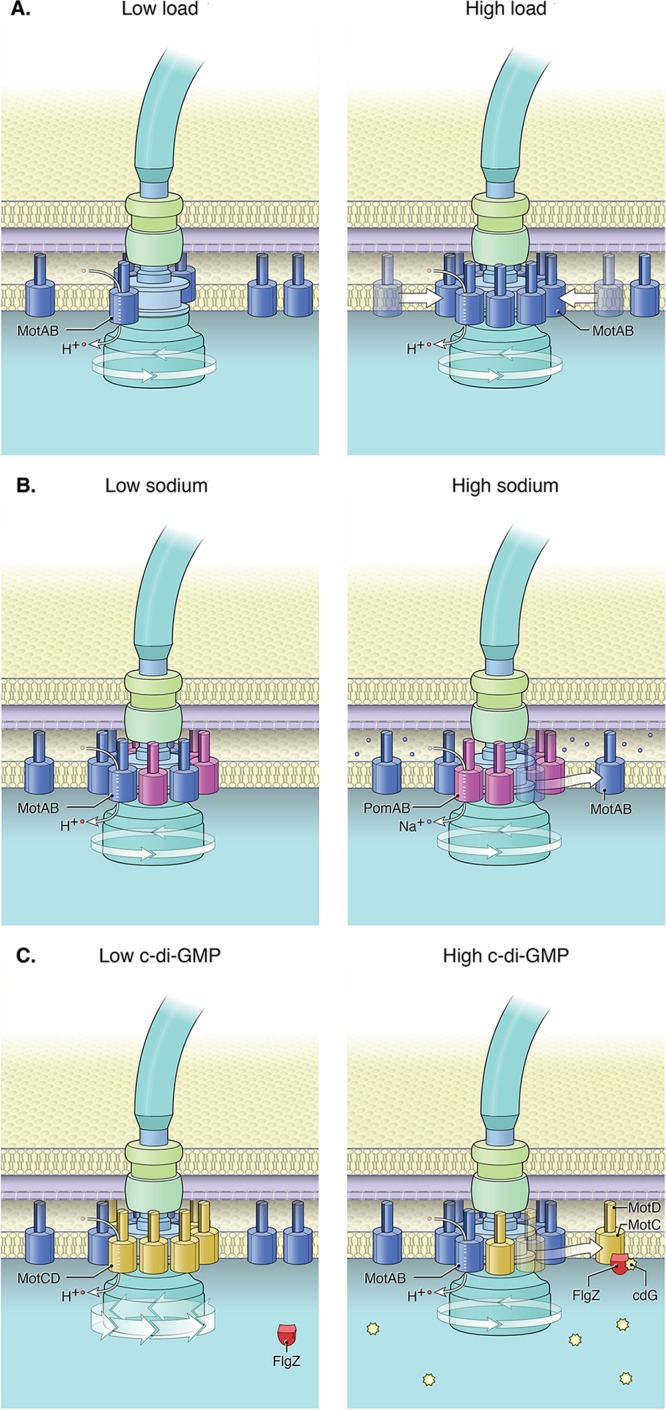
Models of stator dynamics. (A) Additional MotAB stators are incorporated into the motor as the mechanical load on the flagellum increases in E. coli (13, 14). (B) Some bacteria contain both proton-powered MotAB stators and sodium-powered PomAB stators. Work with S. oneidensis has demonstrated that the number of MotAB stators engaged in the motor decreases as the sodium concentration increases (30). (C) P. aeruginosa uses two proton-powered stator sets—MotAB and MotCD. Here we illustrate the current model suggesting that the ratio of the two stator complexes changes at different c-di-GMP levels. In this model, when levels of the signaling molecule c-di-GMP are low, there are increased levels of MotCD in the motor (50). When c-di-GMP levels are high, c-di-GMP (cdG) binds to FlgZ; when bound to c-di-GMP, FlgZ interacts with the MotC component of the MotCD stator complexes (51). We hypothesize that the c-di-GMP-dependent FlgZ-MotC interaction facilitates MotCD disengagement from the motor and decreases motor speed. Here, for ease of illustration, we show a flagellar motor that accommodates 10 stators, but the average number of each stator, as well as the total number of stators in the inner membrane, is unknown for most organisms. The arrow at the base of the motor indicates a motor complex capable of rotating the flagellum; the thicker line indicates the capability of generating relatively greater torque. Copyright, William Scavone; used with permission.
Stator dynamics become more complex in species with multiple stator systems, some of which are driven by different ions (reviewed in reference 12). Proton-dependent stators power most flagella, but motors in several species (especially those in marine environments) depend instead on sodium flux. To regulate motor function in these more complicated systems, bacteria must select and swap between types of stators. Here, we summarize what is known about the factors governing stator arrangement and exchange. We illustrate that stator dynamics are a common mechanism for regulating motor function in response to diverse external and cellular signals.
ENVIRONMENTAL FACTORS DRIVING STATOR EXCHANGE
External load.
As bacterial cells move through heterogeneous environments, they encounter various fluids of different viscosities. Thus, under changing conditions, bacteria experience different levels of viscous drag (or mechanical load) on their flagella. In more viscous environments, motors must generate more force to rotate flagella and do so by recruiting stators to the motor in a load-dependent manner (13, 14) (Fig. 2A). Using E. coli with fluorescently labeled stator proteins, Lele et al. (13) showed that when rotating flagellar stubs are abruptly tethered to latex beads, the motor initially slows because of the increased load. The motor responds to the heavy load by recruiting additional stator units and then increases speed in a stepwise manner. Experiments by Tipping et al. (14) also demonstrated that adding Ficoll to the medium leads to an increasing mechanical load and a corresponding increase in stator incorporation.
These experiments suggest that light loads only require the power of a few stators; as the load increases, the motor recruits additional stators to provide enough torque to support flagellar rotation. The concept that torque increases proportionally with added stators is supported by recent electron cryotomography experiments that show the motor in new detail. These structural studies illustrate diversity in motor architecture among species, in which species known to generate high motor torque have wider stator rings that are likely to accommodate additional stators (15, 16). Clearly, this arrangement and load-dependent regulation of stator incorporation allow the motor to work efficiently and only use as many ions as necessary.
The mechanism of load-dependent stator assembly is not entirely understood, although evidence suggests that load sensing requires motor torque and is independent of motor rotation. For example, sufficient ion motive force is necessary to keep stators bound to the motor (17–19); however, Tipping et al. (14) demonstrated that motors stalled under a heavy load still contain the maximum number of stators, indicating that motor rotation is unnecessary to maintain stator binding. There is also indirect evidence that MotB directly contributes to load sensing; that is, mutational analysis of MotB suggests that the periplasmic domain of MotB is important for maintaining motor torque under light loads (20). Mutation of the critical aspartic acid residue (which binds cations) of Salmonella MotB to a glutamic acid resulted in unexpected speed fluctuations under light loads and stable rotation rates under heavy loads (21). The load-dependent effects of this mutation suggest that the load may impact the coupling between ion translocation and stator conformational changes for torque generation. Thus, load changes may influence the incorporation of stator complexes by triggering conformational changes in MotB.
While it is still not understood how cells sense an external load placed on the flagellum, it is clear that bacteria have exploited this external cue. The concept that cells respond to constraints on flagellar rotation (i.e., flagellar mechanosensing) is widespread in the literature (reviewed in reference 22). Flagellar mechanosensing is one possible mechanism by which bacteria sense surface contact to promote surface-associated behaviors, such as swarming motility and biofilm formation. Surface contact and viscous (i.e., heavy-load) growth conditions result in slowed flagellar rotation and trigger swarmer cell differentiation in Vibrio parahaemolyticus (23) and Proteus mirabilis (24). The flagellar mechanosensing pathway is also induced in V. parahaemolyticus by phenamil, a sodium channel inhibitor. These data indicate that V. parahaemolyticus may be sensing decreased flagellar rotation or decreased Na+ flux through the stators (25). Interestingly, Harshey and Partridge proposed that changes in flagellar rotation speed and/or ion flux might be a direct response to an altered conformation at the rotor-stator interface (26). Such a model would posit that an external change in load is propagated to an intracellular change in the motor shape and thereby serve as a means to initiate a signal transduction event.
Ion availability.
Ion availability can also serve as a signal to modify stator composition. Early experiments with E. coli demonstrated that varying membrane potential via an external voltage source resulted in loss of torque at low membrane potentials and subsequent resurrection upon reestablishment of proton motive force (PMF) (18). Later experiments demonstrated a stepwise stator resurrection upon PMF restoration, along with visualization of motor dissociation upon loss of PMF (17), analogous to what is observed as the load changes. The delayed recovery in speed upon the return of PMF supports the conclusion that stators dissociate upon loss of PMF and take time to return to the motor.
Stators that use sodium motive force (SMF) also exchange depending on ion availability. Extracellular Na+ is required for stator assembly in species including V. alginolyticus (19), and stator engagement (or resurrection) upon restoration of the SMF was observed (27). Notably, these dynamics in response to ions may not be universal across species since there is no evidence of stator loss when PMF is disrupted in Salmonella (28).
Organisms with multiple stator sets sense ion availability to select the appropriate stator set. In Shewanella oneidensis MR-1, an organism with both H+-powered and Na+-powered stators, the exchange rate of stators depends on Na+ levels (29) (Fig. 2B). Higher Na+ levels had no apparent effect on the localization or turnover of the Na+-dependent stator PomAB, but there is evidence of increased incorporation of MotAB at low Na+ concentrations (30). These results imply that this bacterium can sense when low Na+ conditions require it to engage its proton motor.
It has been suggested that the width of the ion channels created by the stators determines ion selectivity (31), but it is still not known how the flagellar motor senses changing ion availability and how the exchange of stators is subsequently controlled. Since ion motive force (which refers, in general, to the force generated by protons, Na+, or any other ion) appears to be critical for stator engagement in most cases, it may be that ion limitation causes stators to more rapidly dissociate from the motor. Whether this dissociation is secondary to loss of ion flow or if other factors are required to displace the stator is an open question. It is clear that for organisms with multiple stators powered by different ions, such as S. oneidensis, the two-stator systems provide distinct motor functions under different environmental conditions. This makes ion sensing and the associated stator exchange a critical regulatory mechanism for bacteria to move efficiently through their varying habitats.
CELLULAR FEATURES DRIVING STATOR EXCHANGE
Flagellum-associated FliL.
FliL is a flagellum-associated protein important for motility and stator function in many bacterial species. For example, FliL is essential for swimming motility in Caulobacter crescentus (32) and Rhodobacter sphaeroides (33). Deletion of fliL has only a small effect on swimming by E. coli, Salmonella, and P. mirabilis but completely abolishes swarming motility (34, 35). Similarly, a fliL mutant of V. alginolyticus, swims more slowly than wild-type cells only in high-viscosity liquids (36). Thus, in some species, FliL only seems to play a significant role under conditions wherein the flagellum experiences a greater external load.
FliL may function via its ability to make direct contact with the stator. For example, bacterial two-hybrid assays suggest an interaction between FliL and MotB in Campylobacter jejuni (37) and between FliL and MotAB complexes in Salmonella (38). Similarly, in V. alginolyticus, localization of FliL to the flagellar pole depends on the stator (36). Electron cryotomography experiments place FliL between the stators and the rotor in Borrelia burgdorferi (39). Furthermore, FliL's impact on motility and its interaction with the stators imply that FliL impacts stator engagement, and this conclusion is consistent with FRAP experiments by Partridge et al. (38), which showed that in Salmonella, recovery of MotA-yellow fluorescent protein was 5 to 10% higher in wild-type cells than in a fliL mutant. It is unknown whether FliL increases stator engagement by influencing the rate of stator incorporation or the stator dwell time in the motor. Mutations in the plug domain of MotB that allow proton leakage restore higher motor speeds to a fliL mutant, suggesting that FliL increases ion flux through stator channels or, alternatively, responds to changes in ion flux by regulating stator insertion/displacement (38). It is also possible that the role of FliL is to sense the load on the flagellum or ion availability.
c-di-GMP.
Cyclic diguanylate (c-di-GMP) is a widespread intracellular signaling molecule that regulates a variety of cellular processes in bacteria, including the motile-to-sessile transition (40). In general, low levels of c-di-GMP promote motility and high c-di-GMP levels stimulate surface attachment and biofilm formation (41, 42). c-di-GMP controls motility by a variety of mechanisms that impinge on flagellar biosynthesis and function (43). PilZ domain proteins are a class of c-di-GMP effectors that share a conserved c-di-GMP binding motif (44). In E. coli and Salmonella enterica, a PilZ domain protein, YcgR, is thought to act as a “backstop brake” on the flagellar motor to slow flagellar rotation and bias the direction of rotation in response to c-di-GMP. It was reported that YcgR impacts flagellar rotation through direct interactions with components of the motor (45–47). Studies by different groups suggest that YcgR interacts with different partners, including MotA (45) and rotor proteins FliG (46, 47) and FliM (47). While YcgR's exact mechanism of action is still unclear, these models all imply disruption of the rotor-stator interface as a mechanism of motility regulation.
One recent addition to c-di-GMP's repertoire of motility control mechanisms is the influence of this signal on stator selection in Pseudomonas aeruginosa. P. aeruginosa has two distinct stator complexes, MotAB and MotCD, to power the rotation of a single flagellum. However, only MotCD supports motility under increased-load conditions—swarming motility on a surface (48–50). Visualization of functional stator proteins fused to GFP in P. aeruginosa reflects dynamic localization, indicating stator exchange. The localization patterns of MotAB and MotCD suggest that these two stator sets coexist in flagellar motors (50). Localization of MotCD to the cell pole decreases in mutants with high c-di-GMP levels, suggesting that c-di-GMP influences the dynamics of MotCD, the “powering stator” required for flagellar rotation during swarming (50) (Fig. 2C). MotCD localization is influenced by c-di-GMP via direct interactions between MotC and FlgZ, a YcgR homolog (51). Another c-di-GMP effector, MapZ, in P. aeruginosa was recently found to control flagellar output by regulating chemotaxis (52). It is likely that c-di-GMP plays a role in flagellar motor dynamics in other species in which a high c-di-GMP level generally leads to reduced motility.
The ability of c-di-GMP to regulate stator exchange offers a mechanism for cells to respond and adapt quickly to changing environments. While c-di-GMP levels clearly influence stator incorporation, the input signals leading to altered c-di-GMP levels are not well understood in many organisms. In V. parahaemolyticus, changes in c-di-GMP occur as a response to surface growth, which then stimulates lateral flagellum formation (53). Although evidence suggests that these changes in c-di-GMP are induced by quorum sensing (54), we speculate that changes in c-di-GMP levels that occur upon surface growth in organisms such as P. aeruginosa may be a response to altered stator engagement.
AN OPEN QUESTION: MECHANISM OF STATOR EXCHANGE
By analyzing bacterial genomic sequences, Thormann and Paulick (12) identified 69 species encoding two or more putative stators for a single flagellar system. How do these organisms that have multiple stator systems regulate stator exchange? In S. oneidensis MR-1, the rate of stator exchange is highest when both the H+-powered (MotAB) and Na+-powered (PomAB) stator sets are present (30). This finding suggests that even with ion availability as a critical factor for stator incorporation, MotAB and PomAB are able to share space in the flagellar motor.
In contrast, for an organism like P. aeruginosa, with multiple stator sets driven by the same ion, we suspect that competition between stators could play an important role in these dynamics. It is not understood why P. aeruginosa has two stator sets both powered by the same ion, while other bacteria (e.g., E. coli) only require one set. It is likely that the two sets in P. aeruginosa are important under different environmental conditions or perhaps even play distinct roles in flagellar function. One stator set (MotCD) seems to generate torque more efficiently than the other (MotAB), and it is apparent that MotCD plays a key role in swarming motility (48–50). We speculate that MotAB may tune the motor for behaviors, such as near-surface swimming or spinning (55), that may be critical for surface attachment or detachment.
CONCLUSIONS
Flagellar motility is a critical survival tool for many motile bacteria. The activity and regulation of flagella contribute to mechanisms that control the transition between the free-swimming and surface-attached lifestyles. As we highlight in this review, stator exchange remains a poorly understood mechanism by which bacteria control their flagellar motility in response to a variety of stimuli. It is apparent that load and ion availability can have diverse effects on the motor among these different species, which experience unique environments. Some bacteria also use flagellum-associated proteins such as FliL and signaling molecules, including c-di-GMP, to facilitate stator remodeling and function. Because stator rearrangement is a common output to extracellular stimuli, we and others speculate that the level of stator engagement within the motor may be actively recognized by cells (26) and could thereby serve as an input for subsequent signal transduction events. Alternatively, modulation of stator occupancy or exchange offers a way for the bacterium to indirectly sense and/or respond to environmental perturbations, such as surface contact and ion availability, by surveying its motor status. The dynamic exchange of stators also provides a means to respond to environmental cues on the order of seconds rather than the longer response times that would be required for the synthesis of new cellular components in response to environmental cues.
A next challenge will be to identify the molecular mechanisms by which the motor modulates its stator exchange dynamics. Understanding how the composition of the flagellar motor is regulated is critical for understanding how bacteria establish and disperse from biofilms and how biofilm establishment can be prevented. Although we focus on stator dynamics, plasticity is common to other parts of the motor as well. For example, components of the rotor—FliN and FliM—also turn over frequently and their exchange is likely important for motility control (56–58). Further molecular analyses in combination with new, ever advancing and evolving microscopy techniques will undoubtedly improve our understanding of motor arrangement and dynamics.
ACKNOWLEDGMENTS
We thank S. L. Kuchma for her critical reading of the manuscript.
This work was supported by National Institutes of Health grant R01 2 R37 AI83256-06 to G.A.O.
REFERENCES
- 1.Berg HC. 2003. The rotary motor of bacterial flagella. Annu Rev Biochem 72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 2.Kojima S, Blair DF. 2004. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry 43:26–34. doi: 10.1021/bi035405l. [DOI] [PubMed] [Google Scholar]
- 3.Hizukuri Y, Kojima S, Homma M. 2010. Disulphide cross-linking between the stator and the bearing components in the bacterial flagellar motor. J Biochem 148:309–318. doi: 10.1093/jb/mvq067. [DOI] [PubMed] [Google Scholar]
- 4.Kojima S, Blair DF. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40:13041–13050. doi: 10.1021/bi011263o. [DOI] [PubMed] [Google Scholar]
- 5.Lloyd SA, Blair DF. 1997. Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J Mol Biol 266:733–744. doi: 10.1006/jmbi.1996.0836. [DOI] [PubMed] [Google Scholar]
- 6.Zhou J, Lloyd SA, Blair DF. 1998. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc Natl Acad Sci U S A 95:6436–6441. doi: 10.1073/pnas.95.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kojima S, Nonoyama N, Takekawa N, Fukuoka H, Homma M. 2011. Mutations targeting the C-terminal domain of FliG can disrupt motor assembly in the Na(+)-driven flagella of Vibrio alginolyticus. J Mol Biol 414:62–74. doi: 10.1016/j.jmb.2011.09.019. [DOI] [PubMed] [Google Scholar]
- 8.Blair DF, Berg HC. 1988. Restoration of torque in defective flagellar motors. Science 242:1678–1681. doi: 10.1126/science.2849208. [DOI] [PubMed] [Google Scholar]
- 9.Block SM, Berg HC. 1984. Successive incorporation of force-generating units in the bacterial rotary motor. Nature 309:470–472. doi: 10.1038/309470a0. [DOI] [PubMed] [Google Scholar]
- 10.Reid SW, Leake MC, Chandler JH, Lo CJ, Armitage JP, Berry RM. 2006. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc Natl Acad Sci U S A 103:8066–8071. doi: 10.1073/pnas.0509932103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leake MC, Chandler JH, Wadhams GH, Bai F, Berry RM, Armitage JP. 2006. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443:355–358. doi: 10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
- 12.Thormann KM, Paulick A. 2010. Tuning the flagellar motor. Microbiology 156:1275–1283. doi: 10.1099/mic.0.029595-0. [DOI] [PubMed] [Google Scholar]
- 13.Lele PP, Hosu BG, Berg HC. 2013. Dynamics of mechanosensing in the bacterial flagellar motor. Proc Natl Acad Sci U S A 110:11839–11844. doi: 10.1073/pnas.1305885110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tipping MJ, Delalez NJ, Lim R, Berry RM, Armitage JP. 2013. Load-dependent assembly of the bacterial flagellar motor. mBio 4:e00551. doi: 10.1128/mBio.00551-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beeby M, Ribardo DA, Brennan CA, Ruby EG, Jensen GJ, Hendrixson DR. 2016. Diverse high-torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold. Proc Natl Acad Sci U S A 113:E1917-1926. doi: 10.1073/pnas.1518952113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin Z, Lin WT, Zhu S, Franco AT, Liu J. 14 November 2016 Imaging the motility and chemotaxis machineries in Helicobacter pylori by cryo-electron tomography. J Bacteriol doi: 10.1128/JB.00695-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tipping MJ, Steel BC, Delalez NJ, Berry RM, Armitage JP. 2013. Quantification of flagellar motor stator dynamics through in vivo proton-motive force control. Mol Microbiol 87:338–347. doi: 10.1111/mmi.12098. [DOI] [PubMed] [Google Scholar]
- 18.Fung DC, Berg HC. 1995. Powering the flagellar motor of Escherichia coli with an external voltage source. Nature 375:809–812. doi: 10.1038/375809a0. [DOI] [PubMed] [Google Scholar]
- 19.Fukuoka H, Wada T, Kojima S, Ishijima A, Homma M. 2009. Sodium-dependent dynamic assembly of membrane complexes in sodium-driven flagellar motors. Mol Microbiol 71:825–835. doi: 10.1111/j.1365-2958.2008.06569.x. [DOI] [PubMed] [Google Scholar]
- 20.Castillo DJ, Nakamura S, Morimoto YV, Che YS, Kami-Ike N, Kudo S, Minamino T, Namba K. 2013. The C-terminal periplasmic domain of MotB is responsible for load-dependent control of the number of stators of the bacterial flagellar motor. Biophysics (Nagoya-shi) 9:173–181. doi: 10.2142/biophysics.9.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Che YS, Nakamura S, Morimoto YV, Kami-Ike N, Namba K, Minamino T. 2014. Load-sensitive coupling of proton translocation and torque generation in the bacterial flagellar motor. Mol Microbiol 91:175–184. doi: 10.1111/mmi.12453. [DOI] [PubMed] [Google Scholar]
- 22.Belas R. 2014. Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol 22:517–527. doi: 10.1016/j.tim.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 23.McCarter L, Hilmen M, Silverman M. 1988. Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell 54:345–351. doi: 10.1016/0092-8674(88)90197-3. [DOI] [PubMed] [Google Scholar]
- 24.Belas R, Suvanasuthi R. 2005. The ability of Proteus mirabilis to sense surfaces and regulate virulence gene expression involves FliL, a flagellar basal body protein. J Bacteriol 187:6789–6803. doi: 10.1128/JB.187.19.6789-6803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawagishi I, Imagawa M, Imae Y, McCarter L, Homma M. 1996. The sodium-driven polar flagellar motor of marine Vibrio as the mechanosensor that regulates lateral flagellar expression. Mol Microbiol 20:693–699. doi: 10.1111/j.1365-2958.1996.tb02509.x. [DOI] [PubMed] [Google Scholar]
- 26.Harshey RM, Partridge JD. 2015. Shelter in a swarm. J Mol Biol 427:3683–3694. doi: 10.1016/j.jmb.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sowa Y, Rowe AD, Leake MC, Yakushi T, Homma M, Ishijima A, Berry RM. 2005. Direct observation of steps in rotation of the bacterial flagellar motor. Nature 437:916–919. doi: 10.1038/nature04003. [DOI] [PubMed] [Google Scholar]
- 28.Morimoto YV, Nakamura S, Kami-ike N, Namba K, Minamino T. 2010. Charged residues in the cytoplasmic loop of MotA are required for stator assembly into the bacterial flagellar motor. Mol Microbiol 78:1117–1129. doi: 10.1111/j.1365-2958.2010.07391.x. [DOI] [PubMed] [Google Scholar]
- 29.Paulick A, Koerdt A, Lassak J, Huntley S, Wilms I, Narberhaus F, Thormann KM. 2009. Two different stator systems drive a single polar flagellum in Shewanella oneidensis MR-1. Mol Microbiol 71:836–850. doi: 10.1111/j.1365-2958.2008.06570.x. [DOI] [PubMed] [Google Scholar]
- 30.Paulick A, Delalez NJ, Brenzinger S, Steel BC, Berry RM, Armitage JP, Thormann KM. 2015. Dual stator dynamics in the Shewanella oneidensis MR-1 flagellar motor. Mol Microbiol 96:993–1001. doi: 10.1111/mmi.12984. [DOI] [PubMed] [Google Scholar]
- 31.Nishihara Y, Kitao A. 2015. Gate-controlled proton diffusion and protonation-induced ratchet motion in the stator of the bacterial flagellar motor. Proc Natl Acad Sci U S A 112:7737–7742. doi: 10.1073/pnas.1502991112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenal U, White J, Shapiro L. 1994. Caulobacter flagellar function, but not assembly, requires FliL, a non-polarly localized membrane protein present in all cell types. J Mol Biol 243:227–244. doi: 10.1006/jmbi.1994.1650. [DOI] [PubMed] [Google Scholar]
- 33.Suaste-Olmos F, Domenzain C, Mireles-Rodriguez JC, Poggio S, Osorio A, Dreyfus G, Camarena L. 2010. The flagellar protein FliL is essential for swimming in Rhodobacter sphaeroides. J Bacteriol 192:6230–6239. doi: 10.1128/JB.00655-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Attmannspacher U, Scharf BE, Harshey RM. 2008. FliL is essential for swarming: motor rotation in absence of FliL fractures the flagellar rod in swarmer cells of Salmonella enterica. Mol Microbiol 68:328–341. doi: 10.1111/j.1365-2958.2008.06170.x. [DOI] [PubMed] [Google Scholar]
- 35.Cusick K, Lee YY, Youchak B, Belas R. 2012. Perturbation of FliL interferes with Proteus mirabilis swarmer cell gene expression and differentiation. J Bacteriol 194:437–447. doi: 10.1128/JB.05998-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu S, Kumar A, Kojima S, Homma M. 2015. FliL associates with the stator to support torque generation of the sodium-driven polar flagellar motor of Vibrio. Mol Microbiol 98:101–110. doi: 10.1111/mmi.13103. [DOI] [PubMed] [Google Scholar]
- 37.Rajagopala SV, Titz B, Goll J, Parrish JR, Wohlbold K, McKevitt MT, Palzkill T, Mori H, Finley RL Jr, Uetz P. 2007. The protein network of bacterial motility. Mol Syst Biol 3:128. doi: 10.1038/msb4100166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Partridge JD, Nieto V, Harshey RM. 2015. A new player at the flagellar motor: FliL controls both motor output and bias. mBio 6:e02367. doi: 10.1128/mBio.02367-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Motaleb MA, Pitzer JE, Sultan SZ, Liu J. 2011. A novel gene inactivation system reveals altered periplasmic flagellar orientation in a Borrelia burgdorferi fliL mutant. J Bacteriol 193:3324–3331. doi: 10.1128/JB.00202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 41.Caiazza NC, Merritt JH, Brothers KM, O'Toole GA. 2007. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189:3603–3612. doi: 10.1128/JB.01685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simm R, Morr M, Kader A, Nimtz M, Romling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol 53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 43.Wolfe AJ, Visick KL. 2008. Get the message out: cyclic-di-GMP regulates multiple levels of flagellum-based motility. J Bacteriol 190:463–475. doi: 10.1128/JB.01418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryjenkov DA, Simm R, Romling U, Gomelsky M. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem 281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 45.Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. 2010. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141:107–116. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 46.Fang X, Gomelsky M. 2010. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol Microbiol 76:1295–1305. doi: 10.1111/j.1365-2958.2010.07179.x. [DOI] [PubMed] [Google Scholar]
- 47.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell 38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doyle TB, Hawkins AC, McCarter LL. 2004. The complex flagellar torque generator of Pseudomonas aeruginosa. J Bacteriol 186:6341–6350. doi: 10.1128/JB.186.19.6341-6350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toutain CM, Zegans ME, O'Toole GA. 2005. Evidence for two flagellar stators and their role in the motility of Pseudomonas aeruginosa. J Bacteriol 187:771–777. doi: 10.1128/JB.187.2.771-777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuchma SL, Delalez NJ, Filkins LM, Snavely EA, Armitage JP, O'Toole GA. 2015. Cyclic di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa PA14 requires the MotAB stator. J Bacteriol 197:420–430. doi: 10.1128/JB.02130-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker AE, Diepold A, Kuchma SL, Scott JE, Ha DG, Orazi G, Armitage JP, O'Toole GA. 2016. PilZ domain protein FlgZ mediates cyclic di-GMP-dependent swarming motility control in Pseudomonas aeruginosa. J Bacteriol 198:1837–1846. doi: 10.1128/JB.00196-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu L, Xin L, Zeng Y, Yam JK, Ding Y, Venkataramani P, Cheang QW, Yang X, Tang X, Zhang LH, Chiam KH, Yang L, Liang ZX. 2016. A cyclic di-GMP-binding adaptor protein interacts with a chemotaxis methyltransferase to control flagellar motor switching. Sci Signal 9:ra102. doi: 10.1126/scisignal.aaf7584. [DOI] [PubMed] [Google Scholar]
- 53.Ferreira RB, Antunes LC, Greenberg EP, McCarter LL. 2008. Vibrio parahaemolyticus ScrC modulates cyclic dimeric GMP regulation of gene expression relevant to growth on surfaces. J Bacteriol 190:851–860. doi: 10.1128/JB.01462-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trimble MJ, McCarter LL. 2011. Bis-(3′-5′)-cyclic dimeric GMP-linked quorum sensing controls swarming in Vibrio parahaemolyticus. Proc Natl Acad Sci U S A 108:18079–18084. doi: 10.1073/pnas.1113790108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Conrad JC, Gibiansky ML, Jin F, Gordon VD, Motto DA, Mathewson MA, Stopka WG, Zelasko DC, Shrout JD, Wong GC. 2011. Flagella and pili-mediated near-surface single-cell motility mechanisms in P. aeruginosa. Biophys J 100:1608–1616. doi: 10.1016/j.bpj.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Delalez NJ, Berry RM, Armitage JP. 2014. Stoichiometry and turnover of the bacterial flagellar switch protein FliN. mBio 5:e01216-14. doi: 10.1128/mBio.01216-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Delalez NJ, Wadhams GH, Rosser G, Xue Q, Brown MT, Dobbie IM, Berry RM, Leake MC, Armitage JP. 2010. Signal-dependent turnover of the bacterial flagellar switch protein FliM. Proc Natl Acad Sci U S A 107:11347–11351. doi: 10.1073/pnas.1000284107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fukuoka H, Inoue Y, Terasawa S, Takahashi H, Ishijima A. 2010. Exchange of rotor components in functioning bacterial flagellar motor. Biochem Biophys Res Commun 394:130–135. doi: 10.1016/j.bbrc.2010.02.129. [DOI] [PubMed] [Google Scholar]


