ABSTRACT
Porcine epidemic diarrhea virus (PEDV) causes enteric disease in pigs, resulting in significant economic losses to the swine industry worldwide. Current vaccination approaches against this emerging coronavirus are only partially effective, though natural infection protects pigs against reinfection and provides lactogenic immunity to suckling piglets. The viral spike (S) glycoprotein, responsible for receptor binding and cell entry, is the major target for neutralizing antibodies. However, knowledge of antibody epitopes, their nature and location in the spike structure, and the mechanisms by which the antibodies interfere with infection is scarce. Here we describe the generation and characterization of 10 neutralizing and nonneutralizing mouse monoclonal antibodies raised against the S1 receptor binding subunit of the S protein. By expression of different S1 protein fragments, six antibody epitope classes distributed over the five structural domains of the S1 subunit were identified. Characterization of antibodies for cross-reactivity and cross-neutralization revealed antigenic differences among PEDV strains. The epitopes of potent neutralizing antibodies segregated into two epitope classes and mapped within the N-terminal sialic acid binding domain and in the more C-terminal receptor binding domain. Antibody neutralization escape mutants displayed single amino acid substitutions that impaired antibody binding and neutralization and defined the locations of the epitopes. Our observations picture the antibody epitope landscape of the PEDV S1 subunit and reveal that its cell attachment domains are key targets of neutralizing antibodies.
IMPORTANCE Porcine epidemic diarrhea virus (PEDV), an emerging porcine coronavirus, causes an economically important enteric disease in pigs. Effective PEDV vaccines for disease control are currently lacking. The spike (S) glycoprotein on the virion surface is the key player in virus cell entry and, therefore, the main target of neutralizing antibodies. To understand the antigenic landscape of the PEDV spike protein, we developed monoclonal antibodies against the spike protein's S1 receptor binding region and characterized their epitopes, neutralizing activity, and cross-reactivity toward multiple PEDV strains. Epitopes of antibodies segregated into six epitope classes dispersed over the multidomain S1 structure. Monoclonal antibodies revealed antigenic variability in B-cell epitopes between PEDV strains. The epitopes of neutralizing antibodies mapped to two distinct domains in S1 that are involved in binding to carbohydrate and proteinaceous cell surface molecules, respectively, indicating the importance of these cell attachment sites on the PEDV spike protein in eliciting a protective humoral immune response.
KEYWORDS: PEDV, coronavirus, monoclonal antibodies, spike protein, virus neutralization
INTRODUCTION
The porcine epidemic diarrhea virus (PEDV), a member of the Coronaviridae family within the Alphacoronavirus genus (1), is an emerging virus that causes severe enteric disease in pigs (2). PEDV is transmitted via the oral-fecal route and replicates in the mature enterocytes of the intestinal epithelium (3). In naive swine herds, PEDV infection is characterized by acute diarrhea and vomiting, with the rate of mortality in neonatal piglets being high (4, 5). The virus gained increased attention in 2010 when outbreaks caused by new strains more pathogenic than the classical strains observed in the 1980s were reported (2, 6). In 2013 such pathogenic strains spread into the Americas (7). During this outbreak two major groups of PEDV strains, named S-Indel and non-S-Indel after a set of insertions and deletions observed in the viral spike (S) glycoprotein, were defined (8). These insertions and deletions appeared to be also present within S proteins of classical strains, including the CV777 strain (8). The S-Indel and non-S-Indel viruses were shown to cause different degrees of mortality in piglets, with the PEDV S-Indel strains representing the less pathogenic viruses (9).
Given the high rates of morbidity and mortality caused by PEDV, particularly in piglets, there is an urgent need to control the infection (10). Both inactivated and live-attenuated virus vaccines are commercially available and have been used in Asia against PEDV (11). However, despite reducing mortality, the currently used vaccines are not able to control virus infection and spread and are considered to be not sufficiently effective (11). Despite the inefficacy of current vaccines, vaccination represents a realistic and viable way of controlling PEDV infection, given that natural infection protects pigs against reinfection and provides lactogenic immunity to suckling piglets (5). The induction of mucosal immunity in the enteric tract is critical for protection against enteric diseases. Although precise correlates of protection are not really known for this virus yet, humoral immunity through the intestinal production of secretory IgA antibodies is likely to be essential (3). Maternal antibodies in pregnant sows are not translocated to the uterus during gestation. Instead, piglets obtain passive lactogenic immunity through the continuous supply of antibodies present in colostrum and milk. Again, IgA antibodies are particularly considered to be important in providing lactogenic immunity because of their relative resistance to proteolytic cleavage in the piglet's digestive tract (12).
As for all coronaviruses, the S protein of PEDV is the key player in virus cell entry and therefore the main target of neutralizing antibodies (13, 14). This ∼200-kDa large glycoprotein assembles into homotrimers which mediate attachment and membrane fusion through its S1 and S2 subunits, respectively (Fig. 1A). Recently elucidated cryo-electron microscopy (cryo-EM) structures of alpha- and betacoronavirus spike trimers revealed a multidomain architecture, particularly of the S1 receptor binding subunit (15–17). This subunit is comprised of four core domains, S1A to S1D, whereas many alphacoronaviruses, including PEDV, contain an additional N-terminal domain that was coined the S10 domain. The S1B domain is known to function as a receptor binding domain for most coronaviruses (15–17) (Fig. 1A and B). The S1B domain of the spike proteins of a number of alphacoronaviruses, including the porcine transmissible gastroenteritis virus (TGEV) and human coronavirus (HCoV) 229E, recruits the aminopeptidase N (APN) protein as a functional receptor, whereas S1B of the human coronavirus NL63 spike protein binds angiotensin-converting enzyme 2 (ACE2) as a receptor. The role of APN as a receptor for PEDV is controversial. PEDV has been reported to utilize APN as a functional cellular receptor (18–22) via its S1B domain (23), but its actual functioning as a receptor is currently disputed (24). Besides interacting with proteinaceous receptors, various coronaviruses also bind to sialoglycoconjugates (25). For the alphacoronavirus TGEV, for instance, the sialic acid (Sia) binding activity was shown to reside in the S10 domain (26) and to be required for enteropathogenicity (27). Recently, Sia binding and hemagglutinating activities were also described for certain PEDV strains; they were mapped to the amino-terminal 246 residues—comprising S10—of the PEDV S protein (23, 28, 29). This N-terminal region in S1 shows a high degree of genetic diversity among PEDV strains and includes the domains in which the insertions and deletions that distinguish S-Indel and non-S-Indel strains occur. The genetic diversity in S10 may also contribute to the variation in Sia binding activity observed among PEDV strains (23, 28, 29).
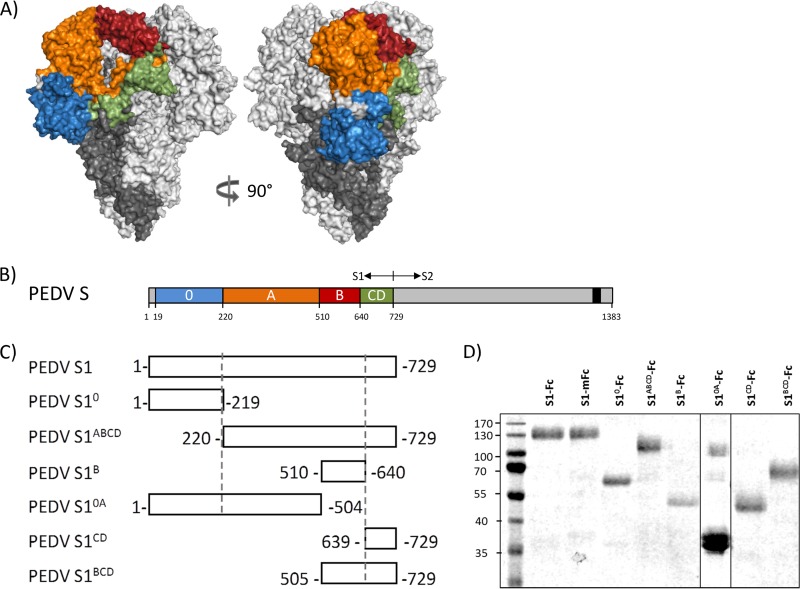
FIG 1.
Domains within the PEDV S protein. (A) Model of the PEDV S trimer based on the HCoV NL63 S structure. The different domains in the S1 subunit of one protomer are colored, with S10 being presented in blue, S1A in orange, S1B in red, and the domains S1CD in green. The S2 subunit of this protomer is marked in dark gray, and the two remaining S protomers are in light gray. (B) Schematic presentation of the PEDV S protein with the S1 part (the domains are colored as described in the legend to panel A) and the S2 part (the C-terminal transmembrane domain is indicated by a black box). (C) Diagram of the different S1 variants used in this study for epitope mapping. All S1 variants were C-terminally tagged with the Fc part of human IgG1 or mouse IgG2a (not shown in the figure). (D) Affinity-purified Fc-tagged S1 variants were separated by SDS-PAGE, and a compilation of Coomassie blue-stained protein gels is shown. Numbers on the left are molecular masses (in kilodaltons).
Receptor binding domains on viruses are often targets for potent neutralizing antibodies. Indeed, most of the neutralizing antibodies that have been described for coronaviruses interfere with receptor binding by targeting the receptor binding domain in the S1 subunit (16, 30). Limited data on the immunogenicity of the PEDV spike protein are available. Polyclonal sera raised against the part of the S1 protein (residues 499 to 638) now known to contain the S1B domain or against a downstream region (residues 636 to 789) spanning the S1-S2 border were shown to have neutralizing activity (14, 31). Only one neutralizing monoclonal antibody (MAb; 8A3A10) has been described; its epitope is located within a broad region of the spike protein (residues 390 to 789) (32). In addition, two B-cell epitopes (SS2 [residues 748 to 755] and SS6 [residues 764 to 771]) targeted by nonneutralizing antibodies were identified downstream of the predicted S1-S2 junction (33).
Despite its relevance for vaccine development, the antigenic landscape of the PEDV spike and the occurrence of neutralizing epitopes within the receptor binding S1 subunit have not been thoroughly explored. To fill this gap, we generated a panel of S1-specific monoclonal antibodies. Using a collection of expressed S1 subdomains, their epitopes were mapped on the PEDV spike. In addition, their neutralizing activity and cross-reactivity toward multiple PEDV strains was evaluated. The observations clearly define the two S1 domains with which the virus interacts with its target cells to be the immunologically most critical.
RESULTS
Generation of monoclonal antibodies targeting PEDV S1 and mapping of their epitopes.
To study the antigenic landscape of the PEDV S1 receptor binding subunit, mouse monoclonal antibodies (MAbs) were generated. Hybridomas were produced from B cells of mice immunized with the spike S1 subunit of the pathogenic strain GDU that was C-terminally extended with a murine Fc tag. Ten monoclonal antibodies targeting PEDV S1 were purified from hybridoma cell culture supernatants. All monoclonal antibodies appeared to be of the IgG1 isotype, with the exception of MAb 71 (IgG2b isotype). In order to map the epitopes of the antibodies to one of the domains within the S1 subunit, we defined a model of the PEDV spike protein based on the cryo-EM structure of the trimeric S protein of the related HCoV NL63 alphacoronavirus and identified five continuous domains (S10 and S1A to S1D) in PEDV S1 (Fig. 1A). Several S1 polypeptides encompassing one or more S1 domains were expressed and purified (Fig. 1B). All S1 polypeptides were checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and shown to migrate according to their expected size. It should be noted that the majority of S10A appeared to be cleaved, which could not be prevented by inclusion of protease inhibitors in the cell culture medium and during the purification procedure. The reactivity of the monoclonal antibodies to these constructs was assessed by enzyme-linked immunosorbent assay (ELISA) (Fig. 2).
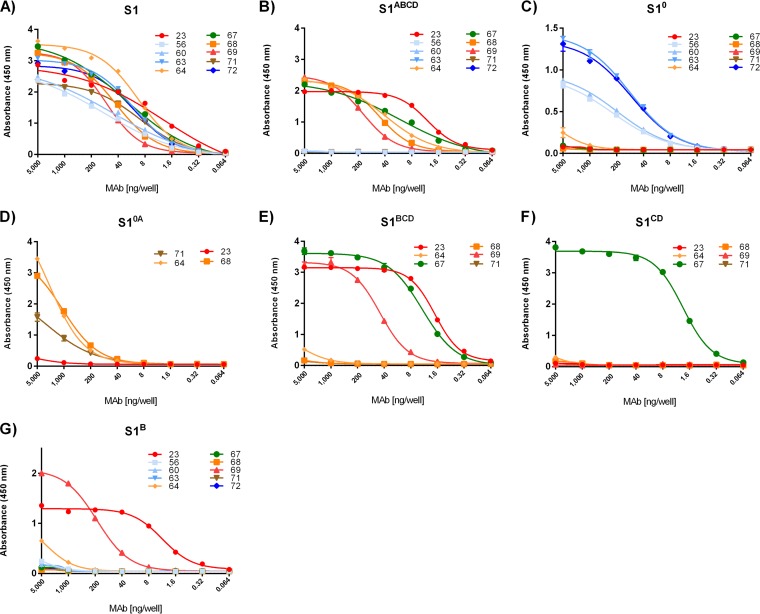
FIG 2.
Mapping of antibody epitopes to S1 domains. (A to G) Antibody binding to human Fc-tagged S1 variants coated onto 96-well plates was tested by ELISA. Antibodies are colored according to the colors given in the legend to Fig. 1 for the single domain to which they bound. S10 binding antibodies are colored in blue, S1A binding antibodies are colored in orange, S1B binding antibodies are colored in red, and S1CD binding antibodies are colored in green; additionally, MAb 71 binding is shown in brown. The S1-Fc variants used for coating are indicated at the top of each panel. All graphs represent the means from three independent experiments. Error bars indicate standard deviations.
Epitopes of the 10 S1-specific MAbs were mapped to S1 domains 0, A, B, and CD (Fig. 2). Antibodies 56, 60, 63, and 72 each bound to S10 (Fig. 2C), whereas MAbs 64 and 68 were found to bind S1A (Fig. 2B to E). Two antibodies, MAbs 23 and 69, bound S1B. MAb 67 was the only antibody binding S1CD (Fig. 2F). MAb 71 showed considerable binding to S10A (Fig. 2D) but not to S10 (Fig. 2C) or S1ABCD (Fig. 2B), suggesting that this antibody targets the boundary between S10 and S1A.
Evaluation of competition of monoclonal antibody binding to PEDV S1 antigen.
After mapping of the antibodies to domains within the S1 subunit, competition in binding between the antibodies was investigated using biolayer interferometry (Fig. 3). PEDV GDU S1-Fc was immobilized on the protein A-coated biosensor tip surface, after which unoccupied sites were blocked using polyclonal feline antibodies. Biosensor tips were then exposed to one antibody until saturation was reached, followed by exposure to a second antibody. Binding of antibodies was recorded by detecting changes in the interference pattern.
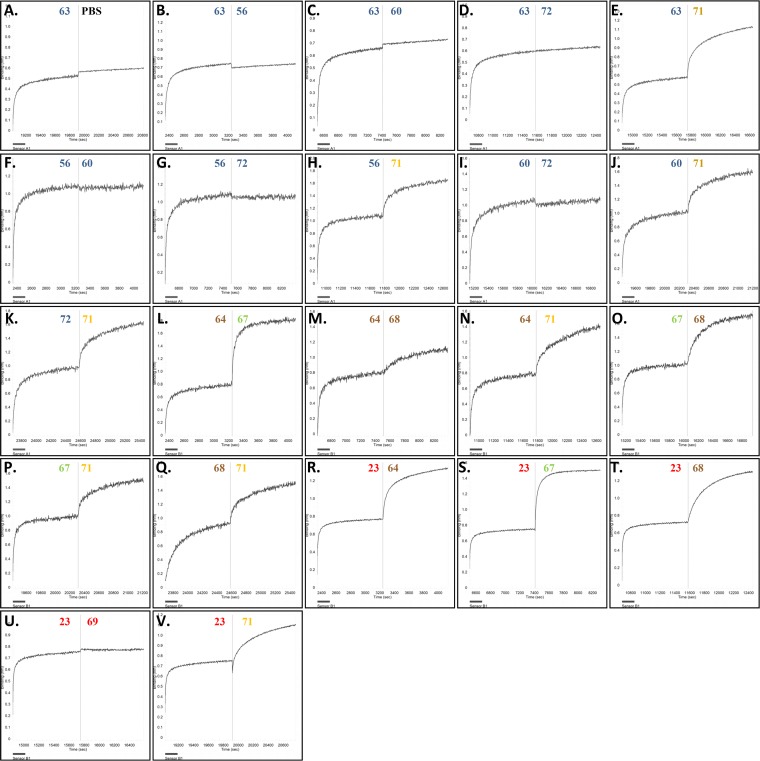
FIG 3.
Evaluation of monoclonal antibody binding competition through biolayer interferometry. To measure competition of binding to an epitope between antibodies, biosensors coated with the S1-Fc protein were exposed to one antibody until saturation in binding was achieved; the sensor was then put in contact with a different antibody in order to measure the competition for an epitope between antibodies. This was done in different combinations. Antibody binding was detected by changes in the light interference pattern. The x axis represents time (in seconds), and the y axis represents the shift in wavelength (in nanometers). The antibody used is indicated above the curve, and the colors are described in the legend to Fig. 1. The experiment was performed twice, and representative binding images are shown.
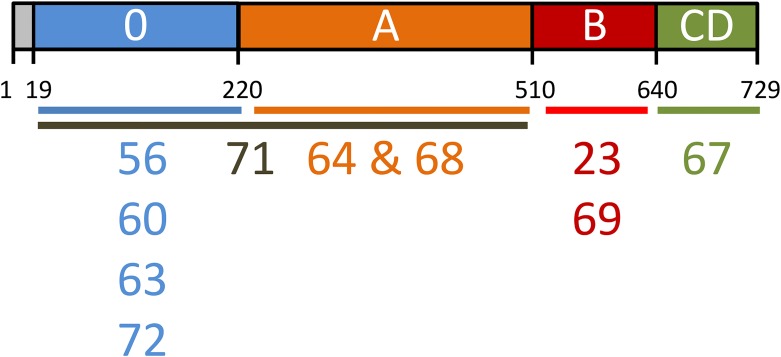
All S10 MAbs (MAbs 56, 60, 63, and 72) competed with each other for binding (Fig. 3A to G and I), indicating that these antibodies share an epitope region within S10. The S1A MAbs 64 and 68 bound distinct sites within domain A (Fig. 3M). MAb 71, which reacts to S10A, did not share epitope regions with either S10 or S1A antibodies (Fig. 3H, J, K, N, and Q). MAbs 23 and MAb 69, which target S1B containing the presumed receptor binding domain, were found to belong to the same epitope class (Fig. 3U). The combined results of antibody epitope mapping to S1 domains and antibody binding competition indicate that the 10 monoclonal antibodies segregated into six nonoverlapping epitope classes within PEDV S1: (i) MAbs 56, 60, 63, and 72 (epitope within S10), (ii) MAb 64 (S1A), (iii) MAb 68 (S1A), (iv) MAb 71 (S10 and S1A boundary), (v) MAbs 23 and 69 (S1B), and (vi) MAb 67 (S1CD) (Fig. 4).
FIG 4.
Monoclonal antibody epitope regions in the PEDV S1 subunit. MAbs and their epitope regions are indicated below the schematic of the PEDV S1 subunit. MAbs sharing an epitope region are aligned vertically.
Cross-reactivity of S1 monoclonal antibodies against PEDV strains.
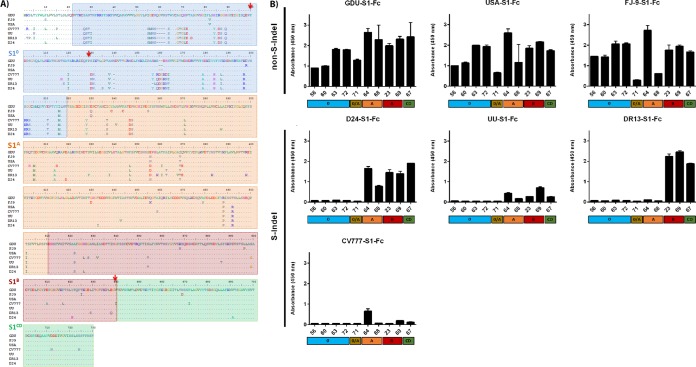
We subsequently tested the cross-reactivity of the S1 monoclonal antibodies against different PEDV strains. Antibody binding to different Fc-tagged PEDV S1 subunits of both non-S-Indel strains (GDU, USA, FJ-9) and S-Indel strains (D24, UU, DR13, CV777) was analyzed by ELISA (Fig. 5). Equal coating of the Fc-tagged antigens was corroborated using a horseradish peroxidase (HRP)-conjugated anti-human IgG antibody (data not shown). All monoclonal antibodies showed moderate to high cross-reactivity to spike protein S1 subunits of non-S-Indel strains, which is not surprising, as they were raised by immunization with the S1 antigen of a non-S-Indel GDU strain.
FIG 5.
Cross-reactivity of monoclonal antibodies to S1 subdomains of non-S-Indel and S-Indel PEDV strains revealed by ELISA. (A) Alignment of S1 subunit sequences of spike proteins of PEDV strains. The amino acid sequences of the S1 subunits of spike protein of PEDV strains were aligned using the Clustal Omega program (http://www.ebi.ac.uk/Tools/msa/clustalo/). The PEDV strains used included GDU (GenBank accession no. KU985230), FJ-9 (GenBank accession no. AGG34696), USA (GenBank accession no. AII20255), CV777 (GenBank accession no. AF353511), UU (GenBank accession no. KU985229), DR13 (GenBank accession no. JQ023162.1), and D24 (GenBank accession no. KY399745). The sequences corresponding to the different S1 domains are indicated by colored boxes, with S10 presented in blue, S1A in orange, S1B in red, and S1CD in green. The positions of mutations F100L, P129L, and V638G, observed in escape mutants resistant to MAbs 72, 63, and 23, respectively, are indicated by arrows. (B) Equal amounts of Fc-tagged S1 proteins of multiple PEDV strains were coated, and antibody binding was measured by ELISA. The S1-Fc proteins tested are indicated at the top of each panel. The absorbance at 450 nm and the MAbs are shown on the y and x axes, respectively. Additionally, the S1 domain targeted by each antibody is indicated at the bottom of each panel. All graphs represent the means from three independent experiments. Error bars indicate standard deviations.
Consistent with the considerable sequence variation found in the N-terminal part of S1 between these two types of viruses (Fig. 5A), none of the S10-specific antibodies (MAbs 56, 60, 63, and 72) showed any reactivity to the S-Indel spike proteins. These differences also eliminated the epitope occurring in GDU S1 at the interface between domains S10 and S1A seen by MAb 71. MAbs targeting the C-terminal part of S1 showed different degrees of cross-reactivity against the S1 subunit of S-Indel strains. Of the two antibodies specific for S1 domain A, MAb 64 bound the most strongly; both had variable cross-reactivity to the S-Indel-type S proteins, with hardly any binding to the DR13 S protein being seen (Fig. 5B). Similarly, both antibodies recognizing the epitope in domain B and the one antibody targeting domain CD were highly cross-reactive with non-S-Indel proteins but variably cross-reactive with S-Indel proteins, but in this case, binding to the S1 proteins of European strains UU and CV777 was very poor (Fig. 5B).
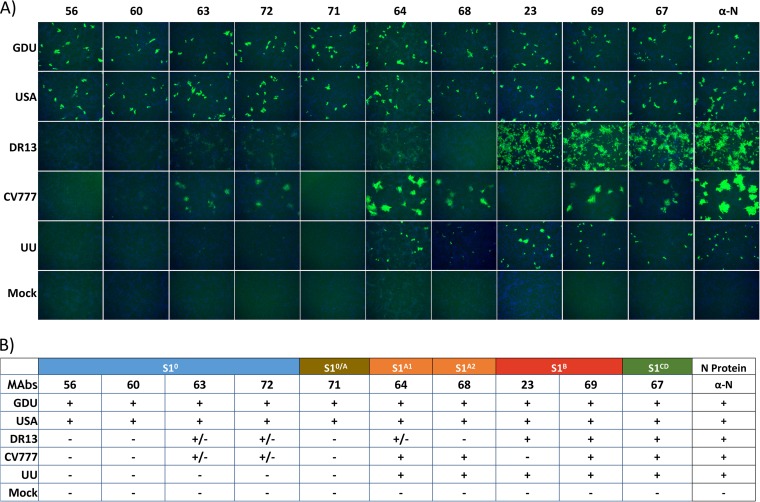
To corroborate the antibody cross-reactivity data obtained by ELISA, an immunofluorescence assay (IFA) was performed using Vero cells infected with three S-Indel strains (CV777, DR13, UU) and two available non-S-Indel strains (GDU, USA) (Fig. 6). The profile of the cross-reactivity of the monoclonal antibodies to PEDV-infected Vero cells observed by IFA was consistent with the cross-reactivity data obtained by ELISA. S10-specific MAbs showed no or poor reactivity against S-Indel viruses. Antibodies targeting the S1 domains A, B, and CD (MAbs 64, 68, 23, 69, and 67) were able to recognize the non-S-Indel viruses, while binding to S-Indel virus-infected cells was generally less efficient and more variable.
FIG 6.
Reactivity of monoclonal antibodies with PEDV-infected cells determined by IFA. (A) IFA pictures showing binding of antibodies to cells infected with different PEDV strains. Binding was visualized with Alexa Fluor 488-coupled goat anti-mouse antibody, while DAPI was used to visualize the cell nuclei. The strain used for cell infection is shown on the left, and the different antibodies used for the assay are indicated at the top. Antibody 3F12 (α-N), which binds to the nucleocapsid protein of PEDV, reacted to all virus strains and was used as a control antibody. (B) Summary of the monoclonal antibody S1 binding reactivity observed in panel A. The experiment was repeated two times, and representative images are shown.
Virus neutralization capacity and mode of action of S1 monoclonal antibodies.
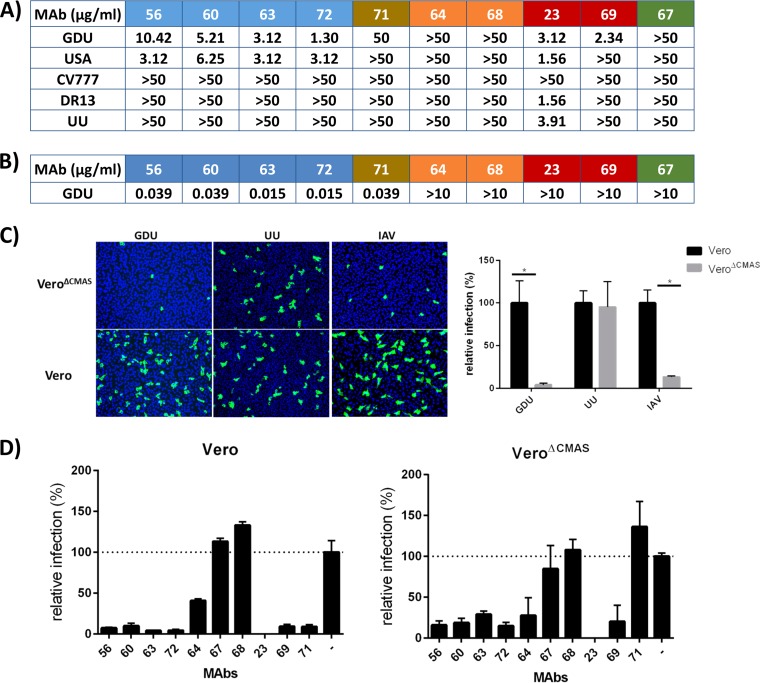
The neutralizing potential of the S1 monoclonal antibodies toward different PEDV strains was determined by virus neutralization assay (Fig. 7A). Consistent with the findings for antibodies raised against the GDU S1 protein, generally strong neutralization of the non-S-Indel strains GDU and USA and poor neutralization of the S-Indel strains CV777, DR13, and UU were observed. Potent neutralization was observed only with antibodies targeting S10 and S1B; MAbs to S1A and S1CD showed no neutralization even at the highest concentration tested (50 μg/ml), while the MAb targeting S0A was moderately neutralizing but only against strain GDU. Significantly, while the 4 MAbs that recognized the same epitope in S10 strongly neutralized the non-S-Indel viruses, they were each inactive against the S-Indel viruses. Of the two antibodies that recognized the same epitope in S1 domain B, MAb 69 strongly neutralized PEDV GDU but did not neutralize any of the other viruses, including the USA strain. In contrast, for MAb 23 broad and strong neutralization of all tested non-S-Indel and S-Indel strains, with the exception of the CV777 strain, was observed. Collectively, the observed neutralization pattern of the monoclonal antibodies is in good accordance with the ELISA- and IFA-based serological cross-reactivity profiles (Fig. 5 and 6).
FIG 7.
Neutralization by monoclonal antibodies and role of Sia binding interference in neutralization. (A) Neutralization of different PEDV strains by S1 MAbs. Serial dilutions of antibodies were mixed with different PEDV strains, and neutralization was scored from the readout of the occurrence of a cytopathic effect at 2 days postinfection. The antibodies and the PEDV strain used are indicated at the top and left, respectively. The minimal antibody concentration required to inhibit a cytopathic effect is shown and is the average from two to three independent experiments. (B) Hemagglutination inhibition (HAI) activity of MAbs. The lowest antibody concentration needed to inhibit the hemagglutination of 160 HA units/ml of PEDV GDU or the PEDV GDU S1-Fc protein is shown. (C) Cells were infected with PEDV or IAV at multiplicities of infection of 0.1 and 1, respectively, and at 14 h postinfection, cells were fixed and infected cells were stained with an anti-PEDV or anti-HA monoclonal antibody. Alexa Fluor 488-conjugated goat anti-rabbit or human IgG (H+L) (Molecular Probes) was used as the secondary antibody, and DAPI was used to visualize the cell nuclei. The percentage of infected cells (relative to the number of PBS-treated cells) was calculated by counting the infected cells in 10 microscopic fields. (D) Interference of Sia binding as a mechanism of virus neutralization. The relative infection of Vero cells (left) or of the Sia-lacking Vero cell clone (VeroΔCMAS; right) by PEDV GDU in the presence of MAbs is shown. PEDV was preincubated with each monoclonal antibody at a final concentration of 25 μg/ml MAb or with phosphate-buffered saline (−) prior to inoculation of cells.
Since the PEDV S1 subunit has been shown to contain Sia binding activity, we next examined the potential inhibitory effect of our S1 antibodies on Sia binding using the hemagglutination (HA) inhibition (HAI) assay. As Fig. 7B shows, the S10 MAbs 57, 60, 63, and 72 as well as S10A MAb 71 inhibited hemagglutination by PEDV GDU S1 at low concentrations. In contrast, the antibodies targeting domain A, B, or CD did not inhibit hemagglutination at any of the concentrations tested.
Binding to cell surface sialoglycoconjugates has been shown to be important for cell entry of PEDV GDU (28). Since S10 antibodies display potent virus neutralization and hemagglutination inhibition activity, we assessed whether the mechanism of action of these antibodies was related to Sia binding interference. Enzymatic depletion of sialic acids from the cell surface has been shown to significantly inhibit infection by the PEDV GDU strain, though infection is not fully abrogated (28). To further assess the antibody-mediated interference of Sia binding as a mechanism of neutralization, we constructed a mutant Vero cell line lacking cell surface expression of Sia that was generated by knockout of the CMP N-acetylneuraminic acid synthetase (CMAS) gene (VeroΔCMAS) through clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 genome engineering. Infection with the Sia-dependent influenza A virus (IAV) and the Sia-independent PEDV UU virus was used to confirm the phenotype of the VeroΔCMAS mutant cell line. Infection of VeroΔCMAS cells with Sia-dependent influenza A virus and PEDV GDU was reduced to ∼5% of that of parental Vero cells, whereas no change in infection efficiency was seen for Sia-independent PEDV UU (Fig. 7C). The residual infection of PEDV GDU on the Sia-lacking VeroΔCMAS allowed us to assess the contribution of Sia binding interference by monoclonal antibodies to neutralization by comparing their PEDV GDU neutralization capacity on wild-type and mutant Vero cells (Fig. 7D). As expected, the extent of inhibition of PEDV GDU infection by domain A, B, and CD antibodies was similar on Vero and VeroΔCMAS cells. However, in contrast to wild-type Vero cells, neutralization by the S10A-targeting MAb 71 was not seen in Sia-lacking VeroΔCMAS cells, indicating that neutralization by MAb 71 on Vero cells is achieved by interference with Sia binding. Puzzlingly, the residual infectivity of PEDV GDU toward VeroΔCMAS cells was still almost as sensitive to S10-targeting MAbs 56, 60, 63, and 72 as its residual infectivity toward wild-type Vero cells.
Selection of neutralizing antibody escape mutants.
Mapping of the mutations of viruses that are resistant to virus neutralization can provide further information about the neutralizing antibody epitopes. Thus, the neutralizing S1B MAb 23 and S10 MAbs 63 and 72 were used to select escape mutants by serial passaging of PEDV GDU on cells with increasing concentrations of antibody. For each antibody we obtained escape mutants of which the spike gene was entirely sequenced. Single nucleotide changes were found in the S gene of all mutants, resulting in single amino acid substitutions. Escape mutants resistant to MAbs 63, 72, and 23 displayed amino acid substitutions in the spike protein at positions 129 (Pro-to-Leu [P129L]), 100 (Phe-to-Leu [F100L]), and 638 (Val-to-Gly [V638G]), respectively, that mapped to the S1 domain to which the selecting antibodies bound.
Binding analysis of monoclonal antibodies to S1 proteins carrying escape mutations.
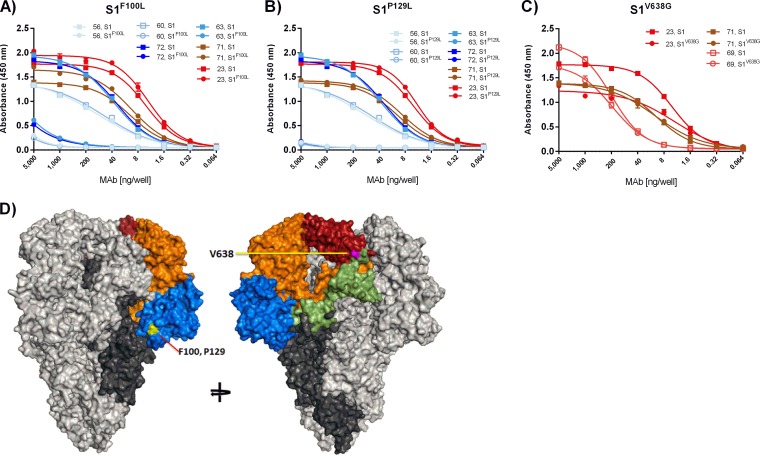
To confirm that the neutralizing antibodies bound at the site of the escape mutations, an ELISA was performed with recombinant Fc-tagged S1 proteins bearing the identified amino acid substitutions (Fig. 8). The P129L substitution in S1 (S1P129L) fully prevented binding of the selecting S10 MAb 63. A similar loss of binding to S1P129L was seen for S10 MAbs 56, 60, and 72, confirming that the epitopes of these four S10-targeting MAbs overlap. As expected, the P129L substitution did not affect the binding of control MAbs (MAb 71 and MAb 23) that bound other epitope classes (Fig. 8A). Likewise, the F100L substitution in S1 (S1F100L) that was observed in the MAb 72-resistant virus almost fully prohibited binding of all four S10 MAbs (MAbs 56, 60, 63, and 72), whereas no significant change in binding was observed for control antibodies MAb 71 and MAb 23 (Fig. 8B). Thus, F100 and P129 are critical residues in the epitope recognized by the S10 MAbs 56, 60, 63, and 72. These residues cluster together at the base of the S10 domain in the predicted three-dimensional PEDV S structure (Fig. 8D). The V638G substitution in S1 (S1V638G) present in the MAb 23 escape mutant significantly but only partially inhibited the binding of MAb 23 as well as that of MAb 69. No change in binding to S1V638G was observed for control MAb 71 (Fig. 8C).
FIG 8.
Binding of MAbs to escape mutant S1 proteins. (A to C) The binding of MAbs to wild-type and mutant S1-Fc proteins containing single amino acid substitutions in the presence of different antibody concentrations was measured by ELISA. The antibody concentration and the absorbance at 450 nm are indicated on the x and y axes, respectively. The coloring of the antibodies is consistent with that described in the legend to Fig. 1. All graphs represent the means from three independent experiments. Error bars indicate standard deviations. (B) Position of amino acids in the PEDV S trimer structure model that are subject to mutation in viruses resistant to MAbs 72 (F100L; yellow), 63 (P129L; yellow), or 23 (V638G, purple).
Quantitation of antibody neutralization resistance of virus escape mutants.
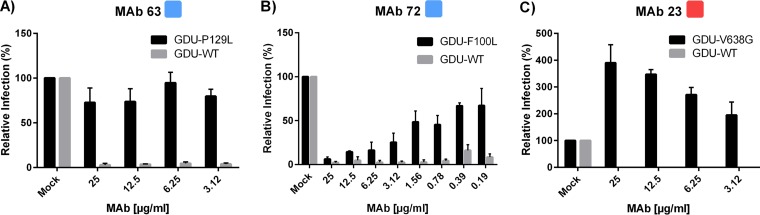
To study the effect of the single-site substitutions in the context of virus infection, antibody neutralization of the escape viruses was quantified (Fig. 9). The PEDV SP129L mutant was fully resistant to MAb 63, which is in accordance with the complete loss of binding to S1P129L by MAb 63 and all other S10 MAbs. PEDVF100L showed only partial resistance to MAb 72, despite the complete loss of binding of this antibody to S1F100L. Surprisingly, enhancement of infection of PEDV SV638G by MAb 23 was observed in an antibody concentration-dependent manner; a 3-fold increase in the level of infection was measured at the antibody concentration used for the selection of this virus mutant.
FIG 9.
Neutralization resistance of PEDV GDU escape mutants bearing single amino acid substitutions in the spike proteins. Wild-type PEDV GDU and PEDV escape mutants isolated under selection pressure with MAbs 63, 72, and 23 were tested for neutralization by the selecting antibody. Infection at different concentrations of antibody was quantified relative to that for mock-treated viruses. The antibody used is indicated at the top of each panel, with the antibody concentration and the percent relative infection being shown on the x and y axes, respectively. For escape the mutants and parental (wild-type [WT]) virus, relative infection is shown in black and gray, respectively. All graphs represent the means from three independent experiments. Error bars indicate standard deviations.
DISCUSSION
PEDV causes significant morbidity in pigs of all ages and high mortality in young piglets, inflicting serious economic losses to the swine industry (10). Vaccination has been used extensively as a means for disease control; however, despite many efforts, vaccine efficiency has not been sufficient for global needs (11, 34). As is known for other coronaviruses, the S1 subunit of the PEDV spike is responsible for target cell attachment and entry and is, hence, a target for neutralizing antibodies (31, 32), yet its antigenic landscape and the location of neutralizing epitopes are poorly defined. To increase our understanding of antibody neutralization of this pathogen relevant to veterinary medicine, a collection of 10 monoclonal antibodies against PEDV S1 was generated, and the MAbs were characterized for their epitope locations and (cross-)neutralizing potential. Six nonoverlapping epitope regions were identified within the S1 subunit. These epitope regions were distributed over the five structural domains of the S1 protein that were recently resolved. Antibodies to two epitope regions were particularly potent in virus neutralization. Epitopes of these neutralizing antibodies mapped to the S10 and S1B structural domains, which are involved in binding to sialoglycoconjugates and to a proteinaceous receptor, respectively.
The receptor binding domain of the coronavirus spike is a key target of potent neutralizing antibodies (35–38). PEDV was reported to bind to a proteinaceous receptor by its S1B domain, analogous to other alphacoronaviruses for which receptors have been identified, although the identity of the host receptor (i.e., porcine APN) has recently been disputed (24). Epitopes of neutralizing antibodies have been mapped to the S1 subunit, which we now know contains the S1B structural domain (31, 32). Here we describe two neutralizing monoclonal antibodies, MAbs 23 and 69, targeting overlapping epitopes in S1B. S1B MAb 23 was broadly reactive; it bound and potently cross-neutralized all tested S-Indel and non-S-Indel viruses, with the exception of CV777. Mapping of the mutation in a MAb 23 neutralization escape virus additionally defined the position of the neutralization epitope. The mutant virus resistant to S1B MAb 23 contained a valine-to-glycine substitution at sequence position 638. The PEDV spike structural model, which we based on the spike structure of the related HCoV NL63, predicts that V638 is a surface-exposed residue close to the boundary of S1B and S1CD (Fig. 8D). Interestingly, in the CV777 strain, an amino acid substitution (V638I) occurs at the same position in S1B, which correlates with the lack of neutralization of this virus by MAb 23. Of note, both S1B antibodies still bound to S1V638G in ELISA, though they did so with a lower affinity than they bound to wild-type S1. Remarkably, infection by the MAb 23 escape mutant virus was enhanced in the presence of the selecting antibody. The underlying mechanism of this antibody-dependent enhancement is difficult to comprehend but may be related to the structural dynamics of the functioning spike. Virus binding to receptors on the target cell surface initiates a series of conformational changes in the metastable spike protein, which culminate in membrane fusion (39, 40). Many viruses conceal critical neutralizing epitope sites (e.g., receptor binding sites) by limiting their exposure to conformational stages subsequent to attachment (41–44). Alphacoronaviruses may utilize a similar immune evasion strategy. The structure of the HCoV NL63 S ectodomain trimer revealed that the receptor-interacting loops in S1B are buried, indicating that a reorientation of S1B is required for receptor engagement (16). It is conceivable that a similar structural rearrangement of S1B is required to expose the receptor binding site of the PEDV spike protein and that antibodies such as MAb 23 that target a hinge region in S1 can alter the prefusion spike stability. We speculate that the V638G escape mutation may counteract such an effect of the bound antibody on the PEDV S protein, yet simultaneously cause dependency on MAb 23 for virus infectivity.
Using the collection of S1-specific monoclonal antibodies, we identified and characterized a novel neutralizing antibody epitope class in the N-terminal domain (S10) of the spike protein. S10 MAbs (MAbs 56, 60, 63, and 72) potently neutralized PEDV GDU infectivity, and analysis of escape mutants further defined the epitope to be a region within S10 comprising F100 and P129. These S10 MAbs prevented binding of PEDV GDU to sialic acids, as demonstrated by their ability to block virus-mediated hemagglutination. Binding to cell surface sialoglycoconjugates has been shown to be important for infection of PEDV GDU (28). However, the relative level of infection in the presence of the S10 antibodies was only slightly higher on Sia-lacking mutant cells than on wild-type cells, suggesting that interference with Sia binding by these S10 antibodies only partially contributes to virus neutralization. Thus, the main mechanism of neutralization by S10 MAbs remains unknown, but it could be related to steric hindrance or interference with binding to other attachment factors, such as heparan sulfate (45). Contrary to the findings for S10 MAbs, the main mechanism of virus neutralization by MAb 71, targeting the S10-S1A domain junction, could be attributed to Sia binding interference, since no neutralization by this antibody was seen on Sia-deficient Vero cells.
S10 antibodies showed binding to and effective neutralization of only closely homologous non-S-Indel strains, while S1B antibodies showed broader reactivity and cross-neutralization toward the more heterologous S-Indel strains. We observed that single-residue changes in S10 MAb escape mutants were already sufficient to generate a loss of antibody binding to and escape from antibody neutralization. Comparison of full-length PEDV spike sequences from field isolates indicated that most sequence variation was found in the N-terminal 350 residues of the S protein, particularly within the S10 region (46). Antigenic diversity in this region may have arisen through a long history of natural selection of variants that escape immune surveillance through mutations in neutralizing epitopes. However, only minor differences in cross-reactivity and cross-neutralization between non-S-Indel and S-Indel strains were seen when polyclonal antisera obtained from non-S-Indel strain- and S-Indel strain-infected pigs were used (47, 48), yet antigenic variation in S1 between non-S-Indel and S-Indel strains was revealed in our study using monoclonal antibodies, and the observed antigenic variability in (neutralizing) epitopes may lower cross-protection in vivo. Consistently, PEDV S-Indel strains were shown to induce only partial cross-protective immunity against the non-S-Indel strain (47, 49, 50). Intriguingly, PEDV strains with spike genes containing large deletions in the S10-encoding region were detected in pig farms in Japan together with PEDV strains with intact spike genes. Whether these spike deletions occurred in viruses to escape from immunity remains to be seen. Antigenic drift in the S10 domain would be in line with the relevance of S10-mediated Sia binding activity in virus entry and the existence of S10-specific neutralizing antibodies that display limited cross-reactivity and a limited cross-neutralizing capacity. The strategy of viruses to escape immune surveillance by antigenic drift imposes a hurdle to vaccine development. The homology of vaccine strains with circulating strains might need to be taken into account, particularly if neutralizing antibodies against the variable S10 domain are to be elicited.
In conclusion, we defined six nonoverlapping neutralizing and nonneutralizing epitopes in the S1 subunit of the PEDV spike protein mapping to different domains. Potent neutralization is achieved by antibodies targeting the Sia binding domain (S10) or the protein receptor binding domain (S1B), further underlining the importance of cell attachment domains on viral surface proteins as major targets of potent neutralizing antibodies (51).
MATERIALS AND METHODS
Cells and viruses.
PEDV UU (GenBank accession no. KU985229), a contemporary Dutch S-Indel strain, was isolated with the help of the Dutch veterinary practice De Oosthof. Qigai He provided two contemporary non-S-Indel Chinese isolates, PEDV FJ-9 (GenBank accession no. AGG34696) and PEDV GDU (GenBank accession no. KU985230). PEDV USA, a contemporary U.S. non-S-Indel isolate, was kindly provided by MSD Animal Health (Boxmeer, The Netherlands). PEDV CV777 (GenBank accession no. AF353511), a classical S-Indel PEDV strain isolated in Belgium, was provided by Kristin van Reeth. PEDV DR13 originated from a commercial vaccine of Green Cross (Republic of Korea). Finally, cDNA of the spike gene of PEDV strain D24 (GenBank accession no. KY399745) was provided by Young Zou of the Shanghai Academy of Agricultural Sciences. Viruses were grown and titrated as described previously (52).
Modeling of trimeric PEDV S protein structure.
A model of the trimeric PEDV S protein structure was generated on the automated protein structure Swiss-Model homology modeling server using the ProMod3 modeling engine (version 1.0.0; https://swissmodel.expasy.org) (53). The PEDV GDU S protein sequence (GenBank accession no. KU985230) was used as a target sequence, and the trimeric HCoV NL63 S structure (PDB accession no. 5SZS) was used as a template. Figures were generated using the PyMOL molecular graphics system (version 1.8; Schrödinger, LLC; http://www.pymol.org).
Expression of recombinant proteins.
Expression of S1 variants of the spike proteins of PEDV strains GDU (GenBank accession no. KU985230.1), UU (GenBank accession no. KU985229.1), CV777 (GenBank accession no. AF353511), DR13 (GenBank accession no. JQ023162.1), D24 (GenBank accession no. KY399745), and FJ-9 (GenBank accession no. AGG34696) was performed as described before (28). Briefly, plasmids expressing the entire PEDV S1 subunit or a subdomain thereof fused to the Fc part of human IgG1 (Fc) or mouse IgG2a (mFc) were transiently expressed in HEK293T cells, as described before (54). Plasmids expressing S1-Fc proteins containing single amino acid substitutions were generated by site-directed mutagenesis. The proteins were purified from tissue culture supernatants by protein A-affinity chromatography (catalog no. 17-0780-01; GE Healthcare), eluted using acid solution (0.1 M citric acid, pH 3.0), and immediately neutralized using Tris, pH 8.8 (final concentration, 0.2 M). Purified proteins were quantified by NanoDrop spectrophotometry, checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and stored at −80°C until use.
Generation of monoclonal antibodies targeting PEDV S1.
PEDV-specific monoclonal antibodies were produced using a previously described method (55) with minor modifications. Briefly, 6-week-old female BALB/c mice were immunized with 7 μg of purified mFc-tagged PEDV S1 (strain GDU) emulsified in n-GNE adjuvant (a nonbacterial oil-water adjuvant; proprietary product; MSD Animal Health, Boxmeer, The Netherlands). Each mouse received three intramuscular (i.m.) injections of PEDV S1-mFc at 4-week intervals. Mice with the highest titers of antibodies against PEDV S1-mFc were further boosted by intravenously injecting 2 μg of purified PEDV S1-mFc in 0.9% saline at 5 weeks after the last i.m. injection. At 5 days after the final injection, the mice were sacrificed, their spleens were collected, and erythrocyte- and monocyte-depleted spleen cell populations were prepared.
The monoclonal antibody-producing hybridomas were prepared by subjecting the spleen cell population or the antigen-specific B cells isolated from those spleen cell populations using paramagnetic beads coated with PEDV S1-mFc to electrofusion with NS-1 myeloma cells. In some cases, antigen-specific B cells were clonally expanded in the presence of human T-cell/macrophage supernatant and irradiated murine thymoma helper cells before subjecting them to electrofusion with NS-1 myeloma cells. At 10 days after electrofusion, the hybridoma cultures were examined for growth and the culture supernatants were screened, using ELISA, for the production of PEDV S1-specific antibodies. The specific antibody-producing hybridoma cultures were cloned by fluorescence-activated cell sorting-based single-cell sorting into 96-well plates. For monoclonal antibody production, hybridoma clones were grown in Dulbecco modified Eagle medium–F-12 medium (catalog no. 11320033; Thermo Fisher Scientific Inc.). IgG was purified from the medium using protein G-conjugated Sepharose beads (catalog no. 6511-100; BioVision). All animal experiments were conducted according to the Guidelines for Animal Experimentation at MSD Animal Health (Boxmeer, The Netherlands).
ELISA.
Microtiter plates (catalog no. 655092; Greiner Bio-One) were coated with the S1 polypeptide (4.0 ng per well diluted in phosphate-buffered saline [PBS]) and incubated overnight at 4°C. After three washes with washing buffer (PBS containing 0.05% Tween 20), the plates were blocked with blocking buffer (PBS containing 3% bovine serum albumin [BSA], 0.1% Tween 20) for 3 h at 37°C and then incubated with different monoclonal antibodies diluted in PBS containing 1% BSA for 1 h at 37°C. After a washing step, 1:3,000-diluted horseradish peroxidase (HRP)-conjugated rabbit anti-mouse IgG (catalog no. P0260; Dako) or HRP-conjugated goat anti-human IgG (catalog no. 109-035-088; Jackson ImmunoResearch) was added, and the mixture was incubated at 37°C for 1 h. The peroxidase reaction was visualized using a tetramethylbenzidine Super Slow one-component HRP microwell substrate (BioFX), and optical densities (OD) were measured at 450 nm.
Competitive binding assay.
A competitive binding assay was performed through biolayer interferometry using an Octet QK system (FortéBio, USA), which measures changes in the interferometry wave pattern of light produced by the binding of molecules to a biosensor layer, according to a previously described method (56). Briefly protein A sensors (FortéBio, USA) were coated with PEDV GDU S1-Fc protein until saturation. The sensor was subsequently blocked with polyclonal cat IgG (catalog no. 002-000-003; Jackson ImmunoResearch). The S1-Fc-loaded sensor was exposed to a first antibody, followed by a brief wash, and was subsequently exposed to a second antibody while the interferometry signal was recorded. Finally, the sensor layer was reconstituted with 10 mM glycine solution (pH 2.0).
Immunofluorescence assay.
Cells were infected with PEDV in the presence of trypsin as described previously (28). Briefly, cells were inoculated for 1 h at 37°C. The inoculum was removed, and the cells were washed and incubated at 37°C in medium supplemented with 1% fetal calf serum and 40 μg/ml soybean protease inhibitor (SBTI; catalog no. T6522; Sigma). At 16 h postinoculation, the cells were fixed and the nuclei were visualized using 4′,6-diamidino-2-phenylindole (DAPI; Molecular Probes). The binding of S1 monoclonal antibodies and of a nucleocapsid protein binding mouse monoclonal antibody 3F12 (BioNote, Republic of Korea) was detected with a goat anti-mouse antibody-Alexa Fluor 488 (Thermo Fisher) conjugate. Nuclei of cells were visualized by DAPI staining.
Virus neutralization assay.
MAbs (start concentration, 100 μg/ml) were serially diluted 2-fold in Eagle's minimum essential medium with the alpha modification (catalog no. 22571-020; Life Technologies) and mixed with an equal volume of PEDV (2,000 50% tissue culture infective doses [TCID50]/ml) at 37°C for 60 min. One hundred microliters of these mixtures was used for inoculation of Vero cells in 96-well plates; for the trypsin-dependent PEDV strains (GDU, USA, CV777, UU), trypsin was added at a final concentration of 15 μg/ml. After 2 days, the cytopathic effect was scored using an inverted microscope, and the neutralizing concentration was defined as the lowest concentration of MAbs that prevented the occurrence of a cytopathic effect.
HAI assay.
The hemagglutination inhibition (HAI) assay for PEDV was used to measure HAI activity by monoclonal antibodies as described before, with slight modifications (28, 57). Briefly, PEDV GDU was diluted in PBS to a final hemagglutination titer of 160 HA units/ml. Serial 2-fold dilutions (25 μl) of the monoclonal antibodies were prepared and mixed with an equal volume of virus solution. An equal volume of a human erythrocyte suspension (0.5% in PBS) was added, and the mixture was incubated for 2 h at 4°C. The specific HAI activity (HAI titer) of the MAb represents the lowest concentration of antibody showing HAI activity.
Generation of the Vero CMAS knockout cell line.
CMP N-acetylneuraminic acid synthetase (CMAS) gene knockout Vero cells were generated using the CRISPR-Cas9-mediated genome editing system as described earlier (58, 59). To knock out the monkey CMAS gene in Vero CCL81 cells, two guide RNAs targeting exon 1 (nucleotides 79 to 98, 5′-CTGCAGCGCAACTCTCGCGG-3′) and intron 1 (nucleotides 971 to 990, 5′-GATACATTGCCAAATTGGTC-3′) were used. Single-cell clones of CMAS knockout cells were obtained by limiting dilution and genotyped by PCR and DNA sequencing. Knockout of CMAS expression in those cells was further confirmed by testing the cells' susceptibility to sialic acid-dependent influenza A virus (strain WSN) infection.
Generation of monoclonal antibody-resistant virus.
Mutant viruses were generated by growing the virus in the presence of increasing concentrations of antibody for 15 passages. Briefly, Vero cells grown in a 48-well plate were infected with virus at a TCID50 of 200 per well in the presence of a nonneutralizing concentration of antibody (a quarter of the minimal concentration required for neutralization in virus neutralization assays). The antibody concentration was increased in subsequent passages, with virus being selected for further passage on fresh Vero cells on the basis of the observed cytopathic effect. Virus was harvested from infected cells by three cycles of freeze-thawing. After 15 passages, virus was harvested and RNA was extracted from the cells using a QIAamp viral RNA minikit (Qiagen) and reverse transcribed into cDNA with random hexamer primers using a SuperScript III reverse transcriptase kit (Invitrogen). The PEDV spike gene was amplified by PCR and entirely sequenced.
ACKNOWLEDGMENTS
We thank Ger Arkestein for his help with the single-cell sorting of hybridoma cells and Peter van Kooten for technical advice on hybridoma culturing and antibody purification.
C.L. was supported by the SAAS training project (grant PG05) and The Science and Technology Commission of Shanghai Municipality Project (grant 13391901500).
REFERENCES
- 1.Gonzalez J, Gomez-Puertas P, Cavanagh D, Gorbalenya A, Enjuanes L. 2003. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch Virol 148:2207–2235. doi: 10.1007/s00705-003-0162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li W, Li H, Liu Y, Pan Y, Deng F, Song Y, Tang X, He Q. 2012. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg Infect Dis 18:1350–1353. doi: 10.3201/eid1808.120002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chattha KS, Roth JA, Saif LJ. 2015. Strategies for design and application of enteric viral vaccines. Annu Rev Anim Biosci 3:375–395. doi: 10.1146/annurev-animal-022114-111038. [DOI] [PubMed] [Google Scholar]
- 4.Saif LJ. 1993. Coronavirus immunogens. Vet Microbiol 37:285–297. doi: 10.1016/0378-1135(93)90030-B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun RQ, Cai RJ, Chen YQ, Liang PS, Chen DK, Song CX. 2012. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg Infect Dis 18:161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Niu B, Yan H, Gao D, Yang X, Chen L, Chang H, Zhao J, Wang C. 2013. Genetic properties of endemic Chinese porcine epidemic diarrhea virus strains isolated since 2010. Arch Virol 158:2487–2494. doi: 10.1007/s00705-013-1767-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oka T, Saif LJ, Marthaler D, Esseili MA, Meulia T, Lin C, Vlasova AN, Jung K, Zhang Y, Wang Q. 2014. Cell culture isolation and sequence analysis of genetically diverse US porcine epidemic diarrhea virus strains including a novel strain with a large deletion in the spike gene. Vet Microbiol 173:258–269. doi: 10.1016/j.vetmic.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vlasova AN, Marthaler D, Wang Q, Culhane MR, Rossow KD, Rovira A, Collins J, Saif LJ. 2014. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013-February 2014. Emerg Infect Dis 20:1620–1628. doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Q, Gauger PC, Stafne MR, Thomas JT, Madson DM, Huang H, Zheng Y, Li G, Zhang J. 2016. Pathogenesis comparison between the United States porcine epidemic diarrhoea virus prototype and S-INDEL-variant strains in conventional neonatal piglets. J Gen Virol 97:1107–1121. doi: 10.1099/jgv.0.000419. [DOI] [PubMed] [Google Scholar]
- 10.Crawford K, Lager KM, Kulshreshtha V, Miller LC, Faaberg KS. 2016. Status of vaccines for porcine epidemic diarrhea virus in the United States and Canada. Virus Res 226:108–116. doi: 10.1016/j.virusres.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Song D, Moon H, Kang B. 2015. Porcine epidemic diarrhea: a review of current epidemiology and available vaccines. Clin Exp Vaccine Res 4:166–176. doi: 10.7774/cevr.2015.4.2.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hurley WL, Theil PK. 2011. Perspectives on immunoglobulins in colostrum and milk. Nutrients 3:442–474. doi: 10.3390/nu3040442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee D-K, Cha S-Y, Lee C. 2011. The N-terminal region of the porcine epidemic diarrhea virus spike protein is important for the receptor binding. Korean J Microbiol Biotechnol 39:140–145. [Google Scholar]
- 14.Sun DB, Feng L, Shi HY, Chen JF, Liu SW, Chen HY, Wang YF. 2007. Spike protein region (aa 636-789) of porcine epidemic diarrhea virus is essential for induction of neutralizing antibodies. Acta Virol 51:149–156. [PubMed] [Google Scholar]
- 15.Walls AC, Tortorici MA, Bosch B, Frenz B, Rottier PJ, DiMaio F, Rey FA, Veesler D. 2016. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature 531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walls AC, Tortorici MA, Frenz B, Snijder J, Li W, Rey FA, DiMaio F, Bosch B, Veesler D. 2016. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat Struct Mol Biol 23:899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirchdoerfer RN, Cottrell CA, Wang N, Pallesen J, Yassine HM, Turner HL, Corbett KS, Graham BS, McLellan JS, Ward AB. 2016. Pre-fusion structure of a human coronavirus spike protein. Nature 531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nam E, Lee C. 2010. Contribution of the porcine aminopeptidase N (CD13) receptor density to porcine epidemic diarrhea virus infection. Vet Microbiol 144:41–50. doi: 10.1016/j.vetmic.2009.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B, Ge J, Li Y. 2007. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virology 365:166–172. doi: 10.1016/j.virol.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh JS, Song DS, Park BK. 2003. Identification of a putative cellular receptor 150 kDa polypeptide for porcine epidemic diarrhea virus in porcine enterocytes. J Vet Sci 4:269–275. [PubMed] [Google Scholar]
- 21.Cong Y, Li X, Bai Y, Lv X, Herrler G, Enjuanes L, Zhou X, Qu B, Meng F, Cong C, Ren X, Li G. 2015. Porcine aminopeptidase N mediated polarized infection by porcine epidemic diarrhea virus in target cells. Virology 478:1–8. doi: 10.1016/j.virol.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shan Z, Yin J, Wang Z, Chen P, Li Y, Tang L. 2015. Identification of the functional domain of the porcine epidemic diarrhoea virus receptor. J Gen Virol 96:2656–2660. doi: 10.1099/vir.0.000211. [DOI] [PubMed] [Google Scholar]
- 23.Deng F, Ye G, Liu Q, Navid MT, Zhong X, Li Y, Wan C, Xiao S, He Q, Fu ZF. 2016. Identification and comparison of receptor binding characteristics of the spike protein of two porcine epidemic diarrhea virus strains. Viruses 8:55. doi: 10.3390/v8030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shirato K, Maejima M, Islam MT, Miyazaki A, Kawase M, Matsuyama S, Taguchi F. 2016. Porcine aminopeptidase N is not a cellular receptor of porcine epidemic diarrhoea virus, but promotes its infectivity via aminopeptidase activity. J Gen Virol 97:2528–2539. doi: 10.1099/jgv.0.000563. [DOI] [PubMed] [Google Scholar]
- 25.Schwegmann-Weßels C, Herrler G. 2006. Sialic acids as receptor determinants for coronaviruses. Glycoconj J 23:51–58. doi: 10.1007/s10719-006-5437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reguera J, Ordono D, Santiago C, Enjuanes L, Casasnovas JM. 2011. Antigenic modules in the N-terminal S1 region of the transmissible gastroenteritis virus spike protein. J Gen Virol 92:1117–1126. doi: 10.1099/vir.0.027607-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shahwan K, Hesse M, Mork A, Herrler G, Winter C. 2013. Sialic acid binding properties of soluble coronavirus spike (S1) proteins: differences between infectious bronchitis virus and transmissible gastroenteritis virus. Viruses 5:1924–1933. doi: 10.3390/v5081924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W, FJ van Kuppeveld, He Q, Rottier PJ, Bosch B. 2016. Cellular entry of the porcine epidemic diarrhea virus. Virus Res 226:117–127. doi: 10.1016/j.virusres.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu C, Tang J, Ma Y, Liang X, Yang Y, Peng G, Qi Q, Jiang S, Li J, Du L, Li F. 2015. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J Virol 89:6121–6125. doi: 10.1128/JVI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reguera J, Mudgal G, Santiago C, Casasnovas JM. 2014. A structural view of coronavirus-receptor interactions. Virus Res 194:3–15. doi: 10.1016/j.virusres.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang SH, Bae JL, Kang TJ, Kim J, Chung GH, Lim CW, Laude H, Yang MS, Jang YS. 2002. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol Cells 14:295–299. [PubMed] [Google Scholar]
- 32.Zhang Y, Yao Y, Gao X, Wang Y, Jia X, Xiao Y, Wang T, Li X, Tian K. 2016. Development of a neutralizing monoclonal antibody against porcine epidemic diarrhea virus S1 protein. Monoclon Antib Immunodiagn Immunother 35:37–40. doi: 10.1089/mab.2015.0049. [DOI] [PubMed] [Google Scholar]
- 33.Sun D, Feng L, Shi H, Chen J, Cui X, Chen H, Liu S, Tong Y, Wang Y, Tong G. 2008. Identification of two novel B cell epitopes on porcine epidemic diarrhea virus spike protein. Vet Microbiol 131:73–81. doi: 10.1016/j.vetmic.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee C. 2015. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol J 12:193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coughlin MM, Babcook J, Prabhakar BS. 2009. Human monoclonal antibodies to SARS-coronavirus inhibit infection by different mechanisms. Virology 394:39–46. doi: 10.1016/j.virol.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mou H, Raj VS, van Kuppeveld FJ, Rottier PJ, Haagmans BL, Bosch BJ. 2013. The receptor binding domain of the new Middle East respiratory syndrome coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J Virol 87:9379–9383. doi: 10.1128/JVI.01277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sune C, Jimenez G, Correa I, Bullido MJ, Gebauer F, Smerdou C, Enjuanes L. 1990. Mechanisms of transmissible gastroenteritis coronavirus neutralization. Virology 177:559–569. doi: 10.1016/0042-6822(90)90521-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sui J, Deming M, Rockx B, Liddington RC, Zhu QK, Baric RS, Marasco WA. 2014. Effects of human anti-spike protein receptor binding domain antibodies on severe acute respiratory syndrome coronavirus neutralization escape and fitness. J Virol 88:13769–13780. doi: 10.1128/JVI.02232-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ristic M, Abou-Youssef MH. 1972. Comments on immunology of transmissible gastroenteritis. J Am Vet Med Assoc 160:549–553. [PubMed] [Google Scholar]
- 40.Boulant S, Stanifer M, Lozach P. 2015. Dynamics of virus-receptor interactions in virus binding, signaling, and endocytosis. Viruses 7:2794–2815. doi: 10.3390/v7062747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis GK, Finzi A, DeVico AL, Pazgier M. 2015. Conformational masking and receptor-dependent unmasking of highly conserved Env epitopes recognized by non-neutralizing antibodies that mediate potent ADCC against HIV-1. Viruses 7:5115–5132. doi: 10.3390/v7092856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bankwitz D, Pietschmann T. 2016. Hepatitis C virus plays hide and seek with neutralizing antibodies. Hepatology 64:1840–1842. doi: 10.1002/hep.28760. [DOI] [PubMed] [Google Scholar]
- 43.Sommerstein R, Flatz L, Remy MM, Malinge P, Magistrelli G, Fischer N, Sahin M, Bergthaler A, Igonet S, Ter Meulen J. 2015. Arenavirus glycan shield promotes neutralizing antibody evasion and protracted infection. PLoS Pathog 11:e1005276. doi: 10.1371/journal.ppat.1005276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.López S, Arias CF. 2004. Multistep entry of rotavirus into cells: a Versaillesque dance. Trends Microbiol 12:271–278. doi: 10.1016/j.tim.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 45.Huan C, Wang Y, Ni B, Wang R, Huang L, Ren X, Tong G, Ding C, Fan H, Mao X. 2015. Porcine epidemic diarrhea virus uses cell-surface heparan sulfate as an attachment factor. Arch Virol 160:1621–1628. doi: 10.1007/s00705-015-2408-0. [DOI] [PubMed] [Google Scholar]
- 46.Jarvis MC, Lam HC, Zhang Y, Wang L, Hesse RA, Hause BM, Vlasova A, Wang Q, Zhang J, Nelson MI. 2016. Genomic and evolutionary inferences between American and global strains of porcine epidemic diarrhea virus. Prev Vet Med 123:175–184. doi: 10.1016/j.prevetmed.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin C, Saif LJ, Marthaler D, Wang Q. 2016. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res 226:20–39. doi: 10.1016/j.virusres.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin C, Gao X, Oka T, Vlasova AN, Esseili MA, Wang Q, Saif LJ. 2015. Antigenic relationships among porcine epidemic diarrhea virus and transmissible gastroenteritis virus strains. J Virol 89:3332–3342. doi: 10.1128/JVI.03196-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin CM, Annamalai T, Liu X, Gao X, Lu Z, El-Tholoth M, Hu H, Saif LJ, Wang Q. 2015. Experimental infection of a US spike-insertion deletion porcine epidemic diarrhea virus in conventional nursing piglets and cross-protection to the original US PEDV infection. Vet Res 46:134. doi: 10.1186/s13567-015-0278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goede D, Murtaugh MP, Nerem J, Yeske P, Rossow K, Morrison R. 2015. Previous infection of sows with a “mild” strain of porcine epidemic diarrhea virus confers protection against infection with a “severe” strain. Vet Microbiol 176:161–164. doi: 10.1016/j.vetmic.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 51.Corti D, Lanzavecchia A. 2013. Broadly neutralizing antiviral antibodies. Annu Rev Immunol 31:705–742. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- 52.Wicht O, Li W, Willems L, Meuleman TJ, Wubbolts RW, FJ van Kuppeveld Rottier PJ, Bosch BJ. 2014. Proteolytic activation of the porcine epidemic diarrhea coronavirus spike fusion protein by trypsin in cell culture. J Virol 88:7952–7961. doi: 10.1128/JVI.00297-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raj VS, Mou H, Smits SL, Dekkers DH, Müller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steenbakkers PG, Hubers HA, Rijnders AW. 1994. Efficient generation of monoclonal antibodies from preselected antigen-specific B cells. Mol Biol Rep 19:125–134. doi: 10.1007/BF00997158. [DOI] [PubMed] [Google Scholar]
- 56.Estep P, Reid F, Nauman C, Liu Y, Sun T, Sun J, Xu Y. 2013. High throughput solution-based measurement of antibody-antigen affinity and epitope binning, p 270–278. In Anonymous MAbs. Taylor & Francis, New York, NY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Coligan J. 2005. Short protocols in immunology: a compendium of methods from current protocols in immunology. John Wiley & Sons, Inc, Hoboken, NJ. [Google Scholar]
- 58.Hsu PD, Lander ES, Zhang F. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langereis MA, Rabouw HH, Holwerda M, Visser LJ, van Kuppeveld FJ. 2015. Knockout of cGAS and STING rescues virus infection of plasmid DNA-transfected cells. J Virol 89:11169–11173. doi: 10.1128/JVI.01781-15. [DOI] [PMC free article] [PubMed] [Google Scholar]